Abstract
This paper reports on the alignment between mouse and human anatomies, a critical resource for comparative science as diseases in mice are used as models of human disease. The two ontologies under investigation are the NCI Thesaurus (human anatomy) and the Adult Mouse Anatomical Dictionary, each comprising about 2500 anatomical concepts. This study compares two approaches to aligning ontologies. One is fully automatic, based on a combination of lexical and structural similarity; the other is manual. The resulting mappings were evaluated by an expert. 715 and 781 mappings were identified by each method respectively, of which 639 are common to both and all valid. The applications of the mapping are discussed from the perspective of biology and from that of ontology.
INTRODUCTION
Comparing gene-related information across model organisms is crucial to understanding similarities and differences among species. The availability of fully-sequenced genomes for an increasing number of model organisms enables comparisons across species. At the sequence level, tools such as BLAST help identify orthologous genes based on sequence similarity. Analogously, functional genomics uses the annotations of gene products (e.g., to the Gene Ontology) to compare genes.
One element shared by all model organisms is anatomy. Anatomical structures can be described at the subcellular (e.g., nucleus) and cellular (e.g., lymphocyte) level as well as in terms of tissues (e.g., adipose tissue), organs and organ parts (e.g., heart, aortic valve), body systems and their components (e.g., central nervous system, brain) and entire organisms. Anatomy is in fact central to the biomedical domain as the description of virtually every biological process makes reference to some anatomical structure. For example, the information stored about microarray experiments routinely includes tissue and cell type. Many representations of anatomy have been developed for various model organisms [1]. While some of them are mere lists of anatomical names for anatomical entities (e.g., Terminologica Anatomica), others are full-fledged ontologies, organizing anatomical entities in hierarchies (isa, part of) in order to support reasoning (e.g., Foundational Model of Anatomy1).
The general framework of this study is that of ontology alignment. For a survey of alignment techniques, the interested reader is referred to [2]. Lexical features (i.e., the names of anatomical entities) and structural features (e.g., the relations described among anatomical entities) can be used to compare representations of anatomies within and across species. In previous research, we have developed methods for comparing two representations of human anatomy [3]. The objective of this study is to align two ontologies of anatomy, for human and mouse anatomy. In other words, we aim at identifying equivalent anatomical entities between the NCI Thesaurus (human anatomy) and the Adult Mouse Anatomical Dictionary (mouse anatomy). The originality of this study lies in using two independent approaches (lexical and manual), followed by a structural validation and a manual evaluation of the results. This study is a contribution to the caBIG project2.
MATERIALS
The two resources under investigation in this study are part of the Open Biomedical Ontologies3 (OBO).
NCI Thesaurus
Published by the National Cancer Institute (NCI), this thesaurus4 contains the working terminology of many data systems in use at NCI [4]. Its scope is broad as it covers vocabulary for clinical care as well as translational and basic research. Among its 37,386 concepts, 4,410 (11.8%) correspond to anatomical entities (anatomic structure, system, or substance hierarchy). For example, the concept liver is identified by C12392 and has several synonyms (e.g., hepatic organ system). Additionally, liver is subsumed by organ and related to abdominal cavity (has_location) and to gastrointestinal system (is_physical_part_of). The version used in this study is version 04.09a (September 10, 2004).
Adult Mouse Anatomical Dictionary
The Adult Mouse Anatomical Dictionary5 has been developed as part of the mouse Gene Expression Database (GXD) project [5] to provide standardized nomenclature for anatomical entities in the postnatal mouse [6]. It will be used to annotate and integrate different types of data pertinent to anatomy, such as gene expression patterns and phenotype information, which will contribute to an integrated description of biological phenomena in the mouse. The ontology contains more than 2,400 unique terms, is structured as a directed acyclic graph (DAG) and is organized hierarchically in both spatial and functional ways. For example, the concept liver is identified by MA:0000358 and is a ‘child’ of (is_a) abdomen organ as well as part_of the liver/biliary system. The version used in this study was downloaded on December 22, 2004 (under the name Mus adult gross anatomy in OBO).
Of the 4,410 NCI Thesaurus terms, over 2,000 terms correspond to entities that are not included in the Adult Mouse Anatomical Dictionary, such as cell types and subcellular components. Thus, only about 2,400 would be expected to be candidates for matching.
METHODS
The method used for aligning the NCI Thesaurus (NCI) and Adult Mouse Anatomical Dictionary (MA) was originally developed by the authors for aligning two ontologies of human anatomy: the Foundational Model of Anatomy (FMA) and GALEN [3] and can be summarized as follows. Concept names and relations are extracted from each ontology. In the lexical approach, additional synonyms are collected. All names are normalized and compared across ontologies. Lexically similar names form the basis for identifying equivalent concepts. Structural similarity (e.g., shared relations to other equivalent concepts) is required for concepts to be aligned.
Independently of the lexical approach, a manual alignment is performed, resulting in a different set of equivalent concepts. Structural similarity is also computed on this set in order to eliminate from the alignment the concepts failing to be supported by structural evidence.
The evaluation consists of comparing the sets of equivalent concepts resulting from each approach. Equivalent concepts are reviewed manually by an expert for accuracy. An overview of the methods is presented in Figure 1.
Figure 1.
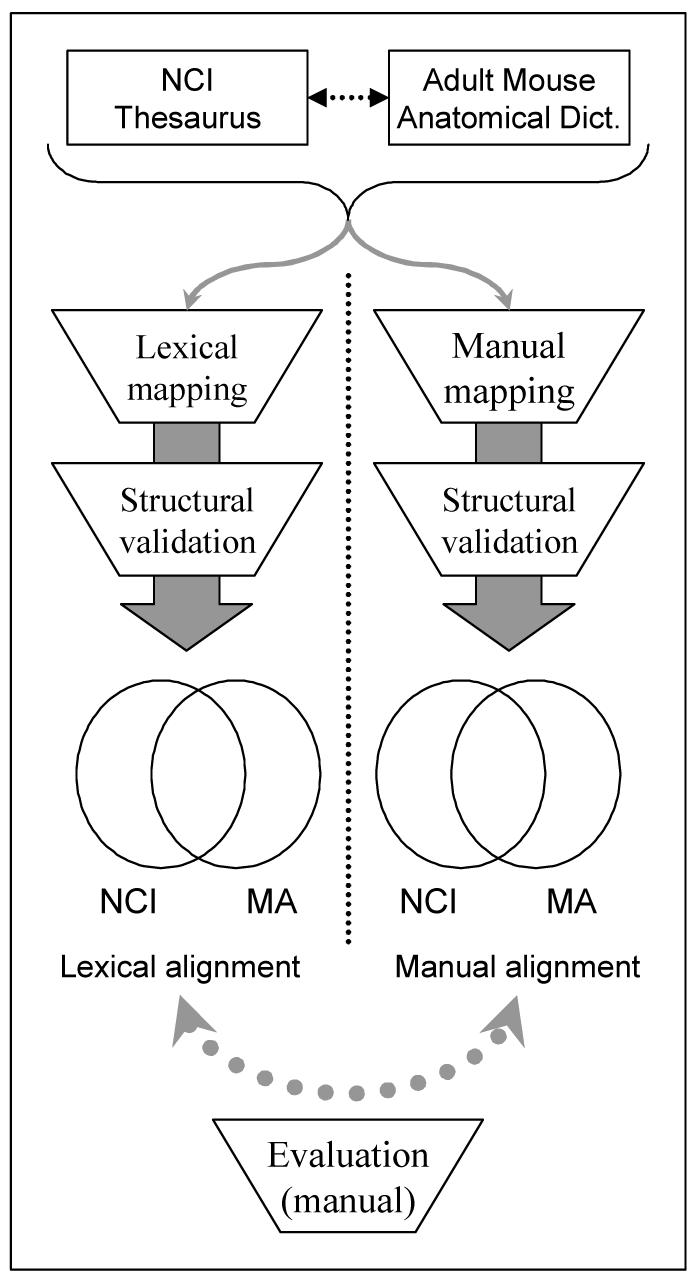
Overview of the alignment methods
Lexical approach
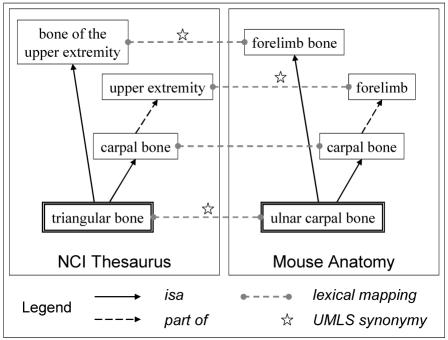
The lexical alignment identifies shared concepts across systems lexically through exact match and after normalization. Concepts exhibiting similarity at the lexical level across systems are called anchors, as they are going to be used as reference concepts in the structural alignment. Additional anchors are identified through UMLS synonymy. Two concepts across systems are considered anchors if their names are synonymous in the UMLS Metathesaurus (i.e., if they name the same UMLS concept) and if the corresponding concept is in the anatomy domain (i.e., has a semantic type related to Anatomy). For example, the NCI term triangular bone and the MA term ulnar carpal bone, although lexically different, were mapped through the synonymy of these two terms in the UMLS Metathesaurus.
Manual approach
This approach utilized the search capabilities of the available web-based ontology browsers for the NCI Thesaurus6 and Adult Mouse Anatomical Dictionary7 to identify potential matching concepts between the two ontologies. A manual comparison of adult mouse anatomy terms against the entire NCI Thesaurus was performed using lexical similarity and synonymy in both sources. This process was repeated in order to match each mouse term against a list of approximately 4,400 human anatomy terms provided by NCI. Additional matches were identified by a domain expert (TH) familiar with both mouse and human anatomy. All matches were then validated using available anatomy reference sources.
Validation by structural similarity
The structural alignment first consists of acquiring the semantic relations explicitly represented in each system. In order to facilitate the comparison of relations across systems, the transitive closure of isa relations is computed in each system, as well as that of part_of relations. With these semantic relations, the structural alignment identifies structural similarity among anchors across systems. Structural similarity, used as positive structural evidence, is defined by the presence of at least one common hierarchical relation among anchors across systems, e.g., <c1, part_of, c2> in one system and <c1’, part_of, c2’> in another where {c1, c1’} and {c2, c2’} are anchors across systems. For example, the anchor concepts triangular bone in the NCI Thesaurus and ulnar carpal bone in Mouse Anatomy, presented earlier, received positive structural evidence because they share hierarchical links to some of the other anchors across systems. As illustrated in Figure 2, triangular bone is related to bone of the upper extremity (isa) and to upper extremity (part of) through carpal bone (isa). These relations from the NCI Thesaurus mirror relations among equivalent concepts in the Mouse Anatomy. The structural validation is performed automatically in both cases, but separately on the set of equivalent concepts identified by each approach (lexical and manual).
Figure 2.
Structural validation following lexical alignment
Evaluation
The evaluation consists of comparing the sets of equivalent concepts obtained by lexical and manual alignment, after eliminating the concepts not supported by structural similarity. An additional manual review of the pairs identified specifically by either lexical or manual alignment was performed by an expert (TH).
RESULTS
Quantitative results
As mentioned earlier, of the 4,410 NCI Thesaurus terms, over 2,000 terms correspond to entities that are not included in the Adult Mouse Anatomical Dictionary. Thus, only about 2,400 NCI Thesaurus terms would be expected to be candidates for matching against the 2,404 terms in the Adult Mouse Anatomical Dictionary.
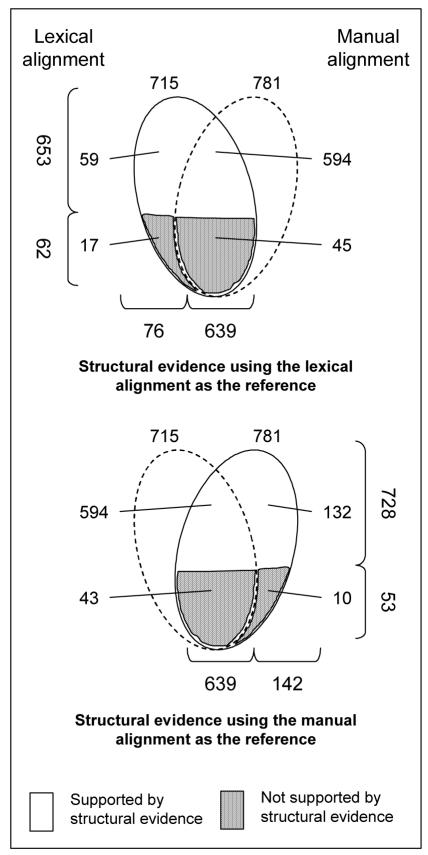
As illustrated in Figure 3, the lexical alignment identified 715 equivalent concepts (anchors), while the manual identified 781. Of these, the structural alignment removed the anchors for which no structural evidence was found: 62 (8.7%) for the concepts aligned lexically and 53 (6.8%) for those aligned manually, leaving 653 pairs of equivalent concepts for the lexical alignment and 728 for the manual alignment.
Figure 3.
Results after structural validation
The intersection of these two sets of concepts contains 639 concepts. In other words, most equivalent concepts are identified by both approaches simultaneously. The vast majority of these 639 mappings is supported by structural evidence. Only 45 of them (7.0%) are rejected for lack of structural evidence when the lexical alignment is used as the reference (43 when the manual alignment serves as the reference). 76 concepts were considered equivalent by the lexical method only, while 142 concepts were identified by the manual approach only.
Qualitative results
Mappings identified by both approaches
All of the 639 mappings identified simultaneously by both approaches were determined to be appropriate matches by a domain expert, including the 43 or 45 for which no structural evidence was found, depending which alignment is used as the reference. Examples of such mappings include the pairs {uterine cervix (MA:000392) and Cervix Uteri (NCI:C12311)}, identified by lexical similarity and supported by structural evidence, and {tendon (MA:0000115) and Tendon (NCI:C13045)}. The latter pair was not supported by structural evidence because it is related to connective tissue (isa) in MA but to Musculoskeletal System (part of) in NCI.
Mappings specific to the lexical approach
Of the 76 mappings identified by the lexical method only, 61 (80.2%) were deemed to be appropriate matches which, in theory, should have been picked up by the manual approach. Use of UMLS synonymy clearly augmented the sensitivity of the lexical approach. The remaining 15 concept pairs were not considered to be valid.
Examples of valid mappings specific to the lexical approach include the pairs {urinary bladder urothelium (MA:0001693) and Transitional Epithelium (NCI:C13318)} (where MA provides transitional epithelium as a synonym for urinary bladder urothelium) and {lienal artery (MA:0001991) and Splenic Artery (NCI:C33597)} (where the synonymy between the two terms comes from the UMLS concept Structure of splenic artery (UMLS:C0037996)).
The following mapping was considered invalid: between cerebellum lobule I (MA:0000998) and Lingula of the Lung (NCI:C40373), both terms having lingula as a synonym. In both cases, lingula refers to a tongue-shaped entity, but one is part of the cerebellum, while the other is part of the lung.
Mappings specific to the manual approach
Of the 142 mappings identified only by the manual approach, 133 (93.7%) were validated as matches. In 6 of the 9 remaining mappings, an operator-generated error was involved; that is, incorrect numerical identifiers had been used for one of the terms. In each of these cases, at least one of the terms in the set was found to have another, more appropriate match; 4 of these were already represented on the lists, while 2 new matches were identified.
Examples of valid mappings specific to the manual approach include the pairs {alveolus epithelium (MA: 0001771), Alveolar Epithelium (NCI: C12867)} and {cervical vertebra 1 (MA:0001421) and C1 Vertebra (NCI:C32239)}. In these cases, the equivalence is easy to determine for an expert. However, the lexical approach failed to identify these mappings because the terms are different and no common synonym is provided by either source or found unambiguously in the UMLS.
DISCUSSION
Applications of the mapping for biologists
The mapping between human and mouse anatomies is one element of a broader framework under development whose objective is to support reasoning about diseases across various model organisms. More precisely, the mapping between human and mouse anatomies together with mappings between diseases in humans and mice will enable comparative science, i.e., will help validate the use of mouse models of human disease. In practice, instead of creating a unique ontology for the anatomies of various species, relations such as Anatomy Equivalent_To Anatomy will be created in the NCI Thesaurus between human and mouse anatomical entities. Analogously, the relationship Disease Similar_To Disease will be used to relate human and mouse diseases. Users of the mapping between mouse and human anatomies will include, for example, researchers from the NCI Mouse Models of Human Cancers Consortium8 (MMHCC).
The following hypothetical use case illustrates the practical application of this framework. Researchers studying small cell lung carcinoma (SCLC) in humans may want to use a mouse model of lung cancer to test a new therapeutic agent. In order to find out whether there is a mouse equivalent of SCLC in humans, they start their search from SCLC (human diseases) and use the link Similar_To to reach diseases in mice. Such a link would lead possibly to Neuroendocrine Carcinoma of the Mouse Pulmonary System. Analogously, the links between anatomies would help determine, for example, correspondences between parts of the mouse lung and the parts of the human lung of interest in their study. The availability of such correspondences would also greatly facilitate the retrieval by clinicians familiar with human anatomy and diseases of scientific information about the neuroendocrine carcinoma models in mouse.
Applications of the mapping for ontologists
Many large-scale alignment experiments lack thorough validation, because it requires a manual review of the mappings by experts, which is a labor intensive task. This study provides a curated mapping between the human and mouse anatomy ontologies. However, one limitation of this study is that evaluation of the alignment was performed by the same expert who also performed the manual alignment. In practice, the results show that the expert reversed some of her initial judgements during the evaluation, which confirms that the bias, if any, is not important.
The large proportion of mappings identified by the fully automatic lexical approach and deemed satisfactory by the expert confirmed that the alignment techniques we developed for large ontologies remain valid on smaller ontologies. The large proportion of valid mappings not supported by structural evidence also indicates that this alignment technique is rather conservative in evaluating validity. Moreover, assuming our definition of structural evidence is correct, the presence of valid mappings not supported by structural evidence essentially reveals a lack of relationships represented in the ontologies or differing modeling choices between them.
The analysis of fine differences between human and mouse anatomies is beyond the scope of this paper but is addressed in [7]. However, a careful analysis of the mappings (and failures) also reveals differences between the two ontologies and thus helps identify issues in the representation of anatomical entities, including missing and inaccurate synonyms and relations. The results obtained will serve as a guide in addressing these weaknesses, and in complementing and harmonizing the anatomical concepts and relations in both ontologies. As the human and mouse ontologies are refined and harmonized, the mapping approaches presented here can also help monitor progress.
Lexical vs. manual
Both approaches showed strengths and weaknesses. The lexical approach performs consistently, at a low cost, but its model of lexical similarity is limited. In contrast, experts may identify mappings in the absence of lexical similarity, but they also sometimes make mistakes, and their knowledge comes at a high price. This study suggests a strategy in which the results of an automatic mapping (combining lexical and structural similarity) could be used to direct the attention of experts on those cases where their knowledge is required. Such a strategy would make it possible to achieve high quality mapping with limited human resources and to bring consistency to the mapping between large ontologies.
The limited overlap found between human anatomy in NCI and mouse anatomy is not due to the lack of power of our methods, but rather to differences in coverage. For example, cell types, developmental stages and subcellular entities are covered by NCI, but not by MA. The presence of mouse-specific anatomical structures in MA also accounts for some of the differences.
Acknowledgements
The research was supported in part by an appointment to the National Library of Medicine Research Participation Program administrated by the Oak Ridge Institute of Science and Education through an interagency agreement between the U.S. Department of Energy and the National Library of Medicine.
Footnotes
References
- 1.Bard JB. Anatomics: the intersection of anatomy and bioinformatics. J Anat. 2005;206(1):1–16. doi: 10.1111/j.0021-8782.2005.00376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noy NF. Tools for mapping and merging ontologies. In: Staab S, Studer R, editors. Handbook on Ontologies: Springer-Verlag; 2004. p. 365–384
- 3.Zhang S, Bodenreider O. Aligning representations of anatomy using lexical and structural methods. AMIA Annu Symp Proc. 2003:753–7. [PMC free article] [PubMed] [Google Scholar]
- 4.De Coronado S, Haber MW, Sioutos N, Tuttle MS, Wright LW. NCI Thesaurus: Using Science-based Terminology to Integrate Cancer Research Results. Medinfo. 2004;2004:33–7. [PubMed] [Google Scholar]
- 5.Hill DP, Begley DA, Finger JH, Hayamizu TF, McCright IJ, Smith CM, et al. The mouse Gene Expression Database (GXD): updates and enhancements. Nucleic Acids Res. 2004;32(Database issue):D568–71. doi: 10.1093/nar/gkh069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayamizu TF, Mangan M, Corradi JP, Kadin JA, Ringwald M. The Adult Mouse Anatomical Dictionary: a tool for annotating and integrating data. Genome Biology. 2005;6(3):R29. doi: 10.1186/gb-2005-6-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Travillian RS, Gennari JH, Shapiro LG. Of mice and men: Design a comparative anatomy information system. AMIA Annu Symp Proc 2005:(in press) [PMC free article] [PubMed]