Abstract
Epsin and related proteins play important roles in various steps of protein trafficking in animal and yeast cells. Many epsin homologs have been identified in plant cells from analysis of genome sequences. However, their roles have not been elucidated. Here, we investigate the expression, localization, and biological role in protein trafficking of an epsin homolog, Arabidopsis thaliana EPSIN1, which is expressed in most tissues we examined. In the cell, one pool of EPSIN1 is associated with actin filaments, producing a network pattern, and a second pool localizes primarily to the Golgi complex with a minor portion to the prevacuolar compartment, producing a punctate staining pattern. Protein pull-down and coimmunoprecipitation experiments reveal that Arabidopsis EPSIN1 interacts with clathrin, VTI11, γ-adaptin-related protein (γ-ADR), and vacuolar sorting receptor1 (VSR1). In addition, EPSIN1 colocalizes with clathrin and VTI11. The epsin1 mutant, which has a T-DNA insertion in EPSIN1, displays a defect in the vacuolar trafficking of sporamin:green fluorescent protein (GFP), but not in the secretion of invertase:GFP into the medium. Stably expressed HA:EPSIN1 complements this trafficking defect. Based on these data, we propose that EPSIN1 plays an important role in the vacuolar trafficking of soluble proteins at the trans-Golgi network via its interaction with γ-ADR, VTI11, VSR1, and clathrin.
INTRODUCTION
After translation in eukaryotic cells, a large number of proteins are transported to subcellular compartments by a variety of different mechanisms. Newly synthesized vacuolar proteins that are delivered to the endoplasmic reticulum (ER) by the cotranslational translocation mechanism are transported to the vacuole from the ER by a process called intracellular trafficking. Trafficking of a protein to the vacuole from the ER occurs through two organelles, the Golgi complex and the prevacuolar compartment (PVC) (Rothman, 1994; Hawes et al., 1999; Bassham and Raikhel, 2000; Griffiths, 2000). Transport of a protein from the ER to the Golgi complex is performed by coat protein complex II vesicles. Transport from the trans-Golgi network (TGN) to the PVC occurs via clathrin-coated vesicles (CCVs) (Robinson et al., 1998; Tang et al., 2005; Yang et al., 2005).
Transport of a protein from the ER to the vacuole/lysosome requires a large number of proteins, including components of vesicles, factors involved in vesicle generation and fusion, regulators of intracellular trafficking, adaptors for the cargo proteins, and other accessory proteins (Robinson and Kreis, 1992; Bennett, 1995; Schekman and Orci, 1996; da Silva Conceição et al., 1997; Kirchhausen, 1999; Sever et al., 1999; Bassham and Raikhel, 2000; Griffiths, 2000; Jin et al., 2001; Robinson and Bonifacino, 2001). Most of these proteins are found in all eukaryotic cells from yeast, animals, and plants, suggesting that protein trafficking mechanisms from the ER to the vacuole/lysosome may be highly conserved in all eukaryotic cells.
Of the large number of proteins involved in intracellular trafficking, a group of proteins that have the highly conserved epsin N-terminal homology (ENTH) domain have been identified as playing a critical role at various trafficking steps in animal and yeast cells (Chen et al., 1998; De Camilli et al., 2002; Wendland, 2002; Overstreet et al., 2003; Legendre-Guillemin et al., 2004). The ENTH domain binds to phosphatidylinositols (PtdIns), although the lipid binding specificity differs with individual members of the epsin family. For example, epsin1 binds to PtdIns(4,5)P2, whereas EpsinR and Ent3p bind to PtdIns(4)P and PdtIns(3,5)P2, respectively (Itoh et al., 2001). The ENTH domain is thought to be responsible for targeting these proteins to specific compartments and also for introducing curvature to the bound membranes to assist in the generation of CCVs (Legendre-Guillemin et al., 2004). However, the exact steps of intracellular trafficking in which ENTH-containing proteins play a role are complex. Epsin homologs can be divided into two groups based on the pathway in which they play a role. One group, which includes epsin1 in animal cells and Ent1p and Ent2p in yeast cells, is involved in endocytosis from the plasma membrane (Chen et al., 1998; De Camilli et al., 2002; Wendland, 2002). The other group, which includes EpsinR/clint/enthoprotin in animal cells and Ent3p and Ent4p in yeast cells, is involved in protein trafficking from the TGN to the lysosome/vacuole as well as retrograde trafficking from the early endosomes to the TGN (Kalthoff et al., 2002; Wasiak et al., 2002; Hirst et al., 2003; Chidambaram et al., 2004; Eugster et al., 2004; Saint-Pol et al., 2004).
Another common feature of epsin-related proteins is that they play a role in CCV-mediated protein trafficking at both the TGN and the plasma membrane. These proteins can bind directly to clathrin through their multiple clathrin binding motifs; thus, they may recruit clathrin to the plasma membrane or the TGN to generate CCVs (Rosenthal et al., 1999; Wendland et al., 1999; Drake et al., 2000). In addition, these proteins interact with many other proteins, such as heterotetrameric clathrin adaptor complexes (APs), monomeric adaptor Golgi-localized, γ-ear–containing Arf binding proteins (GGAs), and soluble NSF attachment protein receptors (SNAREs). Epsin1 interacts with AP-2, Epsin15, and intersectin (Chen et al., 1998; Legendre-Guillemin et al., 2004), whereas EpsinR/enthoprotin/clint and Ent3p interact with SNAREs such as vti1b and vti1p, respectively (Chidambaram et al., 2004) and with adaptor proteins such as GGAs and AP-1 (Duncan et al., 2003; Mills et al., 2003). In addition, epsin homologs have ubiquitin-interacting motifs and are ubiquitinated (Oldham et al., 2002; Shih et al., 2002). Protein ubiquitination acts as a signal for endocytosis from the plasma membrane and trafficking from the TGN through the endosome/PVC to the lysosome/vacuole (Polo et al., 2002; Horak, 2003; Raiborg et al., 2003; Scott et al., 2004). The binding of epsin homologs to ubiquitin raises the possibility that epsin homologs may bind directly to cargo proteins that are destined for the vacuole/lysosome from either the plasma membrane or the TGN (Chen and De Camilli, 2005; Sigismund et al., 2005).
In plant cells, sequence analysis of the entire Arabidopsis thaliana genome reveals several proteins with the highly conserved ENTH domains (Holstein and Oliviusson, 2005). However, their biological roles have not been addressed. In this study, we investigate the functional role of EPSIN1, an Arabidopsis epsin homolog, at the molecular level. In particular, we focus on its possible role in protein trafficking in plant cells. We demonstrate that EPSIN1 interacts with clathrin, AP-1, VSR1, and VTI11 and plays an important role in the vacuolar trafficking of a soluble protein from the Golgi complex to the central vacuole.
RESULTS
EPSIN1, a Member of the Epsin Family, Is Ubiquitously Expressed in Arabidopsis
The Arabidopsis genome encodes three highly similar epsin-related proteins, EPSIN1, EPSIN2, and EPSIN3 (Holstein and Oliviusson, 2005). In this study, we investigated the biological role of EPSIN1. EPSIN1 has the highly conserved ENTH domain at the N terminus. However, the rest of the molecule is less similar to other epsin-related proteins, although it has motifs, such as LIDL and DPF, that may function as clathrin and AP-1 binding motifs, respectively.
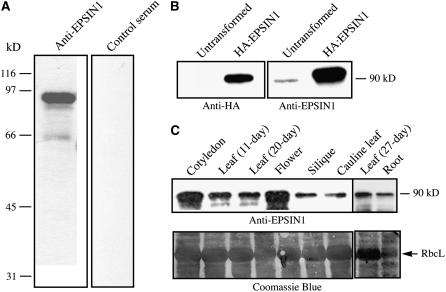
To understand the biological role of EPSIN1, its expression in various plant tissues was examined. An antibody was raised against the middle domain of EPSIN1 (amino acid residues 153 to 337). The antibody recognized a protein band at 90 kD, which was much larger than the expected size, 60 kD, of EPSIN1 (Figure 1A). It was shown previously that epsin-related proteins migrate slower than expected in SDS-PAGE (Chen et al., 1998). The control serum did not recognize any protein bands. This result suggested that the antibody specifically recognized EPSIN1. To confirm this, protoplasts were transformed with EPSIN1 tagged with HA at the N terminus (HA:EPSIN1) and protein extracts from the transformed protoplasts were analyzed by protein gel blotting using anti-HA and anti-EPSIN1 antibodies. The anti-HA antibody specifically recognized a protein band from the transformed protoplasts, but not from the untransformed protoplasts, at 90 kD (Figure 1B). In addition, the 90-kD protein species was recognized by the anti-EPSIN1 antibody, confirming that the 90-kD band was EPSIN1. The expression of EPSIN1 in various tissues was examined using the anti-EPSIN1 antibody. Protein extracts were prepared from various tissues at different stages of plant growth and used for protein gel blot analysis. EPSIN1 was expressed in all of the tissues examined, with the highest expression in cotyledons and flowers (Figure 1C).
Figure 1.
EPSIN1 Is Expressed in Various Arabidopsis Tissues.
(A) Generation of anti-EPSIN1 antibody. The middle domain, corresponding to amino acid residues 153 to 337, was expressed as the Hisx6-tagged form in E. coli and used to raise antibody in a rabbit. Control serum was obtained from the rabbit before immunization. Total protein extracts were obtained from leaf tissues and used to test the anti-EPSIN1 antibody.
(B) Specificity of the anti-EPSIN1 antibody. Protein extracts were obtained from protoplasts expressing EPSIN1 tagged with the HA epitope at the N terminus and used for protein gel blot analysis using anti-HA and anti-EPSIN1 antibodies.
(C) Expression of EPSIN1 in various tissues. Total protein extracts from the indicated tissues were analyzed by protein gel blotting using anti-EPSIN1 antibody. Leaf tissues were harvested 11 and 20 d after germination. Cotyledons were obtained from 5-d-old plants. The membranes were stained with Coomassie blue to control for protein loading. RbcL, large subunit of the ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) complex.
EPSIN1 Produces Both Network and Punctate Staining Patterns
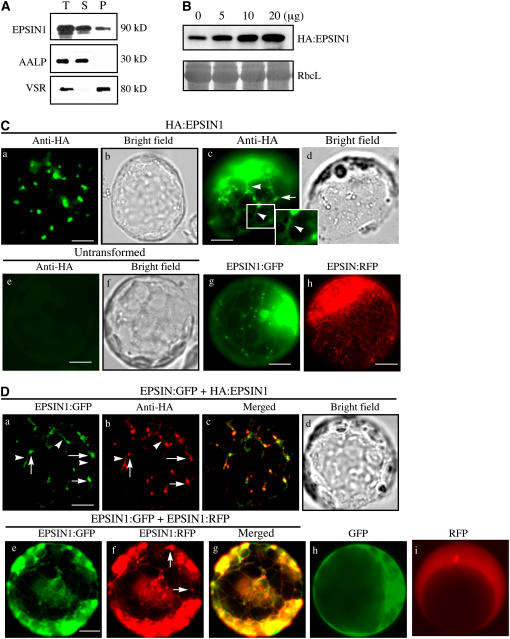
To examine the subcellular distribution of EPSIN1, total protein extracts from leaf tissues were separated into soluble and membrane fractions and analyzed by protein gel blotting using anti-EPSIN1 antibody. EPSIN1 was detected in both membrane (pellet) and soluble fractions (Figure 2A). As controls for the fractionation, Arabidopsis aleurain-like protease (AALP) and Arabidopsis vacuolar sorting receptor (VSR) were detected with anti-AALP and anti-VSR antibodies, respectively (Sohn et al., 2003). AALP is a soluble protein present in the vacuolar lumen, and VSR is a membrane protein that is localized primarily to the PVC with a minor portion to the Golgi complex (da Silva Conceição et al., 1997; Ahmed et al., 2000). As expected, AALP and VSR were detected in the supernatant and pellet fractions, respectively. These results indicated that EPSIN1 localized to multiple locations, consistent with the behavior of other epsin-related proteins (Legendre-Guillemin et al., 2004).
Figure 2.
EPSIN1 Produces Both Network and Punctate Staining Patterns.
(A) Subcellular fractionation of EPSIN1. Total (T) protein extracts of leaf tissues were separated into soluble (S) and pellet (P) fractions and analyzed by protein gel blotting using anti-EPSIN1, anti-AALP, and anti-VSR antibodies.
(B) Expression level of EPSIN1 in transformed protoplasts. Protoplasts were transformed with various amounts of HA:EPSIN1 DNA, and the level of EPSIN1 was determined by protein gel blotting with anti-EPSIN1 antibody. Protein extracts from untransformed protoplasts were used as a control. The membrane was also stained with Coomassie blue to control for loading.
(C) Localization of EPSIN1. Protoplasts were transformed with the indicated constructs (5 to 10 μg), and the localization of EPSIN1 was examined either by immunostaining with anti-HA antibody or by direct detection of the GFP or RFP signal. Untransformed protoplasts were immunostained with anti-HA antibody as a control. Bars = 20 μm.
(D) Colocalization of EPSIN1 proteins. The localization of EPSIN1 protein was examined in protoplasts transformed with HA:EPSIN1 and EPSIN1:GFP or with EPSIN1:GFP and EPSIN1:RFP. As controls, GFP and RFP alone were transformed into protoplasts. Bars = 20 μm.
Next, we defined the subcellular localization of EPSIN1. Our initial attempts to localize the endogenous EPSIN1 with the anti-EPSIN1 antibody failed. Thus, we determined the localization of EPSIN1 protein transiently expressed in protoplasts. EPSIN1 was tagged with the HA epitope, green fluorescent protein (GFP), or red fluorescent protein (RFP). The amount of total EPSIN1 protein was determined using various amounts of HA:EPSIN1 plasmid DNA by protein gel blot analysis with anti-EPSIN1 antibody and was found to be proportional to the amount of plasmid used (Figure 2B). For the localization, we used a minimal amount (5 to 10 μg) of EPSIN1 plasmid DNAs. Protoplasts were transformed with HA:EPSIN1, and localization of EPSIN1 was determined by immunostaining with anti-HA antibody. HA:EPSIN1 produced primarily a punctate staining pattern (Figure 2Ca). In addition to punctate stains, we occasionally observed weakly stained strings that connected punctate stains (Figure 2Cc, arrowheads). By contrast, the nontransformed controls did not produce any patterns (Figure 2Ce). In protoplasts transformed with EPSIN1:GFP and EPSIN1:RFP, both EPSIN1 fusion proteins produced a network pattern with punctate stains (Figures 2Cg and 2Ch), whereas GFP and RFP alone produced diffuse patterns (Figures 2Dh and 2Di), indicating that EPSIN1 produces the network pattern with punctate stains. These results were further confirmed by cotransforming the protoplasts with either EPSIN1:GFP and HA: EPSIN1 or EPSIN1:GFP and EPSIN1:RFP. The punctate staining pattern of EPSIN1:GFP closely overlapped that of HA:EPSIN1 (Figures 2Da to 2Dc). In addition, the network and punctate staining patterns of EPSIN1:GFP closely overlapped those of EPSIN1:RFP (Figures 2De to 2Dg). However, the fine networks revealed by EPSIN1:GFP in the live protoplasts were nearly absent in the fixed protoplasts. Thus, the differences in the staining patterns between fixed and live protoplasts may be attributable to the fact that the network pattern of live protoplasts are not well preserved under the fixing conditions used. In addition, the strings occasionally observed in the fixed protoplasts may represent the remnants of the network pattern revealed by HA:EPSIN1. These results strongly suggest that EPSIN1 is responsible for the network pattern as well as the punctate stains.
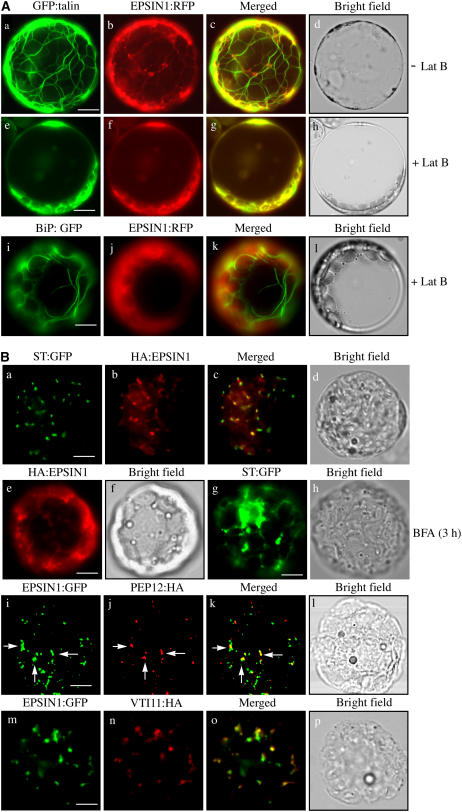
The network pattern was reminiscent of the ER or actin pattern in plant cells (Boevink et al., 1998; Jin et al., 2001; Kim et al., 2005), whereas the punctate staining pattern suggested that EPSIN1 may localize to the Golgi complex or endosomes, as observed previously with epsin homologs in animal and yeast cells (Wasiak et al., 2002; Chidambaram et al., 2004; Saint-Pol et al., 2004). Therefore, protoplasts were cotransformed with EPSIN1:RFP and GFP:talin, a marker for actin filaments consisting of GFP and the actin binding domain of mouse talin (Kost et al., 1998; Kim et al., 2005). As expected, GFP:talin produced the network pattern (Figure 3A) (Kost et al., 1998; Kim et al., 2005). Furthermore, the red fluorescent network pattern of EPSIN1:RFP closely overlapped the green fluorescent network pattern of GFP:talin (Figure 3A), raising the possibility that EPSIN1:GFP bound to the actin filaments rather than to the ER. To confirm this, the EPSIN1:RFP pattern was examined after treatment with latrunculin B (Lat B), a chemical agent known to disrupt actin filaments (Spector et al., 1983). Lat B–treated protoplasts produced the diffuse green fluorescent pattern of GFP:talin (Figure 3A), an indication of solubilized actin filaments, as observed previously (Kim et al., 2005). In addition, the Lat B–treated protoplasts displayed a diffuse red fluorescent pattern of EPSIN1:RFP (Figure 3A), indicating that EPSIN1 is associated with actin filaments but not with the ER. Furthermore, the punctate staining pattern of EPSIN1:RFP also was not observed in the presence of Lat B, indicating that actin filaments played a role in yielding the punctate staining pattern of EPSIN1. In the same conditions, BiP:GFP, an ER marker (Lee et al., 2002), produced a network pattern, indicating that Lat B does not disrupt the ER network patterns (Figure 3Ai).
Figure 3.
Localization of EPSIN1 in Protoplasts.
(A) Colocalization of EPSIN1 with actin filaments. Protoplasts were transformed with the indicated constructs, and the localization of these proteins was examined in the presence (+ Lat B) and absence (− Lat B) of Lat B (10 μM). Bars = 20 μm.
(B) Localization of EPSIN1 to the Golgi complex and the PVC. Protoplasts were transformed with the indicated constructs, and localization of the proteins was examined after immunostaining with anti-HA. The GFP signals were observed directly in the fixed protoplasts. For BFA treatment, BFA (30 μg/mL) was added to the transformed protoplasts at 24 h after transformation and incubated for 3 h. Arrows indicate the overlap between EPSIN1:GFP and PEP12p:HA. Bars = 20 μm.
To identify the organelle responsible for the punctate staining pattern of EPSIN1, its localization was compared with that of ST:GFP and PEP12p/SYP21. ST:GFP, a chimeric protein between rat sialyltransferase and GFP, localizes to the Golgi complex, and PEP12p, a t-SNARE, localizes to the PVC (da Silva Conceição et al., 1997; Boevink et al., 1998; Jin et al., 2001). Protoplasts were cotransformed with HA:EPSIN1 and ST:GFP. The localization of these proteins was examined after staining with anti-HA antibody. ST:GFP was observed directly with the green fluorescent signals. A major portion of the HA:EPSIN1-positive punctate stains closely overlapped with those of ST:GFP (Figures 3Ba to 3Bc). To further confirm the Golgi localization of HA:EPSIN1, protoplasts transformed with HA:EPSIN1 were treated with brefeldin A (BFA), a chemical known to disrupt the Golgi complex (Driouich et al., 1993), and the localization of HA:EPSIN1 was examined. In the presence of BFA, HA:EPSIN1 yielded a largely diffuse pattern with aggregates, but not the punctate staining pattern, indicating that BFA affects EPSIN1 localization (Figure 3Be). In the same conditions, ST:GFP produced a network pattern with large aggregates (Figure 3Bg), confirming that the Golgi complex was disrupted. These results support the notion that EPSIN1 localizes to the Golgi complex. Next, we examined the possibility of EPSIN1 localizing to the PVC. Protoplasts were cotransformed with EPSIN1:GFP and PEP12p:HA. The localization of PEP12p:HA was examined after staining with anti-HA antibody. EPSIN1:GFP was observed directly with the green fluorescent signals. Only a minor portion of the EPSIN1:GFP-positive punctate stains overlapped with the PEP12p:HA-positive punctate stains (Figures 3Bi to 3Bk, arrows). These results indicated that EPSIN1 localized primarily to the Golgi complex with a minor portion to the PVC.
To obtain independent evidence for the localization, we examined the colocalization of EPSIN1 with VTI11, a v-SNARE that is distributed equally to both the TGN and the PVC (Zheng et al., 1999; Bassham et al., 2000; Kim et al., 2005). Protoplasts were cotransformed with EPSIN1:GFP and VTI11:HA, and the localization of these proteins was examined by immunostaining with anti-HA antibody. EPSIN1-positive punctate stains largely colocalized with those of VTI11:HA (Figures 3Bm to 3Bo), confirming that EPSIN1 localizes to both the Golgi complex and the PVC.
EPSIN1 Binds to and Colocalizes with Clathrin
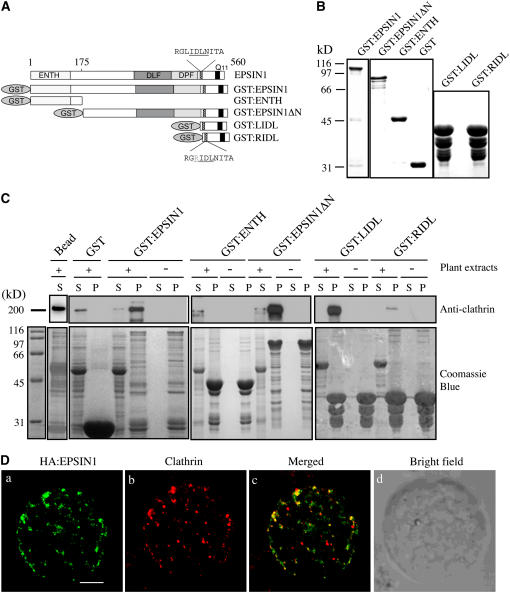
The members of the epsin family have two clathrin binding motifs (Rosenthal et al., 1999; Wendland et al., 1999; Drake et al., 2000). Sequence analysis indicated that EPSIN1 has a potential clathrin binding motif. To explore the possibility that EPSIN1 binds to clathrin, glutathione S-transferase–fused EPSIN1 (GST:EPSIN1) was constructed for a protein pull-down assay (Figure 4A). GST:EPSIN1 was expressed in Escherichia coli and purified from E. coli extracts (Figure 4B). The purified GST:EPSIN1 was mixed with protein extracts obtained from leaf tissues. Proteins pelleted with glutathione–agarose were analyzed by protein gel blotting using anti-clathrin antibody. GST:EPSIN1, but not GST alone, precipitated from the plant extracts a 180-kD protein species that was recognized by anti-clathrin antibody (Figure 4C), indicating that EPSIN1 bound to clathrin.
Figure 4.
EPSIN1 Binds to and Colocalizes with Clathrin.
(A) Constructs. GST was fused to the N terminus. ENTH, the epsin N-terminal homology domain. DLF and DPF motifs are similar to AP-1 and AP-3 binding motifs, respectively. Q11 indicates a stretch of 11 Glu residues. The clathrin binding motif (LIDL) and the Leu-to-Arg substitution in the clathrin binding motif (RIDL) are shown in the C-terminal region. The numbers indicate amino acid positions.
(B) Expression of GST-fused EPSIN1 proteins. Constructs were introduced into E. coli, and their expression was induced by isopropylthio-β-galactoside.GST fusion proteins were purified from E. coli extracts with glutathione–agarose beads. Purified proteins were stained with Coomassie blue.
(C) Interaction of EPSIN1 with clathrin. GST-fused EPSIN1 proteins were mixed with protein extracts from leaf tissues. EPSIN1 binding proteins were precipitated using glutathione–agarose beads and analyzed by protein gel blotting using anti-clathrin antibody. Supernatants also were included in the protein gel blot analysis. Subsequently, the membranes were stained with Coomassie blue. Bead, glutathione–agarose beads alone; P, pellet; S, supernatant (10% of total).
(D) Colocalization of EPSIN1 with clathrin. Protoplasts transformed with HA:EPSIN1 were fixed with paraglutaraldehyde, and the localization of HA:EPSIN1 and clathrin was examined by immunostaining with anti-HA and anti-clathrin antibodies, respectively. Bar = 20 μm.
To further examine its binding to clathrin, EPSIN1 was divided into two regions, the ENTH and the remainder of the molecule (EPSIN1ΔN) (Figure 4A). These regions were expressed in E. coli as GST fusion proteins, GST:ENTH and GST:EPSIN1ΔN, respectively (Figure 4B). Protein pull-down experiments using leaf cell extracts were performed with purified GST:ENTH and GST:EPSIN1ΔN. GST:EPSIN1ΔN, but not GST:ENTH, precipitated clathrin from the plant extracts (Figure 4C). To identify the clathrin binding motif, the C-terminal region containing the putative clathrin binding motif, LIDL (Lafer, 2002), as well as GST:RIDL, which contained an Arg substitution of the first Leu residue in the motif, were expressed as GST fusion proteins in E. coli (Figures 4A and 4B). GST:LIDL, but not GST:RIDL, precipitated clathrin from protein extracts (Figure 4C), indicating that the LIDL motif functioned as a clathrin binding motif.
The in vitro binding of EPSIN1 with clathrin strongly suggested that EPSIN1 was likely to colocalize with clathrin. Therefore, immunohistochemistry for the localization of EPSIN1 and clathrin was performed. Protoplasts were transformed with HA:EPSIN1, and the localization of HA:EPSIN1 and clathrin was examined by staining with anti-HA and anti-clathrin antibodies, respectively. The anti-clathrin antibody produced a punctate staining pattern (Figure 4D). A majority (60 to 70%) of the HA:EPSIN1-positive punctate stains closely overlapped with a pool (40 to 50%) of clathrin-positive punctate stains (Figure 4D), consistent with an interaction between EPSIN1 and clathrin. There was also a pool of clathrin-positive punctate stains that lacked the HA:EPSIN1 signal, suggesting that clathrin also was involved in an EPSIN1-independent process.
To further characterize the interaction between EPSIN1 and clathrin, we examined whether or not EPSIN1 is permanently associated with CCVs. Protein extracts from leaf tissues were first separated into soluble and pellet fractions by ultracentrifugation. The pellet fraction was treated with Triton X-100 and further fractionated by gel filtration, and the fractions were analyzed by protein gel blotting using anti-clathrin, anti-EPSIN, and anti-VSR antibodies. Clathrin was detected in a peak between 443 and 669 kD (see Supplemental Figure 1 online). Interestingly, VSR, the vacuolar cargo receptor, was eluted at the same position with clathrin. By contrast, EPSIN1 was eluted at 90 kD. These results suggest that EPSIN1 is not permanently associated with CCVs.
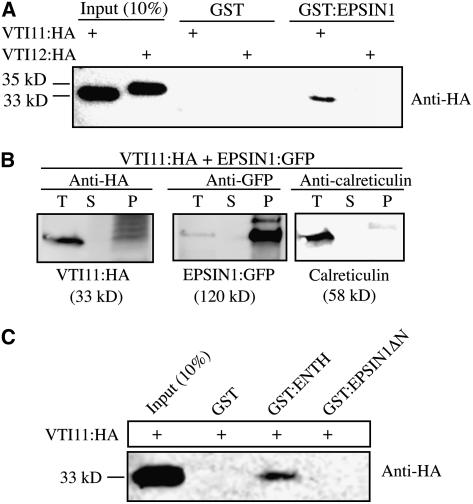
EPSIN1 Interacts with VTI11
Epsin-related proteins in animal and yeast cells are involved in either endocytosis or vacuolar/lysosomal protein trafficking (Chen et al., 1998; De Camilli et al., 2002; Wendland, 2002; Overstreet et al., 2003; Legendre-Guillemin et al., 2004). To elucidate the pathway of EPSIN1 involvement, binding partners of EPSIN1 were examined. In animal and yeast cells, epsin-like proteins have been shown to interact with SNAREs (Chen et al., 1998; Chidambaram et al., 2004). Because EPSIN1 localized to the Golgi complex and the PVC, EPSIN1 interactions with Arabidopsis VTI11 and VTI12 (formerly At VTI1a and At VTI1b, respectively) were examined. VTI11 is a v-SNARE that localizes to the TGN and travels to the PVC (Zheng et al., 1999; Bassham et al., 2000). VTI11 and VTI12 were tagged with HA at the C terminus and introduced into protoplasts. The expression of VTI11:HA and VTI12:HA in protoplasts was confirmed by protein gel blot analysis using anti-HA antibody. The anti-HA antibody detected protein bands at 33 and 35 kD (Figure 5A), the expected positions of VTI11:HA and VTI12:HA, respectively. Purified GST:EPSIN1 from E. coli extracts was mixed with plant extracts from the VTI11:HA- or VTI12:HA-transformed protoplasts, and GST:EPSIN1-bound proteins were precipitated from the mixture using glutathione–agarose beads. The pellet fraction was analyzed by protein gel blotting using anti-HA antibody. VTI11:HA, but not VTI12:HA, was detected from the pellet (Figure 5A). GST alone did not precipitate VTI11:HA from the plant extracts. These results indicated that although VTI11 and VTI12 are highly similar to each other, EPSIN1 specifically binds to VTI11:HA. To further confirm this interaction, we performed a reciprocal protein pull-down experiment (i.e., pull-down of EPSIN1 with VTI11) using protein extracts obtained from protoplasts transformed with VTI11:HA and EPSIN1:GFP. VTI11:HA-bound proteins were immunoprecipitated with anti-HA antibody, and the immunoprecipitates were analyzed by protein gel blotting using anti-HA, anti-GFP, and anti-calreticulin antibodies. Anti-calreticulin antibody was used as a negative control. In addition to VTI11:HA, EPSIN1:GFP was detected in the immunoprecipitates (Figure 5B). However, calreticulin was not detected in the pellet. These results further confirm the interaction between VTI11 and EPSIN1. To determine the VTI11 binding domain of EPSIN1, protein pull-down experiments were performed using GST:ENTH and GST:EPSIN1ΔN. GST:ENTH, but not GST:EPSIN1ΔN, precipitated VTI11:HA from the plant extracts (Figure 5C), indicating that the ENTH domain contained the VTI11 binding motif. Similarly, in animal and yeast cells, EpsinR and Ent3p have been shown to bind to vti1b and vti1p, respectively (Chidambaram et al., 2004).
Figure 5.
EPSIN1 Binds to VTI11.
(A) Protein extracts were prepared from VTI11:HA- and VTI12:HA-transformed protoplasts and mixed with GST alone or GST:EPSIN1. EPSIN1-bound proteins were precipitated from the mixture with glutathione–agarose beads and analyzed by protein gel blotting using anti-HA antibody.
(B) Coimmunoprecipitation of EPSIN1:GFP with VTI11:HA. Protein extracts from protoplasts cotransformed with VTI11:HA and EPSIN1:GFP were used for immunoprecipitation with anti-HA antibody. The immunoprecipitates were analyzed by protein gel blotting with anti-HA, anti-GFP, and anti-calreticulin antibodies. P, immunoprecipitate; S, supernatant; T, total protein extracts (5% of the input).
(C) For binding experiments, protein extracts from protoplasts transformed with VTI11:HA were mixed with GST alone, GST:ENTH, and GST:EPSIN1ΔN. Proteins were precipitated with glutathione-agarose beads and analyzed by protein gel blotting using anti-HA antibody. The amount of the input proteins is indicated.
EPSIN1 Binds to the Arabidopsis Homolog of γ-Adaptin of AP-1
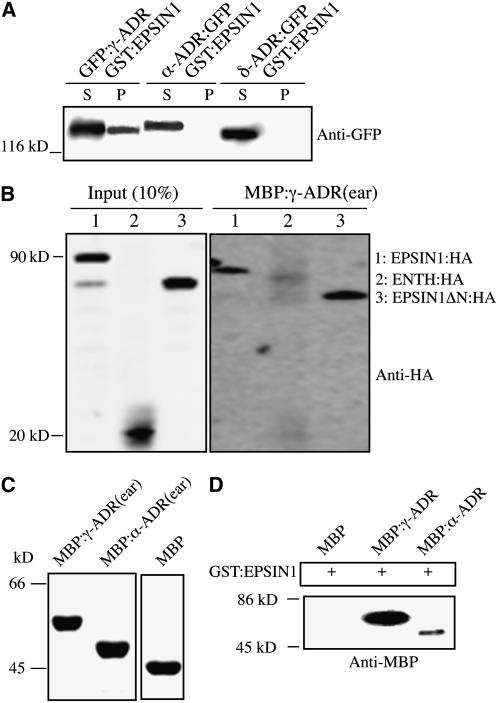
Epsin homologs bind to adaptor proteins (APs) (Duncan et al., 2003; Mills et al., 2003). In animal cells, EPSIN1 binds to the α-adaptin of AP-2 via the DΦF/W (where Φ indicates a hydrophobic amino acid) and FXDXF motifs (Figure 4A) (Brett et al., 2002). Arabidopsis EPSIN1 has three DPF motifs to which α-adaptin of AP-2 could bind. In addition, EPSIN1 has two regions with motifs similar to the acidic Phe motif for binding AP-1 and GGAs (Duncan et al., 2003). Therefore, the interactions of EPSIN1 with AP complexes were examined. We isolated the Arabidopsis proteins γ-adaptin related protein (γ-ADR), α-ADR, and δ-ADR, which were most closely related to γ-adaptin, α-adaptin, and δ-adaptin of AP-1, AP-2, and AP-3, respectively. These Arabidopsis proteins were tagged with GFP and expressed transiently in protoplasts. Protein extracts from the transformed protoplasts were mixed with purified GST:EPSIN1, and the GST:EPSIN1-bound proteins were precipitated. The pellet was analyzed by protein gel blotting using anti-GFP antibody. GFP:γ-ADR, but not α-ADR:GFP or δ-ADR:GFP, was detected in the pellet (Figure 6A). The control for the protein pull-down assay, GST alone, did not precipitate any of these proteins. These results strongly suggested that EPSIN1 interacts with γ-ADR specifically. To further confirm the interaction between EPSIN1 and γ-ADR, we performed a reciprocal protein pull-down experiment (i.e., pull down of EPSIN1 proteins with γ-ADR). The ear domain of γ-ADR was expressed as a maltose binding protein (MBP) fusion protein, MBP:γ-ADR(ear), in E. coli. Purified MBP:γ-ADR(ear) was mixed with protein extracts obtained from protoplasts transformed with full-length EPSIN1:HA or its deletion mutants, and MBP:γ-ADR(ear)-bound proteins were precipitated with amylose resin. The precipitates were analyzed by protein gel blotting using anti-HA antibody. Full-length HA:EPSIN1 and HA:EPSIN1ΔN, but not HA:ENTH, were detected in the pellets (Figure 6B), further confirming the interaction between γ-ADR and EPSIN1. Furthermore, these results indicated that the C-terminal region of EPSIN1 was responsible for the interaction with γ-ADR.
Figure 6.
EPSIN1 Binds to γ-ADR.
(A) GFP-fused γ-ADR, α-ADR, and δ-ADR were constructed and transiently expressed in protoplasts. Protein extracts were prepared from the transformed protoplasts and used for binding assays with GST:EPSIN1. The EPSIN1 binding proteins were precipitated from the reaction mixture and analyzed by protein gel blotting using anti-GFP antibody. P, pellet; S, supernatant (20% of total).
(B) Pull-down of EPSIN1 proteins with MBP:γ-ADR(ear). MBP:γ-ADR(ear) was mixed with protein extracts from protoplasts transformed with the indicated constructs. MBP:γ-ADR(ear) binding proteins were precipitated with amylose resin, and the pellets were analyzed by protein gel blotting using anti-HA antibody.
(C) The ear domains of γ-ADR and α-ADR were expressed as MBP fusion proteins in E. coli and used for binding experiments. Proteins were detected by protein gel blotting using anti-MBP antibody.
(D) GST:EPSIN1 was mixed with MBP alone, MBP:γ-ADR(ear), or MBP:α-ADR(ear). The EPSIN1 binding proteins were precipitated from the reaction mixture and analyzed by protein gel blotting using anti-MBP antibody.
To confirm the interaction between EPSIN1 and γ-ADR and that EPSIN1 binds to γ-ADR directly, the binding of EPSIN1 was examined with MBP-fused ear domains of γ-ADR or α-ADR expressed in E. coli, respectively (Figure 6C). GST:EPSIN1 was incubated with MBP:γ-ADR(ear), MBP:α-ADR(ear), or MBP alone. GST:EPSIN1-bound proteins were precipitated from the reaction mixture and analyzed by protein gel blotting using anti-MBP antibody. MBP:γ-ADR(ear) bound strongly to GST:EPSIN1 (Figure 6D). Interestingly, MBP:α-ADR(ear) also bound weakly to GST:EPSIN1, possibly because of the three DPF motifs in the middle domain of EPSIN1. DPF or DPW motifs are known to bind to the α-ear domain of AP-2 (Owen et al., 1999; Brett et al., 2002). However, the binding affinity of EPSIN1 for the γ-ADR ear domain was >10-fold higher than that for the α-ADR ear domain. These results strongly suggest that γ-ADR and α-ADR are Arabidopsis γ-adaptin of AP-1 and α-adaptin of AP-2, respectively, and that EPSIN1 binds primarily to γ-ADR. It is not clear whether the α-ADR binding of EPSIN1 has a physiological role in vivo.
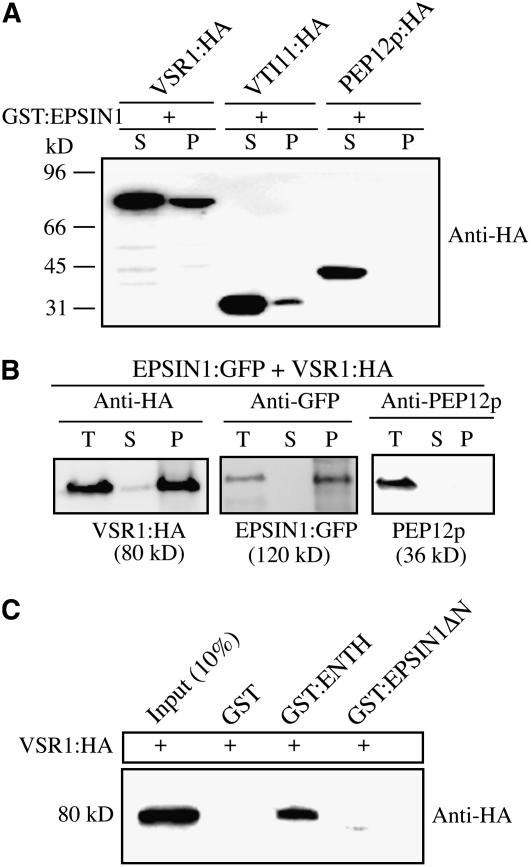
EPSIN1 Interacts with VSR1
To further characterize the binding partners of EPSIN1, EPSIN1 binding to VSR—the Arabidopsis vacuolar sorting receptor that localizes primarily to the PVC but travels to the TGN for cargo binding—was examined (Ahmed et al., 1997, 2000; Tse et al., 2004). VSR1/ATELP1 tagged with HA at the C terminus was expressed transiently in protoplasts. We showed previously that transiently expressed VSR1:HA localized primarily to the PVC with a minor portion to the Golgi complex (Lee et al., 2004). Protein extracts were prepared from the transformed protoplasts and incubated with GST:EPSIN1. As negative and positive controls, protein extracts were prepared from protoplasts expressing PEP12p:HA and VTI11:HA, respectively (da Silva Conceição et al., 1997). EPSIN1-bound proteins were precipitated from the reaction mixture and analyzed by protein gel blotting using anti-HA antibody. VSR1:HA, but not PEP12p:HA, was detected in the pellet (Figure 7A), indicating that EPSIN1 bound specifically to VSR1. To further confirm the interaction, we performed a protein pull-down experiment of EPSIN1:GFP with VSR1:HA from the extracts of protoplasts cotransformed with EPSIN1:GFP and VSR1:HA. VSR1:HA was immunoprecipitated with anti-HA antibody, and the immunoprecipitates were analyzed by protein gel blotting using anti-HA, anti-GFP, and anti-PEP12p antibodies. Anti-PEP12p antibody was used as a negative control. In addition to VSR1:HA, EPSIN1:GFP was detected in the immunoprecipitates (Figure 7B). However, PEP12p was not detected in the pellet. These results further confirm the interaction between EPSIN1 and VSR1. To define the VSR1 binding region of EPSIN1, GST:ENTH and GST:EPSIN1ΔN were used in protein pull-down experiments. GST:ENTH, but not GST:EPSIN1ΔN, brought down VSR1:HA from the reaction mixture (Figure 7C), indicating that the ENTH domain contained the VSR1 binding domain.
Figure 7.
EPSIN1 Binds to VSR1 via the ENTH Domain.
(A) Protein extracts were prepared from protoplasts transformed with the indicated constructs and incubated with GST-fused EPSIN1. EPSIN1-bound proteins were precipitated with glutathione–agarose beads, and the pelleted proteins were analyzed by protein gel blotting using anti-HA antibody. P, pellet; S, supernatant (10% of total).
(B) Protein extracts were prepared from protoplasts cotransformed with VSR1:HA and EPSIN1:GFP and used for immunoprecipitation with anti-HA antibody. The immunoprecipitates were analyzed by protein gel blotting using anti-HA, anti-GFP, and anti-PEP12 antibodies. P, immunoprecipitate; S, supernatant; T, total protein extracts (10% of input).
(C) Protein extracts from protoplasts transformed with VSR1:HA were incubated with GST alone or GST-fused EPSIN1 proteins. EPSIN1 binding proteins were precipitated with glutathione–agarose beads, and the pelleted proteins were analyzed by protein gel blotting using anti-HA antibody.
EPSIN1 Is Involved in Vacuolar Trafficking of Soluble Cargo Proteins
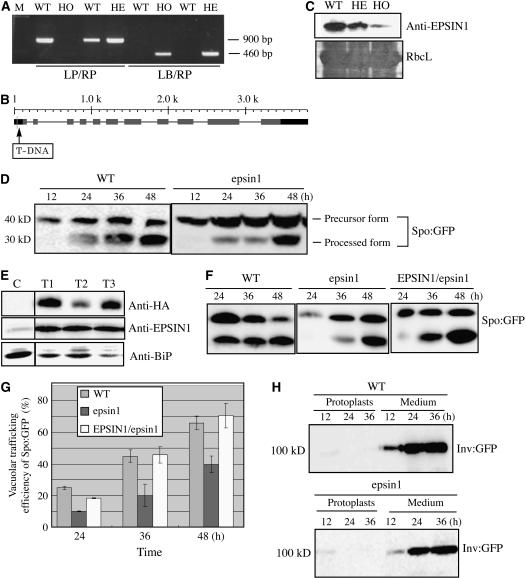
The protein–protein interaction data of EPSIN1 strongly suggested that EPSIN1 was involved in trafficking soluble proteins to the lytic (central) vacuole. To examine the role of EPSIN1, the T-DNA insertion mutant, SALK_049204, from the SALK T-DNA insertion mutant library was used. PCR was used to confirm that this mutant, epsin1, had a T-DNA insertion in the promoter region of EPSIN1 (Figures 8A and 8B). Homozygote mutant plants were screened by PCR, because the epsin1 mutant did not display any morphological alterations. Because the epsin1 mutant had a T-DNA insertion in the promoter region of EPSIN1, protein gel blot analysis with anti-EPSIN1 antibody was used to determine the EPSIN1 level. In the homozygote epsin1 plants, the EPSIN1 level was reduced to 10 to 20% of the wild-type level (Figure 8C).
Figure 8.
EPSIN1 Is Necessary for the Efficient Vacuolar Trafficking of Spo:GFP.
(A) Screen of an epsin1 mutant. T-DNA insertion in EPSIN1 was confirmed by PCR. HE, heterozygote; HO, homozygote; LB, left border primer; LP, left primer; M, size marker; RP, right primer.
(B) Scheme of the epsin1 mutant. The position of the T-DNA insertion site is indicated. The numbers indicate nucleotide positions. Shaded boxes, exons; dark boxes, promoters or 3′ untranslated regions.
(C) Total proteins were prepared from wild-type plants and heterozygotes and homozygotes of epsin1 plants and analyzed by protein gel blotting using anti-EPSIN1 antibody. The large subunit of the Rubisco complex (RbcL) stained with Coomassie blue was used as a loading control.
(D) Trafficking assay of Spo:GFP in epsin1 protoplasts. Protoplasts from wild-type and epsin1 plants were transformed with Spo:GFP. Protein extracts prepared from protoplasts at various time points after transformation were analyzed by protein gel blotting using anti-GFP antibody.
(E) Protein gel blot analysis of epsin1 mutants harboring HA:EPSIN1. Protein extracts were prepared from the T2 generation of epsin1 transgenic plants harboring HA:EPSIN1 and examined by protein gel blot analysis using anti-HA, anti-EPSIN1, and anti-BiP antibodies. C, epsin1 mutant; T1 to T3, independent transgenic plants harboring HA:EPSIN1 on the epsin1 background.
(F) and (G) Protoplasts obtained from wild-type, epsin1, and epsin1 complemented with HA:EPSIN1 (EPSIN1/epsin1; T3) plants were transformed with Spo:GFP.
(F) Protein extracts were obtained from protoplasts and analyzed by protein gel blotting using anti-GFP antibody.
(G) Trafficking efficiency was determined as the ratio of the amount of the processed form to the total amount of Spo:GFP (processed plus precursor forms). Error bars indicate sd (n = 3).
(H) Invertase:GFP secretion was not inhibited. Protoplasts obtained from epsin1 plants were transformed with invertase:GFP, and the secretion of invertase:GFP was examined at various time points after transformation. Protein extracts were prepared from the protoplasts, as well as from the media, and analyzed by protein gel blotting using anti-GFP antibody.
The mutant plants were examined for defects in protein trafficking. Previously, we showed that sporamin:GFP (Spo:GFP), a chimeric protein consisting of GFP and the N-terminal region of sporamin, was targeted to the central vacuole in protoplasts (Kim et al., 2001). It had been shown previously that when Spo:GFP was transported to the central vacuole in protoplasts, it was processed from a 40-kD precursor to a 30-kD form (Sohn et al., 2003). Therefore, the amount of the processed form could be used to measure trafficking efficiency. Protoplasts obtained from the epsin1 mutant were transformed with Spo:GFP, and trafficking of the reporter protein to the central vacuole was examined by protein gel blot analysis using anti-GFP antibody (Figure 8D). Protein extracts were obtained from protoplasts at various time points after transformation. With wild-type protoplasts at 36 and 48 h after transformation, ∼45 and 70% of the Spo:GFP, respectively, was in the processed form (Figure 8D). However, in protoplasts from epsin1 mutants, the amount of 30-kD processed protein was reduced significantly: at 36 and 48 h after transformation, the amount of the processed form was ∼20 and 40%, respectively (Figure 8D). These results clearly indicated that EPSIN1 was involved in the vacuolar trafficking of Spo:GFP. To confirm that the inhibition of vacuolar trafficking was attributable to the mutation, transgenic plants were generated with HA:EPSIN1 on the epsin1 background. The expression of HA:EPSIN1 in transgenic plants (EPSIN1/epsin1) was examined by protein gel blot analysis using protein extracts obtained from three independent lines of the T2 generation. The anti-HA antibody detected a specific band from all three independent lines (Figure 8E). Furthermore, the levels of EPSIN1 proteins were significantly increased in the transgenic plants compared with epsin1 plants (Figure 8E). Next, protoplasts were obtained from the leaf tissues of wild-type epsin1 plants and EPSIN1/epsin1 plants and transformed with Spo:GFP. The trafficking efficiency was recovered to the wild-type level in the EPSIN1/epsin1 plants (Figures 8F and 8G), confirming that the defect in the vacuolar trafficking in epsin1 mutant plants is attributable to the mutation in EPSIN1.
To examine the specificity of EPSIN1 in vacuolar trafficking, the secretion into the medium of invertase:GFP, a chimeric protein consisting of secretory invertase and GFP (Kim et al., 2005), was examined. Protoplasts obtained from epsin1 and wild-type plants were transformed with invertase:GFP. Protein extracts were prepared from the transformed protoplasts and from the incubation medium and analyzed by protein gel blotting using anti-GFP antibody. As expected, wild-type protoplasts efficiently secreted invertase:GFP into the medium. Nor was the secretion of invertase:GFP affected in protoplasts from the epsin1 mutants (Figure 8I), indicating that EPSIN1 is not involved in the secretion of the protein.
DISCUSSION
In this study, we demonstrate that EPSIN1 is involved in the trafficking of soluble proteins to the central (lytic) vacuole. This conclusion is based on three lines of evidence. First, EPSIN1 localizes primarily to the Golgi complex (possibly to the TGN) with a minor portion to the PVC. Second, EPSIN1 interacts specifically with VTI11, VSR1, clathrin, and γ-ADR, all of which play important roles in vacuolar trafficking from the TGN (Ahmed et al., 1997, 2000; Bassham et al., 2000; Surpin et al., 2003). Finally, the vacuolar trafficking of Spo:GFP is inhibited in epsin1 mutant protoplasts, and this defect can be complemented by stably expressed HA:EPSIN1.
Localization of EPSIN1 in Arabidopsis
A large number of epsin homologs have been identified (Chen et al., 1998; De Camilli et al., 2002; Wendland, 2002; Overstreet et al., 2003; Legendre-Guillemin et al., 2004). One group of these proteins, which includes epsin1, has been shown to be involved in endocytosis (Chen et al., 1998; De Camilli et al., 2002; Wendland, 2002). The other group, which includes EpsinR, has been shown to play an important role in lysosomal/vacuolar trafficking in animal and yeast cells (Kalthoff et al., 2002; Wasiak et al., 2002; Hirst et al., 2003; Chidambaram et al., 2004; Eugster et al., 2004; Saint-Pol et al., 2004). Thus, amino acid sequence homology between EPSIN1 and other epsin homologs cannot be used to define the pathways in which EPSIN1 plays a role. Therefore, we examined the localization of transiently expressed HA-, GFP-, and RFP-tagged EPSIN1s. Regardless of the tag, EPSIN1 displays both network and punctate staining patterns. The network pattern results from EPSIN1's association with actin filaments. The punctate staining pattern is attributable to the protein's localization largely to the Golgi complex with a minor portion to the PVC. This was demonstrated by colocalization of EPSIN1 with ST:GFP, PEP12p, and VTI11. At present, EPSIN1's interaction with actin filaments is not understood clearly. Actin filaments recently were shown to play a critical role in vacuolar trafficking from the Golgi complex in plant cells (Kim et al., 2005). Thus, it is possible that actin filaments guide EPSIN1 to the TGN and the PVC. Consistent with this notion is the observation that in the presence of Lat B the punctate staining pattern of EPSIN1 was also not observed. Its localization suggests that EPSIN1 is more closely related functionally to EpsinR in animal cells. EpsinR has been shown to localize to the TGN as well as endosomes (Kalthoff et al., 2002; Saint-Pol et al., 2004).
Inhibition of the Vacuolar Trafficking of Spo:GFP in epsin1 Mutants
One of the most compelling pieces of evidence for the role of EPSIN1 is the result of trafficking assays in protoplasts from epsin1 mutants. An epsin1 mutant with a T-DNA insertion in the promoter region has greatly reduced EPSIN1 protein levels, although the mutants display no visible phenotype. It is possible that the low level of EPSIN1 in the mutants is enough to support vacuolar trafficking. Another possibility is that AP-1 can support vacuolar trafficking at low levels of EPSIN1. However, despite the lack of a visible phenotype, trafficking of Spo:GFP to the central vacuole in protoplasts is inhibited significantly. The defects in vacuolar trafficking in epsin1 mutants are rescued by HA:EPSIN1 expressed stably in transgenic plants with the epsin1 background, further supporting the hypothesis that the trafficking inhibition is attributable to the mutation in EPSIN1. In epsin1 mutants, the secretion into the medium of the reporter protein invertase:GFP (Kim et al., 2005) is normal, indicating that the effects of the mutations are specific for the vacuolar trafficking.
EPSIN1 Interaction Partners
A large numbers of proteins are involved in lysosomal/vacuolar trafficking in eukaryotic cells. Clathrin is believed to function as a coat protein for vesicles derived from the TGN in Arabidopsis (Hohl et al., 1996; Ahmed et al., 1997; Robinson et al., 1998), as has been observed in other eukaryotic cells (Dahms et al., 1989; Pryer et al., 1992). EPSIN1 strongly interacts with clathrin. It has a typical clathrin binding motif, LIDL (Lafer, 2002), and colocalizes with a pool of clathrin. As with most epsin homologs (Rosenthal et al., 1999; Wendland et al., 1999; Drake et al., 2000), EPSIN1 appears to have at least two different clathrin binding motifs, and this study demonstrates that LIDL is one of them. However, at the moment, the other motif is not apparent and does not show any sequence homology with known clathrin binding motifs. The fact that EPSIN1 binds to and colocalizes with clathrin suggests that EPSIN1 may recruit clathrin to the TGN for vesicle formation (Rosenthal et al., 1999; Wendland et al., 1999; Drake et al., 2000).
EPSIN1 also binds to VTI11. EpsinR and Ent3p interact with vti1b and vti1p in animal and yeast cells, respectively (Chidambaram et al., 2004). However, EPSIN1 does not bind to VTI12, which also localizes to the TGN but at a different region than VTI11 (Bassham et al., 2000; Surpin et al., 2003). The two isoforms of VTI are thought to be involved in different pathways: the zig mutant that has a mutation in VTI11 displays a defect in the gravitropic response (Kato et al., 2002), whereas atvti12 mutants appear normal under normal growth conditions (Surpin et al., 2003). However these isoforms appear to be somewhat functionally redundant, because plants carrying either mutation are viable but mutations in both genes are lethal (Surpin et al., 2003). The fact that despite the high degree of similarity between VTI11 and VTI12 EPSIN1 binds specifically to VTI11 but not VTI12 strongly suggests that EPSIN1 may contribute to the functional diversification of these two v-SNAREs in the vacuolar trafficking at the TGN.
EPSIN1 also interacts with VSR1. VSR1 and its homologs that localize to the TGN and the PVC are the best characterized sorting receptors in plants (Ahmed et al., 1997, 2000; Sanderfoot et al., 1998; Paris and Neuhaus, 2002; Tse et al., 2004; daSilva et al., 2005). VSR1 has been shown to interact specifically with the N-terminal propeptide motif that is present in many soluble proteins destined for the central vacuole (Ahmed et al., 2000) and plays a role in the sorting of N-terminal propeptide motif–containing proteins. However, its role has been complicated by the mutant phenotype of atvsr1, which displays a defect in protein trafficking to the protein storage vacuole in seed cells (Shimada et al., 2003). The ENTH domain of EPSIN1 interacts specifically with VSR1, although the exact sites of interaction in the two molecules are unknown. This finding is rather surprising because it had been thought that VSR1 interacts with AP-1 in plant cells (Sanderfoot et al., 1998; Happel et al., 2004). In addition, in animal cells, ENTH-containing proteins are thought to assist in vesicle formation by recruiting clathrin for adaptors, APs, or GGAs (Legendre-Guillemin et al., 2004). However, the interaction between EPSIN1 and VSR1 strongly suggests that EPSIN1 is involved directly in recruiting clathrin for the sorting receptor of vacuolar trafficking.
In addition to the interaction between EPSIN1 and VSR1, EPSIN1 also interacts directly with γ-ADR, a close homolog of γ-adaptin of AP-1 in vitro, but not with α-ADR, a homolog of α-adaptin of AP-2, or δ-ADR, a homolog of δ-adaptin of AP-3. EPSIN1 has a putative acidic Phe motif that has been shown to be the binding site of the heterotetrameric adaptor complex AP-1 and monomeric adaptor GGAs (Duncan et al., 2003; Duncan and Payne, 2003). Protein pull-down experiments clearly demonstrate that γ-ADR binds to the EPSIN1 fragment containing the acidic Phe motif. The fact that EPSIN1 interacts with γ-ADR, a close homolog of γ-adaptin of AP-1, places it clearly in the vacuolar pathway rather than the endocytotic pathway. As in animal cells, plant cells are thought to have four different types of AP complexes, AP-1 to AP-4, although their precise roles have not been demonstrated directly (Kirchhausen, 1999; Robinson and Bonifacino, 2001). In animal and yeast cells, AP-1 functions as the adaptor complex for CCVs and binds to the cargo receptor at the TGN (Kirchhausen, 1999; Robinson and Bonifacino, 2001). It is likely that AP-1 plays a similar role in plant cells (Ahmed et al., 1997). Consistent with this notion are the findings that VSR1/ATELP1 interacts with animal AP-1 and that the μA subunit of AP-1 interacts in vitro with the YXXΦ motif (where Φ is a bulky, hydrophobic residue) that is present in the cytoplasmic C-terminal domain of VSR-PS1, a pea (Pisum sativum) VSR1 (Sanderfoot et al., 1998; Happel et al., 2004). AP-1 is likely to play a role in recruiting clathrin to the TGN through its interaction with VSR1 (Ahmed et al., 1997). The interaction between EPSIN1 and γ-ADR is quite similar to that between EpsinR and γ-adaptin of AP-1 in animal cells (Mills et al., 2003; Legendre-Guillemin et al., 2004). Thus, EPSIN1 may recruit clathrin for AP-1–mediated vesicle formation at the TGN.
We demonstrate here that EPSIN1 is involved in the trafficking of Spo:GFP to the central vacuole. epsin1 mutants display significant inhibition of the vacuolar trafficking of Spo:GFP but do not show a complete block in the vacuolar trafficking. It is possible that both AP-1 and EPSIN1 function independently to recruit clathrin to the TGN for the cargoes that are sorted by VSR1, but they may recruit clathrin to the TGN much more efficiently when they act cooperatively. In addition, the interaction between EPSIN1 and VTI11 suggests that EPSIN1 may incorporate the v-SNARE VTI11 into the vesicle at the TGN.
METHODS
Growth of Plants and Screening of a T-DNA Insertion Mutant
Arabidopsis thaliana (ecotype Columbia) plants were grown on Murashige and Skoog plates in a growth chamber. Leaf tissues were harvested from 10- to 14-d-old plants and used immediately for protoplast isolation as described previously (Jin et al., 2001). In addition, plants were grown on soil at 20 to 25°C in a greenhouse with a 16/8-h light/dark cycle.
A T-DNA insertion mutant of EPSIN1, the SALK_049204 line, was obtained from the ABRC (Ohio State University). To identify homozygotic plants, genomic DNA from 20 individual plants was used for PCR with left border primer 5′-GATGCAATCGATATCAGCCAATTTTAGAC-3′and the gene-specific primer 5′-TCGATAGAGAAAACGGAAAAC-3′.
Construction of Plasmids
The construction of VTI11:HA and GFP:talin was described previously (Kim et al., 2005). EPSIN1 (At5g11710) was isolated by PCR from an Arabidopsis cDNA library using the specific primers Epsin-5 (5′-AAGAATTCAAATGGATTTCATGAAG-3′) and Epsin-3 (5′-TTCTCGAGTCACTGCTTAAAGCCACC-3′). To tag the N terminus of EPSIN1 with the HA epitope, the PCR product was reamplified with the primers 5′-TCTATGGCTTACCCATACGATGTTCCAGATTACGCTATGGATTTCATGAAGGTCTTC-3′ and Epsin-3. To construct EPSIN1:GFP and EPSIN1:RFP, the EPSIN1 cDNA without the termination codon was amplified by PCR using the primers 5′-AAAGAGTACCGCCGAACAATGGATTTCATGAAGGTC-3′ and 5′-GTCCCGGGACTGCTTAAAGCCACCAGATTGGTACC-3′ and ligated in-frame to the 5′ ends of the GFP and mRFP coding regions, respectively (Campbell et al., 2002). EPSIN1:GFP and EPSIN1:RFP were placed between the cauliflower mosaic virus 35S promoter and the nos terminator in a pUC vector.
To generate the GST:EPSIN1 fusion construct, full-length EPSIN1 was ligated in-frame to pGEX-5X-2 using EcoRI and XhoI sites. The EPSIN1 deletion or substitution mutants, ENTH, EPSIN1ΔN, LIDL (the C-terminal region from amino acid position 461), and RIDL (identical to LIDL except for the L-to-R substitution at position 470) were amplified by PCR using the following primers: 5′-AAGAATTCAAATGGATTTCATGAAGGTC-3′ and Epsin-3 for ENTH, 5′-ATGAATTCATGGGTTCCTCAGCTTCA-3′ and Epsin-3 for EPSIN1ΔN, 5′-GAATTCATTTGGGCAGATTCGCTG-3′ and Epsin-3 for LIDL, and 5′-GAATTCATTTGGGCAGATTCGCTGAGCCGTGGAAGGATTGATCTCAATATAACT-3′ and Epsin-3 for RIDL. These PCR products were ligated to pGEX-5X-2 using EcoRI and XhoI sites. All of the PCR products were sequenced.
α-ADR (At5g22770), γ-ADR (At1g23900), and δ-ADR (At1g48760) were amplified by PCR from an Arabidopsis cDNA library using gene-specific primers. The primers were 5′-AAGCTTATGAATCCCTTTTCTTCT-3′ and 5′-CTCGAGTCACAACCCGCGAGGGAA-3′ for γ-ADR, 5′-GAATTCATGACCGGAATGAGAGGTCTCTCCGTA-3′ and 5′-GAATTCTTAAAGTAAGCCAGCGAGCATAGCTCC-3′ for α-ADR, and 5′-GGTACCATGTCGTCGTCTTCCACTTCTATAATG-3′ and 5′-GGTACCTCACAAGAGAAAATCTGGAATTATAAC-3′ for δ-ADR. To construct GFP:γ-ADR, γ-ADR was ligated to the C terminus of the GFP coding region without the termination codon. To construct α-ADR:GFP and δ-ADR:GFP, α-ADR and δ-ADR were ligated to the N terminus of the GFP coding region. The γ-ADR-ear and α-ADR-ear domains were amplified by PCR. The gene-specific primers were 5′-ACCGAATTCCCAGCTTATGCACCTATTGTT-3′ and 5′-TCACAAAAGCTTAGGGAAGTTGCTGACTTG-3′ for the γ-ADR-ear domain and 5′-TTAGAATTCTTTCTTCAACCTCTACAATTG-3′ and 5′-TCAAAGCTTTTGTTCCTTGATAAATTCTTCCAA-3′ for the α-ADR-ear domain. The PCR products were ligated to pMAL-c2 (New England Biolabs) to generate MBP:γ-ADR(ear) and MBP:α-ADR(ear).
Transient Expression and in Vivo Targeting of Reporter Cargo Proteins
Plasmids were introduced into Arabidopsis protoplasts by polyethylene glycol–mediated transformation (Jin et al., 2001; Lee et al., 2002). The expression of the fusion constructs was monitored at various time points after transformation. Images were captured with a cooled CCD camera attached to a Zeiss Axioplan fluorescence microscope. The filter sets used were XF116 (exciter, 474AF20; dichroic, 500DRLP; emitter, 510AF23) and XF33/E (exciter, 535DF35; dichroic, 570DRLP; emitter, 605DF50) (Omega) for GFP/fluorescein isothiocyanate and RFP/tetramethylrhodamine isothiocyanate, respectively. The data were processed using Adobe Photoshop software, and the images were rendered in pseudocolor (Jin et al., 2001).
Generation of Antibody
To raise an antibody to EPSIN1, the DNA fragment encoding the middle domain, amino acid residues 153 to 337, was amplified by PCR using specific primers (5′-GGATCCGACAAAGCAGTAGCAAATCGT-3′ and 5′-GCTCGAGGTCCACTTGGTTGGCACTGCC-3′) and ligated in-frame with the His tag to pRSET-A (Invitrogen). To raise an antibody to calreticulin (At1g56340), the C-terminal region containing amino acid residues 212 to 425 was amplified by PCR using primers 5′-GGATCCAAGTGCTAAGAAGCC-3′ and 5′-TTAGAGCTCGTCATGGGCGGCATC-3′ and ligated in-frame to pRSET-B (digested with BamHI and HindIII). The resulting constructs, PREST:EPSIN-M and pRSET-Cal, were introduced into Escherichia coli. Expression of the recombinant proteins was induced with isopropylthio-β-galactoside and the proteins were purified using a nickel-nitrilotriacetic acid agarose column (Qiagen) according to the manufacturer's instructions. Affinity-purified recombinant proteins were used to immunize rabbits. The antibody was affinity-purified using recombinant protein as described previously (Harlow and Lane, 1988).
Immunohistochemistry
Immunohistochemistry was performed as described previously (Frigerio et al., 1998; Park et al., 2004). The fixed protoplasts were incubated with appropriate primary antibodies, such as rat monoclonal anti-HA (Roche Diagnostics) or rabbit anti-clathrin antibodies, in TSW buffer (10 mM Tris-HCl, pH 7.4, 0.9% NaCl, 0.25% gelatin, 0.02% SDS, and 0.1% Triton X-100) at 4°C overnight and then washed with TSW buffer three times. The protoplasts were incubated with tetramethylrhodamine isothiocyanate– or fluorescein isothiocyanate–conjugated goat anti-rabbit IgG (Sigma-Aldrich) or anti-rat IgG (Zymed) antibodies in TSW buffer and washed three times with TSW buffer.
Protein Preparation, Protein Gel Blot Analysis, and Immunoprecipitation
Cell extracts from protoplasts were prepared as described previously (Jin et al., 2001). To obtain the proteins from the culture medium, cold trichloroacetic acid (100 μL) was added to the medium (1 mL) and protein aggregates were precipitated by centrifugation at 10,000g at 4°C for 5 min. The protein aggregates were dissolved in the lysis buffer. The supernatant was used for protein gel blot analysis with anti-GFP, anti-EPSIN1, anti-VSR, and anti-HA antibodies (Roche Diagnostics), as described previously (Jin et al., 2001). The protein blots were developed with an ECL detection kit (Amersham Pharmacia Biotech), and images were obtained using an LAS3000 image-capture system (Fujifilm).
Immunoprecipitation was performed as described previously (Park et al., 2005). Briefly, protein extracts (100 μg of total protein) in immunoprecipitation buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2.0 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, and 1% [v/v] Triton X-100) supplemented with an EDTA-free protease inhibitor cocktail were first incubated with Protein A–Sepharose (CL-4B; Amersham) for 30 min and centrifuged at 10,000g for 5 min at 4°C for preclearing. Subsequently, 4 μg of anti-HA antibody (12CA5; Roche Diagnostics) was added to the supernatant and incubated for 3 h at 4°C. The immunocomplexes were precipitated by incubating them with Protein A–agarose for 1 h at 4°C. The pellet was then washed three times with immunoprecipitation buffer, suspended in homogenization buffer, and subjected to immunoblot analyses.
Protein Pull-Down Assay
GST and MBP fusion constructs were introduced into the E. coli BL21(DE3)LysS strain, and expression of the fusion proteins was induced by isopropylthio-β-galactoside (1 mM) at 37°C. GST and MBP fusion proteins were purified using glutathione–agarose 4B (Peptron) beads and amylose resin (New England Biolabs), respectively, according to the manufacturers' instructions.
For protein pull-down assays, partially purified recombinant GST or MBP fusion proteins bound to glutathione–agarose beads or amylose resin, respectively, were mixed with total protein extracts in 100 μL of protein pull-down buffer (40 mM HEPES-KOH, pH 7.5, 10 mM KCl, 3 mM MgCl2, 0.4 M sucrose, 1 mM EDTA, 1 mM DTT, and 0.2% Triton X-100) and incubated at 4°C for 1 h with agitation. The beads were then pelleted by centrifugation at 2000g for 1 min at 4°C and washed four times with protein pull-down buffer. Bound proteins were eluted, fractionated by 10% SDS-PAGE, and subjected to protein gel blot analysis using appropriate antibodies.
Generation of Transgenic Plants
The cauliflower mosaic virus 35S promoter and HA:EPSIN1 were inserted into the polylinker region of pBIB-HYG (Becker, 1990), and the resulting construct was introduced into plants by Agrobacterium tumefaciens–mediated transformation (Clough and Bent, 1998).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: EPSIN1, At5g11710; EPSIN2, At2g43170; EPSIN3, At3g59290; AALP, At5g60360; VSR1, At3g52850; PEP12p, At5g16830; α-ADR, At5g22770; γ-ADR, At1g23900; δ-ADR, At1g48760; VTI11, At5g39510; and VTI12, At1g26670.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. EPSIN1 Is Not Permanently Associated with Clathrin-Coated Vesicles.
Supplementary Material
Acknowledgments
The T-DNA insertion mutant was obtained from the ABRC (Ohio State University). This work was supported by a grant from the Creative Research Initiatives Program of the Ministry of Science and Technology (Korea).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Inhwan Hwang (ihhwang@postech.ac.kr).
Online version contains Web-only data.
References
- Ahmed, S.U., Bar-Peled, M., and Raikhel, N.V. (1997). Cloning and subcellular location of an Arabidopsis receptor-like protein that shares common features with protein-sorting receptors of eukaryotic cells. Plant Physiol. 114 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, S.U., Rojo, E., Kovaleva, V., Venkataraman, S., Dombrowski, J.E., Matsuoka, K., and Raikhel, N.V. (2000). The plant vacuolar sorting receptor AtELP is involved in transport of NH(2)-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J. Cell Biol. 149 1335–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham, D.C., and Raikhel, N.V. (2000). Unique features of the plant vacuolar sorting machinery. Curr. Opin. Cell Biol. 12 491–495. [DOI] [PubMed] [Google Scholar]
- Bassham, D.C., Sanderfoot, A.A., Kovaleva, V., Zheng, H., and Raikhel, N.V. (2000). AtVPS45 complex formation at the trans-Golgi network. Mol. Biol. Cell 11 2251–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, D. (1990). Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 18 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M.K. (1995). SNAREs and the specificity of transport vesicle targeting. Curr. Opin. Cell Biol. 7 581–586. [DOI] [PubMed] [Google Scholar]
- Boevink, P., Oparka, K., Santa Cruz, S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15 441–447. [DOI] [PubMed] [Google Scholar]
- Brett, T.J., Traub, L.M., and Fremont, D.H. (2002). Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure 10 797–809. [DOI] [PubMed] [Google Scholar]
- Campbell, R.E., Tour, O., Palmer, A.E., Steinbach, P.A., Baird, G.S., Zacharias, D.A., and Tsien, R.Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., and De Camilli, P. (2005). The association of epsin with ubiquitinated cargo along the endocytic pathway is negatively regulated by its interaction with clathrin. Proc. Natl. Acad. Sci. USA 102 2766–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., Fre, S., Slepnev, V.I., Capua, M.R., Takei, K., Butler, M.H., Di Fiore, P.P., and De Camilli, P. (1998). Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature 394 793–797. [DOI] [PubMed] [Google Scholar]
- Chidambaram, S., Mullers, N., Wiederhold, K., Haucke, V., and von Mollard, G.F. (2004). Specific interaction between SNAREs and epsin N-terminal homology (ENTH) domains of epsin-related proteins in trans-Golgi network to endosome transport. J. Biol. Chem. 279 4175–4179. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Dahms, N.M., Lobel, P., and Kornfeld, S. (1989). Mannose 6-phosphate receptors and lysosomal enzyme targeting. J. Biol. Chem. 264 12115–12118. [PubMed] [Google Scholar]
- da Silva Conceição, A., Marty-Mazars, D., Bassham, D.C., Sanderfoot, A.A., Marty, F., and Raikhel, N.V. (1997). The syntaxin homolog AtPEP12p resides on a late post-Golgi compartment in plants. Plant Cell 9 571–582. [PMC free article] [PubMed] [Google Scholar]
- daSilva, L.L., Taylor, J.P., Hadlington, J.L., Hanton, S.L., Snowden, C.J., Fox, S.J., Foresti, O., Brandizzi, F., and Denecke, J. (2005). Receptor salvage from the prevacuolar compartment is essential for efficient vacuolar protein targeting. Plant Cell 17 132–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Camilli, P., Chen, H., Hyman, J., Panepucci, E., Bateman, A., and Brunger, A.T. (2002). The ENTH domain. FEBS Lett. 513 11–18. [DOI] [PubMed] [Google Scholar]
- Drake, M.T., Downs, M.A., and Traub, L.M. (2000). Epsin binds to clathrin by associating directly with the clathrin-terminal domain. Evidence for cooperative binding through two discrete sites. J. Biol. Chem. 275 6479–6489. [DOI] [PubMed] [Google Scholar]
- Driouich, A., Zhang, G.F., and Staehelin, L.A. (1993). Effect of brefeldin A on the structure of the Golgi apparatus and on the synthesis and secretion of proteins and polysaccharides in sycamore maple (Acer pseudoplatanus) suspension-cultured cells. Plant Physiol. 101 1363–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, M.C., Costaguta, G., and Payne, G.S. (2003). Yeast epsin-related proteins required for Golgi-endosome traffic define a gamma-adaptin ear-binding motif. Nat. Cell Biol. 5 77–81. [DOI] [PubMed] [Google Scholar]
- Duncan, M.C., and Payne, G.S. (2003). ENTH/ANTH domains expand to the Golgi. Trends Cell Biol. 13 211–215. [DOI] [PubMed] [Google Scholar]
- Eugster, A., Pecheur, E.I., Michel, F., Winsor, B., Letourneur, F., and Friant, S. (2004). Ent5p is required with Ent3p and Vps27p for ubiquitin-dependent protein sorting into the multivesicular body. Mol. Biol. Cell 15 3031–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio, L., de Virgilio, M., Prada, A., Faoro, F., and Vitale, A. (1998). Sorting of phaseolin to the vacuole is saturable and requires a short C-terminal peptide. Plant Cell 10 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, G. (2000). Gut thoughts on the Golgi complex. Traffic 1 738–745. [DOI] [PubMed] [Google Scholar]
- Happel, N., Honing, S., Neuhaus, J.M., Paris, N., Robinson, D.G., and Holstein, S.E. (2004). Arabidopsis mu A-adaptin interacts with the tyrosine motif of the vacuolar sorting receptor VSR-PS1. Plant J. 37 678–693. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Hawes, C.R., Brandizzi, F., and Andreeva, A.V. (1999). Endomembranes and vesicle trafficking. Curr. Opin. Plant Biol. 2 454–461. [DOI] [PubMed] [Google Scholar]
- Hirst, J., Motley, A., Harasaki, K., Peak Chew, S.Y., and Robinson, M.S. (2003). EpsinR: An ENTH domain-containing protein that interacts with AP-1. Mol. Biol. Cell 14 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl, I., Robinson, D.G., Chrispeels, M.J., and Hinz, G. (1996). Transport of storage proteins to the vacuole is mediated by vesicles without a clathrin coat. J. Cell Sci. 109 2539–2550. [DOI] [PubMed] [Google Scholar]
- Holstein, S.E., and Oliviusson, P. (2005). Sequence analysis of Arabidopsis thaliana E/ANTH-domain-containing proteins: Membrane tethers of the clathrin-dependent vesicle budding machinery. Protoplasma 226 13–21. [DOI] [PubMed] [Google Scholar]
- Horak, J. (2003). The role of ubiquitin in down-regulation and intracellular sorting of membrane proteins: Insights from yeast. Biochim. Biophys. Acta 1614 139–155. [DOI] [PubMed] [Google Scholar]
- Itoh, T., Koshiba, S., Kigawa, T., Kikuchi, A., Yokoyama, S., and Takenawa, T. (2001). Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science 291 1047–1051. [DOI] [PubMed] [Google Scholar]
- Jin, J.B., Kim, Y.A., Kim, S.J., Lee, S.H., Kim, D.H., Cheong, G.W., and Hwang, I. (2001). A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13 1511–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalthoff, C., Groos, S., Kohl, R., Mahrhold, S., and Ungewickell, E.J. (2002). Clint: A novel clathrin-binding ENTH-domain protein at the Golgi. Mol. Biol. Cell 13 4060–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, T., Morita, M.I., Fukaki, H., Yoshiro, Y., Uehara, M., Nihama, M., and Tasaka, M. (2002). SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell 14 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.H., Eu, Y.-J., Yoo, C.M., Kim, Y.-W., Pih, K.T., Jin, J.B., Kim, S.J., Stenmark, H., and Hwang, I. (2001). Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H., Park, M., Kim, S.J., and Hwang, I. (2005). Actin filaments play a critical role in vacuolar trafficking at the Golgi complex in plant cells. Plant Cell 17 888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen, T. (1999). Adaptors for clathrin-mediated traffic. Annu. Rev. Cell Dev. Biol. 15 705–732. [DOI] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16 393–401. [DOI] [PubMed] [Google Scholar]
- Lafer, E.M. (2002). Clathrin-protein interactions. Traffic 3 513–520. [DOI] [PubMed] [Google Scholar]
- Lee, G.J., Sohn, E.J., Lee, M.H., and Hwang, I. (2004). The Arabidopsis rab5 homologs rha1 and ara7 localize to the prevacuolar compartment. Plant Cell Physiol. 45 1211–1220. [DOI] [PubMed] [Google Scholar]
- Lee, M.H., Min, M.K., Lee, Y.J., Jin, J.B., Shin, D.H., Kim, D.H., Lee, K.H., and Hwang, I. (2002). ADP-ribosylation factor 1 of Arabidopsis plays a critical role in intracellular trafficking and maintenance of endoplasmic reticulum morphology in Arabidopsis. Plant Physiol. 129 1507–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre-Guillemin, V., Wasiak, S., Hussain, N.K., Angers, A., and McPherson, P.S. (2004). ENTH/ANTH proteins and clathrin-mediated membrane budding. J. Cell Sci. 117 9–18. [DOI] [PubMed] [Google Scholar]
- Mills, I.G., Praefcke, G.J., Vallis, Y., Peter, B.J., Olesen, L.E., Gallop, J.L., Butler, P.J., Evans, P.R., and McMahon, H.T. (2003). EpsinR: An AP1/clathrin interacting protein involved in vesicle trafficking. J. Cell Biol. 160 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham, C.E., Mohney, R.P., Miller, S.L., Hanes, R.N., and O'Bryan, J.P. (2002). The ubiquitin-interacting motifs target the endocytic adaptor protein epsin for ubiquitination. Curr. Biol. 12 1112–1126. [DOI] [PubMed] [Google Scholar]
- Overstreet, E., Chen, X., Wendland, B., and Fischer, J.A. (2003). Either part of a Drosophila epsin protein, divided after the ENTH domain, functions in endocytosis of delta in the developing eye. Curr. Biol. 13 854–860. [DOI] [PubMed] [Google Scholar]
- Owen, D.J., Vallis, Y., Noble, M.E., Hunter, J.B., Dafforn, T.R., Evans, P.R., and McMahon, H.T. (1999). A structural explanation for the binding of multiple ligands by the alpha-adaptin appendage domain. Cell 97 805–815. [DOI] [PubMed] [Google Scholar]
- Paris, N., and Neuhaus, J.M. (2002). BP-80 as a vacuolar sorting receptor. Plant Mol. Biol. 50 903–914. [DOI] [PubMed] [Google Scholar]
- Park, M., Kim, S.J., Vitale, A., and Hwang, I. (2004). Identification of the protein storage vacuole and protein targeting to the vacuole in leaf cells of three plant species. Plant Physiol. 134 625–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, M., Lee, D., Lee, G.J., and Hwang, I. (2005). AtRMR1 functions as a cargo receptor for protein trafficking to the protein storage vacuole. J. Cell Biol. 170 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo, S., Sigismund, S., Faretta, M., Guidi, M., Capua, M.R., Bossi, G., Chen, H., De Camilli, P., and Di Fiore, P.P. (2002). A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416 451–455. [DOI] [PubMed] [Google Scholar]
- Pryer, N.K., Wuestehube, L.J., and Schekman, R. (1992). Vesicle-mediated protein sorting. Annu. Rev. Biochem. 61 471–516. [DOI] [PubMed] [Google Scholar]
- Raiborg, C., Rusten, T.E., and Stenmark, H. (2003). Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15 446–455. [DOI] [PubMed] [Google Scholar]
- Robinson, D.G., Hinz, G., and Holstein, S.E. (1998). The molecular characterization of transport vesicles. Plant Mol. Biol. 38 49–76. [PubMed] [Google Scholar]
- Robinson, M.S., and Bonifacino, J.S. (2001). Adaptor-related proteins. Curr. Opin. Cell Biol. 13 444–453. [DOI] [PubMed] [Google Scholar]
- Robinson, M.S., and Kreis, T.E. (1992). Recruitment of coat proteins onto Golgi membranes in intact and permeabilized cells: Effects of brefeldin A and G protein activators. Cell 69 129–138. [DOI] [PubMed] [Google Scholar]
- Rosenthal, J.A., Chen, H., Slepnev, V.I., Pellegrini, L., Salcini, A.E., Di Fiore, P.P., and De Camilli, P. (1999). The epsins define a family of proteins that interact with components of the clathrin coat and contain a new protein module. J. Biol. Chem. 274 33959–33965. [DOI] [PubMed] [Google Scholar]
- Rothman, J.E. (1994). Mechanisms of intracellular protein import. Nature 372 55–63. [DOI] [PubMed] [Google Scholar]
- Saint-Pol, A., Yelamos, B., Amessou, M., Mills, I.G., Dugast, M., Tenza, D., Schu, P., Antony, C., McMahon, H.T., Lamaze, C., and Johannes, L. (2004). Clathrin adaptor epsinR is required for retrograde sorting on early endosomal membranes. Dev. Cell 6 525–538. [DOI] [PubMed] [Google Scholar]
- Sanderfoot, A.A., Ahmed, S.U., Marty-Mazars, D., Rapoport, I., Kirchhausen, T., Marty, F., and Raikhel, N.V. (1998). A putative vacuolar cargo receptor partially colocalizes with AtPEP12p on a prevacuolar compartment in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 95 9920–9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman, R., and Orci, L. (1996). Coat proteins and vesicle budding. Science 271 1526–1533. [DOI] [PubMed] [Google Scholar]
- Scott, P.M., Bilodeau, P.S., Zhdankina, O., Winistorfer, S.C., Hauglund, M.J., Allaman, M.M., Kearney, W.R., Robertson, A.D., Boman, A.L., and Piper, R.C. (2004). GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat. Cell Biol. 6 252–259. [DOI] [PubMed] [Google Scholar]
- Sever, S., Muhlberg, A.B., and Schmid, S.L. (1999). Impairment of dynamin's GAP domain stimulates receptor-mediated endocytosis. Nature 398 481–486. [DOI] [PubMed] [Google Scholar]
- Shih, S.C., Katzmann, D.J., Schnell, J.D., Sutanto, M., Emr, S.D., and Hicke, L. (2002). Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell Biol. 4 389–393. [DOI] [PubMed] [Google Scholar]
- Shimada, T., Fuji, K., Tamura, K., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (2003). Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100 16095–16100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund, S., Woelk, T., Puri, C., Maspero, E., Tacchetti, C., Transidico, P., Di Fiore, P.P., and Polo, S. (2005). Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl. Acad. Sci. USA 102 2760–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn, E.J., Kim, E.S., Zhao, M., Kim, S.J., Kim, H., Kim, Y.W., Lee, Y.J., Hillmer, S., Sohn, U., Jiang, L., and Hwang, I. (2003). Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 15 1057–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector, I., Shochet, N.R., Kashman, Y., and Groweiss, A. (1983). Latrunculins: Novel marine toxins that disrupt microfilament organization in cultured cells. Science 219 493–495. [DOI] [PubMed] [Google Scholar]
- Surpin, M., Zheng, H., Morita, M.T., Saito, C., Avila, E., Blakeslee, J.J., Bandyopadhyay, A., Kovaleva, V., Carter, D., Murphy, A., Tasaka, M., and Raikhel, N. (2003). The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15 2885–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, B.L., Wang, Y., Ong, Y.S., and Hong, W. (2005). COPII and exit from the endoplasmic reticulum. Biochim. Biophys. Acta 1744 293–303. [DOI] [PubMed] [Google Scholar]
- Tse, Y.C., Mo, B., Hillmer, S., Zhao, M., Lo, S.W., Robinson, D.G., and Jiang, L. (2004). Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16 672–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasiak, S., Legendre-Guillemin, V., Puertollano, R., Blondeau, F., Girard, M., de Heuvel, E., Boismenu, D., Bell, A.W., Bonifacino, J.S., and McPherson, P.S. (2002). Enthoprotin: A novel clathrin-associated protein identified through subcellular proteomics. J. Cell Biol. 158 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland, B. (2002). Epsins: Adaptors in endocytosis? Nat. Rev. Mol. Cell Biol. 3 971–977. [DOI] [PubMed] [Google Scholar]
- Wendland, B., Steece, K.E., and Emr, S.D. (1999). Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J. 18 4383–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.D., Elamawi, R., Bubeck, J., Pepperkok, R., Ritzenthaler, C., and Robinson, D.G. (2005). Dynamics of COPII vesicles and the Golgi apparatus in cultured Nicotiana tabacum BY-2 cells provides evidence for transient association of Golgi stacks with endoplasmic reticulum exit sites. Plant Cell 17 1513–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, H., von Mollard, G.F., Kovaleva, V., Stevens, T.H., and Raikhel, N.V. (1999). The plant vesicle-associated SNARE AtVTI1a likely mediates vesicle transport from the trans-Golgi network to the prevacuolar compartment. Mol. Biol. Cell 10 2251–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.