Abstract
In the cyanobacterium Synechococcus elongatus PCC 7942, the KaiA, KaiB and KaiC proteins are essential for generation of circadian rhythms. We quantitatively analyzed the intracellular dynamics of these proteins and found a circadian rhythm in the membrane/cytosolic localization of KaiB, such that KaiB interacts with a KaiA–KaiC complex during the late subjective night. KaiB–KaiC binding is accompanied by a dramatic reduction in KaiC phosphorylation and followed by dissociation of the clock protein complex(es). KaiB attenuated KaiA-enhanced phosphorylation both in vitro and in vivo. Based on these results, we propose a novel role for KaiB in a regulatory link among subcellular localization, protein–protein interactions and post-translational modification of Kai proteins in the cyanobacterial clock system.
Keywords: circadian clock/cyanobacteria/KaiB/KaiC phosphorylation/localization
Introduction
Circadian rhythms, endogenous oscillations of physiological activities with a period of ∼24 h, are found in a wide spectrum of organisms and enhance their fitness in a day/night alternating environment (Bünning, 1973). Among these organisms, cyanobacteria are the simplest organisms that exhibit circadian rhythms (Golden et al., 1997; Iwasaki and Kondo, 2000). Three clock genes kaiA, kaiB and kaiC have been identified in the cyanobacterium, Synechococcus elongatus PCC 7942 as essential timekeeping components. A transcription/translation-based autoregulatory loop of kaiBC gene expression has been proposed to drive circadian rhythms (Ishiura et al., 1998). In this model, KaiA and KaiC are proposed as positive and negative regulators of kaiBC expression, respectively (Ishiura et al., 1998). More recently, we found that KaiA-mediated activation of kaiBC expression is KaiC dependent, suggesting that KaiC also functions in a positive feedback process in the molecular oscillatory mechanism (Iwasaki et al., 2002).
The stability of self-sustained oscillation and the length of the circadian period are key features of circadian rhythms. These properties are likely to be realized by interactions between multiple feedback processes (Somers, 1999; Roenneberg and Merrow, 2002) and multiple post-translational controls of the core feedback process, including subcellular translocation, protein– protein interactions and phosphorylation of some circadian clock proteins (Dunlap, 1999; Young and Kay, 2001). In cyanobacteria, the following biochemical properties of the Kai proteins have been reported to be important for circadian timekeeping: (i) interactions between Kai proteins in various combinations (Iwasaki et al., 1999); (ii) ATP-binding and autophosphorylating activities of KaiC in vitro (Nishiwaki et al., 2000); (iii) amplification of basic oscillation by a KaiC-binding histidine kinase, SasA (Iwasaki et al., 2000); and (iv) circadian accumulation of KaiB and KaiC proteins (Xu et al., 2000). More recently, we found that KaiC is phosphorylated at serine and threonine residues in a circadian manner, and that KaiA enhances this phosphorylation. Biochemical and genetic studies strongly suggested that KaiA-stimulated KaiC phosphorylation is important for circadian timing, and that the level of KaiC phosphorylation affects the rate of kaiBC transcription (Iwasaki et al., 2002).
Even though these biochemical properties of clock proteins are essential for circadian oscillation, they are still not sufficient to explain fully their molecular functions in the circadian oscillator of cyanobacteria. In particular, the biochemical function of KaiB is rather unknown, except that it is somehow crucial for rhythm generation. In this study, we examined the intracellular dynamics of Kai proteins with respect to their cellular amounts, subcellular localization, protein–protein interactions and regulation of KaiC phosphorylation. We propose that a time-specific membrane/cytosolic localization of KaiB protein is important for circadian rhythms in formation of Kai protein complexes, and that KaiB functions as an attenuator of KaiA-enhanced phosphorylation of KaiC.
Results
Absolute cellular amounts of Kai proteins
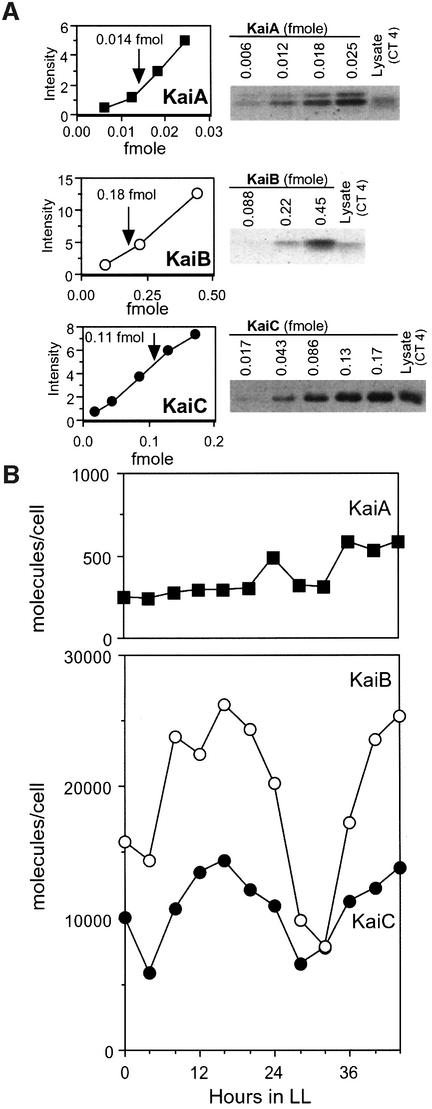
The levels of KaiB and KaiC proteins exhibit robust circadian rhythms, peaking at early circadian night phase (CT 15–16, circadian time), while levels of KaiA are almost constant throughout the circadian cycle (Xu et al., 2000). To understand the behavior of Kai proteins in the cell, we quantitatively assessed the cellular abundance of these proteins. The amount of each Kai protein in Synechococcus cells was estimated by normalization of western blot signals to a dilution series of known amounts of recombinant Kai proteins (see Materials and methods). Densitometric analysis of the western blots indicated that all the signals were within the linear range, which enabled us to calculate the absolute amount of each Kai protein in a single cell (Figure 1A). Then, circadian variation in the cellular amounts of the Kai proteins was determined every 4 h under continuous illumination (LL; Figure 1B). We found that the amount of KaiB and KaiC changed rhythmically, with an ∼2:1 molecular ratio during the circadian cycle. In contrast, KaiA levels were almost constant and relatively low, at 5% of the lowest levels of KaiB and KaiC (Figure 1B). The low level expression of KaiA is presumably due to both transcriptional and translational regulation, as deduced from the following observations: (i) quantitative analysis by a luciferase reporter has demonstrated that the promoter activity of the kaiA gene is ∼10 times less than that of the kaiBC operon (Ishiura et al., 1998); and (ii) the estimated start codon for the KaiA protein is not AUG but GUG, which is often used for poorly expressed genes in bacteria (Kozak, 1983; Reddy et al., 1985).
Fig. 1. Circadian profiles of absolute amounts of Kai proteins. (A) A series of dilutions of known amounts of Kai proteins produced in E.coli was subjected to SDS–PAGE. Synechococcus whole-cell extracts were also loaded on the same gel and then analyzed by western blot so that the signals could be compared directly (right panel, cell extracts; data for the CT 4 sample are shown as representatives). Densitometric analysis (left panels) shows a linear relationship between the input amounts of KaiA (closed square), KaiB (open circle) and KaiC (closed circle) and signal intensity upon western analysis. These data allow absolute quantification of the levels of three Kai proteins in the cell (see Materials and methods). Arrows point to the results at CT 4 for lysate. (B) Circadian profiles of absolute amounts of KaiA (closed square), KaiB (open circle) and KaiC (closed circle) in a single cell under continuous illumination (LL) (50 µE/m2/s). Whole-cell extracts were prepared from Synechococcus cells grown in LL after two 12 h light:12 h dark cycles. The extracts were electrophoresed and analyzed by western blot. The immunoblot signals were normalized with signals of known amounts of purified Kai proteins, as described above.
Intracellular localization of clock proteins during the circadian cycle
In eukaryotic clock systems, a circadian rhythm in the subcellular localization of clock-related proteins, most typically the transport of core Drosophila clock proteins such as Period and Timeless into the nucleus, is important for generating basic molecular oscillation (Young and Kay, 2001). Although prokaryotic cyanobacteria do not contain nuclei, they have developed cytoplasmic membranes around the cell envelope and thylakoid membranes for photosynthesis. Thus, we examined the temporal profile of membrane/cytosolic localization of the Kai proteins.
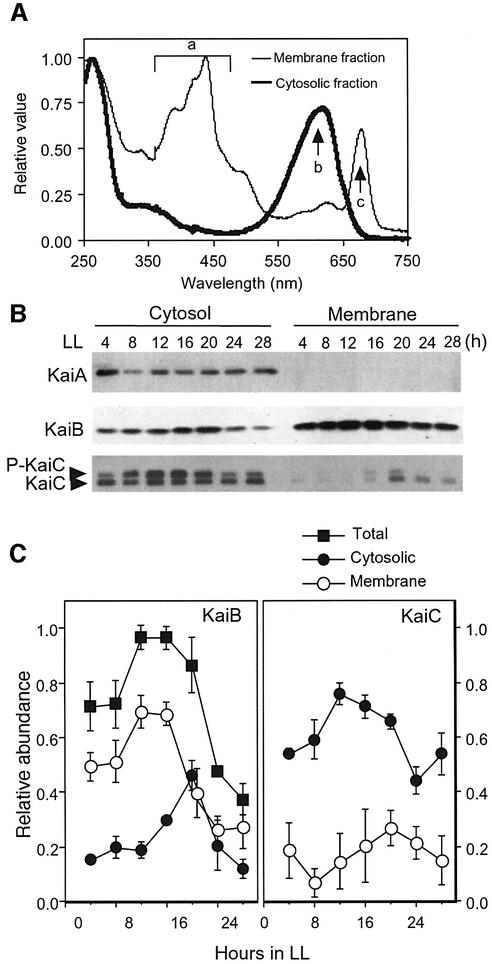
As shown in Figure 2A, we separated crude extract of Synechococcus cells into cytosolic and total membrane fractions (see Materials and methods) and subjected them to western blot analysis. KaiA was found exclusively in the cytosol throughout the entire circadian cycle (Figure 2B). A large amount of KaiC protein (70–90%) was localized in the cytosolic fraction during the day (Figure 2B and C). The upper and lower signals from the doublet bands for cytosolic KaiC on the western blots (Figure 2B) correspond to phosphorylated and non-phosphorylated forms of KaiC, respectively (Iwasaki et al., 2002). As reported previously (Iwasaki et al., 2002), cytosolic KaiC showed a robust circadian rhythm in its phosphorylation, peaking at CT 12–16. We also found that in membrane fractions, most KaiC protein was found in a non-phosphorylated form (Figure 2B; data not shown).
Fig. 2. Subcellular localization of KaiA, KaiB and KaiC proteins. Cells were grown in a continuous culture system in LL (50 µE/m2/s). After two 12 h light:12 h dark cycles, cells were returned to LL, collected at the indicated times, and then separated into cytosolic and total membrane fractions. (A) Absorption spectra of cytosolic and membrane fractions for wavelengths ranging from 250 to 750 nm. Arrows indicate absorption peaks of carotenoids (a), phycobiliproteins (b) and chlorophyll a (c). (B) Cytosolic and total membrane fractions were subjected to SDS–PAGE and western blot. Equal amounts of each sample were loaded (3.0 µg protein per lane) for the cytosolic fraction collected at the indicated time. Isolated membranes were resuspended in an amount of buffer equal to that of the corresponding cytosolic fraction and analyzed at each time point. As a volume equal to the cytosolic fraction was loaded onto the gels, the intensity of immunoblot signals of the membrane and cytosolic fractions are quantitatively comparable. In the bottom panel (KaiC), the upper band corresponds to phosphorylated KaiC and the lower band to the non-phosphorylated form. (C) Rhythmic accumulation profiles of KaiB and KaiC in cytosol (closed circle), membranes (open circle) and total (sum of cytosol and membranes, closed square) were plotted against time under LL conditions. Peak values for the total were set to 1, and the rest of the values were normalized to these values. The results are shown with mean ± SEM (n = 3).
KaiB was present in both cytosolic and membrane fractions, with 20–50% of KaiB molecules localized to the cytosol. Surprisingly, cytosolic KaiB protein levels oscillated, peaking at CT 20, which was ∼4 h delayed relative to the total KaiB accumulation rhythm in whole-cell extracts, which peaked at CT 16 (Figures 1 and 2C). In contrast, the rhythm in the level of membrane-associated KaiB peaked at LL 12–16 (Figure 2C). Thus, in the cytosol, KaiB cycling is delayed by several hours relative to that of KaiC, while total cellular amounts exhibit circadian rhythms with the same phase. In addition, it is notable that the cytosolic levels of KaiB and KaiC are estimated to be approximately equimolar, considering the KaiB:KaiC ratio in the whole cell (Figure 1) and the proportion of KaiB in the cytosol (∼20–50%, Figure 2).
Clock proteins form time-specific complexes in the cytosol
The three Kai proteins interact in all possible combinations in vitro, in the yeast two-hybrid system and in Synechococcus cells (Iwasaki et al., 1999). Therefore, differences in the cytosolic accumulation of KaiA, KaiB and KaiC are expected to play a profound role in controlling the interaction(s) of these proteins.
We examined interactions between Kai proteins by co-immunoprecipitation assays using Synechococcus cytosolic extracts over the circadian cycle. As shown in Figure 3A, we observed a high amplitude rhythm of KaiB–KaiC complex formation which peaked at LL 20 (CT 20). This phase coincides well with the cytosolic fluctuation in KaiB abundance (Figure 2). Moreover, we observed that KaiC rhythmically associated with KaiA prior to binding with KaiB (Figure 3A, LL 16–20). A dramatic increase in KaiB–KaiC complex formation at CT 20 was accompanied by a decrease in KaiC phosphorylation (Figure 3B). These results raise the possibility that KaiB may modulate the state of KaiC phosphorylation.
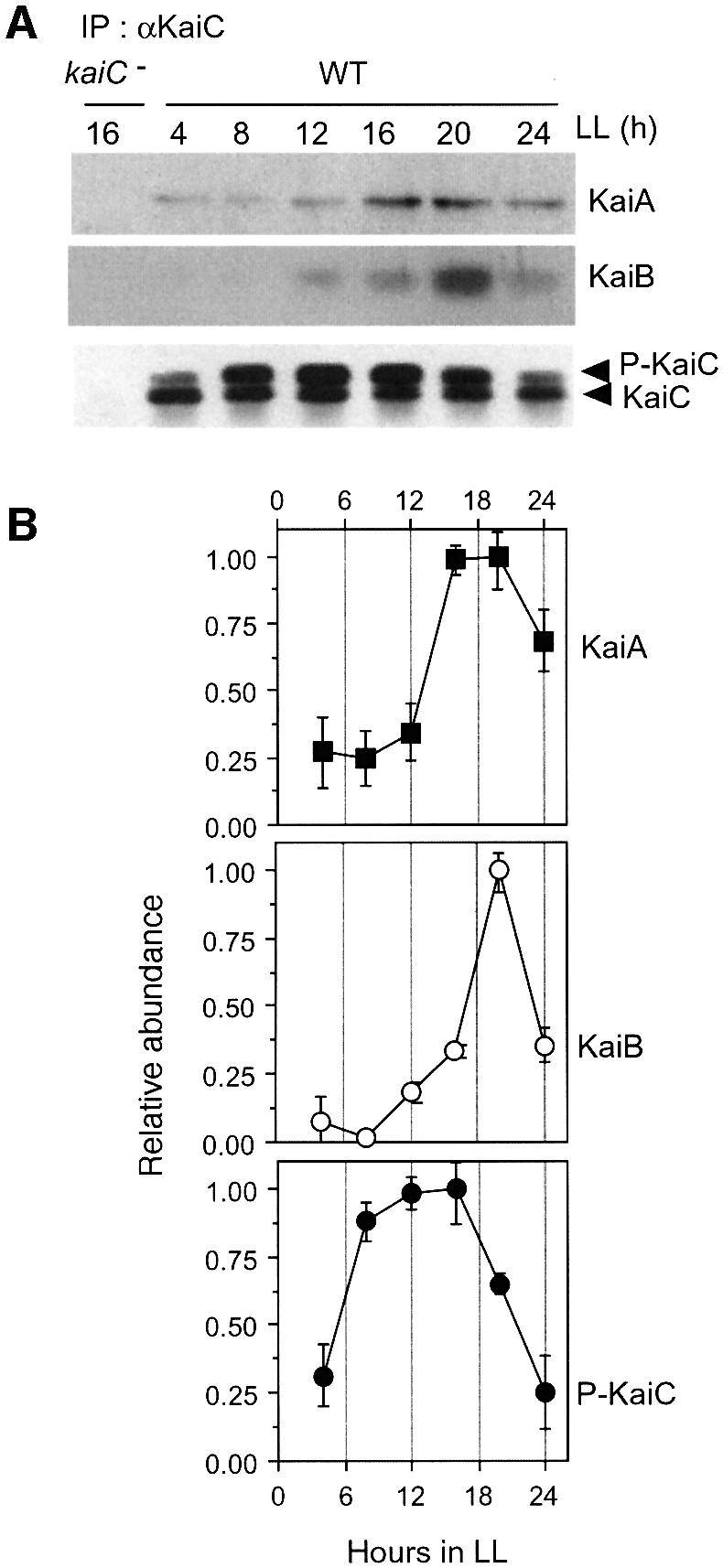
Fig. 3. Circadian rhythms of KaiC-interacting KaiA and KaiB. (A) Wild-type (WT) cells were collected at the indicated times, and kaiC-depleted (kaiC –) cells were harvested after 16 h in LL. Cytosolic fractions were prepared and subjected to co-immunoprecipitation analysis using an antibody against KaiC. The resulting immune complexes were electrophoresed and subjected to immunoblot analysis to detect KaiC-interacting KaiA and KaiB. Eleven percent polyacrylamide gels (acrylamide:N,N′-methylenebisacrylamide 29.8: 0.2) were used to detect KaiC, and 13% gels (acrylamide:N,N′-methylenebisacrylamide 29.2:0.8) were used to detect KaiA and KaiB. In the bottom panel (KaiC), the upper band corresponds to phosphorylated KaiC and the lower band to the non-phosphorylated form. (B) Rhythmic KaiC interaction profiles of KaiA (closed squares) and KaiB (open circles) and rhythmic accumulation profiles of phosphorylated KaiC in the cytosolic fraction (closed circle) were seen. Peak values for each protein were set to 1. The results are shown with mean ± SEM (n = 4 for KaiA and KaiC; n = 3 for KaiB).
KaiB attenuates KaiA-stimulated KaiC phosphorylation
To examine the role of KaiB in the accumulation and phosphorylation of KaiC in vivo, we analyzed expression profiles of Kai proteins in the kaiB-null strain as well as in wild-type, kaiA-null and kaiC-null strains by immunoblotting assays. In contrast to the kaiA-null mutant, in the kaiB-null mutant, the level of phosphorylated KaiC increased, while the non-phosphorylated form was dramatically reduced (Figure 4A). Thus, KaiB is crucial for accumulation of non-phosphorylated KaiC protein.
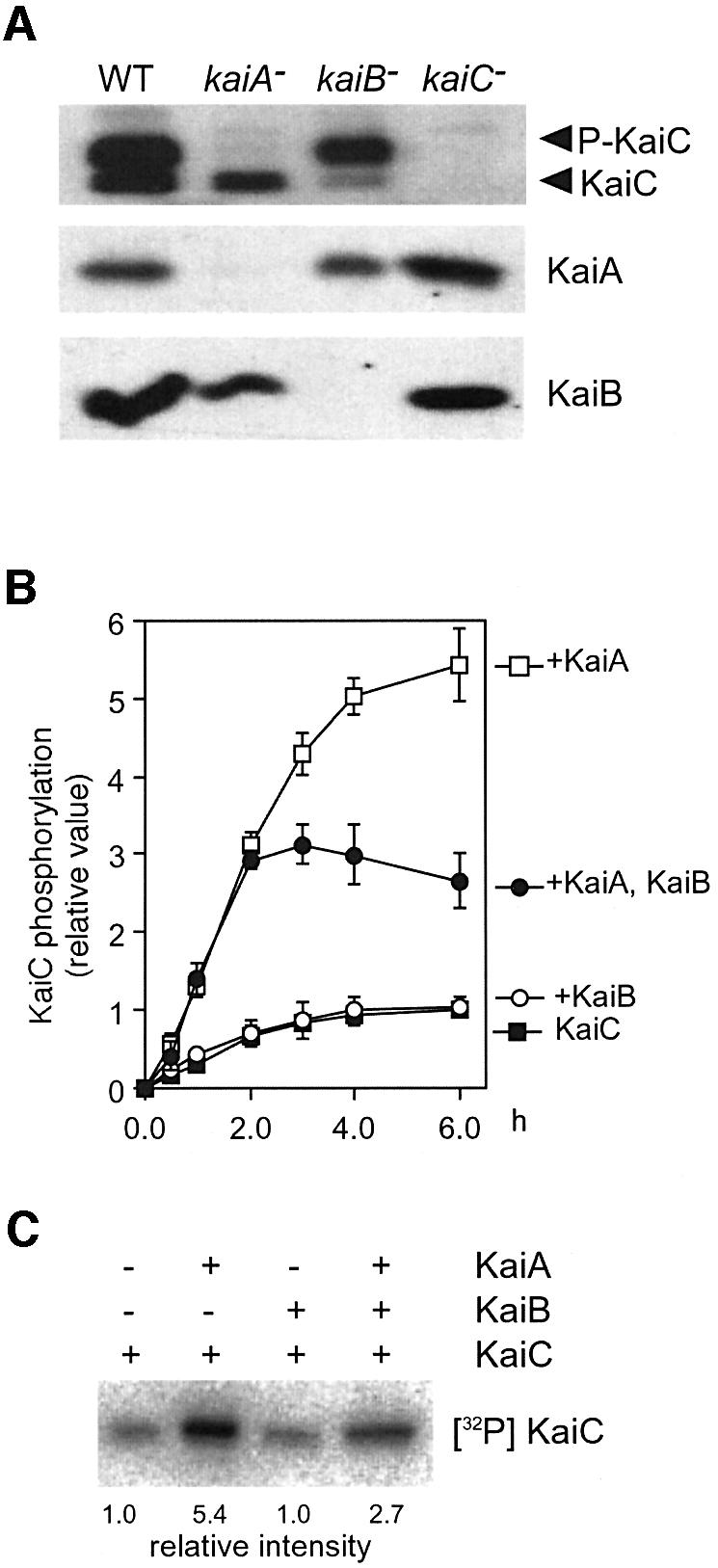
Fig. 4. Attenuation of KaiA-mediated KaiC phosphorylation by KaiB. (A) Accumulation of Kai proteins in wild-type (WT), kaiA-null (kaiA–), kaiB-null (kaiB–) and kaiC-null (kaiC–) strains (immunoblot analysis). Cells were cultured in LL (50 µE/m2/s). After a 12 h dark treatment, cells were returned to LL conditions and then collected. Cells collected at LL 16, when the level of KaiC phosphorylation is maximal in the wild-type strain, were used for western blots shown here. Whole-cell proteins were extracted and subjected to SDS–PAGE, followed by western blot (5.0 µg protein per lane). Note that no circadian fluctuation was observed in any kai gene mutants in terms of the state of KaiC abundance and its phosphorylation throughout the circadian cycle (data not shown). In the top panel (KaiC), the upper band corresponds to phosphorylated KaiC and the lower band to a non-phosphorylated form. (B) In vitro autokinase assay (time course experiments). KaiC (4.6 pmol) was incubated with [γ-32P]ATP in the presence or absence of KaiA (0.23 pmol) and/or KaiB (9.2 pmol) at 30°C for the indicated time. Proteins were analyzed by SDS–PAGE, blotted onto PVDF membranes and then subjected to autoradiography. The results are shown with means ± SEM (n = 4). (C) A representative autoradiograph of in vitro autokinase assay (30°C, 6 h). The numbers at the bottom indicate relative signal intensity which was normalized to that of KaiC without additional protein.
Our previous study indicated that KaiC autophosphorylation is activated by KaiA, but not affected by KaiB alone in vitro (Iwasaki et al., 2002). As KaiB formed a complex with KaiC following KaiA–KaiC interaction in vivo (Figure 3A), we examined whether KaiB would affect KaiA-enhanced KaiC autokinase activity in vitro by incubation of KaiC with [γ-32P]ATP in the presence of KaiA and/or KaiB protein. As the amount of KaiA is very limiting for KaiC in vivo (Figure 1B), we performed the in vitro assay with a similar KaiA:KaiB:KaiC (1:40:20) molecular ratio (see Materials and methods). We found that KaiA strikingly enhanced KaiC autokinase activity, even if it was present at 5% of the KaiC molecules, and also confirmed that KaiB alone did not affect the activity (Figure 4B). When KaiA and KaiB were incubated together with KaiC for 6 h, however, the level of KaiC autophosphorylation was dramatically reduced to ∼50% of that in the presence of KaiA alone (Figure 4B). Thus, KaiB also inhibits or negates KaiA-enhanced KaiC autophosphorylation in vitro. A similar result was obtained previously by Williams et al. (2002), who reported ∼70% attenuation of the initial velocity of KaiA-enhanced KaiC phosphorylation by KaiB when the KaiA:KaiB:KaiC molecular ratio was 3:3:1. In our experimental conditions with a very limited KaiA ratio, the significant effect of KaiB protein on KaiC autophosphorylation was not observed reproducibly over the first 2 h (Figure 4B). This result may be explained by delayed binding of KaiB to a limited concentration of KaiA–KaiC complex, as discussed for the in vivo findings (see Discussion). We also found that the level of 32P-labeled KaiC decreased from 3 h after incubation with KaiA and KaiB (Figure 4B). Thus, it is possible that KaiB activates dephosphorylation of the phosphorylated KaiC protein in the presence of KaiA (see Discussion). Therefore, KaiC hyperphosphorylation in the kaiB-null strain appears to be partly due to the lack of a negative effect of KaiB on KaiA-enhanced KaiC phosphorylation, as demonstrated in vitro. Based on these results, we suggest that KaiB is crucial for rhythmicity by balancing the phosphorylated and non-phosphorylated levels of KaiC.
Effects of kai gene disruption on Kai protein localization
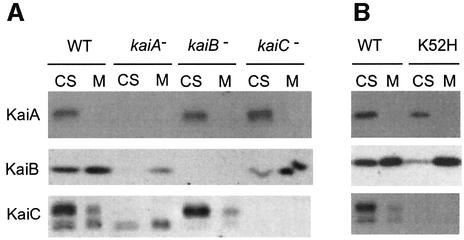
Finally, we examined the effect of kai gene disruption on protein localization to characterize further the relationship between KaiC phosphorylation and subcellular localization of clock proteins. In the kaiA-null mutant strain, KaiC was present in the non-phosphorylated form (Figure 4A) and present predominantly in membrane fractions (Figure 5A). As non-phosphorylated KaiC was more abundant in the membrane fraction in wild-type cells (Figure 2B), it is possible that the KaiC distribution profile in the kaiA-null mutant may be due to accumulation of non-phosphorylated KaiC. We also found that KaiC associated with membranes in the kaiB-null mutant. Thus, membrane association of KaiC is not mediated by KaiB.
Fig. 5. Effects of inactivation of the kai gene on subcellular localization. Cells were grown in LL (50 µE/m2/s). After two 12 h light:12 h dark cycles, cells were returned to LL, separated into cytosolic and total membrane fractions, and then analyzed by immunoblotting assays, as described in Figure 2. Note that no circadian alteration was observed in any kai gene-inactivated mutants in terms of Kai protein distribution throughout the circadian cycle (data not shown). Cells collected at LL 16, when the level of KaiC phosphorylation is maximal in the wild-type strain, were used for western blots. Cytosolic fractions (CS) and total membrane fractions (M) were prepared and analyzed from wild-type (WT), kaiA-null (kaiA–), kaiB-null (kaiB–) and kaiC-null (kaiC–) strains (A), and from wild-type (WT) and kaiC [K52H] strains (B). In the bottom panel (KaiC), the upper and lower bands correspond to phosphorylated and non-phosphorylated forms of KaiC, respectively.
Localization of KaiA did not change in any of the clock mutants (Figure 5), while that of KaiB was affected by kaiA and kaiC inactivation. In both kaiA- and kaiC-null mutants, a larger amount of KaiB associated with membranes than in the wild-type strain, while it still remained in the cytosol (Figure 5A). The same result was obtained in a kaiC [K52H] mutant strain, in which mutant KaiC fails to accumulate with normal accumulation of KaiA (Figure 5B). In the kaiC [K52H] mutant, an ATP-binding domain of KaiC (P-loop) is mutated (Nishiwaki et al., 2000) to abolish its ATP-binding activity and rhythmicity. Thus, KaiC can partly function in translocation of membrane KaiB to the cytosol or interfere with membrane association of KaiB.
Discussion
Our results suggest a novel function for the KaiB protein in a regulatory link controlling subcellular Kai protein localization, clock protein complex formation and KaiC phosphorylation. In this study, we found that the circadian rhythm of KaiB accumulation in the cytosol peaked at CT 20, which is several hours delayed relative to the rhythm of total KaiB abundance in whole-cell extracts (Figure 2). As kaiBC mRNA expression rhythm peaks at CT 8 and total KaiB accumulation peaks at CT16, it is more likely that KaiB is released from membranes to the cytosol than that KaiB is synthesized to remain in the cytosol at CT 20. The mechanism by which KaiB associates with membranes currently is unknown. The KaiB amino acid sequence does not contain a hydrophobic stretch as a transmembrane domain. We performed the phase separation assay with Triton X-114 (Bordier, 1981; Lipman et al., 1998) for the Synechococcus membrane fractions to separate hydrophobic (transmembrane) and hydrophilic (peripheral) proteins. We observed KaiB to be a peripheral membrane-associating protein in the aqueous phase (data not shown). Moreover, KaiB was released from membrane fractions into soluble fractions by exposure to high salts and alkaline pH conditions (data not shown). These results suggest that the association of KaiB with membranes is not a tight one. In cyanobacteria, some associations between soluble or extrinsic proteins and membranes are regulated by the physiological state of the cell. For example, the polypeptide composition of phycobilisomes associating with thylakoid membranes is regulated by nitrogen availability (Duke et al., 1989), and ribosome D1 nascent chain complexes are targeted into thylakoid membranes by exposure to light (Tyystjärvi et al., 2001). We suggest that membrane association of KaiB is similarly flexible.
In contrast, most KaiC proteins remain in the cytosol throughout the circadian cycle. Less KaiC was found in the membrane fraction, with a higher population of the non-phosphorylated form (Figures 2B and 5A). This implies that KaiC is mainly phosphorylated in the cytosol, or that phosphorylation of KaiC reduces its affinity for membranes. As changes in the rate of KaiC phosphorylation have been suggested to affect gene expression in the Synechococcus clock system (Iwasaki et al., 2002), distinct localization of phosphorylated and non-phosphorylated KaiC could be important for circadian rhythm control.
Our results also provide novel insights into the complex formation profile of Kai proteins in the cell. As KaiB and KaiC failed to be co-fractionated by gel filtration analysis in the kaiA-null mutant strain (Kageyama et al., 2003), it is plausible that KaiA mediates KaiB–KaiC association. Our quantitative analysis demonstrated that KaiB and KaiC were present at molecular excess relative to KaiA (Figure 1). Considering that KaiC can form a homo-hexamer in vivo (Kageyama et al., 2003) and in vitro (Mori et al., 2002) and that 20% of KaiC molecules associate with membranes (Figure 2), >80% of KaiC hexamers do not associate physically with KaiA in the cytosol. It is unlikely that KaiC forms a solid complex with KaiA at an ∼20:1 ratio because our previous gel filtration analysis detected KaiC within the 400–600 kDa fractions (the size of KaiC monomer is ∼58 kDa) (Kageyama et al., 2003). Nevertheless, gel filtration analysis demonstrated that most KaiB molecules were co-fractionated with KaiC during the late subjective night (Kageyama et al., 2003). Therefore, we suggest that KaiA binds KaiC in a processive manner and thereby enhnaces KaiB–KaiC interaction, which peaks at CT 20.
Although KaiA is essential for accumulation of phosphorylated KaiC in vivo and enhances its autophosphorylation in vitro (Iwasaki et al., 2002), accumulation of phosphorylated KaiC starts increasing prior to accumulation of the KaiA–KaiC complex (Figure 3). This implies that weak processive association is sufficient for KaiA to enhance KaiC phosphorylation. In addition, increases in the magnitude of KaiC phosphorylation during the late subjective day may also be due to elevated KaiC accumulation enhancing self-association for KaiC intermolecular autophosphorylation. Formation of a KaiA–KaiC complex from CT 16 to 20, which is presumably important for KaiB–KaiC association peaking at CT 20, is probably due to the phosphorylation state and/or an as yet unknown KaiC post-translational state in the late subjective night. A very limited amount of KaiA relative to KaiB and KaiC (Figure 1) would explain the delayed accumulation of the KaiB–KaiC complex relative to the KaiA–KaiC association (Figure 3). We suggest that KaiA can be released from a transient KaiA–KaiB–KaiC complex and then act on another KaiC molecule stimulating interaction with KaiB, thereby maintaining the high level (transient) KaiC–KaiA complex formation and gradually increasing the amount of more stable KaiB–KaiC complex from CT 16 to 20. In addition, the delayed accumulation of cytosolic KaiB peaking at CT 20 (Figure 2) must also be an important process for dramatic increases in the formation of KaiB–KaiC complexes during the late subjective night.
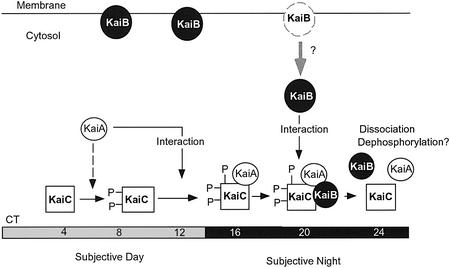
Figure 6 summarizes a possible scheme of dynamic circadian behaviors of Kai proteins in the Synechococcus cell. The rhythms of KaiB and KaiC accumulation in the total cell delay kaiBC mRNA rhythm by several hours, increasing at CT 8 to a peak at CT 16 (Figure 1). KaiB is translocated selectively into the membrane to be present as a temporal reservoir regulating cytosolic KaiB concentration. The cytosolic accumulation of KaiB reaches a peak at CT 20 (Figure 2C). KaiA enhances the accumulation of phosphorylated KaiC protein at CT 8–12 by enhancing its autokinase activity in a processive manner. The resulting phosphorylated KaiC may accelerate its binding to KaiA during the middle subjective night (CT 16). KaiA–KaiC complex formation and elevated KaiB protein accumulation in the cytosol enhance stable KaiB–KaiC complex formation peaking at CT 20. We suggest that this KaiB binding is presumably important in the subsequent dramatic reduction in the magnitude of KaiC phosphorylation.
Fig. 6. A possible role for KaiB in the cyanobacterial clock. In the early morning, the phosphorylation level of KaiC is relatively low. During the subjective day, KaiC gradually accumulates in the cytosol. This increase in KaiC activates its own autokinase activity, which is enhanced by KaiA, even without the formation of a stable KaiA–KaiC complex during CT 8–12 (broken arrow). Phosphorylation of KaiC protein may accelerate its binding to KaiA during the middle subjective night (CT 16). Both the KaiA–KaiC complex and KaiB accumulate in cytosol at CT 20, and then KaiB can form a stable complex with KaiC. Although the whole cellular amount of KaiB peaks at CT 12–16, the cytosolic KaiB accumulation is delayed by several hours to peak at CT 20, most probably due to circadian-gated translocation of KaiB from membranes to the cytosol. This enables KaiB to bind to KaiA-interacting KaiC, which would reduce the magnitude of KaiC phosphorylation and trigger dissociation of the clock protein complex(es). The dynamic regulation of KaiC phosphorylation and Kai protein complex formation could affect the rate of kaiBC expression, which is the basis for generating the basic oscillation.
How does KaiB attenuate KaiC phosphorylation? One possibility is that KaiB enhances degradation of the phosphorylated form(s) of KaiC. Neither kaiB depletion nor kaiB overexpression, however, affected the total amount of KaiC protein (Figure 4A; data not shown). Rather, it is more plausible that KaiB inhibits the effects of KaiA, which enhances KaiC phosphorylation, or that KaiB stimulates dephosphorylation of KaiC. Our results shown in Figure 4B support the latter, since the level of phosphorylated KaiC was reduced gradually from 3 h after incubation with KaiA and KaiB. In contrast, the magnitude of KaiC phosphorylation was increased when KaiC alone or KaiC in the presence of KaiA was incubated with [γ-32P]ATP (Figure 4B). In either case, KaiB affects KaiC phosphorylation dependent on KaiA function. In summary, we suggest that subcellular localization of KaiB is important for KaiABC complex formation and functions as an attenuator of KaiA-enhanced KaiC, thereby changing the rate of kaiBC expression, which forms a basic oscillatory loop for circadian rhythm generation in Synechococcus.
Materials and methods
Bacterial strains
We used the wild-type, kaiA-inactivated and kaiC-depleted Synechococcus strain (Iwasaki et al., 2002) and the kaiC [K52H] strain (Nishiwaki et al., 2000), which harbor the PkaiBC::luxAB reporter gene set. A kaiB-inactivated strain expressing the PkaiBC::luxAB reporter was generated by mutagenesis of a wild-type reporter strain, NUC42 (Nishimura et al., 2002), as described previously (Ishiura et al., 1998).
Expression and purification of recombinant Kai proteins
KaiB and KaiC were produced by induction of GST fusion proteins in Escherichia coli followed by purification with glutathione–Sepharose and removal of the GST tag, as described previously (Iwasaki et al., 2002), followed by further purification with Resource-Q anion exchange chromatography (Amersham Pharmacia). For recombinant KaiA protein, the kaiA open reading frame was cloned into the pGEX-6P-1 vector (Amersham). Escherichia coli DH5α cells harboring the resulting plasmid, pGEX6P-kaiA, were used to produce GST–KaiA fusion protein, with subsequent affinity purification and GST tag removal as described (Iwasaki et al., 2002).
Western blot analysis
Synechococcus whole-cell extracts were prepared as described (Iwasaki et al., 2002), with minor modifications. Cells were disrupted using a Multi-Beads Shocker (Yasui Kikai, Japan) with zirconium beads (0.1 mm diameter; Biospec Products) for 5 min at 30 s intervals at 2°C. After centrifugation, the supernatants were removed to new tubes, and the pellets were disrupted further, as described above. Protein concentration was determined by the BCA method using bovine serum albumin (BSA) as a standard. Whole-cell extracts were subjected to SDS–PAGE and transferred to nitrocellulose membranes. Immunoblots were incubated in the presence of anti-KaiA, anti-KaiB and anti-KaiC antibodies, as described (Iwasaki et al., 1999), and protein was detected with enhanced chemiluminescence (ECL; Amersham).
Quantification of cellular Kai protein
Synechococcus cells were cultured in a continuous culture system, in BG-11 liquid medium to maintain an OD730 of 0.25, corresponding to 2.5 × 108 cells/ml, under LL (50 µE/m2/s) at 30°C. After two 12 h light:12 h dark (LD) cycles, cells were returned to LL conditions and then collected every 4 h. Total proteins were extracted, as described above. Recombinant KaiA, KaiB and KaiC proteins were produced in E.coli and purified, as described above, as standard proteins for quantitative immunoblotting assays. Serial dilutions of the standard protein and of Synechococcus cell extracts were loaded onto the same gel and subjected to SDS–PAGE and immunoblotting assays. The intensity of each appropriate signal was measured with a densitometer. In each experiment, we confirmed that the intensity of the relevant immunoreactive bands for whole-cell extracts and standard proteins yielded a linear dose response (Figure 1A; data not shown). Comparison of the signal intensity of a standard protein band with that of a lysate protein enabled an estimate of the absolute amount of each Kai protein in a single cell. We obtained similar results from two independent experiments.
KaiC autokinase assay
Autophosphorylation assays were performed at 30°C, as described (Iwasaki et al., 2002), with some modification. KaiC protein (4.6 pmol) was incubated in 15 µl of a reaction buffer [20 mM Tris–HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, 5 mM MgCl2, 5 mM [γ-32P]ATP (300 c.p.m./pmol)] in the presence or absence of KaiA (0.23 pmol) and KaiB (9.2 pmol). The solution was incubated at 30°C. The reaction was terminated by adding SDS sample buffer, followed by SDS–PAGE, blotting onto Immobilon-P membranes (Millipore) and autoradiography.
Preparation of total membranes and cytosolic fractions
The total membrane fraction was prepared as described (Ogawa, 1992) with minor modifications. Synechococcus cells were grown in a continuous culture system in BG-11 liquid medium to maintain an optical density (OD730) of 0.2 under LL conditions (50 µE/m2/s). After two LD cycles, cells were returned to LL conditions and then collected every 4 h. Cells (0.12 g) were suspended in 3 ml of TES–NaOH buffer (10 mM TES–NaOH, 5 mM NaCl, 5 mM EDTA pH 7.0) and disrupted twice by a French pressure cell press (SIM-Aminco) at 40 MPa. The cell suspension was centrifuged at 5000 g for 10 min to remove undisrupted cell debris, and the supernatant was again centrifuged at 100 000 g for 1 h at 4°C. The pellet was washed with TES–NaOH buffer, resuspended in 3 ml of 1× SDS sample buffer, and used as a total membrane fraction. The supernatant was used as the cytosolic fraction. Integrity of fractionation was checked by examining the absorption spectra for each fraction over a wavelength range from 250 to 750 nm (Figure 2A).
Co-immunoprecipitation assays
Immunoprecipitation was performed as described by Iwasaki et al. (2000), with modifications. In advance of immunoprecipitation reactions, AffiGel-Hz resin (BioRad) containing immobilized anti-KaiC IgG (Iwasaki et al., 2000) was pre-washed with 0.5% BSA in an IP buffer (50 mM Tris–HCl, 100 mM KCl, 5 mM MgCl2, 0.2% glycerol, 0.1 mM EDTA) to block non-specific interaction. A 150 µg aliquot of cytosolic proteins in 700 µl of TES–NaOH buffer was prepared as described above, and then added to 700 µl of IP buffer (supplemented with 100 mM KCl). A 1400 µl aliquot of the resulting cytosolic extract was incubated with the pre-washed beads coupled to anti-KaiC IgG at 4°C for 2 h. Immune complexes were washed twice with 1 ml each of IP buffer with 0.5% BSA and four times with 1 ml each of IP buffer without BSA, and then resolved in 1× SDS sample buffer for immunoblotting assays.
Triton X-114 phase separation
Triton X-114 partitioning assays were performed as described by Bordier (1981). Briefly, the total membranes prepared above were solubilized in 1% Triton X-114 by agitation for 30 min at 4°C. Insoluble materials were removed by centrifugation, and the supernatant was incubated at 30°C for 3 min. The supernatant was layered onto a pre-incubated sucrose (6%) cushion containing 0.06% Triton X-114, 10 mM Tris–HCl pH 7.4 and 150 mM NaCl, and centrifuged at 300 g for 3 min. The detergent (lower) and aqueous (upper) phases were recovered separately. The detergent phase was extracted three times with the same buffer, while the aqueous phase was rinsed three times with 2% Triton X-114 without a sucrose cushion. Aliquots of the separated phases were analyzed by SDS–PAGE followed by immunoblotting assays.
Acknowledgments
Acknowledgements
We thank Masato Nakajima (Nagoya University) for valuable comments and technical help. We are also grateful to Drs Stanly B.Williams and Susan S.Golden (Texas A & M University) for sharing experimental results and comments. This research was supported in part by Grants-in-aid from the Japanese Ministry of Education, Culture, Sports, Science and Technology (11233203 to T.K.and H.I., and COE 13CE2005 to T.K.), the Japanese Society for Promotion of Science (13680778 to H.I.), the Kurata Memorial Hitachi Science and Technology Foundation (to H.I.), and an Inoue Research Award for Young Scientists from the Inoue Foundation (to H.I.). Y.K. was supported by the JSPS fellowship for young scientists (13001761), and. T.N. was supported by the Hayashi Memorial Foundation for women natural scientists.
References
- Bordier C. (1981) Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem., 256, 1604–1607. [PubMed] [Google Scholar]
- Bünning E. (1973) The Physiological Clock, 3rd edn. Springer-Verlag, New York, NY.
- Duke C.S., Cezeaux,A. and Allen,M.M. (1989) Change in polypeptide composition of Synechocystis sp. strain 6803 phycobilisomes induced by nitrogen starvation. J. Bacteriol., 171, 1960–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap J.C. (1999) Molecular bases for circadian clocks. Cell, 96, 271–290. [DOI] [PubMed] [Google Scholar]
- Golden S.S., Ishiura,M., Johnson,C.H. and Kondo,T. (1997) Cyanobacterial circadian rhythm. Annu. Rev. Plant Physiol. Plant Mol. Biol., 48, 327–354. [DOI] [PubMed] [Google Scholar]
- Ishiura M., Kutsuna,S., Aoki,S., Iwasaki,H., Andersson,C.R., Tanabe,A., Golden,S.S., Johnson,C.H. and Kondo,T. (1998) Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science, 281, 1519–1523. [DOI] [PubMed] [Google Scholar]
- Iwasaki H. and Kondo,T. (2000) The current state and problems of circadian clock studies in cyanobacteria. Plant Cell Physiol., 41, 1013–1020. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Taniguchi,Y., Ishiura,M. and Kondo,T. (1999) Physical interactions among circadian clock proteins KaiA, KaiB and KaiC in cyanobacteria. EMBO J., 18, 1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H., Williams,S.B., Kitayama,Y., Ishiura,M., Golden,S.S. and Kondo,T. (2000) AKaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell, 101, 223–233. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Nishiwaki,T., Kitayama,Y., Nakajima,M. and Kondo,T. (2002) KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc. Natl Acad. Sci. USA, 99, 15788–15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama H., Kondo,T. and Iwasaki,H. (2003) Circadian formation of clock protein complexs by KaiA, KaiB, KaiC and SasA in cyanobacteria. J. Biol. Chem., 278, 2388–2395. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1983) Comparison of initiation of protein synthesis in prokaryote, eukaryotes and organelles. Microbiol. Rev., 47, 1–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman M.L., Panda,D., Bennett,H.P.J., Henderson,J.E., Shane,E., Shen,Y., Goltzman,D. and Karaplis,A.C. (1998) Cloning of human PEX cDNA. Expression, subcellular localization and endopeptidase activity. J. Biol. Chem., 273, 13729–13737. [DOI] [PubMed] [Google Scholar]
- Mori T., Saveliev,S.V., Xu,Y., Stafford,W.F., Cox,M.M., Inman,R.B. and Johnson,C.H. (2002) Circadian clock protein KaiC forms ATP-dependent hexameric rings and binds DNA. Proc. Natl Acad. Sci. USA, 99, 17203–17208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura H., Nakahira,Y., Imai,K., Tsuruhara,A., Kondo,H., Hayashi,H., Hirai,M., Saito,H. and Kondo,T. (2002) Mutations in KaiA, a clock protein, extend the period of circadian rhythm in the cyanobacterium Synechococcus elongatus PCC 7942. Microbiology, 148, 2903–2909. [DOI] [PubMed] [Google Scholar]
- Nishiwaki T., Iwasaki,H., Ishiura,M. and Kondo,T. (2000) Nucleotide binding and autophosphorylation of the clock protein KaiC as a circadian timing process of cyanobacteria. Proc. Natl Acad. Sci. USA, 97, 495–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T. (1992) Identification and characterization of the ictA/ndhL gene product essential to inorganic carbon transport of Synechocystis PCC6803. Plant Physiol., 99, 1604–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P., Peterkofsky,A. and McKenney,K. (1985) Translational efficiency of the Escherichia coli adenylate cyclase gene: mutation of the UUG initiation codon to GUG or AUG results in increased gene expression. Proc. Natl Acad. Sci. USA, 82, 5656–5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roenneberg T. and Merrow,M. (2002) ‘What watch?… such much’: complexity and evolution of circadian clocks. Cell Tissue Res., 309, 3–9. [DOI] [PubMed] [Google Scholar]
- Somers D.E. (1999) The physiology and molecular bases of the plant circadian clock. Plant Physiol., 121, 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyystjärvi T., Herranen,M. and Apo,E. (2001) Regulation of translation elongation in cyanobacteria: membrane targeting of the ribosome nascent-chain complexes controls the synthesis of D1 protein. Mol. Microbiol., 40, 476–484. [DOI] [PubMed] [Google Scholar]
- Williams S.B., Vakonakis,I., Golden,S.S. and LiWang,A.C. (2002) Structure and function from the circadian clock protein KaiA of Synechococcus elongatus reveals a clock input mechanism. Proc. Natl Acad. Sci. USA, 99, 15357–15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Mori,T. and Johnson,C.H. (2000) Circadian clock-protein expression in cyanobacteria: rhythms and phase setting. EMBO J., 19, 3349–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.W. and Kay,S.A. (2001) Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet., 2, 702–715. [DOI] [PubMed] [Google Scholar]