Abstract
Mitochondria require NADPH for anti-oxidant protection and for specific biosynthetic pathways. However, the sources of mitochondrial NADPH and the mechanisms of maintaining mitochondrial redox balance are not well understood. We show here that in Saccharomyces cerevisiae, mitochondrial NADPH is largely provided by the product of the POS5 gene. We identified POS5 in a S.cerevisiae genetic screen for hyperoxia-sensitive mutants, or cells that cannot survive in 100% oxygen. POS5 encodes a protein that is homologous to NAD+ and NADH kinases, and we show here that recombinant Pos5p has NADH kinase activity. Pos5p is localized to the mitochondrial matrix of yeast and appears to be important for several NADPH-requiring processes in the mitochondria, including resistance to a broad range of oxidative stress conditions, arginine biosynthesis and mitochondrial iron homeostasis. Pos5p represents the first member of the NAD(H) kinase family that has been identified as an important anti-oxidant factor and key source of the cellular reductant NADPH.
Keywords: iron homeostasis/hyperoxia/mitochondria/NADPH/reactive oxygen species
Introduction
As a consequence of oxidative metabolism, aerobic organisms are continuously bombarded with reactive oxygen species (ROS), such as superoxide (·O2–), hydrogen peroxide (H2O2) and hydroxyl radicals (·OH), which can damage DNA, proteins and lipids. Most of these highly reactive molecules are generated in the mitochondria as by-products from the reduction of O2 to H2O during oxidative phosphorylation (Boveris and Cadenas, 1982). Cells can also be exposed to exogenous sources of oxidative stress such as hyperoxia.
Hyperoxia is a state in which individual cells or organisms are exposed to oxygen concentrations well above that of atmospheric oxygen. In laboratory animals, hyperoxia exposure (>90% O2) leads to death within 3–7 days (O’Reilly, 2001). Specific hyperoxia-induced disorders in humans include bronchopulmonary dysplasia (Northway et al., 1967) and adult respiratory distress syndrome (Halliwell et al., 1992). In spite of the importance of hyperoxia in health-related issues, little is known regarding the mechanisms by which high concentrations of oxygen can cause damage or the defense systems that guard against hyperoxia damage.
All cells possess numerous anti-oxidant defense systems. Included in this list are the superoxide dismutase enzymes, which disproportionate ·O2– into H2O2 and O2, and peroxidases, which catalyze the reduction of hydroperoxides. Non-enzymatic defense systems include the tripeptide glutathione (GSH) and the small protein thioredoxin (TRX), which can serve either as reductants themselves or as cofactors for anti-oxidant enzymes such as GSH peroxidases, glutaredoxins, TRX peroxidases and methionine sulfoxide reductases (Jamieson, 1998; Carmel-Harel and Storz, 2000; Weissbach et al., 2002). All of these enzymes rely on the reduced forms of GSH and TRX, which are regenerated through the action of NADPH-requiring GSH and TRX reductases. As such, NADPH lies at the heart of many anti-oxidant defenses of the cell.
In the cytosol, NADPH is provided primarily by enzymes in the pentose phosphate pathway, including glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase, which catalyze the reduction of NADP+. Accordingly, these enzymes have been shown to play an important role in protection from oxidative stress in both yeast and mammals (Pandolfi et al., 1995; Juhnke et al., 1996; Slekar et al., 1996). The sources of NADPH in the mitochondria, however, where the bulk of ROS are generated, are less clear. In mammalian cells, mitochondrial NADP+-dependent isocitrate dehydrogenase (NADP-IDHm) has been reported to be an important source of mitochondrial NADPH (Jo et al., 2001). However, a deletion of the corresponding gene in the bakers’ yeast Saccharomyces cerevisiae (IDP1) had no effect on oxidative stress sensitivity or cell growth (Minard et al., 1998). Therefore, NADP-IDHm cannot be the only means by which eukaryotic mitochondria produce NADPH. The other sources have not yet been identified.
In this study, we provide evidence that in bakers’ yeast, the POS5 gene product is a major source of mitochondrial NADPH. Saccharomyces cerevisiae POS5 was identified in a screen for yeast genes that protect against hyperoxia damage. By sequence analysis, the POS5 gene encodes a member of the NAD(H) kinase family. We demonstrate that Pos5p has NADH kinase activity and localizes to the yeast mitochondrial matrix, where it appears to provide the NADPH needed for oxidative stress protection and for specific mitochondrial biogenesis reactions. This is the first demonstration of an NAD(H) kinase acting as a key source of NADPH.
Results
The pos5Δ mutant is sensitive to several types of oxidative stress
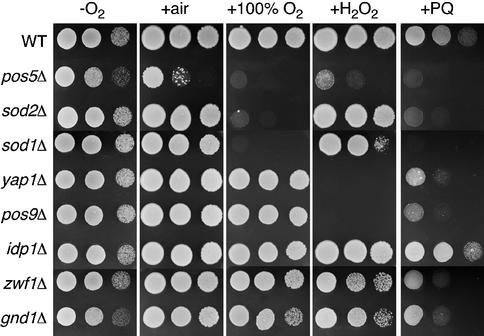
In order to identify anti-oxidant factors that provide protection against hyperoxia-related damage, we developed a genetic screen for yeast mutants that are sensitive to high oxygen conditions. The Research Genetics BY4741 haploid knockout collection was screened for mutants that fail to grow under hyperoxia (100% O2) conditions, but grow well in an oxygen-depleted environment. One of the hyperoxia-sensitive mutants identified in this screen was pos5Δ. To confirm that POS5 alone was responsible, we engineered a pos5Δ gene deletion in strain BY4741 and found that the resultant mutant is likewise hyperoxia sensitive (Figure 1). Along with high oxygen, the pos5Δ mutant is also sensitive to H2O2 (as has been shown previously; Krems et al., 1995) and paraquat, a superoxide-generating agent (Figure 1). Two other mutants isolated from the hyperoxia sensitivity screen were sod1Δ and sod2Δ, encoding the two superoxide dismutases in yeast. As shown in Figure 1, the sod mutants show sensitivity to hyperoxia and paraquat, but are not markedly sensitive to H2O2. We also tested deletion mutants for the two principal oxidative stress transcription factors in yeast, Yap1p and Pos9p/Skn7p, which control induction of the oxidative stress response (Lee et al., 1999). These mutants show hypersensitivity to H2O2 and paraquat, but not to hyperoxia. The strong sensitivity of pos5Δ mutants to all three oxidative stress conditions appears to be unique.
Fig. 1. Comparison of oxidative stress sensitivity of pos5Δ and other mutants with anti-oxidant function. The indicated yeast strains were tested for growth by plating 5 µl of solution at 2.0, 0.2 and 0.02 OD600 units onto YPD plates. –O2 = growth in anaerobic culture jars; +air = aerobic growth; +100% O2 = growth in chambers flushed with 100% O2; +H2O2 = aerobic growth on YPD plates containing 2 mM H2O2; +PQ = aerobic growth on YPD plates containing 1 mM paraquat. Strains utilized: wild-type, BY4741; pos5Δ, CO205; sod1Δ, LJ284; sod2Δ, yap1Δ, pos9Δ, idp1Δ, zwf1Δ and gnd1Δ were obtained from ResGen as kanMX4 deletions in parental strain BY4741.
Pos5p is a mitochondrial matrix protein required for proper mitochondrial function
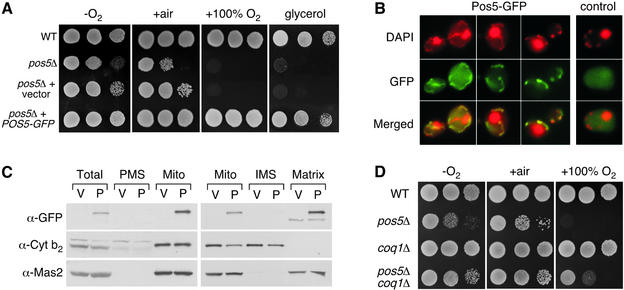
The function of S.cerevisiae Pos5p has not been determined previously. However, the mutant grows poorly on glycerol (Figure 2A), as has been reported previously (Dimmer et al., 2002), suggesting a role in mitochondrial function. In order to determine the subcellular localization of Pos5p, a Pos5–green fluorescent protein (GFP) expression plasmid was constructed with GFP fused to the C- terminus of Pos5p. This fusion protein, under the control of the POS5 promoter, is functional since the plasmid fully complements both the hyperoxia sensitivity and glycerol growth defects of the pos5Δ strain (Figure 2A). The fact that the plasmid complements the slow growth on glycerol of pos5Δ mutants suggests that the apparent mitochondrial defect does not result from secondary mitochondrial DNA mutations. However, when the experiment is repeated with older stocks of pos5Δ mutants that have undergone several generations, only the hyperoxia, but not poor growth on glycerol, is complemented by episomal POS5 (data not shown). This suggests that the pos5Δ yeast mutant does have the propensity to accumulate mitochondrial DNA mutations over time.
Fig. 2. Pos5p is a mitochondrial matrix protein that is needed for proper mitochondrial function. (A) The indicated strains were tested for growth by plating cell dilutions as in Figure 1 onto YPD (–O2, +air, +100% O2) or YPG (glycerol) plates in air. –O2, +air, +100% O2 = same as in Figure 1. Strains utilized: wild-type, BY4741; pos5Δ, CO205; pos5Δ + vector, strain CO205 transformed with vector pAA1; pos5Δ + POS5-GFP, strain CO205 transformed with plasmid pCO101. (B) Strain BY4741 transformed with pCO101 (Pos5–GFP) or pAA1 (control) was prepared for fluorescence microscopy as described in Materials and methods. Merged = merged images of DAPI and GFP fluorescence. (C) Cell lysates were prepared from strain BY4741 transformed with the Pos5–GFP plasmid pCO101 (P) or the vector control pAA1 (V), and 140 µg of total cell protein was fractionated into post-mitochondrial supernatant (PMS) and crude mitochondria (Mito) (left). Where indicated, mitochondria (25 µg of protein) were fractionated further into intermembrane space (IMS) and matrix components (right). All samples were subject to SDS–PAGE and immunoblotting using antibodies directed against either GFP (marking Pos5p), cytochrome b2 (mitochondrial IMS) or Mas2 (mitochondrial matrix). (D) The indicated strains were tested for growth as in (A). Strains utilized: wild-type, BY4741; pos5Δ, BY4741 pos5Δ::kanMX4; coq1Δ, CO217; pos5Δ coq1Δ, CO200.
Cells expressing Pos5–GFP were stained with 4′,6-diamidino-2-phenylindole (DAPI) and examined by fluorescence microscopy. DAPI staining for DNA highlights both the mitochondria (string-like structures) and the nucleus (large, rounded structure) as exemplified in Figure 2B. Pos5–GFP specifically co-localized with DAPI staining of the mitochondrial, but not nuclear DNA, indicating that Pos5p is localized to the mitochondria (Figure 2B). To confirm this result, cellular fractionation experiments were conducted in which crude mitochondria were isolated from cells expressing Pos5–GFP, followed by western blotting with an anti-GFP antibody. As seen in Figure 2C, Pos5–GFP co-localized with mitochondria and was not detected in the post-mitochondrial supernatant (PMS), largely cytosolic fraction. Furthermore, additional separation of the mitochondria into intermembrane space (IMS) and matrix fractions indicated that Pos5p is targeted specifically to the mitochondrial matrix as determined by co-localization with the mitochondrial processing protease Mas2p (Figure 2C).
Deletion of COQ1 partially rescues the hyperoxia sensitivity phenotype of pos5Δ strains
Since Pos5p is targeted to the mitochondria, we wanted to determine whether the respiratory chain may be contributing to the hyperoxia sensitivity of the pos5Δ mutant. Two sites in the respiratory chain are known to contribute to ROS production: the NADH dehydrogenase and the ubi-semiquinone anion derivative of co-enzyme Q (Boveris et al., 1976; Turrens and Boveris, 1980; Turrens et al., 1985; Fang and Beattie, 2003). Saccharomyces cerevisiae expresses three mitochondrial NADH dehydrogenases (encoded by NDE1, NDE2 and NDI1; De Vries et al., 1992; Luttik et al., 1998). Although these are single subunit enzymes that do not form complex I, they still contain a flavin that can reduce oxygen (Fang and Beattie, 2003). However, single and combined deletions in these genes did not suppress the hyperoxia sensitivity of a pos5Δ mutant (data not shown). By comparison, a deletion in COQ1 affecting co-enzyme Q synthesis partially suppressed the hyperoxia sensitivity of pos5Δ mutants (Figure 2D), suggesting that the ubi-semiquinone anion of the respiratory chain may help contribute to high oxygen damage. Together with our mitochondrial localization of the protein, it appears that Pos5p functions in the mitochondria to protect against oxidative damage derived from component(s) in the respiratory chain.
Pos5p is an NADH-specific kinase
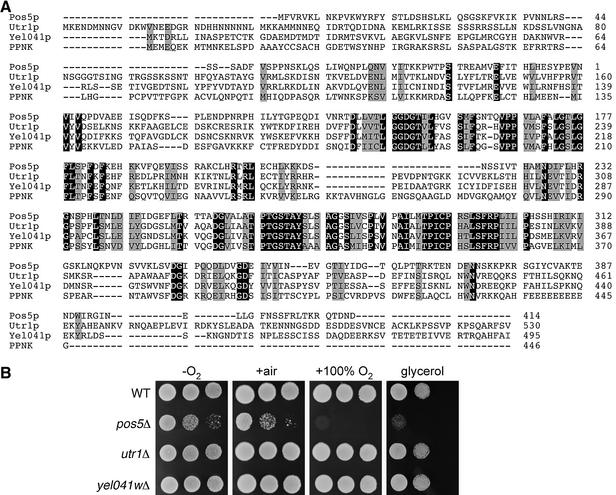
The amino acid sequence of Pos5p is shown in Figure 3A. Pos5p is predicted to be a 46.3 kDa protein with homology to NAD+ and NADH kinases, which are enzymes that catalyze the phosphorylation of NAD+ or NADH, respectively. This family of enzymes is present in organisms from bacteria to humans, including Escherichia coli, Caenorhabditis elegans, Drosophila melanogaster, plants and mice. In S.cerevisiae, NAD+ kinase activity has been reported in the cytosol, mitochondria and microsomes, while NADH kinase activity is found solely in the mitochondria (Bernofsky and Utter, 1968; Apps, 1970; Griffiths and Bernofsky, 1972; Iwahashi et al., 1989). Pos5p has two other homologs identified in the yeast genome, namely Utr1p and Yel041p. Utr1p has recently been identified as an NAD+ kinase (Kawai et al., 2001); however, the function of Yel041p is unknown. An amino acid sequence comparison of Pos5p, Utr1p and Yel041p with their human homolog PPNK [also known as FLJ13052 (accession No. NP_075394), encoding an NAD+ kinase (Lerner et al., 2001)] is given in Figure 3A. Despite the strong sequence similarities between Pos5p and its yeast homologs, only Pos5p is required for protection from hyperoxia and for proper respiratory function. As shown in Figure 3B, deletion of UTR1 or YEL041W does not result in hypersensitivity to high O2 or growth defects on a non-fermentable carbon source.
Fig. 3. Pos5p yeast homologs are not required for protection from hyperoxia or growth on a non-fermentable carbon source. (A) The amino acid sequences of S.cerevisiae Pos5p, Utr1p and Yel041p and human PPNK (accession No. NP_075394) were aligned using Clustal_W 1.5. Identical residues are highlighted in black, and similar residues are outlined in gray. (B) The indicated strains were tested for growth as in Figures 1 and 2A. Strains utilized: wild-type, BY4741; pos5Δ, CO205; utr1Δ, BY4742 utr1Δ::kanMX4; yel041wΔ, BY4741 yel041wΔ::kanMX4.
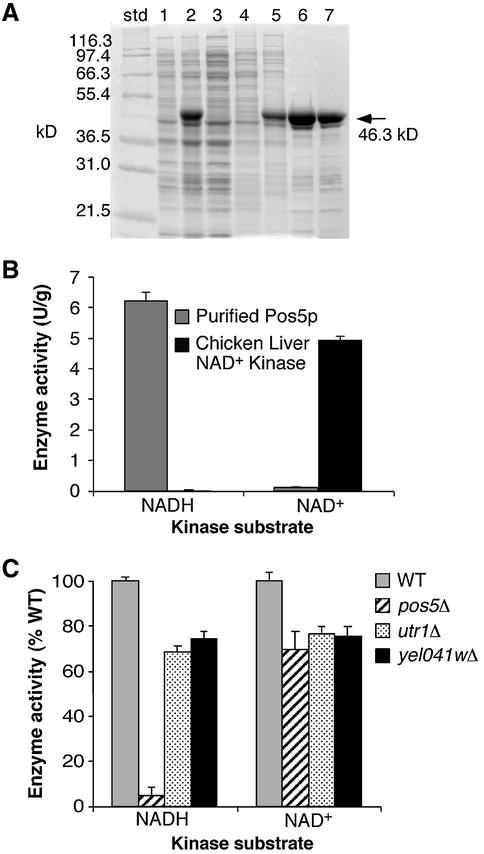
In order to determine if Pos5p has NAD(H) kinase activity, the recombinant protein was overexpressed and purified from E.coli (Figure 4A). The protein was tested for both NAD+ and NADH kinase activity (see Materials and methods) using ATP as a phosphate source. The results, shown in Figure 4B, indicate that recombinant Pos5p is an NADH kinase. The recombinant enzyme also exhibits weak NAD+ kinase activity; however, this activity is ∼50-fold lower than the NADH kinase activity. In comparison, chicken liver NAD+ kinase has the opposite activity profile, with NAD+ kinase activity ∼150-fold higher than NADH kinase activity (Figure 4B). These results demonstrate that Pos5p can phosphorylate NADH using ATP as a phosphate donor and is therefore predicted to catalyze the production of NADPH within yeast mitochondria.
Fig. 4. Recombinant Pos5p is an NADH kinase. (A) SDS– polyacrylamide gel from recombinant Pos5p purification procedures. std, molecular weight standards; lane 1, uninduced cells; lane 2, induced cells; lane 3, sonication supernatant; lane 4, MES/urea extract; lane 5, Tris/urea extract; lane 6, DEAE flow-through; lane 7, 15 µg of refolded Pos5p (see Materials and methods). (B) and (C) NADH and NAD+ kinase activities were assayed as described in Materials and methods using either (B) purified recombinant Pos5p (left hand histogram for both NADH and NAD+) and chicken liver NAD+ kinase (Sigma; right hand histogram for both NADH and NAD+) or (C) crude mitochondria obtained from cells grown to mid-log phase in YPD. Results are presented either in the form of enzyme units (B) where one unit = 1.0 µmol NADPH or NADP+ produced per min, or as a percentage of the total activity of wild-type mitochondria (C). Yeast strains utilized in (C) are the same as in Figure 3B. The reported values in (B) and (C) are the mean of three independent experiments; error bars are standard deviations.
NADH and NAD+ kinase assays were also performed on mitochondrial extracts from various yeast strains. As shown in Figure 4C, mitochondrial NADH kinase activity was greatly reduced in the pos5Δ mutant, while NAD+ kinase activity was largely unaffected. Furthermore, utr1Δ and yel041wΔ gene deletions had little effect on mitochondrial NADH and NAD+ kinase activities. These results indicate that of the three NAD(H) kinase homologs in yeast, Pos5p is the primary NADH kinase in the mitochondria.
Comparison of Pos5p with other potential sources of NADPH
There are a number of enzymes that have been predicted to contribute to cellular production of NADPH in yeast, including mitochondrial NADP+-specific isocitrate dehydrogenase (Idp1p) (Haselbeck and McAlister-Henn, 1991). However, an idp1Δ yeast mutant is not sensitive to hyperoxia, paraquat or H2O2 (Figure 1; Minard et al., 1998). Two other NADPH sources are ZWF1 and GND1, which encode cytosolic enzymes that produce NADPH as part of the pentose phosphate pathway (Juhnke et al., 1996). Deletion of either of these genes also did not produce strong hyperoxia sensitivity, and effects on paraquat and H2O2 resistance were mild in comparison with pos5Δ mutations (Figure 1). Therefore, of all the known potential sources of cellular NADPH, Pos5p seems the most crucial for protection against oxidative damage in yeast.
Mitochondrial iron defects in the pos5Δ mutant
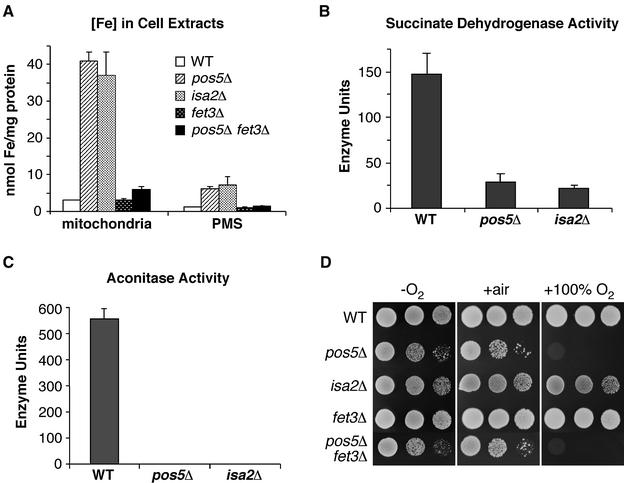
Since defects in metal homeostasis have been linked to oxidative stress (Jamieson, 1998), we examined metal levels in the cytosol and crude mitochondria of the pos5Δ mutant. Surprisingly, the mitochondrial fraction of pos5Δ shows a dramatic accumulation of iron relative to wild-type cells, with an ∼14-fold increase (Figure 5A). The iron levels were also elevated in the PMS fraction, with an ∼6-fold increase. In contrast, the levels of manganese and copper were not considerably affected in pos5Δ cells (data not shown). Since hyperaccumulation of iron in the mitochondria is a hallmark of defects in iron–sulfur cluster biogenesis (for a review see Muhlenhoff and Lill, 2000), we examined the activity of mitochondrial Fe–S enzymes in the pos5Δ mutant. Succinate dehydrogenase (SDH) and aconitase activities were greatly reduced in the pos5Δ mutant relative to wild-type (Figure 5B and C). In fact, the aconitase activity was virtually undetectable under these assay conditions (Figure 5C). Addition of iron to the growth medium did not rescue this defect (data not shown). It is noteworthy that the pos5Δ defects in Fe–S enzyme activity and iron accumulation are comparable to that seen with a S.cerevisiae isa2Δ mutant, known to be defective in iron–sulfur cluster biogenesis (Figure 5A–C) (Jensen and Culotta, 2000; Pelzer et al., 2000).
Fig. 5. Mitochondrial iron homeostasis defects in pos5Δ. (A) The indicated strains were grown to mid-log phase and the iron content measured in mitochondria and post-mitochondrial supernatant fractions by atomic absorption spectroscopy. (B) and (C) Cell lysates were prepared for the indicated yeast strains and tested for succinate dehydrogenase (B) and aconitase (C) activity. One unit of enzyme activity is equivalent to 1.0 nmol of substrate converted/min/mg protein. Substrates were dichlorophenol indophenol (SDH) and cis-aconitate (aconitase). In (A–C), the reported values are the mean of three independent experiments; error bars are standard deviations. (D) The indicated strains were tested for growth as in Figure 2A. Strains utilized: wild-type, BY4741; pos5Δ, CO205; isa2Δ, LJ102; fet3Δ, BY4741 fet3Δ::kanMX4; pos5Δ fet3Δ, CO207.
We addressed whether the pos5Δ defects in iron homeostasis contribute to the hyperoxia sensitivity of this mutant. First, the elevated iron of pos5Δ mutants was reduced by deleting the FET3 gene needed for high affinity iron uptake (Figure 5A). This reduction in mitochondrial iron levels, however, did not reverse the hyperoxia sensitivity of pos5Δ mutants (Figure 5D). Secondly, we noted that the isa2Δ mutant, which accumulates similarly high levels of mitochondrial iron (Figure 5A), is not hyperoxia sensitive (Figure 5D). Evidently, the defects in mitochondrial iron homeostasis of pos5Δ mutants are not connected to the hyperoxia sensitivity of this strain and appear to represent a separate outcome of mitochondrial NADPH depletion (see Discussion).
The requirements for mitochondrial and cytosolic NADPH in amino acid biosynthesis
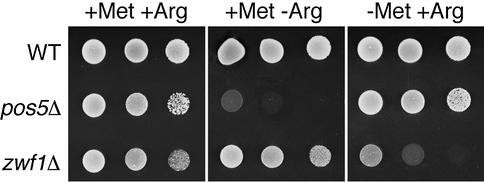
While optimizing growth conditions for the pos5Δ mutant, we discovered that it is auxotrophic for arginine (Figure 6). This observation may be explained by an NADPH-requiring step in arginine biosynthesis that takes place in the mitochondria, specifically the third step in the conversion of glutamate to arginine catalyzed by N-acetylglutamyl phosphate reductase (Jauniaux et al., 1978). While mitochondrial NADPH is required for arginine biosynthesis, cytosolic NADPH is needed for methionine biosynthesis (Thomas et al., 1991; Slekar et al., 1996), and low cytosolic NADPH caused by deletion of ZWF1 results in methionine auxotrophy (Figure 6). The mitochondrial and cytosolic pools of NADPH are kept quite separate, because pos5Δ mutants exhibit no defect in methionine biosynthesis and zwf1Δ mutants are not auxotrophic for arginine. This result is consistent with the fact that the mitochondrial inner membrane is impermeable to pyridine nucleotides, prohibiting exchange of these molecules with the cytosol (von Jagow and Klingenberg, 1970).
Fig. 6. Amino acid auxotrophies of pos5Δ and zwf1Δ strains. The indicated strains were tested for growth by spotting 10 µl of solution at 2.0, 0.2 and 0.02 OD600 units onto complete SD plates with arginine, or SD plates lacking methionine or arginine. Strains utilized: wild- type, BY4742; pos5Δ, BY4742 pos5Δ::kanMX4; zwf1Δ, BY4742 zwf1Δ::kanMX4.
Discussion
Due to the constant generation of respiratory chain ROS, the mitochondria may be particularly prone to oxidative damage. This organelle has therefore evolved with a set of anti-oxidant defense factors that are distinct from that of the cytosol, including a separate system for the generation of the key reductant, NADPH. Until recently, the mito chondrial source for NADPH has been poorly understood. We provide strong evidence herein that in bakers’ yeast, mitochondrial NADPH is generated through the action of an NADH kinase encoded by the POS5 gene.
Mitochondrial NADPH and oxidative stress
In contrast to other anti-oxidant factors in yeast (e.g. superoxide dismutases and oxidative stress transcriptional regulators), Pos5p provides protection from a wide range of oxidative stress insults, including hyperoxia, hydrogen peroxide and superoxide-generating agents. The effect of pos5Δ mutations is consistent with the widespread requirement for NADPH in anti-oxidant defense systems. Both TRX and GSH are maintained in their reduced state through the action of NADPH-requiring reductases. TRX, in turn, serves as cofactor for thioredoxin peroxidases (peroxiredoxins) and methionine sulfoxide reductases. GSH itself can act as a reductant, but is also the cofactor for GSH peroxidases and glutaredoxins (Jamieson, 1998). Yeast express mitochondrial forms of thioredoxin reductase (Trr2p), thioredoxin (Trx3p) and peroxiredoxin (Prx1p) (Pedrajas et al., 1999, 2000), as well as two mitochondrial glutaredoxins, Grx2p and Grx5p (Pedrajas et al., 2002; Rodriguez-Manzaneque et al., 2002). Although the subcellular localization of GSH reductase has not yet been determined for yeast, oxidized glutathione (GSSG) will not cross the mitochondrial membrane (Olafsdottir and Reed, 1988), which necessitates a mitochondrial form of GSH reductase, as has been identified in mammalian cells (Mbemba et al., 1985; Taniguchi et al., 1986). In any case, the numerous GSH- and TRX-requiring systems in the mitochondria may account for the widespread impact of pos5 mutations on oxidative stress resistance.
Mitochondrial NADPH and iron homeostasis
In addition to a strong sensitivity towards oxidative stress, we find that pos5Δ mutants also exhibit defects in iron homeostasis that are suggestive of a deficiency in Fe–S cluster biogenesis. As with all known mutants for Fe–S cluster synthesis (Muhlenhoff and Lill, 2000), pos5Δ cells accumulate high mitochondrial iron and are defective in mitochondrial Fe–S cluster-containing enzymes. Why would NADPH be required for Fe–S cluster biogenesis? As one possibility, NADPH may be required indirectly (via GSH) to maintain the activity of the mitochondrial Grx5p glutaredoxin, which is known to be required for Fe–S cluster assembly (Rodriguez-Manzaneque et al., 2002). Another possibility is that NADPH may be needed for the function of S.cerevisiae Arh1p, an adrenodoxin reductase homolog that can utilize either NADH or NADPH and is needed for Fe–S cluster assembly (Lacour et al., 1998; Li et al., 2001; Muhlenhoff et al., 2002). The possibility also exists that the pos5Δ defect in iron homeostasis reflects an unknown requirement for NADPH in mitochondrial iron metabolism. Finally, we cannot rule out the alternative theory that the loss of Fe–S enzyme activity is not due solely to changes in iron homeostasis, but to elevated levels of mitochondrial ROS in pos5Δ mutants that could directly damage the Fe–S cluster proteins. In any case, our studies show that this defect in Fe–S enzyme activity is not the cause of pos5Δ hyperoxia sensitivity. Clearly, separate NADPH-requiring processes are involved.
Cytosolic versus mitochondrial NADPH
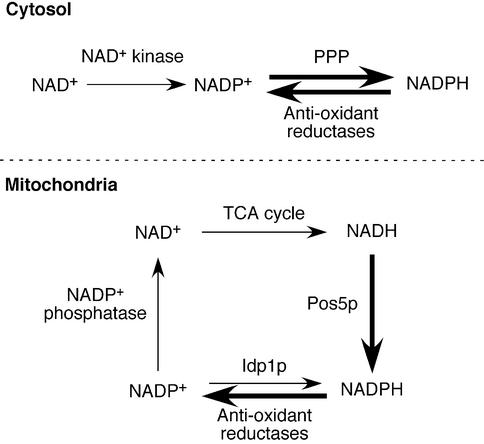
Based on our results, we propose a model depicting two different pathways for the production of NADPH in the cytosol versus the mitochondria of yeast (Figure 7). In the cytosol, NADP+ is converted to NADPH by the pentose phosphate pathway. However, in the mitochondria of S.cerevisiae, NADH, and not NADP+, serves as the substrate for NADPH generation, and this is made possible through the NADH kinase activity of Pos5p. At first glance, it would appear that this method of NADPH generation would deplete mitochondrial pools of NADH. However, NADP+ phosphatase activity has been detected in yeast mitochondria (Bernofsky and Utter, 1968), providing a means of recycling NAD+ back into the citric acid cycle (Figure 7).
Fig. 7. Proposed model for the generation of NADPH in the cytoplasm and mitochondria of yeast. In the cytosol, NAD+ may be phosphorylated to NADP+ by an NAD+ kinase (e.g. S.cerevisiae Utr1p). The resultant NADP+ is then converted to NADPH by enzymes of the pentose phosphate pathway (PPP). NADPH-requiring reductases (e.g. TRX and GSH reductases) can then use NADPH as a cofactor, thereby regenerating NADP+ for the PPP. In the mitochondria, a different pathway exists for the primary generation of NADPH. The NADH generated in the tricarboxylic acid (TCA) cycle apparently can be phosphorylated by the NADH kinase Pos5p, generating NADPH. Following consumption of NADPH via anti-oxidant reductases, an NADP+ phosphatase presumably regenerates NAD+ for recycling back into the TCA cycle. NADP+-IDHm (Idp1p) may also contribute to NADPH regeneration, although to a minimal extent in S.cerevisiae. Arrows shown in bold indicate enzymatic reactions that are critical for oxidative stress resistance.
In the case of mammals, an NADP+-specific isocitrate dehydrogenase has been reported to be a key source of mitochondrial NADPH (Jo et al., 2001). Although it remains possible that mammalian mitochondria also employ an NADH kinase to generate NADPH, no such enzyme has been identified to date. At this point, it is not clear why yeast mitochondria would employ NADH preferentially over NADP+ for the generation of NADPH. This may reflect the oxidation state of NADH. Although mammalian cells are reported to have 10–20% of their total cellular NAD(H) in the reduced form, in yeast cells ∼50% is in the reduced form, and this may also be true of mitochondrial pools (Jacobson and Jacobson, 1976; Ting et al., 1977). Such availability of reduced NADH could help explain the utilization of an NADH kinase for generating NADPH.
Mitochondria and hyperoxia
Finally, our findings highlight the important role that mitochondria play in protection from hyperoxia toxicity. Although a high oxygen level has long been known to cause cellular damage, the pro-oxidant and anti-oxidant mechanisms at play have been poorly understood. Two major mitochondrial anti-oxidant factors, Pos5p and Sod2p, are absolutely required for hyperoxia resistance (Figure 1; and van Loon et al., 1986; Gralla and Kosman, 1992). Sod1p was also found to be important (Gralla and Kosman, 1992), and although this enzyme is largely cytosolic, a fraction of it is known to reside in mitochondria as well (Sturtz et al., 2001). Moreover, a deletion of COQ1 needed for mitochondrial respiration partially alleviated the hyperoxia sensitivity of a pos5Δ mutant, directly implicating the respiratory chain in hyperoxia toxicity. This result is consistent with the observation that hyperoxia conditions increase the rate of ROS generation in mitochondria compared with aerobic conditions (Boveris and Chance, 1973). It is likely that the mitochondria play a key role in hyperoxia damage in mammalian cells as well.
Materials and methods
Yeast strains, media and growth conditions
Yeast strains used in this study were derived from the parental strains BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) (Brachmann et al., 1998). The BY4741 knockout library, a collection of yeast deletion strains created with the kanMX4 cassette, was obtained from Research Genetics (ResGen). Details of this library can be found at ftp://ftp.resgen.com/pub/deletions/mat_a_041902.txt. Construction of LJ102 (isa2Δ::HIS3) was described previously (Jensen and Culotta, 2000). The strain LJ284 (sod1Δ::LEU2) was constructed by introducing the SOD1 gene replacement plasmid pKS1 (Culotta et al., 1995) into the strain BY4741. Strains CO205 (pos5Δ::URA3) and CO207 (pos5Δ::URA3 fet3Δ) were constructed by deleting the POS5 gene of strains BY4741 and BY4741 fet3Δ::kanMX4, respectively, using the POS5 replacement plasmid pCO105. Strains CO217 (coq1Δ::LEU2) and CO200 (pos5Δ coq1Δ::LEU2) were obtained by disruption of the COQ1 gene in the strains BY4741 and BY4741 pos5Δ::kanMX4, respectively, with the coq1Δ::LEU2 plasmid pSG108 (S.Garland, unpublished data). Yeast transformations were performed by electroporation (Becker and Guarente, 1991) or the lithium acetate procedure (Gietz and Schiestl, 1991). All gene deletions, including those from the original ResGen collection, were verified by PCR analysis.
Strains were maintained at 30°C on either enriched yeast extract– peptone-based medium supplemented with 2% glucose (YPD) or 3% glycerol (YPG), or on minimal synthetic defined medium (SD) supplemented with the appropriate amino acids (Sherman et al., 1978). Anaerobic cultures were maintained by growth in an O2-depleted culture jar (BBL Gas Pak) (Liu et al., 1992). For growth in 100% O2, cultures were placed in a modular incubator chamber (Billups-Rothenburg, Inc., Del Mar, CA) and flushed with 100% O2 for 10–30 min before sealing.
Plasmids
To construct the POS5 deletion plasmid pCO105, POS5 sequences from –902 to +71 and +736 to +1293 were amplified by PCR using primers that introduced an EcoRI site at –893, a BamHI site at +64 and a SalI site at +726. The PCR products were digested at these sites and at an internal EcoRI site at +1281, and ligated in a trimolecular reaction to the SalI and BamHI sites of the URA3 integrating vector pRS306 (Sikorski and Hieter, 1989). The resulting plasmid, pCO105, was linearized with EcoRI and used to delete the chromosomal POS5 gene sequences +72 to +735. The Pos5–GFP plasmid pCO101, which contains one copy of GFP fused to the C-terminus of Pos5p, was constructed as follows. The POS5 gene was amplified from –894 to the stop codon with primers that introduced a PstI site at –888 and a NotI site at +1243. The PCR fragment was digested with these enzymes and inserted into the PstI and NotI sites of pAA1 (LEU2 CEN), producing an in-frame fusion with the GFP sequence already present in this vector (Hobbs et al., 2001). To create pCO100, a bacterial overexpression vector for recombinant Pos5p, the POS5 gene was amplified using primers that introduced an NdeI site at the start codon, and a SalI site 16 bp after the stop codon. The fragment was digested with these enzymes and inserted into the NdeI and SalI sites of pET21a (Novagen). The sequence integrity of all plasmids was confirmed by double-stranded DNA sequencing (Synthesis and Sequencing Facility, Johns Hopkins University School of Medicine). Upon sequencing, a polymorphism was detected in POS5 that changed amino acid 180 from serine to leucine (TCA→TTA). The POS5 gene with this single amino acid change fully complemented the hyperoxia sensitivity, arginine auxotrophy and respiratory deficiency phenotypes of the pos5Δ strain.
Hyperoxia sensitivity screen
The fifty-three 96-well plates comprising the ResGen MATa haploid knock-out collection were completely thawed and 3 µl of cells from each well were spotted onto four sets of YPD plates. Two sets were placed in an anaerobic chamber while two sets were placed in a chamber flushed with 100% oxygen. After incubation at 30°C for 48 h, growth under the anaerobic and hyperoxic conditions was compared in order to identify deletion strains that are hyperoxia sensitive.
Fluorescence and immunodetection techniques
For fluorescence studies, the BY4741 parental strain transformed with pCO101 (Pos5–GFP CEN plasmid) or pAA1 (vector control) were grown aerobically to an OD600 of 2.0–2.5 in selecting SD medium. Cells were fixed with formaldehyde then the DNA was stained by incubation with 1 µg/ml DAPI for 5 min. GFP and DAPI were monitored by fluorescence microscopy on a Zeiss Axiovert 135TV microscope (Microscopy Facility, Johns Hopkins Medical Institutions) at a magnification of 1000×.
For western blot analysis, BY4741 strains transformed with pCO101 (Pos5–GFP) or pAA1 (vector control) were grown aerobically to an OD600 of 1.0 in selecting SD medium with 2% galactose. To obtain mitochondrial and PMS fractions, cells were converted to spheroplasts prior to gentle lysis by Dounce homogenization as described previously (Daum et al., 1982; Jensen and Culotta, 2000). Mitochondrial intermembrane and matrix fractions were prepared as previously described by resuspending the isolated mitochondria in a hypotonic buffer (Jensen and Culotta, 2000). All samples were separated by electrophoresis on a 12% SDS–polyacrylamide gel and analyzed by western blotting using an anti-GFP antibody (Molecular Probes) diluted to 1:5000 and a secondary anti-rabbit IgG (Amersham) diluted to 1:12 500. Mitochondrial fractions were monitored by using antibodies (diluted to 1:10 000) directed against cytochrome b2 in the intermembrane space and Mas2 in the matrix (kind gifts of R.Jensen). Detection employed the enhanced chemiluminescence (ECL) kit (Amersham) used according to the manufacturer’s specifications. Protein concentrations were determined using the Bradford method (Bio-Rad) with bovine serum albumin (BSA) as the calibration standard.
Recombinant Pos5p purification
For production of recombinant Pos5p, the bacterial overexpression plasmid pCO100 was transformed into the E.coli strain BL21(DE3) (Novagen). The transformants were grown in 8 l of LB media with shaking at 37°C and induced with 400 µM isopropyl-β-d-thiogalacto pyranoside when OD600 = 0.6. The cells were harvested by centrifugation 2.5 h after induction and then stored at –80°C. A portion of this frozen cell paste (equal to ∼400 ml of cell culture) was thawed and sonicated in 20 ml of 20 mM Tris–HCl pH 8.0. After centrifugation and an additional wash in the sonication buffer, the cell pellet was then extracted twice in 20 ml of MES extraction buffer (20 mM MES-Na, pH 6.0, 6 M urea, 2% Triton X-100) in order to remove contaminants. The cell pellet was then resuspended in 10 ml of Tris extraction buffer [20 mM Tris–HCl pH 8.0, 6 M urea, 1 mM EDTA, 1 mM phenylmethylesulfonyl fluoride (PMSF), 50 mM dithiothreitol (DTT)] and incubated on ice for 1.5–2 h. After centrifugation, the supernatant containing the solubilized protein was loaded onto a DEAE–cellulose (Whatman) column equilibrated with 20 mM Tris–HCl pH 8.0, 6 M urea, 1 mM EDTA and 5 mM DTT. Recombinant Pos5p did not bind to this column and was collected in the flow-through fraction (∼15 ml). The protein solution was concentrated 5-fold to 1 mg/ml and dialyzed in 500 ml of 20 mM MES-Na pH 6.0. After removal of precipitated protein by centrifugation, the solubilized, refolded protein was concentrated to ∼1–2 mg/ml and stored at –80°C. A 400 ml cell culture typically yielded 0.5 mg of purified protein. Despite the addition of protease inhibitors, ∼10–20% of the purified protein is partially proteolyzed during the purification process as evidenced by the presence of several lower molecular weight bands in the SDS–polyacrylamide gel (see Figure 4A, lane 7). However, the protein appeared to be stable upon freezing, since there was no reduction in activity after storage for 2 months at –80°C.
NAD+ and NADH kinase assays
The two-step procedure for the NAD+ and NADH kinase assays was adapted from previous methods (Griffiths and Bernofsky, 1972; Jacobson and Jacobson, 1976; Iwahashi et al., 1989) with some modifications as follows. For the first step, the reaction mixture included 90 mM Tris–HCl pH 7.8, 2 mM NADH (or NAD+), 3 mM ATP, 10 mM MgCl2, 20 mM sodium acetate, 1 mg/ml BSA and 10–15 µg of purified Pos5p or 5–10 µg of mitochondrial extract in a final volume of 100 µl. Purified chicken liver NAD+ kinase (Sigma) was utilized in a parallel reaction as a control. Reactions were initiated by the addition of enzyme and were allowed to proceed for 20 min at 30°C. The NADH and NAD+ kinase assays were terminated by alkalination and acidification, respectively, followed by neutralization as described previously (Iwahashi et al., 1989).
For the second step in the kinase assays, the amount of NADP+ produced from the first step was determined using a cycling assay system. Each assay mixture contained 100 mM Tris–HCl pH 8.0, 10 mM MgCl2, 3.3 mM glucose-6-phosphate, 0.42 mM 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium (MTT), 1.66 mM phenazine ethosulfate (PES), 0.83 mg/ml BSA and 100 µl of the NADH assay mixture or 50 µl of the NAD+ assay mixture from step 1 to a final volume of 1.0 ml. The reaction was initiated by the addition of 2 U of glucose-6-phosphate dehydrogenase from yeast (ICN Biomedicals) and monitored at 570 nm for 5 min to determine the rate of reduction of MTT. This rate was proportional to the amount of NADP(H) present in solution in step 2. NADPH and NADP+ standards were also subjected to the same treatments throughout the two-step procedure in order to develop a standard curve.
Iron accumulation and Fe–S enzyme analyses
For iron accumulation analyses, yeast strains were grown to mid-log phase in YPD. Mitochondrial and PMS fractions were prepared as described above for the western blot. Iron analysis of these fractions was performed on a Perkin-Elmer AAnalyst 600 graphite furnace atomic absorption spectrometer according to the manufacturer’s specifications. For SDH assays, cells were grown to stationary phase in YPD with 0.6% glucose and fractionated into mitochondria and PMS. Aconitase and SDH assays were conducted essentially as described previously (Strain et al., 1998).
Acknowledgments
Acknowledgements
We would like to thank R.Jensen for the cytochrome b2 and Mas2 antibodies, and plasmid pAA1, and L.Jensen for strains LJ102 and LJ284. This work was supported by the JHU NIEHS center and by NIH grant GM 50016 to V.C.C. C.E.O. was supported by the NIEHS training grant ES 07141 and the NIH post-doctoral fellowship GM 66594.
References
- Apps D.K. (1970) The NAD kinases of Saccharomyces cerevisiae. Eur. J. Biochem., 13, 223–230. [DOI] [PubMed] [Google Scholar]
- Becker D.M. and Guarente,L. (1991) High-efficiency transformation of yeast by electroporation. Methods Enzymol., 194, 182–187. [DOI] [PubMed] [Google Scholar]
- Bernofsky C. and Utter,M.F. (1968) Interconversions of mitochondrial pyridine nucleotides. Science, 159, 1362–1363. [DOI] [PubMed] [Google Scholar]
- Boveris A. and Cadenas,E. (1982) Production of superoxide radicals and hydrogen peroxide in mitochondria. In Oberley,L. (ed.), Superoxide Dismutase. CRC Press, Boca Raton, FL, Vol. II, pp. 15–30.
- Boveris A. and Chance,B. (1973) The mitochondrial generation of hydrogen peroxide. General properties and effect of hyperbaric oxygen. Biochem. J., 134, 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveris A., Cadenas,E. and Stoppani,A.O. (1976) Role of ubiquinone in the mitochondrial generation of hydrogen peroxide. Biochem. J., 156, 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C.B., Davies,A., Cost,G.J., Caputo,E., Li,J., Hieter,P. and Boeke,J.D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast, 14, 115–132. [DOI] [PubMed] [Google Scholar]
- Carmel-Harel O. and Storz,G. (2000) Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol., 54, 439–461. [DOI] [PubMed] [Google Scholar]
- Culotta V.C., Joh,H.D., Lin,S.J., Slekar,K.H. and Strain,J. (1995) A physiological role for Saccharomyces cerevisiae copper/zinc superoxide dismutase in copper buffering. J. Biol. Chem., 270, 29991–29997. [DOI] [PubMed] [Google Scholar]
- Daum G., Bohni,P.C. and Schatz,G. (1982) Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem., 257, 13028–13033. [PubMed] [Google Scholar]
- De Vries S., Van Witzenburg,R., Grivell,L.A. and Marres,C.A. (1992) Primary structure and import pathway of the rotenone-insensitive NADH-ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Eur. J. Biochem., 203, 587–592. [DOI] [PubMed] [Google Scholar]
- Dimmer K.S., Fritz,S., Fuchs,F., Messerschmitt,M., Weinbach,N., Neupert,W. and Westermann,B. (2002) Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell, 13, 847–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J. and Beattie,D.S. (2003) External alternative NADH dehydrogenase of Saccharomyces cerevisiae: a potential source of superoxide. Free Radic. Biol. Med., 34, 478–488. [DOI] [PubMed] [Google Scholar]
- Gietz R.D. and Schiestl,R.H. (1991) Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast, 7, 253–263. [DOI] [PubMed] [Google Scholar]
- Gralla E.B. and Kosman,D.J. (1992) Molecular genetics of superoxide dismutases in yeasts and related fungi. Adv. Genet., 30, 251–319. [DOI] [PubMed] [Google Scholar]
- Griffiths M.M. and Bernofsky,C. (1972) Purification and properties of reduced diphosphopyridine nucleotide kinase from yeast mitochondria. J. Biol. Chem., 247, 1473–1478. [PubMed] [Google Scholar]
- Halliwell B., Gutteridge,J.M. and Cross,C.E. (1992) Free radicals, antioxidants and human disease: where are we now? J. Lab. Clin. Med., 119, 598–620. [PubMed] [Google Scholar]
- Haselbeck R.J. and McAlister-Henn,L. (1991) Isolation, nucleotide sequence and disruption of the Saccharomyces cerevisiae gene encoding mitochondrial NADP(H)-specific isocitrate dehydrogenase. J. Biol. Chem., 266, 2339–2345. [PubMed] [Google Scholar]
- Hobbs A.E., Srinivasan,M., McCaffery,J.M. and Jensen,R.E. (2001) Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J. Cell Biol., 152, 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahashi Y., Hitoshio,A., Tajima,N. and Nakamura,T. (1989) Characterization of NADH kinase from Saccharomyces cerevisiae. J. Biochem., 105, 588–593. [DOI] [PubMed] [Google Scholar]
- Jacobson E.L. and Jacobson,M.K. (1976) Pyridine nucleotide levels as a function of growth in normal and transformed 3T3 cells. Arch. Biochem. Biophys., 175, 627–634. [DOI] [PubMed] [Google Scholar]
- Jamieson D.J. (1998) Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast, 14, 1511–1527. [DOI] [PubMed] [Google Scholar]
- Jauniaux J.C., Urrestarazu,L.A. and Wiame,J.M. (1978) Arginine metabolism in Saccharomyces cerevisiae: subcellular localization of the enzymes. J. Bacteriol., 133, 1096–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L.T. and Culotta,V.C. (2000) Role of Saccharomyces cerevisiae ISA1 and ISA2 in iron homeostasis. Mol. Cell. Biol., 20, 3918–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S.H. et al. (2001) Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J. Biol. Chem., 276, 16168–16176. [DOI] [PubMed] [Google Scholar]
- Juhnke H., Krems,B., Kotter,P. and Entian,K.D. (1996) Mutants that show increased sensitivity to hydrogen peroxide reveal an important role for the pentose phosphate pathway in protection of yeast against oxidative stress. Mol. Gen. Genet., 252, 456–464. [DOI] [PubMed] [Google Scholar]
- Kawai S., Suzuki,S., Mori,S. and Murata,K. (2001) Molecular cloning and identification of UTR1 of a yeast Saccharomyces cerevisiae as a gene encoding an NAD kinase. FEMS Microbiol. Lett., 200, 181–184. [DOI] [PubMed] [Google Scholar]
- Krems B., Charizanis,C. and Entian,K.D. (1995) Mutants of Saccharomyces cerevisiae sensitive to oxidative and osmotic stress. Curr. Genet., 27, 427–434. [DOI] [PubMed] [Google Scholar]
- Lacour T., Achstetter,T. and Dumas,B. (1998) Characterization of recombinant adrenodoxin reductase homologue (Arh1p) from yeast. Implication in in vitro cytochrome p45011β monooxygenase system. J. Biol. Chem., 273, 23984–23992. [DOI] [PubMed] [Google Scholar]
- Lee J., Godon,C., Lagniel,G., Spector,D., Garin,J., Labarre,J. and Toledano,M.B. (1999) Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem., 274, 16040–16046. [DOI] [PubMed] [Google Scholar]
- Lerner F., Niere,M., Ludwig,A. and Ziegler,M. (2001) Structural and functional characterization of human NAD kinase. Biochem. Biophys. Res. Commun., 288, 69–74. [DOI] [PubMed] [Google Scholar]
- Li J., Saxena,S., Pain,D. and Dancis,A. (2001) Adrenodoxin reductase homolog (Arh1p) of yeast mitochondria required for iron homeostasis. J. Biol. Chem., 276, 1503–1509. [DOI] [PubMed] [Google Scholar]
- Liu X.F., Elashvili,I., Gralla,E.B., Valentine,J.S., Lapinskas,P. and Culotta,V.C. (1992) Yeast lacking superoxide dismutase. Isolation of genetic suppressors. J. Biol. Chem., 267, 18298–18302. [PubMed] [Google Scholar]
- Luttik M.A., Overkamp,K.M., Kotter,P., de Vries,S., van Dijken,J.P. and Pronk,J.T. (1998) The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J. Biol. Chem., 273, 24529–24534. [DOI] [PubMed] [Google Scholar]
- Mbemba F., Houbion,A., Raes,M. and Remacle,J. (1985) Subcellular localization and modification with ageing of glutathione, glutathione peroxidase and glutathione reductase activities in human fibroblasts. Biochim. Biophys. Acta, 838, 211–220. [DOI] [PubMed] [Google Scholar]
- Minard K.I., Jennings,G.T., Loftus,T.M., Xuan,D. and McAlister-Henn,L. (1998) Sources of NADPH and expression of mammalian NADP+-specific isocitrate dehydrogenases in Saccharomyces cerevisiae. J. Biol. Chem., 273, 31486–31493. [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U. and Lill,R. (2000) Biogenesis of iron–sulfur proteins in eukaryotes: a novel task of mitochondria that is inherited from bacteria. Biochim. Biophys. Acta, 1459, 370–382. [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U., Richhardt,N., Gerber,J. and Lill,R. (2002) Characterization of iron–sulfur protein assembly in isolated mitochondria. A requirement for ATP, NADH and reduced iron. J. Biol. Chem., 277, 29810–29816. [DOI] [PubMed] [Google Scholar]
- Northway W.H. Jr, Rosan,R.C. and Porter,D.Y. (1967) Pulmonary disease following respiratory therapy of hyaline-membrane disease. Bronchopulmonary dysplasia. N. Engl. J. Med., 276, 357–368. [DOI] [PubMed] [Google Scholar]
- Olafsdottir K. and Reed,D.J. (1988) Retention of oxidized glutathione by isolated rat liver mitochondria during hydroperoxide treatment. Biochim. Biophys. Acta, 964, 377–382. [DOI] [PubMed] [Google Scholar]
- O’Reilly M.A. (2001) DNA damage and cell cycle checkpoints in hyperoxic lung injury: braking to facilitate repair. Am. J. Physiol., 281, L291–L305. [DOI] [PubMed] [Google Scholar]
- Pandolfi P.P., Sonati,F., Rivi,R., Mason,P., Grosveld,F. and Luzzatto,L. (1995) Targeted disruption of the housekeeping gene encoding glucose 6-phosphate dehydrogenase (G6PD): G6PD is dispensable for pentose synthesis but essential for defense against oxidative stress. EMBO J., 14, 5209–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrajas J.R., Kosmidou,E., Miranda-Vizuete,A., Gustafsson,J.A., Wright,A.P. and Spyrou,G. (1999) Identification and functional characterization of a novel mitochondrial thioredoxin system in Saccharomyces cerevisiae. J. Biol. Chem., 274, 6366–6373. [DOI] [PubMed] [Google Scholar]
- Pedrajas J.R., Miranda-Vizuete,A., Javanmardy,N., Gustafsson,J.A. and Spyrou,G. (2000) Mitochondria of Saccharomyces cerevisiae contain one-conserved cysteine type peroxiredoxin with thioredoxin peroxidase activity. J. Biol. Chem., 275, 16296–16301. [DOI] [PubMed] [Google Scholar]
- Pedrajas J.R., Porras,P., Martinez-Galisteo,E., Padilla,C.A., Miranda-Vizuete,A. and Barcena,J.A. (2002) Two isoforms of Saccharomyces cerevisiae glutaredoxin 2 are expressed in vivo and localize to different subcellular compartments. Biochem. J., 364, 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelzer W., Muhlenhoff,U., Diekert,K., Siegmund,K., Kispal,G. and Lill,R. (2000) Mitochondrial Isa2p plays a crucial role in the maturation of cellular iron–sulfur proteins. FEBS Lett., 476, 134–139. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Manzaneque M.T., Tamarit,J., Belli,G., Ros,J. and Herrero,E. (2002) Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell, 13, 1109–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink,G.R. and Lawrence,C.W. (1978) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slekar K.H., Kosman,D.J. and Culotta,V.C. (1996) The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J. Biol. Chem., 271, 28831–28836. [DOI] [PubMed] [Google Scholar]
- Strain J., Lorenz,C.R., Bode,J., Garland,S., Smolen,G.A., Ta,D.T., Vickery,L.E. and Culotta,V.C. (1998) Suppressors of superoxide dismutase (SOD1) deficiency in Saccharomyces cerevisiae. Identification of proteins predicted to mediate iron–sulfur cluster assembly. J. Biol. Chem., 273, 31138–31144. [DOI] [PubMed] [Google Scholar]
- Sturtz L.A., Diekert,K., Jensen,L.T., Lill,R. and Culotta,V.C. (2001) A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J. Biol. Chem., 276, 38084–38089. [DOI] [PubMed] [Google Scholar]
- Taniguchi M., Hara,T. and Honda,H. (1986) Similarities between rat liver mitochondrial and cytosolic glutathione reductases and their apoenzyme accumulation in riboflavin deficiency. Biochem. Int., 13, 447–454. [PubMed] [Google Scholar]
- Thomas D., Cherest,H. and Surdin-Kerjan,Y. (1991) Identification of the structural gene for glucose-6-phosphate dehydrogenase in yeast. Inactivation leads to a nutritional requirement for organic sulfur. EMBO J., 10, 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting H.Y., Jacobson,L. and Jacobson,M.K. (1977) Regulation of nicotinamide adenine dinucleotide phosphate levels in yeast. Arch. Biochem. Biophys., 183, 98–104. [DOI] [PubMed] [Google Scholar]
- Turrens J.F. and Boveris,A. (1980) Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem. J., 191, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrens J.F., Alexandre,A. and Lehninger,A.L. (1985) Ubisemiquinone is the electron donor for superoxide formation by complex III of heart mitochondria. Arch. Biochem. Biophys., 237, 408–414. [DOI] [PubMed] [Google Scholar]
- van Loon A.P., Pesold-Hurt,B. and Schatz,G. (1986) A yeast mutant lacking mitochondrial manganese-superoxide dismutase is hypersensitive to oxygen. Proc. Natl Acad. Sci. USA, 83, 3820–3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Jagow G. and Klingenberg,M. (1970) Pathways of hydrogen in mitochondria of Saccharomyces carlsbergensis. Eur. J. Biochem., 12, 583–592. [DOI] [PubMed] [Google Scholar]
- Weissbach H., Etienne,F., Hoshi,T., Heinemann,S.H., Lowther,W.T., Matthews,B., St John,G., Nathan,C. and Brot,N. (2002) Peptide methionine sulfoxide reductase: structure, mechanism of action and biological function. Arch. Biochem. Biophys., 397, 172–178. [DOI] [PubMed] [Google Scholar]