Abstract
The centromere is the chromosomal site for assembly of the kinetochore where spindle fibers attach during cell division. In most multicellular eukaryotes, centromeres are composed of long tracts of satellite repeats that are recalcitrant to sequencing and fine-scale genetic mapping. Here, we report the genomic and genetic characterization of the complete centromere of rice (Oryza sativa) chromosome 3. Using a DNA fiber-fluorescence in situ hybridization approach, we demonstrated that the centromere of chromosome 3 (Cen3) contains ∼441 kb of the centromeric satellite repeat CentO. Cen3 includes an ∼1,881-kb domain associated with the centromeric histone CENH3. This CENH3-associated chromatin domain is embedded within a 3113-kb region that lacks genetic recombination. Extensive transcription was detected within the CENH3 binding domain based on comprehensive annotation of protein-coding genes coupled with empirical measurements of mRNA levels using RT-PCR and massively parallel signature sequencing. Genes <10 kb from the CentO satellite array were expressed in several rice tissues and displayed histone modification patterns consistent with euchromatin, suggesting that rice centromeric chromatin accommodates normal gene expression. These results support the hypothesis that centromeres can evolve from gene-containing genomic regions.
INTRODUCTION
The centromere is the chromosomal site for kinetochore assembly and also plays a major role in sister chromatid cohesion. These functions are conserved among the centromeres from all eukaryotic species. Homologs of several proteins have been found in the centromeres of various eukaryotic species (Houben and Schubert, 2003; Amor et al., 2004a). In particular, a centromere-specific histone 3 variant, referred to as CENH3, appears to be a universal marker for centromeric chromatin (Henikoff et al., 2001). By contrast, the primary DNA sequence that underlies centromeres has no discernable conservation between various model organisms and can be significantly diverged among closely related species. This enigma has evoked extensive studies on the structure and function of the centromeres in several model eukaryotes.
The budding yeast Saccharomyces cerevisiae is one of the simplest model eukaryotic species. The sizes of S. cerevisiae chromosomes range from 250 kb to 2 Mb in length. The centromeres of S. cerevisiae consist of only ∼125 bp of unique sequence (Clarke, 1998). However, in multicellular eukaryotes, centromeres are often embedded within cytologically distinctive heterochromatin and are associated with megabase-sized and highly homogenized satellite DNA (Zhong et al., 2002; Hall et al., 2003; Sun et al., 2003; Rudd and Willard, 2004; Lamb et al., 2005). The highly repetitive nature of centromeres makes them difficult to clone and sequence (Henikoff, 2002). The genomes of several multicellular eukaryotes, including Drosophila melanogaster, human, mouse, and Arabidopsis thaliana, have been sequenced. However, none of the centromeres in these species has been fully sequenced due to the difficult nature of the genomic and genetic characterization of the centromeres in these model species.
Human centromeres are the most extensively studied centromeres among multicellular eukaryotes. The most abundant DNA sequence in human centromeres is the α satellite that consists of multiples of an ∼171-bp monomer repeat (Willard, 1998). The amount of the α satellite in different centromeres varies from ∼250 kb to >4 Mb (Wevrick and Willard, 1989; Oakey and Tyler-Smith, 1990). Human artificial chromosomes were successfully assembled using either synthetic or cloned α satellite DNA as a centromeric component (Harrington et al., 1997; Ikeno et al., 1998; Henning et al., 1999). Structural and functional analyses of DNA in the human X chromosome centromere revealed that the α satellite repeats are more diverged on the edges of the centromere and become homogenized toward the functional core (Schueler et al., 2001), suggesting that the centromeres evolve through selection of new repeats within the functional core and movement of older repeats to the flanking regions (Schueler et al., 2001).
The centromeres of a model plant species, Arabidopsis, have also been studied extensively. The Arabidopsis centromeres were genetically mapped and contain various types of repetitive DNA elements (Copenhaver et al., 1999). The most abundant DNA element within the genetically mapped Arabidopsis centromeres is a 178-bp satellite repeat (Round et al., 1997; Heslop-Harrison et al., 1999; Hall et al., 2003). Fine physical mapping revealed that each Arabidopsis centromere contains several megabases of this 178-bp repeat (Kumekawa et al., 2000, 2001; Hosouchi et al., 2002). Chromatin immunoprecipitation (ChIP) analysis showed that the 178-bp repeat is the only known repetitive DNA element that interacts with CENH3 (Nagaki et al., 2003). In addition, only the middle portion of the multimegabase 178-bp repeat array in each centromere is associated with CENH3 (Shibata and Murata, 2004). Thus, the general DNA structure of Arabidopsis centromeres is similar to that of human centromeres.
Rice (Oryza sativa) centromeres contain two major classes of repetitive DNA elements: the centromeric CRR retrotransposon and a 155-bp satellite repeat CentO (Dong et al., 1998; Miller et al., 1998; Cheng et al., 2002). The CRR retrotransposons are highly enriched in the centromeres and are intermingled with the CentO satellite (Cheng et al., 2002; Nagaki et al., 2005). The amount of the CentO satellite varies from ∼60 kb on chromosome 8 to ∼2 Mb on chromosome 11 (Cheng et al., 2002). The lack of extensive amounts of satellite repeats allowed sequencing of the entire centromere of rice chromosome 8 (Cen8; Nagaki et al., 2004; Wu et al., 2004). An ∼750-kb region within Cen8 is associated with rice CENH3 (Nagaki et al., 2004). Sequence analysis of Cen8 revealed the presence of several active genes within the CENH3 binding domain (Nagaki et al., 2004; Yan et al., 2005). Although genes located in the pericentromeric regions have been reported in several muticellular eukaryotes (Yasuhara and Wakimoto, 2006), only the Cen8 genes represent centromeric genes that are embedded within CENH3-associated chromatin.
We have been interested to know whether Cen8 represents a rare evolutionary case and whether active genes are present within rice centromeres that contain an extensive amount of satellite repeats. In this study, we used genomic and genetic approaches to comprehensively analyze the centromere of rice chromosome 3 (Cen3). We demonstrate that Cen3 contains ∼441 kb of the CentO satellite. ChIP analysis revealed an ∼1881-kb region that is associated with rice CENH3. This CENH3 binding domain is embedded within a 3113-kb recombination-suppressed chromosomal domain. We detected extensive transcription within the CENH3 binding domain. We demonstrate that rice Cen3 and Cen8 share similar genomic and genetic features, although Cen3 contains significantly more centromeric satellite repeats than Cen8. The implications of these results for centromere evolution are discussed.
RESULTS
Cen3 Contains ∼441 kb of the CentO Satellite
The sequence map of rice chromosome 3 spans 36.1 Mb and includes 323 BAC and P1 artificial chromosome clones (Rice Chromosome 3 Sequencing Consortium, 2005). The 36.1-Mb pseudomolecule covers most of the chromosome, with only five physical gaps present in the two arms, gaps at both telomeres, and one gap in the centromere. The most centromere-proximal BAC (b0038E06) on the long arm (Figure 1A), the only BAC clone within this region that is unfinished but is ordered with oriented contigs, contains extensive amounts of CentO repeats (87.2% of the total 21,287-bp CentO in the chromosome 3 pseudomolecule). However, the most centromere-proximal BAC (b0047D08) on the short arm (Figure 1A) contains only 2005 bp of the CentO repeat. These results indicate that the long arm sequence may have already extended into the major CentO array of Cen3. However, it is not known if the short arm sequence has extended into the major CentO array. Therefore, the exact size of the centromeric gap in the current chromosome 3 pseudomolecule is unknown.
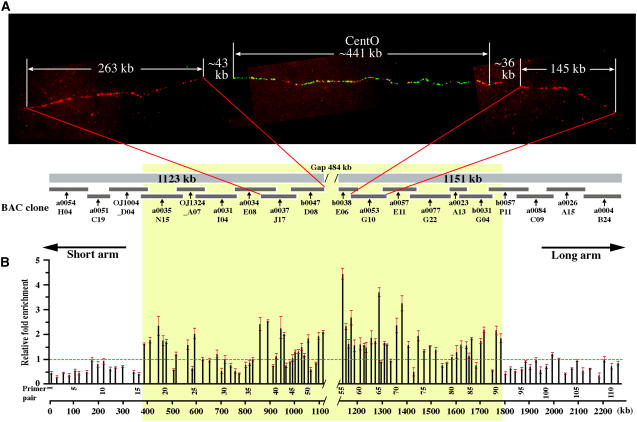
Figure 1.
Mapping of the CENH3 Binding Domain of Cen3.
(A) Physical mapping of Cen3. Top panel: fiber-FISH mapping of the centromeric gap within the chromosome 3 sequence map. Three BAC clones, including a0037J17 and b0047D08 from the short arm side of the gap and a0053G10 from the long arm side, were labeled in red and hybridized together with the centromeric satellite repeat CentO (in green) on an extended DNA fiber. The average lengths of the fiber-FISH signals were converted into kilobases and are labeled on the image. The red signals interspersed with the green signals are derived from the CRR elements contained in the three BAC clones. Bottom panel: physical map of Cen3. Virtual contigs on the short arm side (1123 kb) and long arm side (1151 kb) were assembled from sequences of nine and 10 BAC clones, respectively.
(B) ChIP quantitative PCR assay revealed an ∼1881-kb region (in yellow) between primer pairs 16 and 91, which binds rice CENH3.
A fiber-fluorescence in situ hybridization (fiber-FISH) mapping approach was used to determine the physical coverage of the centromeric region by the current chromosome 3 pseudomolecule. We used two overlapping clones, a0037J17 (AC135226) and b0047D08 (AC137925), which flank the short arm side of Cen3, and a single BAC, a0053G10 (AC091233), which flanks the long arm side of Cen3 (Figure 1A). These three BACs were labeled in red, the CentO repeat was labeled in green, and these probes were cohybridized onto extended DNA fibers (Figure 1A). The lengths of the fiber-FISH signals from the short and long arm sides of the CentO array differ significantly from each other and can be unambiguously identified. Gaps between the BAC-derived and CentO-derived signals were observed at both junctions (Figure 1A). We collected 10 complete fiber-FISH signals derived from these probes (Table 1). The insert size of BAC a0053G10 (145 kb) and the combined sizes of BACs a0037J17 and b0047D08 (263 kb) were used as the reference for estimating the signal lengths of individual fibers to minimize the stretching variation among different fibers. The length (in micrometers) of fiber-FISH signals and the sizes of the gaps were converted into kilobases based on the average resolution (kb/μm) calculated from the signals derived from these three BAC clones. The fiber-FISH mapping revealed a 43 kb ± 12 kb gap between a0037J17/b0047D08 and the CentO array and a 36 kb ± 9 kb gap between a0053G10 and the CentO array (Figure 1A, Table 1). The size of the CentO array was calculated as 441 kb ± 47 kb based on the 10 signals. Collectively, the physical size of the centromeric gap on the sequence map is ∼484 kb (441 kb + 43 kb) (Figure 1A).
Table 1.
Fiber-FISH Measurements of the CentO Array in Cen3
| Fibers | A0037J17 + 0047D08 (μm)a | Gap 1 (kb)b | Gap 1 (μm)b | CentO (kb) | CentO (μm) | Gap 2 (kb)b | Gap 2 (μm)b | a0053G10 (μm)a |
|---|---|---|---|---|---|---|---|---|
| 1 | 106.9 | 61.8 | 23.5 | 446.9 | 169.7 | 36.2 | 13.7 | 48.1 |
| 2 | 83.5 | 48.5 | 15.2 | 420.1 | 132.0 | 43.7 | 13.7 | 44.7 |
| 3 | 69.0 | 41.4 | 11.3 | 446.1 | 121.5 | 26.9 | 7.3 | 42.1 |
| 4 | 79.5 | 40.5 | 12.2 | 388.8 | 117.2 | 37.9 | 11.4 | 43.5 |
| 5 | 77.1 | 40.1 | 11.5 | 455.2 | 130.1 | 30.4 | 8.7 | 39.5 |
| 6 | 78.8 | 23.7 | 6.8 | 448.9 | 127.9 | 27.1 | 7.7 | 37.4 |
| 7 | 81.3 | 50.3 | 15.6 | 437.6 | 136.0 | 27.4 | 8.5 | 45.5 |
| 8 | 81.4 | 29.8 | 9.2 | 369.2 | 114.4 | 43.9 | 13.6 | 45.0 |
| 9 | 79.2 | 54.7 | 15.5 | 450.0 | 127.2 | 54.2 | 15.3 | 36.1 |
| 10 | 68.4 | 35.0 | 8.8 | 547.3 | 137.4 | 31.3 | 7.9 | 34.0 |
| Average | 80.5 | 42.6 | 13.0 | 441.0 | 131.3 | 35.9 | 10.8 | 41.6 |
| sd | 10.6 | 11.6 | 4.8 | 47.2 | 15.4 | 9.1 | 3.1 | 4.6 |
BACs a0037J17 (150 kb; AC135226) and b0047D08(141 kb; AC137925) overlap 28 kb. Therefore, excluding the overlap, these two BACs generate 263 kb of fiber-FISH signals. BAC a0053G10 is 145 kb (AC091233). As an example, the kb/μm conversion rate for fiber 1 is calculated as (263 + 145)/(106.9 + 48.1) = 2.632.
Gap 1 refers to the gap between the signal from BACs a0037J17 and b0047D08 and the signal from CentO; gap 2 refers to the gap between the signal from CentO and the signal from BAC a0053G10.
Cen3 Contains an ∼1881-kb Domain Associated with CENH3
We constructed a 2,275,221-bp virtual contig from the finished sequences of 19 rice BAC clones that flank both sides of the CentO array (Figure 1A). This virtual contig contains the ∼484-kb centromeric gap that is represented by a tract of 974 Ns from position 1,123,027 bp to 1,124,000 bp. We then used ChIP to determine the CENH3 binding region within this virtual contig. Specific primers were designed from the 2.3-Mb Cen3 sequence for real-time PCR analysis. We designed 111 pairs of specific primers, spaced every 8.8 to 47 kb, along the entire 2.3-Mb sequence (see Supplemental Table 1 online).
We plotted the relative enrichment of CENH3 binding of each primer pair over the negative control (a putative manganese transport protein gene, located at 122.8 centimorgan [cM] on chromosome 3) along the Cen3 virtual contig (Figure 1B). This plot illustrated two flanking regions, demarcated by primer pairs 1 to 15 on the left side and 92 to 111 on the right side that did not show relative enrichment above the background. Based on these results, we localized the CENH3 binding region to a portion of this virtual contig starting at primer 16 (389-kb position) and ending at primer 91 (1786-kb position). The size of this functional centromere domain is estimated at 1881 kb, consisting of 1397 kb of assembled sequence and the ∼484-kb physical gap (Figure 1B). We observed that a significant fraction of the primer pairs mapped to this functional domain did not show clear binding with CENH3, which is consist with our earlier finding of noncontinuous binding of CENH3 in rice Cen8 (Nagaki et al., 2004).
Genetic Location of Cen3
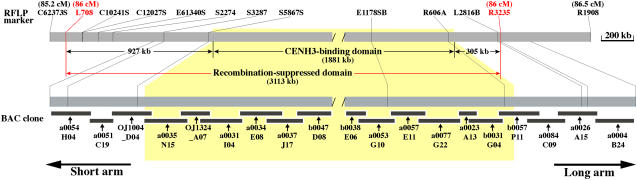
The genetic position of Cen3 was mapped between 80.2 and 87 cM on the linkage map of rice chromosome 3 based on a mapping population that included 186 F2 plants derived from a cross between O. sativa subsp japonica var Nipponbare and O. sativa subsp indica var Kasalath (Harushima et al., 1998). The centromeric position on the linkage map was previously determined by assigning individual genetically mapped DNA markers to the short or long arm using cytogenetic stocks (Singh et al., 1996; Harushima et al., 1998). We analyzed the positions of the restriction fragment length polymorphism (RFLP) markers mapped between 80.2 and 87 cM on the current chromosome 3 pseudomolecule and narrowed down the recombination-suppressed domain to the 86-cM region between RFLP markers L708 and R3235 (Figure 2). A total of 11 cosegregating RFLP markers were mapped within this region, together with the CentO array and the centromeric gap on the sequence map (Figure 1A). We refer to this region with <1/186 recombinants as the recombination-suppressed domain. The size of this recombination-suppressed domain is estimated to be 3113 kb, spanning the entire 1881-kb CENH3 binding domain. The recombination-suppressed domain also included 927 and 305 kb, respectively, flanking the short and long arm sides of the CENH3 binding domain (Figure 2).
Figure 2.
Genetic Position of Cen3.
The genetic position of Cen3 was previously mapped between 80.2 and 87 cM (Harushima et al., 1998) and is narrowed down to 86 cM. The sequenced region corresponding to 86 cM is demarcated by RFLP markers L708 and R3235. This region spans ∼3113 kb, including the centromeric gap, and contains 11 cosegregated RFLP markers. The ∼1881-kb CENH3 binding domain (in yellow) is embedded within this region.
Cen3 Contains a Low Density of Genes Compared with Pericentromeric Regions
We manually annotated protein-coding gene models using publicly available rice ESTs and full-length cDNAs (fl-cDNAs) and proteins from rice and other organisms, in combination with ab initio predictions. We identified 127 protein-coding genes in the 2275-kb virtual contig, excluding genes related to transposable elements, pseudogenes, noncoding genes (protein coding sequence <153 bp), and partial genes. Sixty-five genes showed evidence of expression activity as these genes matched spliced (47) or unspliced (18) cDNA transcripts in GenBank (cutoff criteria of >95% identity and >93% coverage). The putative function can be determined for 36 active genes on the basis of their similarity to entries in the UniRef100 database (http://www.pir.uniprot.org/). The remaining 62 genes are predicted based on similarities to annotated proteins (7) or solely on computational predictions (55) (see Supplemental Figure 1 and Supplemental Table 2 online). Collectively, these genes represent 14% of the sequence in the virtual contig, with a density of 17.9 kb per gene.
Among the 65 genes with cDNA support in the 2275-kb virtual contig, 19 of these expressed genes were detected within the 1881-kb CENH3 binding domain. BLASTP searches against the Arabidopsis proteome (The Institute for Genomic Research [TIGR] V5, http://www.tigr.org/tdb/e2k1/ath1/) identified significant Arabidopsis homologs for nine of these 19 active genes (see Supplemental Table 3 online). Genes Cen3.t01107 (no. 52) and Cen3.t01160 (no. 53), the two genes closest to the centromeric gap, are only 9453 and 9411 bp away from the nearest CentO arrays (see Supplemental Figure 1 online). Both genes match spliced transcripts in the rice EST and fl-cDNA databases; these two genes have protein coding sequences of 768 and 3291 bp, respectively. We conducted RT-PCR analysis to confirm the expression of these two genes. Gene Cen3.t01160 and all three models from gene Cen3.t01107 were expressed in all seven different tissues/treatments (Figure 3), demonstrating that these two genes are transcriptionally competent in spite of their proximity to a major centromeric satellite array.
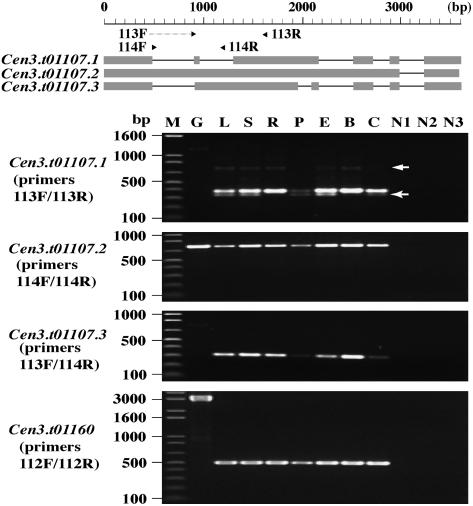
Figure 3.
RT-PCR Verification of Expression for Genes Cen3.t01107 and Cen3.t01160 in Cen3.
Gene Cen3.t01107 covers 3607 bp and has three splicing isoforms (Cen3.t01107.1 to Cen3.t01107.3) as predicted from existing transcript sequences. Two forward and two reverse primers were designed to differentiate these three isoforms; primer 113F was from an exon junction, but 113F/113R and 113F/114R still yielded very weak amplifications on genomic DNA for Cen3.t01107.1 and Cen3.t01107.3. Primer pair 113F/113R amplified the expected 383-bp fragment from all seven cDNA populations that corresponds to gene model Cen3.t01107.1, plus another two weak bands of 741 and ∼320 bp, respectively (arrows), while the 741-bp band represents the amplification product for gene model Cen3.t01107.3; the smaller band of 320 bp that is detected in five cDNA populations (not in cDNA samples from roots and buds) most likely represents a new isoform not yet identified. Gene models Cen3.t01107.2 and Cen3.t01107.3 are both transcribed in all seven tissues/treatments, with the sizes of the amplified products matching the annotation. Gene Cen3.t01160 has 11 exons totaling 7789 bp, and primers designed from exons 2 and 6 yielded amplification products in all seven cDNA samples. M, molecular marker; G, genomic DNA; L, cDNA from leaves; S, cDNA from shoots; R, cDNA from roots; P, cDNA from panicles; E, cDNA from etiolated leaves/shoots; B, cDNA from buds; C, cDNA from calli; N1 (L+S), N2 (R+P), and N3 (E+B+C), negative controls without adding reverse transcriptase (two or three negative controls for each primer pair were loaded to the same lane).
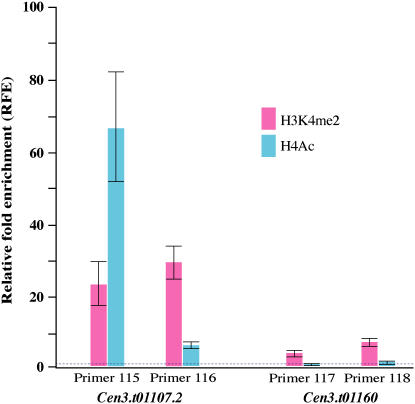
To reveal the chromatin status of these two active genes, we conducted ChIP analysis using antibodies against histone H3 dimethylation at Lys 4 (H3K4me2) and H4 acetylation (H4Ac) that are mostly associated with active euchromatin (Jenuwein and Allis, 2001). Using exon-derived primers in a real-time PCR assay, we detected high levels of enrichment of both H3K4me2 and H4Ac for gene Cen3.t01107.2 and less but significant (∝ < 0.05) enrichment of H3K4me2 for gene Cen3.t01160 (Figure 4). The ChIP results showed that these two active genes are associated with euchromatic features.
Figure 4.
H3K4me2 and H4Ac Associated with Two Cen3 Genes That Are <10 kb Away from the CentO Array.
Two primer sets were designed from exon 1 (primers 115 and 116) of gene Cen3.t01107.2 and from exon 4 (primer 117) and exon 11 (primer 118) of gene Cen3.t01160. Relative fold enrichment (RFE) is calculated from three replicates and indicated by the height of the bar with standard error; the baseline (RFE = 1; dashed line) represented the RFE of reference gene Cen8.t00238 that shows no H3K4me2 and no H4Ac (Yan et al., 2005).
The CENH3 binding domain has a highly reduced density of genes (29.1 kb per gene, with genes corresponding to 8.2% of the sequence) compared with a density of 11.1 kb per gene (23.4% of the sequence) for the regions that flank the CENH3 binding domain in the virtual contig (Table 2). Overall, the gene density in the Cen3 virtual contig is significantly lower than that of the rest of the chromosome, which has one gene every 7.1 kb, or accounts for 38.6% of the sequence (TIGR Osa1 Release 3; Yuan et al., 2005).
Table 2.
Gene Distribution in the CENH3 Binding Domain and Its Flanking Domains
| Short Arm Flanking Domain | CENH3 Binding Domain | Long Arm Flanking Domain | |
|---|---|---|---|
| Coordinates (kb) in the Cen3 virtual contig | 1–389 | 389–1786 | 1786–2275 |
| Size (kb) | 389 | 1397 | 489 |
| Genes with EST/fl-cDNA | 19 | 19 | 27 |
| Genes without EST/fl-cDNA | 9 | 29 | 24 |
| Total number of genes | 28 | 48 | 51 |
| Gene density (kb/gene) | 13.9 | 29.1 | 9.6 |
mRNA and Small RNA Signatures Associated with Cen3
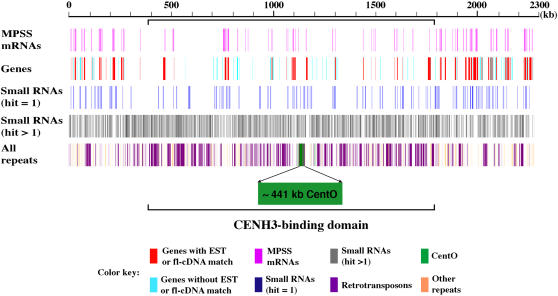
To find additional evidence for transcription within Cen3, we compared the ∼2.3-Mb Cen3 virtual contig to our collection of 17-bp signatures generated by massively parallel signature sequencing (MPSS) (Brenner et al., 2000). These data represent tags derived from the 3′ ends of transcripts of 22 rice mRNA libraries (http://mpss.udel.edu/rice). We found 504 distinct mRNA signatures that have perfect matches in Cen3, including 161 signatures that matched to unique sequences. Comparison between these 161 Cen3-specific signatures and the annotated genes revealed that 81% of the signatures were mapped to genes, with the other 19% mapped to the intergenic regions (Figure 5, Table 3). More than three-quarters of these 161 signatures mapped to genes demonstrated to be transcribed based on fl-cDNA/EST data, including 11 signatures mapped to introns and 28 to the antisense strand of exons (Table 3). In addition, the MPSS data indicated transcriptional activity for an additional four Cen3 genes that were previously not recognized as transcribed based on EST and/or fl-cDNA evidence. The MPSS data also suggested that as many as 17 previously unannotated transcripts may be produced from the intergenic regions that were >500 bp away from the annotated Cen3 genes (Table 3). Combined with the cDNA sequences, these data suggest substantial mRNA transcription in the Cen3 region, although at a lower density than the rest of the chromosome.
Figure 5.
Mapping of MPSS mRNA and Small RNA Signatures to the ∼2.3-Mb Cen3 Virtual Contig.
Most of the single-hit MPSS mRNA signatures were mapped to regions annotated as active genes, while single-hit small RNA signatures were largely found within intergenic regions and tended to cluster to discrete loci. The distribution of small RNAs with at least two genome hits is generally correlated with that of repetitive DNA.
Table 3.
Distribution of Single-Hit MPSS mRNA and Small RNA Signatures in Cen3
| MPSS mRNA Signatures
|
MPSS Small RNA Signatures
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Category | No. of Genes | No. of Signatures | Exon | Intron | No. of Genes | No. of Signatures | Exon | Intron | No. of Genes |
| Genes with EST/fl-cDNA | 65 | 126 | 115 (28)a | 11 (3)a | 52 | 8 | 2 (1) | 6 | 6 |
| Genes without EST/fl-cDNA | 62 | 4 | 4 (1) | 0 | 4 | 16 | 10 (4) | 6 (4) | 13 |
| Intergenic | 31b | 192c | |||||||
| Total signatures | 161 | 216 | |||||||
Numbers in parentheses represent matches to the antisense strand of annotated genes. Note that the data in this table indicate only signatures with a single match in the genome (excluding duplicated signatures).
Two signatures mapped to the antisense strand in exon (1) or intron (1) matched antisense transcripts in public databases.
Nine signatures match noncoding Cen3 transcripts, one matches a transcribed partial gene, and the other 21 do not match any existing Cen3 transcripts, including four located within 150 bp of the 3′ end and 17 located >500 bp away from genes.
The 159 signatures are >500 bp away from annotated genes.
We also surveyed small RNA signatures that match the Cen3 sequence using an MPSS-based method described previously (Lu et al., 2005). From two libraries representing flower and seedling tissues, we detected 7062 distinct small RNA signatures mapped to Cen3; however, only 216 (3%) of these signatures are Cen3 specific, matching nowhere else in the rice genome. In contrast with the mRNA signatures that were mostly located within genes, 89% of these single-hit small RNAs were mapped to the intergenic regions (Table 3). Among the 216 single-hit small RNAs, 116 were >500 bp away from one another and appeared to be dispersed throughout Cen3, while 87 were organized as 32 clusters (a minimum of two signatures overlapped or separated by <150 bp) with signatures from 18 clusters matching both strands of Cen3, suggesting the presence of specific elements as sources and/or targets of small RNAs. Each of the remaining 6846 signatures had perfect matches to two or more locations in the genome, so the exact source of these small RNAs cannot be determined.
To correlate these repetitive signatures to the sequence features in Cen3, we compared the distribution of these 6846 small RNA sequences to predictions of repetitive DNA found using RepeatMasker (see below for details). A total of 5071 small RNA signatures had matches to Cen3 regions annotated as repetitive DNA, with 80% of the matched repetitive sequences identified as retrotransposons (Figure 5). The other 20% of the 5071 small RNAs matched to regions primarily consisting of miniature inverted repeat transposable elements (MITEs), transposons, or unclassified repeats. We found another 1420 signatures that did not match to known repetitive DNA in Cen3 (as defined by RepeatMasker) but matched other repetitive genomic sequences found in >20 copies throughout the rice genome (see below for details). Based on this analysis, we estimated that ∼95% (5071 + 1420) of the 6846 small RNA signatures are likely to be derived from repetitive sequences in the rice genome. Because 95% of the Cen3-specific single-hit small RNAs and 91% of the remaining Cen3-matching small RNAs are found at low abundances (≤10 transcripts per 250,000 in both libraries), it is likely that nearly all of them are functioning as short interfering RNAs (siRNAs) rather than microRNAs. Therefore, most of the small RNAs matching to Cen3 are likely siRNAs that are acting in trans for repeats distributed across the genome, while a small number of sequences unique to Cen3 are both the sources and targets of additional siRNAs.
Repetitive Sequences in Cen3
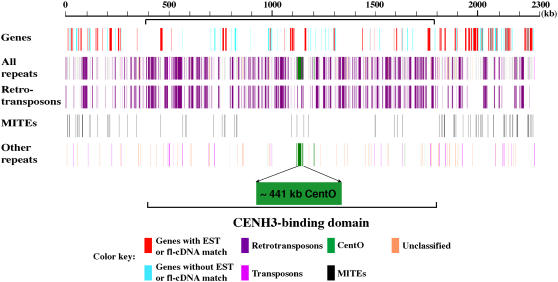
We used RepeatMasker (http://www.repeatmasker.org/) to identify repetitive DNA in the 2.3-Mb Cen3 virtual contig using the TIGR Oryza repeat database (Ouyang and Buell, 2004) as the filter. This analysis identified 857,485 bp of repetitive DNA, accounting for 37.7% of the Cen3 virtual contig (see Supplemental Table 4 online), which is higher than the chromosome 3 average of 21.4% (Rice Chromosome 3 Sequencing Consortium, 2005). The predominant type of repetitive sequences identified was retrotransposons, which account for 30.3% of the Cen3 virtual contig. The Ty3-gypsy class of retrotransposon represents 17.2% of the Cen3 virtual contig, followed by MITEs (2.7%). Approximately 51% of the 1397-kb assembled sequences in the CENH3 binding domain are repetitive in nature. The CENH3 binding domain is clearly more enriched with repetitive DNA sequences than its flanking domains (Figure 6).
Figure 6.
Repetitive DNA Sequences Associated with the ∼2.3-Mb Cen3 Virtual Contig.
All repeats were plotted against their coordinates on the virtual contig, showing that most repetitive DNA sequences reside within intergenic regions. Repetitive DNA, excluding MITEs, is more concentrated in the CENH3 binding domain.
It is likely that the estimated 37.7% of repetitive DNA is an underestimation of the actual content of repetitive DNA in Cen3 because some repetitive DNA elements are likely missing in the TIGR Oryza repeat database. To confirm this hypothesis, we binned the repeat-masked Cen3 sequence into 60-bp fragments, with a sliding window size of 20 bp, and aligned these fragments with the TIGR Osa1 version 3 pseudomolecule. We found that ∼516 kb of Cen3 sequence has ≥20 copies in the genome, based on the cutoff of a minimum aligned length of 40 bp. These newly identified multiple-copy sequences most likely represent highly diverged repetitive DNA sequences undetected by the RepeatMasker program or yet to be defined novel repetitive DNA elements. Indeed, the experimental data provided by the small RNA analysis show many siRNAs with multiple matches to the genome corresponding to regions not detected by RepeatMasker (Figure 5).
We plotted the Cen3 genes and repetitive sequences along the Cen3 virtual contig, which clearly revealed the abundance of repetitive DNA and paucity of genes in the CENH3 binding domain (Figure 6). Repetitive DNA mostly occupies regions devoid of genes, while MITEs are more abundant in the flanking regions that have a much higher density of genes, in agreement with their preferential association with genes (Jiang et al., 2004). MITEs account for 4.95% of the sequences in the flanking regions versus only 1.22% in the CENH3 binding domain.
DISCUSSION
Comparative genome mapping has revealed that the positions of centromeres are dynamic during evolution (Murphy et al., 2005). This centromere repositioning phenomenon has been well documented in mammalian species. Ventura et al. (2001) demonstrated the first such centromere reposition case involved in the X chromosome of mammalian species. The genetic synteny of the X chromosomes of black lemur (Eulemur macaco), ringtailed-lemur (Lemur catta), and humans is highly conserved. However, the centromeres of the three X chromosomes are located in different positions and contain different centromeric satellite repeats (Ventura et al., 2001). This result suggests that emergence of new centromeres, rather than chromosome rearrangements, is the most likely explanation of centromere repositioning during X chromosome evolution. Similar centromere repositioning has since been demonstrated for several other mammalian chromosomes based on the comparative mapping results (Eder et al., 2003; Ventura et al., 2004; O'Neill et al., 2005).
Centromere repositioning is likely achieved via neocentromere activation (Figure 7). More than 70 human neocentromeres have been reported (Amor and Choo, 2002; Warburton, 2004). These neocentromeres are located all over the human genome (Warburton, 2004), indicating that a significant portion of the human genome is competent for neocentromere activation. Human neocentromeres are fully functional and can be stably maintained through both mitosis and meiosis (Tyler-Smith et al., 1999; Amor et al., 2004b). Most importantly, although neocentromere formation results in substantial remodeling of chromatin associated with the functional centromeric domain, a recent study demonstrated that neocentromere formation has no measurable effect on transcriptional competency of the underlying genes or accessibility of the cell transcriptional machinery to this DNA (Saffery et al., 2003). Thus, repositioning of a centromere into a genic region is not detrimental to the transcription of the genes. Neocentromere activation has also been reported recently in a plant species (Nasuda et al., 2005).
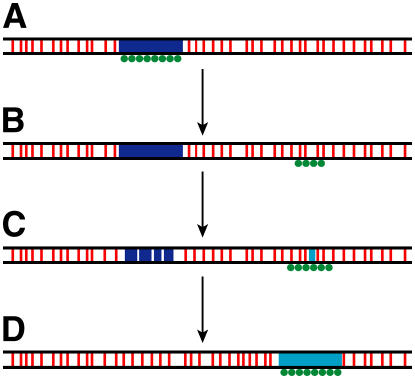
Figure 7.
Model of Centromere Evolution.
(A) The original centromere contains mainly satellite repeats (blue box) that are associated with CENH3 (green circles).
(B) A neocentromere emerges, coupled with the inactivation of the original centromere via unknown epigenetic mechanisms. The CENH3-associated chromatin in the neocentomere may contain active genes (red bars).
(C) A new satellite repeat (turquoise box) emerges in the neocentromere. The satellite array in the inactivated centromere may degrade and eventually be eliminated during evolution.
(D) The new satellite repeat array expands, and the survived neocentromere evolves to eventually become a typical satellite repeat-based mature centromere.
Results of centromere repositioning and neocentromere activation suggest that centromeres are evolved from neocentromeres (Figure 7). Neocentromeres may land in regions that are typical to the rest of the genome and may contain active genes. Coexistence of a non-α satellite–based neocentromere with the α satellite–based but inactivated original centromere has been documented in humans (Tyler-Smith et al., 1999; Amor et al., 2004b). During evolution, the satellite repeats from other centromeres may invade the emergent neocentromeres via currently unknown mechanisms. Alternatively, new satellite repeats may emerge in the neocentromeres (Figure 7). The starting satellite repeat array in the emergent neocentromere may be short, such as the CentO array in rice Cen8, but may ultimately expand via mechanisms that govern tandem repeat evolution (Charlesworth et al., 1986). Expansion of the satellite arrays will be favorable to the survival of the neocentromeres as it may attract more CENH3, thus binding more microtubules during cell division (Henikoff et al., 2001). Rice Cen3 and Cen8 may eventually accumulate megabase-sized CentO arrays. Interestingly, the Cen3 and Cen8 in Oryza punctata, a wild species related to cultivated rice, contain 1.47- and 1.44-Mb arrays of the CentO repeats that almost perfectly overlay with CENH3 (Zhang et al., 2005).
The discovery of the active genes in the CENH3 binding domain of rice Cen8 was surprising since centromeres in most eukaryotes contain only repetitive DNA sequences. In this study, we demonstrate that rice Cen3 also contains active genes. Two of the Cen3 genes reside <10 kb from the centromeric satellite array. These genes are normally expressed in different tissues and show typical euchromatic histone modification patterns (Figures 3 and 4). Profiling of Cen3-derived MPSS mRNA and small RNA signatures revealed the complexity of the transcriptome for this centromere, including antisense transcription, transcription in intergenic regions, and potential transcription of repetitive DNA elements within Cen3. Some of these transcripts are apparently processed into small RNAs. Our findings of transcription in rice centromeres are consistent with several recent reports that the CENH3 binding domains are associated with euchromatic features, such as H3K4 methylation, and are distinct from classical heterochromatin (Cam et al., 2005; Lam et al., 2006). Rice Cen3, similar to Cen8, may also represent an intermediate stage in the evolution from a genic region to a repeat-based mature centromere.
The CENP-A (human CENH3) binding domains of several human neocentromeres in mammalian species have been determined to range from ∼130 to 460 kb (Lo et al., 2001a, 2001b; Alonso et al., 2003). By contrast, the CENP-A binding domain in normal human centromeres was estimated to be between 540 and 1700 kb (Irvine et al., 2004). Quantitative analysis of CENP-A incorporation into centromeres showed that three human neocentromeres contain an average of 32 to 45% CENP-A of the mean of all human centromeres (Irvine et al., 2004). These results indicate that newly emerged neocentromeres may have smaller CENH3 binding domains and/or less effective deposition of CENH3 than the native centromeres. During evolution, the new centromeres will expand the CENH3 binding domains by acquiring large blocks of satellite DNA that are specific to the native centromeres. This would support a similar model in which the accumulation of satellite repeats may be a driving force to achieve the formation of a larger and functionally more stable kinetochore (Irvine et al., 2004).
METHODS
Fiber-FISH
Fiber-FISH was performed according to published protocols (Jackson et al., 1998). DNA probes were labeled with biotin-dUTP or digoxigenin-dUTP (Roche). Images were captured digitally using a Sensys CCD camera (Roper Scientific). The camera control and imaging analysis were performed using IPLab Spectrum v3.1 software (Signal Analytics). Plasmid clone pRCS2 (Dong et al., 1998) was used as a probe to detect the CentO satellite.
DNA Sequence Analysis
We constructed a virtual sequence contig to cover the centromere of rice (Oryza sativa) chromosome 3. The short arm side of this virtual contig consists of a minimal tiling path of nine BAC clones, while the long arm side of the virtual contig contains 10 BAC clones (one clone is located within the interval of 86 to 86.5 cM). We downloaded sequences from GenBank for the nine clones on the short arm side of the gap and 10 clones on the long arm side. We assembled them separately into two virtual contigs of 1,123,026 and 1,151,221 bp, respectively, and merged them into a single virtual contig of 2,275,221 bp by inserting a string of Ns of 974 bp to delineate the gap. Gene annotation and repeat analysis were performed using published procedures (Yan et al., 2005). Repetitive DNA was identified using RepeatMasker with the TIGR Oryza repeat database, http://www.tigr.org/tdb/e2k1/plant.repeats/ (Ouyang and Buell, 2004), as the filter. The cutoff is score ≥225 and divergence ≤35%.
MPSS was performed essentially as described (Brenner et al., 2000). The mRNA data were analyzed using the methods described previously (Meyers et al., 2004). The small RNA libraries were constructed as previously described (Lu et al., 2005). For both data sets, we compared MPSS signatures to the rice centromere sequence and assigned signatures to each location at which a perfect match was found. The number of matches in the centromere and then across the genome was recorded as the hits. All plant material was from O. sativa cv Nipponbare. For the small RNA libraries, inflorescence tissue was harvested from plants grown in soil in a growth chamber with 12-h light and dark cycle, with 80% relative humidity in the day and 60% in the night for 13 weeks at 26°C day and 20°C night temperatures. Floral tissue included the early stage panicles (immature panicles before emergence from shoots). Total RNA was isolated using Trizol reagents (Invitrogen). Seedlings used for the leaf samples were grown under the same conditions and were harvested after 2 weeks.
The raw and normalized MPSS data are available at http://mpss.udel.edu/rice, and these data were deposited in the Gene Expression Omnibus database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/). Our website allows users to query these databases on physical location, gene identifiers, or by sequence.
ChIP
ChIP using the rice anti-CENH3 antibody was performed according to Nagaki et al. (2004). We designed ChIP-PCR primers with amplification products between 154 and 343 bp. Quantitative real-time PCR analysis was used to determine the relative enrichment of CENH3-associated sequences in the bound fraction over the mock control. PCR reactions were performed in duplicates using the DyNAmo HS SYBR Green qPCR kit (MJ Research) and run at 95°C for 15 min, 45 cycles of 95°C for 10 s, 63 to 65°C for 30 s, and 72°C for 30 s. The cycle threshold (Ct) was taken with the baseline of fluorescence intensity manually set at a value between 0.05 and 0.1. For each primer pair, we calculated the RFE of the amplified product using comparative CT method according to Saffery et al. (2003). The RFE value of each sequence was normalized using manganese transport protein (located at 122.8 cM on chromosome 3) as the reference gene, whose RFE value was set at 1. Primer sequences of the reference gene are 5′-GGGGCAACCTAGATCAGGGGTGTCTCC-3′ (forward) and 5′-AAAGCCCTCCTGTTGTGACCCAAGCAG-3′ (reverse). ChIP analysis using antibodies against histone H3 dimethylation at Lys 4 and H4 acetylation were performed as described (Yan et al., 2005). Two active genes, Cen3.t01107 (represented by Cen3.t01107.2) and Cen3.t01160, were tested for these histone modifications. Two sets of primers (see Supplemental Table 1 online) were designed from exons of each of the two genes. The negative control is a NB-ARC domain–containing gene (Cen8.t00238) from the centromere of rice chromosome 8 (Yan et al., 2005). Significance of enrichment difference between antibody binding fraction and mock treatment was tested for each primer set using Student's t test (α = 0.05). RFE was then calculated as 2−ΔΔCT (Yan et al., 2005).
RT-PCR
We used RT-PCR to test the expression of genes Cen3.t01107 and Cen3.t01160. Primers (see Supplemental Table 1 online) were between 24- and 28-bp long, with an annealing temperature from 65.4 to 71.3°C. We isolated mRNA from seven different rice tissues or treatments for RT-PCR, including (1) leaves, (2) shoots, (3) roots collected from 14-d-old plants growing in Biotron greenhouse, (4) etiolated leaves/shoots collected from 14-d-old plants growing in the darkness in the same Biotron greenhouse, (5) panicles collected 3 d after flowering, (6) calli that were induced by keeping seeds on Murashige and Skoog (MS) medium with a supplement of 2 mg/L 2,4-D for 22 d and collected after another 12 d of growing on new MS medium, and (7) buds collected from germinated seeds on quarter-strength MS medium.
Accession Numbers
Sequence data from this article can be found in the Gene Expression Omnibus database at NCBI (http://www.ncbi.nlm.nih.gov/geo/) under the following accession numbers: the platform identifiers are GPL3776 (small RNA) and GPL3777 (mRNA), and sample identifiers are GSM109198 (small RNA) to GSM109200 (mRNA).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Distribution of Genes within the ∼2.3-Mb Cen3 Virtual Contig.
Supplemental Table 1. Primers Used in ChIP Analysis and RT-PCR.
Supplemental Table 2. Gene Models Annotated in the ∼2.3-Mb Cen3 Pseudomolecule.
Supplemental Table 3. Nineteen Transcribed Genes Identified in the CENH3 Binding Domain of Cen3
Supplemental Table 4. Summary of Repetitive Sequences in Cen3.
Supplementary Material
Acknowledgments
We thank Steve Henikoff and Chris Tyler-Smith for their critical comments on the manuscript. This research was supported by Grant FG02-01ER15266 from the Department of Energy (DOE) and by Grant 2006-35604-16649 from the USDA Cooperative State Research, Education, and Extension Service (CSREES) to J.J. and partly by rice chromosome 3 sequencing grants from the USDA-CSREES, the National Science Foundation (NSF), and the DOE to R.A.W., C.R.B., and J.J. The MPSS expression analyses were supported by NSF Award 0321437 (B.C.M. and G.-L.W.), NSF Small Grants for Exploratory Research Award 0439186 (P.J.G. and B.C.M.), and USDA-CSREES 2005-35604-15326 (B.C.M. and P.J.G.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jiming Jiang (jjiang1@wisc.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso, A., Mahmood, R., Li, S., Cheung, F., Yoda, K., and Warburton, P.E. (2003). Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum. Mol. Genet. 12 2711–2721. [DOI] [PubMed] [Google Scholar]
- Amor, D.J., Bentley, K., Ryan, J., Perry, J., Wong, L., Slater, H., and Choo, K.H.A. (2004. b). Human centromere repositioning “in progress”. Proc. Natl. Acad. Sci. USA 101 6542–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor, D.J., and Choo, K.H.A. (2002). Neocentromeres: Role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 71 695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor, D.J., Kalitsis, P., Sumer, H., and Choo, K.H.A. (2004. a). Building the centromere: From foundation proteins to 3D organization. Trends Cell Biol. 14 359–368. [DOI] [PubMed] [Google Scholar]
- Brenner, S., et al. (2000). Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 18 630–634. [DOI] [PubMed] [Google Scholar]
- Cam, H.P., Sugiyama, T., Chen, E.S., Chen, X., FitzGerald, P.C., and Grewal, S.I.S. (2005). Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37 809–819. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., Langley, C.H., and Stephan, W. (1986). The evolution of restricted recombination and the accumulation of repeated DNA sequences. Genetics 112 947–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z.K., Dong, F., Langdon, T., Ouyang, S., Buell, C.B., Gu, M.H., Blattner, F.R., and Jiang, J. (2002). Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14 1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, L. (1998). Centromeres: Proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Curr. Opin. Genet. Dev. 8 212–218. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G.P., et al. (1999). Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286 2468–2474. [DOI] [PubMed] [Google Scholar]
- Dong, F., Miller, J.T., Jackson, S.A., Wang, G.L., Ronald, P.C., and Jiang, J. (1998). Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc. Natl. Acad. Sci. USA 95 8135–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder, V., Ventura, M., Ianigro, M., Teti, M., Rocchi, M., and Archidiacono, N. (2003). Chromosome 6 phylogeny in primates and centromere repositioning. Mol. Biol. Evol. 20 1506–1512. [DOI] [PubMed] [Google Scholar]
- Hall, S.E., Kettler, G., and Preuss, D. (2003). Centromere satellites from Arabidopsis populations: Maintenance of conserved and variable domains. Genome Res. 13 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, J.J., Bokkelen, G.V., Mays, R.W., Gustashaw, K., and Willard, H.F. (1997). Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 15 345–355. [DOI] [PubMed] [Google Scholar]
- Harushima, Y., et al. (1998). A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S. (2002). Near the edge of a chromosome's ‘black hole’. Trends Genet. 18 165–167. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., and Malik, H.S. (2001). The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293 1098–1102. [DOI] [PubMed] [Google Scholar]
- Henning, K.A., Novotny, E.A., Compton, S.T., Guan, X.-Y., Liu, P.P., and Ashlock, M.A. (1999). Human artificial chromosomes generated by modification of a yeast artificial chromosome containing both human alpha satellite and single-copy DNA sequences. Proc. Natl. Acad. Sci. USA 96 592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison, J.S., Murata, M., Ogura, Y., Schwarzacher, T., and Motoyoshi, F. (1999). Polymorphisms and genomic organization of repetitive DNA from centromeric regions of Arabidopsis chromosomes. Plant Cell 11 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosouchi, T., Kumekawa, N., Tsuruoka, H., and Kotani, H. (2002). Physical map-based sizes of the centromeric regions of Arabidopsis thaliana chromosomes 1, 2, and 3. DNA Res. 9 117–121. [DOI] [PubMed] [Google Scholar]
- Houben, A., and Schubert, I. (2003). DNA and proteins of plant centromeres. Curr. Opin. Plant Biol. 6 554–560. [DOI] [PubMed] [Google Scholar]
- Ikeno, M., Grimes, B., Okazaki, T., Nakano, M., Saitoh, K., Hoshino, H., McGill, N.I., Cooke, H., and Masumoto, H. (1998). Construction of YAC-based mammalian artificial chromosomes. Nat. Biotechnol. 16 431–439. [DOI] [PubMed] [Google Scholar]
- Irvine, D.V., Amor, D.J., Perry, J., Sirvent, N., Pedeutour, F., Choo, K.H.A., and Saffery, R. (2004). Chromosome size and origin as determinants of the level of CENP-A incorporation into human centromeres. Chromosome Res. 12 805–815. [DOI] [PubMed] [Google Scholar]
- Jackson, S.A., Wang, M.L., Goodman, H.M., and Jiang, J. (1998). Application of fiber-FISH in physical mapping of Arabidopsis thaliana. Genome 41 566–572. [PubMed] [Google Scholar]
- Jenuwein, T., and Allis, C.D. (2001). Translating the histone code. Science 293 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jiang, N., Feschotte, C., Zhang, X., and Wessler, S.R. (2004). Using rice to understand the origin and amplification of miniature inverted repeat transposable element (MITEs). Curr. Opin. Plant Biol. 7 115–119. [DOI] [PubMed] [Google Scholar]
- Kumekawa, N., Hosouchi, T., Tsuruoka, H., and Kotani, H. (2000). The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 5. DNA Res. 7 315–321. [DOI] [PubMed] [Google Scholar]
- Kumekawa, N., Hosouchi, T., Tsuruoka, H., and Kotani, H. (2001). The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 4. DNA Res. 8 285–290. [DOI] [PubMed] [Google Scholar]
- Lam, A.L., Boivin, C.D., Bonney, C.F., Rudd, M.K., and Sullivan, B.A. (2006). Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc. Natl. Acad. Sci. USA 103 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J.C., Kato, A., and Birchler, J.A. (2005). Sequences associated with A chromosome centromeres are present throughout the maize B chromosome. Chromosoma 113 337–349. [DOI] [PubMed] [Google Scholar]
- Lo, A.W., Craig, J.M., Saffery, R., Kalitsis, P., Irvine, D.V., Earle, E., Magliano, D.J., and Choo, K.H. (2001. a). A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 20 2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, A.W.I., Magliano, D.J., Sibson, M.C., Kalitsis, P., Craig, J.M., and Choo, K.H.A. (2001. b). A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 11 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C., Tej, S.S., Luo, S.J., Haudenschild, C.D., Meyers, B.C., and Green, P.J. (2005). Elucidation of the small RNA component of the transcriptome. Science 309 1567–1569. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C., Tej, S.S., Vu, T.H., Haudenschild, C.D., Agrawal, V., Edberg, S.B., Ghazal, H., and Decola, S. (2004). The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res. 14 1641–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J.T., Dong, F., Jackson, S.A., Song, J., and Jiang, J. (1998). Retrotransposon-related DNA sequences in the centromeres of grass chromosomes. Genetics 150 1615–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, W.J., et al. (2005). Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 309 613–617. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Cheng, Z.K., Ouyang, S., Talbert, P.B., Kim, M., Jones, K.M., Henikoff, S., Buell, C.R., and Jiang, J. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36 138–145. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Neumann, P., Zhang, D., Ouyang, S., Buell, C.R., Cheng, Z., and Jiang, J. (2005). Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice. Mol. Biol. Evol. 22 845–855. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Talbert, P.B., Zhong, C.X., Dawe, R.K., Henikoff, S., and Jiang, J.M. (2003). Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasuda, S., Hudakova, S., Schubert, I., Houben, A., and Endo, T.R. (2005). Stable barley chromosomes without centromeric repeats. Proc. Natl. Acad. Sci. USA 102 9842–9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakey, R., and Tyler-Smith, C. (1990). Y chromosome DNA haplotyping suggests that most European and Asian men are descended from one of two males. Genomics 7 325–330. [DOI] [PubMed] [Google Scholar]
- O'Neill, R.J., Eldridge, M.D.B., and Metcalfe, C.J. (2005). Centromere dynamics and chromosome evolution in marsupials. J. Hered. 95 375–381. [DOI] [PubMed] [Google Scholar]
- Ouyang, S., and Buell, C.R. (2004). The TIGR Plant Repeat Databases: A collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 32 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Chromosome 3 Sequencing Consortium (2005). Sequence, annotation, and analysis of synteny between rice chromosome 3 and diverged grass species. Genome Res. 15 1284–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round, E.K., Flowers, S.K., and Richards, E.J. (1997). Arabidopsis thaliana centromere regions: Genetic map positions and repetitive DNA structure. Genome Res. 7 1045–1053. [DOI] [PubMed] [Google Scholar]
- Rudd, M.K., and Willard, H.F. (2004). Analysis of the centromeric regions of the human genome assembly. Trends Genet. 20 529–533. [DOI] [PubMed] [Google Scholar]
- Saffery, R., Sumer, H., Hassan, S., Wong, L.H., Craig, J.M., Todokoro, K., Anderson, M., Stafford, A., and Choo, K.H.A. (2003). Transcription within a functional human centromere. Mol. Cell 12 509–516. [DOI] [PubMed] [Google Scholar]
- Schueler, M.G., Higgins, A.W., Rudd, M.K., Gustashaw, K., and Willard, H.F. (2001). Genomic and genetic definition of a functional human centromere. Science 294 109–115. [DOI] [PubMed] [Google Scholar]
- Shibata, F., and Murata, M. (2004). Differential localization of the centromere-specific proteins in the major centromeric satellite of Arabidopsis thaliana. J. Cell Sci. 117 2963–2970. [DOI] [PubMed] [Google Scholar]
- Singh, K., Ishii, T., Parco, A., Huang, N., Brar, D.S., and Khush, G.S. (1996). Centromere mapping and orientation of the molecular linkage map of rice (Oryza sativa L.). Proc. Natl. Acad. Sci. USA 93 6163–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X., Le, H.D., Wahlstrom, J.M., and Karpen, G.H. (2003). Sequence analysis of a functional Drosophila centromere. Genome Res. 13 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Smith, C., Gimelli, G., Giglio, S., Floridia, C., Pandya, A., Terzoli, G., Warburton, P.E., Earnshaw, W.C., and Zuffardi, O. (1999). Transmission of a fully functional human neocentromere through three generations. Am. J. Hum. Genet. 64 1440–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura, M., Archidiacono, N., and Rocchi, M. (2001). Centromere emergence in evolution. Genome Res. 11 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura, M., et al. (2004). Recurrent sites for new centromere seeding. Genome Res. 14 1696–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton, P.E. (2004). Chromosomal dynamics of human neocentromere formation. Chromosome Res. 12 617–626. [DOI] [PubMed] [Google Scholar]
- Wevrick, R., and Willard, H.F. (1989). Long-range organization of tandem arrays of a satellite DNA at the centromeres of human chromosomes: High frequency array-length polymorphism and meiotic stability. Proc. Natl. Acad. Sci. USA 86 9394–9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard, H.F. (1998). Centromeres: The missing link in the development of human artificial chromosomes. Curr. Opin. Genet. Dev. 8 219–225. [DOI] [PubMed] [Google Scholar]
- Wu, J.Z., et al. (2004). Composition and structure of the centromeric region of rice chromosome 8. Plant Cell 16 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H.H., Jin, W.W., Nagaki, K., Tian, S., Ouyang, S., Buell, C.R., Talbert, P.B., Henikoff, S., and Jiang, J. (2005). Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell 17 3227–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara, J.C., and Wakimoto, B.T. (2006). Oxymoron no more: The expanding world of heterochromatic genes. Trends Genet. 22 330–338. [DOI] [PubMed] [Google Scholar]
- Yuan, Q., Ouyang, S., Wang, A., Zhu, W., Maiti, R., Lin, H., Hamilton, J., Haas, B., Sultana, R., Cheung, F., Wortman, J., and Buell, C.R. (2005). The TIGR Osa1 rice genome annotation database. Plant Physiol. 138 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Yi, C., Bao, W., Liu, B., Cui, J., Yu, H., Cao, X., Gu, M., Liu, M., and Cheng, Z. (2005). The transcribed 165-bp CentO satellite is the major functional centromeric element in the wild rice species Oryza punctata. Plant Physiol. 138 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C.X., Marshall, J.B., Topp, C., Mroczek, R., Kato, A., Nagaki, K., Birchler, J.A., Jiang, J.M., and Dawe, R.K. (2002). Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.