Abstract
The secretory factor VEGF-C has been directly implicated in various physiological processes during embryogenesis and human cancers. However, the importance of the conversion of its precursor proVEGF-C to mature VEGF-C in tumorigenesis, and vessel formation and the identity of the protease(s) that regulate these processes is/are not known. The intracellular processing of proVEGF-C that occurs within the dibasic motif HSIIRR227SL suggests the involvement of the proprotein convertases (PCs) in this process. In addition, furin and VEGF-C were found to be coordinately expressed in adult mouse tissues. Cotransfection of the furin-deficient colon carcinoma cell line LoVo with proVEGF-C and different PC members revealed that furin, PC5, and PC7 are candidate VEGF-C convertases. This finding is consistent with the in vitro digestions of an internally quenched synthetic fluorogenic peptide mimicking the cleavage site of proVEGF-C (220Q-VHSIIRR↓SLP230). The processing of proVEGF-C is blocked by the inhibitory prosegments of furin, PC5, and PACE4, as well as by furin-motif variants of α2-macroglobulin and α1-antitrypsin. Subcutaneous injection of CHO cells stably expressing VEGF-C into nude mice enhanced angiogenesis and lymphangiogenesis, but not tumor growth. In contrast, expression of proVEGF-C obtained following mutation of the cleavage site (HSIIRR227SL to HSIISS227SL) inhibits angiogenesis and lymphangiogenesis as well as tumor growth. Our findings demonstrate the processing of proVEGF-C by PCs and highlight the potential use of PC inhibitors as agents for inhibiting malignancies induced by VEGF-C.
Introduction
VEGF-C, initially purified from culture medium conditioned by PC3 prostate adenocarcinoma cells, belongs to the PDGF/VEGF family of growth factors (1). VEGF-C is a ligand for the lymphatic endothelial receptor VEGFR-3 (Flt4), but also binds to VEGFR-2, which is the major mitogenic signal transducer for VEGF in blood vessel endothelial cells (1–3). The concomitant expression of VEGF-C and VEGFR-3 in many tissues, including tumors, involves the paracrine action of VEGF-C in angiogenesis of the lymphatic vasculature, whereas its ability to activate VEGFR-2 suggests its functional redundancy with VEGF (1–3). Human VEGF-C cDNA encodes a protein of 419 AA residues with a predicted molecular mass of 59 kDa (1, 4). Newly synthesized VEGF-C is a preproprotein (referred to hereafter as proVEGF-C) consisting of an N-terminal signal sequence (AAs 1–12) followed by an N-terminal propeptide (AAs 13–102), the VEGF homology domain (AAs 103–227), and a cysteine-rich C-terminal segment (AAs 228–419). ProVEGF-C is secreted as a disulfide-bonded homodimer that is proteolytically processed from the precursor polypeptide. The secreted form contains the C-terminal silk domain (4). Upon examination of the AA sequence of the VEGF-C precursor (NCBI sequence data base; NP 005420), a dibasic motif, 220Q-VHSIIRR↓SLP230, resembling those recognized by the proprotein convertases (PCs) (5, 6) is found, suggesting the involvement of these convertases in the maturation of VEGF-C.
The mammalian subtilisin-like PCs constitute a family of seven known dibasic-specific proteinases: furin, PC1, PC2, PC4, PACE4, PC5 (and its isoform PC5B), and PC7, as well as the non–basic-specific convertase SKI-1 (5). The first seven dibasic-specific enzymes are implicated in the processing of multiple protein precursors, including growth factors, receptors, proteases of the coagulation and complement cascades, glycoproteins of viral envelopes, and bacterial exotoxins at multibasic recognition sites exhibiting the general motif (K/R)-(X)n-(K/R)↓, where n = 0, 2, 4, or 6 (5–7), and where K is lysine, R is arginine, and X is any amino acid. PC1 and PC2 are found within dense core secretory granules and process precursors therein. In contrast, furin, PC5B, and PC7 (the only members of the mammalian PCs with a transmembrane domain), together with PC5A and PACE4, are the main enzymes that process precursors sorted to the constitutive secretory pathway (5, 6).
Previously, we demonstrated that inhibition of PCs by the general PC inhibitor α1-PDX (furin-motif variant of α1-antitrypsin [α1-antitrypsin Portland]) blocked the processing of several proteins involved in tumorigenesis, such as MT1-MMP and IGF-1 receptor (6, 8). In the present study, we evaluated the involvement of PCs in the processing of proVEGF-C and assessed the importance of this processing step in tumorigenesis, angiogenesis, and lymphangiogenesis.
Methods
HSIISS227SL mutant VEGF-C, transfections, and cell culture.
The coding region of human VEGF-C precursor was cloned into the Taq-amplified cloning vector pCRII–TOPO (Invitrogen Corp., San Diego, California, USA) by RT-PCR from the human adenocarcinoma PC3 cell line (CRL 1435; American Type Culture Collection, Rockville, Maryland, USA) using the sense primer 5′-TTCCACCATGCACTTGCTG-3′ and the antisense primer 5′-GAAGGGACACAACGACACAC-3′. The VEGF-C product was completely sequenced, compared with the published sequence (1), and cloned again in pCRII-TOPO using the sense primer, but with the following extended antisense primer containing a short segment of missing translated 3′ end and an in-frame XhoI site to allow the addition of C-terminal Flag tag 5′-CTCGAGGCTCATTTGTGGTCTTTTCCAATATGAAGGGACACAACGACACAC-3′. This extended VEGF-C clone was inserted into the EcoRI–XhoI sites in pCMV-Tag4A, and subsequently, VEGF-C-Flag clone was inserted into the EcoRI and KpnI sites of pcDNA3-zeo (Invitrogen Corp.). The final product, completely sequenced, matched the published sequence (1). To generate the mutant VEGF-C (HSIIRR227SL to HSIISS227SL), mutagenesis was carried out by PCR mutagenesis using the sense primer 3′-GACAAACACCTTCTTTAAACCTCC-5′, antisense mutant primer 5′-CAGGGAAGAGCTAATAATGGAATG-3′, sense mutant primer 5′-CCATTATTAGCTCTTCCCTGCCAG-3′, and the same antisense primer described above (5′-GAAGGGACACAACGACACAC-3′).
Furin-deficient LoVo-C5 human colon adenocarcinoma cells (9) were transiently cotransfected with the pIRES2-EGFP (Clontech Laboratories Inc., Palo Alto, California, USA) and pcDNA3-zeo-Flag.cm5 empty vectors, pIRES2-EGFP empty vector, and pcDNA3-zeo-Flag.cm5 containing VEGF-C cDNA, or with the pcDNA3-zeo-Flag.cm5 vector containing VEGF-C and pIRES2-EGFP vectors that express full-length human furin, PACE4, SKI-1, mouse PC5A, PC5B, or rat PC7. Human embryonic kidney (HEK) 293 cells were transiently cotransfected with the pIRES2-EGFP and pcDNA3-zeo-Flag.cm5 empty vectors, pIRES2-EGFP empty vector, and pcDNA3-zeo-Flag.cm5 containing VEGF-C cDNA, or with the pcDNA3-zeo-Flag.cm5 vector containing VEGF-C and pIRES2-EGFP vector that expresses PC inhibitors including p-furin, pPACE4, pPC5, and pPC7, wild-type or mutated α2-macroglobulin, and α1-antitrypsin. In several experiments, the PC3 prostate adenocarcinoma cells were transiently transfected with pIRES2-EGFP vector expressing either PC inhibitors or PCs to assess the endogenous processing of VEGF-C. CHO tumor cells were stably transfected with pcDNA3-zeo-Flag.cm5 empty vector, pcDNA3-zeo-Flag.cm5 vector containing wild-type VEGF-C, or pcDNA3-zeo-Flag.cm5 vector containing mutated VEGF-C (HSIIRR227SL to HSIISS227SL) cDNA’s. Populations of stably transfected cells were selected using zeocin resistance and controlled by Western blotting for wild-type and mutant VEGF-C expression. All the transfections were carried out using Effectene transfection reagent (QIAGEN Inc., Mississauga, Ontario, Canada) as recommended by the manufacturer. Cells were grown in DMEM supplemented with 10% FCS, 100 units/ml penicillin, and 100 mg/ml streptomycin (Life Technologies Inc., Burlington, Ontario, Canada). For the stably transfected CHO cells, 200 μg/ml zeocin was added.
Analysis of furin and VEGF-C mRNA expression by RT-PCR.
Total RNA was extracted using Trizol (Life Technologies Inc.) according to the manufacturer’s instructions. RNA was reverse-transcribed in a buffer containing 50 mM Tris-HCl (pH 8.3), 30 mM KCl, 8 mM MgCl2, 1 mM dNTP’s, and 0.5 units of SuperScript reverse transcriptase (Invitrogen Corp.). The mixture was sequentially incubated for 10 minutes at 25°C, for 60 minutes at 37°C, and for 5 minutes at 95°C. PCR amplification of cDNA’s used the oligonucleotides 5′-ACGCAACAGGGAATTTGATG-3′ and 5′-ACAGACTTGGGCCTCTGTTAC-3′ for VEGF-C (400-bp product), 5′-TGAGCCATTCGTATGGCTACG-3′ and 5′-GGACACAGCTTTTCTGGTGCA-3′ for furin (576-bp product), and 5′-TGGAAATCCCATCACCATCT-3′ and 5′-GTCTTCTGGGTGGCAGTGAT-3′ (520-bp product) for GAPDH. PCR was performed in a buffer supplied by the manufacturer to which the cDNA sample, 5 μM each primer, 200 μM dNTP’s, and 0.5 units Taq polymerase (QIAGEN Inc.) were added. The conditions for the reaction were as follows: 30 seconds at 94°C, 30 seconds at 56°C, and 30 seconds at 72°C for 25 cycles. Amplified PCR products were analyzed on a 1.5% agarose gel.
Biosynthetic labeling and immunoprecipitation.
Two days after transfection, cells were washed and then pulse-incubated for 2–3 hours with 200 μCi/ml [35S]Cys. After the pulse period, cells were lysed in lysis buffer containing 150 mM NaCl, 50 mM Tris-HCl, pH 6.8, 0.5% NP-40, 0.5% sodium deoxycholate, and a protease inhibitor mixture (Roche Molecular Biochemicals, Montreal, Quebec, Canada) and prepared for immunoprecipitations as previously described (7). The monoclonal antibodies directed against Flag (1:500 dilution; Stratagene, La Jolla, California, USA) and anti–VEGF-C (1:200 dilution; Santa Cruz Biotechnology Inc., Santa Cruz, California, USA) were used as primary antibodies. Band intensities of the autoluminographs were quantitated by densitometry.
Western blotting.
Twenty-four hours after transfection, cells were lysed in PBS containing 2% NP-40, lysates were subjected to SDS-PAGE on 8% polyacrylamyde gel, and proteins were blotted onto nitrocellulose membranes. The primary antibodies anti-Flag and anti–VEGF-C were revealed by HPR-conjugated secondary antibodies (at 1:10,000 dilution; Amersham Biosciences Inc., Baie d’Urfé, Quebec, Canada) and enhanced chemiluminescence (ECL Plus; Amersham Biosciences Inc.) according to the manufacturer’s instructions. Band intensities of the autoluminographs were quantitated by densitometry.
Peptide synthesis.
An intramolecularly quenched fluorogenic (IQF) peptide, Q-h-VEGF-C [Abz-220Q-VHSIIRR↓SLP230-Y(NO2)-A-CONH2] containing the processing site of proVEGF-C was synthesized using an automated solid-phase peptide synthesizer (Pioneer model; PerSeptive Biosystems, Framingham, Massachusetts) (10). Abz is 2-(o)-aminobenzoic acid (fluorogenic group) and Y(NO2) is 3-nitrotyrosine (fluorescence quench group). After purification of the crude peptide by reverse-phase HPLC (RP-HPLC) on a Varian instrument (Varian Canada Inc., Ottawa, Ontario, Canada), the peptide purity and concentration were determined by quantitative AA analysis following 24 hours of hydrolysis in the presence of 5.7 N HCl in a vacuum at 110°C on a Beckman 6300 autoanalyzer with a postcolumn ninhydrin detection system (Beckman Coulter Inc., Miami, Florida, USA). The identity of the purified peptide was confirmed by matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-ToF-MS) (Voyager DE-PRO; PerSeptive Biosystems).
In vitro digestions of QVEGF-C with furin, PC5, and PC7.
The recombinant forms of furin, PC5, and PC7 were obtained from culture media of either somatomammotroph GH4C1 or BSC-40 cells following infection of respective cDNA’s with vaccinia viruses. The collected media were concentrated, dialyzed, and partially purified through chromatography on an anion exchange column as described (11). The reaction was conducted in water using 10 mM of Q-h-VEGF-C (2 μl), digested with 10 μl of each recombinant PC separately. The amount of each PC used represents nearly identical levels of enzymatic activity as measured in the presence of the more universal PC substrate, the fluorogenic peptide pERTKR-MCA (100 μM). The reaction was conducted in a total volume of 100 μl containing 25 mM Tris, 25 mM methyl-ethane-sulfonic acid and 2.5 mM CaCl2, pH 7.4 at 37°C using a 96-well flat-bottom plate. Aliquots were removed at various time intervals for RP-HPLC and mass spectral analyses to determine the sites and extent of cleavage. RP-HPLC was performed on an analytical 10-μm 300-Å C18 Jupiter column (0.46 × 25 cm) (Phenomenex, Torrance, California, USA). The conditions used were the same as described earlier except that the flow was maintained at 1.0 ml/min and the elution of the peptides was monitored by both UV absorbency (λ = 210 or 230 nm) and fluorescence measurement (Dynamax FL-2 fluorescence detector; Rainin, Walnut Creek, California, USA), with λex (excitation wavelength) and λem (emission wavelength) fixed at 320 nm and 420 nm, respectively. All fluorometric assays were performed with a spectrofluorometer (Gemini; Molecular Devices Corp., Sunnyvale, California, USA) as described previously (12).
Determination of the kinetic parameters Vmax (apparent) and Km (apparent) for in vitro digestion of QVEGF-C by furin, PC5, and PC7.
The determination of kinetic parameters Vmax (apparent) and Km (apparent) for the cleavage of QVEGF-C by furin, PC5, and PC7 was carried out by incubating varying amounts of QVEGF-C (0.5 –200 μM, depending on the enzyme) with 10 μl of each PC (exhibiting a similar level of activity when measured against 100 μM of pERTKR-MCA). The fluorescence released at various times was measured at λex 320 nm and λem 420 nm. The data collected after either 1 hour or 4 hours of incubation were used to calculate Vmax (apparent) and Km (apparent) using GraFit software (Sigma-Aldrich, Oakville, Ontario, Canada)as described earlier (10–12).
In vitro cell growth and in vivo tumorigenicity assays.
To assess the mitogenic activity of control CHO cells or CHO cells expressing wild-type or mutant VEGF-C (HSIIRR227SL to HSIISS227SL), a [3H]thymidine incorporation assay was carried out as previously described (12). For tumor growth measurement, CHO tumor cells were injected subcutaneously into 4- to 6-week-old male nude mice, and animals were monitored for tumor formation every 3 days. Tumor volume was calculated using the formula previously described (13).
Immunohistochemical analyses.
Fifteen to 20 days after the subcutaneous injection of the populations of CHO control cells and CHO cells stably transfected with either VEGF-C or VEGF-C mutant, the developed tumors were cryosectioned and fixed in 4% PBS/paraformaldehyde. Slides were analyzed for angiogenesis as we described previously (3, 8) using anti-mouse CD-31 monoclonal antibody (Pharmingen, San Diego, California, USA) at 1:50 dilution and for lymphangiogenesis using an anti–VEGFR-3 polyclonal antibody (Santa Cruz Biotechnology Inc.) at 1:200 dilution. Angiogenesis and lymphangiogenesis were quantitated in tumor sections by counting the number of CD-31–positive vascular vessels and VEGFR-3–positive lymphatic vessels, respectively. For each section, only the stained vessels that were clearly separate from others were counted in the ten most densely vasculated areas. Counting was performed at a magnification of ×200. For each tumor, at least four independent sections were analyzed.
Results
Processing of proVEGF-C by members of the PC family.
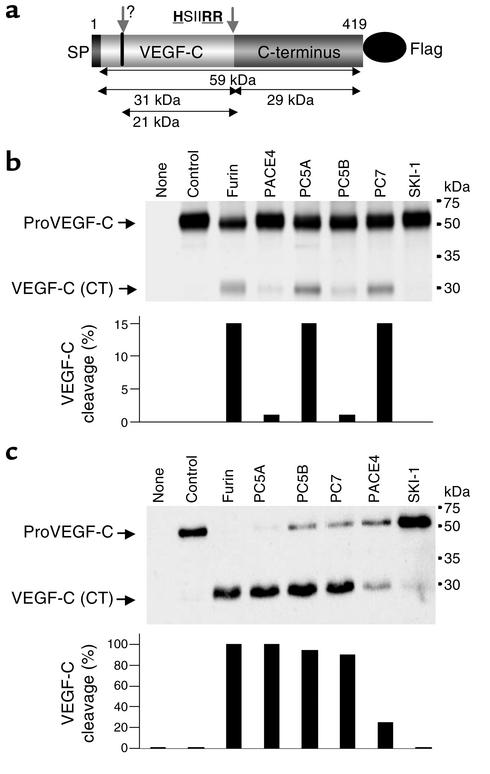
VEGF-C has been identified as a new member of the PDGF/VEGF superfamily that requires specific endoproteolytic cleavage for its activity (1, 4). However, nothing is known about the enzymes involved in its processing and activation. To identify which PC can process proVEGF-C, we cloned the coding regions of human VEGF-C precursor from the human adenocarcinoma PC3 cell line by RT-PCR and tagged its C-terminal domain with a Flag sequence (Figure 1a). The precursor proVEGF-C and each of the PCs were transiently coexpressed in LoVo cells, a furin-deficient cell line (9). After transfection, the cells were pulse-labeled for 2 hours with [35S]Cys, and the media were immunoprecipitated with an anti-Flag antibody (Figure 1b). In parallel, supernatants collected 24 hours after transfection were analyzed for proVEGF-C processing by immunoblotting using an anti–VEGF-C antibody (Figure 1c). As illustrated in Figure 1, b and c, analyses of media derived from LoVo cells cotransfected with vector encoding proVEGF-C and control vector showed only one band with an apparent molecular mass of about 59 kDa, corresponding to the intact VEGF-C precursor. Cotransfection of cells with proVEGF-C and vectors encoding either furin, PC5A, PC5B, PC7, PACE4, or SKI-1 revealed that expression of furin, PC5A, and PC7 in these cells was associated with a reduction in the level of the immunoreactive (∼59 kDa) precursor with a concomitant appearance of a product of approximately 29 kDa, corresponding to the CT domain of proVEGF-C. These data revealed that under steady-state conditions all PCs except SKI-1 were able to mediate cleavage of proVEGF-C (Figure 1c).
Figure 1.
Processing of proVEGF-C by furin, PC5, and PC7. (a) Schematic representation of the primary structure of the 419-AA human proVEGF-C. Shown are the signal peptide (SP), PC-processing site (HSIIRR227SL), an unknown protease site (indicated by question mark) that generates the 21-kDa VEGF-C, and the Flag attached to the C-terminus. ProVEGF-C processing was analyzed by biosynthesis (b) and Western blotting (c) of LoVo-C5–conditioned media obtained from cells transiently transfected with either the empty vectors (None), pIRES2-EGFP vector and pcDNA3-zeo-Flag.cm5 vector containing proVEGF-C (Control), or with the pcDNA3-zeo-Flag.cm5 vector containing proVEGF-C and pIRES2-EGFP vector that expresses full-length human furin, PACE4, or SKI-1; mouse PC5A or PC5B; or rat PC7. The corresponding percentages of proVEGF-C cleavage calculated from the ratio of band intensities of VEGF-C/(proVEGF-C + VEGF-C) are indicated.
In vitro digestions of Q-h-VEGF-C with furin, PC5, and PC7.
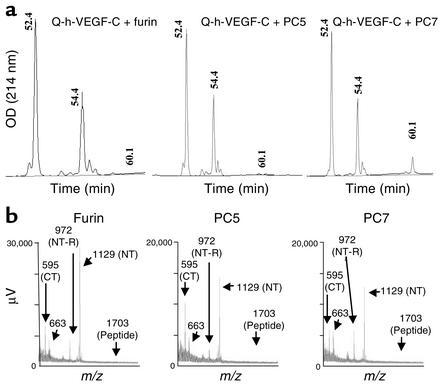
To confirm the processing of VEGF-C by furin, PC5, and PC7, we analyzed the cleavage of an intramolecularly quenched fluorogenic substrate, Q-h-VEGF-C, by these enzymes. As assessed by mass spectrometry and RP-HPLC (Figure 2, a and b), Q-h-VEGF-C was cleaved in vitro by furin, PC5, and PC7 at the correct physiological HSIIRR227↓SL site. Upon incubation with PC5 for 4 hours at room temperature, the RP-HPLC chromatogram of the crude digest (Figure 2a) exhibited, in addition to a small peak of undigested peptide [Rt = 60.1 min, m/z = 1,703.1], two additional peaks at retention time (Rt) = 54.4 and 52.4 minutes. These were identified by MALDI-ToF-MS as the highly fluorogenic N-terminal fragment Abz-220Q-VHSIIRR-OH [m/z = 1,129] and the nonfluorescent C-terminal peptide SLP230-Y(NO2)-A-CONH2 [m/z = 595] (Figure 2b). A nearly identical observation was noted with furin and PC7, although with the latter some undigested peptide remained (Rt = 60.1 min). When digested for a prolonged period of time (24 hours), PC7 also resulted in a complete cleavage of Q-h-VEGF-C (data not shown). In all cases, intense peaks at m/z 1,129 and 595 were detectable, identifying them as the N-terminal and C-terminal fragments, respectively (Figure 2b). An additional peak at m/z 972, which was also present in the wild-type digest of the peptide (run in parallel), was attributed to the peptide fragment Abz-220Q-VHSIIR-OH. This cleavage may be generated by the action of the basic-specific carboxypeptidase present in the enzyme preparation. Another peak at m/z 663 also found in the wild-type digest (not shown) was not PC-related. Thus, no additional cleavage of Q-h-VEGF-C by PCs other than that at the physiological IIRR227↓ site of VEGF-C was detected, even after prolonged incubation of Q-h-VEGF-C with these PCs, indicating the specificity of the PCs for this cleavage.
Figure 2.
In vitro digestions of Q-h-VEGF-C with recombinant furin, PC5, and PC7. (a) RP-HPLC chromatogram of the crude digest following 4 hours of incubation at 37°C of 20 μg of QVEGF-C with furin, PC5, or PC7 in 25 mM Tris, 25 mM Mes, and 2.5 mM CaCl2, pH 7.4. The elution of the peaks was monitored on-line by UV absorbance at 214 nm as well as by fluorescence detectors (λex, 320 nm; λem, 420 nm). (b) MALDI-ToF mass spectra of the crude digests following 24 hours of incubation at 37°C of 20 μg of QVEGF-C with furin, PC5, and PC7 in 25 mM Tris, 25 mM Mes, and 2.5 mM CaCl2, pH 7.4. Note the absence of the peak at m/z 1,703 suggesting complete cleavage of QVEGF-C. The peaks at m/z 1,129 and 595 were attributed to the highly fluorescent N-terminal (NT) (Abz-Q-VHSIIRR-OH) and the nonfluorescent C-terminal (CT) [SLP(NO2)-A-CONH2] fragments, respectively. The peaks at m/z 972 and 663 are not PC-dependent since they are also present in the crude digest of QVEGF-C by wild-type medium (data not shown). NT-R, the N-terminal sequence without Arginine residue.
Kinetic parameters Vmax (apparent) and Km (apparent) for the cleavage of Q-h-VEGF-C by furin, PC5, and PC7.
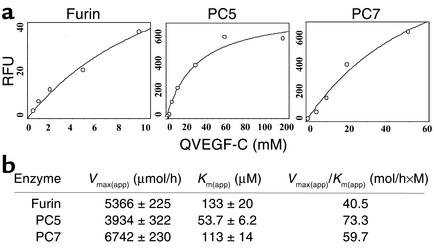
The kinetic parameters Vmax (apparent) and Km (apparent) for the cleavage of Q-h-VEGF-C by the above PCs were determined by Michaelis-Menten plots. The digestion curves of Q-h-VEGF-C for each of these PCs are shown in Figure 3a, and indicate rapid digestion of Q-h-VEGF-C by PC5 compared with furin and PC7. The measurement of the kinetic parameters of this analysis revealed that PC5 was the most active enzyme in processing Q-h-VEGF-C, with a Vmax (apparent)/Km (apparent) ratio of 73.3 (mol/h×M). Second most active was PC7 (59.7 mol/h×M), and then furin (40.5 mol/h×M) (Figure 3b).
Figure 3.
(a) Kinetic parameters Vmax (apparent) and Km (apparent) for the cleavage of QVEGF-C by furin, PC5, and PC7. Various concentrations of QVEGF-C (0.5–200 μM) were incubated in the presence of furin, PC5, or PC7, and the fluorescence released at various times was measured at λex 320 nm and λem 420. (b) The data collected after either 1 hour or 4 hours of incubation were used to calculate Vmax (apparent) and Km (apparent) using GraFit software as described in Methods. The amounts of furin, PC5, and PC7 used in this study were adjusted to show similar levels of enzymatic activity when measured in the presence of the universal PC substrate, the fluorogenic peptide pERTKR-MCA (100 μM). RFU, raw fluorescence unit; app, apparent.
Blockade of proVEGF-C processing by PC inhibitors.
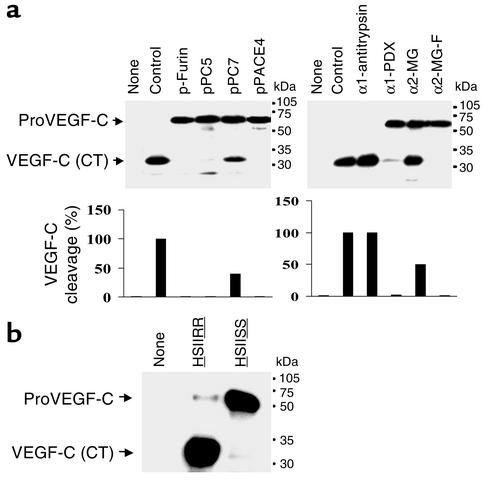
In order to assess the efficiency of PC inhibitors on proVEGF-C processing by PC-like endogenous convertases, we expressed different PC inhibitors in HEK 293 cells, including the PC prosegments (p-furin, pPACE4, pPC5, and pPC7); the furin-motif variants of α2-macroglobulin (α2-MG-F) and of α1-PDX; and wild-type α2-macroglobulin and α1-antitrypsin. Thus, HEK 293 cells were transiently cotransfected with vectors encoding proVEGF-C and one of each of the PC inhibitors. As demonstrated by Western blot analysis (Figure 4a), transfection of HEK 293 cells with the vector encoding proVEGF-C resulted in 100% processing. Cotransfection of cells with PC inhibitors revealed that processing of proVEGF-C is significantly blocked (90–100%) by α2-MG-F and α1-PDX, as well as by p-furin, pPC5, and pPACE4. In contrast, the prosegment pPC7, wild-type α1-antitrypsin, and wild-type α2-macroglobulin failed to significantly inhibit proVEGF-C processing.
Figure 4.
Blockade of proVEGF-C processing. (a) Processing of proVEGF-C was analyzed by Western blotting in HEK 293 cells transiently cotransfected with the empty pIRES2-EGFP vector (None) and pcDNA3-zeo-Flag.cm5 vector containing proVEGF-C cDNA (Control), or with the pcDNA3-zeo-Flag.cm5 vector containing proVEGF-C and pIRES2-EGFP vector that expresses profurin, proPACE4, proPC5, and proPC7, wild-type or mutated α2-macroglobulin (α2-MG and α2-MG-F, respectively), and α1-antitrypsin. The corresponding percentages of proVEGF-C cleavage calculated from the ratio of band intensities of VEGF-C/(proVEGF-C + VEGF-C) is indicated. (b) CHO tumor cells were stably transfected with empty pcDNA3-zeo-Flag.cm5 vector (None), pcDNA3-zeo-Flag.cm5 vector containing wild-type proVEGF-C cDNA (HSIIRR227), or pcDNA3-zeo-Flag.cm5 vector containing mutated proVEGF-C cDNA (HSIISS227). Populations of stably transfected cells were selected using Zeocin resistance and analyzed by Western blot using the VEGF-C antibody.
To determine the processing site in proVEGF-C by the PC family of endoproteases, we analyzed the effect of the mutation of proVEGF-C at HSIIRR227SL to HSIISS227SL on its processing in CHO cells. CHO cells were transfected with wild-type or mutant proVEGF-C, and media derived from these cells were analyzed by Western blotting (Figure 4b). The mutation of HSIIRR227SL to HSIISS227SL prevented the processing of the 59-kDa proVEGF-C into the 29 kDa form, suggesting the importance of the dibasic ArgArg227 in this cleavage.
Endogenous VEGF-C processing by PC-like activity.
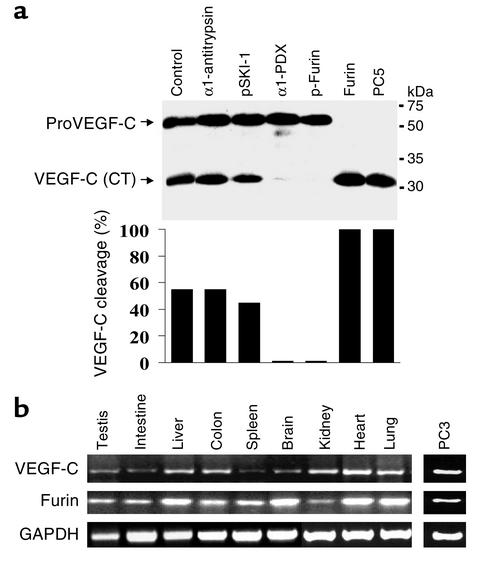
To assess the possibility that endogenous proVEGF-C is proteolytically activated by PC-like endoproteases, we transfected PC3 cells, which are known to endogenously produce VEGF-C, with vectors encoding either α1-PDX or the prosegment p-furin, which inhibits furin activity (11, 14). As demonstrated by Western blot analysis using anti–VEGF-C antibody (Figure 5a), both the unprocessed and the processed form of VEGF-C were detected in these cells (50–60% processing). Expression of α1-PDX or p-furin resulted in complete blockade of proVEGF-C maturation, leading to an accumulation of the 59-kDa unprocessed proVEGF-C. In contrast, the prosegment pSKI-1 and α1-antitrypsin failed to inhibit processing. Semiquantitative RT-PCR analysis of total RNA extracted from PC3 cells using primers specific for furin, PC5, PC7, and VEGF-C revealed the presence of furin (Figure 5b) and the absence of PC5 in these cells, whereas PC7 was weakly expressed (data not shown). Thus, furin is likely to be the major endogenous processing enzyme of VEGF-C in PC3 cells. In addition, overexpression of furin or PC5 in these cells resulted in 100% processing of proVEGF-C into VEGF-C (Figure 5b).
Figure 5.
Endogenous VEGF-C processing by PC-like activity and coexpression of furin and VEGF-C in mouse tissues. (a) Endogenous proVEGF-C processing was analyzed by Western blotting of PC3 cell–conditioned media obtained from cells transiently transfected with either the pIRES2-EGFP empty vectors (Control) or vector expressing α1-antitrypsin, pSKI-1, α1-PDX, p-furin, furin, or PC5. The corresponding percentages of proVEGF-C cleavage calculated from the ratio of band intensities of VEGF-C/(proVEGF-C + VEGF-C) are indicated. (b) Total RNA was extracted from PC3 cells and the indicated tissues and organs, and RT-PCR analysis was performed using primers specific for VEGF-C, furin, and GAPDH (control) under the conditions described in Methods. Results shown are representative of three experiments.
Coexpression of furin and VEGF-C in tissues.
To determine whether VEGF-C and furin are coexpressed in vivo, total RNA from various mouse tissues was analyzed by RT-PCR. We found that both VEGF-C and furin are coexpressed in the tissues examined (Figure 5b). Additionally, the highest expression of VEGF-C mRNA’s was found in heart, lung, and kidney, in agreement with a previous Northern blot analysis report (15). In some of these tissues the level of furin was also higher (Figure 5b).
Role of proVEGF-C processing in tumorigenesis.
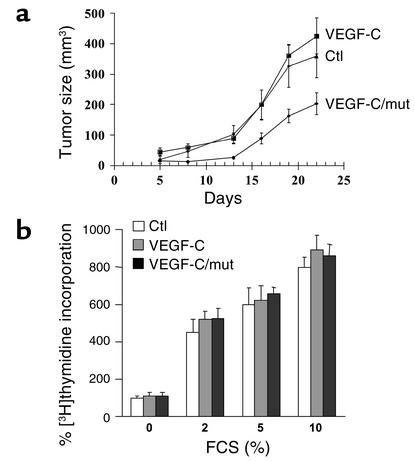
Prior to analysis, pools of CHO tumor cells stably expressing wild-type and mutant VEGF-C were selected, and the efficient production and secretion of these proteins was verified by Western blotting of proteins found in the conditioned media (Figure 4b). To assess the importance of proVEGF-C processing on the tumorigenicity of CHO cells, three groups of male nude mice were inoculated subcutaneously with 3 × 106 to 5 × 106 of control cells, cells expressing wild-type VEGF-C or cells expressing mutant VEGF-C. As illustrated in Figure 6a, the expression of VEGF-C in these tumor cells did not significantly affect tumor growth compared with control cells, whereas mice inoculated with tumor cells expressing mutant VEGF-C developed smaller tumors than did mice inoculated with CHO controls. To determine whether VEGF-C expression directly affected tumor cell proliferation, CHO transfectants were incubated with various concentrations of serum and their ability to incorporate [3H]thymidine was evaluated. As illustrated in Figure 6b, the growth rate of all the transfected cells was unaffected, suggesting the absence of a VEGF-C–mediated autocrine effect.
Figure 6.
Blockade of proVEGF-C processing inhibits in vivo tumor cell growth. (a) Populations of control CHO cells (Ctl) or CHO cells expressing wild-type (VEGF-C) or mutant VEGF-C (VEGF-C/mut) were injected subcutaneously into 4-week-old male nude mice. Tumor size was measured every 3 days. (b) Starved populations of control CHO cells or CHO cells expressing wild-type (VEGF-C) or mutant VEGF-C (VEGF-C/mut) were incubated for 24 hours in medium containing increasing concentrations of serum (0–10% FCS). [3H]thymidine was added for the final 6 hours of incubation. [3H]thymidine incorporation was measured as described previously (9). Data are presented as mean ± SE of four experiments.
Processing of proVEGF-C by PCs is required for VEGF-C–induced tumor angiogenesis and lymphangiogenesis.
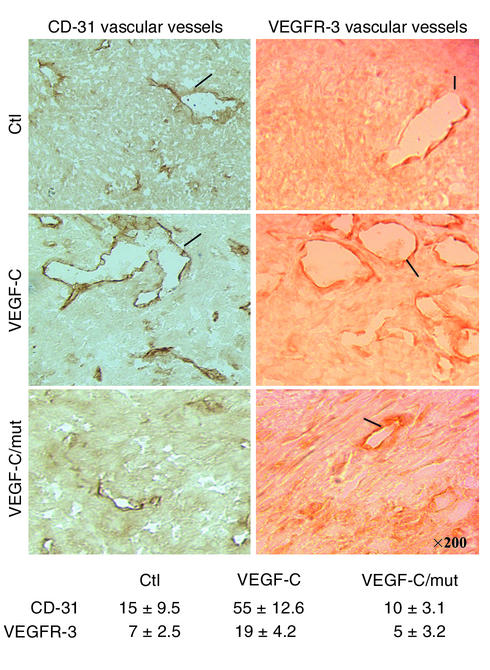
To analyze intratumoral angiogenesis and lymphangiogenesis, tumor sections derived from mice inoculated with control CHO cells or cells expressing proVEGF-C or proVEGF-C mutant were stained with antibodies against mouse CD-31 and VEGFR-3, respectively. Results shown in Figure 7 reveal significantly increased formation of vascular and lymphatic vessels in tumors derived from VEGF-C–transfected cells. In contrast, the relative tumor area covered by vascular and lymphatic vessels and the vessel number were significantly decreased in CHO VEGF-C mutant tumors compared with control CHO tumors.
Figure 7.
Immunohistochemical analyses. Tumors that developed 20 days after subcutaneous injection into nude mice of CHO cells stably transfected with empty vector (Ctl), vector containing wild-type or mutant proVEGF-C cDNA were analyzed for angiogenesis and lymphangiogenesis using an anti-mouse CD-31 monoclonal antibody and an anti–VEG-FR-C polyclonal antibody, respectively. The sections were observed at a magnification of ×200. Arrows indicate vessels. The number of intratumoral vascular and lymphatic vessels was counted in ten different fields for each tumor. Data are presented as mean ± SD.
Discussion
In this study we demonstrate that the convertases furin, PC5, and PC7 are the candidate proteases implicated in the proteolytic processing of proVEGF-C at HSIIRR227↓SL, dividing it into N-terminal (∼31 kDa) and cysteine-rich C-terminal (∼29 kDa) polypeptides (1, 4). Additional processing removes the N-terminal propeptide and generates the 21-kDa (approximate size) VEGF-C. This enzymatic reaction occurs extracellularly and/or at the cell surface via an unknown secretory protease (4). It is believed that the conversion of the 31-kDa VEGF-C to the 21-kDa form results in greatly enhanced affinity for and activating properties toward VEGFR-3 and VEGFR-2 (4).
In our model, the cleavage site of proVEGF-C between Arg227 and Ser228 was confirmed by mutagenesis and by in vitro enzymatic digestion of the fluorogenic substrate Q-h-VEGF-C mimicking this cleavage site. The efficient secretion of proVEGF-C from CHO VEGF-C cells indicated that intracellular proteolytic cleavage was not a prerequisite for VEGF-C secretion. This was further confirmed by the accumulation of proVEGF-C in media derived from VEGF-C–transfected LoVo cells (Figure 1, b and c) and by observed expression of PC inhibitors in HEK 293 (Figure 4a) and PC3 (Figure 5a) cells.
The ubiquitous expression of furin and the presence of a dibasic cleavage motif in proVEGF-C suggested that the PCs are good candidates for VEGF-C processing at this site. Although furin preferentially cleaves substrates containing an R-X-(K/R)R↓X (hydrophobic) sequence with basic residues at P4, P2, and P1 and a hydrophobic residue at P2′ (16), it also processes other precursors in which the P4 Arg is replaced by a basic residue at P6 (16). In particular, certain proteins such as α4-, αv-, and α8-integrins (6, 17, 18) contain a His at P6 and an Ile at P4. These proteins have been shown to be preferentially cleaved by furin and PC5 (18). The putative processing site of proVEGF-C contains all the furin/PC5 recognition hallmarks also found in α4-, αv-, and α8-integrins (8). This suggested that furin and/or PC5 could be the cognate proVEGF-C convertases. To test this hypothesis, we took advantage of the furin-deficient LoVo cells (9), and indeed demonstrated that not only furin and PC5 but also PC7 are the major candidate processing enzymes of proVEGF-C in this cell line (Figure 1, b and c). This finding is in agreement with the in vitro digestions that indicated PC5 to be the most efficient enzyme, followed by furin and then by PC7 (Figure 2 and Figure 3). These conclusions were further corroborated in HEK 293 cells, where the expression of p-furin, pPC5, pPACE4, α1-PDX, and α2-MG-F all gave similar results (Figure 4a). Thus, results from cellular coexpression and in vitro kinetics analysis suggested that furin and PC5 are the most effective proVEGF-C convertases.
In order to extend these conclusions to more physiological situations, we used human prostate cancer PC3 cells as a source of endogenous VEGF-C. Both α1-PDX and p-furin (11, 14) effectively blocked the processing of proVEGF-C (Figure 5a). This suggested that furin and/or PC5, which are the two convertases best inhibited by these agents (11, 14), could be the major processing enzymes of proVEGF-C in PC3 cells. To delineate between their contributions, RT-PCR revealed the presence of furin, but little if any PC5 mRNA in PC3 cells. The observed (approximately 50%) processing of proVEGF-C in PC3 cells (Figure 5a) may be due to either a limiting concentration of furin and/or to the very low PC5 expression. Indeed, overexpression of furin or PC5 in PC3 cells led to complete proVEGF-C processing. In agreement, a recent study of human prostate cancer cell lines LNCaP and PC3 revealed that these cells synthesize and secrete prostate-derived factor (PDF), which is processed by PCs (19). The efficiency of PDF processing was much lower in PC3 cells than in LNCaP cells. This was correlated with low in vitro convertase activity of PC3 versus LNCaP cell lysates (19). Exposure of these cells to a peptide PC inhibitor blocked proPDF processing and induced a loss of epithelial cell differentiation. These data directly implicated PCs in the control of the prostate cancer cell phenotype (19). Analysis of various tissues showed the coexpression of VEGF-C and furin mRNA’s (Figure 5b), reinforcing the functional link between the two. These data are further corroborated by the striking temporal correlation between the expression of furin and VEGF-C in embryos at day 7 and VEGFR-3 at embryonic day 8.5 (15, 20, 21), suggesting a role for convertases in regulating VEGF-C functions during embryonic development.
VEGF-C was originally described as a specific growth factor for lymphatic vessels (2, 3, 22), but later was also found to induce angiogenesis of blood vessels (3, 23–28), raising questions about the mechanism(s) that determine the distinct effects of VEGF-C in vivo. In addition, VEGF-C overexpression was linked to various cancers and metastasizing tumors in which VEGF-C was shown to mediate vascular and lymphatic vessel formation (3, 23, 29). To directly investigate the biological role of proVEGF-C processing in tumors, we took advantage of CHO cells, which do not produce endogenous VEGF-C. Expression of VEGF-C in these cells failed to stimulate their growth in vitro and in vivo (Figure 6). In contrast, VEGF-C expression stimulated both tumor lymphangiogenesis and angiogenesis (Figure 7). Previous reports linked the inability of VEGF-C to stimulate cell proliferation to its weak mitogenic action compared with the other VEGFs. Indeed, VEGF-C is 50- to 100-fold less potent than VEGF in inducing proliferation of endothelial cells (1, 4, 30, 31). In contrast, VEGF-C is a potent angiogenic factor, and acts synergistically with VEGF in angiogenesis in vitro (32). Other angiogenic factors were also reported to synergize with VEGF-C, including basic FGF, angiopoietin-1, angiopoietin-2, TGF-α, and PDGF (32, 33). These observations provide a possible rationale to our observation of enhanced angiogenesis and lymphangiogenesis of CHO cells expressing VEGF-C, probably through its cooperation with VEGF or other factors arising from the tumor microenvironment.
In addition to the cooperative action of VEGF and VEGF-C, their common activation of VEGFR-2 was reported to constitute an important way to mediate in vivo angiogenesis. Thus, VEGF-C and VEGF can both bind VEGFR-2, and probably displace each other (4). Previously, Joukov et al. demonstrated that the unprocessed proVEGF-C can effectively displace mature [125I]VEGF-C from VEGFR-2 and VEGFR-3 (4). On a functional level, analysis of VEGFR-2 and VEGFR-3 autophosphorylation after binding to proVEGF-C and VEGF-C revealed that proVEGF-C had a weaker effect on autophosphorylation of VEGFR-3, and no effect on VEGFR-2 autophosphorylation and vascular permeability (4). Thus proVEGF-C seems to behave as an antagonist to both receptor subtypes and to prevent their activation. In our model, the secretion of proVEGF-C in the microenvironment of CHO-derived tumors may compete with active VEGF-C and VEGF produced by cells surrounding the tumors, such as endothelial and hematopoietic cells (25, 34, 35), and hence antagonize the action of the latter factors on tumor growth and formation of lymphatic and vascular vessels.
VEGF-C has been reported to be involved in the immune response through its ability to induce chemotaxis of macrophages in vitro and in vivo (25, 35). We also observed an increase in macrophage recruitment in CHO VEGF-C–derived tumors compared with control and CHO proVEGF-C–derived tumors (data not shown). Our data showing no significant effect of VEGF-C on tumor growth (Figure 6) may be explained in part by its reported ability to reduce tumor growth through recruitment of macrophages (25). However, VEGF-C can promote lymphangiogenesis and angiogenesis in melanomas, and only lymphangiogenesis in breast cancer (14, 15). This divergence in VEGF-C–mediated tumor angiogenesis or lymphangiogenesis is likely related to the level of the VEGF-C forms accumulated in the developed tumors (14, 15). Thus, whereas in melanomas, fully processed 21-kDa VEGF-C is predominant, breast cancer tumors mostly express the 31-kDa form, which selectively induces lymphangiogenesis through activation of VEGFR-3 (14, 15). Together, these studies emphasize that the biological effect of VEGF-C in tumors is functionally dependent on its in vivo proteolytic processing.
Our findings demonstrate that the blockade of VEGF-C processing by mutagenesis generates predominately the 59-kDa proVEGF-C form. This form was reported to behave as an antagonist of VEGFR-2 and VEGFR-3 (4), and to possibly prevent their optimal oligomerization, which is needed for their signalling functions. Accordingly, the observed reduction in tumor growth, angiogenesis, and lymphangiogenesis of the 59-kDa proVEGF-C–producing tumors (Figure 6 and Figure 7) may be related to the dominant-negative characteristics of this growth factor. Recently, an opposing role of unprocessed and processed PC substrates was reported for other proteins and was demonstrated to be related to their ability to mediate distinct signalling pathways. This includes pro–nerve growth factor (proNGF) (36), the receptor Notch (37, 38), and proBMP-4 (39). In our model, a similar mechanism may explain the divergent action of VEGF-C and proVEGF-C on tumorigenesis. However, further investigations are required to assess the ability of VEGF-C and proVEGF-C to activate different receptors to mediate distinct signalling pathways responsible for their observed divergent functions.
In conclusion, this study demonstrated that furin, PC5, and PC7 are the cognate PCs implicated in the dibasic cleavage and activation of proVEGF-C at the dibasic ArgArg227, and underlined the importance of this processing in VEGF-C–mediated functions. In this context, inhibition of the PC-mediated processing of proVEGF-C may provide a novel potential strategy in cancer therapy (6), in conjunction with other approaches such as using a soluble VEGFR-3 extracellular domain to inhibit tumor angiogenesis and lymphangiogenesis (40–42). VEGF-C expression has been detected in 50% of all the human cancers analyzed, and its expression has been correlated with lymph node dissemination in human cancers (43). The development of specific inhibitors of PCs may lead to treatments of malignancies induced by this growth factor.
Acknowledgments
This work was supported by the Protein Engineering Network of Centres of Excellence, by a group grant from the Canadian Institutes of Health Research (MGP-44363), and by a grant from the Ottawa Health Research Institute start-up fund (to A.M. Khatib). The authors are grateful to JoAnn McDonald for the secretarial work.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: proprotein convertase (PC); furin-motif variant of α1-antitrypsin (α1-antitrypsin Portland) (α1-PDX); human embryonic kidney (HEK); reverse-phase (RP); matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-ToF-MS); furin-motif variant of α2-macroglobulin (α2-MG-F).
References
- 1.Joukov V, et al. A novel vascular endothelial growth factor, VEGF-C, is a ligand for the Flt4 (VEGFR-3) and KDR (VEGFR-2) receptor tyrosine kinases. [erratum 1996, 15:1751] EMBO J. 1996;15:290–298. [PMC free article] [PubMed] [Google Scholar]
- 2.Huang HY, Ho CC, Huang PH, Hsu SM. Co-expression of VEGF-C and its receptors, VEGFR-2 and VEGFR-3, in endothelial cells of lymphangioma. Implication in autocrine or paracrine regulation of lymphangioma. Lab. Invest. 2001;81:1729–1734. doi: 10.1038/labinvest.3780386. [DOI] [PubMed] [Google Scholar]
- 3.Karkkainen MJ, Makinen T, Alitalo K. Lymphatic endothelium: a new frontier of metastasis research. Nat. Cell Biol. 2002;4:E2–E5. doi: 10.1038/ncb0102-e2. [DOI] [PubMed] [Google Scholar]
- 4.Joukov V, et al. Proteolytic processing regulates receptor specificity and activity of VEGF-C. EMBO J. 1997;16:3898–3911. doi: 10.1093/emboj/16.13.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seidah NG, Chretien M. Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res. 1999;848:45–62. doi: 10.1016/s0006-8993(99)01909-5. [DOI] [PubMed] [Google Scholar]
- 6.Khatib AM, Siegfried G, Chretien M, Metrakos P, Seidah NG. Proprotein convertases in tumor progression and malignancy: novel targets in cancer therapy. Am. J. Pathol. 2002;160:1921–1935. doi: 10.1016/S0002-9440(10)61140-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benjannet S, et al. Post-translational processing of beta-secretase (beta-amyloid-converting enzyme) and its ectodomain shedding. The pro- and transmembrane/cytosolic domains affect its cellular activity and amyloid-beta production. J. Biol. Chem. 2001;276:10879–10887. doi: 10.1074/jbc.M009899200. [DOI] [PubMed] [Google Scholar]
- 8.Khatib AM, et al. Inhibition of proprotein convertases is associated with loss of growth and tumorigenicity of HT-29 human colon carcinoma cells: importance of insulin-like growth factor-1 (IGF-1) receptor processing in IGF-1-mediated functions. J. Biol. Chem. 2001;276:30686–30693. doi: 10.1074/jbc.M101725200. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi S, et al. A mutation of furin causes the lack of precursor-processing activity in human colon carcinoma LoVo cells. Biochem. Biophys. Res. Commun. 1993;195:1019–1026. doi: 10.1006/bbrc.1993.2146. [DOI] [PubMed] [Google Scholar]
- 10.Basak A, Zhong M, Munzer JS, Chretien M, Seidah NG. Implication of the proprotein convertases furin, PC5 and PC7 in the cleavage of surface glycoproteins of Hong Kong, Ebola and respiratory syncytial viruses: a comparative analysis with fluorogenic peptides. Biochem. J. 2001;353:537–545. doi: 10.1042/0264-6021:3530537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong M, et al. The prosegments of furin and PC7 as potent inhibitors of proprotein convertases. In vitro and ex vivo assessment of their efficacy and selectivity. J. Biol. Chem. 1999;274:33913–33920. doi: 10.1074/jbc.274.48.33913. [DOI] [PubMed] [Google Scholar]
- 12.Basak A, Chretien M, Seidah NG. A rapid fluorometric assay for the proteolytic activity of SKI-1/S1P based on the surface glycoprotein of the hemorrhagic fever Lassa virus. FEBS Lett. 2002;514:333–339. doi: 10.1016/s0014-5793(02)02394-3. [DOI] [PubMed] [Google Scholar]
- 13.Kyriazis AP, et al. Human pancreatic adenocarcinoma line Capan-1 in tissue culture and the nude mouse: morphologic, biologic, and biochemical characteristics. Am. J. Pathol. 1982;106:250–260. [PMC free article] [PubMed] [Google Scholar]
- 14.Jean F, et al. alpha1-Antitrypsin Portland, a bioengineered serpin highly selective for furin: application as an antipathogenic agent. Proc. Natl. Acad. Sci. U. S. A. 1998;95:7293–7298. doi: 10.1073/pnas.95.13.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kukk E, et al. VEGF-C receptor binding and pattern of expression with VEGFR-3 suggests a role in lymphatic vascular development. Development. 1996;122:3829–3837. doi: 10.1242/dev.122.12.3829. [DOI] [PubMed] [Google Scholar]
- 16.Nakayama K. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J. 1997;327:625–635. doi: 10.1042/bj3270625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seidah NG, Chretien M. Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res. 1999;848:45–62. doi: 10.1016/s0006-8993(99)01909-5. [DOI] [PubMed] [Google Scholar]
- 18.Lissitzky JC, et al. Endoproteolytic processing of integrin pro-alpha subunits involves the redundant function of furin and proprotein convertase (PC) 5A, but not paired basic amino acid converting enzyme (PACE) 4, PC5B or PC7. Biochem. J. 2000;346:133–138. [PMC free article] [PubMed] [Google Scholar]
- 19.Uchida K, Chaudhary LR, Sugimura Y, Adkisson HD, Hruska KA. Proprotein convertases regulate activity of prostate epithelial cell differentiation markers and are modulated in human prostate cancer cells. J. Cell. Biochem. 2003;88:394–399. doi: 10.1002/jcb.10327. [DOI] [PubMed] [Google Scholar]
- 20.Zheng M, Streck RD, Scott RE, Seidah NG, Pintar JE. The developmental expression in rat of proteases furin, PC1, PC2, and carboxypeptidase E: implications for early maturation of proteolytic processing capacity. J. Neurosci. 1994;14:4656–4673. doi: 10.1523/JNEUROSCI.14-08-04656.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lymboussaki A, Olofsson B, Eriksson U, Alitalo K. Vascular endothelial growth factor (VEGF) and VEGF-C show overlapping binding sites in embryonic endothelia and distinct sites in differentiated adult endothelia. Circ. Res. 1999;85:992–999. doi: 10.1161/01.res.85.11.992. [DOI] [PubMed] [Google Scholar]
- 22.Karkkainen MJ, Petrova TV. Vascular endothelial growth factor receptors in the regulation of angiogenesis and lymphangiogenesis. Oncogene. 2000;19:5598–5605. doi: 10.1038/sj.onc.1203855. [DOI] [PubMed] [Google Scholar]
- 23.Skobe M, et al. Induction of tumor lymphangiogenesis by VEGF-C promotes breast cancer metastasis. Nat. Med. 2001;7:192–198. doi: 10.1038/84643. [DOI] [PubMed] [Google Scholar]
- 24.Koolwijk P, et al. Involvement of VEGFR-2 (kdr/flk-1) but not VEGFR-1 (flt-1) in VEGF-A and VEGF-C-induced tube formation by human microvascular endothelial cells in fibrin matrices in vitro. Angiogenesis. 2001;4:53–60. doi: 10.1023/a:1016637700638. [DOI] [PubMed] [Google Scholar]
- 25.Skobe M, et al. Concurrent induction of lymphangiogenesis, angiogenesis, and macrophage recruitment by vascular endothelial growth factor-C in melanoma. Am. J. Pathol. 2001;159:893–903. doi: 10.1016/S0002-9440(10)61765-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh SJ, et al. VEGF and VEGF-C: specific induction of angiogenesis and lymphangiogenesis in the differentiated avian chorioallantoic membrane. Dev. Biol. 1997;188:96–109. doi: 10.1006/dbio.1997.8639. [DOI] [PubMed] [Google Scholar]
- 27.Jeltsch M, et al. Hyperplasia of lymphatic vessels in VEGF-C transgenic mice. Science. 1997;276:1423–1425. doi: 10.1126/science.276.5317.1423. [DOI] [PubMed] [Google Scholar]
- 28.Kaipainen A, et al. Expression of the fms-like tyrosine kinase 4 gene becomes restricted to lymphatic endothelium during development. Proc. Natl. Acad. Sci. U. S. A. 1995;92:3566–3570. doi: 10.1073/pnas.92.8.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karpanen T, et al. Vascular endothelial growth factor C promotes tumor lymphangiogenesis and intralymphatic tumor growth. Cancer Res. 2001;61:1786–1790. [PubMed] [Google Scholar]
- 30.Lee J, et al. Vascular endothelial growth factor-related protein: a ligand and specific activator of the tyrosine kinase receptor Flt4. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1988–1992. doi: 10.1073/pnas.93.5.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Witzenbichler B, et al. Vascular endothelial growth factor-C (VEGF-C/VEGF-2) promotes angiogenesis in the setting of tissue ischemia. Am. J. Pathol. 1998;153:381–394. doi: 10.1016/S0002-9440(10)65582-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pepper MS, Mandriota SJ, Jeltsch M, Kumar V, Alitalo K. Vascular endothelial growth factor (VEGF)-C synergizes with basic fibroblast growth factor and VEGF in the induction of angiogenesis in vitro and alters endothelial cell extracellular proteolytic activity. J. Cell. Physiol. 1998;177:439–452. doi: 10.1002/(SICI)1097-4652(199812)177:3<439::AID-JCP7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 33.Eggert A, et al. High-level expression of angiogenic factors is associated with advanced tumor stage in human neuroblastomas. Clin. Cancer Res. 2000;6:1900–1908. [PubMed] [Google Scholar]
- 34.Wartiovaara U, et al. Peripheral blood platelets express VEGF-C and VEGF which are released during platelet activation. Thromb. Haemost. 1998;80:171–175. [PubMed] [Google Scholar]
- 35.Fielder W, et al. Expression of FLT4 and its ligand VEGF-C in acute myeloid leukemia. Leukemia. 1997;11:1234–1237. doi: 10.1038/sj.leu.2400722. [DOI] [PubMed] [Google Scholar]
- 36.Lee R, Kermani P, Teng KK, Hempstead BL. Regulation of cell survival by secreted proneurotrophins. Science. 2001;294:1945–1948. doi: 10.1126/science.1065057. [DOI] [PubMed] [Google Scholar]
- 37.Mumm JS, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 38.Bush G, et al. Ligand-induced signaling in the absence of furin processing of Notch1. Dev. Biol. 2001;229:494–502. doi: 10.1006/dbio.2000.9992. [DOI] [PubMed] [Google Scholar]
- 39.Cui Y, Jean F, Thomas G, Christian JL. BMP-4 is proteolytically activated by furin and/or PC6 during vertebrate embryonic development. EMBO J. 1998;17:4735–4743. doi: 10.1093/emboj/17.16.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He Y, et al. Suppression of tumor lymphangiogenesis and lymph node metastasis by blocking vascular endothelial growth factor receptor 3 signalling. J. Natl. Cancer Inst. 2002;94:819–825. doi: 10.1093/jnci/94.11.819. [DOI] [PubMed] [Google Scholar]
- 41.Kubo H, et al. Blockade of vascular endothelial growth factor receptor-3 signaling inhibits fibroblast growth factor-2-induced lymphangiogenesis in mouse cornea. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8868–8873. doi: 10.1073/pnas.062040199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makinen T, et al. Inhibition of lymphangiogenesis with resulting lymphedema in transgenic mice expressing soluble VEGF receptor-3. Nat. Med. 2001;7:199–205. doi: 10.1038/84651. [DOI] [PubMed] [Google Scholar]
- 43.Salven P, et al. Vascular endothelial growth factors VEGF-B and VEGF-C are expressed in human tumors. Am. J. Pathol. 1998;153:103–108. doi: 10.1016/S0002-9440(10)65550-2. [DOI] [PMC free article] [PubMed] [Google Scholar]