Abstract
Mono-ADP-ribosylation is the enzymatic transfer of ADP-ribose from NAD+ to acceptor proteins. It is catalysed by cellular ADP-ribosyltransferases and certain bacterial toxins. There are two subclasses of cellular enzymes: the ectoenzymes that modify targets such as integrins, defensin and other cell surface molecules; and the intracellular enzymes that act on proteins involved in cell signalling and metabolism, such as the β-subunit of heterotrimeric G proteins, GRP78/BiP and elongation factor 2. The genes that encode the ectoenzymes have been cloned and their protein products are well characterized, yet little is known about the intracellular ADP-ribosyltransferases, which may be part of a novel protein family with an important role in regulating cell function. ADP-ribosylation usually leads to protein inactivation, providing a mechanism to inhibit protein functions in both physiological and pathological conditions.
Keywords: ADP-ribosylhydrolase/ADP-ribosyltransferase/cell signalling/G proteins/post-translational modifications
Introduction
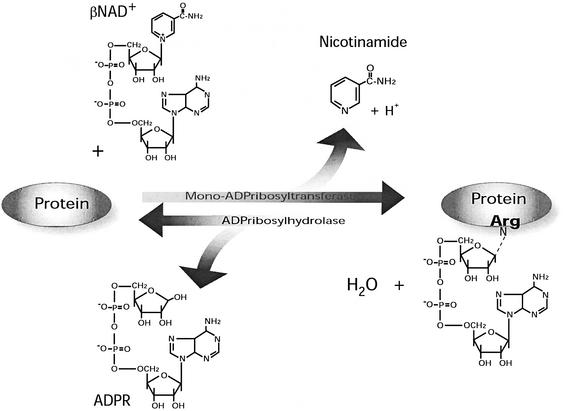
Mono-ADP-ribosylation of cellular proteins can occur through enzymatic and non-enzymatic mechanisms. The enzymatic reaction, originally identified as the mechanism of action of certain bacterial toxins (Ueda and Hayaishi, 1985), is catalysed by mono-ADP-ribosyltransferases (EC 2.4.2.31) that transfer an ADP-ribose residue from βNAD+ to a specific amino acid of the acceptor protein (Figure 1). These processes are distinct from that catalysed by poly(ADP-ribose) polymerases (PARPs) (EC 2.4.2.30), which transfer branched polymers of ADP-ribose to target proteins (for a review see Smith, 2001).
Fig. 1. A schematic representation of the reversible mono-ADP-ribosylation reaction catalysed by an arginine-specific mono-ADP-ribosyltransferase and an ADP-ribosylhydrolase (see text for details). Other amino acid residues that can be modified by this reaction are cysteine, diphtamide and asparagine (Okazaki and Moss, 1999; Table I).
The extent of protein modification by ADP-ribosylation depends on the activity of cellular ADP-ribosylhydrolases that reverse the reaction by hydrolysing the protein–ADP-ribose linkage (Figure 1). For some time, the only gene known to encode a soluble, intracellular ADP-ribosylhydrolase specific for the ADP-ribose–arginine bond was that cloned from mammals (Moss et al., 1992). Recently, two additional genes have been identified by screening the human genome sequence for relatives of the known hydrolase (Glowacki et al., 2002). Although the enzymatic activity and bond specificity of these new gene products have not yet been determined, their identification suggests that the hydrolase family may be larger than originally thought.
The presence of ADP-ribosyltransferase and ADP-ribosylhydrolase activities in the cell suggests that reversible protein mono-ADP-ribosylation acts as a regulatory mechanism for the protein substrates of these reactions (Figure 1). Indeed, recent data are fully consistent with this proposal (see below).
In this review, we will focus mainly on the enzymatic mono-ADP-ribosylation of intracellular proteins and discuss the physiological relevance of this reaction and the recent identification of novel enzymes.
Enzymes and substrates in prokaryotes and viruses
ADP-ribosylation as a mechanism of pathogenesis
ADP-ribosyltransferase activities have been observed in many prokaryotic and eukaryotic species and in viruses. The best characterized mono-ADP-ribosylation reactions are those catalysed by bacterial toxins, such as diphtheria, cholera, pertussis and clostridial toxins, which act by modifying crucial host cell proteins such as the α-subunit of heterotrimeric GTP-binding (G) proteins, the small GTPase Rho, monomeric actin and elongation factor 2 (EF-2), resulting in permanent activation or inactivation of the cell functions modulated by these protein substrates (for a review, see Krueger and Barbieri, 1995; see Table I and Supplementary table I available at The EMBO Journal Online). The mono-ADP-ribosylation reactions induced by these bacterial toxins are a crucial part of the pathogenic mechanisms that cause cholera, botulism and other diseases.
Table I. Mono-ADP-ribosylation: enzymes and substratesa.
| Enzymes | Source | Substrate/amino acid | Effect of the reaction |
|---|---|---|---|
| Viruses | |||
| ALT | Bacteriophage T4 | RNA polymerase/Arg265 | Enhances viral transcription |
| Mod A | Bacteriophage T4 | RNA polymerase/Arg265 | Enhances viral transcription |
| Mod B |
Bacteriophage T4 |
RNA polymerase/Arg265, S1 ribosomal protein/Arg |
Enhances viral transcription |
| Prokaryotes | |||
| Toxins | |||
| Diphtheria | Corynebacterium diphtheriae | EF-2/diphtamide715 | Inhibits protein synthesis |
| Exotoxin A | Pseudomonas aeruginosa | EF-2/diphtamide715 | Inhibits protein synthesis |
| Exotoxin S | Pseudomonas aeruginosa | Ras family/Arg41 | Disrupts actin microfilaments |
| Cholera | Vibrio cholerae | Gαs, Gαt/Arg187 | Inhibits GTPase activity |
| LT1, LT2 | Escherichia coli | Gαs, Gαt/Arg187 | Inhibits GTPase activity |
| Pertussis | Bordetella pertussis | Gαi, Gαo, Gαt/Cys351 | Uncouples receptor and G protein |
| C2, iota t | Clostridium botulinum | Actin | Prevents actin polymerization |
| C3 | Clostridium botulinum | Rho, Rac/Asn41 | Disrupts actin cytoskeleton |
| C3-like | Clostridium limosum | Rho, Rac/Asn41 | Disrupts actin cytoskeleton |
| EDIN | Staphylococcus aureus | Rho/Asn41 | Disrupts Golgi apparatus |
| VIP2 | Bacillus cereus | Rho/Asn41 | Disrupts actin cytoskeleton |
| SpvB | Salmonella enterica | Actin | Prevents actin polymerization |
| Mtx | Bacillus sphaericus | Unknown | Unknown |
| Intracellular | |||
| DRAT |
Rhodospirillum rubrum |
Dinitrogenase reductase/Arg101 |
Inhibits dinitrogenase reductase |
| Eukaryotes | |||
| Ectoenzymes | |||
| ART1 | Human, rat, mouse | Integrin, defensin/Arg | Inhibits substrate activity |
| ART2(A,B) | Rat, mouse | Unknown/Arg | Role in T cell proliferation |
| ART3 | Human, rat, mouse | Unknown | Unknown |
| ART4 | Human, rat, mouse | Unknown | Unknown |
| ART5 | Human | Unknown/Arg | Unknown |
| ART6(A,B) | Chicken | p33/actin/Arg28-206 | Inhibits substrate activity |
| ART7 | Chicken | Unknown | Inhibits substrate activity |
| Intracellular | |||
| Sir2p | Yeast | Histones/Sir2p/acetyl-lysine | Involved in histone deacetylation |
| Sirtuin2 | Human | Albumin/acetyl-lysine | Involved in histone deacetylation |
| Pierisin1,2 | Cabbage butterfly | DNA | Cytotoxic |
| Arginine-specific | Hamster, human | Gβ/Arg129 | Inhibits substrate activity |
| Cysteine-specific | Human | GDH/Cys | Inhibits substrate activity |
aThe table shows some of the well-defined ADP-ribosylation reactions. See text for additional examples, references and details. For a complete list of references to the table, see Supplementary table.
Interestingly, some bacterial toxins are encoded by lysogenic phages that incorporate into the DNA of their host bacteria (Uchida et al., 1971). So far, three different ADP-ribosyltransferases have been discovered that are encoded by the bacteriophage T4: Alt, ModA and ModB. Alt, a structural component of the phage head, ADP-ribosylates Arg265 of the host RNA polymerase (Goff, 1984) with consequent enhancement of viral transcription (Sommer et al., 2000). ModA and ModB both ADP-ribosylate the α-subunit of the bacterial RNA polymerase (the preferred substrate for ModA) and the S1 ribosomal protein (Tiemann et al., 1999), both of which support the phage’s programme to gain control over the infected host cell.
The human genome contains >200 genes of bacterial origin including those encoding ADP-ribosylhydrolases (Lander et al., 2001). The characteristics of the ADP-ribosyltransferase genes (widely present in bacteria and vertebrates, but not in the fully sequenced genomes of Saccharomyces, Arabidopsis, Caenorhabditis and Drosophila) led Pallen and co-workers to propose recently that the human mono-ADP-ribosyltransferases are an example of horizontal gene transfer across species (Pallen et al., 2001; Glowacki et al., 2002). An alternative possibility is that these genes were lost during the evolution of the lower eukaryotes. Recently, however, ADP-ribosylating activities have been found in insects and yeast (see below), suggesting that these activities are conserved.
Endogenous mono-ADP-ribosylation: the DRAT/DRAG cycle
Bacterial mono-ADP-ribosylation reactions occur not only through the secreted toxins but also within the bacteria themselves to control metabolic enzymes. This is the case in the photosynthetic nitrogen-fixing bacterium Rhodospirillum rubrum, where mono-ADP-ribosylation regulates the nitrogenase enzymes responsible for nitrogen fixation (Ludden, 1994); this modulation, which operates only in certain nitrogen-fixing bacteria (Halbleib and Ludden, 2000), was the first clear demonstration of the physiological role of this reaction.
Dinitrogenase reductase (an enzyme that transfers electrons to dinitrogenase, which in turn reduces nitrogen to ammonium) is rapidly and reversibly inactivated by mono-ADP-ribosylation on Arg101, so preventing unproductive nitrogen fixation during energy-limiting or nitrogen-sufficient conditions. This reaction, catalysed by a dinitrogenase reductase arginine-specific mono-ADP- ribosyltransferase (DRAT), is thus very similar to that of cholera toxin, but there is no obvious amino acid sequence homology between these enzymes (Lowery and Ludden, 1988; Bazan and Koch-Nolte, 1997).
The dinitrogenase reductase is fully reactivated by a specific ADP-ribosylarginine hydrolase called dinitrogenase reductase-activating glycohydrolase (DRAG) that hydrolyses the N-glycosidic bond of the ADP-ribosylated protein (Fitzmaurice et al., 1989). The activities of DRAT and DRAG are known to be controlled in vivo (Liang et al., 1991).
Genes and functions in eukaryotes
After the discovery of bacterial ADP-ribosyltransferases, similar enzymatic activities were detected in turkey erythrocytes, rat liver homogenates and Xenopus tissues (Okazaki and Moss, 1999).
ARTs
Members of the vertebrate family of mono-ADP-ribosyltransferases are now known as ARTs. They are all ectoenzymes that modify extracellular substrates (see Table I for a list of known ARTs, their sources and substrates). This relatively small family has been confirmed by recent database searches and structure prediction analyses aimed at identifying all the recognizable human and mouse mono-ADP-ribosyltransferases and ADP-ribosylhydrolases (Glowacki et al., 2002). ART1–ART4 are all glycosylphosphatidylinositol (GPI)-anchored membrane proteins, with an extracellular catalytic domain, whereas ART5, ART6 and ART7 possess a hydrophobic N-terminal signal sequence and they are secreted proteins (reviewed in Bazan and Koch-Nolte, 1997; Okazaki and Moss, 1999).
Despite their limited primary sequence homology, three common domains have been identified in the ART structure: an N-terminal region, characterized by either a conserved histidine or arginine; a central, hydrophobic, NAD+-binding region; and a highly acidic region with a conserved glutamate residue crucial for catalysis (Domenighini et al., 1994; Domenighini and Rappuoli, 1996). Common substrate recognition domains have not yet been identified (reviewed in Bazan and Koch-Nolte, 1997; Han and Tainer, 2002).
The ARTs are expressed preferentially in cells of the immune system and thus have been proposed to participate in the immune response (Haag and Koch-Nolte, 1998; Paone et al., 2002). However, ART substrates also include integrin α7, whose mono-ADP-ribosylation by ART1 is proposed to play a role in myogenesis (Zolkiewska and Moss, 1993). Another important and recently identified ART1 substrate is defensin, an antimicrobial peptide secreted by immune cells, the ADP-ribosylated form of which has been found in the bronchoalveolar lavage fluid of smokers (Paone et al., 2002). This finding points to a role for ADP-ribosylation in the innate immune response in man (Corda and Di Girolamo, 2002; Paone et al., 2002).
Intracellular mono-ADP-ribosylation
ADP-ribosylation, like protein phosphorylation, has all the characteristics of a mechanism crucial for cell regulation. Indeed, its regulatory function has already been demonstrated in some signalling cascades, for example the mono-ADP-ribosylation of the ubiquitous heterotrimeric G protein βγ-subunit (Lupi et al., 2000, 2002). This reaction is catalysed by an arginine-specific, plasma membrane-associated, mono-ADP-ribosyltransferase that has an intracellular catalytic site and specifically modifies Arg129, a crucial residue in the common effector-binding surface of the β-subunit. Mono-ADP ribosylation of this residue prevents β-subunit-dependent modulation of effectors such as type 1 adenylyl cyclase, phosphoinositide 3-kinase and phospholipase C (Lupi et al., 2000, 2002). The modified β-subunit is a substrate for a cytosolic hydrolase that releases the bound ADP-ribose (Lupi et al., 2000). Therefore, these activities are part of a cycle of intracellular ADP-ribosylation and de-ADP-ribosylation regulating the function of the βγ dimer. The physiological role of this reaction has been supported further by the demonstration of its hormonal control exerted by thrombin, serotonin and cholecystokinin (Lupi et al., 2002).
Mono-ADP-ribosylation could, therefore, act as a signal termination mechanism for βγ; when an activated G protein-coupled receptor induces dissociation of the α- and βγ-subunits of the G protein (which both go on to interact with their specific effectors), it also initiates a signal termination process by inducing mono-ADP-ribosylation of the active βγ dimer. The modified dimer no longer interacts with the effector and, in time, will be de-ADP-ribosylated allowing it to reassociate with the α-subunit and form the inactive G protein heterotrimer, ready then for another activation cycle.
Interestingly, there is evidence that the G protein α-subunits can also be ADP-ribosylated by enzymatic activities isolated from various tissues (Di Girolamo and Corda, 2003), suggesting that signalling cascades modulated by α-subunits might be regulated by this modification.
The function of G protein α-subunits is also regulated by mono-ADP-ribosylation of their interacting proteins, as in the case of rod photoreceptor membranes from the amphibian retina, where the interaction between the α-subunit of the G protein called transducin and Pγ (the regulatory subunit of the cGMP phosphodiesterase) is prevented when Pγ is ADP-ribosylated (Bondarenko et al., 1999).
ADP-ribosylation also plays a role in regulating the cell cytoskeleton. Two major cytoskeletal proteins, actin and desmin, can be ADP-ribosylated by arginine-specific ADP-ribosyltransferases in homogenates of various cells and tissues. Mono-ADP-ribosylation of non-muscle actin inhibits its ability to polymerize in chicken heterophils, which might affect actin-mediated functions such as the release of azurophilic granules (Terashima et al., 1995). In HL-60 cells, mono-ADP-ribosylation of actin is involved in the modification of the cytoskeleton leading to apoptosis (Lodhi et al., 2001). Similarly, desmin, the muscle-specific intermediate filament protein, when ADP-ribosylated, cannot assemble into intermediate filaments in vitro, and incubation with ADP-ribosylarginine hydrolase restores the self-assembly properties of the protein (Huang et al., 1993, 1996).
Additional examples of functionally relevant mono-ADP-ribosylation in intact cells include that of GRP78/BiP, a molecular chaperone that resides in the lumen of the endoplasmic reticulum. This specific modification occurs in response to nutritional stress in cells starved of either amino acids or glucose (Leno and Ledford, 1989). Laitusis et al. (1999) recently hypothesized that ADP-ribosylated GRP78/BiP provides a buffering system by which the rates of protein processing can be balanced with that of protein synthesis. In cells with high rates of protein synthesis, the bulk of the GRP78/BiP in the unmodified form would be complexed with protein folding intermediates. Any slowing of protein synthesis relative to protein processing would result in GRP78/BiP inactivation by ADP-ribosylation. Mono-ADP-ribosylation has also been involved in the inhibition of protein synthesis due to the direct modification of EF-2 by cellular ADP-ribosyltransferases present in several tissues (Fendrick and Iglewski, 1989).
A different mechanism of activation of intracellular ADP-ribosylation is by the fungal toxin brefeldin A (BFA), which affects the structure and function of the Golgi complex (De Matteis et al., 1994; Corda et al., 2002). BFA activates an as yet uncharacterized cellular ADP-ribosyltransferase that specifically modifies glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and another protein called BFA-dependent ADP-ribosylation substrate or CtBP3/BARS (De Matteis et al., 1994; Di Girolamo et al., 1995). This protein is one of the cytosolic factors that can counteract the effect of BFA on the Golgi complex, since the addition of purified CtBP3/BARS abolishes the tubulation of the organelle induced by BFA in permeabilized cells (Spano et al., 1999). As with the other ADP-ribosylated proteins, the ADP-ribosylation of CtBP3/BARS results in its inactivation. Once modified, CtBP3/BARS, which has lysophosphatidic acid-specific acyl transferase activity and is a potent activator of the fission of Golgi tubular-reticular domains, loses its catalytic activity and its ability to promote the fission of Golgi tubules (Weigert et al., 1999). Specific inhibitors of BFA-induced ADP-ribosylation largely prevent the toxic effect of BFA on the Golgi complex (Weigert et al., 1997), underpinning the functional role of this reaction.
A growing family
It is becoming clear that mono-ADP-ribosylation is a phylogenetically ancient and crucial mechanism to regulate protein function. ADP-ribosyltransferases (Alt, ModA and ModB) appear to date back to the divergence of bacteriophages during evolution (Goff, 1984; Tiemann et al., 1999). Alt is responsible for the first modification made to the Escherichia coli RNA polymerase after T4 infection (Goff, 1984) that enhances viral transcription (Sommer et al., 2000). The bacterial ADP-ribosylating toxins themselves may derive from the genome of lysogenic phages incorporated into the DNA of their hosts (Pallen et al., 2001). The two classes of bacterial ADP-ribosylating enzymes (the bacterial exotoxins and the DRAT/DRAG system, discussed earlier) are the first known members of growing families. A recent database search (Pallen et al., 2001) discovered 21 putative new toxin-like ADP-ribosyltransferase sequences that fall into the cholera toxin group, characterized by a conserved arginine in the NAD+-binding domain (Domenighini et al., 1994; Domenighini and Rappuoli, 1996). Also, a search for DRAT relatives revealed novel DRAT homologues in the genomes of five different bacteria, but failed to find similarity to the ARTs (Pallen et al., 2001).
Even if the amino acid sequence homology is very limited, there is clearly an evolutionary relationship between prokaryotic and eukaryotic ADP-ribosyltransferases (Domenighini et al., 1994; Domenighini and Rappuoli, 1996) that has been confirmed by recent database searches (Pallen et al., 2001; Glowacki et al., 2002). In eukaryotes, only one family (ART1–7) is well characterized from a molecular point of view, but we predict that distinct families of ADP-ribosylating enzymes are waiting to be found.
All the human and mouse ecto-mono-ADP-ribosyltransferases have been identified (Glowacki et al., 2002), and classified as either GPI-anchored or secretory proteins, implying that in both cases the targets of these enzymes are extracellular proteins. Therefore, it is unlikely that members of this family are involved in mono-ADP-ribosylation of intracellular targets, such as heterotrimeric G proteins. The intracellular ADP-ribosyltransferases probably constitute a different family of proteins with no obvious sequence similarity to the well-known ARTs. Yet their identification may not be easy. Some putative bacterial ADP-ribosyltransferases that were identified by one database search (Pallen et al., 2001) escaped another later search (Glowacki et al., 2002), probably because of their pronounced divergence. This indeed suggests that other surprises are possible when more potent instruments for database searching are developed.
The unique ADP-ribosylating toxins pierisin-1 and pierisin-2 discovered in the cabbage butterfly are a case in point (Watanabe et al., 1999; Matsushima-Hibiya et al., 2000). Only after the characterization of their enzymatic activity did the very remote similarity to bacterial ADP-ribosylating toxins finally emerge (Glowacki et al., 2002). Indeed, the amino acid sequence deduced from the cloned pierisin-1 cDNA has regional sequence similarities to the ADP-ribosylating subunit of cholera toxin, in particular with the conserved catalytic glutamic acid residue (Watanabe et al., 1999). Mutation of this Glu165 residue greatly reduces pierisin’s apoptosis-inducing activity, as do inhibitors of ADP-ribosylating enzymes, thus linking pierisin ADP-ribosylating activity to its ability to induce apoptosis (Watanabe et al., 1999).
Piersins are not the only examples of unusual ADP-ribosyltransferases. Yeast Sir2p (silent information regulator 2 protein) also has ADP-ribosyltransferase activity (Frye, 1999; Tanny et al., 1999). Yeast Sir2p is an NAD+-dependent histone/protein deacetylase involved in gene silencing, chromosomal stability and ageing (for a review, see Grozinger and Schreiber, 2002). Recently, Sir2p was shown to catalyse a unique reaction in which the cleavage of NAD+ and the deacetylation of histones are coupled to formation of the novel metabolite, O-acetyl-ADP-ribose, which blocks oocyte maturation (Tanner et al., 2000; Borra et al., 2002). Moreover, seven human homologues of Sir2p (sirtuins 1–7) able to metabolize NAD+ and endowed with ADP-ribosyltransferase activities have been cloned (Frye, 2000). Again, these Sir2p-like proteins do not share any obvious sequence homology with the well-known ART family and are good candidates for intracellular ADP-ribosyltransferases.
Protein mono-ADP-ribosylation is emerging as a regulatory process in prokaryotes and eukaryotes with features that, due to the lack of information in living cells, are only starting to be established. Clearly, a better understanding of the ADP-ribosylation machinery in living cells will not only increase our understanding of the functional role of this reaction in processes such as signalling, immune responses and membrane traffic, but will also help to define new targets for drug development.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank C.Featherstone (The ELSO Gazette) and M.A.De Matteis (Consorzio Mario Negri Sud, Italy) for their comments on the manuscript in preparation, R.Le Donne for preparation of the figure, and the Italian Association for Cancer Research (AIRC, Milano, Italy), Telethon Italy (no. E.841) and the Italian National Research Council (CNR, Rome, Italy) ctr. no. 01.00027.PF49 for financial support.
References
- Bazan J.F. and Koch-Nolte,F. (1997) Sequence and structural links between distant ADP-ribosyltransferase families. Adv. Exp. Med. Biol., 419, 99–107. [DOI] [PubMed] [Google Scholar]
- Bondarenko V.A., Yamazaki,M., Hayashi,F. and Yamazaki,A. (1999) Suppression of GTP/Tα-dependent activation of cGMP phosphodiesterase by ADP-ribosylation by its γ subunit in amphibian rod photoreceptor membranes. Biochemistry, 38, 7755–7763. [DOI] [PubMed] [Google Scholar]
- Borra M.T., O’Neill,F.J., Jackson,M.D., Marshall,B., Verdin,E., Foltz,K.R. and Denu,J.M. (2002) Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+ dependent deacetylases. J. Biol. Chem., 277, 12632–12641. [DOI] [PubMed] [Google Scholar]
- Corda D. and Di Girolamo,M. (2002) Mono-ADP-ribosylation: a tool for modulating immune response and cell signalling. Sci. STKE, http://www.stke.org/cgi/content/full/sigtrans;2002/163/pe53. [DOI] [PubMed] [Google Scholar]
- Corda D., Hidalgo Carcedo,C., Bonazzi,M., Luini,A. and Spanò,S. (2002) Molecular aspects of membrane fission in the secretory pathway. Cell. Mol. Life Sci., 59, 1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis M.A. et al. (1994) Stimulation of endogenous ADP-ribosylation by brefeldin A. Proc. Natl Acad. Sci. USA, 91, 1114–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Girolamo M. and Corda,M. (2003) Mono-ADP-ribosylation of heterotrimeric G proteins. In Bradshaw,R. and Dennis,E. (eds), Handbook of Cell Signalling. Academic Press, San Diego, CA, in press.
- Di Girolamo M., Silletta,M.G., De Matteis,M.A., Braca,A., Colanzi,A., Pawlak,D., Rasenick,M.M., Luini,A. and Corda,D. (1995) Evidence that the 50-kDa substrate of brefeldin A-dependent ADP-ribosylation binds GTP and is modulated by the G-protein βγ subunit complex. Proc. Natl Acad. Sci. USA, 92, 7065–7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenighini M. and Rappuoli,R. (1996) Three conserved consensus sequences identify the NAD-binding site of ADP-ribosylating enzymes, expressed by eukaryotes, bacteria and T-even bacteriophages. Mol. Microbiol., 21, 667–674. [DOI] [PubMed] [Google Scholar]
- Domenighini M., Magagnoli,C., Pizza,M. and Rappuoli,R. (1994) Common features of the NAD-binding and catalytic site of ADP-ribosylating toxins. Mol. Microbiol., 14, 41–50. [DOI] [PubMed] [Google Scholar]
- Fendrick J.L. and Iglewski,W.J. (1989) Endogenous ADP-ribosylation of elongation factor 2 in polyoma virus-transformed baby hamster kidney cells. Proc. Natl Acad. Sci. USA, 86, 554–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzmaurice W.P., Saari,L.L., Lowery,R.G., Ludden,P.W. and Roberts,G.P. (1989) Genes coding for the reversible ADP-ribosylation system of dinitrogenase reductase from Rhodospirillum rubrum. Mol. Gen. Genet., 218, 340–347. [DOI] [PubMed] [Google Scholar]
- Frye R.A. (1999) Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun., 260, 273–279. [DOI] [PubMed] [Google Scholar]
- Frye R.A. (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun., 273, 793–798. [DOI] [PubMed] [Google Scholar]
- Glowacki G. et al. (2002) The family of toxin-related ecto-ADP-ribosyltransferases in humans and the mouse. Protein Sci., 11, 1657–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff C.G. (1984) Coliphage-induced ADP-ribosylation of Escherichia coli RNA polymerase. Methods Enzymol., 106, 418–429. [DOI] [PubMed] [Google Scholar]
- Grozinger C.M. and Schreiber,S.L. (2002) Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem. Biol., 9, 3–16. [DOI] [PubMed] [Google Scholar]
- Haag F. and Koch-Nolte,F. (1998) Endogenous relatives of ADP-ribosylating bacterial toxins in mice and men: potential regulators of immune cell function. J. Biol. Regul. Homeost. Agents, 12, 53–62. [PubMed] [Google Scholar]
- Halbleib C.M. and Ludden,P.W. (2000) Regulation of biological nitrogen fixation. J. Nutr., 130, 1081–1084. [DOI] [PubMed] [Google Scholar]
- Han S. and Tainer,J.A. (2002) The ARTT motif and a unified structural understanding of substrate recognition in ADP-ribosylating bacterial toxins and eukaryotic ADP-ribosyltransferases. Int. J. Med. Microbiol., 291, 523–529. [DOI] [PubMed] [Google Scholar]
- Huang H.Y., Graves,D.J., Robson,R.M. and Huiatt,T.W. (1993) ADP-ribosylation of the intermediate filament protein desmin and inhibition of desmin assembly in vitro by muscle ADP-ribosyltransferase. Biochem. Biophys. Res. Commun., 197, 570–577. [DOI] [PubMed] [Google Scholar]
- Huang H.Y., Zhou,H., Huiatt,T.W. and Graves,D.J. (1996) Target proteins for arginine-specific mono(ADP-ribosyl)transferase in membrane fractions from chick skeletal muscle cells. Exp. Cell Res., 226, 147–153. [DOI] [PubMed] [Google Scholar]
- Krueger K.M. and Barbieri,J.T. (1995) The family of bacterial ADP-ribosylating exotoxins. Clin. Microbiol. Rev., 8, 34–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitusis A.L., Brostrom,M.A. and Brostrom,C.O. (1999) The dynamic role of GRP78/BiP in the coordination of mRNA translation with protein processing. J. Biol. Chem., 274, 486–493. [DOI] [PubMed] [Google Scholar]
- Lander E.S. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Leno G.H. and Ledford,B.E. (1989) ADP-ribosylation of the 78-kDa glucose-regulated protein during nutritional stress. Eur. J. Biochem., 186, 205–211. [DOI] [PubMed] [Google Scholar]
- Liang J.H., Nielsen,G.M., Lies,D.P., Burris,R.H., Roberts,G.P. and Ludden,P.W. (1991) Mutations in the draT and draG genes of Rhodospirillum rubrum result in loss of regulation of nitrogenase by reversible ADP-ribosylation. J. Bacteriol., 173, 6903–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodhi I.J., Clift,R.E., Omann,G.M., Sweeney,J.F., McMahon,K.K. and Hinshaw,D.B. (2001) Inhibition of mono-ADP-ribosyltransferase activity during the execution phase of apoptosis prevents apoptotic body formation. Arch. Biochem. Biophys., 387, 66–77. [DOI] [PubMed] [Google Scholar]
- Lowery R.G. and Ludden,P.W. (1988) Purification and properties of dinitrogenase reductase ADP-ribosyltransferase from the photosynthetic bacterium Rhodospirillum rubrum. J. Biol. Chem., 263, 16714–16719. [PubMed] [Google Scholar]
- Ludden P.W. (1994) Reversible ADP-ribosylation as a mechanism of enzyme regulation in procaryotes. Mol. Cell. Biochem., 138, 123–129. [DOI] [PubMed] [Google Scholar]
- Lupi R., Corda,D. and Di Girolamo,M. (2000) Endogenous ADP-ribosylation of the G protein β subunit prevents the inhibition of type 1 adenylyl cyclase. J. Biol. Chem., 275, 9418–9424. [DOI] [PubMed] [Google Scholar]
- Lupi R., Dani,N., Dietrich,A., Marchegiani,A., Turacchio,S., Berrie,C.P., Moss,J., Gierschik,P., Corda,D. and Di Girolamo,M. (2002) Endogenous mono-ADP-ribosylation of the free G βγ prevents stimulation of phosphoinositide 3 kinase-γ and phospholipase C-β2 and is activated by G-protein-coupled-receptors. Biochem. J., 367, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima-Hibiya Y., Watanabe,M., Kono,T., Kanazawa,T., Koyama,K., Sugimura,T. and Wakabayashi,K. (2000) Purification and cloning of pierisin-2, an apoptosis-inducing protein from the cabbage butterfly, Pieris brassicae. Eur. J. Biochem., 267, 5742–5750. [DOI] [PubMed] [Google Scholar]
- Moss J., Stanley,S.J., Nightingale,M.S., Murtagh,J.J.,Jr, Monaco,L., Mishima,K., Chen,H.C., Williamson,K.C. and Tsai,S.C. (1992) Molecular and immunological characterization of ADP-ribosylarginine hydrolases. J. Biol. Chem., 267, 10481–10488. [PubMed] [Google Scholar]
- Okazaki I.J. and Moss,J. (1999) Characterization of glycosylphosphatidylinositol-anchored, secreted and intracellular vertebrate mono-ADP-ribosyltransferases. Annu. Rev. Nutr., 19, 485–509. [DOI] [PubMed] [Google Scholar]
- Pallen M.J., Lam,A.C., Loman,N.J. and McBride,A. (2001) An abundance of bacterial ADP-ribosyltransferases: implications for the origin of exotoxins and their human homologues. Trends Microbiol., 9, 302–307. [DOI] [PubMed] [Google Scholar]
- Paone G., Wada,A., Stevens,L.A., Matin,A., Hirayama,T., Levine,R.L. and Moss,J. (2002) ADP-ribosylation of human neutrophil peptide-1 regulates its biological properties. Proc. Natl Acad. Sci. USA, 99, 8231–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. (2001) The world according to PARP. Trends Biochem. Sci., 26, 174–179. [DOI] [PubMed] [Google Scholar]
- Sommer N., Salniene,V., Gineikiene,E., Nivinskas,R. and Ruger,W. (2000) T4 early promoter strength probed in vivo with unribosylated and ADP-ribosylated Escherichia coli RNA polymerase: a mutation analysis. Microbiology, 146, 2643–2653. [DOI] [PubMed] [Google Scholar]
- Spano S. et al. (1999) Molecular cloning and functional characterization of brefeldin A-ADP-ribosylated substrate. A novel protein involved in the maintenance of the Golgi structure. J. Biol. Chem., 274, 17705–17710. [DOI] [PubMed] [Google Scholar]
- Tanner K.G., Landry,J., Sternglanz,R. and Denu,J.M. (2000) Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl Acad. Sci. USA, 97, 14178–14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny J.C., Dowd,G.J., Huang,J., Hilz,H. and Moazed,D. (1999) An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell, 99, 735–745. [DOI] [PubMed] [Google Scholar]
- Terashima M., Yamamori,C. and Shimoyama,M. (1995) ADP-ribosylation of Arg28 and Arg206 on the actin molecule by chicken arginine-specific ADP-ribosyltransferase. Eur. J. Biochem., 231, 242–249. [PubMed] [Google Scholar]
- Tiemann B., Depping,R. and Ruger,W. (1999) Overexpression, purification and partial characterization of ADP-ribosyltransferases modA and modB of bacteriophage T4. Gene Expr., 8, 187–196. [PMC free article] [PubMed] [Google Scholar]
- Uchida T., Gill,D.M. and Pappenheimer,A.M.,Jr (1971) Mutation in the structural gene for diphtheria toxin carried by temperate phage. Nature, 233, 8–11. [DOI] [PubMed] [Google Scholar]
- Ueda K. and Hayaishi,O. (1985) ADP-ribosylation. Annu. Rev. Biochem., 54, 73–100. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Kono,T., Matsushima-Hibiya,Y., Kanazawa,T., Nishisaka,N., Kishimoto,T., Koyama,K., Sugimura,T. and Wakabayashi,K. (1999) Molecular cloning of an apoptosis-inducing protein, pierisin, from cabbage butterfly: possible involvement of ADP-ribosylation in its activity. Proc. Natl Acad. Sci. USA, 96, 10608–10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigert R. et al. (1997) Characterization of chemical inhibitors of brefeldin A-activated mono-ADP-ribosylation. J. Biol. Chem., 272, 14200–14207. [DOI] [PubMed] [Google Scholar]
- Weigert R. et al. (1999) CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature, 402, 429–433. [DOI] [PubMed] [Google Scholar]
- Zolkiewska A. and Moss,J. (1993) Integrin α7 as substrate for a glycosylphosphatidylinositol-anchored ADP-ribosyltransferase on the surface of skeletal muscle cells. J. Biol. Chem., 268, 25273–25276. [PubMed] [Google Scholar]