Abstract
Surgical procedures can be viewed as a process composed of a sequence of steps performed on, by, or with the patient’s anatomy. This sequence is typically the pattern followed by surgeons when generating surgical report narratives for documenting surgical procedures. This paper describes a methodology for semi-automatically deriving a model of conducted surgeries, utilizing a sequence of derived Unified Medical Language System (UMLS) concepts for representing surgical procedures. A multiple sequence alignment was computed from a collection of such sequences and was used for generating the model. These models have the potential of being useful in a variety of informatics applications such as information retrieval and automatic document generation.
Keywords: Medical concept representation, Unified Medical Language System (UMLS), natural language processing, surgical procedures
Introduction
Surgical reports contain much potentially valuable knowledge which can be utilized for various informatics applications. As such, this paper describes a methodology to semi-automatically create a model of surgeries by analyzing the various tasks conducted within a collection of dictated surgical report narratives. We chose to target radical retropubic prostatectomy (RRP) because it is a relatively straightforward procedure with a manageable degree of variation. There are several major steps that must be performed for this procedure under most circumstances, with variations in other steps depending on the patient’s particular situation (e.g., anatomical features, other abnormalities, etc.).
In general, surgical procedures can be viewed as a process composed of a sequence of steps performed on, by, or with the patient’s anatomy. Following previous work in defining ontologies for surgeries, we call an anatomical concept with its corresponding action concept a surgical deed11. The MetaMap tool was utilized to extract relevant Unified Medical Language System (UMLS) concepts from free-text surgical reports (e.g., anatomical terms) to track these steps within each report. As a result, a surgical procedure was represented as a sequence of these surgical deeds. A multiple sequence alignment algorithm was then applied to a collection of surgical deed sequences to derive a model which depicts the various steps and variations of the RRP. The goal of the resulting surgical model is to provide researchers with information such as:
The steps were taken during one or more surgeries
The order of the steps performed
The frequency of the performed steps in a given collection of surgeries
This information has the potential to inform several clinical applications that can offer new insights and efficiencies to epidemiologists, surgeons and administrators. First, a model based on the alignment of data from surgeries of a given type can deliver useful statistics on the number of variations performed within a collection of surgeries (eg., “What steps are conducted most frequently? In how many prostatectomies is step Y conducted?”). Second, this model can act as a template for automated surgical report generation, allowing surgeons to more easily generate text by choosing the appropriate sequence of steps through the model. Finally, such a model offers a new dimension for querying a collection of surgical reports. With input from a clinical expert the most frequently conducted surgical steps, which are automatically derived from the surgical model, can be categorized into the various phases of a surgery (eg., patient preparation, initial incision, etc.). Therefore, processed surgeries can be automatically indexed and searched by standardized concepts and by steps within the process of a given surgery - a potentially significant approach to the retrieval information stored in narrative texts. For example, a surgeon interested in retrieving cases in which profuse bleeding occurred during the lymph node dissection of a prostatectomy might otherwise be limited to a term-matching search for heavy bleeding.
This study also presents the results of our work to measure the performance of a surgical model-building system in terms of both identifying the relevant surgical deeds as well as the ability to accurately place them in sequences.
Background
The identification of standardized concepts from medical free-text reports has been used in several indexing efforts1, 2, 3, 4. As part of the National Library of Medicine’s (NLM) Semantic Knowledge Representation (SKR) program, MetaMap5, 6 maps free text to the UMLS Metathesaurus, the largest thesaurus in the biomedical domain. MetaMap returns a ranked set of UMLS concepts extracted from a body of text. In order to do so, a five-step process is employed. First, the SPECIALIST minimal commitment parser is used to identify simple noun phrases. Second, for each phrase, lexical variants are generated, including abbreviations, synonyms, plurality, etc. Next, a set of candidate matches is returned which is then scored based on lexical principles. Finally, a complete mapping is made by combining candidate matches from disjoint parts of the phrase and again calculating a score based on lexical principles.
Most medical ontologies make provisions for classifying and categorizing surgical procedures. One notable work is the Galen-In-Use project7 which extends the European Committee for Standardization (CEN)’s standard schema for surgical procedures8. Central to defining a surgical procedure is the concept of a surgical deed, which is an action performed on the patient’s anatomy by the surgeon during the procedure9. Though the semantic structure is provided for describing procedures, there was no mention of work on automatically building a model which represents the sequence of steps which comprise a typical procedure.
Multiple sequence alignments have been used within the natural language processing community for several purposes. This work includes methodologies for automatically generating paraphrases for sentences10 and recognizing certain discourse structures using the alignment as a data mining tool11. Similarly, our work uses multiple sequence alignment to determine patterns across a group of surgical reports which we use as the basis for building our model.
Methodology
A Model for Surgical Procedures
Surgical procedures can be modeled using a directed graph, where the nodes represent an action being performed with anatomy (i.e., the surgical deed) and the links represent the temporal relationships between the deeds. In the case of RRPs, for instance, besides patient preparations and the administration of anesthesia, the first significant step is the opening of the patient, which deals with an action performed on anatomy. Further actions on anatomy can then be followed to trace the procedure step by step (e.g., incising the pubis, or separating the rectus muscles). We chose to model procedures based on surgical deeds because, from a medical standpoint, the way a patient’s anatomy is affected by the intervention is the fundamental outcome of a procedure. Second, from a natural language processing perspective, terms representing anatomy or actions on anatomy are generally less difficult to isolate within free-text documents because they usually follow standard terminology.
Identifying Surgical Deeds
For our experiment, we developed an application to utilize MetaMap’s Java API, which has been made available by the NLM to researchers. By doing so, we were able to exercise a more granular level of control over the output of the application to further disambiguate target anatomical concepts that might be referred to in a variety of forms and contexts in free-text. In this preliminary study, we identified several concepts related to anatomy from UMLS as representative of surgical deeds. In a later section, we describe how we incorporate context words for differentiating between cases where the same anatomical concept is used but within different contexts. Future work will include using syntactic structure to identify the action concept which best corresponds to each anatomical concept.
To limit the results of MetaMap to only relevant concepts for use as surgical deeds in our sequence alignment we limited the selection of surgical deeds to derived concepts of the following anatomy-related semantic types from the UMLS Semantic Net Hierarchy (which we call anatomical concepts):
| T023 | Body Part, Organ or Organ Component |
| T024 | Tissue |
| T029 | Body Location or Region |
| T030 | Body Space or Junction |
| T061 | Therapeutic or Preventative Procedure |
To utilize the ranking performed by MetaMap, we considered only the top concept returned and we ignored the phrase entirely if the first concept returned was not of the semantic type listed above. To facilitate the integration of the results with our alignment algorithm, the concepts, the original phrases they were derived from, and their indexes within the reports were packaged as an XML document.
Computing the Multiple Sequence Alignment
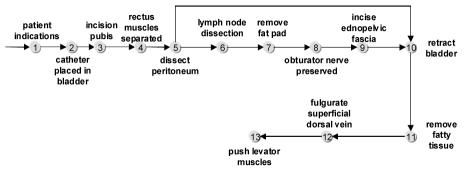
The result of the mapping step is a sequence of anatomical concepts representing each document in the corpus, where the ordering of these concepts reflects the order that they occurred in the original narratives (Figure 1). We have found that it is rather difficult, even for domain experts, to determine the important steps and the inherent variations of a procedure by manually examining a corpus of reports. We propose to gather a large group of these sequences and perform a multiple sequence alignment on them, which generates a model of the surgery that reflects the various surgical steps and their relative ordering.
Figure 1.
Sequence of anatomical concepts
Multiple sequence alignments (MSAs) have been readily used in bioinformatics and data mining for determining patterns given a set of sequences. To compute the MSA, we used a simple bottom-up clustering algorithm12. Bottom-up clustering algorithms create tree structures, where clusters of sequences are merged into larger ones as the tree is built up towards the root. Clustering algorithms can be readily adapted to perform iterative pair-wise alignments to incrementally build an MSA. We used the Needleman-Wunsch global alignment algorithm12 to compute the MSA using the bottom-up clustering approach. Needleman-Wunsch was configured such that gaps were favored over mismatches, which results in an alignment where the columns contain only one type of element along with any gaps introduced by the alignment algorithm.
The clustering algorithm for generating the MSA proceeds as follows: initialization is performed by placing each sequence within its own cluster. For each iteration of the algorithm, a search is conducted for the pair of clusters which currently are the most similar to one another. We define the degree of similarity between two clusters A and B as the highest Needleman-Wunsch score between a sequence in A with a sequence in B.
If it turns out that A and B are currently the two most similar clusters among all the clusters, then we merge A and B and form a new cluster C. Cluster C is generated using sequences from clusters A and B, but with the sequences aligned against each other using Needleman-Wunsch. The creation of a larger cluster from two smaller clusters results in a global MSA of the sequences contained in both smaller clusters.
This process continues until there is only one final cluster remaining. Because clusters of sequences are aligned against each other whenever a new cluster is formed, the final single cluster (the root of the tree) contains an alignment of all the original sequences. The resulting alignment contained in this final cluster is the MSA of our original collection of sequences.
Context Words
The alignment of two sequences is dependent upon how similarity is computed between surgical deeds. The basic approach is to use an all-or-nothing matching scheme, where deeds which match exactly receive a score of 1, for instance, and those that do not receive a score of 0. Because the anatomical concept which represents a surgical deed such as “prostate” may be used in several different contexts, a better approach would be to allow for partial matches. This can be done by factoring context words into the match score. We define context words to be the group of words which surround the anatomical concept in the original report. Context words were determined by gathering all words which co-occur within the same sentence as the anatomical concept of the surgical deed. If two deeds match, meaning they both are represented by the same anatomical concept, then partial matching of the deeds depends on the percentage of context words they have in common. The context word score, scorecw can be calculated as:
Here, Wi and Wj are the set of context words for anatomical concepts i and j respectively. We also attempted to normalize context words by using their corresponding UMLS concept names instead of using the actual words themselves. As mentioned in the results section, using context word scoring slightly improved the alignment over using just the anatomical concepts representing the surgical deeds.
Generating the Model
Given the alignment of document sequences, the model was generated by first determining the significant columns within the alignment. The significance of a column was defined as the number of non-gaps divided by the total number of sequences in the alignment. Those columns which did not meet a preset significance level were not included in the final model. Once the significant columns were determined, the model was created as follows: each significant column in the alignment becomes a surgical deed in the final model. A link between two deeds was added to the model if there was a link from one deed’s corresponding column in the alignment to the other either directly or through any number of gaps. Figure 2 shows an example alignment and its resulting model, where the significance level used was 50% or above. In the figure, nodes with letters represent surgical deeds and those with a dash represent gaps. Link weights are determined by the number of times the deeds’ corresponding columns within the alignment are connected.
Figure 2.
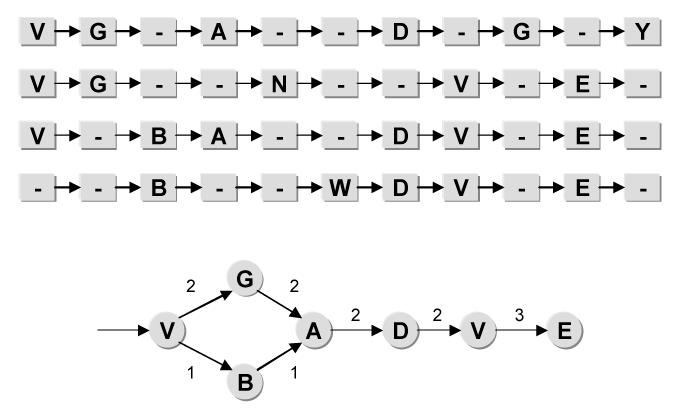
Multiple sequence alignment and corresponding model
Evaluation
Identification of Anatomical Concepts
In order to evaluate the system’s effectiveness at correctly identifying all anatomical concepts which represent surgical deeds, the authors first manually identified all terms of interest in the surgical reports in order to measure the recall and precision of the results. For a term to be considered representative of a relevant surgical deed, we required that it describe an anatomical part that was being affected by some surgical action or an action that was being conducted on the anatomical part. Identified concepts of the appropriate semantic type that did not actually describe a surgical action performed (e.g., “we chose not to conduct a nerve-sparing procedure”) were counted as false positives. Multiple occurrences of the same concept also impacted precision in a negative way. Reasons for missed concepts and false positives were tracked in order to inform future refinement of the system.
Recall of the system was 63% with a substantial proportion of missed concepts caused by our decision to limit the inclusion of an anatomical concept to the first returned (highest ranked concept). An analysis of the effect of including both the first and second ranked concepts showed an estimated increased recall to approximately 75%. Iteration through all concepts returned would have increased recall further, but the negative impact of this on the precision of the system needs further investigation. The majority (83%) of the anatomical concepts of interest that were not ranked as the first concept which negatively affected recall was of type T061 (“Therapeutic or Preventative Procedure”). Similarly, the majority of recall errors (62%) caused by no matching concept were verbs such as “encircled,” “prepped,” or “retracted,” which were considered relevant in context but were not included in semantic type T061.
The precision of the returned results was 86%, with fewer than anticipated errors caused by multiple occurrences within reports or by the presence of negation. Instead, incorrectly identified terms that were repeated consistently across the corpus had a noticeable affect on precision. The term “incision,” for example, was ranked first as the body part, “incisor” and appeared over 120 times in the collection of 52 reports. The term “given” was also consistently identified as “Gamete Intrafallopian Transfer” several times across multiple reports.
Multiple Sequence Alignment
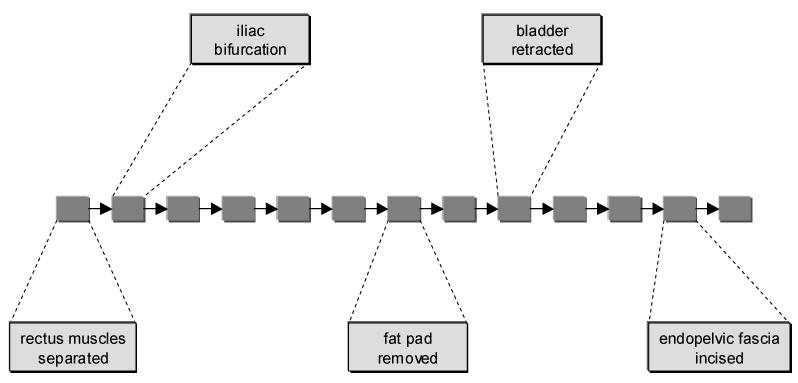
To measure the success of the MSA algorithm, we used two evaluations: the consistency of each column in the alignment, and how well the resulting model for RRP matches with one created by a urologist. A portion of this urologist-created model is shown in Figure 3.
Figure 3.
Urologist-created RRP model (partial)
To measure the consistency of each column in the alignment, we compared each anatomical concept’s corresponding sentence within which the concept was found. For instance, a concept representing the term “prostate” may have a corresponding sentence “The prostate was incised” because it was found in that sentence. A score of 1.0 represents a column where all the anatomical concepts are used in the same context. The average column consistency score for a model generated with a significance level of 25% was .39, whereas the average column score for the same alignment without using context words was .37, exhibiting a slight decrease in consistency. The resulting model generated using the anatomical concepts extracted from 52 RRP reports and using a 25% column significance threshold had 102 nodes. When compared with the urologist-created model, out of the 102 nodes in our generated model, 6 nodes were ambiguous (5.9%). This means that the context in which the anatomical concept was being used was not easily identifiable by inspection. A total of 3 nodes (2.9%) were found to be out of order when compared against the urologist-created model. Also, 8 nodes which were included in the urologist’s model were not included in the generated one.
Discussion
The number of multiple occurrences of the same type of error when identifying anatomical concepts spread across several reports suggests that further techniques may need to be developed to augment the capabilities of MetaMap. Some errors are the result of curious classification in UMLS while others stem from consistent use of a term in ways inconsistent with what is contained in UMLS (e.g.,“Denonvillier’s fascia” rather than the recognized “Denonvilliers’ fascia”). Repeated occurrences of these types of errors indicate the need to consider the use of custom rules to improve performance.
Concepts of type T061 clearly presented the greatest challenge to the system’s recall and precision performance. In response, we are creating a controlled vocabulary of surgical verbs or tasks to enhance our use of T061. Additional consideration will be given to the inclusion of spatial concepts (e.g., “left side of”), as well as inclusion of concepts of type T074, “Medical Device.”
Errors in the multiple sequence alignment were due mainly to the match scoring between two surgical deeds. The context words technique was designed to alleviate some of the problems involved with matching terms out of context. We are exploring better approaches to represent contexts, such as utilizing parse trees to determine words associated with the surgical deed’s corresponding phrase based on syntactic structure.
We plan to present the preliminary models generated by our multiple sequence alignment methodology to domain experts within a graphical user interface for further refinement and error correction. Gathering and analyzing surgical reports for commonalities is a tedious and time-consuming task. We are also planning to implement a surgical report generation system using models of surgical procedures as the backbone. This system would allow surgeons to create documents based on the answers to a small number of questions regarding the specifics of a particular procedure to be documented.
Conclusion
We presented a methodology for generating a model for surgical procedures by first mapping terms from surgical reports to UMLS concepts. These concepts were used to represent surgical deeds and sequences of surgical deeds were used to represent a particular surgical procedure. A multiple sequence alignment algorithm was used to create a model based on a collection of these sequences. Using MetaMap as a basis, we found that mapping to anatomy performed at a high level of precision and recall. MetaMap was less successful, howeverl, in mapping surgical tasks or verbs. Multiple sequence alignment was used to generate our model and it was found to be comparable to an expert-created model for RRPs. Future work includes refinement of the model and more rigorous evaluation of the system with a larger collection of reports.
Acknowledgements
This work was supported in part by grants from the National Institute of Biomedical Imaging and Bioengineering (NIBIB) PO1-EB00216 and RO1-EB002247. The authors would especially like to thank Dr. Kangarloo for his vision, guidance, and continued support.
References
- 1.Elkins JS, Friedman C, Boden-Albala B, Sacco RL, Hripcsak G. Coding neuroradiology reports for the Northern Manhattan Stroke study: a comparison of natural language processing and manual review. Comput Biomed Res. 2000;33(1):1–10. doi: 10.1006/cbmr.1999.1535. [DOI] [PubMed] [Google Scholar]
- 2.Zou Q, Chu WW, Morioka C, Leazer GH, Kangarloo H. Index-Finder: a method of extracting key concepts from clinical texts for indexing. Proc AMIA Symp. 2003:763–7. [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srinivasan P, Rindflesch T. Exploring text mining from MEDLINE. Proc AMIA Symp. 2002:722–6. [PMC free article] [PubMed] [Google Scholar]
- 5.National Library of Medicine Semantic Knowledge Representation Program, http://skr.nlm.nih.gov/papers/index.shtml#MetaMap
- 6.Aronson AR. Effective mapping of biomedical text to the UMLS metathesaurus: the MetaMap program. 2001. AMIA Symposium Hanley & Belfus, 2001:17–21. [PMC free article] [PubMed]
- 7.J-M Rodrigues, B Trombert-Paviot, R Baud, J Wagner et al. Galen-In-Use: An E.U Project applied to the development of a new national coding system for surgical procedures: NCAM. In Medical Informatics Europe '97 pp897–901 (Thessaloniki, Greece) IOS Press Vol 43 ISSN:0926–9630. [PubMed]
- 8.EN 1828 Health informatics-categorical structure for classifications and coding systems of surgical procedures. CEN Brussels 2002.
- 9.Rossi Mori A, Gangemi A, Steve G, Consorti F, Galeazzi E. An Ontological Analysis of Surgical Deeds. in Keravnou E et al (eds), Artificial Intelligence In Medicine, Springer Verlag, 1997, 361–372.
- 10.Barzilay R, Lee L. Learning to paraphrase: an unsupervised approach using multiple sequence alignment. in Proceedings of HLT-NAACL 2003, pp. 16–23.
- 11.Alonso L, Castellón I, Escribano J, Messeguer X, Padró L. Multiple sequence alignment for characterizing the linear structure of revision. In 4th International Conference on Language Resources and Evaluation (LREC 2004).
- 12.Durbin R Eddy S Krogh A and Mitchison G. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press, 1998.