Abstract
We report on the use of medical facts to support the enhancement of natural language processing of biomedical text. Inferencing in semantic interpretation depends on a fact repository as well as an ontology. We used statistical methods to construct a repository of drug-disorder co-occurrences from a large collection of clinical notes, and this resource is used to validate inferences automatically drawn during semantic interpretation of Medline citations about pharmacologic interventions for disease. We evaluated the results against a published reference standard for treatment of diseases.
INTRODUCTION
Natural language processing is increasingly used in the medical domain to support innovative information management applications. A variety of approaches are being pursued to address clinical text. Some recent examples include a semantic grammar for automatic coding of patient data [1], a definite clause grammar to connect the research literature with patient records [2], and machine learning to determine patient medication status [3]. Focusing on the research literature, SemRep [4] uses underspecified semantic interpretation to recover semantic predications from Medline citations. For example, SemRep identifies (2) from (1).
(1) OBJECTIVE: To evaluate the efficacy of donepezil in patients with early-stage Alzheimer disease.
(2) donepezil TREATS Patients
Alzheimer’s Disease OCCURS_IN Patients
Such output is being used to support automatic summarization of the literature on treatment of disease [5] as well as the extraction of information on the genetic basis of disease [6]. However, a class of linguistic structures impedes SemRep’s ability to identify useful information in medical text. In such structures the complete intent of the author is not overtly expressed. For example, from (1) the author intends that we understand (3), which is not asserted, but can be inferred.
(3) donepezil TREATS Alzheimer’s Disease
It is important to address inferencing in natural language processing systems, since not doing so impairs recall performance. However, drawing inferences correctly is not straightforward. For example, it is not valid to infer (5) from (4).
(4) Are beta-blockers efficacious in patients with diabetes mellitus?
(5) Adrenergic beta-Antagonists TREATS Diabetes Mellitus
Felicitous inferencing depends on knowledge beyond that needed for interpreting asserted predications. The reader is expected to know, for example, that donepezil is used to treat Alzheimer’s disease and that adrenergic beta-antagonists are not used for diabetes. In natural language processing a fact repository can be exploited to provide this kind of information. In this paper we describe the construction of such a repository about drug-disease interactions and test its use to support valid inferencing in SemRep processing.
We extracted facts about specific drugs used to treat particular diseases from more than 16 million patient records at the Mayo Clinic. Statistical methods were used to insure that statements about each drug are valid. We then exploit these facts to assign a “medical validity” value to inferences drawn by SemRep. Inferences having a validity value below an empirically determined cut-off are eliminated. To evaluate the effectiveness of this processing we applied SemRep to Medline citations on the treatment of three disorders and evaluated the final results in comparison to curated synopses of interventions for these same diseases.
BACKGROUND
Semantic Interpretation
SemRep relies on domain knowledge in the Unified Medical Language System® (UMLS)® and underspecified linguistic analysis to provide partial semantic interpretation of biomedical text. Input is assigned a syntactic structure based on the SPECIALIST Lexicon [7] and the MedPost tagger [8], which resolves part-of-speech ambiguities. Each noun phrase in this analysis is mapped to a concept in the Metathesaurus using MetaMap [9]. The syntactic structure for (6) enhanced with Metathesaurus concepts is given schematically as (7). UMLS concepts, followed by semantic types (abbreviated), are enclosed in double quotes.
(6) Glucantime for the treatment of cutaneous leishmaniasis
(7) [[“Glucantime:phsu”]NP [for the “treatment(therapeutic aspects):ftcn”]NP [of “Leishmaniasis, Cutaneous:dsyn”]NP]
In identifying semantic predications, this structure is used in conjunction with two further resources: rules that map syntactic “indicators” to semantic predicates and relationships in the UMLS Semantic Network (enhanced for natural language processing). Indicators include verbs, nominalizations, and prepositions. In (7) the nominalization treatment is mapped to (or indicates) the relation TREATS in the Semantic Network. SemRep then uses a Semantic Network relationship (8) to sanction UMLS concepts “Glucantime” (with semantic type ‘Pharmacologic Substance’) and “Leishmaniasis, Cutaneous” (‘Disease or Syndrome’) as arguments. The final interpretation of (6) is (9).
(8) Pharmacologic Substance (phsu) TREATS Disease or Syndrome (dsyn)
(9) Glucantime TREATS Leishmaniasis, Cutaneous
Inferencing
Reasoning is a way of extending current knowledge, and inferencing is a kind of reasoning controlled by a particular type of rule. In artificial intelligence research, inferencing is implemented with rules of the form: IF <condition> THEN <consequent>. Such rules have been used extensively in constructing expert systems [10], including MYCIN [11] in the medical domain.
Language understanding depends on interpreting assertions as well as inferencing based on those assertions [12]. The condition on inferencing rules in language understanding is stated as already asserted semantic predications and the consequent is a new predication. Currently, inferencing in SemRep concentrates on treatment of disease and is controlled by the rule given in (11).
(11) X TREATS Y & Z OCCURS_IN Y
→ X TREATS Z
The variables in (11) refer to semantic types corresponding to arguments of the relevant predicates from the UMLS Semantic Network. For example, X covers such semantic types as ‘Pharmacologic Substance’ and ‘Medical Device’, while Y includes the semantic type ‘Disease or Syndrome’. Predications satisfying this rule must have arguments whose semantic types match the variables in the rule.
The rule in (11) applies to predications derived from text such as (12), namely the first two predications in (13). These satisfy the conditions in (11) and the third predication in (13) is produced by the consequent.
(12) Glucantime for patients with cutaneous leishmaniasis
(13)Glucantime TREATS Patients
Leishmaniasis, Cutaneous OCCURS_IN Patients
Glucantime TREATS(INFER) Leishmaniasis, Cutaneous
In addition to linguistic knowledge, the interpretation of an assertion depends on ontological information [13], while inferencing also requires factual information, as noted earlier. The thrust of the research reported here is to provide SemRep with medical facts sufficient for the felicitous construction of inferences about pharmacologic interventions for disease.
MATERIALS AND METHODS
Clinical Text to Support a Fact Repository
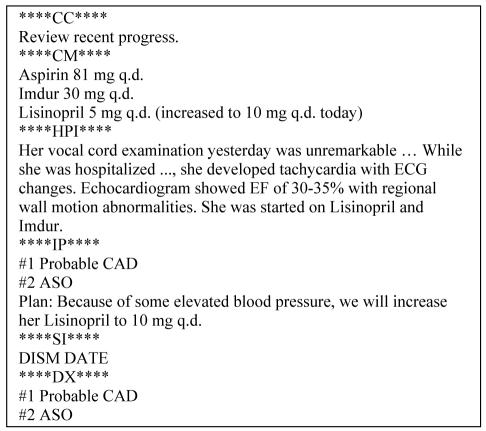
A clinical fact repository was generated from a corpus of over 16 million (de-identified) clinical notes recorded at the Mayo Clinic. These notes follow the HL7 Clinical Document Architecture guidelines [14] and are semi-structured. An example appears in Figure 1.
Figure 1.
Composite Clinical Note from the Mayo Clinic
The Current Medications (CM) and Final Diagnosis (DX) sections are of particular interest to this study because they contain medication and disorder information relevant to constructing a probabilistic fact repository. We automatically annotated the drugs and disorders in these sections using dictionary lookup with RxNorm and SNOMED-CT serving as terminological resources. The former provides standard names for drugs, while the latter has broad coverage of concepts in clinical medicine. We also flagged negated entities using a generalization of the NegEx algorithm [15], which takes advantage of cues such as no evidence of and denies, among others.
We then computed the frequency of each drug-disorder cooccurrence in the corpus as well as the frequency with which each drug and disorder occurred alone. (Negated entities were not counted). In order to reduce noise, drug-disorder pairs that occurred fewer than 4 times were excluded from calculations, as is common in statistical natural language processing.
Hierarchical Concept Matching
In order to account for discrepancies between drug naming conventions in Medline citations and in Mayo clinical text, we exploited hierarchical structure in the UMLS Metathesaurus. We used MetaMap to process both drugs and disorders in the Mayo repository and in SemRep predications. Mayo terms were mapped to Metathesaurus concepts, and the preferred name was kept along with the input repository term. We also used the UMLS Knowledge Source Server [16] to retrieve Metathesaurus hierarchical contexts for drugs and disorders in SemRep predications (which are Metathesaurus preferred names). For example, “Benzodiazepines” has children “Bromazepam,” “Chlordiazepoxide,” “Chlordiazepoxide” “hydrochloride,” “Clobazam,” “Clonazepam,” “Clorazepate,” among many others.
In subsequent comparison between SemRep drugs and disorders and those from the Mayo repository, a match was allowed if the two terms occurred in the same hierarchical context. For example the term “Benzodiazepines” found in a SemRep predication does not occur in the repository. Hierarchical context processing, however, allowed a match with “clonazepam,” the term used at the Mayo Clinic.
If a match between a SemRep term and one in the Mayo repository occurs at a level other than that of a sibling or a child, we assign a score that grows geometrically with the distance of the matched concept from the original. For example, “panic disorder” is a three-level child of “neurotic disorder” and will match to that concept with a score of 33 greater than to “panic disorder.”
Predicting “Medical Validity”
The hierarchical matching score is then used to adjust the ranking of the drug-disorder cooccurrences: The higher the score, the lower the ranking. Some examples of the final values assigned to drug-disorder cooccurrences in the Mayo clinical notes are given in (15). It can be determined from (15), for example, that of all the disorders cooccurring with nifedipine in the same clinical note, scleroderma ranks 114th in frequency.
(15) nifedipine|scleroderma|114
omeprazole|gastroesophageal reflux disease|4
omeprazole|angina pectoris|102
ranitidine|gastroesophageal reflux disease|7
We then established a threshold for this value in predicting whether the cooccurrence is medically valid. We currently use a threshold of 80, with the consequence that a drug-disorder cooccurrence ranked higher than 80 indicates that the drug is probably used to treat the disorder. This score is used to validate SemRep inferences.
For example, the predications in (16) were determined to be true by this method, while those in (17) were not. That is, the inference that omeprazole treats gastroesophageal reflux disease is valid because the drug occurs very frequently with the disease in Mayo clinical notes (4th most frequent cooccurrence).
(16) Omeprazole TREATS(INFER) Gastroesophageal reflux disease
Ranitidine TREATS(INFER) Gastroesophageal reflux disease
(17) Omeprazole TREATS(INFER) Chest Pain
Nifedipine TREATS(INFER) Diffuse Scleroderma
Evaluation
We performed two evaluations to assess the effectiveness of the Mayo fact repository for supporting inferencing in SemRep. The first compared validated inferences against a test collection of 202 inferences marked as true or false by one of the authors (MF). The second evaluation compared drugs obtained from SemRep output to a curated reference standard for treatments of disease [17].
The task-oriented evaluation was centered around drug therapies for three diseases, acute myocardial infarction, Alzheimer’s disease, and panic disorder. For each disease a PubMed query on the name of disease was issued using a methodological filter for therapy [18] and limited to studies on humans with abstracts in English. The most recent 500 citations up to the date of the reference standard were processed by SemRep (with inferencing) and inferences were validated by the Mayo repository. We then created three sets of TREATS predications for each disease. These include SemRep predications with out inferencing (noninferences) as well as both validated and unvalidated inferences. The three sets are: (a) noninferences only, (b) noninferences and unvalidated inferences, and (c) noninferences and validated inferences (only). Drugs were extracted from these groups and compared manually (by MF) to the reference standard for each of the three diseases. For this evaluation we considered the following categories in the reference: Beneficial, Likely to be beneficial, Trade-off between benefits and harms, and Unknown effectiveness.
Assessments of accuracy were made based on the following considerations. If the reference standard refers to an intervention class, members of the class extracted from SemRep output were considered correct, as, for example, paroxetine from SemRep and serotonin uptake inhibitors in the standard. However, a class from SemRep (acetylcholinesterase inhibitors, for example) compared to a member in the standard (donepezil) was marked as an error. Synonyms, such as acetylcholinesterase inhibitors and cholinesterase inhibitors, were allowed.
After marking false positives and negatives, precision and recall were calculated and combined into an F-score. Precision was figured as the total number of drugs retrieved for a disorder that matched the four categories of the reference standard divided by the total number of interventions retrieved. Recall was the total number of interventions for a disorder that matched the four categories of the reference standard divided by the total number of interventions in the reference standard categories. The F score was calculated with β=1; F = (2 x P x R) / (P + R).
RESULTS
In the first test, which evaluated validated inferences directly against a test collection, 81% of the unvalidated SemRep inferences were correct when compared to the test collection, while precision for the inferences validated by the Mayo repository was 89.5%, for an increase of 8%. In this evaluation, inferences were not categorized by disease.
Results of the second evaluation are given in Table 1. The three categories for each disease correspond to the three sets of TREATS predications noted earlier. The category labeled “Inferences” in the table includes noninferences and unvalidated inferences, while “With facts” includes noninferences and validated inferences. It should be noted that this test is more challenging than the first. The inference must correctly reflect the intent of the text, but, further, must agree with the reference standard [17].
Table 1.
Performance Measures. P=Precision, R=Recall, F= F-score, N=Number of drugs retrieved by SemRep.
| Disease | SemRep | P | R | F | N |
|---|---|---|---|---|---|
| Acute myocardial infarction | No inferences | 69% | 79% | 0.73 | 16 |
| Inferences | 47% | 88% | 0.61 | 32 | |
| With facts | 67% | 88% | 0.76 | 21 | |
| Alzheimer's Disease | No inferences | 21% | 57% | 0.30 | 63 |
| Inferences | 18% | 65% | 0.28 | 85 | |
| With facts | 22% | 63% | 0.33 | 68 | |
| Panic disorder | No inferences | 53% | 95% | 0.68 | 40 |
| Inferences | 45% | 100% | 0.62 | 49 | |
| With facts | 54% | 100% | 0.70 | 41 | |
| Overall | No inferences | 38% | 76% | 0.51 | 119 |
| Inferences | 31% | 84% | 0.46 | 166 | |
| With facts | 39% | 82% | 0.53 | 130 |
DISCUSSION
The direct evaluation suggests that a fact repository of drug-disorder cooccurrences generated from clinical notes can validate inferences produced during automatic semantic interpretation with SemRep. Inference validation of the type discussed here could also be supported (perhaps more effectively) by compiled sources of drug indications. However, publicly accessible, comprehensive, online resources of this kind are not readily available.
We are not discouraged by the absolute values in the task-oriented evaluation. We conducted this test to assess the use of a fact repository for validating inferences, and the results suggest that using the Mayo repository consistently improves SemRep output, both without inferencing as well as with unvalidated inferencing.
The F-score for Alzheimer’s disease is notably lower than the results for the other two disorders. This may in part be due to the large number of drugs exhibiting a complex interaction of characteristics that are relevant to Alzheimer’s disease. The perhaps more focused pharmacopoeia for acute myocardial infarction may allow SemRep to produce better results. A further phenomenon encountered in the task-oriented evaluation is that 31% of drugs produced by SemRep were from predications actually asserted in Medline, but not in the reference standard.
Clearly there are curation issues that go beyond accuracy in semantic interpretation, and we are not suggesting that automatic methods can replace the human expertise required to produce a reference standard for treatment of disorders. However, the method we propose might assist human curation by making initial suggestions.
Based on the method we are developing, we suggest that semantic interpretation enhanced with validated inferences (and perhaps supplemented with automatic summarization [5]) could be used for other applications as well. For example, the information extracted from Medline citations by this process might be used to compile profiles on selected drugs, including indications, effectiveness and adverse effects.
CONCLUSION
We propose a method for determining the likelihood that an inference produced by a semantic interpreter, SemRep, has medical validity. We use statistical techniques to process drug-disorder cooccurrences in clinical notes and then use this information to validate SemRep inferences about drug treatment for disease. The method was evaluated against a published reference standard for disease treatments and shows considerable promise for enhancing natural language processing to support biomedical applications.
References
- 1.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mendonça EA, Johnson SB, Seol YH, Cimino JJ. Analyzing the semantics of patient data to rank records of literature retrieval. Proc NAACL Workshop on Natural Language Processing in the Biomedical Domain. 200:269–76. [Google Scholar]
- 3.Pakhomov SV, Ruggieri A, Chute CG. Maximum entropy modeling for mining patient medication status from free text. Proc AMIA Symp. 2002:587–91. [PMC free article] [PubMed] [Google Scholar]
- 4.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;36(6):462–77. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Fiszman M, Rindflesch TC, Kilicoglu H. Abstraction summarization for managing the biomedical research literature. Proc HLT-NAACL Workshop on Computational Lexical Semantics. 2004:76–83. [Google Scholar]
- 6.Libbus B, Kilicoglu H, Rindflesch TC Mork JG, Aronson AR. Using natural language processing, Locus Link, and the Gene Ontology to compare OMIM to MEDLINE. Proc HLT-NAACL Workshop on Linking the Biological Literature, Ontologies and Databases: Tools for Users 2004;69–76.
- 7.McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proc Annu Symp Comput Appl Med Care. 1994:235–9. [PMC free article] [PubMed] [Google Scholar]
- 8.Smith L, Rindflesch T, Wilbur WJ. MedPost: a part-of-speech tagger for biomedical text. Bioinformatics. 2004;20(14):2320–1. doi: 10.1093/bioinformatics/bth227. [DOI] [PubMed] [Google Scholar]
- 9.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 10.Barr A, Feigenbaum EA. The handbook of artificial intelligence. Los Altos, CA: William Kaufman Inc.; 1981.
- 11.Shortliffe E. Computer-based medical consultations: MYCIN. New York: Elsevier; 1976.
- 12.Wilson D, Sperber D. Linguistic form and relevance. Lingua. 1993;90:1–25. [Google Scholar]
- 13.Nirenburg S, Raskin, V. Ontological semantics. Cambridge, MA and London: The MIT Press; 2004.
- 14.http://hl7.org/library/standards.cfm
- 15.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001 Oct;34(5):301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 16.Bangalore A, Thorn KE, Tilley C, Peters L. The UMLS Knowledge Source Server: An object model for delivering UMLS data. Proc AMIA Symp. 2003:51–55. [PMC free article] [PubMed] [Google Scholar]
- 17.Clinical Evidence concise. British Medical Journal.
- 18.Wong SS, Wilczynski NL, Haynes RB. Developing Optimal Search Strategies for Detecting Clinically Relevant Qualitative Studies in MEDLINE. Medinfo. 2004:311–6. [PubMed] [Google Scholar]