Abstract
Synechocystis sp PCC 6803 Slr1471p, an Oxa1p/Alb3/YidC homolog, is an essential protein for cell viability for which functions in thylakoid membrane biogenesis and cell division have been proposed. Using a fusion of green fluorescent protein to the C terminus of Slr1471p, we found that the mutant slr1471-gfp is photochemically inhibited when light intensities increase to 80 μmol·m−2·s−1. We show that photoinhibition correlates with an increased redox potential of the reaction center quinone QA− and a decreased redox potential of QB−. Analysis reveals that membrane integration of the D1 precursor protein is affected, leading to the accumulation of pD1 in the membrane phase. We show that Slr1471p interacts directly with the D1 protein and discuss why the accumulation of pD1 in two reaction center assembly intermediates is dependent on Slr1471p.
INTRODUCTION
It is generally accepted that chloroplasts of higher plants descended from cyanobacteria (Herrmann, 1997; McFadden and van Dooren, 2004). During endosymbiosis, most of the cyanobacterial genes were transferred to the nucleus of the host cell. As a result, most of the chloroplast-localized proteins are nucleus-encoded and have to be imported (Abdallah et al., 2000; Martin et al., 2002). Upon import, the proteins are targeted to the soluble (stroma and the thylakoid lumen) and membrane (inner envelope and thylakoid membrane) compartments of the chloroplast. To date, four different pathways (the spontaneous and the bacteria-related cpSec, cpTAT/ΔpH, and cpSRP pathways) have been described for the targeting of imported proteins into and across the thylakoid membrane, which require specific sets of protein factors and energy sources (Keegstra and Cline, 1999; Eichacker and Henry, 2001; Mori and Cline, 2001; Schleiff and Klösgen, 2001).
By contrast, little is known about the targeting of plastid-encoded proteins to the thylakoid membrane. Recent results suggest that targeting, membrane export, and assembly of the D1 reaction center protein of photosystem II (PSII) might be performed by components of the cpSRP and cpSec pathways: cpSRP54, Alb3p, and cpSecY (Nilsson et al., 1999; Zhang et al., 2001; Ossenbühl et al., 2004). Specifically, cpSRP54 was found to interact early with the nascent D1 protein (D1 fragments smaller than 17 kD) (Nilsson et al., 1999), whereas nascent D1 fragments between 17 and 25 kD were found in interaction with the translocase cpSecY (Zhang et al., 2001). Recently, Alb3.1p of Chlamydomonas reinhardtii was shown to selectively interact with the full-length reaction center protein (RC) D1 during the assembly of D1 into PSII (Ossenbühl et al., 2004).
Alb3p belongs to a widespread protein family, the Oxa1p/Alb3/YidC family. Members of this protein family are found in bacteria, mitochondria, and chloroplasts. They constitute a group of evolutionarily conserved proteins that appear to be involved in the integration and/or assembly of membrane protein complexes. The mitochondrial Oxa1p and the bacterial YidCp are the best analyzed examples (Kuhn et al., 2003). Oxa1p is localized at the inner membrane of mitochondria and facilitates the insertion of both nucleus- and mitochondria-encoded proteins into the inner mitochondrial membrane (Hell et al., 2001). YidC is an essential inner membrane protein of Escherichia coli and is involved in the cotranslational insertion of inner membrane proteins both in connection with the Sec translocase and on its own. Recent models proposed that the YidC protein might also be required for the assembly of inserted membrane proteins by clearing the SecYEG translocase (Chen et al., 2002). Interestingly, the ΔyidC mutant of E. coli can be complemented with ALB3 from Arabidopsis thaliana, thus indicating a common function (Jiang et al., 2002). The protein ALB3p was originally identified by complementation of an albino3 (alb3) mutant of Arabidopsis (Sundberg et al., 1997). The phenotype of the alb3 mutant was described as very severe with strongly reduced thylakoid membranes, suggesting an additional function of Alb3 besides the described integration of light-harvesting complex proteins (LHCPs) (Moore et al., 2000, 2003).
In C. reinhardtii, two nuclear genes encoding ALB3p homologs, Alb3.1p and Alb3.2p, were identified by complementation of a nuclear mutant of the Alb3.1 gene, ac29 (Bellafiore et al., 2002; Göhre et al., 2006). The phenotype of ac29, which accumulates ∼10% LHCPs compared with the wild type, revealed a less pronounced decrease in PSII than the alb3 phenotype in Arabidopsis. In addition to the reduced LHCP levels, a retarded D1 assembly into PSII was found, suggesting that Alb3.1p is involved in the integration and assembly of both nucleus- and plastid-encoded proteins in the thylakoid membranes (Ossenbühl et al., 2004). In the unicellular cyanobacterium Synechocystis sp PCC 6803 (hereafter called Synechocystis), Slr1471p (encoded by the slr1471 gene) was found to be involved in cell division by an unknown mechanism. However, the protein Slr1471p lacks an N-terminal receptor domain, which in Arabidopsis was implicated in organellar division (Fulgosi et al., 2002). Because only the single gene slr1471 was found in all cyanobacterial genome projects, Slr1471p may have an even broader spectrum of substrates and modes of operation than in chloroplasts. This idea is supported by the fact that it is by now not possible to fully segregate a deletion mutant of slr1471. Previous results with partially segregated knockdown mutants suggested that Slr1471p plays a role not only in cell division but in membrane biogenesis as well (Spence et al., 2004). Here, we show that Slr1471p function is significantly impaired by the fusion of green fluorescent protein (GFP) to its C terminus. Specifically, mutant light sensitivity was found to be mediated by an alteration in the redox potential of reaction center quinones QA− and QB−, leading to the photoinhibition of PSII and a concomitant decrease in the mutant growth rate. These results suggest that Slr1471p is important for the de novo assembly of the D1 precursor protein into the PSII reaction center.
RESULTS
Fusion of GFP to the C Terminus of Slr1471p Causes a Light-Sensitive Growth Defect
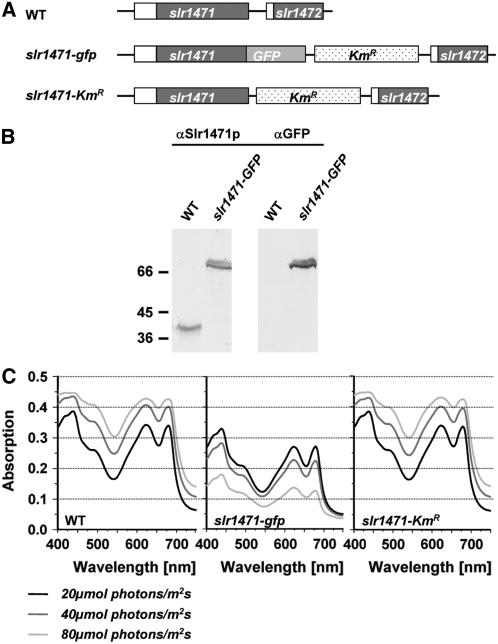
To characterize the function of Slr1471p in cyanobacteria, we fused the C terminus of Slr1471p with the globular and soluble GFP (Figure 1A). Replacement of all copies of the wild-type chromosome in the mutant was verified by PCR and expression of the fusion protein by gel blot analysis using antibodies against Slr1471p (Figure 1B). Surprisingly, during analysis of the photoautotrophic growth of slr1471-gfp cells, we found that the growth of mutant cells grown under a light intensity of 10 μmol·m−2·s−1 was impaired when light intensities were shifted to 20, 40, and 80 μmol·m−2·s−1 (wild-type cells versus slr1471-gfp cells; Figure 1C). When light intensities were increased, wild-type cells responded with growth to higher densities (cell densities in different light conditions for the wild type were as follows: 20 μmol·m−2·s−1, 0.292 × 10−6 OD730/h; 40 μmol·m−2·s−1, 0.958 × 10−6 OD730/h; 80 μmol·m−2·s−1, 1.472 × 10−6 OD730/h), whereas slr1471-gfp cells grew to lower densities (cell densities in different light conditions for slr1471-gfp were as follows: 20 μmol·m−2·s−1, 0.007 × 10−6 OD730/h; 40 μmol·m−2·s−1, 0.000 × 10−6 OD730/h; 80 μmol·m−2·s−1, −0.167 × 10−6 OD730/h).
Figure 1.
Construction and Growth Comparison of slr1471-gfp and slr1471-KmR Mutants.
(A) Genomic region of the slr1471 and slr1472 genes in wild-type, slr1471-gfp, and slr1471-KmR cells. Genes are represented by boxes.
(B) Complete replacement of Slr1471p with the fusion protein Slr1471-GFPp. Total membrane proteins extracted from wild-type and slr1471-gfp cells were separated by 12% SDS-PAGE, transferred to nitrocellulose membranes, and probed with polyclonal anti-Slr1471p antiserum (αSlr1471p) or monoclonal anti-GFP antiserum (αGFP). The calculated molecular masses of Slr1471p and Slr1471-GFPp are 40 and 70 kD, respectively.
(C) Absorption spectra of wild-type, slr1471-gfp, and slr1471-KmR cells. Cultures were inoculated at OD730 = 0.05 and incubated for 3 d at 30°C and the indicated light intensities of 20, 40, and 80 μmol·m−2·s−1.
For segregation of the mutant, GFP was inserted together with a kanamycin resistance (KmR) gene cartridge. To ensure that the light depression of growth in slr1471-gfp was not caused by KmR, we generated the mutant slr1471-KmR (Figure 1A). We found that the growth of slr1471-KmR cells at different light intensities was comparable to that of the wild type (Figure 1C) (cell densities in different light conditions for slr1471-KmR were as follows: 20 μmol·m−2·s−1, 0.319 × 10−6 OD730/h; 40 μmol·m−2·s−1, 0.972 × 10−6 OD730/h; 80 μmol·m−2·s−1, 1.486 × 10−6 OD730/h). We conclude that the fusion of GFP specifically alters the function of Slr1471p, causing the inhibition of photoautotrophic growth at higher light intensity.
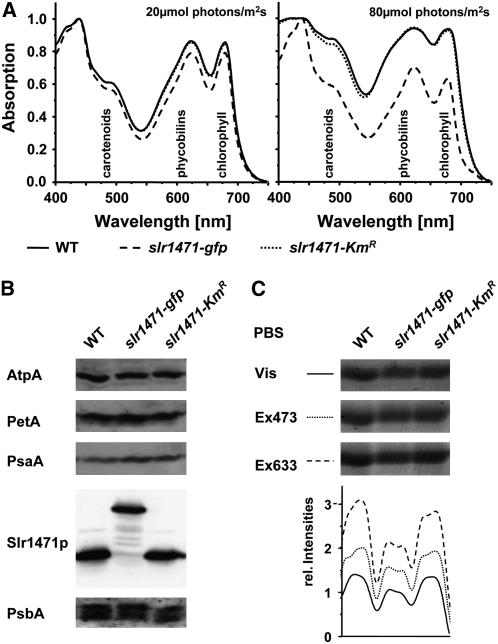
Interestingly, absorption spectra of slr1471-gfp cells grown at 80 μmol·m−2·s−1 indicated that at this increased light intensity, pigmentation was also selectively changed in the mutant (Figure 1C). To compare the absorption spectra of the wild type, slr1471-gfp, and slr1471-KmR grown at low (20 μmol·m−2·s−1) and high (80 μmol·m−2·s−1) light intensity, the spectra of all three strains shown in Figure 1C were normalized to the highest peak at 436 nm (Figure 2A). slr1471-KmR spectra revealed a small decrease in carotenoid content only at the highest light intensity. In slr1471-gfp, overall small decreases of carotenoid (470 to 500 nm), phycobilin (550 to 650 nm), and chlorophyll (670 nm) contents correlated with decreased growth at low light intensity; however, at increased light intensity, pigment contents were selectively reduced, in particular carotenoids and phycobilins (Figure 2A). We also compared the relative amounts of the major thylakoid membrane complexes and of phycobilins bound to phycobilisomes in all three strains grown under high light by immunoblot analysis using antibodies against central subunits (Figure 2B) and by fluorescence and visible scans of the phycobiliproteins on the gel (Figure 2C). These analyses revealed no significant differences in the amount of the major thylakoid membrane complexes PSI, PSII, ATP synthase, and cytochrome b6f between the wild type, slr1471-gfp, and slr1471-KmR (Figure 2B). By contrast, phycobiliproteins showed a reduced fluorescence signal in slr1471-gfp compared with the wild type and slr1471-KmR (Figure 2C), corroborating the reduced amount of phycobilins in slr1471-gfp (Figure 2A). These results indicate in particular that the accumulation of functional proteins from the light-harvesting antenna was affected. Therefore, we analyzed the fluorescence properties of the different antenna systems of PSI and PSII by 77K fluorescence spectroscopy using excitation wavelengths at 440 nm (chlorophyll a) (Figure 3A) and 570 nm (phycobilisomes) (Figure 3B), respectively.
Figure 2.
Pigmentation and Protein Composition of Different Synechocystis Strains.
(A) Absorption spectra from wild-type, slr1471-gfp, and slr1471-KmR cells grown at 20 and 80 μmol·m−2·s−1 (see Figure 1B) were normalized to the absorption at 436 nm. Absorption areas for carotenoids, phycobilins, and chlorophyll are indicated.
(B) Total membrane proteins isolated from high-light-grown (80 μmol·m−2·s−1) wild-type, slr1471-gfp, and slr1471-KmR cells were separated by SDS-PAGE and analyzed on gel blots. Blots were immunodecorated with antibodies against D1 (PsbA), PsaA, AtpA, cytochrome f (PetA), and Slr1471p (Oxa1).
(C) Visible phycobiliproteins (PBS) on the SDS gel shown in (B) were detected with a normal light scanner (Vis) and with a Fuji TLA-3000 fluorescence scanner. Laser excitation wavelengths of 473 nm (Ex473) and 633 nm (Ex633) with emission filter wavelengths of 570 and 675 nm, respectively, were used. Signal intensities of the different scans were analyzed with AIDA software (Fuji) and are plotted under each lane.
Figure 3.
77K Fluorescence Emission Spectra of Wild-Type, slr1471-gfp, and slr1471-KmR Cells.
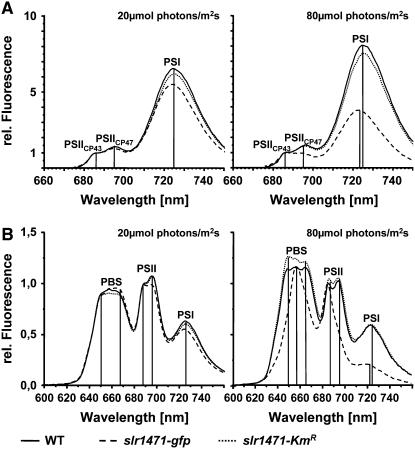
(A) 77K fluorescence emission spectra of wild-type, slr1471-gfp, and slr1471-KmR cell cultures (excitation at 440 nm) grown under 20 and 80 μmol·m−2·s−1 illumination were normalized to the emission at 686 nm (PSIICP43). Fluorescence emission maxima of PSI and PSII (PSIICP43 and PSIICP47) are indicated.
(B) 77K fluorescence emission spectra of wild-type, slr1471-gfp, and slr1471-KmR cell cultures (excitation at 570 nm) grown under 20 and 80 μmol·m−2·s−1 illumination were normalized to the emission at 686 nm (PSIICP43). Fluorescence emission maxima of PSI, PSII (PSIICP43 and PSIICP47), and free phycobilisomes (PBS) are indicated.
Under both low- and high-light conditions, slr1471-KmR cells showed no significant change in PSI antenna fluorescence from chlorophyll a (excitation wavelength, 440 nm) compared with wild-type cells. In addition, low-light-grown slr1471-gfp showed 77K fluorescence spectra comparable to wild type cells, with emission peaks at 686, 695, and 725 nm corresponding to CP43, CP47, and PSI, respectively (Figure 3A; see Supplemental Figure 1 online; Shen and Vermaas, 1994). However, high-light-grown cells of slr1471-gfp showed a selectively reduced fluorescence emission of CP47 at 695 nm and a shifted PSI emission at 723.5 nm (Figure 3A). These results were confirmed by the 77K fluorescence spectra obtained after excitation at 570 nm. Besides the described PSII and PSI peaks, emission spectra showed additional peaks at 650 and 668 nm caused by the fluorescence of the phycobilisome pigments phycocyanin and allophycocyanin, respectively (Figure 3B; see Supplemental Figure 2 online). As after excitation at 440 nm, the emission spectra of low-light-grown slr1471-gfp- and slr1471-KmR cells excited at 570 nm were comparable to the wild-type spectra, whereas high-light-grown cells of slr1471-gfp showed a selectively and strongly reduced fluorescence emission of CP47 at 695 nm and a shifted PSI emission at 723.5 nm (Figure 3B). In addition, the phycocyanin emission peak of high-light-grown slr1471-gfp appeared shifted to 658 nm instead of 650 nm in high-light-grown wild-type and slr1471-KmR cells (Figure 3B; see Supplemental Figure 2 online). PSI fluorescence in high-light-grown slr1471-gfp was reduced to half compared with the wild type, even though the amount of PSI proteins was not changed (Figures 2B and 2C). Furthermore, we detected a blue shift of the PSI fluorescence by 1.5 nm in high-light-grown slr1471-gfp (Figures 3A and 3B).
We conclude that in slr1471-gfp under high-light conditions, the quantum transfer between phycobilisomes, which operate as the outer and inner antenna of the reaction center core complexes, may be affected as a result of reduced amounts of phycobilisomes. This was corroborated by our finding that the protein contents of the major thylakoid membrane complexes in the wild type, slr1471-gfp, and slr1471-KmR grown under different light did not reveal any significant differences in PSI, PSII, cytochrome b6f, and ATPase content by immunoblot analysis but showed a decreased amount of phycobiliproteins (Figures 2B and 2C). Together, these data indicate an altered pigmentation in slr1471-gfp that may result in changes of quantum or electron transfer.
Electron Transfer within PSII of slr1471-gfp Is Altered
To reveal whether changes in quantum transfer within PSII and PSI or electron transfer between the photosystems were responsible for the light sensitivity of slr1471-gfp, we performed oxygen evolution and pulse amplitude–modulated room temperature chlorophyll fluorescence (PAM) measurements. For oxygen evolution, strains were grown and measured at 20 and 80 μmol·m−2·s−1. The relative photosynthetic yield remained unchanged at low light; however, at the increased light intensity, in slr1471-gfp only ∼60% could be obtained (Table 1). For PAM, strains were grown under low- and high-light conditions, and after dark adaptation for 2 min, measurements were performed as described in Methods with an actinic light intensity of 20 μmol·m−2·s−1. Low-light-grown cells showed a maximum photochemical efficiency of PSII in the dark-adapted state (Fv/Fm) of 0.43 ± 0.02 for both the wild type and slr1471-KmR and of 0.40 ± 0.02 for slr1471-gfp (see Supplemental Figure 3 online). Under actinic light, photochemical quenching was comparable in all three strains grown under low light (see Supplemental Figure 3 online). When grown under high light, the wild type and slr1471-KmR performed similarly, whereas slr1471-gfp displayed clear differences (see Supplemental Figure 3 online). The Fv/Fm was decreased to 0.20 ± 0.02 and the actinic light–induced quenching resulted in a steady state chlorophyll fluorescence lower than the initial (minimum) PSII fluorescence in the dark-adapted state (F0). Furthermore, the chlorophyll fluorescence did not recover to F0 even after 15 min of dark incubation. This indicated that under actinic light, the functionality of PSII was decreased as a result of chlorophyll a bleaching.
Table 1.
Oxygen Evolution of Wild-Type, slr1471-gfp, and slr1471-KmR Cells Grown and Measured under Different Light Conditions
| 20 μmol·m−2·s−1
|
80 μmol·m−2·s−1
|
|||
|---|---|---|---|---|
| Strain | nmol Oxygen·h−1·μg−1 Chlorophyll | Relative Oxygen Evolution | μmol Oxygen·h−1·μg−1 Chlorophyll | Relative Oxygen Evolution |
| Wild type | 97 ± 11 | 100 | 165 ± 22 | 100 |
| slr1471-gfp | 99 ± 14 | 102 | 102 ± 21 | 62 |
| slr1471-KmR | 90 ± 12 | 93 | 167 ± 33 | 101 |
To confirm that the decrease of chlorophyll fluorescence is caused by a malfunction of PSII, cells were grown under low light and the chlorophyll fluorescence was compared under actinic light intensities of 20 and 200 μmol·m−2·s−1 in the presence of the herbicide DCMU. Under these experimental conditions, chlorophyll fluorescence was expected to remain high under actinic light, because DCMU blocks electron transport between PSII and cytochrome b6f (Figure 4). Under low actinic light, the wild type, slr1471-gfp, and slr1471-KmR revealed no significant differences (Figure 4). However, high actinic light treatments led to a decrease of the steady state chlorophyll a fluorescence in slr1471-gfp only, demonstrating chlorophyll a bleaching. Switching off the high actinic light again for slr1471-gfp demonstrated that the dark chlorophyll fluorescence F0′ value remained below the starting F0 value, whereas the wild type and slr1471-KmR remained at values above the initial F0 level and also comparable to wild-type and slr1471-KmR values after low actinic illumination (Figure 4). In addition, F0′ levels in slr1471-gfp cells did not recover after high actinic light treatment even after 20 min of dark incubation. Because DCMU blocks the QB binding site in D1, inhibiting electron efflux from QA, we concluded that rapid chlorophyll a bleaching in slr1471-gfp resulted from an altered QA function or redox potential.
Figure 4.
PSII Function Is Impaired in slr1471-gfp.
PAM measurements were performed in the presence of 50 μM DCMU with wild-type, slr1471-gfp, and slr1471-KmR cells grown under 20 μmol·m−2·s−1 illumination and with saturating flashes of 7000 μmol·m−2·s−1 and actinic light of 20 and 200 μmol·m−2·s−1. The start (up arrows) and the end (down arrows) of the actinic light phase are indicated.
Redox Potentials of QA− and QB− Are Altered in slr1471-gfp
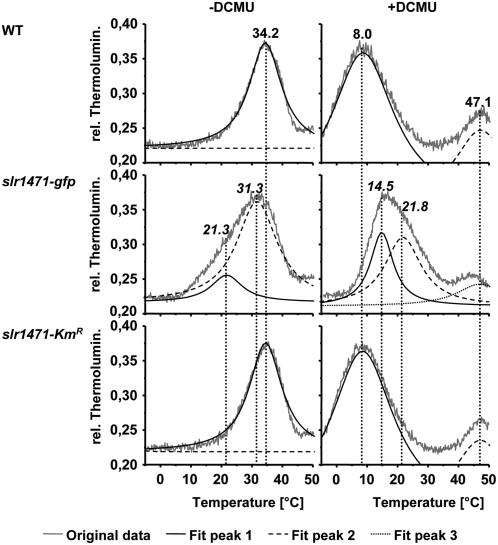
To measure the QA− and QB− redox potentials in wild-type, slr1471-gfp, and slr1471-KmR cells, we performed thermoluminescence measurements (Figure 5; see Supplemental Figure 4 online). No significant changes were found for the wild type and slr1471-KmR. In both cases, the highest thermoluminescence emission revealed a B-band emission maximum at 34.2°C for recombinations from the S2+P680QAQB− state, indicating an unaltered redox potential for QB−. However, for slr1471-gfp, B-band emission was extended to lower temperatures, with a maximum at 31.3°C, indicating that the redox potential for QB− is decreased in the mutant (Figure 5, −DCMU). Fitting of the thermoluminescence spectra revealed the described B-bands for the wild type and slr1471-KmR, but besides the described B-band at 31.3°C, there was an additional significantly smaller peak for slr1471-gfp with an emission maximum temperature of 21.3°C, indicating the emission of a small pool of S2+P680QA− states.
Figure 5.
QA and QB Redox Potentials Are Altered.
Thermoluminescence measurements were performed with wild-type, slr1471-gfp, and slr1471-KmR cells grown under 20 μmol·m−2·s−1 illumination in the absence (−) or presence (+) of 50 μM DCMU. Recorded thermoluminescence curves were mathematically fitted and deconvoluted into the components. Shown are the original data as well as the fitted curves for every peak of each strain. Temperatures for the highest emission of the fitted peaks are given for the B-bands (−DCMU), C-bands, and Q-bands (+DCMU).
In the presence of DCMU, a competitive inhibitor of QB, thermoluminescence measurements with wild-type and slr1471-KmR cells resulted in a Q-band emission maximum of 8.0°C for charge recombination from the S2+P680QA− state. By contrast, in slr1471-gfp cells, an increased Q-band emission maximum at 15.7°C for the S2+P680QA− state indicated an increase in the redox potential of the QA− state (Figure 5, +DCMU). In addition to the Q-band, all spectra showed C-bands at ∼47°C, which seemed similar in all three strains. We performed a curve fit again, revealing two already described peaks at 8.0 and 47.1°C for both the wild type and slr1471-KmR (Figure 5). However, curve fitting of the slr1471-gfp thermoluminescence spectrum in the presence of DCMU showed two peaks at 14.5 and 21.8°C, which appeared merged in the peak at 15.7°C of the original curve, as well as a peak at 47.1°C corresponding to the wild type and slr1471-KmR. It is noteworthy that the intensity of the peaks at 14.5 and 21.8°C was nearly identical, indicating the presence of two pools of S2+P680QA− states in slr1471-gfp. We conclude that in slr1471-gfp, the redox potentials of QA− and QB− are altered, leading to the pronounced light sensitivity and photoinhibition of PSII. These results clearly indicate an altered structure of the PSII reaction center protein D1 and most likely also D2.
D1 Integration into Thylakoid Membranes Is Impaired in slr1471-gfp
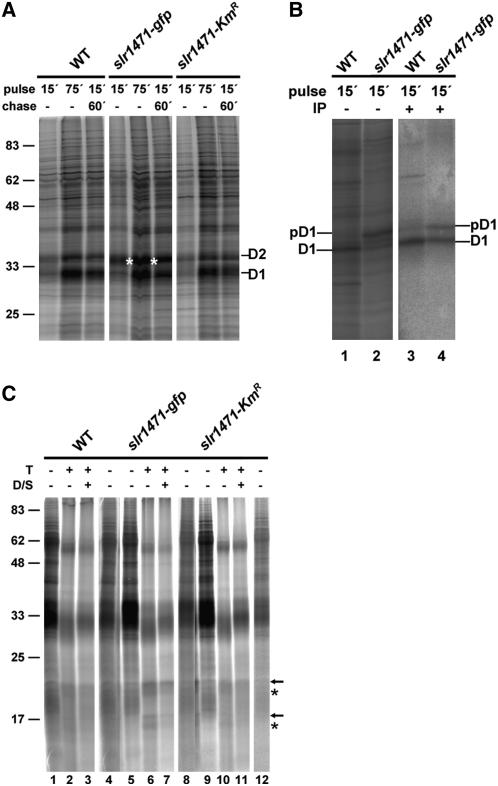
For photosynthetic function, QA and QB have to be assembled with PSII reaction center proteins D2 and D1, respectively. Therefore, we addressed the function of Slr1471p for membrane integration or assembly of D1 and/or D2 by in vivo labeling analysis of wild-type, slr1471-gfp, and slr1471-KmR cells at 30°C and 20 μmol·m−2·s−1. Cells grown under low light were incubated for 15 and 75 min in radiolabeling pulse reactions or were pulse-labeled for 15 min followed by a 60-min chase with l-Met. Isolated and solubilized total membranes were then separated on SDS gels (Figure 6A). In all three strains, the PSII reaction center proteins D1 and D2 were labeled; but in slr1471-gfp, a strong signal with a molecular mass close to that of the D2 protein appeared. To differentiate between radiolabel accumulation in D2 and precursor D1 (pD1), we immunoprecipitated D1 using a D1-specific antibody after denaturing of the isolated membranes with SDS. The results revealed that D1 accumulated dominantly in the pD1 form but also as a mature D1 in slr1471-gfp (Figure 6B). Interestingly, the presence of pD1 was independent of the light conditions but specific for slr1471-gfp cells, indicating that in the mutant, PSII assembly was affected by an impaired processing of pD1. Because QA and QB are bound to transmembrane helices 4 and 5 of the PSII reaction center proteins D2 and D1, respectively, we speculated that either membrane integration or folding of D1 or D2, or both, could be altered in slr1471-gfp. To study the integration status of D1, pD1, and D2 in the membrane, we incubated total membranes of all three strains with trypsin (Figure 6C).
Figure 6.
Integration of D1 into Thylakoid Membrane Is Impaired in slr1471-gfp.
(A) Wild-type, slr1471-gfp, and slr1471-KmR cells were pulse-radiolabeled in vivo with [35S]Met for 15 and 75 min or pulse-labeled for 15 min and chased for an additional 60 min. Isolated total membranes were separated by SDS-PAGE. Radiolabeled D1 and D2 are indicated. Standard protein masses at left are given in kilodaltons.
(B) Wild-type and slr1471-gfp cells were pulse-radiolabeled in vivo with [35S]Met for 15 min. Isolated total membranes (lanes 1 and 2) and isolated and SDS-denatured total membranes subjected to immunoprecipitation with an antibody against D1 (lanes 3 and 4) were separated by SDS-PAGE. Radiolabeled D1 and pD1 are indicated. Lanes 3 and 4 show a longer exposure of the same gel shown in lanes 1 and 2.
(C) Total membranes from wild-type, slr1471-gfp, and slr1471-KmR cells pulse radiolabeled for 15 min were incubated in the absence (lanes 1, 4, 5, 8, 9, and 12) or presence (lanes 2, 6, and 10) of trypsin or trypsin and β-dodecylmaltoside (lanes 3, 7, and 11) for 30 min at room temperature. The samples in lanes 3, 7, and 11 were sonicated briefly before adding the detergent. Samples in lanes 4, 8, and 12 are 1:10 dilutions of the samples in lanes 1, 5, and 9, respectively. Specific degradation products are marked with arrows and asterisks. Standard protein masses at left are given in kilodaltons.
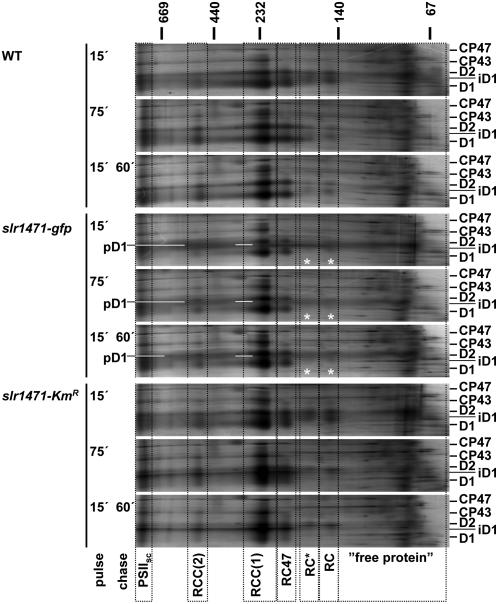
The proteolytic pattern for slr1471-gfp revealed four specific signals, which were either hardly detectable (Figure 6C, arrows) or present only as much weaker signals (Figure 6C, asterisks) in the wild type and slr1471-KmR (Figure 6C, lanes 6 and 7 compared with 2, 3, 10, and 11). Furthermore, during total membrane isolation, D1, pD1, and D2 were not released to the soluble fractions and could not be extracted from the membrane phase by carbonate. We conclude that accumulation of pD1 in the membrane phase in slr1471-gfp increased the amount of degradation product most likely derived from pD1 that was not properly or not at all inserted into the membrane. Given that the precursor protein was not or not properly inserted, an assembly kinetics analysis should reveal difficulties during the assembly of PSII in slr1471-gfp cells. Therefore, we performed two-dimensional blue native (BN)/SDS-PAGE with the isolated total membranes and analyzed the assembly kinetics of D1, pD1, and D2 in the three different strains. For PSII, six assembly states could be readily identified by radiolabeling of the D2 and D1 proteins in wild-type and slr1471-KmR cells [Figure 7, PSIIsc, RCC(2), RCC(1), RC47, RC*, and RC]. In slr1471-gfp cells, the assembly of PSII revealed a major difference (Figure 7). Assembly intermediates corresponding to reaction center complexes (RC and RC*) were not detectable (Figure 7, white asterisks). This finding indicated that in slr1471-gfp, processing of pD1 at the level of the RC assembly intermediate either was very rapid or the assembly step was omitted in the mutant. Furthermore, we noted that radiolabeled pD1 accumulated in slr1471-gfp was present at apparently all molecular mass levels in the first BN dimension gel, indicating nonspecific aggregation of pD1.
Figure 7.
Assembly of PSII Is Altered in slr1471-gfp.
Wild-type, slr1471-gfp, and slr1471-KmR cells (2 × 106) were pulse- and pulse/chase-labeled with [35S]Met in vivo as described for Figure 5. Total membranes were separated by two-dimensional BN/SDS-PAGE. Complexes containing radiolabeled proteins in the SDS-PAGE size range from 25 to 50 kD are presented in a molecular mass window as indicated in kilodaltons for BN-PAGE. Frames (dotted lines) highlight the positions of PSII assembly complexes [RC and RC*, reaction center complexes of PSII including D1 and D2; RC47, reaction center complex of PSII including CP47; RCC(1) and RCC(2), monomeric and dimeric reaction center core complexes of PSII; PSIIsc, PSII supercomplexes; “free protein,” unassembled protein solubilized from the membrane phase]. Radiolabeled proteins CP47, CP43, D2, pD1, intermediate D1 (iD1), and mature D1 (D1) are indicated. Asterisks mark areas of missing reaction center complexes in slr1471-gfp.
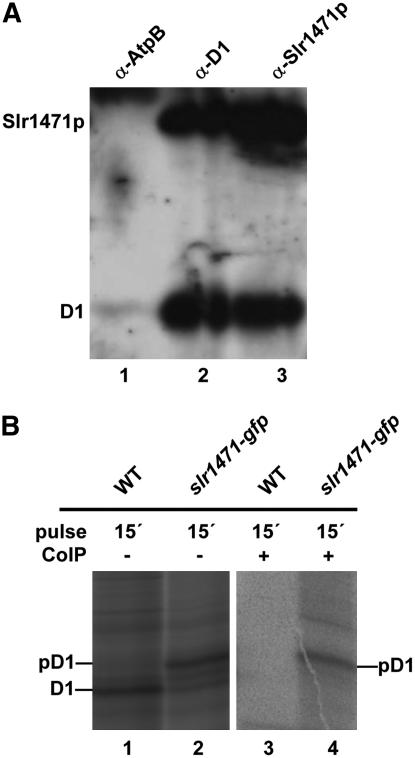
Thus, slr1471-gfp cells only partially accumulated or did not integrate pD1 at all. At least the last transmembrane helix (number 5) appeared not to be integrated properly during assembly, because processing of pD1 requires the C-terminal end to be exported into the thylakoid lumen. In addition, a mature D1 that assembled into PSII complexes displayed altered QA− and QB− redox potentials. These results suggested an impaired function of the Slr1471-GFP fusion protein for the integration and folding of D1 in the thylakoid membrane of Synechocystis. This implies a direct interaction of Slr1471p with D1 and maybe an altered interaction of Slr1471-Gfp with D1. Therefore, we next analyzed whether Slr1471p interacts directly with the D1 protein in the wild-type strain. We used total membranes from the wild type and polyclonal antibodies specific for Slr1471p and D1 and investigated the SynOxa1–D1 interaction by coimmunoprecipitation (Figure 8A). After immunoprecipitation with D1 antibody, coimmunoprecipitated proteins were examined by gel blot analysis with antibodies against D1 and Slr1471p. Both proteins were clearly detectable (Figure 8A, column 2). Conversely, a stronger Slr1471p and a weaker D1 gel blot signal were obtained for immunoprecipitation with an Slr1471p-specific antibody (Figure 8A, column 3). By contrast, immunoprecipitation with an antibody against AtpB could not effectively pull down D1 or Slr1471p when analyzed with antibodies against D1 and Slr1471p (Figure 8A, column 1). Therefore, we conclude that Slr1471p interacts with D1 in Synechocystis.
Figure 8.
Interaction of Slr1471p with D1.
(A) Total membranes from wild-type cells were solubilized with β-dodecylmaltoside. Immunoprecipitation was performed using antisera against AtpB (α-AtpB), D1 (α-D1), and Slr1471p (α-Slr1471p). Immunoprecipitated proteins were analyzed by immunoblotting with antisera against D1 and Slr1471p.
(B) Wild-type and slr1471-gfp cells were pulse-radiolabeled in vivo with [35S]Met for 15 min. Isolated total membranes (lanes 1 and 2) and isolated total membranes subjected to coimmunoprecipitation with an antibody against GFP (lanes 3 and 4) were separated by SDS-PAGE. Radiolabeled D1 and pD1 are indicated. Lanes 3 and 4 show a longer exposure of the same gel shown in lanes 1 and 2.
We then performed coimmunoprecipitation using thylakoid membranes isolated from radioactively labeled wild-type and slr1471-gfp cells and a GFP-specific antibody coupled to microbeads (see Methods). As shown in Figure 8B, pD1 accumulating in slr1471-gfp (column 2) was specifically copurified with Slr1471-Gfp but lacked mature D1 (column 4), whereas no D1 protein could be purified from the wild type (Figure 8B, column 3). These data suggest an interaction of mature D1 with Slr1471p in the wild type, which seemed to be impaired in slr1471-gfp, because only pD1 interacts with Slr1471-Gfp.
DISCUSSION
We found that fusion of GFP to the C terminus of Slr1471p in Synechocystis resulted in light sensitivity and an altered growth rate of mutant slr1471-gfp. Analysis of PSII revealed an altered redox potential for QA− and QB−. In combination with our finding that membrane integration and assembly of reaction center protein D1 was altered in the mutant, we conclude that Slr1471p, the only member of the Alb3/Oxa1/YidC protein family in Synechocystis, operates as a membrane integral chaperone essential for the correct membrane integration, folding, and assembly of PSII reaction center precursor protein pD1.
Analyzing the C-terminal fusion of GFP to Slr1471p, it was intriguing that Slr1471p not only acted as an assembly factor but also catalyzed the proper folding and correct integration of pD1. In higher plant chloroplasts, the D1 protein has been proposed to cotranslationally integrate into the thylakoid membrane or to cotranslationally assemble directly with preassembled reaction center proteins (Zhang et al., 2001). We show here that full-length D1 at least transiently interacts with Slr1471p. Hence, in Synechocystis, assisted membrane integration precedes the functional assembly of D1.
The accumulation of pD1 in slr1471-gfp was caused by the Slr1471-GFP fusion protein, which seemed to be unable or too slow to integrate and fold pD1 correctly. This can be concluded from the interaction of pD1, but not mature D1, with Slr1471-GFP. Interestingly, a pool of mature D1 was integrated completely into thylakoid membranes of slr1471-gfp, but these mature D1 proteins were not integrated and assembled properly into PSII complexes, as shown by altered redox potentials of QA and QB. We speculate that in the mutant, improperly integrated D1 may fail to correctly bind QB during folding, which in turn may alter the correct assembly of RC intermediates, leading to a structural alteration in the binding of QA to the D2 protein. Alternatively, Slr1471-GFP may also fail to integrate and fold D2 correctly into thylakoid membranes, leading to an altered binding of QA.
Interestingly, deconvolution of the original thermoluminescence curves measured in the presence of DCMU revealed two peaks of similar intensities at 14.5 and 21.8°C for the recombination of the S2+P680QA− state in slr1471-gfp, indicating the presence of two nearly identically sized pools of QA with different redox potentials or energy levels. A small peak of 21.3°C was also calculated for slr1471-gfp without DCMU, most likely representing a recombination from the S2+P680QA− state in the absence of a QB− inhibitor. We hypothesize that electron transfer between QA and QB is altered by the closer energetic level (QA significantly lower at two levels, QB slightly higher compared with the wild-type level), which would be more pronounced for the lower QA pool emitting at 21.3°C, leading to the presence of a recombination of the lower S2+P680QA− state even in the absence of DCMU. This alteration of electron flow from QA to QB might also explain the photosensitive phenotype of slr1471-gfp.
Previous attempts to generate a deletion mutant for the slr1471 gene encoding Slr1471p were not successful, suggesting essential functions (Fulgosi et al., 2002; Spence et al., 2004). To alter the gene function, we used the alternative approach of fusing the globular and soluble GFP, which has a molecular mass of ∼30 kD, to the membrane-integral Slr1471p. For yeast mitochondrial Oxa1p, an interaction of the C terminus with ribosomes suggested an important role for Oxa1p function in translation (Jia et al., 2003). When we fused GFP to the C terminus of Slr1471p, we obtained a fully segregated mutant, slr1471-gfp, which exhibited light sensitivity leading to a significantly slower growth rate already at light intensities of 40 to 80 μmol·m−2·s−1. The decreased growth rate was accompanied by a reduced amount of pigment and an altered energy transfer between the antenna systems and the reaction centers of PSI and PSII. In this respect, we interpret our findings that in high-light-grown slr1471-gfp, PSI fluorescence was reduced to half of the PSI fluorescence of the wild type by an effective reduction of phycobilisome antenna and that PSI fluorescence was blue-shifted by 1.5 nm by the relatively higher PSI monomer content relative to PSI trimers in slr1471-gfp. This interpretation is strengthened given that in Spirulina platensis, PSI trimers emitted 77K fluorescence at higher wavelengths than PSI monomers (Kruip et al., 1999), and we confirmed the relatively higher PSI monomer content in slr1471-gfp grown under high light by BN-PAGE [see Supplemental Figure 6 online, PSI (1) versus PSI (3)].
At increased light intensity, PSII fluorescence in slr1471-gfp showed a reduced fluorescence of the inner antenna protein CP47. This result indicated a lower energy transfer between phycobilisomes to CP47 than to the inner antenna protein CP43. Our data suggest a specific, yet unidentified, protein to be essential for the proper attachment and/or energy transfer between phycobilisomes and CP47 that appears to be depleted or ineffective in slr1471-gfp under high-light conditions. This resembles the situation in ac29, the Alb3.1 deletion mutant of C. reinhardtii, in which the loss of Alb3.1p function led to a depletion of CP26, which was found to be essential for the connection between the outer antenna and CP43 (Ossenbühl et al., 2004).
The PSII complex can be damaged if exposed to strong light, a phenomenon known as photoinhibition. In vivo, light damage of D1 leads to nonfunctional PSII and a subsequent D1 exchange. As a result, D1 turnover is very high and D1 synthesis increases in parallel with light intensity (Andersson and Aro, 2001). The degree of photoinhibition in vivo reflects the level of equilibrium between PSII photodamage and PSII repair. If the repair process is inhibited or too slow relative to the damage rate, photodamaged PSII accumulates, leading to slower or completely stopped growth (Allakhverdiev et al., 2005). At light intensities up to 100 μmol·m−2·s−1, the rate of photodamage is balanced by the fast repair of PSII. Significant photoinhibition in higher plants becomes obvious at high light intensities of 1500 to 2000 μmol·m−2·s−1, which is thus widely used as the light intensity for the in vivo analysis of photoinhibition (Trebst et al., 2002; Allakhverdiev et al., 2005; Hakala et al., 2005). Therefore, the growth defect of slr1471-gfp at light intensities of 40 to 80 μmol·m−2·s−1 is remarkable. Light sensitivity in slr1471-gfp relates to a decreased repair rate and an increased photodamaging rate of D1, because the altered D1 state in PSII could not be balanced by a higher rate of repair to replenish functional D1.
In vivo labeling of the mutant revealed that integration and assembly of D1 into the thylakoid membrane and PSII were altered, because an unintegrated or incompletely integrated unprocessed pD1 accumulated. This alteration was light-independent, suggesting that defective D1 membrane integration is the primary effect of the GFP fusion to Slr1471p. Several lines of evidence support the idea that pD1 was at maximum only partially integrated in slr1471-gfp cells: (1) defective C-terminal pD1 processing indicated that the essential transfer of the C terminus into the periplasmic space and/or the thylakoid lumen was affected, so at least the last transmembrane domain needs catalysis to integrate properly into the membrane; (2) although pD1 could not be extracted with carbonate from the membrane, the radiolabeled membrane proteins revealed a different protease digestion pattern (Anbudurai et al., 1994; Klinkert et al., 2004); and (3) after BN-PAGE, pD1 was detectable as a smear over the whole native dimension and not as part of a defined complex, indicating unspecific aggregation of pD1. Besides pD1, a fully integrated and processed D1 accumulated in slr1471-gfp that assembled into PSII complexes. However, the accumulation of two important PSII assembly intermediates characterized as reaction center complexes containing D1 and D2 was not detectable in the mutant (Klinkert et al., 2004; Komenda et al., 2004). This finding suggests that the assembly pathway of RCs is altered in the mutant or that the biogenesis of RC complexes is the rate-limiting step for PSII assembly in slr1471-gfp.
In slr1471-gfp cells, PSII function is impaired, which was demonstrated by changed redox potentials of the primary and secondary electron acceptors, QA and QB, which are bound to transmembrane helices 4 and 5 of D2 and D1, respectively. Similar changes of the QA− and QB− redox potentials were detected in mutants of the D-de loop between transmembrane helices 4 and 5 of D1 (Mäenpää et al., 1995; Nixon et al., 1995; Minagawa et al., 1999). Therefore, changed redox potentials of QA and QB in slr1471-gfp appeared to be caused by structural changes of D1, although we cannot exclude the possibility that the structure of D2 is also altered in slr1471-gfp. Based on the data for the altered D1 integration and assembly in slr1471-gfp, we speculate that the fusion of GFP to the C terminus of Slr1471p impairs the catalysis of the integration and folding of at least the fifth transmembrane helix of D1. This impaired integration is reflected by the accumulated and aggregated pD1, and the impaired folding is reflected in the changed redox potentials of QA and QB. Furthermore, Slr1471p interacts with the full-length D1 in wild-type cells, as revealed by coimmunoprecipitation. Data support the model that Slr1471p is a membrane-integral chaperone essential for the correct integration, folding, and assembly of D1 into PSII, most likely during PSII reaction center biogenesis taking place during de novo assembly as well as during repair of PSII. For this chaperone function, the C terminus of Slr1471p seems to be essential, because fusion of the globular GFP to the C terminus could impair Slr1471p function, possibly by hampering the integration of pD1 sterically. This hypothesis is supported by the fact that Slr1471-Gfp interacts solely with pD1, but not with mature D1, whereas Slr1471p interacts mostly with mature D1 in wild-type cells.
METHODS
Strains and Culture Conditions
The wild-type strain (generously provided by N. Murata, National Institute for Basic Biology, Okazaki, Japan) and generated mutant strains of Synechocystis sp PCC 6803 were grown photoautotrophically at 30°C under illumination from incandescent lamps at the indicated light intensities in BG-11 medium (Stanier et al., 1971) supplemented with 20 mM HEPES/NaOH, pH 8.0. The growth of cells was monitored in terms of OD730. Wild-type and mutant strains were inoculated to the same density (OD730 = 0.05) from precultures grown at a light intensity of 10 μmol·m−2·s−1 and were further incubated for 3 d under light conditions of 20, 40, and 80 μmol·m−2·s−1.
Generation of Synechocystis Mutants
All DNA techniques were performed according to standard procedures (Sambrook et al., 1989). Plasmid pIGA (Kunert et al., 2000) carrying a gfp gene was kindly provided by M. Hagemann (Universität Rostock).
Various portions of a 2.0-kb segment that covers the coding sequence of the slr1471 gene and its flanking regions were amplified by PCR with wild-type genomic DNA as template. Oligonucleotides used were as follows: forward primer M (5′-CATCGTCTATGGCGAAGTGG-3′; corresponding to nucleotides –299 to –280 relative to the AUG of the slr1471 gene) and reverse primer P (5′-CTGACTCGAGTTACGAGGTTTTTTCCTTCTTTTTAC-3′; corresponding to nucleotides +1155 to +1130 of slr1471) to amplify a 1.5-kb fragment that was cloned into the NotI-KpnI site of pBluescript II KS– (Stratagene), yielding plasmid 5UTR-slr1471_pBS containing the 0.3-kb upstream sequence and coding sequence of the slr1471 gene; forward primer K (5′-CAGTTGGAAATTCAGAGCACCGA-3′; corresponding to nucleotides +538 to +560 of slr1471) and reverse primer T (5′-GACTCTGCAGCGAGGTTTTTTCCTTCTTTTTAC-3′; corresponding to nucleotides +1152 to +1130 of slr1471) to amplify the 0.6-kb 3′ half of the slr1471 gene, which was cloned into the HindIII-PstI site of pBluescript II KS–, resulting in slr1471c_pBS; and forward primer 14 (5′-AGTCGGTACCCGGGATTTTTAAACCATAAGTCTTCACC-3′; corresponding to nucleotides –89 to –60 of slr1472) and reverse primer 15 (5′-AGTCGGTACCGATATCCTAACGTCGAGGGCGAATCA-3′; corresponding to nucleotides +495 to +475 of slr1472) to amplify a 0.6-kb fragment covering the sequence containing the stop codon of the slr1471 gene to the stop codon of the slr1472 gene (fragment Kp-slr1472-Kp). For construction of a plasmid used to express the Slr1471-GFP fusion, the gfp gene was amplified with pIGA as template and the forward primer 1 (5′-AGTCCTGCAGATGAGTAAAGGAGAAGAACTTTTCAC-3′; corresponding to nucleotides +1 to +26 relative to the AUG of the gfp gene) and reverse primer 2 (5′-GAGCTCAGATCTCTATTTGTATAGTTCATCCATGCCA-3′; corresponding to nucleotides +716 to +693 of the gfp gene), restricted by PstI and Ecl136II, and inserted into the PstI-SmaI site of slr1471c_pBS, resulting in plasmid slr1471-gfp_pBS. The 3′ half of the slr1471 gene in 5UTR-slr1471_pBS was removed by digestion with HindIII and BamHI, and the corresponding fragment that had been excised from slr1471-gfp_pBS by HindIII and BglII was inserted, resulting in 5UTR-slr1471-gfp_pBS. The KpnI fragment Kp-slr1472-Kp was inserted into the KpnI site of 5UTR-slr1471-gfp_pBS, resulting in plasmid 5UTR-slr1471-gfp-slr1472_pBS. The KmR gene cartridge was excised from pUC4K with HincII and inserted into the SmaI site located downstream of the slr1471-gfp gene in 5UTR-slr1471-gfp-slr1472_pBS. The resulting plasmid was designated pslr1471-gfp. For construction of a plasmid used to generate a kanamycin-resistant control strain, which carried the KmR gene cartridge, slr1471-KmR, just after the coding sequence of the intact slr1471 gene, the 3′ terminus of the slr1471-gfp gene in 5UTR-slr1471-gfp-slr1472_pBS was removed by digestion with HindIII and BamHI and replaced by the corresponding fragment from 5UTR-slr1471_pBS, resulting in plasmid 5UTR-slr1471-slr1472_pBS. The KmR gene cartridge from pUC4K was excised by BamHI and cloned into plasmid 5UTR-slr1471-slr1472_pBS digested with BamHI. The resulting plasmid was designated pslr1471-KmR. Wild-type cells of Synechocystis were transformed with the individual plasmids described previously (Tasaka et al., 1996). For selection of mutant cells, kanamycin was included in the medium at 20 μg/mL.
Generation of Polyclonal Anti-Slr1471 Antibodies
A DNA fragment encoding the C terminus of Slr1471p (Arg-117 to Ser-384) was amplified by PCR from wild-type genomic DNA with the primers 5′-CGGGGATCTCCCTTTTCCGA-3′ (corresponding to nucleotides +349 to +368 relative to the AUG) and 5′-TACGAGGTTTTTTCCTTCTTTTTAC-3′ (corresponding to nucleotides +1152 to +1130). The fragment was cloned into the TA cloning site of pCR T7/NT-TOPO (Invitrogen) fused to a sequence encoding an N-terminal His6 tag. The resulting plasmid, his6-slr1471_pT7/NT, was transformed into Escherichia coli BL21. Recombinant His6-Slr1471 was purified from 100-mL cultures by two rounds of Ni2+ affinity chromatography purification according to the manufacturer's instructions (Qiagen). The eluted fractions containing His6-Slr1471 were concentrated with Biomax-5K centrifugal concentrators (Millipore) and supplemented with SDS to 2% for injection of rabbits. Production of anti-Slr1471 antiserum was examined by protein gel blot analysis (data not shown).
Spectroscopy and Thermoluminescence Measurements
Absorption spectra of 1 mL of Synechocystis culture were recorded with a UV-2401 spectrophotometer equipped with an Ulbrichts sphere (ISR-240A) and the supplied software UV-probe using 0.5-nm step width (Shimadzu Deutschland). 77K fluorescence spectroscopy was performed as described (Ossenbühl et al., 2004) using excitation wavelengths of 440 and 570 nm. Emission spectra were recorded with 0.5-nm step width. PAM measurements were performed as described (Ossenbühl et al., 2004). In Synechocystis, a yield of ∼0.4 to 0.5 in PAM measurements is attributable to the high amount of PSI and corresponds to the yield of 0.8 measured in higher plants.
Thermoluminescence measurements were performed using a thermoluminescence device together with a ThermoRegulator TR 2000, a dual-modulation kinetic fluorometer, and the supplied Fluorwin version 3.0 software (Photon Systems Instruments). For thermoluminescence measurements, dark-adapted cells, which were incubated with or without 50 μM DCMU, were frozen at −10°C, and two saturating flashes were given to drive charge separation within PSII trapped in the S2+P680QAQB− or S2+P680QA− state. Charge recombination of the trapped charges was then measured as luminescence as a function of temperature (Ducruet, 2003).
Oxygen Evolution Measurements
Oxygen evolution measurements were recorded with Chlorolab2 and the supplied Oxylab software (H. Saur Laborbedarf). Synechocystis cells were grown under low (20 μmol·m−2·s−1) and high (80 μmol·m−2·s−1) light. Cells corresponding to 2 μg of chlorophyll a were suspended in 1 mL of culture medium enriched with 20 μL of 0.1 M NaHCO3 and were dark-adapted for 2 min. The amount of oxygen consumption and oxygen evolution was then measured in the presence of 20 μmol·m−2·s−1 for the low-light-grown cells and 80 μmol·m−2·s−1 for the high-light-grown cells.
In Vivo Labeling of Synechocystis, Gel Electrophoresis, Coimmunoprecipitation, and Protein Gel Blot Analysis
The synthesis, stability, and assembly of D1 and D2 into PSII were studied by in vivo labeling of Synechocystis cells, coimmunoprecipitation of protein complexes, and gel electrophoresis of total membranes containing thylakoid membranes as described (Klinkert et al., 2004; Komenda et al., 2004; Ossenbühl et al., 2004). In brief, Synechocystis cells were incubated with [35S]Met for the indicated times and harvested by centrifugation. Washed cells were disrupted by vortexing with glass beads, and total membranes were isolated by centrifugation.
Isolated membranes were solubilized with 1% β-dodecylmaltoside and loaded on a BN-PAGE device in the presence of Coomassie Brilliant Blue G 250.
For coimmunoprecipitation, native solubilized total membranes of wild-type and radiolabeled wild-type and slr1471-gfp cells were incubated in the presence of antibodies against D1 (kindly provided by E. Pistorius, University of Bielefeld), Slr1471p and AtpB (kindly provided by R.G. Herrmann, Ludwig-Maximilians-University), or α-GFP-μMACS (Miltenyi Biotec) for 1 h at 4°C. Protein A–Sepharose (5 mg; Sigma-Aldrich) or Protein A–μMACS (Miltenyi Biotec) was added, and incubation was prolonged for another 1 h. Coimmunoprecipitates were isolated by centrifugation or by purification over μMACS columns (Miltenyi Biotec), washed at least three times, and loaded onto a SDS-PAGE device.
For identification of pD1, total membranes of radiolabeled wild-type and slr1471-gfp cells were solubilized with SDS. Immunoprecipitation was then performed as described above using the antibody against D1 and purification with Protein A–μMACS (Miltenyi Biotec). For gel blot analysis, total membranes of wild type, slr1471-gfp, and slr1471-KmR cells grown at 80 μmol·m−2·s−1 corresponding to 3 μg of chlorophyll a were loaded onto a SDS-PAGE device.
Accession Number
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number BA000022 (Slr1471p).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Resolution of 77K Fluorescence Spectra (Excitation at 440 nm) into Single Components.
Supplemental Figure 2. Resolution of 77K Fluorescence Spectra (Excitation at 570 nm) into Single Components.
Supplemental Figure 3. PSII Function Is Impaired in slr1471-gfp.
Supplemental Figure 4. PSI Complexes in the Wild Type and slr1471-gfp.
Supplementary Material
Acknowledgments
We thank Norio Murata for providing a Synechocystis wild-type strain and Martin Hagemann for the plasmid pIGA. We further thank Cristina DalBosco for help with thermoluminescence and Giusy Canino for discussion. This work was supported by the Deutsche Forschungsgemeinschaft (Grants SFB TR1, SFB 594, and Os192/2).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Lutz A. Eichacker (lutz.eichacker@lrz.uni-muenchen.de).
Online version contains Web-only data.
References
- Abdallah, F., Salamini, F., and Leister, D. (2000). A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci. 5 141–142. [DOI] [PubMed] [Google Scholar]
- Allakhverdiev, S.I., Nishiyama, Y., Takahashi, S., Miyairi, S., Suzuki, I., and Murata, N. (2005). Systematic analysis of the relation of electron transport and ATP synthesis to the photodamage and repair of photosystem II in Synechocystis. Plant Physiol. 137 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbudurai, P.R., Mor, T.S., Ohad, I., Shestakov, S.V., and Pakrasi, H.B. (1994). The ctpA gene encodes the C-terminal processing protease for the D1 protein of the photosystem II reaction center complex. Proc. Natl. Acad. Sci. USA 91 8082–8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, B., and Aro, E.M. (2001). Photodamage and D1 protein turnover in photosystem II. In Regulation of Photosynthesis, Vol. 11, E.M. Aro and B. Andersson B, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 377–393.
- Bellafiore, S., Ferris, P., Naver, H., Gohre, V., and Rochaix, J.D. (2002). Loss of Albino3 leads to the specific depletion of the light-harvesting system. Plant Cell 14 2303–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., Xie, K., Jiang, F., Yi, L., and Dalbey, R.E. (2002). YidC, a newly defined evolutionarily conserved protein, mediates membrane protein assembly in bacteria. Biol. Chem. 383 1565–1572. [DOI] [PubMed] [Google Scholar]
- Ducruet, J.M. (2003). Chlorophyll thermoluminescence of leaf discs: Simple instruments and progress in signal interpretation open the way to new ecophysiological indicators. J. Exp. Bot. 54 2419–2430. [DOI] [PubMed] [Google Scholar]
- Eichacker, L.A., and Henry, R. (2001). Function of a chloroplast SRP in thylakoid protein export. Biochim. Biophys. Acta 1541 120–134. [DOI] [PubMed] [Google Scholar]
- Fulgosi, H., Gerdes, L., Westphal, S., Glockman, C., and Soll, J. (2002). Cell and chloroplast division requires ARTEMIS. Proc. Natl. Acad. Sci. USA 99 11501–11506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhre, V., Ossenbühl, F., Crèvecoeur, M., Eichacker, L.A., and Rochaix, J.-D. (2006). One of two Alb3 proteins is essential for the assembly of the photosystems and for cell survival in Chlamydomonas. Plant Cell 18 1454–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakala, M., Tuominen, I., Keranen, M., Tyystjarvi, T., and Tyystjarvi, E. (2005). Evidence for the role of the oxygen-evolving manganese complex in photoinhibition of photosystem II. Biochim. Biophys. Acta 1706 68–80. [DOI] [PubMed] [Google Scholar]
- Hell, K., Neupert, W., and Stuart, R.A. (2001). Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 20 1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, R.G. (1997). Eukaryotism, towards a new interpretation. In Eukaryotism and Symbiosis, H.E.A. Schenk, R.G. Herrmann, K.W. Jeon, N.E. Müller, and W. Schwemmler, eds (Heidelberg, Germany: Springer), pp. 73–118.
- Jia, L., Dienhart, M., Schramp, M., McCauley, M., Hell, K., and Stuart, R.A. (2003). Yeast Oxa1 interacts with mitochondrial ribosomes: The importance of the C-terminal region of Oxa1. EMBO J. 22 6438–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, F., Yi, L., Moore, M., Chen, M., Rohl, T., van Wijk, K.J., de Gier, J.W., Henry, R., and Dalbey, R.E. (2002). Chloroplast YidC homolog Albino3 can functionally complement the bacterial YidC depletion strain and promote membrane insertion of both bacterial and chloroplast thylakoid proteins. J. Biol. Chem. 277 19281–19288. [DOI] [PubMed] [Google Scholar]
- Keegstra, K., and Cline, K. (1999). Protein import and routing systems of chloroplasts. Plant Cell 11 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkert, B., Ossenbühl, F., Sikorski, M., Berry, S., Eichacker, L., and Nickelsen, J. (2004). PratA, a periplasmic tetratricopeptide repeat protein involved in biogenesis of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 279 44639–44644. [DOI] [PubMed] [Google Scholar]
- Komenda, J., Reisinger, V., Müller, B.C., Dobakova, M., Granvogl, B., and Eichacker, L.A. (2004). Accumulation of the D2 protein is a key regulatory step for assembly of the photosystem II reaction center complex in Synechocystis PCC 6803. J. Biol. Chem. 279 48620–48629. [DOI] [PubMed] [Google Scholar]
- Kruip, J., Karapetyan, N.V., Terekhova, I.V., and Rögner, M. (1999). In vitro oligomerization of a membrane protein complex. Liposome-based reconstitution of trimeric photosystem I from isolated monomers. J. Biol. Chem. 274 18181–18188. [DOI] [PubMed] [Google Scholar]
- Kuhn, A., Stuart, R., Henry, R., and Dalbey, R.E. (2003). The Alb3/Oxa1/YidC protein family, membrane-localized chaperones facilitating membrane protein insertion? Trends Cell Biol. 13 510–516. [DOI] [PubMed] [Google Scholar]
- Kunert, A., Hagemann, M., and Erdmann, N. (2000). Construction of promoter probe vectors for Synechocystis sp. PCC 6803 using the light-emitting reporter systems GFP and LuxAB. J. Microbiol. Methods 48 185–194. [DOI] [PubMed] [Google Scholar]
- Mäenpää, P., Miranda, T., Tyystjärvi, T., Govindjee, Ducruet, J.M., Etienne, A.L., and Kirilovsky, D. (1995). A mutation in the D-de loop of D1 modifies the stability of the S2QA− and S2QB− states of photosystem II. Plant Physiol. 107 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, W., Rujan, T., Richly, E., Hansen, A., Cornelsen, S., Lins, T., Leister, D., Stoebe, B., Hasegawa, M., and Penny, D. (2002). Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA 99 12246–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden, G.I., and van Dooren, G.G. (2004). Evolution: Red algal genome affirms a common origin of all plastids. Curr. Biol. 13 514–516. [DOI] [PubMed] [Google Scholar]
- Minagawa, J., Narusaka, Y., Inoue, Y., and Satoh, K. (1999). Electron transfer between QA and QB in photosystem II is thermodynamically perturbed in phototolerant mutants of Synechocystis sp. PCC 6803. Biochemistry 38 770–775. [DOI] [PubMed] [Google Scholar]
- Moore, M., Goforth, R.L., Mori, H., and Henry, R. (2003). Functional interaction of chloroplast SRP/FtsY with the ALB3 translocase in thylakoids: Substrate not required. J. Cell Biol. 162 1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M., Harrison, M.S., Peterson, E.C., and Henry, R. (2000). Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J. Biol. Chem. 275 1529–1532. [DOI] [PubMed] [Google Scholar]
- Mori, H., and Cline, K. (2001). Post-translational protein translocation into thylakoids by the Sec and DeltapH-dependent pathways. Biochim. Biophys. Acta 1541 80–90. [DOI] [PubMed] [Google Scholar]
- Nilsson, R., Brunner, J., Hoffman, N.E., and van Wijk, K.J. (1999). Interactions of ribosome nascent chain complexes of the chloroplast-encoded D1 thylakoid membrane protein with cpSRP54. EMBO J. 18 733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon, P.J., Komenda, J., Barber, J., Deak, Z., Vass, I., and Diner, B.A. (1995). Deletion of the PEST-like region of photosystem two modifies the QB-binding pocket but does not prevent rapid turnover of D1. J. Biol. Chem. 270 14919–14927. [DOI] [PubMed] [Google Scholar]
- Ossenbühl, F., Göhre, V., Meurer, J., Krieger-Liszkay, A., Rochaix, J.D., and Eichacker, L.A. (2004). Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3. Plant Cell 16 1790–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schleiff, E., and Klösgen, R.B. (2001). Without a little help from ‘my’ friends: Direct insertion of proteins into chloroplast membranes? Biochim. Biophys. Acta 1541 22–33. [DOI] [PubMed] [Google Scholar]
- Shen, G., and Vermaas, W.F. (1994). Chlorophyll in a Synechocystis sp. PCC 6803 mutant without photosystem I and photosystem II core complexes. Evidence for peripheral antenna chlorophylls in cyanobacteria. J. Biol. Chem. 269 13904–13910. [PubMed] [Google Scholar]
- Spence, E., Bailey, S., Nenninger, A., Moller, S.G., and Robinson, C. (2004). A homolog of Albino3/OxaI is essential for thylakoid biogenesis in the cyanobacterium Synechocystis sp. PCC6803. J. Biol. Chem. 279 55792–55800. [DOI] [PubMed] [Google Scholar]
- Stanier, R.Y., Kunisawa, R., Mandel, M., and Cohen-Bazire, G. (1971). Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 35 171–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg, E., Slagter, J.G., Fridborg, I., Cleary, S.P., Robinson, C., and Coupland, G. (1997). ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell 9 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaka, Y., Gombos, Z., Nishiyama, Y., Mohanty, P., Ohba, T., Ohki, K., and Murata, N. (1996). Targeted mutagenesis of acyl-lipid desaturases in Synechocystis: Evidence for the important roles of polyunsaturated membrane lipids in growth, respiration and photosynthesis. EMBO J. 15 6416–6425. [PMC free article] [PubMed] [Google Scholar]
- Trebst, A., Depka, B., and Holländer-Czytko, H. (2002). A specific role for tocopherol and of chemical singlet oxygen quenchers in the maintenance of photosystem II structure and function in Chlamydomonas reinhardtii. FEBS Lett. 516 156–160. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Paakkarinen, V., Suorsa, M., and Aro, E.M. (2001). A SecY homologue is involved in chloroplast-encoded D1 protein biogenesis. J. Biol. Chem. 276 37809–37814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.