Abstract
The balance between cell proliferation, cell cycle arrest, and differentiation needed to maintain the organogenetic program depends on the coordination of gene expression, posttranslational modification, and specific proteolysis of cell cycle regulators. The G1/S and G2/M transitions are critical checkpoints controlled, in part, by cyclin-dependent kinases in the retinoblastoma (RBR)/E2F/DP pathway. Arabidopsis thaliana DPB is regulated by phosphorylation and targeted to proteasome-mediated proteolysis by the SCFSKP2A complex. In addition, DPB interacts in vivo with E2FC, because ectopic coexpression of E2FC and DPB produces severe developmental defects. To understand E2FC/DPB heterodimer function, we analyzed the effect of reducing E2FC mRNA levels with RNA interference. The e2fc-R plants developed organs with more but smaller cells and showed increased cell cycle marker gene expression and increased proliferative activity in developing leaves, meristems, and pericycle cells. This last feature produces plants with more lateral roots, consistent with an E2FC role in restricting lateral root initiation. The e2fc-R plants also show marked reductions in ploidy levels of mature leaves. These results indicate that the transition from cell division to the endocycle is sensitive to different pathways, E2FC/DPB being one of them. Our results show that E2FC/DPB is a key factor in controlling the balance between cell proliferation and the switch to the endocycle program.
INTRODUCTION
The development and growth of multicellular organisms depend on the correct spatiotemporal coordination of cell proliferation, cell differentiation, and subsequent cell specialization (Coffman, 2004). Cell division is a tightly regulated process that is influenced by innate genetic cues and external environmental signals that, ultimately, control gene expression, posttranslational modifications, and proteolysis of multiple regulatory proteins. The retinoblastoma (RB)-E2F pathway is one of the most important regulatory pathways that control and couple cell division and cell differentiation in both animals and plants (Gutierrez et al., 2002; Stevaux and Dyson, 2002). The E2F and DP proteins interact to form active transcription factors that bind to different gene promoters and regulate the expression of genes required for cell cycle progression. The RB protein, known as RB-related (RBR) in plants, binds to E2F proteins, masking the transactivation region and blocking transcriptional activity. This repression can be eliminated by the activity of cyclin-dependent kinases (CDKs), which phosphorylate the RB protein, releasing a functional E2F-DP heterodimer (Cooper and Shayman, 2001; Gutierrez et al., 2002; De Veylder et al., 2003; Dewitte and Murray, 2003; Murray, 2004).
The completion of the Arabidopsis thaliana genome has made possible the identification of the majority of the core cell cycle genes in plants (Arabidopsis Genome Initiative, 2000; Vandepoele et al., 2002). The E2F/DP family in Arabidopsis, formed by six E2F and two DP proteins, can be divided into two subfamilies based on their domain structure (Gutierrez et al., 2002; Shen, 2002). E2FA to E2FC possess the typical organization found in mammalian E2F1 to E2F5, with a DNA binding domain, a DP heterodimerization domain, a RBR binding domain, and a transactivation domain. On the contrary, E2FD to E2FF, which are homologues to the recently identified E2F7 and E2F8 in mammals (Di Stefano et al., 2003; Christensen et al., 2005; Maiti et al., 2005), do not have the DP heterodimerization and RBR binding domains; instead, they have a duplicated DNA binding domain that allows DNA binding without the collaboration of the DP proteins (Gutierrez et al., 2002; Kosugi and Ohashi, 2002a; Shen, 2002; Ramirez-Parra et al., 2004). It appears that E2Fa and E2Fb function preferentially with DPa, based on the use of plants with specific combinations (De Veylder et al., 2002; Sozzani et al., 2006). Previous studies of E2FC have shown that it does not have transcriptional activation properties in transient expression experiments (Kosugi and Ohashi, 2002b). We have shown that overexpression of a nondegradable form of E2FC protein delays cell division and represses the expression of S-phase genes (del Pozo et al., 2002). We also showed that the activity of E2FC is regulated at the level of transcription, CDK-dependent phosphorylation, and specific protein degradation through the SCFSKP2A complex (del Pozo et al., 2002). The SCFSKP2A complex is an E3 ligase of ubiquitin that is composed of a common scaffold (CUL1, ASK, and RBX) (del Pozo and Estelle, 2000) and the F-box subunit SKP2A that specifically interacts with the targets. Furthermore, E2FC degradation was triggered during the transition from dark to light growth conditions, suggesting that E2FC plays an important role in this transition.
In this work, we show that DPB is regulated by phosphorylation and by specific degradation through the ubiquitin pathway, involving the activity of the SCFSKP2A complex. In vivo data show that E2FC and DPB form a heterodimer. Overexpression of DPB and a stable form of E2FC (del Pozo et al., 2002) produces severe defects in cotyledon and leaf development. On the contrary, plants with reduced levels of E2FC show lower levels of DNA endoreplication and increased levels of E2F target gene expression. These molecular alterations are accompanied by the development of more but smaller cells in leaves and cotyledons. Furthermore, partial loss of E2FC is sufficient to highly increase the amount of cyclin B1;1 expression in dividing and differentiated tissues, whereas overexpression of E2FC reduces the number of cells in G2/M in the dividing areas. Another striking feature of these plants is that the pericycle cells are more responsive to auxin and develop more lateral root primordia (LRP). Overexpression of E2FC dramatically reduces the number of cells in the root meristem that express mitotic markers and increases DNA content. Together, these data demonstrate that E2FC/DPB restricts cell division and is one of the components in the coordination between cell proliferation and endoreplication during Arabidopsis development.
RESULTS
DPB and E2FC Form a Heterodimer in Vivo
To understand the role of DPB, we decided to study the expression of this gene by RT-PCR from different organs. DPB is expressed ubiquitously at similar levels in all organs analyzed (see Supplemental Figure 1A online). This is consistent with microarray data (https://www.genevestigator.ethz.ch/) (Zimmermann et al., 2004). This search revealed that DPB and E2FC have similar expression patterns and levels in the different organs analyzed, except in the stamen, where E2FC mRNA is more abundant than DPB (see Supplemental Figure 1B online).
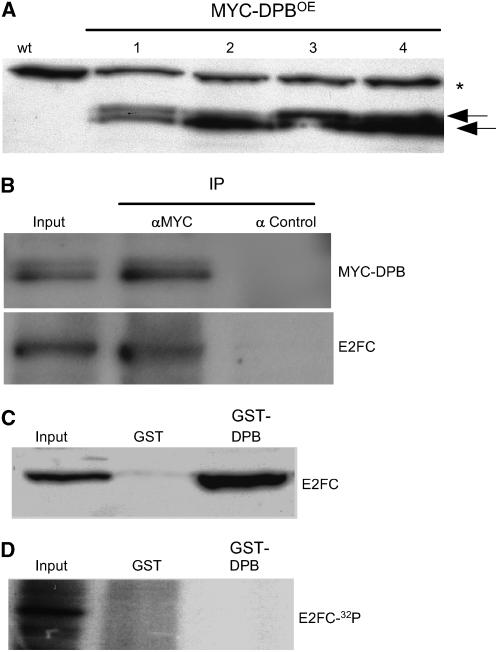
Previous in vitro binding experiments suggested that DPB interacts with E2FC (del Pozo et al., 2002; Mariconti et al., 2002). To evaluate this in vivo, we generated transgenic plants that overexpress DPB protein fused to the MYC epitope (MYC-DPBOE) (Figure 1A). Immunoprecipitation assays showed that E2FC interacts with MYC-DPB in vivo (Figure 1B). We also found that DPB interacts with the nonphosphorylated E2FC produced in bacteria (del Pozo et al., 2002) (Figure 1C). Because CDK activity plays an important role in regulating the cell cycle and E2FC was phosphorylated in vitro (del Pozo et al., 2002), we tested whether CDK phosphorylation affects the DPB–E2FC interaction. As shown in Figure 1D, recombinant His-E2FC, phosphorylated in vitro with CDKA;1/CYCA2;2 as described (del Pozo et al., 2002), does not interact with GST-DPB, whereas the nonphosphorylated His-E2FC form does (Figure 1C), suggesting that CDK activity negatively regulates the formation of E2FC/DPB heterodimers.
Figure 1.
E2FC and DPB Proteins Form a Heterodimer in Vivo.
(A) Protein gel blot analysis of several independent lines (MYC-DPBOE) that overexpress the MYC-DPB protein using the antibody against the MYC epitope. The arrows indicate the two specific bands identified. The asterisk indicates an unspecific cross-reacting band.
(B) Total protein extract from MYC-DPBOE plants were immunoprecipitated using agarose beads coupled to IgGs against the MYC epitope (IP αMYC) or agarose beads coupled to IgGs against the hemagglutinin (HA) epitope as a control (IP α Control). The precipitated proteins were examined by protein gel blot analysis using the antibody against MYC (MYC-DPB) or the affinity-purified anti-E2FC (E2FC) IgGs.
(C) Recombinant E2FC protein was used in pull-down assays with GST or GST-DPB. The pulled-down proteins were examined by protein gel blot analysis using the IgGs against E2FC.
(D) CDKA-phosphorylated radiolabeled E2FC protein was used in pull-down assays with GST or GST-DPB. Bound proteins were fractionated by SDS-PAGE, and the dried gel was exposed to detect the radiolabeled E2FC (E2FC-32P).
DPB Is Regulated by Phosphorylation and the Ubiquitin-Proteasome Pathway
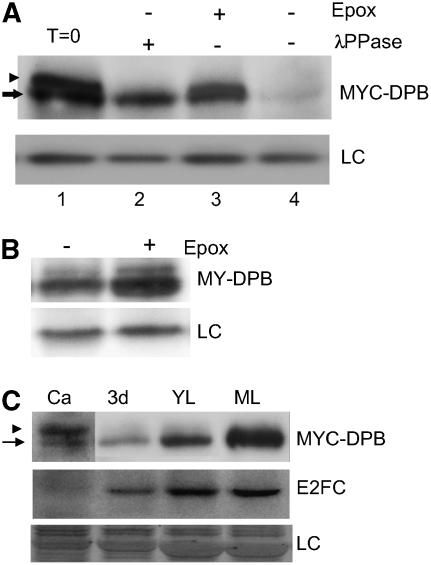
The presence of two specific bands detected in MYC-DPBOE plants (Figure 1A) led us to investigate whether the slower migrating band corresponded to a phosphorylated form of MYC-DPB protein. When total protein extracts of MYC-DPBOE plants were incubated with λ-phosphatase, the slower migrating band disappeared, suggesting that this band corresponded to a phosphorylated form of DPB (Figure 2A, lanes 1 and 2). It is noteworthy that when MYC-DPBOE protein extracts were incubated at 30°C but not treated with λ-phosphatase, the levels of the MYC-DPB protein were reduced dramatically (Figure 2A, lane 4). However, this reduction was prevented by the addition of the highly specific proteasome inhibitor epoxomicin in the reaction, suggesting that MYC-DPB degradation occurred through the proteasome (Figure 2A, lane 3). To confirm this in vivo, we treated MYC-DPBOE seedlings with or without epoxomicin for 4 h and then analyzed the levels of MYC-DPB. As shown in Figure 2B, MYC-DPB accumulated after treatment with the proteasome inhibitor.
Figure 2.
DPB Is Regulated by Phosphorylation and Proteolysis.
(A) Immunoblot analysis of MYC-DPBOE plant extracts treated with or without λ-phosphatase (λPPase). Where indicated, epoxomicin (Epox) was included in the reaction. The arrow indicates the MYC-DPB protein, and the arrowhead points to the phosphorylated MYC-DPB form. LC, loading control, corresponding to an unspecific cross-reacting protein; T=0, fresh extracts without any treatment.
(B) Immunoblot of protein extracts from MYC-DPBOE seedlings treated with (+) or without (−) the proteasome inhibitor epoxomicin. LC, loading control, corresponding to an unspecific cross-reacting protein.
(C) MYC-DPB and E2FC protein phosphorylation patterns at different stages of development. Ca, calli cells; 3d, 3-d-old seedlings; YL, young rosette leaves; ML, mature rosette leaves of 21-d-old plants. Top panel, anti-MYC; middle panel, anti-E2FC. Note that the Ca lane hybridized with anti-MYC was exposed five times longer than the others. LC, loading control, corresponding to the same gel stained with Coomassie blue. The arrow points to the MYC-DPB protein, and the arrowhead indicates phosphorylated MYC-DPB.
Analysis of MYC-DPB protein of overexpressing plants at different stages of development showed that DPB is differentially phosphorylated. As shown in Figure 2C, the phosphorylated DPB isoform is more abundant in dividing cells (calli) than in seedlings and mature rosette leaves. The same blot was analyzed with the IgGs against E2FC, showing that in calli cells the level of E2FC is much lower than in mature leaves (Figure 2C), following a pattern similar to MYC-DPB.
DPB Is Targeted for Proteolysis by the SCFSKP2A Complex
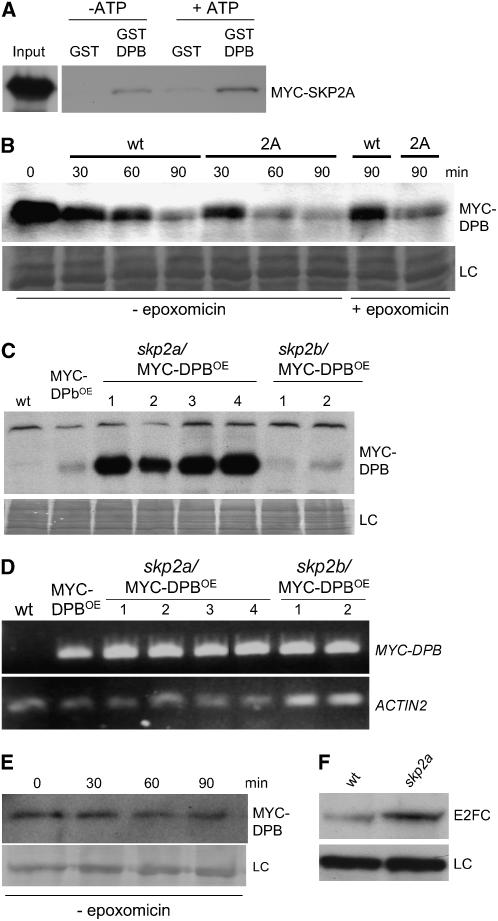
Previously, we showed that the SCFSKP2A complex targets E2FC for proteasome degradation (del Pozo et al., 2002). To test whether the SCFSKP2A complex might also be responsible for targeting DPB protein, we performed pull-down experiments using plant extracts prepared from MYC-SKP2A–overexpressing plants and recombinant GST or GST-DPB. As shown in Figure 3A, GST-DPB, but not GST alone, interacts with MYC-SKP2A. Furthermore, the amount of MYC-SKP2A recovered in the pull-down experiments increased when ATP was added to the reaction (see Methods), suggesting that a higher phosphorylation of DPB likely stimulates its recognition by SKP2A. Consistent with this notion, using an in vitro degradation assay, we found that MYC-DPB was degraded in a proteasome-dependent manner and that such degradation is accelerated by the addition of protein extract from MYC-SKP2AOE plants (Figure 3B).
Figure 3.
SCFSKP2A Targets DPB/E2FC for Ubiquitin-Dependent Degradation.
(A) MYC-SKP2A interacts with GST-DPB but not with GST in pull-down experiments. The interaction was favored in the presence of high levels of ATP (+ATP) during the incubation.
(B) Time-course degradation of the MYC-DPB protein. Protein extracts from MYC-DPBOE plants were incubated with an extract of wild-type plants or with an extract of MYC-SKP2A–overexpressing plants (2A). Where indicated, the proteasome inhibitor (epoxomicin) was added. LC, loading control, corresponding to the same blot stained with Coomassie blue; min, incubation time in minutes.
(C) Protein extracts of wild type, MYC-DPBOE, and different lines (1 to 4) of skp2a/MYC-DPBOE and skp2b/MYC-DPBOE plants were examined by protein gel blot analysis using the anti-MYC antibody to detect MYC-DPB. LC, loading control, corresponding to the same blot stained with Coomassie blue.
(D) The expression level of the MYC-DPB transgene was analyzed by RT-PCR in the same plants used in (C). As a control, the level of the ACTIN2 gene was analyzed.
(E) Time-course degradation of the MYC-DPB protein. Protein extract from skp2a/MYC-DPBOE (line 1) plants was incubated for different times in a degradation assay. MYC-DPB was detected by immunoblotting. LC, unspecific cross-reacting protein used as a loading control.
(F) E2FC protein level in 5-d-old wild-type and skp2a seedlings.
We have identified T-DNA insertion mutants for the SKP2A and SKP2B genes in the Gabi-Kat and SALK collections, respectively, and the structures of these T-DNA insertion lines will be described elsewhere (H. Ren, J.C. del Pozo, J.H. Murray, and M. Estelle, unpublished data). In short, visual analyses of the single and double mutants did not reveal any gross macroscopic alterations. To address whether mutation on SKP2 genes affects the stability of DPB in vivo, the MYC-DPB transgene was introduced into the mutants. As shown in Figure 3C, MYC-DPB accumulates to high levels in the skp2a but not in the skp2b mutant. We analyzed the expression level of the transgene by RT-PCR and confirmed that the mRNA expression level was equivalent in the control and mutant backgrounds (Figure 3D). To test the degradation rate of MYC-DPB in the skp2a mutant, an in vitro degradation assay was performed. The MYC-DPB protein remained very stable in extracts from the skp2a mutant at different times, whereas MYC-DPB was almost degraded in wild-type extracts (Figures 3B and 3E). In addition, we also found higher levels of E2FC protein in the skp2a mutant (Figure 3F). Together, these results indicate that the E2FC/DPB heterodimer is targeted for degradation, in a phosphorylation-dependent manner, by the SCFSKP2A complex.
Overexpression of E2FC and DPB Interferes with Plant Development
The amount of E2FC (del Pozo et al., 2002) and DPB (this work) seems to be regulated by selective proteolysis. Phenotypic analysis of MYC-DPBOE plants did not reveal significant morphological changes with respect to wild-type plants (see Supplemental Figure 2 online). We have generated transgenic plants that ectopically express a genomic region that contains the full-length E2FC gene. Although we have identified plants that overexpress the E2FC mRNA, we did not find plants that accumulate significantly higher levels of E2FC protein. Phenotypic analyses of these plants did not reveal differences between wild-type and E2FCOE plants (data not shown). Furthermore, we crossed plants that overexpress MYC-DPB and full-length E2FC, but these E2FC/DPBOE plants did not show gross macroscopic differences from wild-type plants (data not shown). Plants that overexpress MYC-DPB and a stable form of E2FC, which accumulated to a high level (ΔE2FC) (del Pozo et al., 2002), did show severe defects in plant development (see Supplemental Figure 2 online). Other transgenic ΔE2FC/MYC-DPBOE plants exhibited a milder phenotype, and they have much lower E2FC transgene expression. We determined the ploidy profile in leaves of 14-d-old plants with the mild phenotype and found a significant stimulation of endocycle occurrence. Similarly, the severe dwarf phenotype showed increased polyploidy, in this case analyzed in seedlings (see Supplemental Figure 2 online). These results, together with the in vivo interaction data, show that DPB and E2FC form an active heterodimer in planta that promotes the occurrence of endoreplication cycles.
Reduction of E2FC Levels Affects Cell Proliferation and Nuclear DNA Content
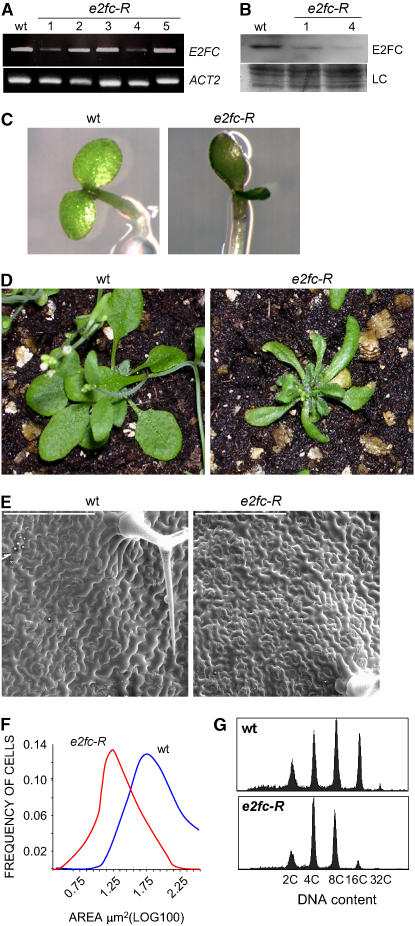
To better define the function of E2FC, we analyzed the phenotype of plants that have lower levels of E2FC. To this end, we generated transgenic plants (e2fc-R) harboring an RNA interference (RNAi) construct with the N-terminal region of the E2FC cDNA, a sequence that did not show significant homology with other members of the Arabidopsis E2F family. The level of E2FC mRNA was analyzed by RT-PCR and shown to be reduced in several independent lines (Figure 4A). Two of these lines (1 and 4) also showed highly reduced levels of E2FC protein (Figure 4B). The e2fc-R lines identified that have lower levels of E2FC mRNA and E2FC protein showed similar phenotypic alterations, which are described below.
Figure 4.
Reduction of E2FC Level Affects Plant Development and Reduces Ploidy Level.
(A) E2FC expression level was analyzed in several independent plant lines (1 to 5) that express an E2FC RNAi (e2fc-R) by semiquantitative RT-PCR of mRNA extracted from 6-d-old seedlings. The ACTIN2 (ACT2) gene was amplified as a control.
(B) E2FC protein levels were analyzed by immunoblotting in the wild type and two lines of e2fc-R (lines 1 and 4) with reduced E2FC mRNA levels. LC, loading control, corresponding to the same blot stained with Coomassie blue.
(C) Phenotypes of 4-d-old wild-type and e2fc-R seedlings.
(D) Phenotypes of 24-d-old wild-type and e2fc-R plants.
(E) Scanning electron micrographs showing epidermal cells of the fourth leaf of 24-d-old wild-type and e2fc-R plants. Bars = 200 μm.
(F) Frequency distribution of epidermal cells with different areas. Measurements were taken using the fourth rosette leaf of 24-d-old wild-type and e2fc-R plants.
(G) Ploidy distribution of fourth rosette leaf nuclei of 24-d-old wild-type and e2fc-R plants.
During the early stage of seedling development, we found, in a high percentage, seedlings with slightly cup-shaped cotyledons of asymmetric size (Figure 4C). Older e2fc-R plants developed smaller and curled rosette leaves (Figure 4D). Analysis at the cellular level showed that these leaves also contained more but smaller epidermal cells than wild-type leaves (Figures 4E and 4F). To analyze the nuclear ploidy level, we chose mature rosette leaves, because these mature organs reflect the changes accumulated during development. We found that the e2fc-R plants had a significantly lower DNA ploidy than wild-type plants. In these e2fc-R leaves, the 16C peak almost disappeared, with a concomitant increase in the amount of 4C nuclei (Figure 4G). In addition, cross-sections of these rosette leaves showed that the e2fc-R plants developed a complete cellular organization, although the sizes of the palisade cells as well as the intercellular air spaces in the spongy mesophyll layer were reduced significantly (see Supplemental Figure 3 online). A similar cellular phenotype was found in e2fc-R cotyledons, which showed a higher number of epidermal cells per unit of area, but these cells were smaller than in the wild type (see Supplemental Figure 3 online). This phenotype was also accompanied by a reduction in the ploidy of cotyledon nuclei, although this was smaller than in leaves (see Supplemental Figure 3 online).
Reduction of E2FC Level Increases Cell Cycle Gene Expression and Cell Division Potential
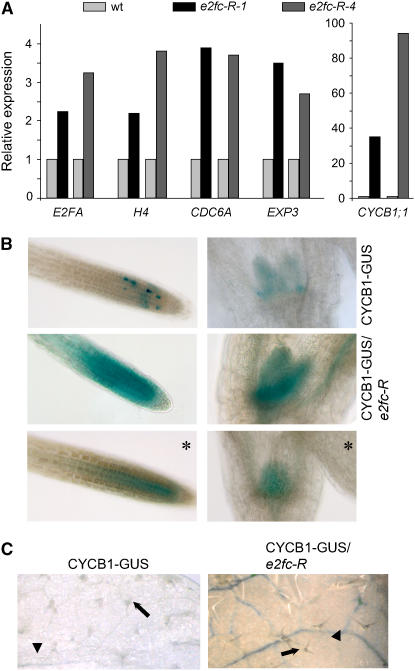
Plant E2F/DP proteins are transcription factors that regulate the expression of cell cycle and differentiation genes (De Veylder et al., 2002; Ramirez-Parra et al., 2003, 2004; Vlieghe et al., 2003; Vandepoele et al., 2005). This, together with the phenotype described above, led us to investigate the expression of a set of cell division genes in each of the two e2fc-R plants (lines 1 and 4) by RT-PCR, using RNA isolated from young dividing or mature rosette leaves. As shown in Figure 5A, the level of E2Fa (De Veylder et al., 2002) was increased in the mature leaves of e2fc-R plants compared with wild-type plants. In addition, we found that the levels of two S-phase markers, histone H4 (Reichheld et al., 1998) and CDC6a (Castellano et al., 2001), were also increased significantly in mature leaves (Figure 5A). Recently, Ramirez-Parra et al. (2004) showed that the expression of some expansin genes, required for cell wall growth during cell differentiation, was regulated by E2Ff. Analysis by RT-PCR showed that the level of EXP3, which contains an E2F site in its promoter, was increased significantly in e2fc-R plants. It is noteworthy that significant changes in gene expression were found in mature leaves but not in young leaves (data not shown), suggesting that E2FC acts by repressing E2F-regulated genes in mature differentiated cells. The level of the CYCB1;1 gene, a G2/M marker (Doerner et al., 1996), increased very significantly in the mature leaves of e2fc-R plants (Figure 5A).
Figure 5.
E2FC Regulates Cell Cycle Gene Expression and Cell Division.
(A) Expression level of different marker genes determined by real-time RT-PCR in mature rosette leaves of 21-d-old wild-type and e2fc-R plants. The two e2fc-R lines (1 and 4) were included in the analysis to show that they exhibit a comparable response to reduction in E2FC level. The expression level of each gene was normalized against the expression of the ACTIN2 gene.
(B) Seven-day-old CYCB1-GUS and CYCB1-GUS/e2fc-R plants stained for GUS activity for 14 h. The photographs correspond to root tips (left panels) and developing leaves (right panels). Panels with asterisks correspond to samples of e2fc-R seedlings stained for only 5 h.
(C) Twenty-one-day-old rosette leaves of CYCB1-GUS and CYCB1-GUS/e2fc-R were stained for GUS activity. Arrowheads indicate vascular strands, and arrows indicate trichome cells.
To further explore the spatial regulation of cell division mediated by E2FC, we analyzed the expression and/or stability of the mitotic cyclin B1;1 in e2fc-R plants. We crossed plants that harbor the CYCB1∷CYCB1-GUS construct, which contains the promoter and the destruction box (D-box) of CYCB1;1 fused to the uidA gene (Colón-Carmona et al., 1999), with e2fc-R plants (CYCB1-GUS/e2fc-R). When we analyzed the β-glucuronidase (GUS) staining pattern in the CYCB1-GUS and in the CYCB1-GUS/e2fc-R lines, we observed that the reduction of E2FC transcripts leads to a striking accumulation of CYCB1-GUS not only in zones of normal cell division, such as root meristems, leaf primordia, and young developing leaves (Figure 5B), but also in the vascular tissue of mature leaves (Figure 5C). This is consistent with the RT-PCR data shown above in mature leaves (Figure 5A). It is remarkable that the increase in GUS activity observed in the CYCB1-GUS/e2fc-R lines spatially correlates with the areas where E2FC is expressed normally (del Pozo et al., 2002), suggesting a direct effect of E2FC in repressing cell proliferation in differentiated cells. Trichomes, specialized cells that undergo several endoreplication cycles, did not accumulate GUS activity in CYCB1-GUS/e2fc-R plants (Figure 5C), although they express E2FC (del Pozo et al., 2002), reinforcing the idea that other components, besides E2FC, are important for cyclin B1 expression, as demonstrated recently (Li et al., 2005).
E2FC Regulates Cell Division of Pericycle Cells
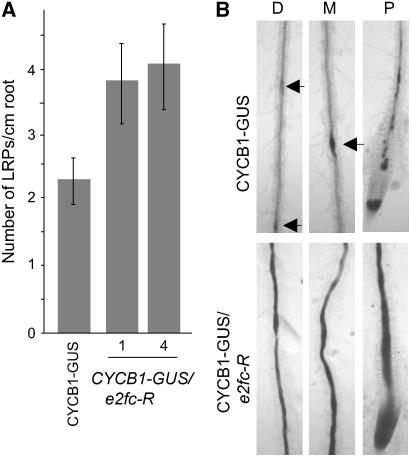
LRP development is a suitable process in which to analyze cell cycle arrest and reactivation, because they are formed from the founder pericycle cells that are arrested at G1 (Himanen et al., 2004). CYCB1-GUS/e2fc-R plants develop ∼40% more LRP (counted as GUS activity; see Methods) per unit length of principal root than control plants (Figure 6A). It is well known that a local increment of auxin induces cell division in pericycle cells, stimulating the formation of LRP (Marchant et al., 2002; Benkova et al., 2003). Several cell cycle genes are induced early during lateral root development, including the CYCB1;1 gene (Himanen et al., 2002, 2004). As shown in Figure 6B, CYCB1;1 levels were strikingly increased in pericycle cells of the CYCB1-GUS/e2fc-R roots when treated with auxin, either close to the root tip or in distal areas from the tip, whereas a discrete increment of activity was detected in CYCB1-GUS roots. These results are fully in agreement with the proposed role of E2FC as a cell cycle repressor and suggest that E2FC is one of the components that maintain pericycle cells arrested in G1 and/or restrict their reentering into the division cycle to form LRP.
Figure 6.
E2FC Negatively Regulates Lateral Root Formation.
(A) Mean values of the numbers of lateral roots developed per centimeter of the principal root in wild-type and two e2fc-R lines (1 and 4) grown for 10 d in Murashige and Skoog (MS) medium. Data are means of 40 seedlings in two independent experiments ± sd.
(B) GUS staining of auxin-treated roots of CYCB1-GUS and CYCB1-GUS/e2fc-R seedlings. D, area distal from the root tip; M, area in the middle of the root; P, area proximal to the root tip. Arrowheads indicate GUS staining of young LRP.
E2FC Regulates the Balance between G2/M and DNA Endoreplication
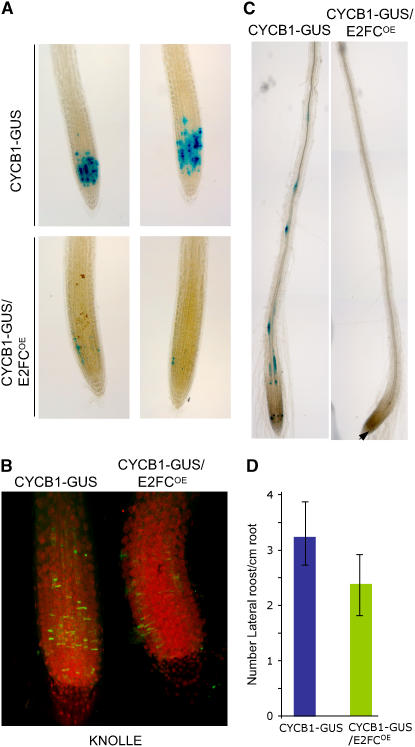
Because overexpression of full-length E2FC does not macroscopically alter the development of Arabidopsis plants, we searched for possible effects at the molecular level. Thus, we crossed plants that overexpress the full-length E2FC (E2FCOE) with plants harboring the CYCB1-GUS cell cycle marker. As shown in Figure 7A, overexpression of E2FC provoked a dramatic reduction of cells that accumulated CYCB1;1 protein in the root meristem or in the shoot meristems and primary leaves (data not shown). To determine whether increased E2FC levels affected the G2/M transition, we identified the KNOLLE protein, which accumulates in the division plane of mitotic cells (Lauber et al., 1997), and found a reduction of mitotic cells in the E2FCOE root meristems (Figure 7B). These results indicate that ectopic expression E2FC reduces cell division potential in the meristems. In addition, CYCB1-GUS/E2FCOE plants treated with auxin did not increase CYCB1-GUS activity in root pericycle cells (Figure 7C). This finding is consistent with a slight reduction of LRP formation compared with wild-type plants in the absence of auxin (Figure 7D).
Figure 7.
Overexpression of E2FC Reduces the Level of CYCB1;1.
(A) GUS staining of CYCB1-GUS and CYCB1-GUS/E2FCOE roots of seedlings grown in MS medium for 7 d.
(B) KNOLLE protein immunolocalization (green) and propidium iodide staining (red) of CYCB1-GUS and CYCB1-GUS/E2FCOE roots. The image shows a stack of multiple confocal layers.
(C) GUS staining of roots of CYCB1-GUS and CYCB1-GUS/E2FCOE seedlings treated with auxin. The arrowhead indicates a GUS-stained cell in the meristem.
(D) Mean values of the numbers of LRP formed per centimeter of the main root in CYCB1-GUS and CYCB1-GUS/E2FCOE seedlings grown for 7 d in MS medium. Data are means of 40 seedlings in two independent experiments ± sd.
DISCUSSION
Plant growth requires a high degree of coordination between cell division and differentiation. Cell division is a greatly regulated process that involves the integrated activity of diverse proteins such as CDK, cyclins, and CDK inhibitors that modulate the RB-E2F pathway, among others (Gutierrez et al., 2002; Dewitte and Murray, 2003). In animals, the RB-E2F pathway regulates cell cycle progression (Harbour and Dean, 2000) and also participates in non-cell-cycle events, although through mechanisms still poorly understood (Harbour and Dean, 2000; Müller et al., 2001; Stevaux and Dyson, 2002; Dimova et al., 2003). In plants, an analogous pathway that involves the RBR protein and E2F/DP factors play important roles in cell proliferation, cell differentiation, and macromolecular biosynthesis (del Pozo et al., 2002; De Veylder et al., 2002; Gutierrez et al., 2002; Ramirez-Parra et al., 2003, 2004; Vlieghe et al., 2003, 2005; Ebel et al., 2004; Magyar et al., 2005; Park et al., 2005; Vandepoele et al., 2005; Desvoyes et al., 2006; Sozzani et al., 2006). Here, we show that the SCFSKP2A complex targets both DPB and E2FC for degradation through the ubiquitin pathway. In addition, the DPB protein is also regulated by phosphorylation, a modification that seems to be important for its turnover. In vivo data show that DPB and E2FC form a heterodimer that likely negatively regulates cell proliferation and lateral root initiation and positively regulates the endocycle program, at least in cotyledons and leaves. Therefore, E2FC/DPB, together with other E2F factors, contributes to the coupling of cell division potential with development.
DPB, an E2FC Partner, Is Regulated by Phosphorylation and Ubiquitin-Dependent Degradation
We have found that hyperphosphorylated DPB is predominant in actively dividing cells, whereas the hypophosphorylated form seems to be more abundant in differentiated tissues. This is opposite what is seen in mammalians cells, where DP-1 was phosphorylated during cell differentiation and the hypophosphorylated DP-1 was associated with cell proliferation (Bandara et al., 1994; Tilli et al., 2003). In these cells, the PP2A phosphatase seems to be implicated in dephosphorylating DP-1 (Tilli et al., 2003). At present, and like the human DP-1, the kinase(s) that phosphorylates DPB is unknown. However, although DPB contains the minimal S/TP consensus CDK phosphorylation sites, we were unable to obtain DPB phosphorylation in vitro with CDKA/CYCA2 or CDKA/CYCD2, complexes that efficiently phosphorylate most of the E2F proteins (data not shown). This suggests that DPB likely is not phosphorylated by the CDKA/CYC complexes, although we cannot disprove that other combinations of CDKA with CYC can phosphorylate DPB.
The programmed proteolysis of cell cycle regulators is of crucial importance in the control of mammalian and yeast cell division. In plants, the ubiquitin-26S pathway regulates multiple processes of development. Recent reports have shown that the ubiquitin-26S pathway also controls cell division in plants, destroying different regulators such as E2FC (del Pozo et al., 2002), cyclin D3 (Planchais et al., 2004), KRP2 (Verkest et al., 2005), and KRP1 (H. Ren, J.C. del Pozo, J.H. Murray, and M. Estelle, unpublished data). The recruitment of target proteins for ubiquitination and subsequent degradation through the proteasome is performed by the E3 ubiquitin ligases (Fang and Weissmann, 2004; Moon et al., 2004). One type of E3 is the SCF complex, which is composed of three scaffold subunits and one exchangeable subunit, the F-box protein, that confers the specificity for target recognition (Patton et al., 1998). Previously, we reported that E2FC is regulated transcriptionally and by CDK-dependent, proteasome-mediated degradation, a process that involves the SCFSKP2 complex (del Pozo et al., 2002). Human SKP2 targets for degradation different cell cycle regulators, such as p27 (Tsvetkov et al., 1999), cyclin E (Nakayama et al., 2000), E2F1 (Marti et al., 1999), CDK9 (Kiernan et al., 2001), RB-like p130 (Tedesco et al., 2002), p57kip2 (Kamura et al., 2003), and CDT1 (Li et al., 2003). Human DP-1 is also regulated by ubiquitin-mediated degradation (Magae et al., 1999), although the pathway involved in this proteolysis is still unknown. Here, we showed that Arabidopsis DPB is degraded by the proteasome in a phosphorylation-dependent manner. In this work, we present strong evidence indicating that SKP2A interacts with DPB, an interaction favored by ATP, and most likely recruits DPB for ubiquitination and degradation, because MYC-DPB degradation is faster in the presence of higher levels of SCFSKP2A. Finally, MYC-DPB accumulates to high levels in the skp2a, but not in the skp2b, mutant. It is remarkable that E2FC also accumulates in the skp2a mutant. Together, the biochemical and genetic data support the conclusion that the SCFSKP2A complex is the E3 that targets the E2FC/DPB transcription factor for degradation through the ubiquitin-proteasome pathway.
E2FC-DPB Restricts Cell Division
In Arabidopsis, in which one RBR, six E2F, and two DP proteins have been identified, the RBR-E2F pathway regulates the cell cycle, cell differentiation, and endoreplication (del Pozo et al., 2002; De Veylder et al., 2002; Ramirez-Parra et al., 2003, 2004; Vlieghe et al., 2003; Ebel et al., 2004; Magyar et al., 2005; Park et al., 2005; Desvoyes et al., 2006; Sozzani et al., 2006). Although E2Fd to E2Ff bind DNA as monomers (Kosugi and Ohashi, 2002a), E2Fa to E2Fc require DP partners to heterodimerize. Several pieces of evidence demonstrate that both E2Fa and E2Fb function as cell cycle activators (De Veylder et al., 2002; Magyar et al., 2005; Sozzani et al., 2006), whereas E2FC acts as a transcriptional repressor (del Pozo et al., 2002). Coexpression experiments in tobacco (Nicotiana tabacum) cells showed that E2Fa and E2Fb induced the expression of an E2F-regulated marker when coexpressed with DPa but only very marginally with DPB (Kosugi and Ohashi, 2002b). Overexpression of E2Fa/DPa in Arabidopsis led to ectopic cell division, increases in DNA endoreplication, and a concomitant increase in the mRNA level of cell cycle genes (De Veylder et al., 2002). E2Fb, in collaboration with DPa, activates cell division in cultured BY2 cells in the absence of auxin in the medium, accelerates both G1/S and G2/M transitions (Magyar et al., 2005), and controls cell cycle progression and development in Arabidopsis (Sozzani et al., 2006).
We showed that E2FC binds to DPB in planta, in agreement with coexpression experiments in yeast and tobacco cells. However, we did not observe gross developmental changes in plants that ectopically express DPB and full-length E2FC mRNA. A likely possibility is that this is caused by their regulation by active proteolysis, because coexpression of DPB together with a stable form of E2FC (ΔE2FC), which accumulates to high levels, has a dramatic effect on the development of Arabidopsis plants. E2FC was proposed to function as a repressor of cell cycle genes and, therefore, a negative regulator of cell proliferation. Sustaining this view, ectopic expression of ΔE2FC delayed cell division and reduced the mRNA level of CDC6, an E2F target gene critical for DNA replication (Castellano et al., 2001; del Pozo et al., 2002). By contrast, the epidermis of leaves and cotyledons, as well as palisade and mesophyll cells, of e2fc-R plants contain more, but smaller, cells than those of the controls, further supporting the idea of E2FC as a repressor of cell division. A similar small-cell phenotype was found in leaves of plants that overexpress cell division activators such as E2Fa (De Veylder et al., 2002) or CYCD3;1 (Dewitte et al., 2003). We also observed that a reduction of E2FC levels leads to an increased expression of cell proliferation markers involved in transcriptional control (E2Fa [De Veylder et al., 2002]), DNA replication (histone H4 [Fobert et al., 1996] and CDC6 [Castellano et al., 2001]), and G2/M (CYCB1;1 [Colón-Carmona et al., 1999]). Together, these results provide compelling evidence that E2FC acts as a negative regulator of cell division.
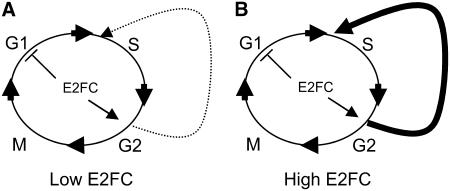
It is remarkable that high levels of CYCB1;1 are found in mature, fully differentiated rosette leaves of e2fc-R plants. This increase of cyclin B1;1 occurs concomitantly with a reduction in DNA endoreplication. The promotion of G2-to-M progression by E2F factors is well known in Drosophila (Neufeld et al., 1998) and in Arabidopsis (Boudolf et al., 2004; Magyar et al., 2005). Our results indicate that E2FC plays a role in this transit, because overexpression of E2FC dramatically reduces the number of cells in mitosis, whereas a reduction of E2FC levels strongly increases the number of CYCB1-containing cells in the root meristem. In a simple view, we think that E2FC negatively regulates the entry into G1/S and positively regulates the switch to the endocycle program (Figure 8). This model explains the low number of cells in G2/M when E2FC is overexpressed, because many of the dividing cells are forced to enter the endoreplication program. Therefore, E2FC-DPB seems to cooperate with E2Fa/DPa and E2Fe/DEL1 (De Veylder et al., 2001; Boudolf et al., 2004; Verkest et al., 2005; Vlieghe et al., 2005) in maintaining the balance between proliferation and the endocycle program.
Figure 8.
Model of E2FC Action.
(A) E2FC negatively regulates the G1/S phase and positively regulates, when the conditions are optimal, the entry of some cells in the endoreplication program.
(B) When E2FC is overexpressed, there is stronger G1/S inhibition and many of the dividing cells are forced into the endoreplication program, thus reducing the proportion of cells undergoing the G2/M transition.
E2FC Functions during LRP Formation
The formation of LRP occurs from G1-arrested pericycle founder cells (Casimiro et al., 2003; Himanen et al., 2004). It is well known that auxin is the signal that triggers lateral root initiation, a very well-suited developmental process in which to study the reactivation of cell proliferation and cell cycle control (Himanen et al., 2002; Casimiro et al., 2003). The lateral root phenotype of e2fc-R plants further supports a role of E2FC as a cell cycle transcriptional repressor, because a reduction of E2FC levels makes pericycle cells more sensitive to auxin and stimulates the formation of LRP. Therefore, these results suggest that E2FC might be one of the components that function as rate-limiting repressors of this process.
Concluding Remarks
Although the majority of the components of the cell cycle machinery are conserved in plants (Vandepoele et al., 2002), we are just getting a glimpse of how cell division is regulated and connected to cell differentiation in plants (Gutierrez, 2005). In particular, the role of the RBR-E2F pathway is crucial for cell division and differentiation (del Pozo et al., 2002; De Veylder et al., 2002; Ramirez-Parra et al., 2003, 2004; Vlieghe et al., 2003, 2005; Ebel et al., 2004; Park et al., 2005; Desvoyes et al., 2006; Sozzani et al., 2006). Based on our results, we think that E2FC/DPB functions at two stages during the cell cycle: (1) at G0, during which E2FC actively represses E2F target genes; and (2) later in the cell cycle, likely in G2/M, when E2FC might participate in the decision to progress through mitosis or to switch to the endocycle and differentiation programs. The available data strongly suggest a complex interplay between activators and repressor factors. The specific degradation of E2FC and its partner DPB through the proteasome provides a temporally controlled mechanism to remove the repression of certain E2F-regulated genes, thus permitting the incorporation of positive regulatory E2F/DP factors into specific promoters of genes required for proper cell cycle progression, such as E2Fa/DPa into G1/S (De Veylder et al., 2002) or G2/M (Boudolf et al., 2004) genes. Furthermore, our work strongly supports an active role of SCFSKP2 in controlling cell proliferation in plants.
METHODS
Plant Material and Transgenic Lines
All analyses were performed using Arabidopsis thaliana ecotype Columbia. The skp2a (GABI_293D12) and skp2b (SALK_028396) mutants were identified in the Arabidopsis Stock Center and Gabi-Kat collections, respectively. To generate MYC-DPB–overexpressing (MYC-DPBOE) and MYC-SKP2A–overexpressing (MYC-SKP2AOE) plants, six copies of the MYC epitope were fused in-frame to the cDNA of DPB (At5g03415) or SKP2A (At1g21410), respectively. These constructs were transferred into the pROK2 binary vector (Baulcombe et al., 1986) and introduced into Agrobacterium tumefaciens (C58C1 strain). To express the full-length E2FC, the genomic clone that contains all of the exons and introns from the ATG to the stop codons was cloned into the pPILY vector (Ferrando et al., 2000) and afterward into the binary vector pBIN19. Arabidopsis plants were transformed by the floral dip method (Clough and Bent, 1998), and independent stable transgenic lines that overexpress MYC-DPB, MYC-SKP2A, or E2FC were identified. To generate the RNAi-E2FC (e2fc-R) lines, we cloned the fragment from the ATG (+1) to the nucleotide at position +450 of the E2FC cDNA into the pHANNIBAL vector (Wesley et al., 2001). To identify and select lines with reduced levels of E2FC, we analyzed its expression by RT-PCR in >100 transgenic lines using the one-step RT-PCR kit from Innogenetics, following the manufacturer's instructions. The MYC-DPB transgene was introduced into the skp2a and skp2b mutants by agroinfiltration. We selected different lines of skp2a/MYC-DPBOE and skp2b/MYC-DPBOE that express similar levels of MYC-DPB transgene to the MYC-DPBOE line used in this work and examined them by protein gel blot analysis.
For the auxin treatment, 6-d-old seedlings were transferred to fresh medium containing 10−7 M 2,4-D for 12 h and then stained for GUS activity as described below.
Pull-Down and Immunoprecipitation Assays
For the pull-down experiments, 0.5 μg of GST or GST-DPB (del Pozo et al., 2002) proteins bound to beads were incubated with 6xHistidine-E2FC (His-E2FC) and expressed in bacteria as described by del Pozo et al. (2002). Where indicated, His-E2FC was phosphorylated by CDKA/CYCA (del Pozo et al., 2002) and incubated with GST or GST-DPB in pull-down assays.
MYC-DPBOE and MYC-SKP2AOE seedlings were grown for 4 d under long-day (16 h of light) conditions. Seedlings were ground in liquid nitrogen, and total proteins were extracted in TBS-0.5 buffer (100 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1× plant protease inhibitor cocktail [Sigma-Aldrich]). The extracts were sonicated three times for10 s each on ice and then incubated for 15 min on ice. Afterward, the extracts were cleared by centrifugation and the supernatants were filtered through 0.22-μm filters before they were used for pull-down or immunoprecipitation experiments. In both cases, after 3 h of incubation, the beads were washed four times for 15 min each with the extraction buffer. For the MYC-DPB immunoprecipitation, we used agarose beads coupled to IgGs against MYC epitope (Sigma-Aldrich). To release the immunopurified proteins, the beads were incubated with the MYC peptide at 0.5 μg/μL (Sigma-Aldrich) for 90 min and then analyzed by immunoblotting. For the pull-down experiments, 2 μg of GST or GST-DPB (del Pozo et al., 2002) proteins bound to beads was incubated with 1 mg of total protein extracted from MYC-SKP2AOE seedlings grown for 4 d. The MYC-SKP2A extracts were incubated under low-phosphorylation (−ATP; extraction buffer alone) or high-phosphorylation (+ATP; extraction buffer plus 5 mM ATP, 0.2 mM DTT, 0.05 unit/μL creatine kinase, and 10 mM phosphocreatine) conditions for 1 h at 25°C and 2 h at 4°C. The washing steps were similar to those described above, and the precipitated proteins were released by boiling in SDS-Laemmli sample buffer. MYC-SKP2A was detected by immunoblotting using the antibody 9E10 (Santa Cruz Biotechnology).
Phosphatase and Degradation Assays
Total proteins were extracted in 50 mM Tris-HCl, pH 8.0, 20 mM NaCl, 0.1% Tween 20, 1 mM PMSF, and 1× plant protease inhibitor cocktail (Sigma-Aldrich). Afterward, the samples were adjusted to phosphatase buffer (extraction buffer plus 0.1 mM Na2EDTA, 5 mM DTT, and 0.01% Brij 35). Approximately 20 μg of total protein was treated with 200 units of λ-phosphatase (New England Biolabs) for 30 or 60 min at 25°C. Where indicated, 4 nM epoxomicin (Affinity) was included during the reaction.
To test in vivo the effect of the proteasome inhibitor on the stability of the MYC-DPB protein, 5-d-old MYC-DPBOE seedlings grown on MS liquid medium were treated with or without 6 nM epoxomicin for 4 h, and then total protein was extracted in the presence of epoxomicin in the buffer and analyzed by immunoblotting with 9E10 antibody. To analyze the stability of the MYC-DPB protein, total protein from 4-d-old MYC-DPBOE plants was extracted in buffer A (50 mM Tris-HCl, pH 7.0, 50 mM NaCl, 0.5% Nonidet P-40, 1 mM PMSF, and 1× plant protease inhibitor cocktail [Sigma-Aldrich]). Twenty micrograms of MYC-DPBOE total protein was mixed with 20 μg of wild-type total protein or with 20 μg of MYC-SKP2AOE total protein. These samples were incubated at 30°C in a degradation assay containing 5 mM ATP, 0.2 mM DTT, 1 μM ubiquitin, 0.01 unit/μL creatine kinase, and 5 mM phosphocreatine in buffer A. Where indicated, 4 nM epoxomicin or the control solvent DMSO was added to the reactions. The reactions were stopped by adding SDS-Laemmli sample buffer and boiling the sample for 4 min. The MYC-DPB protein was detected by immunoblotting using 9E10 antibody.
GUS Assays
The CYCB1-GUS transgenic line harbors the CYCB1;1 promoter and the destruction box fused to the reporter uidA gene (Colón-Carmona et al., 1999). The CYCB1-GUS line was crossed with e2fc-R plants to generate the CYCB1-GUS/e2fc-R line. GUS staining was performed as described by Donnelly et al. (1999).
Real-Time RT-PCR Analysis
Total RNA was extracted using the Trizol reagent (Invitrogen), and RT-PCRs were performed with the ThermoScript RT system (Invitrogen). The LightCycler system with FastStart DNA Master SYBR Green I (Roche) was used for real-time quantitative RT-PCR. The amount of ACTIN2 mRNAs in each sample was determined to normalize for differences in total RNA amount. To avoid amplification of contaminating genomic DNA, primers were designed to scan exon–intron junctions. Primer sequences used are available upon request.
Scanning Electron Microscopy
Fresh plant samples were frozen in liquid nitrogen and analyzed immediately by cryoscanning electron microscopy using the LTSEM method (CryoTrans Oxford CT1500). Area measurements were done using NIH Image 1.61 software, which was previously calibrated to calculate the area in square micrometers using the correlation between micrometers and the pixels of the image. The values represent the frequency distribution of cells with different areas (n ≥ 400 cells) from three different images that correspond to the same areas of three different leaves.
Flow Cytometry Analyses
After the main vein was removed, the plant tissue was chopped out with a razor blade and 0.5 mL of cold Galbraith buffer was added (Galbraith et al., 1983). The extracts were filtered through 60- and 40-μm filters. Afterward, the isolated nuclei were stained with 50 μg/mL propidium iodide (Sigma-Aldrich) for 45 min. Approximately 104 nuclei were measured, and DNA histograms were generated in a FACScalibur flow cytometer (BD Bioscience).
LRP Measurements
CYCB1-GUS and CYCB1-GUS/e2fc-R were grown on MS medium for 10 d. CYCB1-GUS and CYCB1-GUS/E2FCOE seedlings were grown on MS medium for 7 d. In both experiments, seedlings were stained for GUS activity, LPR were counted as spots of GUS signal, and emerged lateral roots were counted using a Leica MC2000 microscope. For these measurements, ∼40 seedlings were counted in two independent experiments. The numbers of lateral roots and LRP were divided by the length of the principal root. The results shown in Figures 6 and 7 correspond to averages and sd values from all measurements.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: E2FC (At1g47870), DPB (At5g03415), SKP2A (At1g21410), and SKP2B (At1g77000).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression Levels of DPB and E2FC in Different Organs.
Supplemental Figure 2. Overexpression of a Truncated ΔE2FC/DPB Drastically Affects Plant Development.
Supplemental Figure 3. Phenotypes of e2fc-R Plants.
Supplementary Material
Acknowledgments
We are indebted to S. Llorens-Berzosa for technical assistance. We also thank P. Doerner for the CYCB1-GUS line, G. Jürgens for the KNOLLE antibodies, and C. Ascaso and F. Pinto for technical help with the LTSEM analysis. This work was partially supported by grants BMC2003-2131, BMC2001-2292, and BIO2004-01749 (Spanish Ministry of Science and Technology), Grant 07B-53-2002 (Comunidad Autonoma de Madrid), and an institutional grant from Fundación Ramon Areces.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Crisanto Gutierrez (cgutierrez@cbm.uam.es).
Online version contains Web-only data.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Bandara, L.R., Lam, E.W., Sorensen, T.S., Zamanian, M., Girling, R., and La Thangue, N.B. (1994). DP-1: A cell cycle-regulated and phosphorylated component of transcription factor DRTF1/E2F which is functionally important for recognition by pRb and the adenovirus E4 orf 6/7 protein. EMBO J. 13 3104–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D.C., Saunders, G.R., Bevan, M.W., Mayo, M.A., and Harrison, B.D. (1986). Expression of biologically-active viral satellite RNA from the nuclear genome of transformed plants. Nature 321 446–449. [Google Scholar]
- Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602. [DOI] [PubMed] [Google Scholar]
- Boudolf, V., Vlieghe, K., Beemster, G.T., Magyar, Z., Acosta, J.A., Maes, S., Van Der Schueren, E., Inze, D., and De Veylder, L. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16 2683–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro, I., Beeckman, T., Graham, N., Bhalerao, R., Zhang, H., Casero, P., Sandberg, G., and Bennett, M.J. (2003). Dissecting Arabidopsis lateral root development. Trends Plant Sci. 8 165–171. [DOI] [PubMed] [Google Scholar]
- Castellano, M.M., del Pozo, J.C., Ramirez-Parra, E., Brown, S., and Gutierrez, C. (2001). Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell 13 2671–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, J., Cloos, P., Toftegaard, U., Klinkenberg, D., Bracken, A.P., Trinh, E., Heeran, M., Di Stefano, L., and Helin, K. (2005). Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Res. 33 5458–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Coffman, J.A. (2004). Cell cycle development. Dev. Cell 6 321–327. [DOI] [PubMed] [Google Scholar]
- Colón-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Spatiotemporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20 503–508. [DOI] [PubMed] [Google Scholar]
- Cooper, S., and Shayman, J.A. (2001). Revisiting retinoblastoma protein phosphorylation during the mammalian cell cycle. Cell. Mol. Life Sci. 58 580–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.C., Boniotti, M.B., and Gutierrez, C. (2002). Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(SKP2) pathway in response to light. Plant Cell 14 3057–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.C., and Estelle, M. (2000). F-box proteins and protein degradation: An emerging theme in cellular regulation. Plant Mol. Biol. 44 123–128. [DOI] [PubMed] [Google Scholar]
- Desvoyes, B., Ramirez-Parra, E., Xie, Q., Chua, N.-M., and Gutierrez, C. (2006). Cell type specific roles of the retinoblastoma/E2F pathway during Arabidopsis thaliana leaf development. Plant Physiol. 140 67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T., de Almeida-Engler, J., Ormenese, S., Maes, S., Naudts, M., Van Der Schueren, E., Jacqmard, A., Engler, G., and Inze, D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T.S., Krols, L., Terras, F., Landrieu, I., van der Schueren, E., Maes, S., Naudts, M., and Inzé, D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13 1653–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Joubes, J., and Inze, D. (2003). Plant cell cycle transitions. Curr. Opin. Plant Biol. 6 536–543. [DOI] [PubMed] [Google Scholar]
- Dewitte, W., and Murray, J.A. (2003). The plant cell cycle. Annu. Rev. Plant Physiol. Plant Mol. Biol. 54 235–264. [DOI] [PubMed] [Google Scholar]
- Dewitte, W., Riou-Khamlichi, C., Scofield, S., Healy, J.M., Jacqmard, A., Kilby, N.J., and Murray, J.A. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova, D.K., Stevaux, O., Frolov, M.V., and Dyson, N.J. (2003). Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 17 2308–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano, L., Jensen, M.R., and Helin, K. (2003). E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 22 6289–6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerner, P., Jorgensen, J.E., You, R., Steppuhn, J., and Lamb, C. (1996). Control of root growth and development by cyclin expression. Nature 380 520–523. [DOI] [PubMed] [Google Scholar]
- Donnelly, P.M., Boneta, D., Tsukaya, H., Dengler, R.E., and Dengler, N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215 407–419. [DOI] [PubMed] [Google Scholar]
- Ebel, C., Mariconti, L., and Gruissem, W. (2004). Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429 776–780. [DOI] [PubMed] [Google Scholar]
- Fang, S., and Weissmann, A.M. (2004). A field guide to ubiquitylation. Cell Mol. Life Sci. 61 1546–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando, A., Farras, R., Jasik, J., Schell, J., and Koncz, C. (2000). Intron-tagged epitope: A tool for facile detection and purification of proteins expressed in Agrobacterium-transformed plant cells. Plant J. 22 553–560. [DOI] [PubMed] [Google Scholar]
- Fobert, P.R., Gaudin, V., Lunes, P., Coen, E.S., and Donan, J.H. (1996). Distinct classes of cdc2-related genes are differentially expressed during the cell division cycle in plants. Plant Cell 8 1465–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith, D.W.H.K., Maddox, J.M., Ayres, N.M., Sharma, D.P., and Firoozabadi, E. (1983). Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220 1049–1051. [DOI] [PubMed] [Google Scholar]
- Gutierrez, C. (2005). Coupling cell proliferation and development in plants. Nat. Cell Biol. 7 535–541. [DOI] [PubMed] [Google Scholar]
- Gutierrez, C., Ramirez-Parra, E., Castellano, M.M., and del Pozo, J.C. (2002). G(1) to S transition: More than a cell cycle engine switch. Curr. Opin. Plant Biol. 5 480–486. [DOI] [PubMed] [Google Scholar]
- Harbour, J.W., and Dean, D.C. (2000). The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Dev. 14 2393–2409. [DOI] [PubMed] [Google Scholar]
- Himanen, K., Boucheron, E., Vanneste, S., de Almeida Engler, J., Inze, D., and Beeckman, T. (2002). Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14 2339–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen, K., Vuylsteke, M., Vanneste, S., Vercruysse, S., Boucheron, E., Alard, P., Chriqui, D., Van Montagu, M., Inze, D., and Beeckman, T. (2004). Transcript profiling of early lateral root initiation. Proc. Natl. Acad. Sci. USA 101 5146–5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura, T., Hara, T., Kotoshiba, S., Yada, M., Ishida, N., Imaki, H., Hatakeyama, S., Nakayama, K., and Nakayama, K.I. (2003). Degradation of p57Kip2 mediated by SCFSkp2-dependent ubiquitylation. Proc. Natl. Acad. Sci. USA 100 10231–10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan, R.E., Emiliani, S., Nakayama, K., Castro, A., Labbe, J.C., Lorca, T., Nakayama, K., and Benkirane, M. (2001). Interaction between cyclin T1 and SCF(SKP2) targets CDK9 for ubiquitination and degradation by the proteasome. Mol. Cell. Biol. 21 7956–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2002. a). E2Ls, E2F-like repressors of Arabidopsis that bind to E2F sites in a monomeric form. J. Biol. Chem. 277 16553–16558. [DOI] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2002. b). Interaction of Arabidopsis E2F and DP proteins confers their concomitant nuclear translocation and transactivation. Plant Physiol. 128 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber, M.H., Waizenegger, I., Steinmann, T., Schwarz, H., Mayer, U., Hwang, I., Lukowitz, W., and Jurgens, G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., Potuschak, T., Colon-Carmona, A., Gutierrez, R.A., and Doerner, P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 102 12978–12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Zhao, Q., Liao, R., Sun, P., and Wu, X. (2003). The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J. Biol. Chem. 278 30854–30858. [DOI] [PubMed] [Google Scholar]
- Magae, J., Illenye, S., Chang, Y.C., Mitsui, Y., and Heintz, N.H. (1999). Association with E2F-1 governs intracellular trafficking and polyubiquitylation of DP-1. Oncogene 18 593–605. [DOI] [PubMed] [Google Scholar]
- Magyar, Z., De Veylder, L., Atanassova, A., Bakó, L., Inzé, D., and Bögre, L. (2005). The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17 2527–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti, B., Li, J., de Bruin, A., Gordon, F., Timmers, C., Opavsky, R., Patil, K., Tuttle, J., Cleghorn, W., and Leone, G. (2005). Cloning and characterization of mouse E2F8, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 280 18211–18220. [DOI] [PubMed] [Google Scholar]
- Marchant, A., Bhalerao, R., Casimiro, I., Eklof, J., Casero, P.J., Bennett, M., and Sandberg, G. (2002). AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariconti, L., Pellegrini, B., Cantoni, R., Stevens, R., Bergounioux, C., Cella, R., and Albani, D. (2002). The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. J. Biol. Chem. 277 9911–9919. [DOI] [PubMed] [Google Scholar]
- Marti, A., Wirbelauer, C., Scheffner, M., and Krek, W. (1999). Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat. Cell Biol. 1 14–19. [DOI] [PubMed] [Google Scholar]
- Moon, J., Parry, G., and Estelle, M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16 3181–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, H., Bracken, A.P., Vernell, R., Moroni, M.C., Christians, F., Grassilli, E., Prosperini, E., Vigo, E., Oliner, J.D., and Helin, K. (2001). E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15 267–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, A.W. (2004). Recycling the cell cycle: Cyclins revisited. Cell 116 221–234. [DOI] [PubMed] [Google Scholar]
- Nakayama, K., et al. (2000). Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 19 2069–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld, T.P., de la Cruz, A.F., Johnston, L.A., and Edgar, B.A. (1998). Coordination of growth and cell division in the Drosophila wing. Cell 93 1183–1193. [DOI] [PubMed] [Google Scholar]
- Park, J.A., Ahn, J.W., Kim, Y.K., Kim, S.J., Kim, J.K., Kim, W.T., and Pai, H.S. (2005). Retinoblastoma protein regulates cell proliferation, differentiation, and endoreduplication in plants. Plant J. 42 153–163. [DOI] [PubMed] [Google Scholar]
- Patton, E.E., Willems, A.R., and Tyers, M. (1998). Combinatorial control in ubiquitin-dependent proteolysis: Don't Skp the F-box hypothesis. Trends Genet. 14 236–243. [DOI] [PubMed] [Google Scholar]
- Planchais, S., Samland, A.K., and Murray, J.A. (2004). Differential stability of Arabidopsis D-type cyclins: CYCD3;1 is a highly unstable protein degraded by a proteasome-dependent mechanism. Plant J. 38 616–625. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra, E., Frundt, C., and Gutierrez, C. (2003). A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J. 33 801–811. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra, E., López-Matas, M.A., Fründt, C., and Gutierrez, C. (2004). Role of an atypical E2F transcription factor in the control of Arabidopsis cell growth and differentiation. Plant Cell 16 2350–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld, J.P., Gigot, C., and Chaubet-Gigot, N. (1998). Multilevel regulation of histone gene expression during the cell cycle in tobacco. Nucleic Acids Res. 26 3255–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W.H. (2002). The plant E2F-Rb pathway and epigenetic control. Trends Plant Sci. 7 505–511. [DOI] [PubMed] [Google Scholar]
- Sozzani, R., Maggio, C., Varotto, S., Canova, S., Bergonioux, C., Albani, D., and Cella, R. (2006). Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol. 140 1355–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaux, O., and Dyson, N.J. (2002). A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 14 684–691. [DOI] [PubMed] [Google Scholar]
- Tedesco, D., Lukas, J., and Reed, S.I. (2002). The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2). Genes Dev. 16 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilli, M.T., Hudgins, S.L., Frech, M.S., Halama, E.D., Renou, J.P., and Furth, P.A. (2003). Loss of protein phosphatase 2A expression correlates with phosphorylation of DP-1 and reversal of dysplasia through differentiation in a conditional mouse model of cancer progression. Cancer Res. 63 7668–7673. [PubMed] [Google Scholar]
- Tsvetkov, L.M., Yeh, K.H., Lee, S.J., Sun, H., and Zhang, H. (1999). p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr. Biol. 9 661–664. [DOI] [PubMed] [Google Scholar]
- Vandepoele, K., Raes, J., De Veylder, L., Rouze, P., Rombauts, S., and Inze, D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele, K., Vlieghe, K., Florquin, K., Hennig, L., Beemster, G.T., Gruissem, W., Van de Peer, Y., Inze, D., and De Veylder, L. (2005). Genome-wide identification of potential plant E2F target genes. Plant Physiol. 139 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest, A., Manes, C., Vercruysse, S., Maes, S., Van Der Schueren, E., Beeckman, T., Genschik, P., Kuiper, M., Inzé, D., and De Veylder, L. (2005). The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17 1723–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe, K., Boudolf, V., Beemster, G.T., Maes, S., Magyar, Z., Atanassova, A., de Almeida Engler, J., De Groodt, R., Inze, D., De Veylder, L. (2005). The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr. Biol. 15 59–63. [DOI] [PubMed] [Google Scholar]
- Vlieghe, K., Vuylsteke, M., Florquin, K., Rombauts, S., Maes, S., Ormenese, S., Van Hummelen, P., Van de Peer, Y., Inze, D., and De Veylder, L. (2003). Microarray analysis of E2Fa-DPa-overexpressing plants uncovers a cross-talking genetic network between DNA replication and nitrogen assimilation. J. Cell Sci. 116 4249–4259. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27 581–590. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.