Abstract
Drug-drug interaction systems exhibit low signal-to-noise ratios because of the amount of clinically insignificant or inaccurate information they contain. MEDLINE represents a respected source of peer-reviewed biomedical citations that potentially might serve as a valuable source of drug-drug interaction information, if relevant articles could be pinpointed effectively and efficiently. We evaluated the classification capability of Support Vector Machines as a method for locating articles about drug interactions. We used a corpus of “positive” and “negative” drug interaction citations to generate datasets composed of MeSH terms, CUI-tagged title and abstract text, and stemmed text words. The study showed that automated classification techniques have the potential to perform at least as well as PubMed in identifying drug-drug interaction articles.
Introduction
Improving the coverage and accuracy of drug-drug interactions can improve delivery of safe and cost-effective patient care[1]. The incidence of drug-drug interactions (DDIs) in patients may range from 4.7% to 8.8%[2], yet many knowledge bases fail to include important drug interactions and contain outdated, irrelevant, or even incorrect information [3–5].
Inadequate techniques for collecting, filtering, and maintaining drug interaction information may be responsible for the incomplete and unreliable information in these knowledge sources. Current pharmaceutical research produces approximately 300,000 MEDLINE citations per year that are labeled with MeSH terms from the “Chemicals and Drugs Category,” and a representative reference publication, Physician’s Desk Reference, has grown from 2,787 pages in 1995 to 3,440 pages in 2005. Unfortunately, filtering articles for new drug information remains predominantly a manual task [6]. Pharmacists responsible for maintaining DDI databases cannot easily review every published article; consequently, both new and revised drug interaction data may be overlooked.
The authors intend to investigate the feasibility of automating portions of the DDI identification process. This report details the first step in this series, and discusses improving the yield from MEDLINE, a potential source of DDI information. Later steps will potentially include tools for extracting interacting drug pairs, evaluating the type and severity of the interaction, and applying this knowledge to drug database creation and maintenance.
This study developed and evaluated an automated method for identifying articles about drug-drug interactions using MEDLINE, a publicly accessible and potentially rich source of DDI information [7]. MEDLINE’s appeal lies not only in its status as a major repository of biomedical literature references [8], but also in that it is an underutilized tool by drug database developers [9].
We hypothesized that text processing and machine learning techniques could identify a set of DDI articles more readily than queries through the PubMed MEDLINE interface. In particular, Support Vector Machines (SVMs), a broadly applicable machine learning technique, have already proven successful at text classification [10] and identifying MEDLINE references according to their quality and usefulness in a clinical setting [11]. SVMs attempt to classify data by projecting vectors of variables into a higher feature space and finding the hyperplane that maximally separates them.
We hope to enhance MEDLINE’s value as a source of DDI information by facilitating identification of new pharmaceutical publications. This effort ultimately will assist in constructing more complete and relevant drug-drug interaction databases.
Methods
Defining a Drug-Drug Interaction Article
Our objective was to locate MEDLINE articles that provide drug-drug interaction information worthy of inclusion in a DDI knowledge base. We define “drug-drug interaction article” as referring only to publications that contain instructive information about the effects of two drugs on each other and on the patient. We excluded (as irrelevant to the study) publications that only discussed the impact of general drug interactions (ie. effect on hospital length-of-stay, dangers of polypharmacy, financial consequences.)
Constructing a Corpus
We first hand-classified a set of 500 MEDLINE citations in order to estimate the prevalence of DDI references in MEDLINE. This process revealed a 1% prevalence of these drug-drug interaction citations in MEDLINE’s collection. We elected to develop a corpus that had an enriched prevalence (10%) of DDI articles for this feasibility study. Therefore, we manually created a corpus of 2000 MEDLINE references, with publication dates between 1985 and 2002, inclusive. The publication era was restricted to reduce the temporal bias resulting from yearly changes in MEDLINE indexing techniques – indexing may have produced significant discrepancies in the classification of an article published in 2004 and from a similar one published in 1970.
To generate a sufficient number of positives for our dataset, we required a reliable and recognized source of influential drug-drug interaction articles. We began by composing a list of recently verified drug-drug interactions. We identified our institution’s care provider order entry system as a good source of well-maintained and expert-reviewed drug interactions: its drug interaction database has been manually curated by expert hospital pharmacists for more than two decades and the database of over 500 significant interactions has evolved over time to exclude false-positive warnings [12]. We randomly selected 150 DDIs from this list to serve as a collection of expert-validated drug-drug interactions.
Next, we transformed the list of 150 pairs of interacting medications into a set of corresponding MEDLINE references from the pre-defined study period. We selected eFacts Online’s Drug Interaction Facts database as a reputable and comprehensive source of drug information with high-quality references to support each of its drug-drug interaction fact sheets [13]. Each reference in eFacts is listed by author, journal name, publication date, volume number and page numbers – providing sufficient information to locate the article in MEDLINE. For each of the 150 DDIs, we included every reference from eFacts that fell within our timeframe. This method identified 200 DDI citations.
To balance our dataset with non-DDI articles, we randomly selected 1800 different articles from MEDLINE and labeled these as negatives. The number of these DDI- articles chosen from each year had to reflect the proportion of that year’s articles in the positive set. If 10% of the positive articles were from the year 2000, for example, we selected 10% of the negatives from the same publication year.
Since preliminary experiments suggested a 1% prevalence of true DDI articles in MEDLINE, we reviewed the titles and abstracts of all 1800 randomly selected references in order to eliminate any true drug-drug interaction articles that may have been randomly included in the set. When a positive DDI article was found (we found 16), it was removed from the set of negatives and replaced with a neighboring article from the sampling frame. This process minimized the number of false negatives and improved the quality of the corpus.
All citations were downloaded in both text and XML format using EFetch, an article retrieval tool provided by PubMed [14]. Each file was marked with its unique PubMed ID and its drug-drug interaction status (DDI+ or DDI-). The final reference dataset was composed of 1800 hand-sorted negatives and 200 expert-reviewed positives, producing a corpus of 2000 unique citations with a 10% prevalence of drug-drug interaction articles.
Evaluating PubMed
The National Library of Medicine (NLM) PubMed interface allows users to run complex queries against the MEDLINE database, and presents an organized set of hyperlinked results. The major search fields include the title (text words), author, abstract (text words), journal name, and publication date of a paper, as well the controlled vocabulary Medical Subject Headings (MeSH) chosen by MEDLINE indexers to characterize a paper’s content [15].
We first conducted a test of PubMed’s ability to extract relevant drug-drug interaction articles from MEDLINE, and evaluated this performance using sensitivity (recall) and positive predictive value (precision). We chose these measures because the value of the query results is dependent on the user’s information needs. A DDI database curator looking for every publication mentioning an uncommon drug, for example, may desire high sensitivity, producing a set that may contain many irrelevant articles, but will not have missed any of the pertinent documents. On the other hand, a second user searching for information about a common drug might not want to retrieve every relevant reference (since that might be excessive), and would therefore prefer a query with high PPV (positive predictive value).
With these two information needs in mind, we worked with expert librarian MEDLINE searchers from Vanderbilt’s Eskind Biomedical Library to develop two queries. The first query aimed to return a set with high sensitivity; the second query focused on maximizing PPV. The details of these two queries are presented in Table 1.
Table 1.
Two PubMed queries
| PubMed Query | |
|---|---|
| Query 1 (maximize sensitivity) | ("drug interactions"[TIAB] NOT Medline[SB]) OR "drug interactions"[MeSH Terms] OR drug interaction[Text Word] |
| Query 2 (maximize PPV) |
Query 1 AND ("Toxicity Tests"[MeSH] OR "Adverse Drug Reaction Reporting Systems"[MeSH] OR "Drug Hypersensitivity"[MeSH] OR "Drug Antagonism"[MeSH] OR "drugs, investigational"[MeSH] OR "Drug evaluation"[MeSH] OR "adverse effects"[Subheading] OR "toxicity"[Subheading] OR "poisoning"[Subheading] OR "chemically induced"[Subheading] OR "contraindications"[Subheading]) |
We executed these queries through PubMed’s MEDLINE interface and intersected the resulting MEDLINE-wide citation set with our study dataset of 2000 references. This identified the true and false positives returned by the PubMed queries, restricted to the study DDI dataset.
Processing the Citation Content
In contrast to the PubMed queries, the experiments involving automated classification techniques required preprocessing of titles and abstracts. We tested two separate methods of text preprocessing, producing two different datasets. We named these datasets CUI and TERMS.
To generate the first dataset (CUI), we applied a text filtering scheme utilizing the UMLS-based MMTx helper application (ver. AA2003) from NLM. To simplify the natural language of our documents’ titles and abstracts, we used MMTx to map free text to UMLS concepts. Each of the 2000 citations was processed separately with MMTx, using the “-a” and “-u” flags to limit acronym processing and the “-I” flag to include each concept’s (numeric) Concept Unique Identifier, or CUI. A binary (present/absent) vector of CUIs was used to represent the text (abstract and title) content of every citation. The set of these binary vectors for all 2000 documents constituted the “CUI dataset” that served as input for automated classification methods.
The second dataset (TERMS) was generated by extracting the title and abstract text of all 2000 corpus documents, removing all stop words (as defined by PubMed), converting text to lowercase, and replacing all punctuation with white space[11]. The remaining terms were reduced to their word stems using a publicly available Perl implementation of the Porter stemming algorithm[16; 17]. This process has been useful for preparing text for machine learning tasks[11], and is considered standard for such work.
Unlike the CUI dataset, TERMS also included the MeSH Headings and Subheadings (also known as Descriptor and Qualifier terms) associated with each MEDLINE. We did not split and stem MeSH terms because they were multi-word phrases representing information content from the full text of the article. To produce the TERMS dataset, we assembled binary present/absent vectors of these stemmed text words and MeSH terms for each of the 2000 documents in the corpus.
Classifying the References
We used the LIBSVM implementation of the SVM algorithm and conducted experiments using Matlab with a freeware SVM API[18; 19]. We tested both polynomial kernels (degree 1–4) with misclassification costs of {0.001, 0.01, 0.1, 1, 10, 100} on both datasets.
The CUI and the TERMS datasets were processed independently using the same methods. We put aside 33.3% of each dataset as a test set, and retained the remaining 66.7% as training data, which we in turn divided into 10 mutually exclusive sets (“folds”). The 10% prevalence of positives was maintained across all the resulting sets. We used a 10-fold cross-validation to obtain an unbiased performance estimate. By repeatedly using nine folds for training and the remaining fold as a validation set, we attempted to prevent overfitting of the data. Performance was measured by maximizing the area under the receiver operating curve (AUC). The kernel and cost parameters that produced – in a cross-validated fashion – the best AUC were used to develop a model that was tested on the previously identified and untouched test set.
For each dataset we also performed feature selection to identify terms with high discriminatory power. HITON is a recently developed algorithm that has shown excellent performance on feature selection tasks with text words [11; 20]. In particular, we used the HITON-MB algorithm, which seeks Markov Blanket variables in a Bayesian network, and the HITON-PC algorithm, which identifies a set of parents and children variables in the Bayesian network. We tested both HITON-PC and HITON-MB with and without a wrapping step that attempts to further reduce the number of features. These feature selection techniques were also performed 10 times in a cross-validated fashion.
Results
PubMed Queries
The results of both PubMed queries are presented in Table 2. For each query, we list the total number of MEDLINE articles returned, the number of those articles present in our corpus, the number of those which were true positives, and the query’s sensitivity and positive predictive value.
Table 2.
PubMed query performance
| PubMed Query | articles returned | Return in corpus | True DDI+ | Sens. | Spec. |
|---|---|---|---|---|---|
| Query1 (Sens.) | 101819 | 167 | 150 | 0.7500 | 0.9906 |
| Query2 (PPV) | 32937 | 78 | 76 | 0.3800 | 0.9989 |
Query 1 retrieved citations from our corpus with a sensitivity of 0.75 and a specificity of 0.9906. Query 2 identified drug-drug interaction documents with a much lower sensitivity (0.38), but a higher specificity (0.9989).
Text Classification Methods
The results of the automated classification experiments are presented for each dataset and feature selection method. For each combination, Tables 3 and 4 list the number of features in the final model (built from the entire training set) and the AUC performance on both the training and testing sets. The training set’s AUC is averaged across all 10 folds of the data. Models with the highest AUC on the test set are highlighted. Table 3 displays the results of the CUI dataset.
Table 3.
CUI dataset results
| Feature Selection Method: | # Features | AUC (train) | AUC (test) |
|---|---|---|---|
| None | 13187 | 0.9504 | 0.9795 |
| HITON-PC | 32 | 0.9050 | 0.9675 |
| HITON-PCW | 30 | 0.9116 | 0.9705 |
| HITON-MB | 152 | 0.9081 | 0.9616 |
| HITON-MBW | 149 | 0.9052 | 0.9474 |
Table 4.
TERMS dataset results
| Feature Selection Method: | # Features | AUC (train) | AUC (test) |
|---|---|---|---|
| None | 22586 | 0.9892 | 0.9887 |
| HITON-PC | 13 | 0.9552 | 0.9893 |
| HITON-PCW | 12 | 0.9577 | 0.9860 |
| HITON-MB | 34 | 0.9633 | 0.9900 |
| HITON-MBW | 24 | 0.9668 | 0.9821 |
The SVM classifier using the full set of 13187 CUIs showed the best performance, producing an AUC of 0.9795. The model identified by HITON-PCW (Parents and Children with wrapping) also scored very highly, but was simpler (only 30 features) and computationally much less costly. The latter model was generated using a linear classifier with a misclassification cost of 10.
While the CUI models were developed from text-to-UMLS mappings, the TERMS data included stemmed text words and MeSH terms. The results of experiments using the TERMS dataset are presented in Table 4.
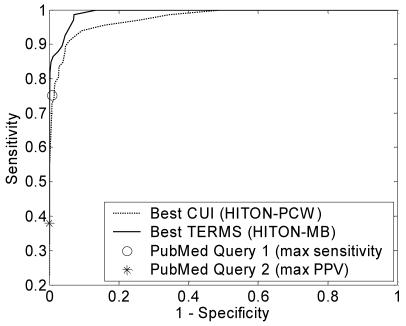
The full TERMS dataset included 22586 distinct word stems and MeSH terms. Of the four varieties of HITON we applied to reduce the number of features, HITON-PC (13 variables) and HITON-MB (34 variables) had the best classification performance. Both models were generated using a linear SVM with misclassification cost of 1. The AUC performance of the SVM classifiers is graphically displayed in Figure 1. The single-point performances of the two PubMed queries are annotated.
Figure 1.
ROC curves for best models
Across all data points, the best TERMS model showed equal or better performance than the best model derived from the CUI dataset. Both models were able to match the performance of PubMed Query 2 (marked with an * in the graph above), which was the query designed to optimize PPV. The TERMS model outperformed PubMed Query 1 as well.
Discussion
The results of this study indicate that SVM classifiers, when trained on a dataset of stemmed text words and MeSH terms, may be at least as good as PubMed queries at identifying articles about drug-drug interactions. We conjecture that the performance of the CUI-trained SVM classifiers resulted from the data-set content rather than the learning method, since the CUI dataset did not include MeSH terms (which are known to have high discriminatory power.) The PubMed queries relied mostly on matching text words and MeSH terms for citation retrieval.
One advantage of the SVM classification model is the ability to easily tune performance based on a user's particular information retrieval needs, adjusting it towards either sensitivity (minimizing false negatives) or PPV (minimizing false positives). The concept of a precision/recall “slider” may prove useful in document retrieval tasks, allowing a user to retrieve either a large, comprehensive set or a small, precise set of articles. In settings where multiple methods are used to retrieve information, for example, users may prefer tools that deliver a reliably useful set of articles from MEDLINE to complement their other strategies. This approach is often cast as a “relevance” measure in typical information retrieval tasks, and may be worth investigating further.
The work presented here represents a feasibility test for quasi-automated drug-drug interaction reference identification. More extensive and applied experiments will be required before we can generate a practical classifier for identifying drug-drug interaction articles in MEDLINE. We are currently working on constructing a wider range of more robust PubMed queries and experimenting with the automated classification of modified datasets that involve word frequencies and weights. In addition, decision trees have proven useful at mapping complex classifiers such as SVMs to Boolean queries[21]. Further work will explore their application to drug-drug interaction article classification.
Acknowledgements
This research was supported by a Training Grant from the National Library of Medicine (T15 LM 007450-03). The authors wish to thank George Robinson and Christine Sommer for their insights into drug database maintenance, and Patricia Lee for help with PubMed searches.
References
- 1.eHealth Initiative. Electronic Prescribing: Toward Maximum Value and Rapid Adoption; 2004
- 2.Stockley IH, editor. Stockley's Drug Interactions, 6 edn. London: Pharmaceutical Press; 2002.
- 3.Enders SJ, Enders JM, Holstad SG. Drug-information software for Palm operating system personal digital assistants: breadth, clinical dependability, and ease of use. Pharmacotherapy. 2002;22 (8):1036–40. doi: 10.1592/phco.22.12.1036.33601. [DOI] [PubMed] [Google Scholar]
- 4.Fulda TR, Baluck R, Vander Zanden J, et al. Disagreement among drug compendia on inclusion and ratings of drug-drug interactions. Curr Ther Res Clin Exp. 2000;61:540–8. [Google Scholar]
- 5.Hazlet TK, Lee TA, Hansten PD, et al. Performance of community pharmacy drug interaction software. J Am Pharm Assoc (Wash) 2001;41 (2):200–4. doi: 10.1016/s1086-5802(16)31230-x. [DOI] [PubMed] [Google Scholar]
- 6.Robinson G, VP of Knowledge Base Development, First DataBank. [personal communication] April 23, 2004.
- 7.Barillot MJ, Sarrut B, Doreau CG. Evaluation of drug interaction document citation in nine on-line bibliographic databases. Ann Pharmacother. 1997;31 (1):45–9. doi: 10.1177/106002809703100106. [DOI] [PubMed] [Google Scholar]
- 8.http://www.nlm.nih.gov/pubs/factsheets/pubmed.html
- 9.Sommer C, Clinical Pharmacist, First DataBank. [personal communication] May 12, 2004.
- 10.Joachims T. Learning to classify text using support vector machines. Boston: Kluwer Academic Publishers; 2002. xvi, 205 p. p. (Kluwer international series in engineering and computer science; SECS 668).
- 11.Aphinyanaphongs Y, Tsamardinos I, Statnikov A, et al. Text categorization models for high-quality article retrieval in internal medicine. J Am Med Inform Assoc. 2005;12 (2):207–16. doi: 10.1197/jamia.M1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potts AL, Barr FE, Gregory DF, et al. Computerized physician order entry and medication errors in a pediatric critical care unit. Pediatrics. 2004;113 (1 Pt 1):59–63. doi: 10.1542/peds.113.1.59. [DOI] [PubMed] [Google Scholar]
- 13.Kupferberg N, Jones Hartel L. Evaluation of five full-text drug databases by pharmacy students, faculty, and librarians: do the groups agree? J Med Libr Assoc. 2004;92 (1):66–71. [PMC free article] [PubMed] [Google Scholar]
- 14.http://eutils.ncbi.nlm.nih.gov/entrez/query/static/efetch_help.html
- 15.http://www.nlm.nih.gov/mesh/
- 16.http://www.tartarus.org/~martin/PorterStemmer/
- 17.Porter M. An algorithm for suffix stripping. Program. 1980;14 (3):130–7. [Google Scholar]
- 18.http://www.csie.ntu.edu.tw/~cjlin/libsvm
- 19.Chang C-C, Lin C-J. LIBSVM: a library for Support Vector Machines.
- 20.Aliferis CF, Tsamardinos I, Statnikov A. HITON: a novel Markov Blanket algorithm for optimal variable selection. AMIA Annual Symp Proc. 2003:21–5. [PMC free article] [PubMed] [Google Scholar]
- 21.Aphinyanaphongs Y, Aliferis CF. Learning boolean queries for article quality filtering. Medinfo. 2004;2004:263–7. [PubMed] [Google Scholar]