Abstract
Cyclotides are peptides from plants of the Rubiaceae and Violaceae families that have the unusual characteristic of a macrocylic backbone. They are further characterized by their incorporation of a cystine knot in which two disulfides, along with the intervening backbone residues, form a ring through which a third disulfide is threaded. The cyclotides have been found in every Violaceae species screened to date but are apparently present in only a few Rubiaceae species. The selective distribution reported so far raises questions about the evolution of the cyclotides within the plant kingdom. In this study, we use a combined bioinformatics and expression analysis approach to elucidate the evolution and distribution of the cyclotides in the plant kingdom and report the discovery of related sequences widespread in the Poaceae family, including crop plants such as rice (Oryza sativa), maize (Zea mays), and wheat (Triticum aestivum), which carry considerable economic and social importance. The presence of cyclotide-like sequences within these plants suggests that the cyclotides may be derived from an ancestral gene of great antiquity. Quantitative RT-PCR was used to show that two of the discovered cyclotide-like genes from rice and barley (Hordeum vulgare) have tissue-specific expression patterns.
INTRODUCTION
Naturally occurring circular proteins are becoming increasingly well known, with examples in bacteria, plants, and animals discovered in recent years (Trabi and Craik, 2002; Craik, 2006). These topologically interesting proteins have a continuous cycle of peptide bonds in their backbone and, accordingly, are devoid of N or C termini. Such proteins were unknown a decade ago, and they differ from previously known cyclic peptides, such as the immunosuppressant cyclosporin and other cyclic peptides found in microorganisms, in that they are conventional gene products rather than the output of nonribosomal synthetic processes. Cyclization of a protein backbone has the potential to provide stabilization relative to conventional proteins, both in a thermodynamic sense (Zhou, 2004) and biologically through their resistance to enzymic hydrolysis (Felizmenio-Quimio et al., 2001). Furthermore, because the termini of conventional proteins are often flexible and the degree of flexibility can be reduced by cyclization, entropic factors can lead to improved receptor binding affinities of circular proteins over corresponding acyclic proteins. Thus, circular proteins potentially have a range of advantages over conventional proteins and may have interesting applications in protein engineering, agriculture, and drug design (Clark et al., 2005; Craik et al., 2006a). Synthetic methods for producing circular proteins have become available in recent years (Camarero and Muir, 1997; Iwai and Pluckthun, 1999; Deechongkit and Kelly, 2002; Williams et al., 2002; Kimura et al., 2006), further stimulating interest in the field.
The cyclotides are the largest known family of circular proteins (Craik et al., 1999). They contain ∼30 amino acids, including six conserved Cys residues that are linked in pairs to form three disulfide bonds. The three disulfide bonds are connected in a knotted topology in which one disulfide threads a loop formed by the two other disulfides and the connecting backbone residues. The structural motif composing this cystine knot and a cyclic peptide backbone has been named the cyclic cystine knot, and based on sequence homology, it has been suggested that this motif is common to all cyclotides (Craik et al., 1999). This unique motif appears to be the main factor that contributes to the remarkable stability of the cyclotides, which retain bioactivity after boiling and are resistant to chemical, thermal, and enzymic treatments that would denature most proteins (Colgrave and Craik, 2004).
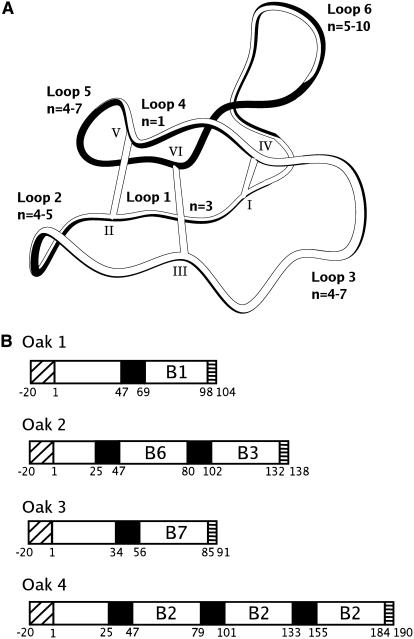
Figure 1 shows the cyclic cystine knot framework characteristic of cyclotides. The peptide backbone comprises six loops linking the six Cys residues, with different loops of different sizes and degrees of sequence conservation. The cyclotides are derived from precursor proteins that contain one, two, or three mature cyclotide domains. The mechanism by which a given precursor is processed into circular peptides is not yet known, but based on an alignment of gene sequences, the ligation point where the N and C termini of the precursor are joined has been localized to loop 6 (Jennings et al., 2001; Ireland et al., 2006). Each precursor protein contains an endoplasmic reticulum signal sequence, a propeptide region, one or more cyclotide domains, and a small hydrophobic tail.
Figure 1.
Summary of Topological, Structural, and Genetic Information on Cyclotides.
(A) Scheme of a cyclotide structure with the circular peptide backbone and knotted arrangement of disulfide bonds. The loop nomenclature, Cys numbering scheme, and range of amino acids (n) that constitute each loop for known cyclotides are also indicated.
(B) Gene organization for the Oak cDNA clones from O. affinis. Each clone contains a classical endoplasmic reticulum signal sequence (diagonal hatching), a precursor region (white), a short hydrophobic tail at the end of the precursor (horizontal hatching), and one to three mature cyclotide domains (labeled B1, B2, B3, B6, and B7) that are each preceded by an N-terminal repeat domain (black).
Approximately 50 cyclotide sequences have been published to date (Craik et al., 2004). Original discoveries were based on observations from native medicine practices or from bioassay-directed screening programs, but cyclotides may also be identified in plant extracts based on their characteristic late elution using reverse-phase HPLC. In recent years, our group (Craik et al., 1999; Trabi et al., 2004) and others (Göransson et al., 1999; Gustafson et al., 2000; Hallock et al., 2000; Hernandez et al., 2000; Bokesch et al., 2001; Broussalis et al., 2001) have screened a range of plants with the aim of discovering and identifying new peptides belonging to the cyclotide family. To date, cyclotides have been identified in every Violaceae plant screened as well as in a few Rubiaceae species. The Rubiaceae and Violaceae are not closely related phylogenetically, with the branch point for the two lineages encompassing the majority of the core eudicots. If the cyclotides did not evolve independently in the Rubiaceae and Violaceae and a common ancestral gene once existed, we might expect the cyclotides to be distributed throughout this varied class of plants. The failure of screening programs to detect cyclotides in a more diverse range of plants may be a function of the reliance on late elution in reverse-phase HPLC for screening, with less hydrophobic members of the group escaping detection. Alternatively, the cyclotides may have been inactivated or lost in the majority of plants within the core eudicots.
The major aim of this study was to determine whether the cyclotides may be more broadly distributed than has been realized and how they might have evolved from a presumably linear ancestral protein. A bioinformatics approach was adopted with searches based on both sequence homology and, to account for inter-Cys sequence divergence, Cys spacing patterns. An added complexity in searching for cyclic sequences is the possibility of variation in the processing point of a protein altering the order of residues in the linear precursor sequence; to address this possibility, a series of permuted searches were conducted.
Here, we report the finding that cyclotide-like sequences are present in a much wider range of plants than has been reported previously. In particular, we demonstrate their presence in some important graminaceous and other monocot species. Furthermore, the possible function of the genes was explored by examining the expression patterns of the discovered barley (Hordeum vulgare) and rice (Ozyza sativa) genes. These new sequences raise the possibility that the cyclotides evolved from an ancient gene common to many members of the plant kingdom. This possibility poses new questions about the evolutionary history of the cyclotides, not the least of which is, when and why did plants evolve the capacity to produce circular proteins?
RESULTS
Homology Searches Using BLAST
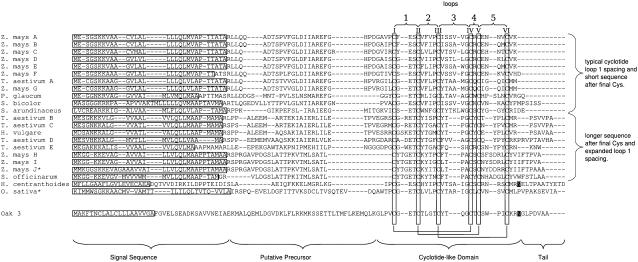
BLAST searches were automated and administered using a Web-based interface developed in our laboratory. Over 2 years, ∼1205 hits were recorded using this system. Each of these hits was examined manually, and those with characteristic Cys spacing were further analyzed using the GENSCAN (Burge and Karlin, 1997) and SignalP (Nielsen et al., 1997) programs. In total, 265 (23.61%) hits were predicted to contain an open reading frame (ORF) with a signal sequence and six-Cys domain, including four hits from known cyclotides (Jennings et al., 2001). From the significant hits, 22 nonredundant sequences were identified from the Poaceae and are shown in Figure 2. Compared with the predicted protein for Oak3, the sequence similarities and identities for the cyclotide-like sequences ranged from 39 to 56% and 14 to 26%, respectively. Maize (Zea mays) contained 10 novel sequences, wheat (Triticum aestivum) had 5 sequences, and single sequences were identified in pearl millet (Pennisetum glaucum), sorghum (Sorghum bicolor), tall fescue (Schedonorus arundinaceus), barley, sugarcane (Saccharum officinarum), and rice. In addition, a sequence was discovered from the Rubiaceae family member Hedyotis centranthoides. A partial sequence similar to the sequence found in H. centranthoides was also found in Hedyotis terminalis. After these novel genes were identified, they were used as the basis of fresh TBLASTN searches that produced 194 new hits but no new sequences.
Figure 2.
Novel Sequences Identified by Database Searching, Showing the Conserved Structure of the Genes in Poaceae Species.
Each sequence contains a signal sequence (horizontal boxes), a precursor region, the six-Cys domain, and a short C-terminal tail region. The numbering of the loops connecting each Cys residue is indicated at top. For comparison, the sequence of Oak3 from O. affinis is included at the bottom of the alignment, and the Asn that is the likely C-terminal processing point is highlighted in white letters on a black background. An Asn residue in a similar location in the cyclotide from H. centranthoides is similarly highlighted.
Regular Expression Searching
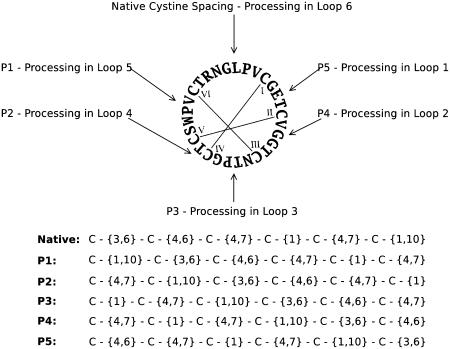
In parallel with the BLAST searching, regular expression (RE) searching was used to examine the completed genome sequences of rice and Arabidopsis thaliana. An RE was generated that coded for six Cys residues with spacings for each loop corresponding to the range observed in characterized cyclotides (Figure 3), and this was used to search DNA databases translated in six reading frames. The search of the Arabidopsis genome yielded no significant hits using the cyclotide Cys spacing. However, the search of rice produced a significant hit for a gene that encoded a six-Cys domain very similar to that in the cyclotides (Figure 2). In addition to the genomic searches, a variety of plant EST databases were searched using the same method. A total of 973 hits were recorded from these databases, and 252 were deemed significant. One sequence from maize was not obtained in the TBLASTN searches. Both of the novel sequences found in these searches were in turn used as queries in a fresh round of TBLASTN searches that produced 23 new hits but no new sequences.
Figure 3.
Sequence and Cys Connectivity of the Prototypical Cyclotide Kalata B1.
The RE corresponding to processing in each loop is indicated with an arrow. The actual REs used are shown at bottom.
Description of Identified Genes
Each of the genes identified in Poaceae species encodes a standard endoplasmic reticulum signal sequence and a putative precursor protein of at most 70 amino acids that includes a C-terminal domain of 26 to 36 amino acids that in turn contains six Cys residues spaced similarly to known cyclotides. Every sequence possesses a short tail after the C-terminal Cys residue that varies in length between 2 and 12 amino acid residues, similar to what is seen for the Oak cyclotide cDNAs shown in Figure 1, although in that case the tail is typically defined as the C-terminal segment following a conserved Asn residue implied in the excision of the cyclic mature peptide from its precursor. No multidomain sequences were discovered, in contrast with cyclotide precursors from Oldenlandia affinis, which often have multiple cyclotide domains encoded by a single mRNA transcript (Jennings et al., 2001), although single cyclotide domain genes are also found.
With the exception of the gene from rice, the gene products from the Poaceae fall into two broad groups (Figure 2). One group displays significant sequence identity in the region upstream from the first Cys residue and generally possesses short tails after the final Cys residue. Members of the second group have longer tails after the C-terminal Cys residue that possess a hydrophobic aspect attributable to the presence of conserved Val and Ala residues, show less interspecies conservation upstream of the six-Cys domain, and also display an expansion in the number of amino acids between the first and second Cys residues. The sequence from rice shares less identity with the other sequences in both the upstream region and in the loops of the six-Cys domain. The sequence also possesses an unusually long, 12–amino acid tail after the C-terminal Cys residue; only T. aestivum D possesses a tail of comparable (11 amino acids) length.
The sequence from H. centranthoides (Rubiaceae) has the organization of a typical cyclotide precursor. It possesses a short signal sequence, a 39–amino acid precursor region preceding the mature domain, and a tail region. It also has an Asn residue in loop 6, which is, after the six-Cys residues and the Glu residue in loop 1, one of the most conserved residues in the cyclotide family. This Asn residue has been suggested to be the C-terminal processing point in other cyclotides (Jennings et al., 2001), and in this case excision of the cyclotide domain would leave an unusually long C-terminal propeptide. Based on cyclotide nomenclature (Craik et al., 1999), the absence of a conserved Pro in loop 5 makes this a member of the bracelet subfamily. The sequence and organization of this clone is shared by the partial clone identified in H. terminalis.
Cyclic Permutant Searches
A consequence of backbone cyclization is that the order in which the Cys residues are arranged in the linear precursor cannot be determined by examination of the mature cyclic product. To take into account the possibility of alternative processing points in cyclic peptides related to the cyclotides, and to provide an independent check on the specificity of the search results, two additional strategies were used to search for linear precursors with circularly permuted Cys arrangements in the linear transcript. First, a series of TBLASTN searches was conducted using the original query sequences with the Cys residues and intervening amino acids systematically shuffled via circular permutation. Similarly, the native spacing RE was permuted to account for alternative processing points (Figure 3) and the same databases searched as above.
In total, five new TBLASTN searches were conducted with the query sets corresponding to the five possible permuted Cys organizations and 891 unique hits were recorded, none of which encoded for peptides with the necessary Cys spacing. Permuted RE searches of plant sequence databases yielded 11,119 hits for all five permutants of the RE. Six hits showed the correct Cys spacing and predicted signal sequence. Five of these hits came from the permutant 3 search of sugarcane and coded for the same sequence. The remaining hit was from the permutant 2 search of beetroot (Beta vulgaris). In both cases, ORF prediction software did not predict an ORF that included the target sequence. The results of the permutant searches were thus extremely definitive. The only permutant that leads to valid hits corresponds to the Cys framework of known cyclotides.
Expression Analysis
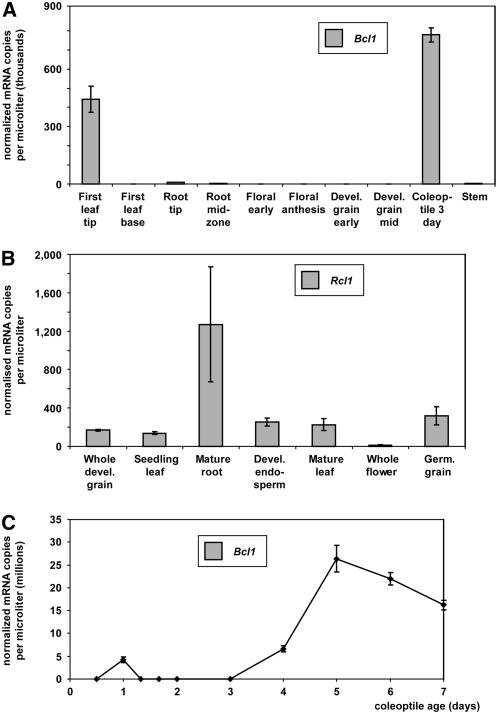
Transcript levels of the identified barley cyclotide-like gene, referred to hereafter as Bcl1 (for Barley cyclotide-like1), and the rice gene identified from a genomic database, referred to hereafter as Rcl1 (for Rice cyclotide-like1), were determined with quantitative PCR using gene-specific primers designed to include 3′ untranslated regions. These genes represented an example from each of the two groups of discovered genes, with Rcl1 containing typical cyclotide loop 1 spacing and Bcl1 containing an expanded loop 1. The transcript levels of the genes of interest were normalized against a series of internal controls (Burton et al., 2004), and the expression levels in a range of tissues were determined (Figure 4). Bcl1 showed relatively low expression levels in all tissues except for the first leaf tip and the 3-d coleoptile (Figure 4A). In these tissues, expression was at least 50 times that of the other tissues. In rice, low transcript levels were detected in all tissues, but the greatest amount was present in mature root and was absent from mature flowers (Figure 4B). Coleoptile libraries were not available for rice, but over a 7-d time course of barley coleoptile growth, expression of Bcl1 increased from 3 d from the start of imbibition and peaked at 5 d (Figure 4C). These results show that expression of Bcl1 is highest in fully expanded tissues.
Figure 4.
Normalized Expression Levels.
(A) Bcl1 from barley in a range of tissues.
(B) Rcl1 from rice in a range of tissues.
(C) Temporal expression of Bcl1 in coleoptile over the course of 7 d. Levels of mRNA are shown as the number of copies per microliter after normalization.
For (A) and (C), the data are averages of four repetitions; for (B), the data are averages of three repetitions. In all cases, the error shown is ±sd.
Mapping of Bcl1 and Rcl1
Bcl1 was mapped to the distal end of the short arm of chromosome 1 using the Clipper × Sahara 3771 DH population (Karakousis et al., 2003). It cosegregates with the restriction fragment length polymorphism marker ksuD14 and is ∼18 centimorgan distal to Hor1. No phenotypic or barley quality quantitative trait loci were present in the vicinity of the Bcl1 gene. Analysis of a 600-bp window of DNA containing Rcl1 with the program GENSCAN revealed the presence of an upstream promoter region as well as the presence of an ORF containing the putative gene. A BLAST search at Gramene (http://www.gramene.org/) revealed that the gene mapped to segment AP003920 136373 1 of chromosome 8.
Structural Modeling of Cyclotide-Like Peptides
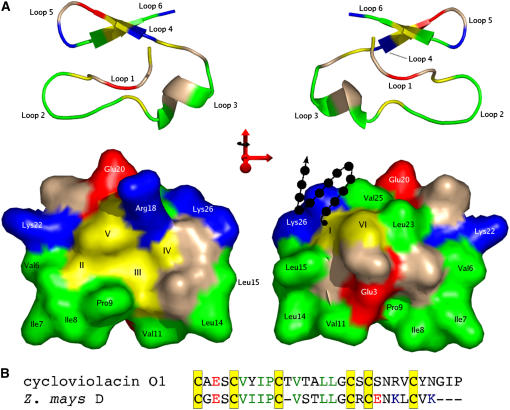
A homology model of one of the putative cyclotide sequences was constructed to determine the surface properties of the molecule if the six Cys residues were connected in a knotted topology. Of the 12 cyclotide structures published (Clark et al., 2006; Craik et al., 2006b; Daly et al., 2006), the sequences discovered in this study share the greatest degree of identity with circulin A and cycloviolacin O1. As the structure of cycloviolacin O1 was of higher resolution (Rosengren et al., 2003), the homology model was calculated using this structure as a template. The sequence identity of each of the new sequences with cycloviolacin O1 was calculated using BLASTP, and Z. mays D possessed the greatest identity (64%). The sequences were aligned using ClustalW (Thompson et al., 1994), and this alignment was used as the basis of the homology model using MODELLER (Fiser and Sali, 2003). The stereochemical quality of the model was analyzed using PROCHECK-NMR (Laskowski et al., 1996), and the covalent geometry of the modeled structures was in agreement with the template structure, with 90.5% of the residues occupying the most favored region of the Ramachandran plot and the remaining 9.5% in additionally allowed regions. Figure 5 shows the homology model. One of the defining characteristics of the cyclotides is the existence of one or more solvent-exposed hydrophobic patches, and it is clear that if the cyclotide-like sequences discovered in this work do contain a cystine knot, then they will also possess a similar pattern of solvent-exposed hydrophobic residues. Given that it is rather unusual for proteins to have a significant proportion of their surface made up of hydrophobic residues, this finding further strengthens the evidence that the sequences discovered in this study are directly related to the cyclotides.
Figure 5.
Homology Model of the Z. mays D Sequence.
(A) In the surface representations, positively charged residues are colored blue, negatively charged residues are shown in red, Cys residues are colored yellow, and hydrophobic residues are shown in green. The ribbon representations are colored in the same way, and the loop nomenclature is indicated. The orientation of the ribbon diagram is shared by the surface model immediately below, and each pair is related by a 180° rotation about the y axis. The arrow-and-circle motif on the surface representation is used to denote the possibility of additional N-terminal residues belonging to the mature peptide. Cys residues are marked with Roman numerals corresponding to their order in the linear precursor.
(B) The numbering of the non-Cys residues corresponds to the alignment shown used to generate the model. The alignment is color-coded in the same manner as the surface and ribbon representations. Molecule representations were prepared using the program PyMOL.
DISCUSSION
Here, we have shown that sequences with high identity to those of the cyclotide family of circular proteins are present in a variety of socially and economically important crop plants within the Poaceae, including rice, maize, and wheat. Furthermore, their existence in EST databases is consistent with expression in a wide variety of tissues and indicates that they may produce mature products with significant sequence identity to the cyclotides. This finding is potentially of great significance because cyclotide sequences have now been found in the monocots as well as both the asterid and rosid lineages of the eudicots (Daly et al., 2001), which suggests that the cyclotides may be derived from an ancestral gene in existence before the divergence of the monocot and dicot lineages ∼150 million years ago (Chaw et al., 2004).
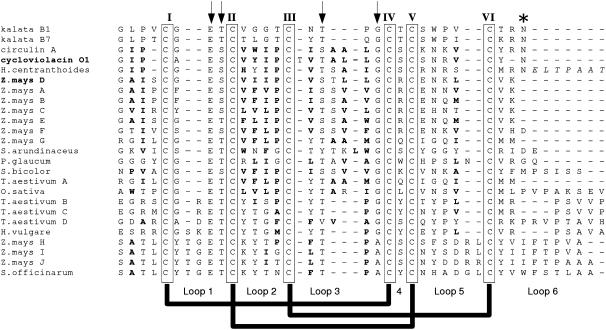
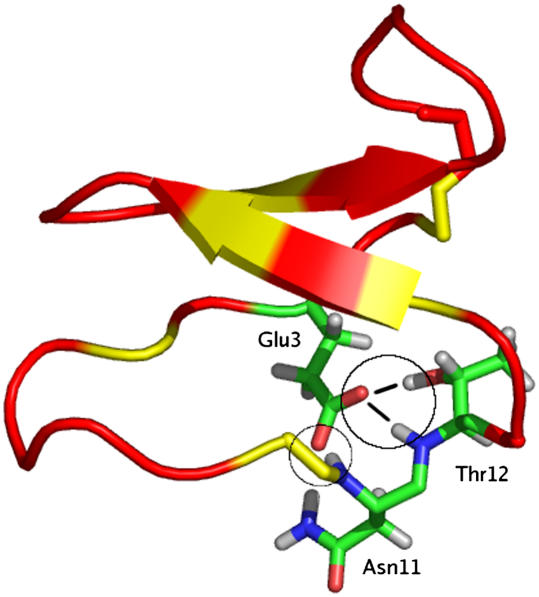
Although the amino acid composition and structures of the mature peptides encoded by these newly analyzed sequences have not yet been determined, the genes discovered in this study are remarkable for their similarity to the cyclotides. The similarities include a number of key residues that are important for the compact structure of the cyclotides (Rosengren et al., 2003). Figure 6 shows an alignment of the putative cyclotide domain of the grass sequences with a small set of known cyclotides. The most striking similarity is in loop 1, which contains a Glu residue that is absolutely conserved across all cyclotides and is present in all sequences discovered in this study. This residue is important in stabilizing the structure of cyclotides by forming a hydrogen bonding network with a hydroxyl-containing residue conserved as the second or third residue of loop 3, as illustrated in Figure 7 (Rosengren et al., 2003). Inspection of Figure 6 shows that this hydroxyl-containing residue is also conserved in all Poaceae sequences discovered except T. aestivum D. Other striking homologies include the conservation of another hydroxyl-bearing (Thr or Ser) residue immediately following the Glu residue in loop 1. This residue has been implicated in intraresidual hydrogen bonding (Rosengren et al., 2003). Also noteworthy is the strong conservation of the last residue in loop 3: in most cases Gly and in some cases Ala. In the cyclotides, this residue is almost always a Gly with a positive φ angle necessary for linking loop 3 to the cystine knot (Rosengren et al., 2003). The embedded ring of the cystine knot in the cyclotides is the smallest that would allow passage of a penetrating disulfide bond, and it is likely that these conserved residues are necessary to allow such a compact fold. Accordingly, their conservation in the sequences discovered here may reflect similarities in the three-dimensional structure of the mature peptides.
Figure 6.
Comparison of Cyclotide-Like Domains of Discovered Sequences with Mature Cyclotide Sequences.
Cys residues are boxed, and the absolutely conserved Glu in loop 1, the conserved hydroxyl-containing residues in loops 1 and 2, and the conserved Gly/Ala in the last position of loop 3 are highlighted with arrows. The conserved Asn residue in loop 6 of the known cyclotides, thought to be the C-terminal processing point, is marked with an asterisk. The loops between Cys residues and the disulfide connectivity of characterized cyclotides are shown at bottom. Boldface residues indicate hydrophobic residues that may be solvent-exposed based on sequence identity with circulin A and cycloviolacin O1. Italicized residues are part of the cyclotide precursor but are not likely to be present in the mature peptide. Z. mays D and cycloviolacin O1 are shown in boldface to denote their use in homology modeling.
Figure 7.
Ribbon Diagram of the Prototypical Cyclotide Kalata B1 Highlighting the Hydrogen Bond Network Formed by Glu-3.
The location of the Cys residues on the ribbon are denoted with a yellow tint. For the stick representations of Glu-3, Asn-11, and Thr-12, oxygen atoms are colored red, hydrogen atoms are gray, nitrogen atoms are blue, and carbon is green. Actual atoms involved in the hydrogen bonding are circled. The diagram was prepared using the program PyMOL based on the Protein Data Base coordinates (code 1NB1) for kalata B1.
All published cyclotide solution structures exhibit a similar fold that forces the exposure of a hydrophobic patch to the surface. The homology model shown in Figure 5 suggests that, at least for Z. mays D, if a cystine knot is present then a hydrophobic surface will be exposed to the surface. It can be seen that the hydrophobic residues from loops 2 and 3 form a hydrophobic patch along one face of the molecule, with charged residues and additional hydrophobic residues arranged on the other face, as is the case in the bracelet cyclotides (Felizmenio-Quimio et al., 2001). It can be seen in Figure 6 that, at least for the sequences with typical loop 1 spacing, the conservation of hydrophobic residues suggests that a similar situation could occur for these proteins. The importance of the solvent-exposed hydrophobic patches for the natural function of the cyclotides is not known, but the high degree of sequence similarity between the Poaceae sequences and cyclotides may indicate that they are necessary for a natural function shared by the two groups of molecules.
The cyclotides exhibit a range of bioactivities. Apart from their uterotonic activity, they display hemolytic (Schöpke et al., 1993; Gustafson et al., 1994; Witherup et al., 1994; Craik et al., 2001; Barry et al., 2003), anti–human immunodeficiency virus (Gustafson et al., 1994; Hallock et al., 2000; Bokesch et al., 2001), neurotensin inhibitory (Witherup et al., 1994), antimicrobial (Tam et al., 1999a), insecticidal (Jennings et al., 2001), antifouling (Göransson et al., 2004), and antitumor/cytotoxic (Tam et al., 1999b; Lindholm et al., 2002) activities. Neither the mechanisms of action nor the functional significance to the plant has been definitively established, although their potent insecticidal activity suggests that they have a role in plant defense. The quantitative PCR data presented here demonstrate that the cyclotide-like sequences found in the Poaceae are expressed in a tissue-specific manner. With so little data on the tissue specificity of cyclotide-like genes available, it is difficult to use this information to interpret possible functions for these genes. However, Basse (2005) found a gene with similar sequence, Umi11 (described in this work as Z. mays I), to be upregulated in basal shoot tissue and in leaf blade tumors formed after challenge with the smut fungus Ustilago maydis. These data suggest that the role of plant defense postulated for cyclotides (Jennings et al., 2001) may be extended to include cyclotide-like genes from the Poaceae.
The distribution of the cyclotides in every Violaceae species but apparently in only a few plants from the Rubiaceae poses questions about the evolution of these macrocyclic peptides. Previous explanations for this distribution have either relied on the failure to detect peptides in particular species using current screening methods or, conversely, indicated that the cyclotides are descended from an ancestral gene that was subsequently lost or inactivated in most eudicot lineages, including the majority of the Rubiaceae (Craik et al., 2004). The discovery of sequences with significant identity to the cyclotides in the Poaceae, and the absence of these sequences in the Arabidopsis genome, adds to the evolutionary picture. If these sequences and the cyclotides are related, then the ancestral gene is even more ancient than the divergence of Rubiaceae and Violaceae. The question becomes why cyclotides or cyclotide-like sequences have been retained in only three phylogenetically dispersed plant families and not in the large number of plants that share similar ancestry. Furthermore if these sequences represent a linear ancestral gene, as discussed below, then the evolution of cyclization either occurred before the branching of the Rubiaceae and Violaceae and was subsequently lost in plants with shared ancestry that do not contain cyclotides, or the cyclizing mechanism evolved separately in the Rubiaceae and Violaceae lineages. Given the interest in cyclization as a method of stabilizing bioactive peptides (Craik et al., 2002; Clark et al., 2005), the elucidation of this process is of great interest. The lack of significant hits in the permutated searches is particularly important because it indicates that the only sequences found that share a resemblance to the cyclotides have the same organization of Cys residues in the linear precursor. This finding suggests that such an organization is an absolute requirement for cyclization.
The cyclization mechanism of the cyclotides has not yet been elucidated. However, by analysis of mRNA transcripts from O. affinis (Jennings et al., 2001) and Viola odorata (Dutton et al., 2004), the C-terminal processing point has been identified in loop 6 immediately following a conserved Asn residue. Apart from the sequence from H. centranthoides, which retains the Asn residue, the sequences reported here do not contain an Asn in loop 6. Indeed, in many of the Poaceae sequences, loop 6 is greatly reduced in size; combined with the lack of the conserved Asn, this suggests that the corresponding mature peptides are not cyclic or that, if they are, a different mechanism of cyclization is involved. Interestingly, those Poaceae sequences with a longer sequence after the C-terminal Cys residue show a prevalence of Val and Ala residues that are reminiscent of the short hydrophobic tails found in the cyclotide precursors (see Figure 2 for the sequence of Oak3). If these sequences are related to the cyclotides, the development of a tail such as this may have been a necessary step toward a cyclic molecule.
The evolution of a cyclic backbone in plant proteins is a particularly intriguing phenomenon. On the one hand, a macrocyclic backbone introduces a new paradigm into the field of protein topology; on the other hand, cyclic proteins can be regarded as just one step away from conventional proteins (i.e., via the addition of just one more peptide bond to the n − 1 that are already present in a protein of n residues). Because the termini of proteins are often structurally disordered, and because the addition of one final tethering peptide bond between the termini has the potential to decrease this disorder, we refer to cyclization as the completion of nature's unfinished business. Head-to-tail cyclization leads to a seamless circle of n peptide bonds for n amino acids. With the field of circular proteins still very much in its infancy, the discovery of potential ancestral proteins to the large family of cyclotides reported here represents an important step in understanding these topologically intriguing molecules.
METHODS
Query Sequences and RE
Protein query sequences used during the BLAST searches consisted of 49 mature cyclotide sequences identified in our own screening programs as well as those characterized by other groups (Craik et al., 1999; Göransson et al., 1999; Gustafson et al., 2000; Hallock et al., 2000; Hernandez et al., 2000; Bokesch et al., 2001; Broussalis et al., 2001; Trabi et al., 2004). The initial native spacing RE was generated by determining the maximum and minimum number of residues in each loop of the 49 sequences (shown in Figure 1). These parameters were used as non-Cys amino acid spacing intervals for each of the loops in the RE. Based on early positive BLAST hits, the RE spacings were amended from the precise spacing of known cyclotides by increasing the maximum length of loop 1 to 6 and decreasing the minimum length of loop 6 to 1.
BLAST Searches
The BLAST searches were automated using a Perl script that submitted a 49-sequence query set to the publicly accessible National Center for Biotechnology Information (NCBI) BLAST servers on a weekly basis. The script searched the NCBI nonredundant database using each query sequence in a TBLASTN search using the BLOSUM62 scoring matrix, with the expected score set to 100. Returned searches were parsed by the script, and novel hits were extracted and stored. A Web-based interface was written using the scripting language PHP that allowed the novel hits to be inspected manually. Those hits that were not obvious nonspecific hits, that possessed the characteristic cystine knot spacing, that did not have large numbers of Cys residues outside of the putative cystine knot domain, and that did not contain long runs of a single amino acid or repeated amino acid patterns were retained for further analysis.
For potentially novel hits, the complete sequence was obtained from the NCBI database and analyzed with GENSCAN for ORFs (Burge and Karlin, 1997) and with SignalP for signal sequences (Nielsen et al., 1997). The parameter set optimized for maize was used for the prediction of ORFs in all monocot databases searched, and the set optimized for Arabidopsis thaliana was used in all eudicot species. If an ORF containing the BLAST hit was predicted, then the translated sequence was analyzed further using SignalP. If a signal sequence was predicted using the eukaryotic organism group, then the sequence was entered into the database. BLAST hits were aligned using the program ClustalW (Webba da Silva et al., 2001), and consensus sequences were derived by choosing the most conserved sequence over the range of hits. These consensus sequences were then used in new TBLASTN searches using the methodology described above.
RE Searches
A script was written using the scripting language Perl that searched flat file databases of DNA sequences in FASTA format for protein patterns corresponding to the REs described above. The script used the Bioperl modules to translate each sequence into six reading frames, and these translations were searched for the particular RE. If a match was made, then the script output the pattern found as well as 20 residues on either side of the pattern. Additional scripts filtered the output of the search. If the database being searched contained EST sequences, then the results were filtered by examining the preceding 20 amino acids for stop codons and discarding hits if these elements were present. Similarly, if the succeeding 20 amino acids did not contain a stop codon or contained additional Cys residues before the stop codon, then the hit was also discarded. Finally, the gi number of the hit was compared with a database of characterized BLAST hits, and if present it was disregarded. The remaining hits were analyzed manually using GENSCAN and SignalP in the manner described above. If a genomic database was being examined, the contig containing the hit was retrieved and analyzed using GENSCAN, and if the hit was recognized as an ORF it was further analyzed with SignalP. Using this procedure, searches were made using the native RE and each of the five permutants (Figure 3) on the complete genome sequence databases of rice (Oryza sativa), Arabidopsis, and various plant EST databases. Redundant hits were identified in the manner described above.
Databases
All TBLASTN searches were conducted using the NCBI BLAST server (http://www.ncbi.nlm.nih.gov/BLAST/) and the nonredundant database. For RE searches, EST databases for individual plants were downloaded from the PlantGDB website (http://www.plantgdb.org/). The complete genome sequence for rice was downloaded from the Rice GD website (http://btn.genomics.org.cn:8080/rice/), and the genome sequence database for Arabidopsis was downloaded from The Arabidopsis Information Resource website (http://www.arabidopsis.org/index.jsp).
Homology Modeling
Each of the Poaceae sequences containing only three residues between the first and second Cys residues was compared with the sequence of cycloviolacin O1 using the program BLASTP (Altschul et al., 1997) and the default settings. The sequence showing the greatest identity, Zea mays D, was used as the basis of a model based on the lowest energy structure of the cycloviolacin O1 structure. The program MODELLER (Fiser and Sali, 2003) was used to calculate 10 homology models by optimizing the objective function from 10 different initial conformations. The model with the lowest objective function was chosen as the representative model. The stereochemical quality of the model was analyzed using PROCHECK-NMR (Laskowski et al., 1996). The ribbon and surface diagrams were produced using the program PyMOL.
Expression Analysis
Tissues for the rice series were collected from mature plants (cv Nippon Bare) grown at 28°C day and 22°C night temperatures under high humidity, a photointensity of 300 μmol·m−2·s−1, and an 11/13-h day/night regime. Mature leaf tissue was fully expanded, whereas young leaf tissue was not. The young developing grain represents the whole grain, and developing endosperm was dissected away from the other tissues of the young grain. All tissues were snap-frozen in liquid nitrogen and used for total RNA extraction and cDNA synthesis as described by Burton et al. (2004).
The preparation of the barley (Hordeum vulgare) tissue series was as described by Burton et al. (2004). The barley coleoptile time course was prepared from grains of the barley cv Sloop, which were soaked in oxygenated water overnight at 20°C, transferred to damp vermiculite, and germinated in the dark at 20°C. The start of the time course was the point at which imbibition began. Coleoptiles were collected for 7 d, all contaminating adherent scutellum or leaf tissue was removed, and the tissue was snap-frozen in liquid nitrogen and stored at −80°C. RNA was extracted, and cDNA was prepared using standard procedures as described by Burton et al. (2004).
Quantitative PCR was performed according to Burton et al. (2004), including the primers for the rice control genes as described by Kim et al. (2003). For details of the primers used, see Supplemental Table 1 online.
Map Location
The barley Clipper × Sahara 3371 doubled haploid mapping population (Karakousis et al., 2003) was used to map the Bcl1 gene. DNA gel blot hybridization analyses were performed on genomic DNA filters kindly supplied by Peter Langridge (Australian Centre for Plant Functional Genomics, Adelaide, Australia), and the locus was positioned using Map Manager QT version b29ppc (Manly and Olson, 1999).
Accession Numbers
Representative sequences for each cyclotide-like gene can be found in GenBank under accession numbers CF060985, CF014141, CK369406, BM379838, CF630454, CF013901, CN070702, BI674581, BM080572, and CK368015 for Z. mays A to J; CA617438, CK154330, CK154890, CA595705, and BE591233 for T. aestivum A to E; BE125990 for S. bicolor; CD725989 for P. glaucum; AL450615 for H. vulgare; CK803164 for S. arundinaceus; CA274667 for S. officinarum; and 2CB084585 for H. centranthoides. There is no EST identifier for the O. sativa gene because it was discovered in a genomic database. A complete list of sequences can be located in the GenBank data libraries using the accession numbers listed in Supplemental Table 2 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. PCR Primers and PCR Product Sizes in Base Pairs, Together with Optimal Acquisition Temperatures for the Genes Analyzed.
Supplemental Table 2. EST Sequences Discovered during BLAST and RE Searches.
Supplementary Material
Acknowledgments
This work was funded by grants from the Australian Research Council (to D.J.C. and M.A.A.) and the Grains Research and Development Corporation (to G.B.F.). We thank Margaret Pallotta for her mapping expertise, Andrew Harvey for primer design assistance, and Ute Baumann.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: David J. Craik (d.craik@imb.uq.edu.au).
Online version contains Web-only data.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, D.G., Daly, N.L., Clark, R.J., Sando, L., and Craik, D.J. (2003). Linearization of a naturally occurring circular protein maintains structure but eliminates hemolytic activity. Biochemistry 42 6688–6695. [DOI] [PubMed] [Google Scholar]
- Basse, C.W. (2005). Dissecting defense-related and developmental transcriptional responses of maize during Ustilago maydis infection and subsequent tumor formation. Plant Physiol. 138 1774–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokesch, H.R., Pannell, L.K., Cochran, P.K., Sowder, R.C., II, McKee, T.C., and Boyd, M.R. (2001). A novel anti-HIV macrocyclic peptide from Palicourea condensata. J. Nat. Prod. 64 249–250. [DOI] [PubMed] [Google Scholar]
- Broussalis, A.M., Göransson, U., Coussio, J.D., Ferraro, G., Martino, V., and Claeson, P. (2001). First cyclotide from Hybanthus (Violaceae). Phytochemistry 58 47–51. [DOI] [PubMed] [Google Scholar]
- Burge, C., and Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268 78–94. [DOI] [PubMed] [Google Scholar]
- Burton, R.A., Shirley, N.J., King, B.J., Harvey, A.J., and Fincher, G.B. (2004). The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol. 134 224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarero, J., and Muir, T. (1997). Chemoselective backbone cyclization of unprotected peptides. Chem. Commun. 15 1369–1370. [Google Scholar]
- Chaw, S.-M., Chang, C.-C., Chen, H.-L., and Li, W.-H. (2004). Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J. Mol. Evol. 58 424–441. [DOI] [PubMed] [Google Scholar]
- Clark, R.J., Daly, N.L., and Craik, D.J. (2006). Structural plasticity of the cyclic-cystine-knot framework: Implications for biological activity and drug design. Biochem. J. 394 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, R.J., Fischer, H., Dempster, L., Daly, N.L., Rosengren, K.J., Nevin, S.T., Meunier, F.A., Adams, D.J., and Craik, D.J. (2005). Engineering stable peptide toxins by means of backbone cyclization: Stabilization of the alpha-conotoxin MII. Proc. Natl. Acad. Sci. USA 102 13767–13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgrave, M.L., and Craik, D.J. (2004). Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: The importance of the cyclic cystine knot. Biochemistry 43 5965–5975. [DOI] [PubMed] [Google Scholar]
- Craik, D.J. (2006). Seamless proteins tie up their loose ends. Science 311 1563–1564. [DOI] [PubMed] [Google Scholar]
- Craik, D.J., Anderson, M.A., Barry, D.G., Clark, R.J., Daly, N.L., Jennings, C.V., and Mulvenna, J. (2002). Discovery and structures of the cyclotides: Novel macrocyclic peptides from plants. Lett. Pept. Sci. 8 119–128. [Google Scholar]
- Craik, D.J., Cĕmažar, M., and Daly, N.L. (2006. a). The cyclotides and related macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Dev. 9 251–260. [PubMed] [Google Scholar]
- Craik, D.J., Cĕmažar, M., Wang, C.K.L., and Daly, N.L. (2006. b). The cyclotide family of circular miniproteins: Nature's combinatorial peptide template. Biopolymers 84 250–266. [DOI] [PubMed] [Google Scholar]
- Craik, D.J., Daly, N.L., Bond, T., and Waine, C. (1999). Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J. Mol. Biol. 294 1327–1336. [DOI] [PubMed] [Google Scholar]
- Craik, D.J., Daly, N.L., Mulvenna, J., Plan, M.R., and Trabi, M. (2004). Discovery, structure and biological activities of the cyclotides. Curr. Protein Pept. Sci. 5 297–315. [DOI] [PubMed] [Google Scholar]
- Craik, D.J., Daly, N.L., and Waine, C. (2001). The cystine knot motif in toxins and implications for drug design. Toxicon 39 43–60. [DOI] [PubMed] [Google Scholar]
- Daly, D.C., Cameron, K.M., and Stevenson, D.W. (2001). Plant systematics in the age of genomics. Plant Physiol. 127 1328–1333. [PMC free article] [PubMed] [Google Scholar]
- Daly, N.L., Clark, R.J., Plan, M.R., and Craik, D.J. (2006). Kalata B8, a novel antiviral circular protein, exhibits conformational flexibility in the cystine knot motif. Biochem. J. 393 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deechongkit, S., and Kelly, J.W. (2002). The effect of backbone cyclization on the thermodynamics of beta-sheet unfolding: Stability optimization of the PIN WW domain. J. Am. Chem. Soc. 124 4980–4986. [DOI] [PubMed] [Google Scholar]
- Dutton, J.L., Renda, R.F., Waine, C., Clark, R.J., Daly, N.L., Jennings, C.V., Anderson, M.A., and Craik, D.J. (2004). Conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins. J. Biol. Chem. 279 46858–46867. [DOI] [PubMed] [Google Scholar]
- Felizmenio-Quimio, M.E., Daly, N.L., and Craik, D.J. (2001). Circular proteins in plants: Solution structure of a novel macrocyclic trypsin inhibitor from Momordica cochinchinensis. J. Biol. Chem. 276 22875–22882. [DOI] [PubMed] [Google Scholar]
- Fiser, A., and Sali, A. (2003). MODELLER: Generation and refinement of homology-based protein structure models. Methods Enzymol. 374 461–491. [DOI] [PubMed] [Google Scholar]
- Göransson, U., Luijendijk, T., Johansson, S., Bohlin, L., and Claeson, P. (1999). Seven novel macrocyclic polypeptides from Viola arvensis. J. Nat. Prod. 62 283–286. [DOI] [PubMed] [Google Scholar]
- Göransson, U., Sjögren, M., Svangård, E., Claeson, P., and Bohlin, L. (2004). Reversible antifouling effect of the cyclotide cycloviolacin O2 against barnacles. J. Nat. Prod. 67 1287–1290. [DOI] [PubMed] [Google Scholar]
- Gustafson, K.R., Sowder, R.C.I., Henderson, L.E., Parsons, I.C., Kashman, Y., Cardellina, J.H.I., McMahon, J.B., Buckheit, R.W.J., Pannell, L.K., and Boyd, M.R. (1994). Circulins A and B: Novel HIV-inhibitory macrocyclic peptides from the tropical tree Chassalia parvifolia. J. Am. Chem. Soc. 116 9337–9338. [Google Scholar]
- Gustafson, K.R., Walton, L.K., Sowder, R.C.I., Johnson, D.G., Pannell, L.K., Cardellina, J.H.I., and Boyd, M.R. (2000). New circulin macrocyclic polypeptides from Chassalia parvifolia. J. Nat. Prod. 63 176–178. [DOI] [PubMed] [Google Scholar]
- Hallock, Y.F., Sowder, R.C.I., Pannell, L.K., Hughes, C.B., Johnson, D.G., Gulakowski, R., Cardellina, J.H.I., and Boyd, M.R. (2000). Cycloviolins A-D, anti-HIV macrocyclic peptides from Leonia cymosa. J. Org. Chem. 65 124–128. [DOI] [PubMed] [Google Scholar]
- Hernandez, J.F., Gagnon, J., Chiche, L., Nguyen, T.M., Andrieu, J.P., Heitz, A., Trinh Hong, T., Pham, T.T., and Le Nguyen, D. (2000). Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry 39 5722–5730. [DOI] [PubMed] [Google Scholar]
- Ireland, D.C., Colgrave, M.L., Nguyencong, P., Daly, N.L., and Craik, D.J. (2006). Discovery and characterization of a linear cyclotide from Viola odorata: Implications for the processing of circular proteins. J. Mol. Biol. 357 1522–1535. [DOI] [PubMed] [Google Scholar]
- Iwai, H., and Pluckthun, A. (1999). Circular beta-lactamase: Stability enhancement by cyclizing the backbone. FEBS Lett. 459 166–172. [DOI] [PubMed] [Google Scholar]
- Jennings, C., West, J., Waine, C., Craik, D., and Anderson, M. (2001). Biosynthesis and insecticidal properties of plant cyclotides: The cyclic knotted proteins from Oldenlandia affinis. Proc. Natl. Acad. Sci. USA 98 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakousis, A., Barr, A.R., Kretschmer, J.M., Manning, S., Jefferies, S.P., Chalmers, K.J., Islam, A.K.M., and Langridge, P. (2003). Mapping and QTL analysis of the barley population Clipper × Sahara. Aust. J. Agric. Res. 54 1137–1140. [Google Scholar]
- Kim, B.-R., Nam, H.-Y., Kim, S.-U., Kim, S.-I., and Chang, Y.-J. (2003). Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol. Lett. 25 1869–1872. [DOI] [PubMed] [Google Scholar]
- Kimura, R.H., Tran, A.-T., and Camarero, J.A. (2006). Biosynthesis of the cyclotide kalata B1 by using protein splicing. Angew. Chem. Int. Ed. Engl. 45 973–976. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R., and Thornton, J.M. (1996). AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8 477–486. [DOI] [PubMed] [Google Scholar]
- Lindholm, P., Goransson, U., Johansson, S., Claeson, P., Gulbo, J., Larsson, R., Bohlin, L., and Backlund, A. (2002). Cyclotides: A novel type of cytotoxic agents. Mol. Cancer Ther. 1 365–369. [PubMed] [Google Scholar]
- Manly, K.F., and Olson, J.M. (1999). Overview of QTL mapping software and introduction to Map Manager QT. Mamm. Genome 10 327–334. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10 1–6. [DOI] [PubMed] [Google Scholar]
- Rosengren, K.J., Daly, N.L., Plan, M.R., Waine, C., and Craik, D.J. (2003). Twists, knots, and rings in proteins. Structural definition of the cyclotide framework. J. Biol. Chem. 278 8606–8616. [DOI] [PubMed] [Google Scholar]
- Schöpke, T., Hasan Agha, M.I., Kraft, R., Otto, A., and Hiller, K. (1993). Hämolytisch aktive komponenten aus Viola tricolor L. und Viola arvensis Murray. Sci. Pharm. 61 145–153. [Google Scholar]
- Tam, J.P., Lu, Y.A., Yang, J.L., and Chiu, K.W. (1999. a). An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc. Natl. Acad. Sci. USA 96 8913–8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam, J.P., Lu, Y.-A., and Yu, Q. (1999. b). Thia zip reaction for synthesis of large cyclic peptides: Mechanisms and applications. J. Am. Chem. Soc. 121 4316–4324. [Google Scholar]
- Thompson, J., Higgins, D., and Gibson, T. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabi, M., and Craik, D.J. (2002). Circular proteins—No end in sight. Trends Biochem. Sci. 27 132–138. [DOI] [PubMed] [Google Scholar]
- Trabi, M., Svangård, E., Herrmann, A., Göransson, U., Claeson, P., Craik, D.J., and Bohlin, L. (2004). Variations in cyclotide expression in Viola species. J. Nat. Prod. 67 806–810. [DOI] [PubMed] [Google Scholar]
- Webba da Silva, M., Sham, S., Gorst, C.M., Calzolai, L., Brereton, P.S., Adams, M.W.W., and La Mar, G.N. (2001). Solution NMR characterization of the thermodynamics of the disulfide bond orientational isomerism and its effect of cluster electronic properties for the hyperthermostable three-iron cluster ferredoxin from the archaeon Pyrococcus furiosus. Biochemistry 40 12575–12583. [DOI] [PubMed] [Google Scholar]
- Williams, N.K., Prosselkov, P., Liepinsh, E., Line, I., Sharipo, A., Littler, D.R., Curmi, P.M., Otting, G., and Dixon, N.E. (2002). In vivo protein cyclization promoted by a circularly permuted Synechocystis sp. PCC6803 DnaB mini-intein. J. Biol. Chem. 277 7790–7798. [DOI] [PubMed] [Google Scholar]
- Witherup, K.M., Bogusky, M.J., Anderson, P.S., Ramjit, H., Ransom, R.W., Wood, T., and Sardana, M. (1994). Cyclopsychotride A, a biologically active, 31-residue cyclic peptide isolated from Psychotria longipes. J. Nat. Prod. 57 1619–1625. [DOI] [PubMed] [Google Scholar]
- Zhou, H.X. (2004). Loops, linkages, rings, catenanes, cages, and crowders: Entropy-based strategies for stabilizing proteins. Acc. Chem. Res. 37 123–130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.