Abstract
Expression of α-amylase genes during cereal grain germination and seedling growth is regulated negatively by sugar in embryos and positively by gibberellin (GA) in endosperm through the sugar response complex (SRC) and the GA response complex (GARC), respectively. We analyzed two α-amylase promoters, αAmy3 containing only SRC and αAmy8 containing overlapped SRC and GARC. αAmy3 was sugar-sensitive but GA-nonresponsive in both rice (Oryza sativa) embryos and endosperms, whereas αAmy8 was sugar-sensitive in embryos and GA-responsive in endosperms. Mutation of the GA response element (GARE) in the αAmy8 promoter impaired its GA response but enhanced sugar sensitivity, and insertion of GARE in the αAmy3 promoter rendered it GA-responsive but sugar-insensitive in endosperms. Expression of the GARE-interacting transcription factor MYBGA was induced by GA in endosperms, correlating with the endosperm-specific αAmy8 GA response. αAmy8 became sugar-sensitive in MYBGA knockout mutant endosperms, suggesting that the MYBGA–GARE interaction overrides the sugar sensitivity of αAmy8. In embryos overexpressing MYBGA, αAmy8 became sugar-insensitive, indicating that MYBGA affects sugar repression. α-Amylase promoters active in endosperms contain GARE, whereas those active in embryos may or may not contain GARE, confirming that the GARE and GA-induced MYBGA interaction prevents sugar feedback repression of endosperm α-amylase genes. We demonstrate that the MYBGA–GARE interaction affects sugar feedback control in balanced energy production during seedling growth and provide insight into the control mechanisms of tissue-specific regulation of α-amylase expression by sugar and GA signaling interference.
INTRODUCTION
In plants, sugars have hormone-like activity and modulate nearly all fundamental processes throughout the entire life cycle (Smeekens, 2000; Halford and Paul, 2003). In cereals, the process of seedling development is divided into three stages: imbibition, germination, and seedling elongation (Thomas and Rodriguez, 1994). Sugars tightly regulate this process by controlling gibberellin (GA) biosynthesis and α-amylase expression, which is essential for the degradation of starch to provide a nonphotosynthetic carbon source for germination and seedling development (Yu et al., 1996). After imbibition of seeds, sugars in the embryo are rapidly consumed, leading to sugar depletion and subsequent activation of α-amylase gene expression during germination (Yu et al., 1996). Meanwhile, the embryo synthesizes GAs that are released to aleurone cells surrounding the starchy endosperm to activate the synthesis and secretion of α-amylases and other hydrolases. The stored starch and other nutrients in the endosperm are digested by these hydrolases to small molecules that are taken up by the embryo to support seedling elongation (Jacobsen et al., 1995). Sugars transported to the embryo in turn repress α-amylase expression and GA biosynthesis in the embryo (Yu et al., 1996; Perata et al., 1997). Another plant hormone, abscisic acid, antagonizes GA action and represses the expression of α-amylases, providing a mechanism for preventing precocious germination (Jacobsen et al., 1995). Accordingly, the expression of α-amylases in germinating cereal grains is subject to multiple modes of regulation by sugars and hormones: induced by GA and sugar depletion and repressed by sugars and abscisic acid (Yu et al., 1992, 1996; Perata et al., 1997).
Sugar repression of α-amylase genes has also been observed in cultured rice (Oryza sativa) suspension cells (Yu et al., 1991, 1992; Chan et al., 1994), with the mechanism extensively studied. αAmy3 and αAmy8 are two α-amylase genes highly induced upon sucrose starvation in rice suspension cells (Sheu et al., 1996). The mechanism of sugar repression involves the regulation of both transcription rate and mRNA stability (Chan et al., 1994; Sheu et al., 1994, 1996; Chan and Yu, 1998a, 1998b). Upon sucrose starvation, αAmy3 has the highest and αAmy8 the next highest transcription rate among eight rice α-amylase genes analyzed (Sheu et al., 1996). Mechanisms of sugar regulation may be similar in rice embryos and suspension cells, as the same cis-acting elements identified in the αAmy3 promoter are regulated similarly in these two tissues (Hwang et al., 1998; Lu et al., 1998; Toyofuku et al., 1998).
In rice and barley (Hordeum vulgare), the accumulation of α-amylase mRNA or protein is regulated negatively by sugars in embryos (Perata et al., 1997; Chen et al., 2002) and positively by GA in endosperms/aleurone cells (Yu et al., 1996; Perata et al., 1997; Loreti et al., 2000). Embryo-specific sugar repression was further demonstrated by the fusion of a 230-bp αAmy8 promoter sequence with a reporter gene and an assay in transgenic rice seeds (Chen et al., 2002). In that study, αAmy8 promoter activity was repressed by sucrose in embryos but was insensitive to sucrose repression and activated by GA in endosperms. This suggested that the tissue-specific dominant regulation of the αAmy8 promoter by sugar and GA is most likely mediated at the transcriptional level.
In vivo functional analyses of α-amylase and other hydrolase gene promoters isolated from barley, wheat (Triticum aestivum), and rice have been performed extensively, with several promoter cis-acting elements responsive to GA or sugar identified. These elements form regulatory complexes and act in concert for GA- or sugar depletion–induced high-level α-amylase gene promoter activity. The GA response complex (GARC) includes the O2S/W box, the pyrimidine box (C/TCTTTT), the GA response element (GARE; C/TAACC/GG/AA/CC/A), and the TA/Amy box (TATCCA) (Lanahan et al., 1992; Gubler et al., 1999; Sun and Gubler, 2004; Zhang et al., 2004). The sugar response complex (SRC) includes the GC box, the G box (CTACGTGG), and the TA box (Lu et al., 1998; Chen et al., 2002). Several transcription factors interacting with these cis-acting elements have been identified and their functions analyzed: barley MYBGA (also called GAMyb) with the GARE (Gubler et al., 1995); the DNA binding protein with one finger (Dof) from rice (DOF) or barley (SAD) with the pyrimidine box (Washio, 2001, 2003; Isabel-LaMoneda et al., 2003); and rice MYBS with the TA box (Lu et al., 2002), WRKY with the W box (Zhang et al., 2004), and BZ8 with the G box (Lee et al., 2003). The GARE and TA box are the key elements in the GARC and SRC, respectively, with other elements acting cooperatively with these two elements for high-level α-amylase gene promoter activity induced by GA and sugar depletion (Lanahan et al., 1992; Rogers and Rogers, 1992; Rogers et al., 1994; Lu et al., 1998; Gomez-Cadenas et al., 2001). Both GARC and SRC require the TA box for their full functions. Consequently, rice MYBS1, which interacts with the TA box, plays dual functions in GA and sugar regulation of α-amylase gene promoters (Lu et al., 2002).
Quantitative expression of α-amylases controls the rates of starch degradation in embryos and endosperms, which profoundly affects seed germination and seedling development. Particularly, the expression of α-amylases during seedling development represents a primary factor contributing to seedling vigor, which is an important agronomic trait (Karrer et al., 1993). A precise understanding of the mechanism of the tissue-specific differential regulation of α-amylase gene expression by sugar and GA is still lacking. Unraveling the mechanism underlying this process may provide important information regarding how to improve the seedling vigor of cereals. In this study, we show that the expression of α-amylase genes in rice embryos and endosperms during germination and seedling development is differentially regulated as a result of the tissue-specific sensitivity of these genes to sugar and GA. By gain- and loss-of-function analyses, we demonstrate that the TA box is essential for sugar regulation in embryos and that the GARE is essential for GA activation and sugar insensitivity in endosperms. Studies with a MYBGA knockout rice mutant and MYBGA-overexpressing transgenic rice seeds further confirmed that the interaction between MYBGA and GARE affects the sugar sensitivity of α-amylases.
RESULTS
GA-Nonresponsive αAmy3 SRC Is Glucose-Sensitive in Both Embryos and Endosperms, Whereas GA-Responsive αAmy8 SRC/GARC Is Glucose-Sensitive Only in Embryos
Rice αAmy3 and αAmy8 were used to study the mechanism of tissue-specific differential regulation of α-amylase genes. Our studies found that the expression of αAmy3 and αAmy8 was synchronized and inversely correlated with sugar concentrations in embryos during germination. On the other hand, expression of αAmy3 and αAmy8 was nonsynchronized in endosperms: αAmy3 expression inversely correlated with sugar concentration, whereas αAmy8 expression positively correlated with GA and MYBGA mRNA levels during seedling elongation (see Supplemental Figure 1 and Supplemental Table 2 online).
To determine whether, and if so how, sugar and GA differentially regulate αAmy3 and αAmy8 in embryos and endosperms, cis-acting elements responding to sugar and GA in αAmy3 and αAmy8 promoters were studied further. The 105-bp SRC of the αAmy3 promoter, containing a GC box, a G box, and two tandem repeats of the TA box (Figure 1), functions as a transcriptional enhancer for sugar depletion–induced promoter activity (Lu et al., 1998; Chen et al., 2002). The 230-bp SRC/GARC of the αAmy8 promoter, containing a putative GC box, a GARE, and a single TA box (Figure 1), functions as a transcriptional enhancer for sucrose depletion– and GA-induced promoter activity (Chen et al., 2002). To determine whether the αAmy3 SRC and αAmy8 SRC/GARC are responsible for tissue-specific sugar and GA regulation in embryos and endosperms, the promoters were fused to a luciferase cDNA (Luc) (Figure 1). The GA and sugar response of the promoters was analyzed through the particle bombardment–mediated transient expression assay system.
Figure 1.
Schemes of Constructs for αAmy3 and αAmy8 Promoter Analysis.
The αAmy3 SRC (–186 to –82 relative to the transcription start site) and αAmy8 SRC/GARC (–318 to –89) were fused upstream of a cauliflower mosaic virus (CaMV) 35S minimal promoter (35Smp)–alcohol dehydrogenase1 (Adh1) intron (In)–Luc–Nos 3′ chimeric gene. Relative positions of cis-acting elements, including GC, G, the GARE, and TA boxes, in promoters are indicated.
Plasmids containing the αAmy3 SRC-Luc or αAmy8 SRC/GARC-Luc construct were transfected into rice embryos and endosperms and incubated with or without glucose or GA, and luciferase activity was analyzed. αAmy8 SRC/GARC was activated by GA in both embryos and endosperms, whereas GA had no effect on αAmy3 SRC activity (see Supplemental Figure 2A online). On the other hand, αAmy3 SRC was repressed by glucose in both embryos and endosperms, whereas αAmy8 SRC/GARC was repressed by glucose in embryos but not in endosperms (see Supplemental Figure 2B online). Similar results were obtained with stable transgenic expression assays (see Supplemental Figure 3 online). These results are summarized in Table 1.
Table 1.
Summary of the Tissue-Dependent Sensitivity of α-Amylase Gene Promoters to GA and Glucose in Rice Seeds
| Sensitive to Glucose
|
Responsive to GA
|
|||
|---|---|---|---|---|
| Promoter | Embryo | Endosperm | Embryo | Endosperm |
| αAmy3 SRC | + | + | − | − |
| αAmy8 SRC/GARC | + | − | + | + |
In Addition to GA Responses, GARE Confers Glucose Insensitivity to αAmy8 SRC/GARC in Endosperms
To investigate the mechanism of the tissue-specific differential regulation of αAmy3 and αAmy8 promoters, the cis-acting elements of the two promoters were compared. One obvious difference between αAmy3 and αAmy8 promoters is the presence of GARE in αAmy8 SRC/GARC but its absence in αAmy3 SRC (Figure 1). αAmy8 SRC/GARC contains the GARE and was responsive to GA (see Supplemental Figure 2A online). To determine whether the GARE plays a role in the glucose insensitivity of the αAmy8 promoter, the GARE in αAmy8 SRC/GARC was mutated (Figure 2A), generating αAmy8 SRC/GARC(mGARE). The sensitivity of the wild type and αAmy8 SRC/GARC(mGARE) to GA induction and glucose repression was examined by the transient expression assay. In endosperms, the activity of αAmy8 SRC/GARC was induced 8.9-fold, but the activity of αAmy8 SRC/GARC(mGARE) was not induced significantly by GA (Figure 2B), indicating that the GA-responsive function of GARE had been impaired by mutation. In endosperms, the activity of αAmy8 SRC/GARC was not altered by glucose, but the activity of αAmy8 SRC/GARC(mGARE) was repressed 2.5-fold by glucose (Figure 2C). This gain of sugar sensitivity resulting from the GARE mutation suggested that GARE is responsible for the sugar insensitivity of αAmy8 SRC/GARC in endosperms.
Figure 2.
Gain-of-Function Analysis Demonstrates That GARE Confers GA Responsiveness and Glucose Insensitivity to the αAmy8 Promoter in Endosperms.
(A) Comparison of nucleotide sequences between the wild-type αAmy8 GARE and the mutated GARE (mGARE). Nucleotides for the GARE are underlined, and substituted nucleotides are shown in lowercase letters.
(B) and (C) Rice endosperms were cotransfected with plasmids containing the αAmy8 SRC/GARC-Luc or αAmy8 SRC/GARC(mGARE)-Luc construct, incubated for 3 d in a buffer containing GA (+GA) or lacking GA (−GA) (B) or for 24 h in a buffer containing glucose (+Glc) or mannitol (−Glc) (C), and luciferase activity was determined. X indicates fold induction or repression of luciferase activity by GA or glucose. Error bars indicate se of three replicate experiments for each construct.
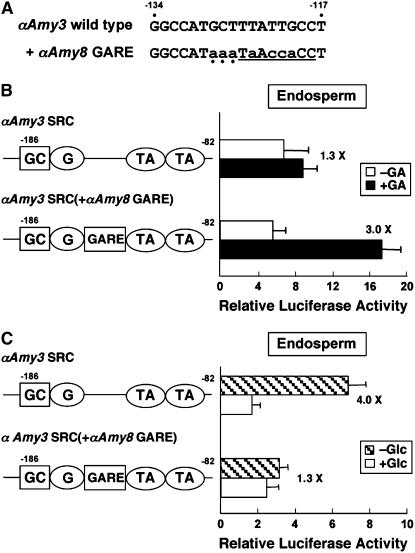
To further confirm that GARE confers endosperm-specific glucose insensitivity to a promoter, αAmy3 SRC was modified to contain an αAmy8 GARE, generating αAmy3 SRC(+αAmy8 GARE) (Figure 3A). This modified construct was analyzed for GA response and sugar sensitivity using the transient expression assay. In endosperms, wild-type αAmy3 SRC was found to be nonresponsive to GA, but αAmy3 SRC(+αAmy8 GARE) was enhanced 3.0-fold by GA (Figure 3B), indicating that the GARE in the modified αAmy3 was functional for GA response. By contrast, the activity of the wild-type αAmy3 SRC was repressed 4.0-fold by glucose, whereas αAmy3 SRC(+αAmy8 GARE) activity was not repressed by glucose (Figure 3C). This loss of sugar sensitivity resulting from the insertion of GARE suggested that GARE can convert glucose-sensitive but GA-nonresponsive αAmy3 SRC to the glucose-insensitive but GA-responsive promoter.
Figure 3.
Loss-of-Function Analysis Demonstrates That the GARE Confers GA Responsiveness and Glucose Insensitivity to the αAmy3 Promoter in Endosperms.
(A) Comparison of nucleotide sequences among the wild-type αAmy3 SRC (region between the G box and the duplicated TA box) and αAmy3 SRC containing the αAmy8 GARE (+αAmy8 GARE). Nucleotides for the GARE are underlined, substituted sequences are shown in lowercase letters, and flanking sequences 5′ of αAmy8 GARE are indicated by dots.
(B) and (C) Rice endosperms were cotransfected with plasmids containing the αAmy3 SRC-Luc or αAmy3 SRC(+αAmy8 GARE)-Luc construct, incubated for 3 d in a buffer containing GA (+GA) or lacking GA (−GA) (B) or for 24 h in a buffer containing glucose (+Glc) or mannitol (−Glc) (C), and luciferase activity was determined. X indicates fold repression or induction of luciferase activity by glucose or GA. Error bars indicate se of three replicate experiments for each construct.
Glucose Repression Overrides GA Activation of αAmy8 in Embryos, Whereas GA Activation Overrides Glucose Repression of αAmy8 in Endosperms
To determine whether the glucose and GA signal interaction plays a role in the tissue-specific regulation of the αAmy8 promoter, the response of αAmy8 SRC/GARC to glucose in the presence or absence of GA was analyzed. Rice embryos and endosperms were transfected with plasmids containing the αAmy8 SRC/GARC-Luc construct and divided into four groups, each group was incubated with or without glucose plus or minus GA, and luciferase activities were determined. In embryos, αAmy8 SRC/GARC activity was repressed by glucose regardless of the presence or absence of GA; by contrast, in endosperms, αAmy8 SRC/GARC activity was induced by GA regardless of the presence or absence of glucose (see Supplemental Figure 4 online). This result was consistent with our previous study using sucrose as a signaling molecule in a stable transgenic expression assay (Chen et al., 2002).
To determine whether endogenous αAmy3 and αAmy8 are subject to tissue-specific regulation by glucose and GA, mRNAs were extracted from rice embryos and endosperms, which were prepared as for use in transient expression assays, and subjected to gel blot analysis. In embryos, the accumulation of αAmy3 and αAmy8 mRNA was repressed by glucose, regardless of the presence or absence of GA (Figure 4, left panel). In endosperms, the accumulation of αAmy3 mRNA was also repressed by glucose regardless of the presence or absence of GA; by contrast, the accumulation of αAmy8 mRNAs was barely detectable in the absence of GA but was significantly induced by GA in spite of the presence of glucose (Figure 4, right panel). The reduced αAmy3 mRNA accumulation in endosperms in the absence of glucose but in the presence of GA (Figure 4, compare lanes 5 and 7) was likely attributable to the repression by endogenous sugars derived from starch hydrolysis by α-amylases in endosperms. These results indicate that the presence of GA interferes with sugar signaling in endosperms.
Figure 4.
Glucose Repression Overrides GA Activation of αAmy8 in Embryos, Whereas GA Activation Overrides Glucose Repression of αAmy8 Expression in Endosperms.
Rice embryos and endosperms were prepared as for use in the transient expression assays and divided into four groups. Each group was incubated in the presence (+) or absence (−) of glucose with or without GA for 24 h for embryos and for 2 d for endosperms. Total RNA was purified from embryos and endosperms and subjected to gel blot analysis using αAmy3 and αAmy8 gene-specific DNAs (3′ untranslated regions [UTRs]) as probes. Ethidium bromide staining of rRNA was used as the RNA loading control.
Elimination of MYBGA Expression Results in Glucose-Responsive αAmy8 Activity in Endosperms
In endosperms, we found that the expression of MYBGA and αAmy8 was coordinately activated by GA (see Supplemental Figure 1C online, panels 2 and 3). Because GARE confers sugar insensitivity to αAmy8 SRC/GARC in endosperms, we speculated that MYBGA might also be involved in such regulation. To explore this possibility, the glucose sensitivity of αAmy8 in endosperms was studied in a rice mutant, gamyb-2, in which the retrotransposon Tos17 had been inserted into the fourth exon of MYBGA (Kaneko et al., 2004). MYBGA function in gamyb-2 is lost, as determined by two major mutant phenotypes (e.g., impaired floral organ development and GA-dependent expression of α-amylases in endosperms) (Kaneko et al., 2004).
A rice mutant carrying homozygous Tos17-inserted alleles is defective in floral organ development (Kaneko et al., 2004) and therefore cannot be propagated by seeds; however, homozygous mutant seeds can be obtained by self-pollination of a mutant carrying a heterozygous Tos17-inserted allele. A PCR-based screen for the identification of homozygous and heterozygous Tos17 mutant seeds was performed using three primers, 5′ and 3′ primers specific to MYBGA and another 3′ primer specific to Tos17. The embryo and endosperm from each individual seed from the mutant population were collected separately. Each isolated embryo was allowed to germinate, and genomic DNA was then extracted from the seedling and used for PCR analysis. Results of some representative samples are shown in Figure 5A. Samples that produced only a 150-bp band, containing only rice DNA but no rice-Tos17 junction DNA, were wild type (+/+) (Figure 5A, lane 8). Samples that produced only a 250-bp band, containing the rice-Tos17 junction DNA, were homozygous for the Tos17 insertion (−/−) (Figure 5A, lanes 2, 3, 6, and 7). Samples that produced both 150- and 250-bp bands were heterozygous for the Tos17 insertion (+/−) (Figure 5A, lanes 4 and 5).
Figure 5.
Elimination of Rice MYBGA Expression Permits αAmy8 to Respond to Glucose in Endosperms.
(A) Screening of the Tos17-tagged rice mutant gamyb-2 identifies homozygous and heterozygous mutants. PCR with primers Myb12-5′ and Myb11-3′ produced a product of 150 bp from wild-type (wt) rice genomic DNA, and PCR with primers Myb12-5′ and LTR4A produced a product of 250 bp from the rice genomic DNA–Tos17 junction region (mt). M, molecular weight marker; +/+, wild type; +/−, heterozygous for the Tos17 tag; −/−, homozygous for the Tos17 tag.
(B) RT-PCR analysis of the expression of αAmy3, αAmy8, MYBGA, and Act1 (as an internal control) in endosperms of wild-type and gamyb-2 mutant rice (cv Nipponbare). Endosperms from the wild type or the gamyb-2 mutant (−/−) were divided into fours groups. Each group containing three endosperms was incubated in the presence (+) or absence (−) of 200 mM glucose with or without 10 μM GA for 2 d. Total RNA was purified from endosperms and subjected to RT-PCR analysis. The number of PCR cycles is indicated at right. ND, not determined.
(C) Quantitative (real-time) RT-PCR analysis of αAmy3 and αAmy8 expression using total RNAs prepared as described for (B). RNA levels of αAmy3 and αAmy8 were quantified and normalized to the level of 18S rRNA. The highest αAmy3 mRNA level was assigned as 100, and levels of expression in other samples were calculated relative to this value. The αAmy8 mRNA levels in gamyb-2 endosperms could be 4 orders of magnitude lower than in wild-type endosperms; therefore, relative mRNA levels are shown on log plots. Error bars indicate se for three replicate experiments.
Isolated endosperms corresponding to wild-type seedlings or Tos17 homozygous seedlings were then treated with or without glucose plus or minus GA and their total RNAs purified. Because of the limited quantity of mutant seeds available for classification into different categories (wild type, homozygous, and heterozygous) for RNA purification and the low abundance of αAmy8 mRNA present in endosperms of mutant seeds, RT-PCR was used to detect αAmy3, αAmy8, and MYBGA mRNAs. As shown in Figure 5B, the accumulation of MYBGA mRNA was detectable in wild-type endosperms (lanes 1 to 4) but undetectable in gamyb-2 endosperms (lanes 5 to 8) by RT-PCR over 25 cycles, indicating the complete knockout of MYBGA expression. In wild-type endosperms, the accumulation of αAmy3 mRNA was repressed by glucose independent of the presence of GA, and the accumulation of αAmy8 and MYBGA mRNAs was enhanced by GA independent of the presence of glucose, as detected by RT-PCR over 25 cycles (Figure 5B, left panel). These results were similar to those observed in Figure 4 (right panel), except that RT-PCR was sensitive enough to detect the accumulation of αAmy8 mRNA in wild-type endosperms in the absence of GA. In mutant endosperms, the accumulation of αAmy3 mRNA was still repressed by glucose, as detected by RT-PCR over 25 cycles (Figure 5B, right panel). The accumulation of αAmy8 mRNA was reduced significantly in the homozygous mutant, as it was not detected by RT-PCR over 25 cycles; however, it was detected and found to be repressed by glucose with RT-PCR over 45 cycles (Figure 5B, right panel). The relative levels of αAmy3 and αAmy8 mRNA in the wild-type and mutant endosperms, under glucose and/or GA treatments, were further confirmed with quantitative (real-time) RT-PCR analyses (Figure 5C). Therefore, αAmy8 was normally highly induced by GA and insensitive to glucose repression in the wild-type endosperm, but it became GA-nonresponsive and glucose-repressible in the mutant endosperm. The reduced αAmy3 mRNA accumulation in wild-type endosperms in the absence of glucose but in the presence of GA was likely attributable to repression by endogenous sugars derived from starch hydrolysis by α-amylases in endosperms (similar to what was observed in Figure 4, right panel); such a phenomenon was not observed in mutant endosperms.
α-Amylase Gene Promoters Highly Active in Endosperms Contain the GARE
Insensitivity to sugar repression could be important for high-level α-amylase expression in endosperms. To determine whether the GARE is necessary for high-level α-amylase expression in endosperms, mRNA accumulation of several α-amylase genes in endosperms was determined. One day after imbibition is critical for embryos, as that is the time when soluble sugar levels decrease rapidly and the expression of α-amylase genes is induced; by contrast, 4 to 5 d after the onset of imbibition is important for endosperms, as that is the time when the expression of most α-amylase genes reaches peak levels (see Supplemental Figure 1 online) (Yu et al., 1996). To compare the relative mRNA abundance of individual α-amylase genes at these two time points, excess amounts of rice rRNA gene, actin cDNA, and α-amylase gene-specific DNA were spotted onto a membrane and hybridized with a 32P-labeled single-stranded cDNA probe transcribed from the total mRNA of embryos collected on day 1 and of endosperms collected on day 5 after seed imbibition. The relative mRNA levels corresponding to eight rice α-amylase genes in a given population of mRNA were then compared. In embryos, levels of αAmy3, αAmy8, and αAmy10 mRNAs were higher than those of other α-amylase genes (Figure 6, panel 1). In endosperms, levels of αAmy6, αAmy7, αAmy8, and αAmy10 were significantly higher than those of other α-amylase genes (Figure 6, panel 2). cis-acting elements on promoters of these α-amylase genes were identified (Figure 6, panel 3). This study shows that the promoters of α-amylase genes more abundantly expressed in embryos may or may not contain the GARE, whereas those highly expressed in endosperms were found to contain the GARE.
Figure 6.
α-Amylase Gene Promoters Actively Expressed in Endosperms Contain the GARE.
Total RNAs were isolated from embryos collected on day 1 (panel 1) and from endosperms collected on day 5 (panel 2) after seed imbibition and used for the synthesis of 32P-labeled cDNA probes. In the slot-blot analysis, 5 μg of plasmid DNAs containing each α-amylase gene was applied in each slot and hybridized with the cDNA probes. The cis-acting elements in individual α-amylase gene promoters are illustrated in panel 3. These cis-acting elements were numbered relative to the translation start site (ATG) of individual α-amylase genes, and the distances between the GARE and the TA box are indicated (panel 4). G, ACGT core–containing G box; Pyr, pyrimidine box.
Overexpression of MYBGA Renders αAmy8 Insensitive to Glucose in Embryos
The studies described above indicate that GA activation, through the interaction between the GARE and MYBGA, overrides the sugar repression of αAmy8 in endosperms. Expression of MYBGA was also detectable in embryos, but the levels were lower than in endosperms and did not correlate with GA levels (see Supplemental Figure 1C online, panel 3). Questions were thus raised regarding why sugar repression overrides GA activation irrespective of the presence of MYBGA in embryos. To determine whether the level of MYBGA affects sugar regulation, MYBGA was fused downstream of the Ubi promoter and used as an effector construct, and the wild type αAmy8 SRC/GARC, αAmy8 SRC/GARC(mTA), αAmy8 SRC/GARC(mGARE), and αAmy3 SRC were used as reporter constructs. The embryo transient expression assays were performed. As shown in Figure 7, without overexpression of MYBGA, activities of all α-amylase promoter constructs were repressed by glucose, but activities of the TA- and GARE-mutated αAmy8 SRC/GARC were reduced significantly to ∼50% of the wild-type αAmy8 SRC/GARC even in the absence of glucose. This finding indicated that the TA box and the GARE act synergistically for αAmy8 SRC/GARC activity in the absence of glucose. Overexpression of MYBGA derepressed glucose repression of the wild-type and the TA-mutated, but not the GARE-mutated, αAmy8 SRC/GARC. Overexpression of MYBGA did not affect glucose repression of αAmy3 SRC. These results indicated that an increase in MYBGA in embryos renders αAmy8 SRC/GARC insensitive to glucose repression, specifically through the interaction between MYBGA and the GARE on the αAmy8 promoter.
Figure 7.
MYBGA Renders αAmy8 SRC/GARC Insensitive to Glucose in Embryos Specifically through the GARE.
Rice embryos were cotransfected with effector, reporter, and control plasmids. The effector construct contained the Ubi-MYBGA chimeric gene. The reporter constructs contained αAmy8 SRC/GARC-Luc, αAmy8 SRC/GARC(mTA)-Luc, αAmy8 SRC/GARC(mGARE)-Luc, or αAmy3 SRC-Luc chimeric genes. Transfected embryos were incubated for 24 h in a buffer containing glucose (+Glc) or mannitol (−Glc), and their luciferase activities were determined. X indicates fold repression of luciferase activity by glucose. Error bars indicate se of three replicate experiments for each construct.
To further examine the effectiveness of MYBGA in conferring sugar insensitivity to αAmy8 in embryos, MYBGA was also overexpressed under the control of the Ubi promoter in transgenic rice seeds. A starch agar plate α-amylase assay method was used for the identification of transgenic rice seeds overexpressing MYBGA. The embryo and endosperm of each individual seed from several T1 transgenic lines were collected separately. Sixteen isolated endosperms of each line were placed on starch agar plates, incubated for 3 d, and stained with iodine. Clear zones appeared after staining when isolated wild-type endosperms were incubated in starch agar containing GA, indicating expression and secretion of α-amylases that hydrolyzed starch (Figure 8A, panel 1). No clear zone was detected with wild-type endosperms incubated in starch agar lacking GA (Figure 8A, panel 2). Large and small clear zones were detected with transgenic endosperms incubated in starch agar lacking GA (Figure 8A, panel 3). Endosperms giving rise to large clear zones might indicate higher levels of MYBGA and α-amylase expression than endosperms with smaller clear zones. A reduced number of large clear zones was detected, compared with small clear zones, which could be attributable to the lethality of seeds in lines expressing high levels of MYBGA, as has been reported in transgenic barley in which the higher the amount of MYBGA overexpressed, the greater the occurrence of male sterility (Murray et al., 2003).
Figure 8.
Overexpression of MYBGA Renders αAmy8 Insensitive to Glucose in Embryos.
(A) Starch agar plate α-amylase activity assays identified transgenic rice endosperms overexpressing MYBGA. Sixteen isolated endosperms from wild-type or transgenic rice were placed on one starch agar plate. Panel 1, wild-type endosperms expressed α-amylases in starch agar containing 1 μM GA; panel 2, wild-type endosperms did not express α-amylases in starch agar lacking GA; panel 3, endosperms from one transgenic line containing Ubi-MYBGA construct: five endosperms expressed low levels of α-amylases, and one endosperm expressed a high level of α-amylases.
(B) RT-PCR analysis of the expression of αAmy3, αAmy8, and endogenous and overexpressed MYBGA. Embryos from the wild type and a transgenic line were incubated at 30°C in the presence (+) or absence (−) of 100 mM glucose for 24 h. Total RNA was purified from embryos and subjected to RT-PCR analyses.
(C) Quantitative (real-time) RT-PCR analysis of the expression of αAmy3 and αAmy8 using total RNAs prepared as described for (B). RNA levels of αAmy3 and αAmy8 were quantified and normalized to the level of 18S rRNA. The highest αAmy3 or αAmy8 mRNA level was assigned as 100, and other levels of expression were calculated relative to this value. Error bars indicate se for three replicate experiments.
Isolated embryos corresponding to wild-type or transgenic endosperms with large clear zones were then collected and treated with or without glucose, and total RNAs were purified. RT-PCR analysis was used for the detection of relative mRNA levels of different genes. As shown in Figure 8B, the accumulation of endogenous MYBGA mRNA was detected in both wild-type and transgenic embryos, whereas the overexpressed MYBGA mRNA was detected only in transgenic embryos. The accumulation of αAmy3 mRNA was repressed by glucose in both wild-type and transgenic embryos, whereas the accumulation of αAmy8 mRNA was repressed by glucose in wild-type embryos but not in transgenic embryos (Figure 8B). The relative levels of αAmy3 and αAmy8 mRNA in wild-type and Ubi:MYBGA embryos, under glucose treatment, were further confirmed with quantitative (real-time) RT-PCR analyses (Figure 8C). These results indicated that overexpression of MYBGA rendered αAmy8 insensitive to glucose in embryos.
DISCUSSION
Interaction between the GARE and MYBGA Causes Endosperm-Specific Sugar Insensitivity of α-Amylase Gene Promoters
Questions have been raised regarding why some rice and barley α-amylase genes are sensitive to sugar repression only in embryos but not in endosperms (Yu et al., 1996; Perata et al., 1997; Chen et al., 2002). The expression of α-amylase in cultured rice suspension cells is repressed by glucose, fructose, and sucrose (Yu et al., 1991), and that in rice and barley embryos is repressed by glucose, fructose, maltose, raffinose, and sucrose (Perata et al., 1997; Umemura et al., 1998). Consequently, it is generally agreed that metabolizable sugars repress α-amylase expression. Starch in endosperms is hydrolyzed to glucose, maltose, and other polysaccharides, which then are absorbed by the scutellum and transported to the growing points of seedlings. Glucose repression of α-amylase gene promoter activity in cereal embryos is well documented (Karrer and Rodriguez, 1992; Perata et al., 1997; Hwang et al., 1998; Morita et al., 1998; Toyofuku et al., 1998; Umemura et al., 1998; Loreti et al., 2000); therefore, glucose was used as the signaling molecule in studying the mechanism of tissue-dependent sugar sensitivity of α-amylase gene promoters.
In this study, both αAmy3 SRC and αAmy8 SRC/GARC were glucose-repressible in embryos; however, only αAmy3 SRC was glucose-repressible in endosperms (see Supplemental Figures 2B and 3B online). One explanation for this tissue-dependent sugar sensitivity could be that the cis-acting element(s) conferring glucose sensitivity in both embryos and endosperms is present only in αAmy3 SRC. Alternatively, an element(s) conferring glucose insensitivity in endosperms may be present only in αAmy8 SRC/GARC.
αAmy3 SRC contains a duplicated TA box, whereas αAmy8 SRC/GARC contains only a single TA box. Duplication of the TA box enhanced, and mutation of the TA box impaired, the promoter activity and sugar sensitivity of αAmy8 SRC/GARC in embryos (see Supplemental Figure 5 online). However, duplication of the TA box did not confer glucose sensitivity of αAmy8 SRC/GARC in endosperms. αAmy3 SRC contains another important cis-acting element, the G box, which is lacking in αAmy8 SRC/GARC. Insertion of a G box adjacent to the GC box in αAmy8 SRC/GARC, similar to what exists in αAmy3 SRC, did not enhance the sugar sensitivity of αAmy8 SRC/GARC in transgenic endosperms (see Supplemental Figure 6 online). These studies indicate that the lack of a duplicated TA box or a G box is not the cause of the glucose insensitivity of αAmy8 SRC/GARC in endosperms.
Both gain- and loss-of-function analyses in endosperms, by insertion of a GARE into the sugar-sensitive αAmy3 SRC and mutation of a GARE in the sugar-insensitive αAmy8 SRC/GARC, showed that the GARE was necessary and sufficient to confer glucose insensitivity to an α-amylase gene promoter in endosperms. Because MYBGA is known to interact with the GARE, its function was examined with the MYBGA knockout mutant gamyb-2. Although GA-dependent accumulation of α-amylases was impaired (Kaneko et al., 2004), αAmy3 was still repressed by glucose in the gamyb-2 (−/−) mutant endosperm (Figures 5B and 5C), indicating that factors required for sugar regulation were maintained in the mutant endosperm. Although αAmy8 was highly GA-inducible and glucose-nonrepressible in the wild-type endosperm, it was GA-noninducible and glucose-repressible, and expressed at very low levels, in the gamyb-2 homozygous mutant (−/−) endosperm (Figures 5B and 5C). These studies further suggest that an interaction between the GARE and MYBGA is required for high-level αAmy8 expression. MYBGA appears to interfere with the sugar repression of α-amylase gene promoters containing a GARE. The proposed role of MYBGA in the regulation of α-amylase expression during germination and seedling development is illustrated in Figure 9.
Figure 9.
Schemes Illustrating the Interactions among Sugars, GAs, MYBGA, and α-Amylases in Rice during Germination and Seedling Development.
(A) In the wild-type endosperm, sugar repression of α-amylase expression is inhibited by MYBGA, which interacts with the GARE in α-amylase gene promoters.
(B) In the gamyb-2 mutant endosperm, the absence of MYBGA leads to the sugar repression of α-amylase expression.
In wild-type endosperms, the accumulation of both MYBGA and αAmy8 mRNA was coordinately induced by exogenous GA (Figure 5B, left panel). Although the expression of MYBGA was significantly lower without exogenous GA, the expression of αAmy8 was still not repressed by glucose (Figure 5B, lanes 1 and 2), suggesting that the basal level of MYBGA in endosperms is sufficient to override sugar repression.
Sugar and GA Signals Compete for Tissue-Specific Regulation of α-Amylase Genes
The phenomena of sugar repression overriding GA activation in embryos, and GA activation overriding sugar repression in endosperms, and thereby regulating α-amylase gene expression, have been observed consistently in rice and barley (Karrer and Rodriguez, 1992; Perata et al., 1997; Morita et al., 1998). In this study, we demonstrate the dominant regulation of αAmy8 SRC/GARC activity and endogenous αAmy8 expression by glucose in embryos and by GA in endosperms. Our study with the MYBGA knockout mutant demonstrated that MYBGA is a major factor overriding the glucose repression of αAmy8 in endosperms (Figure 5).
The distance between the GARE and the TA box of αAmy promoters varies, ranging from 2 to 16 bp in rice αAmy promoters (Figure 6), 12 bp in the high-pI barley Am(-174)IGN promoter (Gubler and Jacobsen, 1992), 6 bp in the low-pI barley Amy32b promoter (Sutliff et al., 1993), and 6 to 12 bp in several other barley and wheat αAmy promoters (Huang et al., 1990). The consensus GARE bound by MYBGA is 8 bp (Gubler et al., 1999). It is known that MYBGA binds to the GARE (Gubler et al., 1995) and MYBS binds to the TA box (Lu et al., 2002) for high-level GA-activated α-amylase expression. These two MYB activators may interact with each other because their binding sites are close to each other. A DNase I footprinting assay demonstrated GA-dependent interaction of partially purified barley aleurone nuclear proteins with several regions of the Amy32b promoter, including the two adjacent GARE and TA box regions separated by six nucleotides (Sutliff et al., 1993). This study suggests that MYBGA and MYBS may interact with each other during GA activation. Mutation of either the TA box or the GARE significantly reduced αAmy8 promoter activity, which further suggests that both MYBS and MYBGA cooperate for high αAm8 promoter activity (Figure 7).
The ability to perceive GA signals and induce MYBGA expression appears to be the cause of the sugar insensitivity of the αAmy8 promoter in endosperms. Levels of MYBGA mRNA in embryos were significantly lower than in endosperms (see Supplemental Figure 1C online), and overexpression of MYBGA rendered αAmy8 promoter activity and mRNA accumulation insensitive to sugar repression (Figures 7 and 8), indicating that high-levels of MYBGA may favor the binding of MYBS to the TA box and may not favor the interaction of MYBS with a repressor. A threshold MYBGA concentration could be required for the competition of MYBS with the repressor. The interaction between MYBGA and MYBS activators and their competition with a repressor may serve as a starting point for the study of how sugar and GA signal interference leads to the tissue-specific regulation of α-amylase expression.
Physiological Significance of Dominant Sugar Regulation in Embryos and Dominant GA Regulation in Endosperms
In several cereals, α-amylase expression and promoter activity are initiated at the scutellar epithelium and then gradually spread over aleurone layers during the progression from germination to seedling elongation (Okamoto and Akazawa, 1979; Okamoto et al., 1980; Ranjhan et al., 1992; Itoh et al., 1995). Thus, starch hydrolysis starts from the scutellar epithelium in the embryo and proceeds into the proximal subaleurone region in the endosperm. Sugars produced from starch hydrolysis by α-amylases in the endosperm are absorbed by the scutellar epithelium and transported to the embryonic axis as the prominent carbon source for seedling growth (Akazawa and Hara-Mishimura, 1985; Beck and Ziegler, 1989; Jacobsen et al., 1995). The absorption of sugars by the scutellar epithelium leads to an increase in sugar concentration and an inhibition of αAmy3 and αAmy8 expression in this tissue during the transition from germination to seedling elongation. The concentration of sugars also builds up significantly in endosperms as the expression of α-amylases increases, which might lead to feedback repression of α-amylase expression in aleurone cells and slow starch degradation in endosperms. From this study, however, we have shown that as a result of the presence of the GARE in promoters and their interaction with MYBGA, α-amylase genes are sugar-insensitive and highly GA-inducible in endosperms. This would explain why αAmy3 expression (which has no GARE) is negatively regulated by glucose but αAmy8 expression is positively regulated by GA and MYBGA in endosperms during seedling elongation. The GARE, therefore, would not only be responsible for GA activation but would also function in preventing the sugar feedback repression of α-amylase genes in endosperms, which would ensure a continuous supply of sugars to embryos during active seedling growth.
All α-amylase genes in the monocot lineage are derived from duplication of a single ancestor gene (Huang et al., 1992). The GARE is present in some α-amylase gene promoters but absent in others throughout evolution. On the other hand, all α-amylase gene promoters active in embryos contain a TA box, indicating that they are regulated by sugar and play an important role in starch hydrolysis in embryos during germination. α-Amylase gene promoters highly active in endosperms were found to contain a GARE, indicating that they are regulated by GA and play an important role in starch hydrolysis in endosperms during seedling development. It is not clear why the Ramy1B promoter also contains a GARE, but it was expressed at a low level in endosperms.
In conclusion, our studies indicate that tight temporal and spatial regulation of α-amylase expression controls rates of sugar production in embryos and endosperms, which is balanced between energy supply (source) and seedling development (sink). Our studies also provide a new insight into the control mechanism of the tissue-specific dominant regulation of α-amylase gene expression by sugar and GA signaling interference. Our finding that the interaction between the GARE and MYBGA prevents the sugar feedback repression of α-amylase genes in endosperms, which contributes significantly to seedling vigor, has advanced our knowledge about how cereal growth and development are controlled at the beginning of the life cycle.
METHODS
Plant Materials
Rice (Oryza sativa cv Tainung 67) was used for all experiments, except that the Tos17-tagged mutant gamyb-2 was derived from cv Nipponbare. The latter was a gift from the National Institute of Agrobiological Resources (Tsukuba, Japan).
Primers
Nucleotide sequences of all primers used for plasmid construction, PCR, quantitative RT-PCR, and RT-PCR analyses are listed in Table 2.
Table 2.
Primers Used for Plasmid Construction, PCR, Quantitative RT-PCR, and RT-PCR Analyses
| Primer | Sequence | Gene/Plasmid |
|---|---|---|
| Promoter construction | ||
| Amy8F | 5′-GC CCGGGTGCGTGATCGGTGATCG-3′ CCGGGTGCGTGATCGGTGATCG-3′ |
pAmy8SRC/GARC |
| Amy8R | 5′-GC GATATCAACAATCAATGATGTTGC-3′ GATATCAACAATCAATGATGTTGC-3′ |
pAmy8SRC/GARC |
| 2TAF | 5′-CCTTATCCATATCCATTATCCGTGAATTGCAACAGC-3′ | pαAmy8SRC/GARC(2TA) |
| 31KpF | 5′-TAACCACCTTTCGAACTGTTGCTTATCCGTG-3′ | pαAmy8SRC/GARC(mTA) |
| 34SacF | 5′-CCGTTGGAGAAAG CTTTATCCATGTTGC-3′ CTTTATCCATGTTGC-3′ |
pαAmy8SRC/GARC(mGARE) |
| 8GARE | 5′-CCTACGTGGCCATAAATAACCACCTTATCCATATCCA-3′ | pαAmy3SRC(+Amy8GARE) |
| Quantitative RT-PCR | ||
| 3RT25A | 5′-GTAGGCAGGCTCTCTAGCCTCTAGG-3′ | αAmy3 |
| 3RTR | 5′-AACCTGACATTATATATTGCACC-3′ | αAmy3 |
| 8RT1 | 5′-CTCAGGGTTCCTGCCGGTAGAAAGCA-3′ | αAmy8 |
| 8RTB | 5′-CGAAACGAACAGTAGCTAG-3′ | αAmy8 |
| GARTF | 5′-CAGACGCTAAAGCAGATTC-3′ | MYBGA |
| GARTR | 5′-GGCTTATCTCCATGCAC-3′ | MYBGA |
| 18SF | 5′-CCTATCAACTTTCGATGGTAGGATA-3′ | 18S rRNA |
| 18SR | 5′-CGTTAAGGGATTTAGATTGTACTCATT-3′ | 18S rRNA |
| RT-PCR | ||
| 7RT1 | 5′-TGAGCGCACGATGACGAGACTCTCA-3′ | αAmy7 |
| 7RT2 | 5′-AATTGCATCCGTAATTCGGA-3′ | αAmy7 |
| S1RTF | 5′-ATGGACGGACATGAGC-3′ | MYBS1 |
| S1RTR | 5′-GCTTTCACCGGGTGTA-3′ | MYBS1 |
| ART1 | 5′-CTGATGGACAGGTTATCACC-3′ | Actin1 |
| ART3 | 5′-CAGGTAGCAATAGGTATTACAG-3′ | Actin1 |
| Genotyping | ||
| Myb12-5′ | 5′-TCAGCTCTCCAAAGTTTCCC-3′ | MYBGA |
| Myb11-3′ | 5′-CAGGTTCATATTTAGGCCCC-3′ | MYBGA |
| LTR4A | 5′-ACTGTATAGTTGGCCCATGTCCAG-3′ | Tos17 |
| GAMYB5′ | 5′-CGC GCCATGTATCGGGTGAAG-3′ GCCATGTATCGGGTGAAG-3′ |
MYBGA cDNA |
| GAMYB3′ | 5′-CCG TCATTTGAATTCTGACAT-3′ TCATTTGAATTCTGACAT-3′ |
MYBGA cDNA |
Plasmids
Plasmid pαAmy3SRC-Luc (p3Luc.18) contains αAmy3 SRC (−186 to −82) fused to CaMV 35S minimal promoter (−46 bp from the transcription start site)–Adh1 intron–Luc coding sequence–nopaline synthase gene (Nos) terminator (Lu et al., 1998). Plasmid pcRAc1.3 contains a 1.4-kb rice actin gene (Act1) cDNA insert in pBluescript II KS+ (McElroy et al., 1990). Plasmid pRY18 carries a 3.8-kb DNA fragment that contains a rice genomic rDNA cluster, including the 3′ half portion of the 18S rRNA gene, the complete 5.8S rRNA gene, and the 5′ half portion of the 25S rRNA gene in pUC13 (Sano and Sano, 1990). Plasmid pUG contains β-glucuronidase (GUS) cDNA fused between a Ubi promoter and a Nos terminator (Christensen and Quail, 1996). Rice α-amylase gene-specific DNAs were obtained as described (Sheu et al., 1996).
Plasmid Construction
The αAmy8 SRC/GARC (−318 to −89) was amplified by PCR using Amy8F and Amy8R as forward and reverse primers and pAG8 (Lu et al., 1998), which contains the 1.2-kb αAmy8 promoter, as the DNA template. This 230-bp DNA fragment with an EcoRI site at both ends was inserted into pBluescript SK II+ (Stratagene), generating p8SRC/GARC. The αAmy8 SRC/GARC was excised from p8SRC/GARC with ApaI and PstI and subcloned into p35mALuc (Lu et al., 1998), generating pαAmy8SRC/GARC-Luc (Figure 2).
For modification or mutation of cis-acting elements in SRC or SRC/GARC, a PCR-based oligonucleotide-directed mutagenesis approach was used (Picard et al., 1994). For construction of mutant αAmy8 SRC/GARC, the TA box in αAmy8 SRC/GARC was duplicated or mutated by PCR amplification in a two-stage PCR. In the first PCR, 2TAF or 31KpF primer was used as the forward primer, T7 primer (Stratagene) was used as the reverse primer, and p8SRC/GARC was used as the DNA template. The PCR product was then used as a reverse megaprimer for a second PCR with T3 primer (Strategene) as the forward primer and p8SRC/GARC as the DNA template. The DNA fragment containing the duplicated or mutated TA box of αAmy8 SRC/GARC with ApaI and PstI sites at both ends was then subcloned into p35mALuc, generating pαAmy8SRC/GARC(2TA)-Luc and pαAmy8SRC/GARC(mTA)-Luc, respectively. The GARE in αAmy8 SRC/GARC was also mutated by two-stage PCR as described above, using 34SacF and T7 primers as forward and reverse primers, respectively, p8SRC/GARC as the DNA template in the first PCR, and then the PCR product as a reverse megaprimer and T3 primer as the forward primer in the second PCR. The DNA fragment containing the mutated GARE with ApaI and PstI sites at both ends was then subcloned into p35mALuc, generating pαAmy8SRC/GARC(mGARE)-Luc.
For the construction of mutant αAmy3 SRC, αAmy8 GARE was amplified by PCR using 8GARE and T7 primers as forward and reverse primers, respectively, and p3Luc.18 as the DNA template in the first PCR. The PCR product was then used as a reverse megaprimer with T3 primer as the forward primer and p3Luc.18 as the DNA template in the second PCR. The DNA fragment containing the αAmy8 GARE with ApaI and PstI sites at both ends was then subcloned into p35mALuc, generating pαAmy3SRC(+αAmy8GARE)-Luc.
For the construction of MYBGA overexpression vector, an 85-bp DNA fragment containing duplicated c-myc epitope (EQKLISEEDL) and residues 410 to 419 of the human c-myc protein (Evan et al., 1985) was synthesized and inserted into the EcoRV site in pBluescript KS+, generating p2cmyc. MYBGA cDNA was amplified by RT-PCR from RNA collected from endosperms of rice seeds germinated for 2 d and cloned into pBluescript, generating pOsMYBGA. The MYBGA coding region was excised from pOsMYBGA with BamHI and XbaI sites and subcloned into the same sites in pLAm (Chan and Yu, 1998b), generating pUbi-OsMYBGA-Amy. A DNA fragment containing the duplicated c-myc tag was excised from p2cmyc with BamHI and inserted into the same site of pUbi-OsMYBGA-Amy, generating pBS-Ubi-myc-OsMYBGA. This fusion protein contains two additional amino acids (Gly and Ser) between c-myc and MYBGA. pBS-Ubi-myc-OsMYBGA was linearized with HindIII and inserted into the same site of the binary vector pSMY1H (Ho et al., 2000), generating pUbi-myc-OsMYBGA.
Purification of RNA from Wild-Type Rice Seeds
Rice seeds were dehulled, sterilized with 3% NaOCl for 30 min, washed extensively with distilled water, and germinated in distilled water at 30°C in the dark for various lengths of time. Young shoots and roots were removed, and total RNA was purified from embryos and endosperms separately as described (Yu et al., 1991).
Real-Time Quantitative RT-PCR Analysis
Five micrograms of purified RNA was treated with 1 unit of RNase-free DNase I (Promega) at 37°C for 15 min. First-strand cDNA synthesis was primed with random hexamers (Promega) and catalyzed with Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen) at 37°C for 1.5 h. Ten-fold dilution of the reaction products was then subjected to real-time quantitative RT-PCR analysis (SYBR Green, Light-Cycler; Roche) using gene-specific primers. PCR amplification of the 112-bp 3′ UTR of αAmy3 cDNA was performed using 3RT25A and 3RTR as forward and reverse primers, respectively, and PCR amplification of the 202-bp 3′ UTR of αAmy8 cDNA (Sheu et al., 1996) was performed using 8RT1 and 8RTB as forward and reverse primers, respectively. PCR amplification of the 182-bp 3′ UTR of MYBGA cDNA (Gubler et al., 1995) was performed using GARTF and GARTR as forward and reverse primers, respectively. Amplification of the 229-bp 18S rRNA amplicon was performed using 18SF and 18SR as forward and reverse primers, respectively. Quantitative RT-PCR was analyzed by Light-Cycler data-analysis software (Roche). Crossing points were measured using the second derivative maximum method. PCR efficiencies were established for each pair of primers (see Supplemental Table 1 online), and relative expression levels were calculated by RelQuant relative quantification software version 1.01 (Roche). After PCR amplification, all samples were electrophoresed on agarose gels to verify the correct molecular weight of amplification products.
Semiquantitative RT-PCR Analysis
The analysis of αAmy3 and αAmy8 mRNA was performed using the same sets of primers described for real-time quantitative RT-PCR analyses. RT-PCR amplification of a 120-bp 3′ UTR of αAmy7 cDNA was performed using 7RT1 and 7RT2 as forward and reverse primers, respectively. RT-PCR amplification of the 234-bp 3′ UTR of MYBS1 cDNA was performed using S1RTF and S1RTR as forward and reverse primers, respectively. RT-PCR amplification of the 494-bp 3′ UTR of MYBGA cDNA was performed using Myb12-5′ and GARTR as forward and reverse primers, respectively. RT-PCR amplification of the 566-bp 3′ UTR of Act1 cDNA was performed using ART1 and ART3 as forward and reverse primers, respectively. RT-PCR amplification of the 564-bp junction region between MYBGA cDNA and the αAmy3 terminator of overexpressed MYBGA was performed using Myb12-5′ and 3RTR as forward and reverse primers, respectively. All amplifications were performed with Taq DNA polymerase (Promega). The resulting PCR products were resolved by 2% agarose gel electrophoresis, visualized with ethidium bromide staining, and quantified with a luminescent image analyzer (model LAS-1000 Plus; Fujifilm) using its Image Gauge program version 3.2.
Rice Embryo and Endosperm Transient Expression Assays
The particle bombardment–mediated transient expression assay system has been used successfully to study sugar and GA signal transduction and the regulation of barley (Hordeum vulgare) and rice α-amylase gene promoter activity (Lanahan et al., 1992; Rogers and Rogers, 1992; Gubler et al., 1997; Morita et al., 1998; Toyofuku et al., 1998; Umemura et al., 1998; Cercos et al., 1999; Gomez-Cadenas et al., 2001; Lu et al., 2002; Lee et al., 2003; Washio, 2003) using barley or rice endosperms/aleurone layers or embryos as hosts. Because transfection efficiency varies from sample to sample using this method, the enzyme activity of an internal control (Ubi-GUS) was used to normalize the reporter enzyme activity.
For embryo transient expression assays, rice seeds (cv Tainung 67) were dehulled, sterilized with 3% NaOCl for 30 min, washed extensively with sterile water, and placed on liquid Murashige and Skoog (MS) medium containing 2 mg/L 2,4-D. Embryos germinated on this medium showing enlarged scutella (where α-amylase is expressed in vivo) would allow an accurate targeting of gold particles into the scutellum. After incubation at 30°C for 8 d, embryos were dissected away from endosperms and placed on MS medium solidified with 0.3% (w/v) Phytagel (Sigma-Aldrich) with the scutellar side up. Each plate contained 16 embryos, arranged in a small circle (∼2.5 cm in diameter), for one bombardment. The particle bombardment–mediated rice embryo transient expression assay was performed as described (Umemura et al., 1998). Sixteen bombarded embryos were divided into two halves, each incubated in liquid MS medium containing 100 mM glucose or mannitol, or medium containing or lacking 10 μM GA3, at 30°C for 24 h.
For endosperm transient expression assays, embryoless half seeds (endosperms) were sterilized as for embryos. Each plate contained 16 endosperms, also arranged in a small circle, for one bombardment. The rice endosperm transient expression assay was performed as described (Lanahan et al., 1992; Cercos et al., 1999; Washio, 2003) with slight modifications. Sixteen bombarded endosperms were divided into two halves, each incubated in a buffer (20 mM CaCl2, 20 mM sodium succinate, pH 5.0, and 10 μM chloramphenicol) containing 200 mM glucose or mannitol, or buffer containing or lacking 10 μM GA3, at 30°C for the indicated times.
Plasmid pUG was used as an internal control for the αAmy3 SRC-Luc or αAmy8 SRC/GARC-Luc construct. The ratio of test DNA to internal control plasmid DNA was 4:1. Each independent experiment consisted of three replicates, with eight embryos or endosperms for each treatment, and was repeated three to four times with similar results.
Luciferase and GUS Activity Assay
Total proteins were extracted from eight bombarded rice embryos with 0.5 mL, or from four bombarded rice endosperms with 0.8 mL, of CCLR buffer [100 mM KH2(PO4), pH 7.8, 1 mM EDTA, 10% glycerol, 1% Triton X-100, and 7 mM β-mercaptoethanol], and 100 and 50 μL of extracted samples were used for luciferase and GUS activity assays, respectively, with methods as described (Lu et al., 1998). The protein concentration was determined with a Coomassie Brilliant Blue R 250 protein assay reagent (Pierce). Luciferase activity was normalized by dividing it against GUS activity.
RNA Gel Blot Analysis
Total RNA was purified from embryos and endosperms as described (Yu et al., 1991). RNA gel blot analysis using α-32P–labeled probes was performed as described (Sheu et al., 1996).
Purification of RNA from Tos17-Tagged Rice Mutant Seeds
Seeds of the Tos17-tagged rice mutant gamyb-2 were propagated in the field for one season. To screen for mutant seeds containing heterozygous or homozygous Tos17 insertion alleles, embryos and endosperms were collected separately and placed on two 96-well plates. Each isolated endosperm and its corresponding embryo were labeled with the same identification number. Embryos were sterilized with 1% NaOCl for 15 min, washed extensively with distilled water, and germinated in MS medium at 30°C in the dark for 1 week. Genomic DNA was individually isolated from each seedling as described (Sheu et al., 1996). The genotyping of each seedling was performed by PCR using transposon-specific primer LTR4A and MYBGA-specific primer Myb12-5′ as forward primers and Myb11-3′ as the reverse primer (Kaneko et al., 2004). Seedlings with homozygous Tos17 insertion alleles and with homozygous wild-type alleles were identified (examples shown in Figure 9B), and their corresponding endosperms were collected and sterilized with 0.5% NaOCl for 20 min, washed extensively with distilled water, and divided into four groups. Each group containing three endosperms was incubated with 200 mM glucose or mannitol with or without 10 μM GA at 30°C for 2 d. Total RNAs were purified from treated endosperms and subjected to RT-PCR analyses.
Synthesis of cDNA Probes and DNA Slot-Blot Analysis
The α-32P–labeled, single-stranded cDNA probe was prepared from total RNA using an oligo(dT) primer and M-MLV reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Excess rRNA, actin cDNA, and α-amylase gene-specific DNAs were denatured with 0.4 N NaOH at room temperature for 30 min and neutralized with a ninefold volume of 6× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate; containing 0.1% DNA agarose gel loading dye). The denatured DNA was blotted onto a Magna nylon membrane (Micron Separations) using a suction slot-blotter (Life Technologies) and hybridized with the cDNA probe.
Rice Transformation
Plasmid pUbi-myc-OsMYBGA was introduced into Agrobacterium tumefaciens strain EHA101, and rice transformation was performed as described (Chen et al., 2002).
Starch Agar Plate α-Amylase Assay
Several transgenic rice lines carrying the Ubi-OsMYBGA construct were generated and T1 seeds collected. To screen for transgenic seeds overexpressing MYBGA, 32 seeds from each independent transgenic rice line were randomly selected. Embryos and endosperms were collected separately and placed on two 96-well plates. Each isolated endosperm and its corresponding embryo were labeled with the same identification number. Isolated endosperms were subjected to the α-amylase activity assay as described (Yamaguchi, 1998). Endosperms were sterilized with 1% NaOCl for 15 min, washed with distilled water, and placed cutting side down on starch agar plates containing 0.2% starch and 2% agar (Duchefa). Sixteen endosperms per plate were incubated at 30°C in the dark for 3 d. Endosperms were removed, and α-amylase activity was examined by staining agar with a solution containing 0.1% I2 and 1% KI. Clear zones appear if α-amylases are expressed in endosperms and secreted into the starch agar. Transgenic endosperms giving rise to large clear zones were assumed to have expressed high levels of MYBGA, and their corresponding embryos were collected. Embryos were sterilized with 1% NaOCl for 15 min, washed with distilled water, placed in a buffer (20 mM CaCl2, 20 mM sodium succinate, pH 5.0, and 10 μM chloramphenicol) containing 100 mM glucose or mannitol, and incubated at 30°C for 24 h. Total RNAs were purified from treated embryos and subjected to RT-PCR analyses.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers M59351 (αAmy3), M59352 (αAmy8), X98355 (MYBGA), X16280 (Act1), and M26461(18S rRNA).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Conditions and Efficiency of Real-Time Quantitative RT-PCR.
Supplemental Table 2. Determination of αAmy3, αAmy8, and MYBGA mRNA Levels in Rice Embryos and Endosperms during Germination and Seedling Development by Real-Time Quantitative RT-PCR Analysis.
Supplemental Figure 1. Expression of αAmy3 and αAmy8 Correlates with Glucose and GA Levels in Germinating Seeds and Developing Seedlings.
Supplemental Figure 2. Transient Expression Assays Demonstrated That GA-Nonresponsive αAmy3 SRC Is Glucose-Sensitive in Both Embryos and Endosperms and GA-Responsive αAmy8 SRC/GARC Is Glucose-Sensitive Only in Embryos.
Supplemental Figure 3. In Transgenic Rice Seeds, GA-Nonresponsive αAmy3 Is Glucose-Sensitive in Both Embryos and Endosperms and GA-Responsive αAmy8 SRC/GARC Is Glucose-Sensitive Only in Embryos.
Supplemental Figure 4. Glucose Repression Overrides GA Activation of αAmy8 SRC/GARC in Embryos, Whereas GA Activation Overrides Glucose Repression of the Same Promoter in Endosperms.
Supplemental Figure 5. The TA Box Enhances the Promoter Activity and Glucose Sensitivity of αAmy8 SRC/GARC in Embryos Only.
Supplemental Figure 6. In Transgenic Endosperms, the G Box Does Not Enhance the Glucose Sensitivity of αAmy8 SRC/GARC.
Supplementary Material
Acknowledgments
We thank Hirohiko Hirochika and Makoto Matsuoka for sharing gamyb-2 mutant seeds, Tuan-Hua David Ho, Kenrick Deen, and Kuo-Wei Lee for critical review of the manuscript, and Lin-Chih Yu and Ku-Ting Chen for technical assistance. This work was supported by grants from the Academia Sinica and the National Science Council (Grant NSC92-2311-B001-037) of the Republic of China.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Su-May Yu (sumay@imb.sinica.edu.tw).
Online version contains Web-only data.
References
- Akazawa, T., and Hara-Mishimura, I. (1985). Topographic aspects of biosynthesis, extracellular secretion and intracellular storage of proteins in plant cells. Annu. Rev. Plant Physiol. 70 441–472. [Google Scholar]
- Beck, E., and Ziegler, P. (1989). Biosynthesis and degradation of starch in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40 95–117. [Google Scholar]
- Cercos, M., Gomez-Cadenas, A., and Ho, T.H. (1999). Hormonal regulation of a cysteine proteinase gene, EPB-1, in barley aleurone layers: Cis- and trans-acting elements involved in the co- ordinated gene expression regulated by gibberellins and abscisic acid. Plant J. 19 107–118. [DOI] [PubMed] [Google Scholar]
- Chan, M.T., Chao, Y.C., and Yu, S.M. (1994). Novel gene expression system for plant cells based on induction of alpha-amylase promoter by carbohydrate starvation. J. Biol. Chem. 269 17635–17641. [PubMed] [Google Scholar]
- Chan, M.T., and Yu, S.M. (1998. a). The 3′ untranslated region of a rice alpha-amylase gene functions as a sugar-dependent mRNA stability determinant. Proc. Natl. Acad. Sci. USA 95 6543–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, M.T., and Yu, S.M. (1998. b). The 3′ untranslated region of a rice alpha-amylase gene mediates sugar-dependent abundance of mRNA. Plant J. 15 685–695. [DOI] [PubMed] [Google Scholar]
- Chen, P.-W., Lu, C.-A., Yu, T.-S., Tseng, T.-H., Wang, C.-S., and Yu, S.-M. (2002). Rice alpha-amylase transcriptional enhancers direct multiple mode regulation of promoters in transgenic rice. J. Biol. Chem. 277 13641–13649. [DOI] [PubMed] [Google Scholar]
- Christensen, A.H., and Quail, P.H. (1996). Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5 213–218. [DOI] [PubMed] [Google Scholar]
- Evan, G.I., Lewis, G.K., Ramsay, G., and Bishop, J.M. (1985). Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5 3610–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cadenas, A., Zentella, R., Walker-Simmons, M.K., and Ho, T.H. (2001). Gibberellin/abscisic acid antagonism in barley aleurone cells: Site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13 667–679. [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., and Jacobsen, J.V. (1992). Gibberellin-responsive elements in the promoter of a barley high-pI alpha-amylase gene. Plant Cell 4 1435–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Kalla, R., Roberts, J.K., and Jacobsen, J.V. (1995). Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell 7 1879–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Raventos, D., Keys, M., Watts, R., Mundy, J., and Jacobsen, J.V. (1999). Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 17 1–9. [DOI] [PubMed] [Google Scholar]
- Gubler, F., Watts, R.J., Kalla, R., Matthews, P., Keys, M., and Jacobsen, J.V. (1997). Cloning of a rice cDNA encoding a transcription factor homologous to barley GAMyb. Plant Cell Physiol. 38 362–365. [DOI] [PubMed] [Google Scholar]
- Halford, N.G., and Paul, M.J. (2003). Carbon metabolite sensing and signalling. Plant Biotechnol. J. 1 381–398. [DOI] [PubMed] [Google Scholar]
- Ho, S.L., Tong, W.F., and Yu, S.M. (2000). Multiple mode regulation of a cysteine proteinase gene expression in rice. Plant Physiol. 122 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, N., Stebbins, G.L., and Rodriguez, R.L. (1992). Classification and evolution of alpha-amylase genes in plants. Proc. Natl. Acad. Sci. USA 89 7526–7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, N., Sutliff, T.D., Litts, J.C., and Rodriguez, R.L. (1990). Classification and characterization of the rice alpha-amylase multigene family. Plant Mol. Biol. 14 655–668. [DOI] [PubMed] [Google Scholar]
- Hwang, Y.S., Karrer, E.E., Thomas, B.R., Chen, L., and Rodriguez, R.L. (1998). Three cis-elements required for rice alpha-amylase Amy3D expression during sugar starvation. Plant Mol. Biol. 36 331–341. [DOI] [PubMed] [Google Scholar]
- Isabel-LaMoneda, I., Diaz, I., Martinez, M., Mena, M., and Carbonero, P. (2003). SAD: A new DOF protein from barley that activates transcription of a cathepsin B-like thiol protease gene in the aleurone of germinating seeds. Plant J. 33 329–340. [DOI] [PubMed] [Google Scholar]
- Itoh, K., Yamaguchi, J., Huang, N., Rodriguez, R.L., Akazawa, T., and Shimamoto, K. (1995). Developmental and hormonal regulation of rice alpha-amylase (RAmy1A)-gusA fusion genes in transgenic rice seeds. Plant Physiol. 107 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, J.V., Gubler, F., and Chandler, P.M. (1995). Gibberellin action in germinated cereal grains. In Plant Hormones: Physiology, Biochemistry, and Molecular Biology, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 246–271.
- Kaneko, M., Inukai, Y., Ueguchi-Tanaka, M., Itoh, H., Izawa, T., Kobayashi, Y., Hattori, T., Miyao, A., Hirochika, H., Ashikari, M., and Matsuoka, M. (2004). Loss-of-function mutations of the rice GAMYB gene impair alpha-amylase expression in aleurone and flower development. Plant Cell 16 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer, E.E., Chandler, J.M., Foolad, M.R., and Rodriguez, R.L. (1993). Correlation between alpha-amylase gene expression and seedling vigor in rice. Euphytica 66 163–169. [Google Scholar]
- Karrer, E.E., and Rodriguez, R.L. (1992). Metabolic regulation of rice alpha-amylase and sucrose synthase genes in planta. Plant J. 2 517–523. [PubMed] [Google Scholar]
- Lanahan, M.B., Ho, T.H., Rogers, S.W., and Rogers, J.C. (1992). A gibberellin response complex in cereal alpha-amylase gene promoters. Plant Cell 4 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.-C., Lu, C.-A., Chen, P.-W., Casaretto, J., and Yu, S.-M. (2003). An ABA-responsive bZIP protein, OsBZ8, mediates sugar repression of alpha-amylase gene expression. Physiol. Plant. 119 78–86. [Google Scholar]
- Loreti, E., Matsukura, C., Gubler, F., Alpi, A., Yamaguchi, J., and Perata, P. (2000). Glucose repression of alpha-amylase in barley embryos is independent of GAMYB transcription. Plant Mol. Biol. 44 85–90. [DOI] [PubMed] [Google Scholar]
- Lu, C.A., Ho, T.H., Ho, S.L., and Yu, S.M. (2002). Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of alpha-amylase gene expression. Plant Cell 14 1963–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C.A., Lim, E.K., and Yu, S.M. (1998). Sugar response sequence in the promoter of a rice alpha-amylase gene serves as a transcriptional enhancer. J. Biol. Chem. 273 10120–10131. [DOI] [PubMed] [Google Scholar]
- McElroy, D., Rothenberg, M., Reece, K.S., and Wu, R. (1990). Characterization of the rice (Oryza sativa) actin gene family. Plant Mol. Biol. 15 257–268. [DOI] [PubMed] [Google Scholar]
- Morita, A., Umemura, T., Kuroyanagi, M., Futsuhara, Y., Perata, P., and Yamaguchi, J. (1998). Functional dissection of a sugar-repressed alpha-amylase gene (RAmy1 A) promoter in rice embryos. FEBS Lett. 423 81–85. [DOI] [PubMed] [Google Scholar]
- Murray, F., Kalla, R., Jacobsen, J., and Gubler, F. (2003). A role for HvGAMYB in anther development. Plant J. 33 481–491. [DOI] [PubMed] [Google Scholar]
- Okamoto, K., and Akazawa, T. (1979). Enzymic mechanism of starch breakdown in germinating rice seeds. 7. Amylase formation in the epithelium. Plant Physiol. 63 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, K., Kitano, H., and Akazawa, T. (1980). Biosynthesis and excretion of hydrolases in germinating cereal seeds. Plant Cell Physiol. 21 201–204. [Google Scholar]
- Perata, P., Matsukura, C., Vernieri, P., and Yamaguchi, J. (1997). Sugar repression of gibberellin-dependent signalling pathway in barley embryos. Plant Cell 9 2197–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard, V., Ersdal-Badju, E., Lu, A., and Bock, S.C. (1994). A rapid and efficient one-tube PCR-based mutagenesis technique using Pfu DNA polymerase. Nucleic Acids Res. 22 2587–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjhan, S., Karrer, E.E., and Rodriguez, R.L. (1992). Localizing alpha-amylase gene expression in germinated rice grains. Plant Cell Physiol. 33 73–79. [Google Scholar]
- Rogers, J.C., Lanahan, M.B., and Rogers, S.W. (1994). The cis-acting gibberellin response complex in high pI alpha-amylase gene promoters. Requirement of a coupling element for high-level transcription. Plant Physiol. 105 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, J.C., and Rogers, S.W. (1992). Definition and functional implications of gibberellin and abscisic acid cis-acting hormone response complexes. Plant Cell 4 1443–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, Y., and Sano, R. (1990). Variation of the intergenic spacer region of ribosomal DNA in cultivated and wild rice species. Genome 33 209–218. [Google Scholar]
- Sheu, J.-J., Jan, S.-P., Lee, H.-T., and Yu, S.-M. (1994). Control of transcription and mRNA turnover as mechanisms of metabolic repression of alpha-amylase gene expression. Plant J. 5 655–664. [Google Scholar]
- Sheu, J.-J., Yu, T.-S., Tong, W.-F., and Yu, S.-M. (1996). Carbohydrate starvation stimulates differential expression of rice alpha-amylase genes that is modulated through complicated transcriptional and posttranscriptional processes. J. Biol. Chem. 271 26998–27004. [DOI] [PubMed] [Google Scholar]
- Smeekens, S. (2000). Sugar-induced signal transduction in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 49–81. [DOI] [PubMed] [Google Scholar]
- Sun, T.p., and Gubler, F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55 197–223. [DOI] [PubMed] [Google Scholar]
- Sutliff, T.D., Lanahan, M.B., and Ho, T.H. (1993). Gibberellin treatment stimulates nuclear factor binding to the gibberellin response complex in a barley alpha-amylase promoter. Plant Cell 5 1681–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, B.R., and Rodriguez, R.L. (1994). Metabolite signals regulate gene expression and source/sink relations in cereal seedlings. Plant Physiol. 106 1235–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku, K., Umemura, T., and Yamaguchi, J. (1998). Promoter elements required for sugar-repression of the RAmy3D gene for alpha-amylase in rice. FEBS Lett. 428 275–280. [DOI] [PubMed] [Google Scholar]
- Umemura, T., Perata, P., Futsuhara, Y., and Yamaguchi, J. (1998). Sugar sensing and alpha-amylase gene repression in rice embryos. Planta 204 420–428. [DOI] [PubMed] [Google Scholar]
- Washio, K. (2001). Identification of Dof proteins with implication in the gibberellin-regulated expression of a peptidase gene following the germination of rice grains. Biochim. Biophys. Acta 1520 54–62. [DOI] [PubMed] [Google Scholar]
- Washio, K. (2003). Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol. 133 850–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, J. (1998). Analysis of embryo-specific α-amylase using isolated mature rice embryos. Breed. Sci. 48 365–370. [Google Scholar]
- Yu, S.M., Kuo, Y.H., Sheu, G., Sheu, Y.J., and Liu, L.F. (1991). Metabolic derepression of alpha-amylase gene expression in suspension-cultured cells of rice. J. Biol. Chem. 266 21131–21137. [PubMed] [Google Scholar]
- Yu, S.M., Lee, Y.C., Fang, S.C., Chan, M.T., Hwa, S.F., and Liu, L.F. (1996). Sugars act as signal molecules and osmotica to regulate the expression of alpha-amylase genes and metabolic activities in germinating cereal grains. Plant Mol. Biol. 30 1277–1289. [DOI] [PubMed] [Google Scholar]
- Yu, S.M., Tzou, W.S., Lo, W.S., Kuo, Y.H., Lee, H.T., and Wu, R. (1992). Regulation of alpha-amylase-encoding gene expression in germinating seeds and cultured cells of rice. Gene 122 247–253. [DOI] [PubMed] [Google Scholar]
- Zhang, Z.L., Xie, Z., Zou, X., Casaretto, J., Ho, T.H., and Shen, Q.J. (2004). A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 134 1500–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.