Abstract
A deeper understanding of the mechanisms that underlie plant growth and development requires quantitative data on three-dimensional (3D) morphology and gene activity at a variety of stages and scales. To address this, we have explored the use of optical projection tomography (OPT) as a method for capturing 3D data from plant specimens. We show that OPT can be conveniently applied to a wide variety of plant material at a range of scales, including seedlings, leaves, flowers, roots, seeds, embryos, and meristems. At the highest resolution, large individual cells can be seen in the context of the surrounding plant structure. For naturally semitransparent structures, such as roots, live 3D imaging using OPT is also possible. 3D domains of gene expression can be visualized using either marker genes, such as β-glucuronidase, or more directly by whole-mount in situ hybridization. We also describe tools and software that allow the 3D data to be readily quantified and visualized interactively in different ways.
INTRODUCTION
A major aim of developmental biology is to understand how the three-dimensional (3D) morphology of organisms arises through molecular and cellular mechanisms. However, traditional anatomical and molecular studies of plant development have mainly relied on two-dimensional (2D) images. 3D properties of plant structures are often inferred indirectly by semiquantitative extrapolations from 2D information. Although this may be sufficient for some aspects of plant biology, understanding of plant growth and function increasingly requires quantitative 3D data. This presents two major challenges. First, we need convenient methods to acquire 3D digital images. Second, software tools are needed that allow 3D images to be readily visualized, interrogated, and quantified.
Various approaches have been used to gain 3D information from plants with a variety of resolutions along the different axes (x, y, and z). Several of these techniques use optical methods. An example is 3D reconstructions from a series of microtome sections (Fiala, 2005). Resolution along the x and y axes is limited by the wavelength of visible light (∼0.5 μm), whereas resolution along the z axis depends on the thickness of the sections (typically 5 to 10 μm). Serial sectioning has the advantage that gene activity and histological stains can be readily viewed. However, the procedure is laborious and prone to alignment errors. Confocal microscopy addresses many of these issues and has a resolution limit of 0.3 μm × 0.3 μm × 0.5 μm (Liu and Chiang, 2003; Jönsson et al., 2006). However, this method is limited by the overall thickness of the specimen that can be visualized: image quality decreases rapidly for optical sections deeper than 60 to 80 μm (Haseloff et al., 1997). The depth can be extended at the expense of spatial resolution using optical coherence microscopy (OCM). OCM was used to obtain 3D images of whole growing Arabidopsis thaliana plants and single cells at a resolution of 3 μm × 3 μm × 5 μm and a maximum specimen depth of 2 to 3 mm. OCM provides a means of continuously monitoring plants and plant cells throughout development and in response to exogenous stimuli (Hettinger et al., 2000; Reeves et al., 2002). More recently, OCM has been used in animal systems to obtain a resolution of 1 μm × 1 μm × 1 μm, with a depth of 10 mm (Greive et al., 2005). Selective plane illumination microscopy also overcomes depth restrictions using a sheet of laser light 2 to 8 μm thick to section a sample optically. Protein expression patterns within living embryos were captured with a resolution of 6 μm × 6 μm × 6 μm, to a depth of 500 μm, using this method (Huisken et al., 2004). Saturated structured-illumination microscopy has been used to obtain 2D images beyond diffraction limitations with resolutions to 50 nm (Gustafsson, 2005). 3D images of the actin cytoskeleton of a human neutrophil were obtained using this technique (http://msg.ucsf.edu:8100/∼mats/).
Depth restrictions can also be overcome using methods based on magnetic resonance or types of radiation that penetrate deeper into specimens. However, this is at the expense of resolution. Magnetic resonance imaging (MRI) offers live imaging without depth restriction and can resolve down to 20 μm × 20 μm × 1.5 mm (Holbrook et al., 2001). Although it is possible to view gene expression patterns by MRI using contrast agents that can indicate reporter expression in living animals (Louie et al., 2000), this process is not well suited for general applications (Ruijter et al., 2004). The x-ray high-resolution computed tomography (HRCT) allows live imaging to a resolution of 13 μm (http://www.mouseimaging.ca/research/gallery_microct_placenta.html). However HRCT depends on the contrast between the x-ray absorption coefficient of the material and does not allow gene expression to be captured (Stuppy et al., 2003). Gene expression in living animals has also been visualized using positron emission tomography; however, this method only achieves resolutions of 1.8 mm × 1.8 mm × 1.8 mm (Gambhir et al., 1999). MRI, HRCT, and positron emission tomography do not allow the visualization of existing histological stains commonly used for light microscopy.
Another method, not previously used with plants, is optical projection tomography (OPT). OPT has the advantages of optical methods, but with greater penetration and the ability to generate 3D images of nonfluorescent signals (such as the blue precipitate from X-Gal). This allows it to capture gene expression and histology using traditional tissue stains to a resolution of 5 μm × 5 μm × 5 μm within the 3D context of whole embryos and organs between 0.5 and 15 mm in depth (Sharpe et al., 2002; Sharpe, 2003). A further advantage of OPT is that the scanner is relatively inexpensive (currently approximately one-quarter of the price of a confocal microscope). A disadvantage of optical techniques, including OPT, is that large specimens typically need to be cleared with an organic solvent, precluding live imaging in most cases. The 3D information about a specimen can be collected in two ways. In fluorescence OPT, the specimen is illuminated with UV light and the signal depends on the fluorescence of stains and markers or the autofluorescence of the tissue. In transmission OPT, visible light is shone through the specimen and the signal depends on the amount of light absorbed. To date, OPT has been applied to numerous vertebrate systems, including mouse (Sharpe et al., 2002; Sharpe, 2003, 2004; Wilkie et al., 2004; Walls et al., 2005; Takeuchi et al., 2005), human (Kerwin et al., 2004; Lickert et al., 2004; Lindsay and Copp, 2005; Lindsay et al., 2005; Sarma et al., 2005), zebrafish (Bryson-Richardson and Currie, 2004), chick (Tickle, 2004), and frog (Tyszka et al., 2005).
One general problem in dealing with volumetric data obtained from OPT and other methods is the need to visualize and analyze the information. When creating software tools, there is a trade-off between flexibility and ease of use. One approach is to provide a flexible software environment that tries to address most issues found in the visualization and analysis of volumetric data, independent of the field of science. A disadvantage is the time taken for users to develop packages that are appropriate for their problem. Successful examples of such software packages are AMIRA (http://www.tgs.com/), VTK (http://public.kitware.com/VTK/), and AVS (http://www.avs.com/). The second approach is to produce solutions tailored to each type of volumetric data. For example, MAPaint has been devised to allow manual painting of regions of interest in 3D data sets (Baldock et al., 2003). We use a combination of these approaches to develop generally applicable software that allows interactive visualization and interrogation of 3D data of the kind generated by OPT.
Here, we explore the use of OPT for obtaining 3D data from plant material. We show that plant structures at a range of scales provide ideal material for visualization by OPT. The method provides a convenient means to examine 3D morphology and gene expression patterns of Arabidopsis and Antirrhinum majus plant structures. OPT allows the 3D analysis of large, thick specimens, with visualization of large cells. However subcellular structures and small cells are not resolvable. In the case of Arabidopsis roots, it is also possible to use OPT for live imaging. We also present QtVolView as a general software solution particularly suited for the interactive visualization and quantitative analysis of OPT data sets
RESULTS
Visualization of OPT Data from Plants
To explore the possibility of using OPT on plant material, specimens were cleared and photographed using the OPT device. Specimens were illuminated with visible light (transmission OPT) or UV light with associated filters (fluorescence OPT). In most cases, illumination with UV light was most effective for revealing tissue structure. The specimen was rotated about its main axis in steps of 0.9° and an image was taken at each step, giving 400 images per scan. This allowed the information for the entire specimen to be reconstructed as a 3D digital image (Sharpe et al., 2002). A 3D digital image consists of a set of volume units, called voxels (just as a 2D digital image comprises a set of pixels). Collectively, these voxels carry quantitative information for the entire volume of the specimen, from its internal structures to the outer surface. Voxel size is measured along three directions (x, y, and z). Software was developed to facilitate viewing and quantification of the OPT data in various ways.
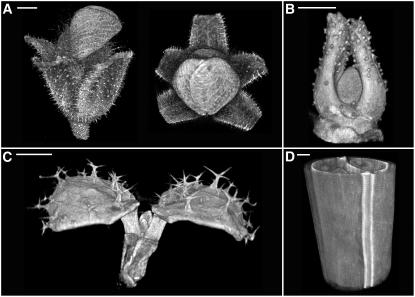
The specimen could be viewed interactively without significant time delays, at 10 to 15 frames per second, from any position in 3D space. Examples of such volume views for floral buds, apices, seedlings, and fruits are shown in Figures 1A to 1D. The viewing point could be varied continuously, improving the user's overall sense of the 3D structure of the specimen. Two sample views are shown for a flower bud in Figure 1A. The QtVolView program was used to calculate voxel sizes obtained by OPT. A range of scales was obtained, from 28 to 1 μm3/voxel.
Figure 1.
OPT Data Reconstructed and Displayed as Volumes.
(A) OPT volume views of an Antirrhinum flower. The image is based on a combination of transmission and fluorescence OPT (Leica TXR filter and GFP1 filters). Bar = 525 μm.
(B) Antirrhinum vegetative meristem imaged by fluorescence OPT (GFP1 filter). Bar = 85 μm.
(C) First true leaves of an Arabidopsis seedling showing trichome cells on the adaxial leaf surface (cotyledons removed). The image was taken with fluorescence OPT (GFP1 filter). Bar = 285 μm.
(D) Part of an Arabidopsis silique imaged by fluorescence OPT (TXR filter). Bar = 42 μm.
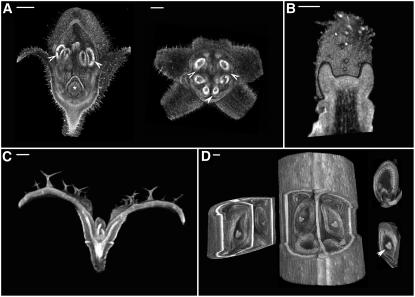
Volume views have the disadvantage that structures nearer to the viewing point can occlude those farther away. One way of circumventing this problem is to cut away some of the structure using standard clipping planes. This can reveal more detail about internal structure. For example, by cutting away part of the flower bud, the internal organs (stamens, carpels) were revealed (Figure 2A). Similarly, primordia, vasculature, and ovules were revealed by clipping an apex, seedling, and silique, respectively (Figures 2B to 2D).
Figure 2.
Clipping to Reveal Internal Morphology.
(A) Virtual dissection using clipping planes of the Antirrhinum flower shown in Figure 1A reveals internal floral structures such as anther lobes (arrowheads) and the ovary at the base of the carpel (asterisk). Pollen is more autofluorescent than the surrounding tissues, making the anthers appear brighter. Bars = 200 μm (left) and 365 μm (right).
(B) Clipping plane through the center of the Antirrhinum shoot apical meristem. Bar = 55 μm.
(C) An Arabidopsis seedling clipped to display vasculature. Vasculature is more autofluorescent than the surrounding tissue, making it appear brighter. Bar = 100 μm.
(D) An Arabidopsis silique (from Figure 1D) clipped to reveal internal structure. A piece was removed using three clipping planes to show the seeds developing within. The removed piece is shown at left. Individual seeds were also dissected out using six clipping planes to display the heart-stage embryo (arrowhead) and endosperm within (two examples shown at right). Bar = 35 μm.
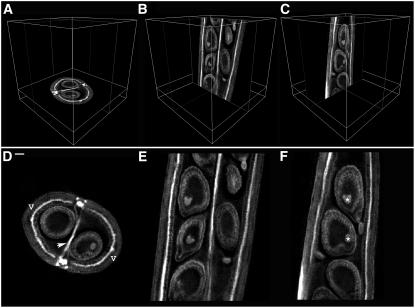
It was also possible to generate slices through the specimen, equivalent to histological sections. Sections of any thickness could be generated using two parallel clipping planes separated by the required thickness. Sections one voxel thick could also be generated by specifying a single plane and visualizing the voxels that intersect it (Figures 3A to 3F).
Figure 3.
Virtual Sectioning of an Arabidopsis Silique.
(A) to (C) Orthogonal sections in their original orientations: xy section (A); zx section (B); and yz section (C).
(D) to (F) Orthogonal sections oriented parallel to the viewing plane: xy section showing two valves (v) surrounding the developing seeds and the septum (arrowhead) (D); zx section (E); and yz section with heart-stage embryos labeled (asterisks) (F). Bar = 32 μm.
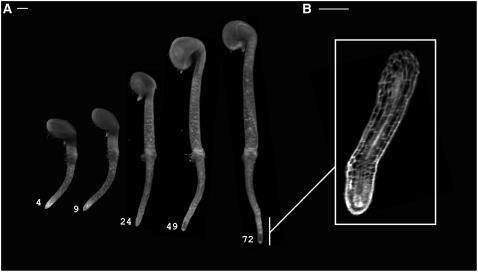
Light does not easily pass through most living plant specimens, so it is usually necessary to clear them in an organic solvent for good-quality optical imaging. However, for structures such as roots that are naturally semitransparent, there is the possibility of collecting OPT data from living tissue without fixing and clearing. To achieve this, an Arabidopsis seed was embedded in agarose, allowed to germinate, and transferred into the OPT device, where it grew submerged in water. OPT images of the germinating seedling were collected several times during a 72-h growth period. The seedling was transparent enough to allow reliable 3D reconstruction without fixation and clearing (see volume views in Figure 4A). Thus, it is possible to use OPT for live 3D imaging for naturally semitransparent material. At higher magnifications, larger individual cells within the root could be identified, raising the possibility of using OPT to extract detailed growth dynamics (Figure 4B).
Figure 4.
Live OPT Imaging.
(A) Volume views of an Arabidopsis seedling growing within the OPT device. A series of OPT scans in water collected at 4, 9, 24, 49, and 72 h after the start of the experiment is shown (transmission OPT channel). Bar = 100 μm.
(B) yz section view of the same root as in (A) captured at higher magnification (8×) collected 30 h after the start of the experiment. Bar = 60 μm.
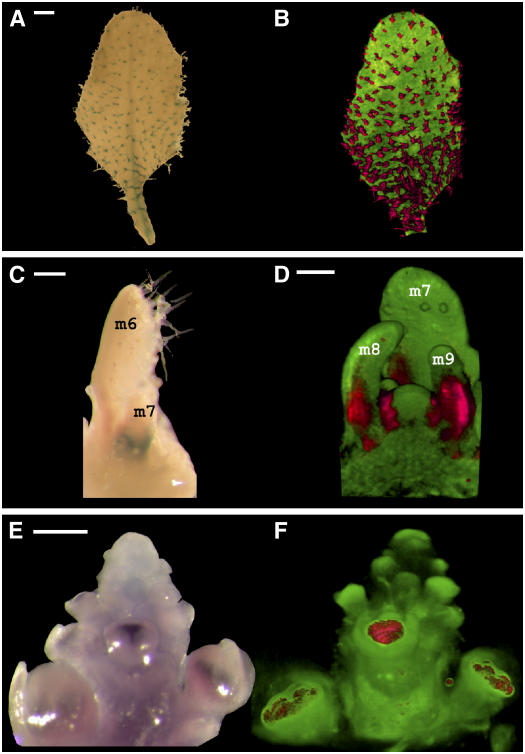
OPT could also be used to capture gene expression patterns in 3D. For example, histochemical staining of Arabidopsis plants containing GLABRA2:β-glucuronidase (GL2:GUS) gives strong signal in trichomes, reflecting GL2 promoter activity in these cells (Figure 5A). GUS-expressing regions could be identified as dark areas by transmission OPT with visible light, whereas the background tissue of the same specimen could be visualized by fluorescence OPT. The information from both channels could then be superimposed to give a combined 3D image with GUS highlighted in red and background tissue in green (Figure 5B). This allowed gene expression in individual leaves or entire seedlings to be visualized. As another example, OPT of LEAFY:GUS (LFY:GUS) plants allowed the 3D pattern of gene activity in meristems to be captured (Figure 5D).
Figure 5.
Visualization of Gene Expression in 3D.
(A) GL2:GUS Arabidopsis leaf showing GUS staining in the trichomes (metamer 5; 13 d from sowing). Bar = 250 μm.
(B) Combined volume view of the leaf shown in (A). Transmission OPT highlights GUS-stained trichomes, shown in red. Fluorescence OPT highlights the remaining leaf tissue, shown in green (GFP1 filter). The two channels were combined and displayed in the same 3D space. Data were collected with OPT Scanner 3001.
(C) LFY:GUS Arabidopsis seedling, after leaf removal and staining to reveal GUS activity (metamers 1 to 5 were removed; 8 d from sowing). Metamers 6 and 7 (m6 and m7) are labeled. Bar = 170 μm.
(D) Combined volume view of the seedling shown in (C). The view was clipped to remove metamer 6 and reveal LFY:GUS staining (red) in emerging leaves (m8 and m9). Signal is also seen in the flanks of the shoot apical meristem. Red signal is from transmission OPT, and green signal is from fluorescence OPT (GFP1 filter). Bar = 45 μm.
(E) DEF expression in stamen and petal primordia of developing flowers in an Antirrhinum inflorescence meristem revealed by whole-mount RNA in situ hybridization. Bar = 350 μm.
(F) Volume view of the Antirrhinum inflorescence shown in (E) with DEF expression highlighted in red and the surrounding tissue in green. This image was obtained by transmission OPT, with a threshold used to distinguish signal from background.
3D gene expression patterns could also be obtained from whole-mount RNA in situ hybridization. For example, whole-mount in situ hybridization with the DEFICIENS (DEF) gene of Antirrhinum (Zachgo et al., 2000) gave dark-staining regions in whorls 2 and 3 of floral meristems (Figure 5E). The 3D expression pattern could be captured by OPT, as shown in Figure 5F.
Segmentation and Quantification of OPT Data
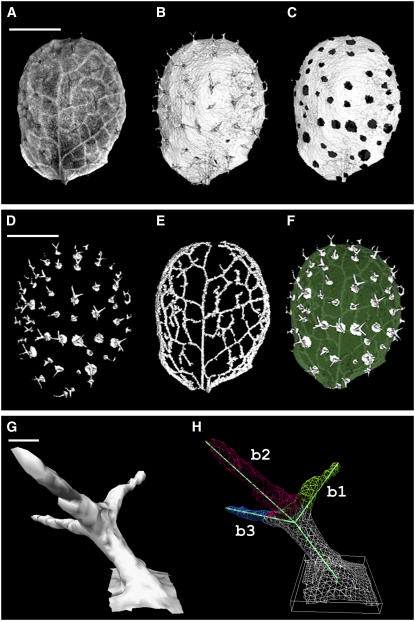
Because only a subset of 3D information can be viewed at any one time, it is helpful to have methods for automatically extracting and measuring features such as 3D domains and their outlines. These domains can correspond, for example, to morphological features such as leaves, veins, and trichomes. Various types of quantitative information can be obtained from domains, such as their position, volume, surface area, or length. For example, the outer surface of a leaf could be extracted from OPT data sets (Figure 6B). Once this surface domain was identified, it was possible to extract individual trichomes as subdomains. This allowed the distribution of trichomes in the leaf to be visualized by showing the leaf surface without trichomes (Figure 6C) or the trichomes without the leaf (Figure 6D).
Figure 6.
Extraction of Morphological Landmarks.
(A) OPT fluorescence volume view of an Arabidopsis leaf (metamer 3; 12 d from sowing) (GFP1 filter). Although the volume data contain information about trichomes and venation pattern, it cannot be readily seen with this view.
(B) Surface extracted from the same data used for (A). Trichomes are now clearly visible on the leaf surface.
(C) to (H) Using information from (A) and (B), it is possible to retrieve lamina and trichomes separately. Each component is surface rendered for clearer visualization.
(C) Leaf lamina without trichomes.
(D) Trichomes without lamina.
(E) Veins alone.
(F) Combined lamina, trichomes, and veins.
(G) Surface rendering of a single trichome.
(H) Main axes of the trichome shown in (G) with angles computed between each branch (b1 to b2, 89°; b2 to b3, 101°; and b3 to b1, 100°).
Bars = 1140 μm in (A) to (F) and 40 μm (G) and (H).
As well as external surfaces, internal domains such as veins could be retrieved using region-growing algorithms (Figure 6E). Views could also be combined to show the trichomes and veins highlighted in the context of the leaf as a whole (Figure 6F). One advantage of identifying trichome domains in this way is that their position and orientation could be automatically quantified down to the cellular scale (Figures 6G and 6H). Moreover, it was possible to measure the angles of trichome branches, with angles computed between each branch. For example, the trichome shown in Figure 6H had angles of 89° between branch 1 and branch 2, 101° between branch 2 and branch 3, and 100° between branch 3 and branch 1.
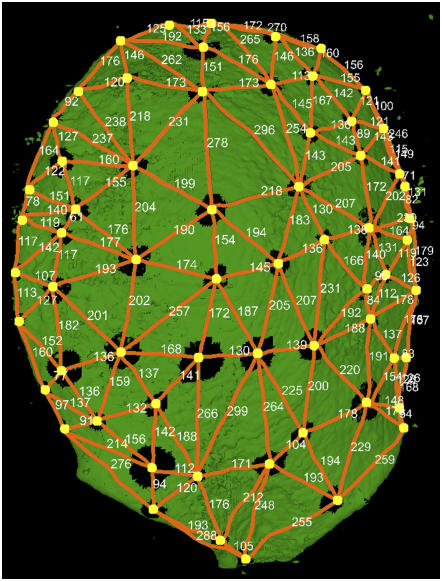
It was also possible to measure automatically trichome spacing along the leaf surface (Figure 7). Trichome positions were extracted for the leaf shown in Figure 6A. The positions of trichome bases were recorded, and distances in micrometers were retrieved along the 3D surface of the leaf (Figure 7). Trichomes were found to be spaced, with distances ranging from 54 to 299 μm and a mean distance of 164 μm.
Figure 7.
Trichome Distribution Map.
Extracted trichome pattern for the leaf shown in Figure 6A. The trichome bases were retrieved (yellow dots), and their separation along the leaf surface is indicated (orange lines). Distances were computed along the 3D surface of the leaf (see Figure 6B) in micrometers. Distances between trichome bases ranged from 54 to 299 μm, with a mean distance of 164 μm.
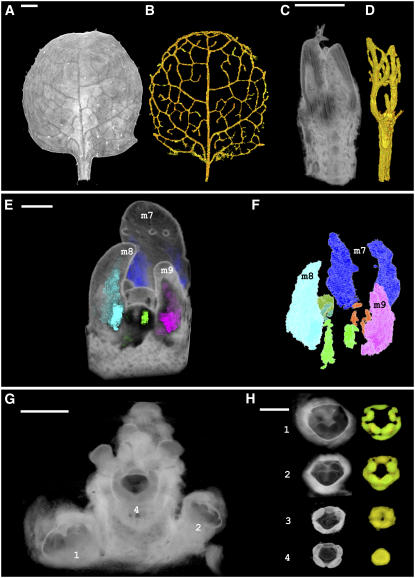
Domains can also correspond to gene expression patterns. For example, regions expressing the vein marker gene Arabidopsis thaliana Homeobox Gene8 (ATHB8) could be extracted using region-growing algorithms (Figures 8A to 8D). This gave a similar result to extracting veins from unstained tissue (Figure 6E) but allowed earlier stages of vein development to be retrieved (Figure 8D). The volume of the vein domains could be quantified at both developmental stages. The vein domain for one of the young leaves shown in Figure 6D was 3.2 × 104 μm3 and occupied 8% of the leaf volume. By comparison, the vein domain of the older leaf shown in Figure 6B was 5 × 107 μm3 and occupied 8.7% of the leaf volume. Total leaf volume increased 1425 times from the young leaf (4 × 105 μm3) to the old leaf (5.7 × 108 μm3). Thus, the proportion of the leaf occupied by the veins remained similar, even though the total volume of the mature leaf increased greatly. This indicates that vein and leaf growth are tightly coupled. Similarly, regions expressing the LFY gene could be extracted and interactively viewed as domains of expression. This allowed expression domains to be classified and color-coded according to metamer position, revealing domains deep within the sample and previously occluded by other tissues (Figures 8E and 8F). Volumes and linear dimensions of domains could also be computed. For example, the LFY expression domain shown in metamer 7 had a volume of 7.5 × 104 μm3 (Figure 8F). Regions of DEF expression in whole-mount in situ images could also be extracted at various stages of development. Domains of DEF signal with different intensities could then be displayed separately for each developmental stage (Figures 8G and 8H).
Figure 8.
Extraction of Gene Expression Patterns from 3D Data Sets.
(A) OPT volume view of a mature ATHB8:GUS Arabidopsis leaf stained to reveal GUS activity (metamer 2; 9 d from sowing). Combined transmission and fluorescent (GFP1) OPT channels were used. Bar = 200 μm.
(B) GUS signal extracted using semiautomatic segmentation tools revealing the expression of ATHB8:GUS in veins.
(C) OPT volume view of a young ATHB8:GUS dissected seedling (metamers 2 and 3; 4 d from sowing). Combined transmission and fluorescent (GFP1) channels were used. Cotyledons were removed and first true leaves are visible. Bar = 60 μm.
(D) ATHB8 venation pattern extracted from (C).
(E) OPT volume view of a LFY:GUS Arabidopsis meristem with expression domains highlighted. Gene expression was segmented as separate domains for each metamer and color-coded accordingly. Bar = 45 μm.
(F) Domains shown in (E) displayed without surrounding tissues. LFY:GUS staining situated deeper within the volume is revealed. Color-coding is as follows: m7, blue; m8, turquoise; m9, pink; m10, dark green; m11, bright green; m12, orange.
(G) OPT volume view of DEF expression in an Antirrhinum inflorescence. Flower buds are labeled consecutively, with flower 1 being the most mature in this series. Flowers are arranged with spiral phyllotaxy; flower 3 being occluded by flower 4 in this view. Bar = 350 μm.
(H) Flowers from (G) were clipped and segmented, with intense DEF expression color-coded in green and weaker expression displayed in yellow. Flower 1, late stage 5 (without sepals). DEF is expressed in petal and stamen primordia. Flower 2, mid stage 5. DEF is strongly expressed in emerging petal and stamen primordia. Flower 3, early stage 5. DEF expression is in the ring of the emerging petal and stamen primordia. Flower 4, stage 4. DEF is expressed in the area where petals and stamens will later develop. Bar = 350 μm.
DISCUSSION
OPT is a convenient method for visualizing plant morphology and gene activity in 3D. Data from a range of specimens including embryos, seeds, seedlings, meristems, leaves, and flowers can be readily obtained. The data can be acquired and visualized at a range of scales, from whole Arabidopsis plants down to large individual cells. In some cases, it is also possible to generate OPT images from live specimens as they grow. Using an appropriate combination of light channels, markers, and stains, it is possible to highlight particular morphological, histological, and gene expression domains. These domains can be easily identified and interrogated, allowing quantitative statistical analysis in 3D of features such as trichomes and gene expression patterns using software developed for OPT image analysis.
The ability to image plants and expression patterns at various scales in 3D opens up new areas to experimental attack. First, it is possible to obtain accurate statistics on the shapes and positions of structures and expression domains at different stages of development. This can provide essential quantitative data for analyzing the relationship between growth and patterning. For example, although there have been extensive molecular genetic studies on trichome and stomatal spacing (Nadeau and Sack, 2002; Geisler et al., 2003; Kirik et al., 2004), we do not have quantitative data on distributions over the leaf as a whole at different developmental stages for wild-type or mutant plants. Such information would allow proposed mechanisms for patterning and growth to be assessed rigorously. Moreover, relationships between different domains, such as the positioning of trichomes and veins, could be readily investigated and quantified.
Second, it is possible to quantify gene expression and morphological domains much more easily and accurately. Quantifying and comparing domains based on 2D sections depends on being able to recreate the same planes of a section, a time-consuming task. For example, a previous study on vascular patterning in Arabidopsis involved manual alignment and tracing of numerous serial sections of ATHB8:GUS plants (Kang et al., 2003). With OPT, similar information can be obtained from a single 3D image. Moreover, it is possible to obtain quantitative data, such as vein and leaf volumes, that allow the growth of different structures to be related. A further advantage is that each metamer can be readily identified, allowing standardization of developmental stages for each leaf and bud along the main axis of the plant.
Finally, the use of OPT for live imaging opens up the possibility of monitoring and quantifying the 3D growth of tissues and cells over time. Combined with imaging of expression domains with markers such as GFP, this would allow growing domains to be quantified from the cellular to the organ and plant level, complementing studies that have recently been performed using confocal microscopy (Grandjean et al., 2004; Heisler et al., 2005).
METHODS
Plant Material
Antirrhinum majus plants were grown in a greenhouse. Arabidopsis thaliana plants were grown in a continuous-light growth cabinet at 25°C.
Plant Nomenclature and Staging
Plant age is described as days from sowing. An Arabidopsis plant was considered as a series of metamers, each comprising three modules: an axillary meristem, a subtending leaf (if present), and a supporting internode. In the main axis of the plant, one cotyledon was defined as m0, the other as m1, and the metamers above this were numbered sequentially m2, m3, to mi, irrespective of the organ at that position (Mundermann et al., 2005). Antirrhinum flowers were staged as described previously (Vincent and Coen, 2004).
GUS Staining
The GUS reporter gene (uidA encoding Escherichia coli β-glucuronidase [Jefferson et al., 1987]) was used for the histochemical localization of GUS activity. LEAFY:GUS (Blazquez et al., 1997), ATHB8:GUS (Kang et al., 2003), and GL2:GUS (Masucci et al., 1996) constructs were used. GL2:GUS seeds were obtained from John Schielefbein. Fresh tissue was dissected and incubated in the dark at 37°C overnight in 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid with 0.5 mM potassium ferrocyanide and 0.5 mM potassium ferricyanide. Stained specimens were washed in deionized water and soaked in 95% ethanol to clear tissue of chlorophyll.
Whole-Mount in Situ Hybridization
Antirrhinum floral inflorescences ∼1 cm long were collected and dissected to remove bracts and sepals. In situ hybridization was performed as described by Coen et al. (1990) and adapted by Desmond Bradley (John Innes Centre) for whole-mount specimens. Fixed tissue in 100% ethanol was rehydrated through an ethanol series and incubated in acetone for 30 min, in PBS for 1 h, in acetic anhydride/triethanolamine for 30 min, and for two washes in PBS for 10 min. The probe to detect the DEF transcript was a 370-bp BamHI-HindIII fragment of DEF A (Sommer et al., 1990) linearized with HindIII. Probe was prepared and hybridized overnight at 50°C as described by Coen et al. (1990). After hybridization, specimens were given three washes of 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 50% formamide at 50°C for 2 h each, two washes of NTE (1× NTE is 0.5 M NaCl, 10 mM Tris-HCl, pH 7.5, and 10 mM EDTA) at 37°C for 30 min each, one wash of NTE and RNase at 37°C for 1 h, two washes of NTE at room temperature for 30 min each, one wash of 2× SSC and 50% formamide at 50°C for 2 h, one wash of 1× SSC at room temperature for 5 min, and one wash of PBS at room temperature for 30 min. Blocking was as described by Coen et al. (1990), with an extended incubation period of up to 4 h. After blocking, specimens were incubated overnight with anti-digoxigenin-alkaline phosphatase at 4°C. They were then washed at room temperature once with 100 mM Tris-HCl, 150 mM NaCl, 1% BSA, and 0.3% Triton for 20 min, once with 100 mM Tris-HCl and 150 mM NaCl for 20 min, and once with 100 mM Tris-HCl, 150 mM NaCl, and 50 mM MgCl2, pH 9.5, for 30 min. Staining for the antibody was as described by Coen et al. (1990) with incubations of 5 to 12 h.
Specimen Preparation for OPT
Specimens for OPT analysis were usually fixed in 4% paraformaldehyde and 0.1% Tween/0.1% Triton followed by dehydration steps of 3 h each through an ethanol/0.85% saline series (50, 70, 85, 95, and finally 100% ethanol, after which the specimen was left overnight at 4°C). Alternatively, specimens were incubated overnight in 100% ethanol without fixation. Arabidopsis seedlings younger than 4 d from sowing and specimens <300 μm in size were stained with a cell wall–specific stain, saffranin (0.3% in 100% ethanol), at 4°C overnight to enhance tissue fluorescence, followed by destaining for up to 1 h in 100% ethanol. Specimens were rehydrated through an ethanol series (70, 50, 30, and 10% ethanol for 30 min each at room temperature). Rehydrated specimens were embedded in 1% low-melting-point agarose, as described previously (Sharpe et al., 2002). Embedded specimens were dehydrated overnight in methanol and cleared in a 1:2 mixture of benzyl alcohol and benzyl benzoate (Sigma-Aldrich) for 2 to 24 h until almost transparent. The strong signal and sharp boundaries of expression observed around trichomes indicate that the diffusion of stains in the benzyl alcohol and benzyl benzoate clearing agent is limited.
For live OPT, Arabidopsis seeds were embedded in 1% low-melting-point agarose, stratified for 3 d at 4°C, and allowed to germinate in a growth room with 16 h of light at 20°C. When the seedlings were 3 d old, they were placed in the OPT scanner in which further growth occurred.
OPT Scanning
Images of embedded plant tissues were collected using a prototype OPT device constructed at the Human Genetics Unit of the Medical Research Council (Edinburgh, UK) (Sharpe et al., 2002) and installed at the John Innes Centre. Specimens were rotated through 360° using a stepper motor with 0.9° steps on a Leica MZ FLIII microscope using either a planapo 0.63× M-series or a plan 0.5× M-series objective, via a 1× C-mount adaptor, with a 100-W mercury-vapor burner for fluorescence imaging (Leica Microsystems). Images were recorded using a Coolsnap CF camera (Roper Scientific, Photometrics) controlled by IPLab imaging software (Scanalytics) (Sharpe et al., 2002). The following Leica filters were used: TXR, 560/40 nm excitation, 610 LP nm emission, for saffranin-stained tissue and autofluorescence; GFP1, 425/60 nm excitation, 480 nm emission; Green, 546/10 nm excitation, 590 nm barrier; and GFP2, 480/40 nm excitation, 510 nm emission for tissue autofluorescence. Visible light transmission images were collected using a 20-W halogen lamp connected to the OPT device. Infrared path filters (700, 750, and 800 nm) reduced dark signal in strongly stained specimens. Voxel size was calculated based on the microscope objective and magnification used and verified with reconstructions of a calibration pin of known size. Voxel sizes at each magnification were obtained and linear measurements and volume measurements of domains were collected with QtVolView. Data for the specimen shown in Figure 5A were obtained using an OPT Scanner 3001 (Bioptonics, MRC Technology). Reconstructions of OPT images were performed as described previously (Sharpe et al., 2002).
For live OPT, germinating Arabidopsis seedlings were scanned in water. Visible light transmission images were collected at several intervals over a 72-h period. The quality of 3D images from water-scanned Arabidopsis roots was superior to 3D data obtained from roots fixed and treated with clearing agent, although internal detail through dense regions, such as the seed coat, was reduced.
Visualization and Segmentation
A software package, called QtVolView, was developed to display combined volume, surface, and section-rendered images. The program exploits graphics hardware to allow more rapid and interactive viewing of 3D data. Interactive viewing of the specimens was achieved by approximating the volumes as series of parallel view-aligned quadrilateral slices. The individual slices were texture-mapped with the corresponding volumetric data and blended together to achieve the 3D volume view, as described by Blythe (1998). The visibility of the renderings was improved with user-specified transfer functions, edge enhancement (Jain, 1989), and per-voxel lighting (Ebert and Rheingans, 2001). Gaussian blurring and thresholding were also used to remove noise in the scans. Up to three OPT scan filter channels were combined within the same 3D space. Each channel could be independently clipped and interrogated. A basic version of the program is presented with the supplemental data online, and the full version of QtVolView is available on request.
Segmentation of the trichomes for the Arabidopsis leaf was achieved using a semiautomatic seeded region-growing algorithm (separate software from QtVolView). This was applied to a surface representation of the leaf. To retrieve the surface representation, the marching-cubes algorithm was applied (Lorenson and Cline, 1987). A surface normal clustering algorithm based on mean shift was then applied to the surface (Yamauchi et al., 2005). The trichomes were then differentiated from the rest of the surface using a region-growing algorithm. To initialize the region-growing algorithm, the user selected a nontrichome vertex as the seed point. The algorithm was then allowed to propagate, by edge traversal and a curvature term, defined as the angle between the normals of two edge vertices. By specifying this curvature term, the region-growing algorithm was able to achieve the desired segmentation. Further processing of individual trichomes using principal component analysis (http://www.cs.otago.ac.nz/cosc453/student_tutorials/principal_components.pdf) (Pearson, 1901) and k-means clustering was used to retrieve the trichome branches (http://fconyx.ncifcrf.gov/∼lukeb/kmeans.html) (MacQueen, 1967).
Distances between trichomes were obtained by first performing Delaunay triangulation on the positions of the trichome bases (www.qhull.org). Points on the leaf surface lying nearest to the triangle edges were then obtained by sampling along the edges at constant intervals. These points were fitted with a B-spline curve to get a smooth path along the leaf surface for each neighboring trichome base. The lengths of the B-spline curves were then calculated to obtain the distances between trichome bases.
For the segmentation of gene expression and veins, a region-growing algorithm combined with information gain was incorporated into QtVolView (Jain, 1989; Singh et al., 2004). Figures were composed with Adobe Photoshop CS.
QtVolView Movies
Movies of images illustrated in Figures 1 to 8 are provided as Supplemental Movies 1 to 14 online. All movies were obtained from QtVolView with an image size of 256 × 256 × 256 pixels unless stated otherwise.
QtVolViewLITE Program
The QtVolViewLITE program is provided with the reconstructed OPT data set illustrated in Figures 1A and 2A to allow the visualization and direct interaction with the 3D data (http://www.cmp.uea.ac.uk/Research/cbg/downloads.htm). Minimum system requirements for the QtVolViewLITE program are a 3.0-GHz CPU, 2 Gb of RAM, and a 256-Mb graphic card. The full QtVolView program is available on request.
Accession Numbers
LEAFY:GUS (Blazquez et al., 1997) seeds were obtained from Nottingham Arabidopsis Stock Centre (NASC) reference N6297. ATHB8:GUS (Kang et al., 2003) seeds were obtained from NASC reference N296.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Movie 1. Movie for Figures 1A and 2A.
Supplemental Movie 2. Movie for Figures 1C and 2C.
Supplemental Movie 3. Movie for Figures 1D and 2D.
Supplemental Movie 4. Movie for Figure 3F.
Supplemental Movie 5. Movie for Figure 4A.
Supplemental Movie 6. Movie for Figure 4B.
Supplemental Movie 7. Movie for Figure 5B.
Supplemental Movie 8. Movie for Figure 5D.
Supplemental Movie 9. Movie for Figure 6A.
Supplemental Movie 10. Movie for Figures 8A and 8B.
Supplemental Movie 11. Movie for Figures 8C and 8D.
Supplemental Movie 12. Movie for Figures 8E and 8F.
Supplemental Movie 13. Movie for Figure 8G.
Supplemental Movie 14. Movie for Figure 8H, Flower 1.
Supplementary Material
Acknowledgments
We thank Richard Baldock, Duncan Davidson, Bill Hill, and Allyson Ross from the Edinburgh Mouse Atlas Group for their invaluable support and for providing us with Mouse Atlas software and expertise. We also thank the Bioptonics team for the use of a prototype OPT Scanner 3001, to obtain the OPT scan shown in Figure 5B. Steve Rawsthorne (John Innes Centre) provided the Arabidopsis silique shown in Figures 1 to 3. We also thank Desmond Bradley for advice on whole-mount in situ techniques and Paul Derbyshire for advice on root growth.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Enrico Coen (enrico.coen@bbsrc.ac.uk).
Online version contains Web-only data.
References
- Baldock, R.A., et al. (2003). EMAP and EMAGE: A framework for understanding spatially organized data. Neuroinformatics 1 309–325. [DOI] [PubMed] [Google Scholar]
- Blazquez, M.A., Soowal, L.N., Lee, I., and Weigel, D. (1997). LEAFY expression and flower initiation in Arabidopsis. Development 124 3835–3844. [DOI] [PubMed] [Google Scholar]
- Blythe, D. (1998). Advanced Graphics Programming Techniques Using OpenGL. SIGGRAPH 1998 Course. (New York: ACM Press).
- Bryson-Richardson, R.J., and Currie, P.D. (2004). Optical projection tomography for spatio-temporal analysis in the zebrafish. Methods Cell Biol. 76 37–50. [DOI] [PubMed] [Google Scholar]
- Coen, E.S., Romero, J.M., Doyle, S., Elliott, R., Murphy, G., and Carpenter, R. (1990). Floricaula: A homeotic gene required for flower development in Antirrhinum majus. Cell 63 1311–1322. [DOI] [PubMed] [Google Scholar]
- Ebert, D., and Rheingans, P. (2001). Volume illustration: Nonphotorealistic rendering of volume models. IEEE Trans. Vis. Comput. Graph. 7 253–264. [Google Scholar]
- Fiala, J.C. (2005). Reconstruct: A free editor for serial section microscopy. J. Microsc. 218 52–61. [DOI] [PubMed] [Google Scholar]
- Gambhir, S.S., et al. (1999). Imaging adenoviral-directed reporter gene expression in living animals with positron emission tomography. Proc. Natl. Acad. Sci. USA 96 2333–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler, M.J., Deppong, D.O., Nadeau, J., and Sack, F.D. (2003). Stomatal neighbour cell polarity and division in Arabidopsis. Planta 216 571–579. [DOI] [PubMed] [Google Scholar]
- Grandjean, O., Vernoux, T., Laufs, P., Belcram, K., Mizukami, M., and Trass, J. (2004). In vivo analysis of cell division, cell growth and differentiation at the shoot apical meristem in Arabidopsis. Plant Cell 16 74–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greive, K., Dubois, A., Simonutti, M., Paques, M., Sahel, J., Le Gargasson, J.-F., and Boccara, C. (2005). In vivo anterior segment imaging in the rat eye with high speed white light full field optical coherence tomography. Opt. Express 13, 6286–6295. [DOI] [PubMed] [Google Scholar]
- Gustafsson, M.G.L. (2005). Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proc. Natl. Acad. Sci. USA 102 13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler, M.G., Ohno, C., Das, P., Sieber, P., Reddy, G.V., Long, J.A., and Meyerowitz, E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15 1899–1911. [DOI] [PubMed] [Google Scholar]
- Hettinger, J.W., de la Pena Mattozzi, M., Myers, W.R., Williams, M.E., Reeves, A., Parsons, R.L., Haskell, R.C., Petersen, D.C., Wang, R., and Medford, J.I. (2000). Optical coherence microscopy. A technology for rapid, in vivo, non-destructive visualization of plants and plant cells. Plant Physiol. 123 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook, N.M., Ahrens, E.T., Burns, M.J., and Zwieniecki, M.A. (2001). In vivo observation of cavitation and embolism repair using magnetic resonance imaging. Plant Physiol. 126 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J., and Stelzer, E.H.K. (2004). Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305 1007–1009. [DOI] [PubMed] [Google Scholar]
- Jain, A.K. (1989). Fundamentals of Digital Image Processing. (Upper Saddle River, NJ: Prentice-Hall).
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson, H., Heisler, M., Shapiro, B., Meyerowitz, E., and Mjolsness, E. (2006). An auxin-driven polarized transport model for phyllotaxis. Proc. Natl. Acad. Sci. USA 103, 1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J., Tang, J., Donnelly, P., and Dengler, N. (2003). Primary vascular pattern and expression of ATHB-8 in shoots of Arabidopsis. New Phytol. 158 443–454. [DOI] [PubMed] [Google Scholar]
- Kerwin, J., Scott, M., Sharpe, J., Puelles, L., Robson, S.C., Martinez-de-la-Torre, M., Ferran, J.L., Feng, G., Baldock, R., Strachan, T., Davidson, D., and Lindsay, S. (2004). 3 dimensional modelling of early human brain development using optical projection tomography. BMC Neurosci. 5 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik, V., Simon, M., Huelskamp, M., and Schiefelbein, J. (2004). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268 506–513. [DOI] [PubMed] [Google Scholar]
- Lickert, H., Takeuchi, J.K., von Both, I., Walls, J.R., McAuliffe, F., Adamson, S.L., Henkelman, R.M., Wrana, J.L., Rossant, J., and Bruneau, B.G. (2004). Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 432 107–112. [DOI] [PubMed] [Google Scholar]
- Lindsay, S., and Copp, A.J. (2005). MRC-Wellcome Trust Human Developmental Biology Resource: Enabling studies of human trends in genetics developmental gene expression. Trends Genet. 21 586–590. [DOI] [PubMed] [Google Scholar]
- Lindsay, S., Sarma, S., Martinez-De-La-Torre, M., Kerwin, J., Scott, M., Ferran, J.L., Baldock, R., and Puelles, L. (2005). Anatomical and gene expression mapping of the ventral pallium in a three-dimensional model of developing human brain. Neuroscience 136 625–632. [DOI] [PubMed] [Google Scholar]
- Liu, Y.C., and Chiang, A.S. (2003). High-resolution confocal imaging and three-dimensional rendering. Methods 30 86–93. [DOI] [PubMed] [Google Scholar]
- Lorenson, W.E., and Cline, H.E. (1987). Marching cubes: A high resolution 3D surface construction algorithm. In Proceedings of the 14th Annual Conference on Computer Graphics and Interactive Techniques. (New York: ACM Press), pp. 163–169.
- Louie, A.Y., Huber, M.M., Ahrens, E.T., Rothbacher, U., Moats, R., Jacobs, R.E., Fraser, S.E., and Meade, T.J. (2000). In vivo visualization of gene expression using magnetic resonance imaging. Nat. Biotechnol. 18 321–325. [DOI] [PubMed] [Google Scholar]
- MacQueen, J. (1967). Some methods for classification and analysis of multivariate observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability. (Berkeley, CA: University of California Press), pp. 281–297.
- Masucci, J.D., Rerie, W.G., Foreman, D.R., Zhang, M., Galway, M.E., Marks, M.D., and Schiefelbein, J.W. (1996). The homeobox gene GLABRA 2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122 1253–1260. [DOI] [PubMed] [Google Scholar]
- Mundermann, L., Erasmus, Y., Lane, B., Coen, E., and Prusinkiewicz, P. (2005). Quantitative modeling of Arabidopsis development. Plant Physiol. 139 960–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau, J.A., and Sack, F.D. (2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296 1697–1700. [DOI] [PubMed] [Google Scholar]
- Pearson, K. (1901). On lines and planes of closest fit to systems of points in space. Philos. Mag. 2, 559–572. [Google Scholar]
- Reeves, A., Parsons, R.L., Hettinger, J.W., and Medford, J.I. (2002). In vivo three-dimensional imaging of plants with optical coherence microscopy. J. Microsc. 208 177–189. [DOI] [PubMed] [Google Scholar]
- Ruijter, J.M., Soufan, A.T., Hagoort, J., and Moorman, A.F. (2004). Molecular imaging of the embryonic heart: Fables and facts on 3D imaging of gene expression patterns. Birth Defects Res. Part C Embryo Today Rev. 72 224–240. [DOI] [PubMed] [Google Scholar]
- Sarma, S., Kerwin, J., Puelles, L., Scott, M., Strachan, T., Feng, G., Sharpe, J., Davidson, D., Baldock, R., and Lindsay, S. (2005). 3D modelling, gene expression mapping and post-mapping image analysis in the developing human brain. Brain Res. Bull. 66 449–453. [DOI] [PubMed] [Google Scholar]
- Sharpe, J. (2003). Optical projection tomography as a new tool for studying embryo anatomy. J. Anat. 202 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe, J. (2004). Optical projection tomography. Annu. Rev. Biomed. Eng. 6 209–228. [DOI] [PubMed] [Google Scholar]
- Sharpe, J., Ahlgren, U., Perry, P., Hill, B., Ross, A., Hecksher-Sorensen, J., Baldock, R., and Davidson, D. (2002). Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science 296 541–545. [DOI] [PubMed] [Google Scholar]
- Singh, H., Crawford, M., Curtin, J., and Zwiggelaar, R. (2004). Automated 3D segmentation of lung airway tree using gain based region growing approach. Lect. Notes Comput. Sci. 3217 975–982. [Google Scholar]
- Sommer, H., Beltràn, J.-P., Huijser, P., Pape, H., Lönnig, W., Saedler, H., and Schwarz-Sommer, Z. (1990). Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 9 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuppy, W.H., Maisano, J.A., Colbert, M.W., Rudall, P.J., and Rowe, T.B. (2003). Three-dimensional analysis of plant structure using high-resolution X-ray computed tomography. Trends Plant Sci. 8 2–6. [DOI] [PubMed] [Google Scholar]
- Takeuchi, J.K., et al. (2005). Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development 132 2463–2474. [DOI] [PubMed] [Google Scholar]
- Tickle, C. (2004). The contribution of chicken embryology to the understanding of vertebrate limb development. Mech. Dev. 121 1019–1029. [DOI] [PubMed] [Google Scholar]
- Tyszka, J.M., Ewald, A.J., Wallingford, J.B., and Fraser, S.E. (2005). New tools for visualization and analysis of morphogenesis in spherical embryos. Dev. Dyn. 234 974–983. [DOI] [PubMed] [Google Scholar]
- Vincent, C.A., and Coen, E.S. (2004). A temporal and morphological framework for flower development in Antirrhinum majus. Can. J. Bot. 82 681–690. [Google Scholar]
- Walls, J.R., Sled, J.G., Sharpe, J., and Henkelman, R.M. (2005). Correction of artefacts in optical projection tomography. Physiol. Med. Biol. 50 4645–4665. [DOI] [PubMed] [Google Scholar]
- Wilkie, A.L., Jordan, S.A., Sharpe, J.A., Price, D.J., and Jackson, I.J. (2004). Widespread tangential dispersion and extensive cell death during early neurogenesis in the mouse neocortex. Dev. Biol. 267 109–118. [DOI] [PubMed] [Google Scholar]
- Yamauchi, H., Lee, S., Lee, Y., Ohtake, Y., Belyaev, A., and Hans-Peter, S. (2005). Feature sensitive mesh segmentation with mean shift. In Shape Modeling International Conference 2005 (Cambridge, MA: IEEE), pp. 236–243.
- Zachgo, S., Perbal, M.-C., Saedler, H., and Schwarz-Sommer, Z. (2000). In situ analysis of RNA and protein expression in whole mounts facilitates detection of floral gene expression dynamics. Plant J. 23, 697–702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.