Abstract
Plant invasion by pathogenic fungi involves regulated growth and highly organized fungal morphological changes. For instance, when the smut fungus Ustilago maydis infects maize (Zea mays), its dikaryotic infective filament is cell cycle arrested, and appressoria are differentiated prior to plant penetration. Once the filament enters the plant, the cell cycle block is released and fungal cells begin proliferation, suggesting a tight interaction between plant invasion and the cell cycle and morphogenesis control systems. We describe a novel factor, Biz1 (b-dependent zinc finger protein), which has two Cys2His2 zinc finger domains and nuclear localization, suggesting a transcriptional regulatory function. The deletion of biz1 shows no detectable phenotypic alterations during axenic growth. However, mutant cells show a severe reduction in appressoria formation and plant penetration, and those hyphae that invade the plant arrest their pathogenic development directly after plant penetration. biz1 is induced via the b-mating–type locus, the key control instance for pathogenic development. The gene is expressed at high levels throughout pathogenic development, which induces a G2 cell cycle arrest that is a direct consequence of the downregulation of the mitotic cyclin Clb1. Our data support a model in which Biz1 is involved in cell cycle arrest preceding plant penetration as well as in the induction of appressoria.
INTRODUCTION
Many phytopathogenic fungi are known to differentiate specific infection structures, called appressoria, that facilitate the penetration of the plant epidermal cell layers (Emmett and Parbery, 1975; Deising et al., 2000). In Magnaporthe grisea, Colletotrichum species, and many other plant pathogens, appressoria are visible as discrete, lobed, or dome-shaped cells that are separated from the germ tube by a septum. This dome-shaped cell generates enormous turgor pressure and physical force, allowing the fungus to breach the host cuticle and to invade the plant tissue (Mendgen et al., 1996; Talbot, 2003). In smut and rust fungi, appressoria are inconspicuous swellings of the germ tube; it is generally assumed that in these fungi cell wall–degrading enzymes, rather than force, aid the penetration of the invading hypha (Gold and Mendgen, 1991). Appressorial development and the penetration step are still poorly understood processes that require the integration of several environmental signals to produce the appropriate differentiation (Dean, 1997). Like other developmental decisions in fungi, appressorium formation must involve regulated growth, control of the cell cycle progression, and highly organized morphological changes.
One of the model systems to investigate the connections between cell cycle and morphogenesis in fungi during plant penetration is Ustilago maydis, a basidiomycete causing smut disease on maize (Zea mays) plants (Basse and Steinberg, 2004). Haploid cells (sporidia) of this fungus are unicellular and grow saprophytically by budding. The pathogenic form, the filamentous dikaryon, is established after fusion of two sporidia that have to harbor different alleles of the a- and b-mating–type loci of U. maydis. The a locus controls the cell fusion via a pheromone receptor–based system. Upon pheromone stimulation, cells arrest budding growth and start the formation of conjugation tubes (Spellig et al., 1994). These mating filaments undergo directed tip growth toward the pheromone source (Snetselaar et al., 1996) followed by cell fusion and the formation of dikaryotic hyphae. The subsequent steps in filament formation and pathogenic development are controlled by the multiallelic b-locus that encodes two distinct homeodomain transcription factors, bE and bW. A heterodimeric complex of the two proteins is formed when they are derived from different alleles, and the presence of this complex is sufficient to initiate pathogenic development (Kahmann and Kämper, 2004).
The dikaryon formed after the fusion of compatible sporidia is arrested in the G2 phase of the cell cycle; for further propagation, it requires plant signals that have not been identified yet. On the plant surface, the filaments differentiate appressoria and penetrate the cuticule (Snetselaar and Mims, 1992, 1993). In contrast with appressoria from other phytopathogenic fungi, such as M. grisea or Colletotrichum species (Bechinger et al., 1999; Talbot, 2003), appressoria of U. maydis are unmelanized, rather small swellings of the hyphal tip that form penetration structures that are less constricted (Snetselaar and Mims, 1993; Snetselaar et al., 2001). Since it is unlikely that entry of U. maydis occurs by mechanical force, it is believed that appressoria simply mark the point at which the growth direction changes. Once the filament enters the plant, the cell cycle is reactivated and the fungal cells proliferate to a network of filaments with septated cell compartments each containing a pair of nuclei (Snetselaar and Mims, 1992; Banuett and Herskowitz, 1996).
The morphological changes of U. maydis cells during the pathogenic process advocate for tight control of the cell cycle. Previous research efforts have defined networks of regulatory genes that control cell cycle progression (Castillo-Lluva et al., 2004; García-Muse et al., 2004; Castillo-Lluva and Pérez-Martín, 2005; Sgarlata and Pérez-Martín, 2005a, 2005b). As in other eukaryotic organisms, in U. maydis cyclin-dependent protein kinases (Cdks) are key regulators of the cell division cycle. Two distinct Cdk-cyclin complexes are responsible for the different cell cycle transitions in U. maydis. While Cdk1-Clb1 is required for the G1/S and the G2/M transitions, the Cdk1-Clb2 complex is specific for the G2/M transition (García-Muse et al., 2004). A third Cdk1-cyclin complex composed of Cdk1 and the G1-like cyclin Cln1 regulates the G1/S transition and has additional functions at the morphogenetic level (Castillo-Lluva and Pérez-Martín, 2005). Regulation of Cdk-cyclin activity throughout the different cell cycle phases is crucial. Inhibitory phosphorylation and activating dephosphorylation of the Cdk1 catalytic subunit are one of the major controls, and we have recently shown that this kind of regulation is crucial for cell cycle progression in U. maydis (Sgarlata and Pérez-Martín, 2005a, 2005b). In addition, proteolysis of cyclins is pivotal for cell cycle control. U. maydis mitotic cyclins are targeted by the anaphase promoting complex in association with different adaptor proteins, such as Cru1 (Castillo-Lluva et al., 2004) and Cdc20 (J. Torreblanca and J. Pérez-Martín, unpublished data).
Recently, the role of cell cycle regulators for pathogenic development of U. maydis has been addressed. Manipulation of the levels of mitotic cyclins either by affecting their rate of degradation or by alteration of their transcriptional regulation produce fungal cells that are unable to infect the plant (Castillo-Lluva et al., 2004; García-Muse et al., 2004; Castillo-Lluva and Pérez-Martín, 2005). These results stress the importance of a tight regulation of the Cdk-cyclin complexes through the pathogenic development of U. maydis.
In this work, we describe a novel transcriptional regulator, Biz1 (b-dependent zinc finger protein), that functions as a repressor for the mitotic cyclin Clb1. biz1 is induced after initiation of pathogenic development. The deletion of biz1 does not lead to any detectable phenotypic alterations during axenic growth. However, strains deleted for biz1 are completely apathogenic. The mutant cells show a low frequency of appressoria formation, and those that produce appressorium and invade the plant arrest their pathogenic development directly after plant penetration.
RESULTS
High Levels of biz1 Expression Induce a G2 Cell Cycle Arrest
To isolate novel regulators for the control of cell cycle and morphogenesis during pathogenic development, we performed a genetic screen devoted to the isolation of mutants affected in cell cycle arrest and/or polarity induction in response to pheromone stimulation (see Supplemental Results for a description of such a genetic screen). Serendipitously, in this screen we found that an open reading frame (ORF) of 783 amino acids, which corresponds to the predicted hypothetical protein um02549 annotated at the MIPS Ustilago maydis Database (http://mips.gsf.de/genre/proj/ustilago/), was able to induce cell cycle arrest and strong polar growth in cells expressing it (see below). We called the gene biz1 (for b-induced zinc finger; see below).
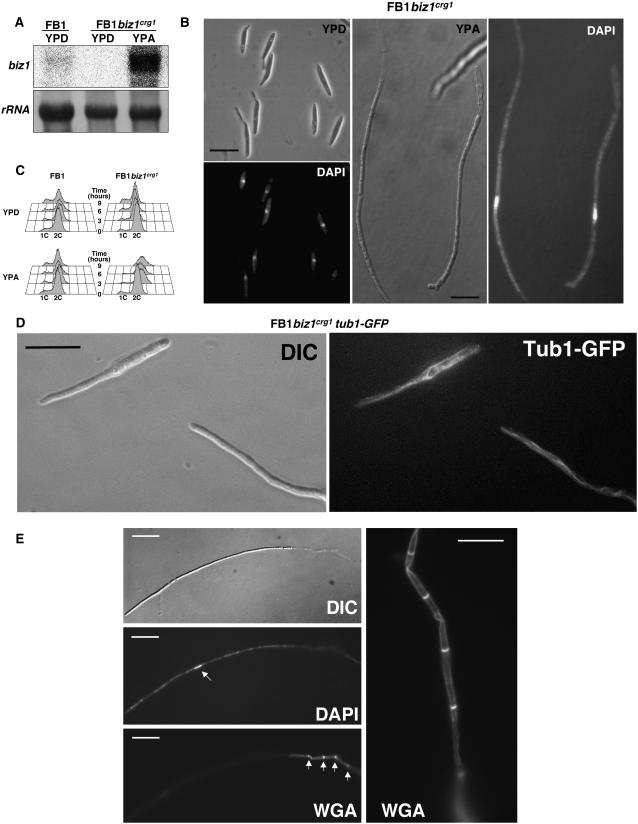
To analyze the functions of Biz1, we examined the effects of increased expression of biz1. For this, we replaced the endogenous promoter within the biz1 locus with the carbon source–regulated crg1 promoter (which is repressed by glucose and induced by arabinose; Bottin et al., 1996). The resulting biz1crg1 allele shows a clear carbon source–dependent pattern of expression (Figure 1A). Under repressing conditions (YPD), cells harboring the biz1crg1 allele were morphologically indistinguishable from wild-type strains. However, after transfer to arabinose-containing medium (YPA), cells expressing high levels of biz1 started to elongate (Figure 1B). We found that these elongated cells contained a single nucleus and long microtubules that reached to the tip of the growing pole (Figure 1D). This polar growth phenotype has been described for U. maydis cells arrested in the G2 phase (Steinberg et al., 2001; Banuett and Herskowitz, 2002). Consistently, fluorescence-activated cell sorter (FACS) analysis revealed that the cells have a 2C DNA content (Figure 1C). We did not observe cell divisions after induction of biz1crg1. Prolonged incubation (>24 h) under inducing conditions resulted in long filaments in which only the tip cell was filled with cytoplasm and the remaining part of the hyphae consisting of empty sections separated by septa (Figure 1E). Strikingly, these hyphae are reminiscent of the growth mode of the dikaryotic hyphae produced after mating. Under these conditions, cell division is arrested and cells show permanent polarized growth (Steinberg et al., 1998). We believe that this growth mode of U. maydis could be a default response to an arrested cell division. For instance, the conditional removal of clb1, encoding a mitotic cyclin, led to the same phenotype (García-Muse et al., 2004).
Figure 1.
Expression of biz1 Arrests the Cell Cycle in G2 Phase.
(A) Arabinose-induced expression of biz1. Wild-type FB1 and conditional UMN53 (FB1biz1crg1) strain were grown for 6 h in noninducing conditions (yeast extract-peptone medium amended with glucose [YPD]) or inducing conditions (yeast extract-peptone medium amended with arabinose [YPA]). The RNA was extracted and analyzed by RNA gel blotting, loading 10 μg total RNA per lane. A probe specific for 18s rRNA was used to control for loading.
(B) Micrographs showing the cell morphology of UMN53 cells after 12 h of growth in YPD (noninducing conditions) or YPA (inducing conditions) liquid cultures. Note the elongated shape and the presence of a single nucleus (4′,6-diamidino-2-phenylindole [DAPI] staining). Bars = 20 μm.
(C) FACS analysis of FB1 and UMN53 (FB1biz1crg1) to assess DNA content in noninducing (YPD) and inducing conditions (YPA). Samples were taken at 0, 3, 6, and 9 h after transfer to inducing conditions. Cells expressing biz1 arrest their growth at G2 and thereby accumulate with a 2C DNA content. The shift to DNA content higher than 2C observed in UMN53 cells incubated in YPA for 9 h was due to mitochondrial DNA staining.
(D) Microtubule network in UMN68 cells carrying an α-tubulin-GFP fusion and expressing high levels of biz1 after 8 h in YPA (Tub1-GFP; epifluorescence). Bar = 20 μm. DIC, differential interference contrast.
(E) UMN53 cells incubated for 24 h in inducing conditions display a distinct phenotype characterized by an extensive polarized growth (top panel; DIC) and single nuclei content (DAPI; arrow points to the nucleus), and empty sections behind that are separated by septa (stained with wheat germ agglutinin [WGA]; arrows) generated by formation of basal vacuoles as result of a permanent G2 cell cycle arrest. The vertical panel shows a detailed view of the basal area of the hypha where the septa-separated empty sections can be observed. Bars = 50 μm in horizontal panels and 15 μm in the vertical panel.
Biz1, a Putative Zinc Finger Transcription Factor in U. maydis
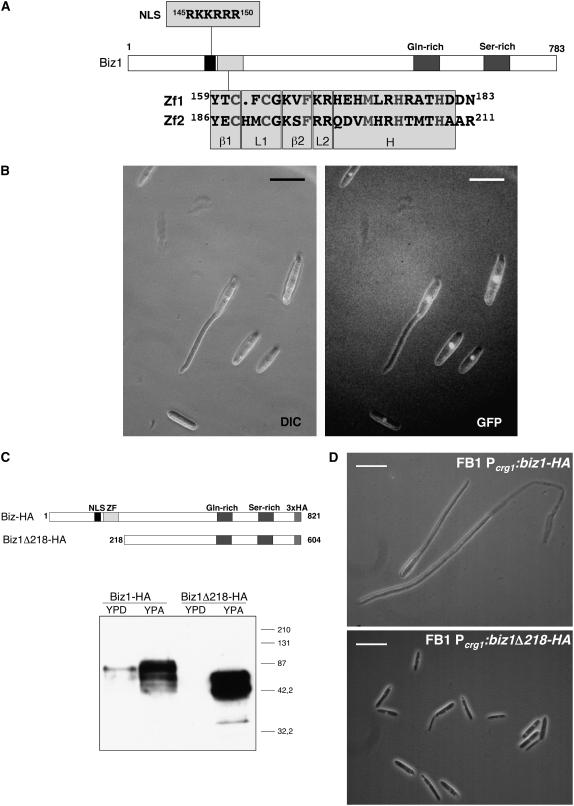
The predicted Biz1 protein contains two putative zinc finger domains of the Cys2His2 class, one of the most common DNA binding motifs found in Eukaryota. The two zinc fingers were designated Zf1 (from residues 159 to 183) and Zf2 (from residues 186 to 211). Both are predicted to be composed of two short β-strands (β1 and β2) separated by a Cys-containing short loop (L1), followed by a second loop (L2) and an α-helix (H) (Figure 2A). While Zf2 comprises the CysX2-4CysX12HisX3-5His consensus for Cys2His2 zinc fingers (Jacobs, 1992), the two Cys residues in Zf1 are separated only by a single amino acid (Figure 2A). Both zinc fingers contain a conserved hydrophobic residue, a Met, at position 4 of the proposed α-helix. The Phe residue at position 3 in the second β-strand (β2) is a frequent but not essential feature of Cys2His2 zinc fingers (Suzuki et al., 1994). We have been unable to find any significant sequence similarity between the Biz1 protein and other entries in the databases outside the zinc finger region. Analysis of the amino acid sequence using PROSITE indicated a Gln-rich region located between residues 507 and 526, a Ser-rich region located between residues 655 and 702, and a putative nuclear localization signal (NLS) located upstream of the first zinc finger (residues 145 to 150). PSORT (Nakai and Horton, 1999) predicts for Biz1 a high probability for nuclear localization (P = 94.1). To verify the subcellular location, we expressed a Biz1–green fluorescent protein (GFP) fusion under the control of the arabinose-inducible crg1 promoter (Bottin et al., 1996) in the U. maydis haploid wild-type strain FB1 (a1b1). Fluorescence microscopy revealed bright signals in the nucleus upon induction of the fusion protein (Figure 2B). Induction resulted in filament formation comparable to that of the cells expressing wild-type Biz1, indicating that the GFP fusion did not render the protein inactive.
Figure 2.
biz1 Encodes a Zinc Finger–Containing Protein.
(A) Domain structure of Biz1. Domains indicated were identified using PROSITE. Biz1 contains an NLS, a Gln-rich region, a Ser-rich region, and two zinc fingers. The bottom part shows the sequence of the two putative Cys2His2 zinc finger domains; amino acids predicted to be involved in the formation of the β-strands (β1 and β2), the loops (L1 and L2), and the α-helix (H) are indicated. Residues corresponding to the Cys2His2 zinc finger consensus sequence are in gray.
(B) The Biz1-GFP fusion protein localizes to the nucleus. Cells harboring an ectopic copy of a biz1-gfp fusion under the control of the arabinose-inducible crg1 promoter (UMN68 strain) were grown for 4 h under inducing conditions (complete medium supplemented with 1% arabinose as carbon source). Bars = 15 μm.
(C) In the top part, a scheme of the C-terminal-tagged Biz1 protein and its derivative lacking the NLS and zinc fingers, Biz1Δ218-HA, is shown. In the bottom part, a protein gel blot assay to detect Biz1 proteins is shown. Extracts were prepared from FB1 Pcrg:Biz1HA and FB1 Pcrg:Biz1Δ218HA grown in inducing conditions (YPA) or repressive conditions (YPD) at an OD600 of 0.5. An equal amount of total protein (50 μg) was loaded into the gel. Anti-HA-peroxidase (Roche) antibodies were used to detect the fusion proteins. The bars at right indicate the molecular masses of the protein marker ladder. Biz1-HA and Biz1Δ218HA have an estimated molecular mass of 88 and 64 kD, respectively.
(D) Morphology of cells producing the Biz1-HA or the Biz1Δ218HA protein, growing in inducing conditions (YPA). Bars = 20 μm.
The presence of two conserved Cys2His2 class zinc finger domains in the predicted sequence of Biz1 suggests a role as transcriptional factor. However, we cannot rule out other functions, as it has been shown that zinc-centered domains can be involved in protein–protein interaction in other proteins than transcriptional regulators (Matthews and Sunde, 2002). To gain support for a role of Biz1 as a transcription factor, first we tried to show that the NLS and zinc fingers are required for the function of Biz1. For this, we expressed an N-terminal truncated protein lacking the NLS and zinc finger–containing region fused to a triple HA tag at the C terminus (Figure 2C). As control, we also expressed a C-terminal-tagged full-length protein. Both proteins were detected by protein gel blot assay using anti-HA antibodies (Figure 2C). The addition of a C-terminal triple HA tag did not affect the activity of Biz1, as high levels of the full-length protein induced the already observed cell cycle arrest and hyperpolarized growth (Figure 2D). Strikingly, the N-terminal deleted mutant produced no effect, supporting a role of the N-terminal end carrying the NLS and the zinc fingers in the activity of the Biz1 protein.
Taken together, the presence of two Cys2His2 zinc fingers and a Gln-rich region, two features that frequently associate with transcription factors (Escher et al., 2000), and the nuclear localization suggest that Biz1 probably functions as a transcription factor.
Biz1 Downregulated the Expression of the Mitotic Cyclin clb1
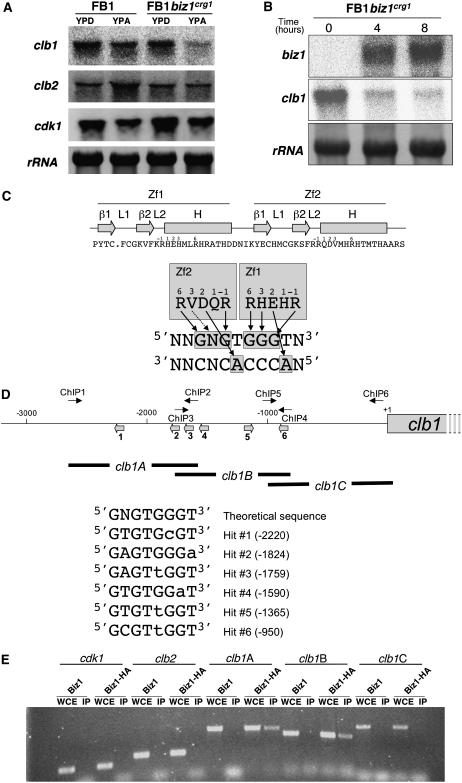
In U. maydis cells, entry into mitosis from G2 phase requires the activity of two B-type cyclins, Clb1 and Clb2, that form a complex with the catalytic subunit Cdk1 (García-Muse et al., 2004). Since biz1 overexpression inhibits the G2/M transition, we wondered whether Biz1 could control the expression of these cell cycle regulators via a function as transcriptional regulator. For this, we analyzed the expression of cdk1, clb1, and clb2 in biz1crg1 cells and in wild-type cells after 5 h growth under noninducing (YPD) and inducing (YPA) conditions. Remarkably, we observed a clear decrease in clb1 expression under conditions of biz1 overexpression (Figure 3A). Furthermore, a time-course analysis of clb1 expression after biz1 induction showed a strict inverse correlation between the biz1 and clb1 mRNA levels (Figure 3B).
Figure 3.
Biz1 Downregulates clb1 mRNA Levels.
(A) Expression level of the genes encoding the components of the mitotic Cdk in U. maydis: cdk1, clb1, and clb2. FB1 and UMN53 (FB1biz1crg1) cells were grown in noninducing (YPD) and inducing (YPA) conditions for biz1 overexpression in liquid cultures for 8 h, and the RNA was extracted and analyzed by RNA gel blotting using specific probes for each gene, as indicated at left. Ten micrograms of total RNA was loaded per lane. A probe specific for 18s rRNA was used to control for loading.
(B) Induction of biz1 and its effect on clb1 expression. UMN53 (FB1biz1crg1) cells growing in glucose-containing YPD were washed three times and transferred to arabinose-containing YPA, and samples were taken at the indicated times (in hours). RNA was isolated and submitted to RNA gel blot analysis as above.
(C) The sequence of the two zinc fingers of Biz1 is shown in the top panel. The secondary structure prediction is also schematized (arrows indicate β-zsheet, while the box indicates the α-helix). The numbers indicate the position with respect to the putative recognition α-helix. At the bottom, a scheme showing the proposed key amino acid–base contacts from positions −1, 2, 3, and 6 of each α-helix is shown.
(D) Scheme of the promoter region of clb1 depicting the location of the primers used in chromatin immunoprecipitation and the location of the different hits of the theoretical recognition sequence for Biz1. The different hits are shown in the bottom panel.
(E) Association of Biz1-HA with the clb1 promoter as measured by chromatin immunoprecipitation. Strains producing nontagged Biz1 and Biz1-HA–tagged proteins were submitted to immunoprecipitation. Control lanes show DNA amplified from extracts without tagged protein (Biz1) or prior to immunoprecipitation (whole-cell extract [WCE]). The following primer pairs were used: cdk1, CDK1-5/CDK1-7; clb2, CYC2-1/CYC2-3; clb1A, ChIP1/ChIP2; clb1B, ChIP3/ChIP4; clb1C, ChIP5/ChIP6.
Each zinc finger binds a single zinc ion that is sandwiched between the two-stranded β-sheet and the α-helix, producing a compact fold. In this fold, the comparison of the known structures between zinc finger and DNA target reveals that the vast majority of the base-specific contacts in the zinc finger–DNA complexes are made from positions −1, 2, 3, and 6 of the α-helix (presumably because these residues are the most prominently exposed on the surface of the helix). The zinc fingers present in Biz1 have a good level of conservation in these residues with respect to well-known zinc fingers, which structures of protein-DNA complexes have been described (Wolfe et al., 1999). In addition, the linker region that connects neighboring zinc fingers, which is an important structural element that controls the spacing of the fingers along the DNA site, shows the most common arrangement composed of five residues between the final His of one finger and the first conserved aromatic of the next finger. These similarities made it possible to assign a putative sequence recognized by the zinc fingers of Biz1 (Figure 3C) in basis of the described docking arrangements of previously known zinc finger–DNA structures (Wolfe et al., 1999; Paillard et al., 2004). Search of putative recognition sites in the regulatory upstream sequence of clb1 indicated six hits with a single mismatch (Figure 3D). We sought to analyze whether Biz1 is associated with the clb1 regulatory region. For this, we made use of chromatin immunoprecipitation, a method that measures the extent to which certain genomic DNA regions can be cross-linked to a specific protein under in vivo conditions (Hecht et al., 1999). Biz1 was provided as a C-terminal HA epitope-tagged protein or untagged protein as control. Cross-linked protein-DNA complexes were immunoabsorbed, and selected stretches of coprecipitated DNA were amplified by PCR and analyzed by gel electrophoresis. We used primers that amplified promoter regions of cdk1 and clb2 as negative controls (since their mRNA levels were unaffected by high dose of Biz1) and three different primer pairs to amplify the clb1 upstream region. We observed that amplification of the upstream clb1 region can be obtained, while a weak or no amplification at all was obtained from control promoters (Figure 3E). Strikingly, the amplification signal of clb1 promoter was obtained only with the distal regions, where the putative Biz1 recognition sites were located.
Downregulation of clb1 Expression Accounts for the Cell Cycle Arrest Induced by biz1 Overexpression
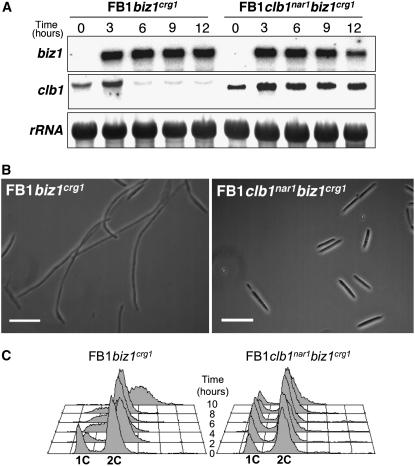
Clb1 forms a complex with Cdk1 and, together with the Clb2-Cdk1 complex, is required for the G2/M transition in U. maydis (García-Muse et al., 2004). Therefore, the observed G2 arrest after biz1 expression could be well explained by the decrease in clb1 mRNA levels. Alternatively, the decrease of clb1 mRNA level could be a consequence and not the cause of the G2 cell cycle arrest. We reasoned that if ectopic clb1 expression circumvents the Biz1-induced cell cycle arrest, it would imply that transcriptional downregulation of clb1 is the cause of the cell cycle arrest. Alternatively, a cell cycle arrest regardless of the promoter driving clb1 expression would suggest that the observed downregulation of clb1 is simply a consequence of the Biz1-induced cell cycle arrest. To address this question, we took advantage of a U. maydis clb1nar1 strain, in which the native clb1 promoter was replaced with the nar1 promoter (García-Muse et al., 2004). The cells of this strain show a wild-type phenotype when grown under conditions where the clb1nar1 gene is expressed (minimal medium amended with nitrate as nitrogen source; García-Muse et al., 2004). We introduced the biz1crg1 allele into the clb1nar1 strain and grew these cells under conditions where clb1nar1 is expressed (nitrate) and either noninduced (glucose) or induced (arabinose) conditions for biz1crg1 expression. As expected, biz1 induction led to the repression of the native clb1 gene, while the expression of the clb1nar1 gene was not altered (Figure 4A). In the latter case, no cell cycle arrested cells were observed, even after prolonged incubation (Figures 4B and 4C). These data indicate that downregulation of clb1 expression by Biz1 is the cause of the observed cell cycle arrest.
Figure 4.
Biz1-Mediated Downregulation of clb1 Induces Cell Cycle Arrest.
(A) Expression of clb1 under a heterologous promoter avoids the downregulation mediated by high levels of Biz1. UMN53 (FB1biz1crg1) and UMN56 (FB1clb1nar1biz1crg1) cells growing in MM-NO3 with glucose as carbon source, conditions in which clb1nar1 is expressed but biz1crg1 is repressed, were transferred to arabinose-containing MM-NO3 to allow the expression of biz1crg1, and samples were taken at the indicated times (in hours). RNA was isolated and submitted to RNA gel blot analysis as above, using either biz1 or clb1 probes, as indicated at left. A probe for the 18S rRNA was used as loading control.
(B) Expression of clb1 under a heterologous promoter avoids the cell cycle arrest mediated by high levels Biz1. Morphology of UMN53 (FB1biz1crg1) and UMN56 (FB1clb1nar1biz1crg1) cells grown for 10 h in MM-NO3 and arabinose as carbon source. Observe the arrested phenotype of UMN53 cells while UMN56 cells showed a wild-type budding pattern. Bars = 20 μm.
(C) FACS analysis of UMN53 (FB1biz1crg1) and UMN56 (FB1clb1nar1biz1crg1) cells growing in MM-NO3 with arabinose as carbon source to assess DNA content in inducing conditions. Samples were taken at 0, 2, 4, 6, 8, and 10 h after transfer to inducing conditions. Cells expressing biz1 arrest their growth at G2 and thereby accumulate with a 2C DNA content; however, in cells carrying the clb1nar1 allele, cells do not accumulate in G2 phase. The shift to DNA content higher than 2C observed in UMN53 cells was due to mitochondrial DNA staining.
Biz1 Is Required for Virulence
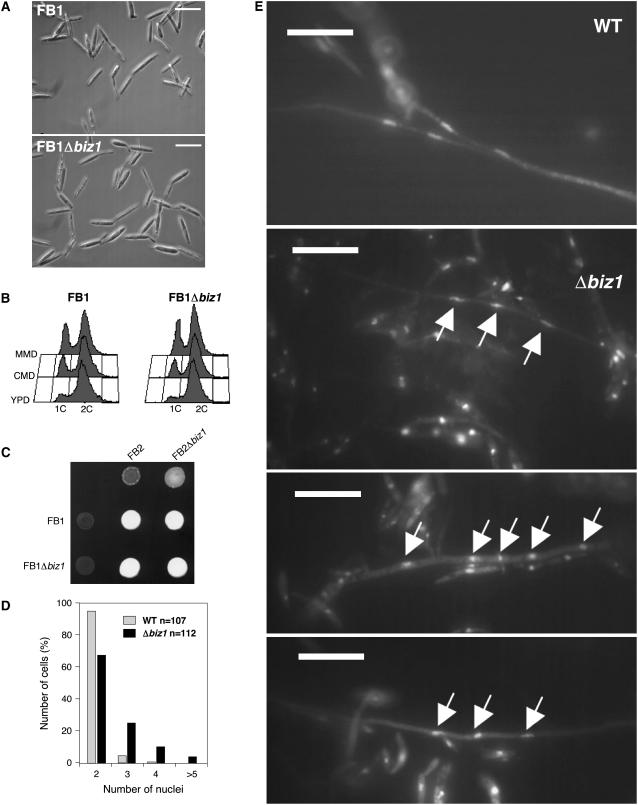
To investigate the physiological role of biz1, null mutants were constructed by one-step replacement in the haploid wild-type strains FB1 (a1b1) and FB2 (a2b2). When grown in liquid culture, the deletion strains showed no obvious phenotypic alterations to wild-type cells with respect to morphology (Figure 5A) and cell cycle profile (Figure 5B) or growth properties in various media (data not shown). To assay mating competence, FB1Δbiz1 and FB2Δbiz1 were cocultivated on charcoal-containing agar plates. Under these conditions, compatible wild-type strains fuse and develop filamentous dikaryotic hyphae that can be recognized as white fuzzy mycelium (Day and Anagnostakis, 1971). In wild-type strains, these filaments are cell cycle arrested and consist of a single binucleated tip cell that leave empty, septated cell compartments behind (Steinberg et al., 1998). biz1 mutant crosses formed filaments similar to those observed in wild-type crosses (Figure 5C), and microscopy observation showed no difference in the morphology of the filaments. However, nuclear staining with DAPI revealed a higher frequency of filaments carrying more than two nuclei in the mutant crosses, suggesting a defect in the ability to arrest mitosis during the formation of the infective hyphae (Figures 5D and 5E).
Figure 5.
Phenotype of U. maydis Δbiz1 Strains in Axenic Culture.
(A) Morphology of wild-type (FB1) and FB1Δbiz1 cells (UMN40) cells growing in nutrient-rich medium (YPD). Bars = 20 μm.
(B) Flow cytometry analysis of the DNA content of a cell population of wild-type (FB1) and FB1Δbiz1 (UMN40) strains growing in different media (YPD, 1% glucose yeast peptone extract; CMD, 1% glucose complete medium; MMD, 1% glucose minimal medium). In different media, the 1C versus 2C DNA content changes similarly in both strains.
(C) Mating assays of biz1 deletion strains. Strains indicated were spotted alone or in combinations on CM-charcoal plates and incubated at 24°C for 48 h. The appearance of white filaments indicates formation of infective hyphae.
(D) Quantitative analysis of nuclei distribution in wild-type and mutant infective hyphae formed by a cross of the wild-type strains FB1 and FB2 or the Δbiz1 mutant strains UMN40 (FB1Δbiz1) and UMN41 (FB2Δbiz1) after 24 h on charcoal-containing agar plates.
(E) Wild-type and mutant infective hyphae stained with DAPI visualizing the nuclei content. Both wild-type and mutant hyphae have the same appearance, although the mutant cross resulted in filaments with an increased frequency of multiple nuclei. Arrows point to multiple nuclei in the mutant hyphae. Bars = 10 μm.
To assess the role of biz1 in pathogenesis, plants were inoculated with a mixture of the compatible FB1Δbiz1 and FB2Δbiz1 strains. Infection with the respective wild-type strains led to tumor production in 89% of the inoculated plants. By contrast, in infections with the mutant strains, no plant tumors were obtained (Table 1). Only on a few occasions was chlorosis around the inoculation point observed.
Table 1.
Pathogenicity Assays
| Tumor Formation
|
|||
|---|---|---|---|
| Inoculum | Genotype | Total | Percentage |
| FB1 × FB2 | a1 b1 × a2 b2 | 62/69 | 89 |
| UMN40 × UMN41 | a1 b1 Δbiz1 × a2 b2 Δbiz1 | 0/74 | 0 |
| SG200 | a1mfa2 bW2bE1 | 59/70 | 84 |
| SG200Δbiz1 | a1mfa2 bW2bE1 Δbiz1 | 0/82 | 0 |
| UMN69 | a1mfa2 bW2bE1 Pdik6:clb1 | 29/62 | 46 |
Taken together, these results illustrate that Biz1 is a factor specifically required for pathogenic development of U. maydis.
Absence of biz1 Impairs Appressorium Formation and Proliferation of the Fungus in Plant Tissue
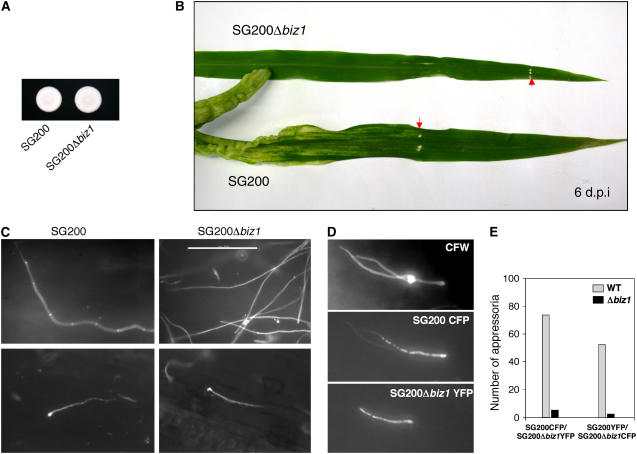
To elucidate at which stage pathogenic development of the mutant strain is blocked, the early infection process was followed using a Δbiz1 derivate of strain SG200. SG200 harbors a compatible combination of the bE1 and bW2 genes and the mfa2 gene inserted in the a1 locus and is thus able to infect plants without the need for a mating partner (Bölker et al., 1995). Similar to the cross of FB1Δbiz1 and FB2Δbiz1, SG200Δbiz1 forms filaments on charcoal plates (Figure 6A) and is apathogenic (Figure 6B, Table 1). After plant inoculation, fungal cells on the plant surface were stained with Calcofluor. We observed vigorous filament formation on the plant surface both in SG200 and SG200Δbiz1 infections (Figure 6C, top row). SG2000Δbiz1 could form appressoria with morphology comparable to that of the wild-type SG200 filaments (Figure 6C, bottom row). However, we noticed a decrease in the number of appressoria produced by the mutant strain. A decrease in the ability to form appressoria has also been noted in infections with mixtures of compatible haploid biz1 mutants versus wild-type strains (data not shown).
Figure 6.
Biz1 Is Required for Plant Infection.
(A) Filament formation of the solopathogenic strain SG200 (a1mfa2bE1bW2) and its derivative SG200Δbiz1. Strains indicated were spotted on CM-charcoal plates and incubated at 24°C for 48 h. The white colony morphology indicates the formation of filaments.
(B) Influence of biz1 deletion on pathogenicity. Maize seedlings were inoculated either with SG200 or SG200Δbiz1. Six days after infection (d.p.i.), tumors were formed on plants infected with SG200, while in infection with the mutant strain, no symptoms were observed. Arrows point to the injection punctures. For a quantitative analysis, see Table 1.
(C) Infection of young maize plants with SG200 or SG200Δbiz1 results in a network of hyphae (top panels) and the production of appressoria (bottom panels) that can be detected on the plant surface after 1 d by Calcofluor staining of the fungal cell wall. Bar = 100 μm.
(D) Expression of yellow- and cyan-shifted derivatives of GFP (YFP and CFP) allows the identification of SG200 and SG200Δbiz1 strains in the same infection. The top panel shows two hyphae with appressoria (Calcofluor staining). One of the appressoria is formed by SG200∷CFP (fluorescence in CFP channel; middle panel) and the second by the SG200Δbiz1∷YFP mutant (fluorescence in YFP channel; bottom panel).
(E) SG200Δbiz1 hyphae are impaired in appressorium formation. Equal numbers of SG200 and SG200Δbiz1 cells tagged with either YFP or CFP were inoculated in young maize seedlings. After 1 d, appressoria were scored with respect to specific CFP and YFP fluorescence. A decrease in the frequency of appressorium formation was observed in SG200Δbiz1, regardless of the fluorescence marker used for tagging.
To allow the quantitative comparison of appressoria formation, we coinfected plants with equal numbers of SG200 and SG200Δbiz1 cells that were tagged with constitutively expressed genes for cyan fluorescent protein (CFP) or yellow fluorescent protein (YFP), respectively (Figure 6D). We found that irrespective of the combination used (i.e., SG200∷CFP and SG200Δbiz1∷YFP or SG200∷YFP and SG200Δbiz1∷CFP), appressoria formation was reduced at least 10-fold in Δbiz1 cells compared with wild-type cells (Figure 6E). These results indicate a role of Biz1 in appressorial structure formation on the plant surface.
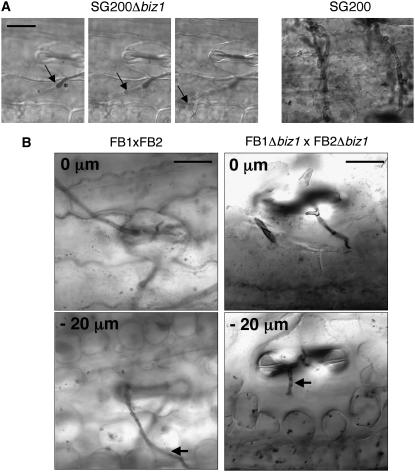
In the few cases where appressoria were produced in SG200Δbiz1 cells, Chlorazole Black E staining was used to visualize invading hyphae (Brachmann et al., 2003). Strikingly, mutant hyphae arrested growth after penetration of the plant surface and did not extend growth beyond the epidermial cell layer (Figure 7A). Supporting the defect in the progression through plant tissue, we found that hyphae entering via the natural opening provided by stomata were unable to progress through the plant tissue (Figure 7B).
Figure 7.
Δbiz1 Cells Arrest Growth after Plant Penetration.
(A) Left panels: Series of z axis projections showing the infection of a SG200Δbiz1 filament. Right panel: Infection with SG200. Plants were infected with SG200 and SG200Δbiz1, respectively, and after 2 d, fungal material was visualized by staining with Chlorazole Black E. In SG200Δbiz1, the appressorium (asterisk) and a short hyphae (arrows) that has penetrated the plant cuticle is visible. In SG200, massive proliferation of hyphae can be observed. Bar = 15 μm.
(B) Series of z axis projections showing a top view of the infection of FB1Δbiz1 × FB2Δbiz1 hyphae that grow through the stomatal opening. Note that even at 7 d after infection, the Δbiz1 hypha (arrow) fails to invade the host tissue and remains in the stomatal cavity. The range of z axis projections (as distance in micrometers) are indicated at the top left in each microphotograph. At the right, a wild-type hypha (arrow) penetrating through stomata is shown. Note that the wild-type hypha can progress through the plant tissue. Bars = 25 μm.
biz1 Is a b-Regulated Gene That Is Highly Expressed during the Infection Process
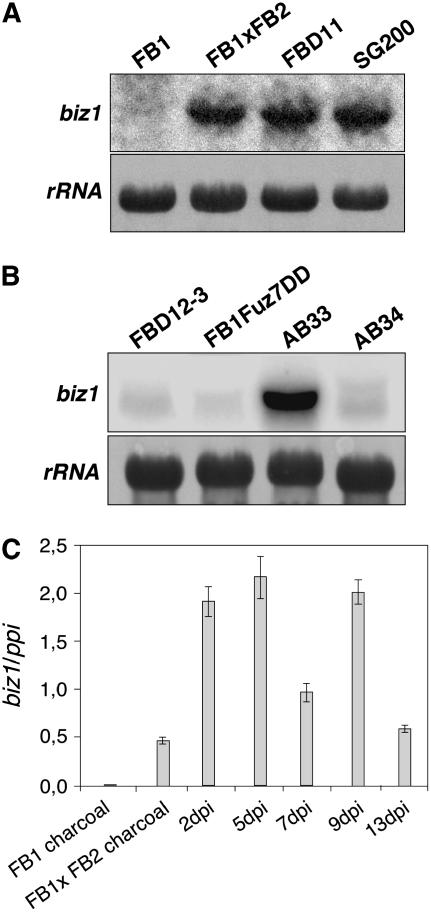
In agreement with the absence of phenotypic alterations in Δbiz1 cells under axenic conditions, biz1 expression was barely detectable in haploid strains growing in liquid culture (for instance, see Figure 1A). Since biz1 is required for virulence, it was likely that its expression would be activated under conditions that induce the pathogenicity program. Thus, we analyzed the biz1 expression in strains with activated a- and b-pathways that were grown on charcoal-containing agar plates, a condition that mimics the plant surface with respect to the ability to induce the mating determinants (Holliday, 1974). We used the wild-type haploid strain FB1 (a1b1), mixtures of FB1 (a1b1) and FB2 (a2b2), the solopathogenic haploid strain SG200 (a1mfa2bW2bE1), and the solopathogenic diploid strain FBD11 (a1ab1b2). RNA gel blot analysis confirmed a low level of biz1 expression in the wild-type haploid strain. However, all strains with compatible a- and b-mating–type combinations and a thus activated pathogenic program (mixture of FB1 and FB2, FBD11, and SG200) displayed a clearly induced biz1 expression (Figure 8A). In the diploid strain FBD12-3 (a1a2b1b1), where pheromone signaling is constitutive and an active b complex is missing, no biz1 expression was present. Furthermore, haploid cells expressing an activated form of the mitogen-activated protein kinase kinase Fuz7 (Fuz7DD; Müller et al., 2003), therefore mimicking an active pheromone signaling, do not express biz1 either (Figure 8B). On the contrary, in the haploid strain AB33 (Brachmann et al., 2001), harboring the compatible bW1/bE2 genes under the control of the nitrate-inducible nar1 promoter (Brachmann et al., 2001), a strong biz1 induction is observed in nitrate-containing medium, whereas in the control strain AB34, harboring the noncompatible bE2/bW2 combination, only a faint signal is detectable under the same conditions (Figure 8B). Thus, we conclude that biz1 expression is independent from the a-locus–mediated pathway and strictly dependent on the presence of an active bE/bW heterodimer.
Figure 8.
Expression of biz1 Depends on the b Locus.
(A) RNA gel blot analysis of biz1 in different genetic backgrounds. Haploid cells (FB1 and FB2), a mixture of FB1 and FB2, diploid (FBD11), and the solopathogenic strain SG200 were grown for 36 h on charcoal-containing plates and total RNA extracted. Ten micrograms of total RNA was loaded per lane. The same filter was hybridized in succession with probes for biz1 and 18s rRNA as loading control.
(B) biz1 expression is a independent but b dependent. In conditions of a locus activation without the presence of compatible b loci, such as the diploid FBD12-3 (a1 a2 b1 b1), or the presence of an active allele of the pheromone-induced mitogen-activated protein kinase kinase Fuz7 in a haploid cell (FB1FuzDD), nearly undetectable expression of biz1 was apparent. By contrast, conditions that produced an active bE/bW heterodimer, such as the AB33 strain growing, resulted in a high level of biz1 expression. AB34 is a control strain carrying noncompatible bE and bW alleles. FBD12-3 cells were grown for 24 h in charcoal-containing plates; FB1 Fuz7DD cells were grown in arabinose-containing liquid complete medium for 8 h; AB33 and AB34 were grown for 12 h in nitrate-containing liquid minimal medium. In all cases 10 μg of total RNA was loaded per lane. The same filter was hybridized in succession with probes for biz1 and 18s rRNA as loading control.
(C) Expression of biz1 in planta. Total RNA was extracted from plants infected with SG200 solopathogenic U. maydis cells at the indicated points. As control of expression out of the plant, RNA was extracted from SG200 growing in charcoal-containing plates. Data represent means of four experiments ± sd. dpi, days after infection.
To analyze biz1 expression during pathogenic development, maize plants were inoculated with a mixture of FB1 and FB2 cells, infected leaf areas were collected at different time points, and RNA was extracted and submitted to RT-PCR analysis (Figure 8C). We found that biz1 is expressed at all stages during pathogenic development. Although Δbiz1 mutants arrest early during infection, the observed expression at later stages suggests additional roles for Biz1 beyond the penetration step.
High Level of Clb1 Interferes with Infective Filament Formation
We wondered whether the biz1-dependent regulation of clb1 could account for the penetration defect observed in Δbiz1 strains. Constitutive high levels of mitotic cyclins interfere with pheromone signaling and, as a consequence, hinder the induction of the bE and bW genes, resulting in decreased virulence (Castillo-Lluva et al., 2004; Castillo-Lluva and Pérez-Martín, 2005). In addition, the inability to control cyclin levels inside the plant affects the infection process (Castillo-Lluva et al., 2004). To circumvent these interferences, we took advantage of the dik6 promoter. dik6 expression is dependent on the bE/bW heterodimer (Brachmann et al., 2001); the gene is strongly expressed in hyphae growing on the plant surface, but it is downregulated once the hyphae have penetrated the plant (G. Weinzierl and J. Kämper, unpublished data). Therefore, to achieve high clb1 mRNA levels specifically during the stage of infection where the appressoria are formed, we introduced clb1 under the control of the dik6 promoter (Pdik6:clb1) into SG200. In this strain, the Pdik6:clb1 fusion should lead to the reversion of the negative regulation of clb1 by biz1 once the b-dependent program is activated. In SG200 Pdik6:clb1 cells grown on charcoal-containing agar plates (conditions in which the bE and bW genes are highly expressed), a strong induction of clb1 was observed. SG200 cells grown under the same conditions show only a weak clb1 expression (Figure 9A). This decrease is not totally dependent on biz1, since in SG200Δbiz1 cells there is still some level of clb1 downregulation in charcoal-containing plates with respect to CMD plates, indicating that additional factors are involved in the regulation of clb1 during the initial steps of pathogenic development. Since clb2 expression seems not to be affected by biz1 expression, we also checked the level of clb2 mRNA as a control (Figure 9A).
Figure 9.
High Levels of clb1 Expression Interfere with Filament Formation.
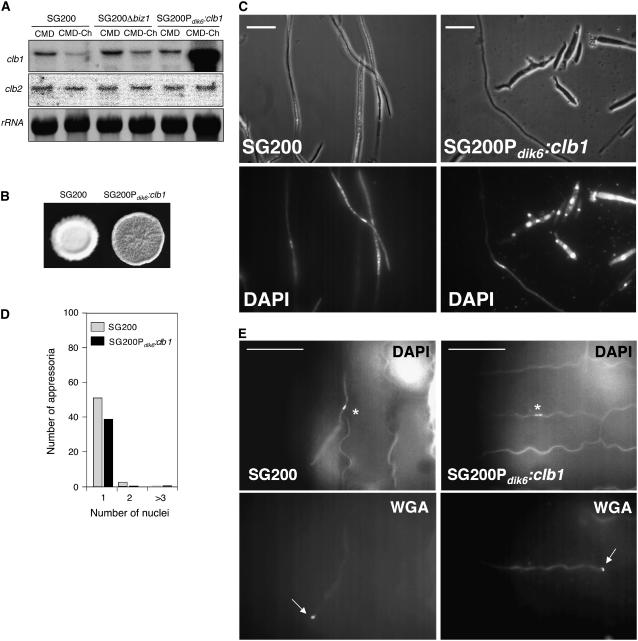
(A) Reprogramming clb1 expression after induction of the pathogenic program. clb1 was placed under the control of the dik6 promoter and ectopically integrated into the solopathogenic strain SG200 (a1mfa2bE1/bW2) to produce the UMN69 strain. The dik6 promoter is induced in response to the active bE1/bW2 heterodimer present in SG200. The expression of bE1/bW2 in SG200 is high on charcoal-containing complete medium (CMD-Ch), while on YPD plates, only low bE1/bW2 levels are observed. As a consequence, dik6p:clb1 is highly expressed in UMN69 on CMD-Ch medium. We also included SG200Δbiz1 as control. Total RNA (10 μg) extracted from SG200 and UMN69 (SG200Pdik6:clb1) cells was used for RNA gel blot analysis using clb1, clb2, and 18S rRNA-specific probes.
(B) clb1-reprogrammed SG200 cells are impaired in filament formation. SG200 and UMN69 (SG200Pdik6:clb1) cells were spread in charcoal-containing complete medium plates. After 48 h, the SG200Pdik6:clb1 colonies displayed significantly less filament formation than SG200.
(C) Analysis of SG200Pdik6:clb1 hyphae from CMD-Ch medium. Cells were spotted on charcoal-containing CMD-Ch plates, and cells were recovered from the surface after 24 h. Observe the presence of cells with aberrant morphology, containing more than one nuclei in strain SG200Pdik6:clb1. Bars = 20 μm.
(D) Appressorium formation is associated with cell cycle–arrested cells. Maize plants were infected with SG200 and SG200Pdik6:clb1. One day after infection, appressorium formation was visualized by staining with wheat germ agglutinin, and nuclear content was visualized by DAPI. Number of appressoria and number of nuclei per appressorium were counted. In SG200Pdik6:clb1, we did not observe appressoria with more than one nucleus.
(E) Examples of appressorium containing a single nucleus in SG200 and SG200Pdik6:clb1 cells. WGA, wheat germ agglutinin staining (arrow indicates the appressorium); DAPI staining marks the nucleus (asterisk indicates the nucleus). Bars = 100 μm.
Strikingly, cells carrying the ectopic Pdik6:clb1 transgene showed a clear decrease in the ability to form filaments on charcoal plates: aerial hyphae were observed as a much thinner layer of filaments in the colony (Figure 9B). Microscopy analysis of the cells within the colony revealed that ∼80% of these cells displayed an aberrant morphology and were carrying several nuclei (Figure 9C), suggesting a defect in the cell cycle arrest associated with the formation of infective filaments. However, it was also possible to detect filaments with wild-type morphology and single nuclei. Despite the clear impairment in filament formation, SG200 Pdik6:clb1 cells were able to infect maize plants, although with lower efficiency (Table 1). Therefore, we analyzed the ability of SG200 Pdik6:clb1 cells to form appressoria on maize leaves. Similar to the situation on CM-charcoal plates, the majority of the cells in planta displayed an aberrant morphology, and a minority of filaments exhibited wild-type morphology (data not shown). Remarkably, appressorium formation on the plant surface was only observed for the filaments with wild type–like appearance and a single nucleus (Figures 9D and 9E), suggesting that only cells able to arrest the cell cycle can produce appressoria.
DISCUSSION
In this study, we have identified a new factor, Biz1, which is required for pathogenic development of U. maydis. Biz1 also has impacts on cell cycle control via the downregulation of the gene for the mitotic cyclin Clb1.
The predicted Biz1 protein contains two conserved nucleic acid binding Cys2His2 zinc finger domains and a Gln-rich region that is known from several eukaryotic transcription factors, such as Sp1 or Oct1, to mediate transcriptional activation (Escher et al., 2000). Consistent with the prediction from PSORT, Biz1 shows a nuclear localization. We found that the N-terminal half of Biz1, carrying the zinc fingers and the NLS, is required for its activity. In addition, using chromatin immunoprecipitation assays we have been able to show a physical interaction between Biz1 and the upstream regulatory region of clb1. Thus, we favor that Biz1 is a transcription factor. A limited number of transcriptional regulators have been described associated with virulence in phytopathogenic fungi. In Fusarium oxysporum, the Cys2His2 zinc finger transcription factor PacC, a conserved regulator involved in pH sensing, is required for virulence (Caracuel et al., 2003). In Claviceps purpurea, the CREB-like transcription factor CPTF1 is involved in oxidative stress response, and deletion of CPTF1 leads to reduced virulence (Nathues et al., 2004). ToxE is a pathway-specific transcription factor essential for the expression of HC-toxin required for pathogenicity of Cochliobolus carbonum (Pedley and Walton, 2001). Finally, in M. grisea, Mst12, a Ste12 homologue, was found to be essential for penetration and invasive growth (Park et al., 2002). However, Biz1 function cannot be related to that of any of these known transcription factors, although the biz1 deletion, similar to the phenotype observed for the MST12 deletion in M. grisea, leads to a penetration defect. However, while an MST12Δ strain in M. grisea is not impaired in appressoria formation, in U. maydis Δbiz1 strains, the frequency of appressoria formation is dramatically reduced.
The biz1 gene is not expressed in haploid wild-type cells, and deletion analysis revealed no function for biz1 during saprophytic growth. biz1 expression is strictly dependent on the presence of a bE/bW heterodimer; induction of the a-pathway either by pheromone or by a constitutive active mitogen-activated protein kinase kinase cascade (Fuz7DD) does not show any effects. It has been proposed that the bE/bW heterodimer triggers a regulatory cascade in which class 1 genes are directly regulated by the binding of the bE/bW heterodimer to a conserved DNA motif termed b binding sequence (bbs) in the upstream region of b-responsive genes, and class 2 genes are indirectly regulated by b via yet unidentified regulatory proteins that are encoded by class 1 genes (Romeis et al., 2000; Brachmann et al., 2001). In the promoter region of biz1, we could not detect a bbs motif, and we observed a delayed expression of biz1 after bE/bW induction during time-course experiments (data not shown). Furthermore, compared with the expression observed after b-induction in charcoal-containing plates, biz1 expression increased at least fourfold after the fungus has penetrated the plant, suggesting additional factors favoring biz1 expression specific for in planta conditions. Therefore, we favor the idea that biz1 is regulated indirectly via a b-dependent signaling cascade that allows the input of additional environmental cues for its regulation. It is conceivable that such cues must be perceived already on the plant surface.
In wild-type cells, after cell fusion, the assembly of a compatible bE/bW heterodimer induces the formation of a cell cycle–arrested dikaryotic filament that is characterized by tip growth and the generation of empty sections in the distal parts of the filament (Steinberg et al., 1998). Mutants lacking Biz1 were able to form this infectious filament, although an increase in filaments carrying more than two nuclei (i.e., not cell cycle arrested) was observed. Strikingly, ectopic expression of biz1 in axenic conditions resulted in a G2 cell cycle arrest. We showed that clb1, encoding a mitotic cyclin essential for G2/M transition, was downregulated by Biz1. We also demonstrated that the cell cycle arrest is a direct consequence of the downregulation of clb1 expression. Since Biz1 seems to be positively regulated by the b locus and since high levels of Biz1 protein arrest the cell cycle, it is tempting to speculate that Biz1-mediated downregulation of clb1 expression is responsible for the b-dependent cell cycle arrest. Moreover, the results obtained after reprogramming clb1 expression during the induction of the infective filament using the dik6 promoter supports the importance of clb1 downregulation for the correct formation of such a structure. However, additional control systems ensuring a cell cycle arrest must exist, since deletion of biz1 or overexpression of clb1 do not completely abolish the formation of the typical cell cycle–arrested hyphae. Such additional controls may well involve inhibitory phosphorylation of the Cdk1 kinase by the Wee1 kinase. We recently reported that overexpression of Cdc25, a phosphatase that counteracts the Wee1-mediated inhibition of Cdk1, impairs the formation of b-dependent filaments (Sgarlata and Pérez-Martín, 2005b). We believe that complementary and somehow redundant mechanisms, such as downregulation of cyclin expression and direct inhibition of kinase activity (i.e., by inhibitory phosphorylation), could be responsible of the b-induced cell cycle arrest. Biz1 is one of the factors responsible for the cyclin downregulation, although not the only one. We based this conclusion on two results: first, the dramatic difference between SG200Δbiz1 and SG200 Pdik6:clb1 cells to form cell cycle–arrested filaments advocates for additional factors that could downregulate clb1 expression during pathogenic development; second, in SG200Δbiz1 cells growing on charcoal medium (and thus having an elevated level of b-expression), clb1 is downregulated (although to a lesser extend than in the wild-type situation). Further research efforts will be needed to define the nature of these putative additional mechanisms of b-dependent cell cycle control.
Is cell cycle arrest connected to appressorium formation? We observed that in the reprogrammed SG200 Pdik6:clb1 strain appressorium formation is always associated cell cycle–arrested filaments but never to aberrant nonarrested cells. On the other hand, SG200Δbiz1 could produce cell cycle–arrested filaments, but only low numbers of appressoria were detected. Taken together, these two observations suggest that cell cycle arrest is necessary but not sufficient for appressorium formation. It is conceivable that during appressorium development the response to external cues (i.e., signals from plant surface) and internal cues (i.e., cell cycle arrest) must be timely coordinated. A delay in the sensing of either external or internal cues (for instance, a delay in cell cycle arrest) could therefore lead to an inefficient response and thus result in impaired appressorium formation. The importance of precise connections between cell cycle regulation and appressorium formation has been recently highlighted in the rice blast fungus M. grisea, where it has been proposed that either a G2/M or a postmitotic checkpoint may regulate appressorium formation (Veneault-Fourrey et al., 2006).
The arrested cell cycle of the infective hyphae is released once the fungus enters the plant. Consistently, we found that clb1 is expressed inside the plant (I. Flor-Parra and M. Vranes, unpublished data). As biz1 is expressed during the biotrophic phase, there must be additional factors that uncouple the clb1 downregulation from Biz1 once the fungus penetrates the plant. These still uncharacterized factors are expected to play an essential role during the infection process.
Finally, we believe that cell cycle arrest and appressorium formation are not the only processes controlled by Biz1 during the infection process. Since biz1 is expressed during the entire biotrophic phase, it is conceivable that the protein plays additional roles at this stage. We found that in the rare occasions that biz1-defective cells penetrate the plant surface either by producing appressoria or by using the natural opening provided by stomata, the fungus was unable to proliferate inside the plant, corroborating the importance of Biz1 for fungal development after penetration. In the future, the characterization of genes regulated by Biz1 will provide insights to understand these proposed downstream roles.
METHODS
Strains and Growth Conditions
Ustilago maydis strains are derived from FB1 (a1b1) (Banuett and Herskowitz, 1989) and are listed in Table 2. Cells were grown at 28°C in YPD (Sherman et al., 1986), complete medium (CM), or minimal medium (MM) (Holliday, 1974). Controlled expression of genes under the crg1 and nar1 promoters were performed as described previously (Brachmann et al., 2001; García-Muse et al., 2004). FACS analysis was described previously (García-Muse et al., 2003). Mating assays and plant infections were performed as described (Gillissen et al., 1992).
Table 2.
U. maydis Strains Used in This Study
| Strain | Relevant Genotype | Reference |
|---|---|---|
| FB1 | a1 b1 | Banuett and Herskowitz (1989) |
| FB2 | a2 b2 | Banuett and Herskowitz (1989) |
| FBD11 | a1a2 b1b2 | Banuett and Herskowitz (1989) |
| FBD12-3 | a1 a2 b1 b1 | Banuett and Herskowitz (1989) |
| SG200 | a1 mfa2 bW2 bE1 | Bölker et al. (1995) |
| AB33 | a2 Pnar1:bW2 Pnar1:bE1 | Brachmann et al. (2001) |
| AB34 | a2 Pnar1:bW2 Pnar1:bE2 | Brachmann et al. (2001) |
| FB1Pcrg1:fuz7DD | a1 b1 Pcrg1:fuz7DD | Müller et al. (2003) |
| UMN40 | a1 b1 Δbiz1 | This work |
| UMN41 | a2 b2 Δbiz1 | This work |
| UMN45 | a1 b1Pcrg1:biz1-3HA | This work |
| UMN47 | a1 b1Pcrg1:biz1 | This work |
| UMN52 | a1 b1Pcrg1:biz1-gfp | This work |
| UMN53 | a1 b1 biz1crg1 | This work |
| UMN56 | a1 b1 clb1nar1 biz1crg1 | This work |
| UMN68 | a1 b1 biz1crg1 tub1-GFP | This work |
| UMN69 | a1 mfa2 bW2 bE1 Pdik6:clb1 | This work |
| UMN75 | a1 b1Pcrg1:biz1Δ218-3HA | This work |
| SG200Δbiz1 | a1 mfa2 bW2 bE1 Δbiz1 | This work |
| SG200CFP | a1 mfa2 bW2 bE1 POMA:CFP | This work |
| SG200YFP | a1 mfa2 bW2 bE1 POMA:YFP | This work |
| SG200Δbiz1CFP | a1 mfa2 bW2 bE1 Δbiz1 POMA:CFP | This work |
| SG200Δbiz1YFP | a1 mfa2 bW2 bE1 Δbiz1 POMA:YFP | This work |
DNA and RNA Analysis
U. maydis DNA isolation was performed as previously described (Tsukuda et al., 1988). RNA isolation from axenic cultures, charcoal plates, and plant tissues was performed as described (Basse et al., 2002). RNA gel blot analysis was performed as described previously (Garrido and Pérez-Martín, 2003). A 579-bp DNA fragment spanning the sequence coding the first 193 amino acids of Biz1 was used as probe. The cdk1, clb1, and clb2 probes were already described (García-Muse et al., 2004). A 5′-end labeled oligonucleotide complementary to the U. maydis 18S rRNA (Bottin et al., 1996) was used as loading control in RNA gel blot analyses. A phosphor imager (Molecular Imager FX; Bio-Rad) and the suitable program (Quantity One; Bio-Rad) were used for visualization and quantification of radioactive signals.
First-strand cDNA synthesis was performed using the SuperScript III first-strand synthesis SuperMix assay (Invitrogen) according to the manufacturer's protocol. One microgram of total RNA was used as a template for the reaction. Samples were incubated at 50°C for 50 min. Real-time PCR was performed on a Bio-Rad iCycler system using the Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) according to the manufacturer's protocol. Cycling conditions were as follows: 95°C for 2 min, 45 cycles of 30 s at 95°C, 30 s at 62°C, and 30 s at 72°C. After each PCR, the specificity of the amplifications was verified, and the threshold cycle above background was calculated using the Bio-Rad iCycler software. The gene encoding peptidylprolyl isomerase (ppi) was used as a constitutive control for normalization. Relative expression values were calculated using the Bio-Rad Gene Expression Macro. Forward (F) and reverse (R) primers were as follows: for biz1, Rt-biz1-F3 (5′-GGATCAGCCAAATGATGGACAG-3′) and Rt-biz1-R3 (5′-TACTCTCGCATCTCTTCCACTC-3′); for ppi, 5′Rt-ppi-F2 (5′-ACATCGTCAAGGCTATCG-3′) and Rt-ppi-R2 (5′-AAAGAACACCGGACTTGG-3′).
Plasmid and Strain Constructions
Plasmid pGEM-T easy (Promega) was used for cloning, subcloning, and sequencing of genomic fragments and fragments generated by PCR. Plasmid pRU11 (Brachmann et al., 2001) was used to express genes under the control of the Pcrg1 promoter. Sequence analysis of fragments generated by PCR was performed with an automated sequencer (ABI 373A; Applied Biosystems) and standard bioinformatic tools. To construct the different strains, transformation of U. maydis protoplasts with the indicated plasmids was performed as described previously (Tsukuda et al., 1988). Homologous recombination of gene replacement into the corresponding loci was verified by diagnostic PCR and subsequent DNA gel blot analysis.
Deletion of biz1 in FB1 and FB2 was done by homologous replacement following the protocols of Kämper (2004) and Brachmann et al. (2004). Briefly, a pair of DNA fragments flanking the biz1 ORF was amplified and ligated to a hygromicin resistance cassette via SfiI sites. The 5′ fragment spans from nucleotide –1000 to nucleotide –1 (considering the adenine in the ATG as nucleotide +1), and it was produced by PCR amplification using the primers UST12-2 (5′-CTGCATGTTCGGACGCCAACAATGGGC-3′) and UST12-3 (5′-ATGGCCATCTAGGCCGCTGTTTCAAGCACACTGGCCCTC-3′). The 3′ fragment spans from nucleotide +2352 to nucleotide +3357, and it was produced by PCR amplification using the primers UST12-6 (5′-TAGGCCTGAGTGGCCAGCATGGGCGAAAGCTGACTCGAC-3′) and UST12-7 (5′-CAGCTAGCGATCCTTGCACGCCGTTCC-3′). To delete biz1 in SG200, we followed the same strategy: the 5′ fragment spans the region from −7 to − 981 (with respect to the ATG), and the 3′ fragment spans 1057 bp 3′of the stop codon. Primers used were Biz1-lbn (5′-CAATGGGCATGTGCTCTTG-3′), Biz1-lb2 (5′-GTTGGCCTGAGTGGCCCAAGCACACTGGCCCTCG-3′), Biz1-rbn (5′-GAAGGGCGTTGAAGGATG-3′), and Biz1-rb2 (5′-GTTGGCCATCTAGGCCGCTGACTCGACTGCCTGC-3′).
To produce a conditional biz1crg1 allele, we constructed a plasmid by ligation of two fragments into pRU11 digested with NdeI and EcoRI. The 5′ fragment (flanked by EcoRI and BamHI) was produced by PCR using the primers USTA (5′-CGGGATCCTCTTGCGCATCAAAGCTGCGCTG-3′) and USTB (5′-CGGAATTCATGCCAGGTAGTCGAGAGCCATC-3′). This fragment spans from nucleotide −967 to nucleotide −256 (considering the adenine in the ATG as nucleotide +1). The 3′ fragment (flanked by NdeI and BamHI) was obtained by PCR amplification with primers UST12-4 (5′-CATATGTCGATGCTTAGCACACGGGCA-3′) and UST12-5 (5′-CCGGAATTCCCAACGACGGCTGTGGTGACC-3′) and spans from nucleotide +1 to nucleotide +2360. The resulting plasmid pBIZ1crg was integrated, after digestion with BamHI, by homologous recombination into the biz1 locus.
To construct the Pdik6:clb1 allele, a 2.1-kb NdeI-NotI fragment from pRU11-CLB1 (García-Muse et al., 2004) carrying the Pcrg1 promoter was exchanged with a PCR-amplified 1.1-kb NdeI-NotI fragment carrying the Pdik6 promoter (C. Sgarlata and J. Pérez-Martín, unpublished data). The resulting plasmid, pCLB1dik6, was integrated after digestion with SspI by homologous recombination into the cbx locus as described previously (Brachmann et al., 2001).
To produce a biz1-gfp fusion, a PCR-generated 2.3-kb NdeI fragment carrying the biz1 ORF without the stop codon was inserted at the single NdeI site of pRU11 (Brachmann et al., 2001), allowing the fusion at the C-terminal end of Biz1 with GFP and an expression under control of crg1 promoter. This construction was integrated, after digestion with XcmI, by homologous recombination into the cbx locus as described previously (Brachmann et al., 2001).
To produce the biz1-HA and biz1Δ218-HA alleles under the control of the crg1 promoter, the biz1 ORF was amplified using UST12-4 (which amplifies from the first ATG; 5′-CATATGTCGATGCTTAGCACACGGGCA-3′) or Biz1ΔZF (which amplifies from nucleotide +652; 5′-CATATGCGAACCACGGCTTCCATGTC-3′) and UST12-5 (which amplifies until the end of the ORF without the stop codon; 5′-GAATTCCCAACGACGCCTGGTGTGACC-3′). The corresponding fragments were cloned as NdeI-EcoRI into pGNB-HA (a pGEX-2T derivate that carries three copies of the HA epitope; J. Pérez-Martín, unpublished data), and the resulting tagged alleles were cloned as NdeI-AflII into pRU11 (Brachmann et al., 2001). The pRU11-Biz1HA and pRU11-BizΔ218HA plasmids were integrated, after digestion with SspI, by homologous recombination into the cbx as described previously (Brachmann et al., 2001).
Plasmids allowing the constitutive expression of cfp and yfp genes (pOMA-CFP and pOMA-YFP) are based on plasmid pCU4 (Loubradou et al., 2001). In pCU4, the o2tef promoter was replaced with a 1.3-kb SpeI-XmaI from plasmid pSL1180-OMA with the OMA promoter (A. Hartmann, unpublished data) to yield pOMA-GFP. The OMA promoter consists of the U. maydis mfa1 minimal promoter fused to eight enhancer elements from the U. maydis prf1 gene promoter and leads to strong constitutive expression (A. Hartmann, unpublished data). The NotI site in the vector backbone in pOMA-GFP was deleted with Klenow polymerase and religation; subsequently, the gfp gene was replaced with XmaI-NotI fragments harboring the yfp and cfp genes from plasmids pOY and pOC (Weber et al., 2003), respectively. The resulting plasmids pOMA-CFP and pOMA-YFP were integrated into the cbx locus after linearization with SspI as described previously (Brachmann et al., 2001).
Chromatin Immunoprecipitation Assays
Chromatin immunoprecipitation PCR assays were performed as described (Hecht et al., 1999). For cross-linking, U. maydis cells producing nontagged or tagged Biz1 protein were treated with 1% formaldehyde for 15 min at room temperature. Chromatin was sheared by sonication to an average size of 1000 bp. Primer sequences used are described in Supplemental Table 1 online.
Microscopy Observations
Microscopy was performed either using a Leica DMLB microscope or a Zeiss Axioplan 2. Phase contrast or standard fluorescein isothiocyanate and DAPI filter sets were used for epifluorescence analysis of nuclear staining with DAPI (García-Muse et al., 2003), wheat germ agglutinin, and Calcofluor, performed as described (Castillo-Lluva and Pérez-Martín, 2005). Photomicrographs were obtained with a Leica 100 or Axiocam HrM camera, and the images were processed with Axiovision (Zeiss) and Photoshop (Adobe Systems).
Sequence Analyses
Predicted amino acid sequences were analyzed using the programs BLAST (Altschul et al., 1990) and PSORT (Nakai and Horton, 1999).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AAP94019 (Um Clb1). The sequence information of Um Biz1 was obtained from the public MIPS Ustilago maydis Database (http://mips.gsf.de/proj/ustilago/) under accession number um02549.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Results. Genetic Screening for biz1.
Supplemental Table 1. Primers Used in Chromatin Immunoprecipitation Assays.
Supplemental Figure 1. Description of the Genetic Screening Where biz1 Was Found.
Supplemental Figure 2. Identification of biz1, Encoding a Zinc Finger–Containing Protein.
Supplementary Material
Acknowledgments
We thank William K. Holloman (Cornell University, Ithaca, NY) for the pCM54-based U. maydis gene library and Jan Schirawski (Max-Planck-Institut) for the wild-type fungus infecting the plant through stomata. This work was supported by a grant from the Ministerio de Ciencía y Tecnologia (MCyT) (BIO2005-02998) and a European Union contract (MRTN-CT-2005-019277) to J.P.-M. and by a grant from the German Bunderministerium für Bildung und Forschung to J.K. I.F.-P. was a recipient of an FPI fellowship from the MCyT. M.V. was supported by a grant from the International Graduate School GRK767 funded by the German Research Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: José Pérez-Martín (jperez@cnb.uam.es).
Online version contains Web-only data.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Banuett, F., and Herskowitz, I. (1989). Different a alleles are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86 5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett, F., and Herskowitz, I. (1996). Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis. Development 122 2965–2976. [DOI] [PubMed] [Google Scholar]
- Banuett, F., and Herskowitz, I. (2002). Bud morphogenesis and the actin and microtubule cytoskeletons during budding in the corn smut fungus, Ustilago maydis. Fungal Genet. Biol. 37 149–170. [DOI] [PubMed] [Google Scholar]
- Basse, C.W., Kolb, S., and Kahmann, R. (2002). A maize-specifically expressed gene cluster in Ustilago maydis. Mol. Microbiol. 43 75–93. [DOI] [PubMed] [Google Scholar]
- Basse, C.W., and Steinberg, G. (2004). Ustilago maydis, model system for analysis of the molecular basis of fungal pathogenicity. Mol. Plant Pathol. 5 83–92. [DOI] [PubMed] [Google Scholar]
- Bechinger, C., Giebel, K.F., Schnell, M., Leiderer, P., Deising, H.B., and Bastmeyer, M. (1999). Optical measurement of invasive forces exerted by appressoria of a plant pathogenic fungus. Science 285 1896–1899. [DOI] [PubMed] [Google Scholar]
- Bölker, M., Genin, S., Lehmler, C., and Kahmann, R. (1995). Genetic regulation of mating and dimorphism in Ustilago maydis. Can. J. Bot. 73 S320–S325. [Google Scholar]
- Bottin, A., Kämper, J., and Kahmann, R. (1996). Isolation of a carbon source-regulated gene from Ustilago maydis. Mol. Gen. Genet. 25 342–352. [DOI] [PubMed] [Google Scholar]
- Brachmann, A., Konig, J., Julius, C., and Feldbrügge, M. (2004). A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 272 216–226. [DOI] [PubMed] [Google Scholar]
- Brachmann, A., Schirawski, J., Mülller, P., and Kahmann, R. (2003). An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J. 9 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, A., Weinzierl, G., Kämper, J., and Kahmann, R. (2001). Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42 1047–1063. [DOI] [PubMed] [Google Scholar]
- Caracuel, Z., Roncero, M.I., Espeso, E.A., Gonzalez-Verdejo, C.I., García-Maceira, F.I., and Di Pietro, A. (2003). The pH signaling transcription factor PacC controls virulence in the plant pathogen Fusarium oxysporum. Mol. Microbiol. 48 765–779. [DOI] [PubMed] [Google Scholar]
- Castillo-Lluva, S., García-Muse, T., and Pérez-Martín, J. (2004). A member of the Fizzy-related family of APC activators is required at different stages of plant infection by Ustilago maydis. J. Cell Sci. 117 4143–4156. [DOI] [PubMed] [Google Scholar]
- Castillo-Lluva, S., and Pérez-Martín, J. (2005). The induction of the mating program in the phytopathogen Ustilago maydis is controlled by a G1 cyclin. Plant Cell 17 3544–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, P.R., and Anagnostakis, S.L. (1971). Corn smut dikaryon in culture. Nat. New Biol. 231 19–20. [DOI] [PubMed] [Google Scholar]
- Dean, R.A. (1997). Signal pathways and appressorium morphogenesis. Annu. Rev. Phytopathol. 35 211–234. [DOI] [PubMed] [Google Scholar]
- Deising, H.B., Werner, S., and Wernitz, M. (2000). The role of fungal appressoria in plant infection. Microbes Infect. 2 1631–1641. [DOI] [PubMed] [Google Scholar]
- Emmett, R.W., and Parbery, D.G. (1975). Appressoria. Annu. Rev. Phytopathol. 13 147–167. [Google Scholar]
- Escher, D., Bodmer-Glavas, M., Barberis, A., and Schaffner, W. (2000). Conservation of Glutamine-rich transactivation function between yeast and humans. Mol. Cell. Biol. 20 2774–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Muse, T., Steinberg, G., and Pérez-Martín, J. (2003). Pheromone-induced G2 arrest in the phytopathogenic fungus Ustilago maydis. Eukaryot. Cell 2 494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Muse, T., Steinberg, G., and Pérez-Martín, J. (2004). Characterization of B-type cyclins in the smut fungus Ustilago maydis: Roles in morphogenesis and pathogenicity. J. Cell Sci. 117 487–506. [DOI] [PubMed] [Google Scholar]
- Garrido, E., and Pérez-Martín, J. (2003). The crk1 gene encodes an Ime2-related protein that is required for morphogenesis in the plant pathogen Ustilago maydis. Mol. Microbiol. 47 729–743. [DOI] [PubMed] [Google Scholar]
- Gillissen, B., Bergemann, J., Sandmann, C., Schrör, B., Bölker, M., and Kahmann, R. (1992). A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 68 647–657. [DOI] [PubMed] [Google Scholar]
- Gold, R.E., and Mendgen, K. (1991). Rust basidiospore germlings and disease initiation. In Initiation in Plants and Animals, G.T. Cole and H.C. Hoch, eds (New York: Plenum Press), pp. 67–99.
- Hecht, A., Strahl-Bolsinger, S., and Grunstein, M. (1999). Mapping DNA interaction sites of chromosomal proteins. Crosslinking studies in yeast. Methods Mol. Biol. 119 469–479. [DOI] [PubMed] [Google Scholar]
- Holliday, R. (1974). Ustilago maydis. In Handbook of Genetics, R.C. King, ed (New York: Plenum Press), pp. 575–595.
- Jacobs, G.H. (1992). Determination of the base recognition positions of zinc fingers from sequence analysis. EMBO J. 11 4507–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahmann, R., and Kämper, J. (2004). Ustilago maydis: How its biology relates to pathogenic development. New Phytol. 164 31–42. [DOI] [PubMed] [Google Scholar]
- Kämper, J. (2004). A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 271 103–110. [DOI] [PubMed] [Google Scholar]
- Loubradou, G., Brachmann, A., Feldbrügge, M., and Kahmann, R. (2001). A homologue of the transcriptional repressor Ssn6 anatagonizes cAMP signaling in Ustilago maydis. Mol. Microbiol. 40 719–730. [DOI] [PubMed] [Google Scholar]
- Matthews, J.M., and Sunde, M. (2002). Zinc fingers—Folds for many occasions. IUBMB Life 54 351–355. [DOI] [PubMed] [Google Scholar]
- Mendgen, K., Hahn, M., and Deising, H. (1996). Morphogenesis and mechanisms of penetration in plant pathogenic fungi. Annu. Rev. Phytopathol. 34 367–386. [DOI] [PubMed] [Google Scholar]
- Müller, P., Weinzierl, G., Brachmann, A., Feldbrügge, M., and Kahmann, R. (2003). Mating and pathogenic development of the smut fungus Ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot. Cell 2 1187–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, K., and Horton, P. (1999). PSORT: A program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem. Sci. 24 34–35. [DOI] [PubMed] [Google Scholar]
- Nathues, E., Joshi, S., Tenberge, K.B., von der Driesch, M., Oeser, B., Baumer, N., Mihlan, M., and Tudzynski, P. (2004). CPTF1, a CREB-like transcription factor, is involved in the oxidative stress response in the phytopathogen Claviceps purpurea and modulates ROS level in its host Secale cereale. Mol. Plant Microbe Interact. 17 383–393. [DOI] [PubMed] [Google Scholar]
- Paillard, G., Deremble, C., and Lavery, R. (2004). Looking into DNA recognition: Zinc finger binding specificity. Nucleic Acids Res. 32 6673–6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, G., Xue, C., Zheng, L., Lan, S., and Xu, J.R. (2002). MST12 regulates infectious growth but not appressoria formation in the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 15 183–192. [DOI] [PubMed] [Google Scholar]
- Pedley, K.F., and Walton, J.D. (2001). Regulation of cyclic peptide biosynthesis in a plant pathogenic fungus by a novel transcription factor. Proc. Natl. Acad. Sci. USA 98 14174–14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis, T., Brachmann, A., Kahmann, R., and Kämper, J. (2000). Identification of a target gene for the be-bW homeodomain protein complex in Ustilago maydis. Mol. Microbiol. 37 54–66. [DOI] [PubMed] [Google Scholar]
- Sgarlata, C., and Pérez-Martín, J. (2005. a). Inhibitory phosphorylation of a mitotic cyclin-dependent kinase regulates the morphogenesis, cell size and virulence of the smut fungus Ustilago maydis. J. Cell Sci. 15 3607–3622. [DOI] [PubMed] [Google Scholar]
- Sgarlata, C., and Pérez-Martín, J. (2005. b). The Cdc25 phosphatase is essential for the G2/M phase transition in the basidiomycete Ustilago maydis. Mol. Microbiol. 58 1482–1496. [DOI] [PubMed] [Google Scholar]
- Sherman, F., Fink, G.R., and Hicks, J.B. (1986). Laboratory Course Manual for Methods in Yeast Genetics. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Snetselaar, K.M., Bölker, M., and Kahmann, R. (1996). Ustilago maydis mating hyphae orient their growth toward pheromone sources. Fungal Genet. Biol. 20 299–312. [DOI] [PubMed] [Google Scholar]
- Snetselaar, K.M., Carfioli, M.A., and Cordisco, K.M. (2001). Pollination can protect maize ovaries from infection by Ustilago maydis, the corn smut fungus. Can. J. Bot. 79 1390–1399. [Google Scholar]
- Snetselaar, K.M., and Mims, C.W. (1992). Sporidial fusion and infection of maize seedlings by the smut fungus Ustilago maydis. Mycologia 84 193–203. [Google Scholar]
- Snetselaar, K.M., and Mims, C.W. (1993). Infection of maize stigmas by Ustilago maydis: Light and electron microscopy. Phytopathology 83 843–850. [Google Scholar]
- Spellig, T., Bölker, M., Lottspeich, F., Frank, R.W., and Kahmann, R. (1994). Pheromones trigger filamentous growth in Ustilago maydis. EMBO J. 13 1620–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg, G., Schliwa, M., Lehmler, C., Bölker, M., Kahmann, R., and McIntosh, J.R. (1998). Kinesin from the plant pathogenic fungus Ustilago maydis is involved in vacuole formation and cytoplasmic migration. J. Cell Sci. 111 2235–2246. [DOI] [PubMed] [Google Scholar]
- Steinberg, G., Wedlich-Söldner, R., Brill, M., and Schulz, I. (2001). Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J. Cell Sci. 114 609–622. [DOI] [PubMed] [Google Scholar]
- Suzuki, M., Gerstein, M., and Yagi, N. (1994). Stereochemical basis of DNA recognition by Zn fingers. Nucleic Acids Res. 22 3397–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot, N.J. (2003). On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57 177–202. [DOI] [PubMed] [Google Scholar]
- Tsukuda, T., Carleton, S., Fotheringham, S., and Holloman, W.K. (1988). Isolation and characterization of an autonomously replicating sequence from Ustilago maydis. Mol. Cell. Biol. 8 3703–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneault-Fourrey, C., Barooah, M., Egan, M., Wakley, G., and Talbot, N.J. (2006). Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 312 580–583. [DOI] [PubMed] [Google Scholar]
- Weber, I., Gruber, C., and Steinberg, G. (2003). A class V myosin required for mating, hyphal growth and pathogenicity in the dimporphic plant pathogen Ustilago maydis. Plant Cell 15 2826–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, S.A., Nekludova, L., and Pabo, C.O. (1999). DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 3 183–212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.