Abstract
We previously reported that laboratory reference strains of Chlamydia trachomatis differing in infection organotropism correlated with inactivating mutations in the pathogen’s tryptophan synthase (trpBA) genes. Here, we have applied functional genomics to extend this work and find that the paradigm established for reference serovars also applies to clinical isolates — specifically, all ocular trachoma isolates tested have inactivating mutations in the synthase, whereas all genital isolates encode a functional enzyme. Moreover, functional enzyme activity was directly correlated to IFN-γ resistance through an indole rescue mechanism. Hence, a strong selective pressure exists for genital strains to maintain a functional synthase capable of using indole for tryptophan biosynthesis. The fact that ocular serovars (serovar B) isolated from the genital tract were found to possess a functional synthase provided further persuasive evidence of this association. These results argue that there is an important host-parasite relationship between chlamydial genital strains and the human host that determines organotropism of infection and the pathophysiology of disease. We speculate that this relationship involves the production of indole by components of the vaginal microbial flora, allowing chlamydiae to escape IFN-γ–mediated eradication and thus establish persistent infection.
Introduction
Chlamydiae are obligate intracellular gram-negative eubacteria that exhibit a highly specialized biphasic developmental cycle (1). Currently, the genus is divided into four species: Chlamydia trachomatis, C. psittaci, C. pneumoniae, and C. pecorum (2). C. trachomatis and C. pneumoniae are important human pathogens, whereas C. psittaci and C. pecorum are primarily pathogens of animals and birds, rarely infecting humans. C. trachomatis is classified into 15 distinct serovars based on antigenic variation of the ompA genes that encode the organism’s major outer membrane protein (MOMP). The 15 different serovars display well-documented and unique tissue tropisms (3). Serovars A, B, Ba, and C are the causative agents of trachoma, the most common infectious cause of blindness worldwide (2, 4, 5). With the exception of serovar B variants, which have been associated with a very low incidence of urogenital disease (6–8), the trachoma serovars are rarely isolated from the genital tract (2). In contrast, serovars D–K are a major cause of urogenital tract infection worldwide but are not associated with blinding trachoma (2). However, serovars D–K can cause ocular infection when newborn infants acquire the organism during passage through the infected birth canal or when adults secondarily inoculate the eye with infected genital secretions. Infections with either ocular (A–C) or genital (D–K) serovars are noninvasive — that is, they are restricted to epithelial surfaces, whereas the LGV strains (L1–L3) are invasive and proliferate in monocytes within lymphatic tissues. In humans, progression from uncomplicated mucosal infections to serious sequelae such as blindness or tubal factor infertility has been linked epidemiologically to either reinfection or persistent infection (2, 9–12). Re-exposure or persistent infection is thought to drive an immunopathological inflammatory response resulting in tissue fibrosis and scarring (13, 14).
Despite this diversity in tissue tropism and disease manifestations, complete genomic sequencing of several Chlamydiaceae has shown that the gene order and content among the different species is remarkably conserved, with the exception of a region termed the plasticity zone (PZ) (15). This observation led to the suggestion that host, organ, and cellular tropism may be attributed to the few genes localized in the PZ. Consistent with this hypothesis, our recent observation using C. trachomatis laboratory reference strains showed that the tryptophan synthase operon (trpRBA) that resides in the PZ are closely linked with genital and ocular tropism (16).
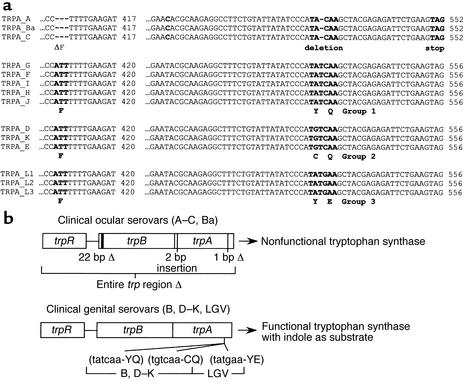
Tryptophan synthase is a tetramer consisting of two α subunits (TrpA) and two β subunits (TrpB) (17–19). The subunits form a bifunctional enzyme that catalyzes the final two reactions in the biosynthesis of tryptophan — namely, the cleavage of indole glycerol 3-phosphate (IGP) to indole by TrpA and the TrpB-mediated conversion of indole to L-tryptophan. Comparative DNA sequencing of the PZ of all 15 human C. trachomatis reference strains showed that 14 of 15 serovars encoded homologues of trpB and trpA (16, 20). The chlamydial TrpB proteins retained conserved residues identified as essential for enzyme activity in other microorganisms (17, 18). In contrast, the chlamydial TrpA protein was shown to have several changes in its primary structure that differed considerably from its trpA orthologue in E. coli. Specifically, the trpA of ocular but not genital serovars had a common consensus triplet nucleotide deletion (nucleotides 408–410) that resulted in the loss of Phe-136 from the polypeptide. Furthermore, a polymorphic mutational “hot spot” was identified in the trpA of both ocular and genital serovars. In ocular serovars, this region contained a single-nucleotide (531) deletion that resulted in a frame shift and expression of a truncated nonfunctional TrpA polypeptide. Interestingly, the trpA of all genital serovars contained two point mutations at this same site giving rise to codon changes resulting in amino acid substitutions that differed among the serovars: YE in LGV serovars; CQ in D, E, and K serovars; and YQ in G, F, H, I, and J serovars (Figure 1a). These amino acids in the Salmonella TrpA are found in loop 6, a region identified in the tryptophan synthase crystal structure as being important for subunit-subunit interactions between TrpB and TrpA, metabolite channeling, and substrate binding (17, 18). Consistent with this conclusion was our finding by genetic complementation studies that the TrpBA of genital serovars was not capable of using IGP but could use exogenous indole for the synthesis of tryptophan (16). The significance of these findings in terms of pathogenesis was that genital but not ocular serovars were readily rescued under conditions of tryptophan starvation by the addition of exogenous indole (16).
Figure 1.
(a) Alignment of partial sequences from the trpA gene from the14 human C. trachomatis reference serovars. A ClustalW alignment of the nucleotide regions containing sequence polymorphisms is illustrated. As compared with genital serovars, ocular serovars have a three-base (nucleotides 408–410) deletion that results in the loss of a phenylalanine. The various serovars have been grouped, in accordance with their nucleotide mutational “hot-spot” sequence. The ocular serovars have a single-base deletion (nucleotide 528) resulting in a nonfunctional truncated TrpA protein. Genital serovar specific missense mutations (nucleotides 530 and 532) that result in amino acid changes in loop 6 of TrpA are shown below the nucleotide sequence. See Fehlner-Gardiner et al. (16) for details. (b) Schematic summary of the tryptophan synthase inactivating mutations identified in clinical ocular serovars and the missense mutations identified in the clinical genital serovars.
Chlamydial infection stimulates an intense cellular immune response (CMI) that is important in protective immunity against mucosal infection and is mediated predominantly by CD4+ type 1 lymphocytes and IFN-γ (21–24). Although the effector mechanism(s) of IFN-γ in vivo are not completely understood, its antichlamydial inhibitory activity for cultured cells in vitro has been comprehensively studied. It is known that IFN-γ eradicates chlamydiae mainly through the induction of the tryptophan-degrading enzyme indoleamine 2,3-dioxygenase (IDO) (21, 25, 26). IDO catabolizes intracellular pools of L-tryptophan, thereby starving chlamydiae of this essential amino acid. Chlamydiae deprived of tryptophan fail to complete secondary differentiation into infectious elementary bodies (EBs), resulting in the loss of infectivity and cell-to-cell transmission. These findings argue that the ability to synthesize tryptophan in a IFN-γ–rich environment would be an important virulence factor for C. trachomatis because it would provide a mechanism for the pathogen to evade host defenses.
Because of the potential implications of our earlier work in understanding chlamydial organotropism and pathogenesis, we have extended our studies to include analyses of more recently isolated ocular and genital isolates from diverse geographical locations. We report here that the paradigm of a functional tryptophan synthase in genital serovars versus a nonfunctional synthase in ocular serovars that we established for laboratory reference strains also holds true for this larger, more heterogeneous population of clinical isolates. The importance of these findings with regard to chlamydial organotropism, evasion of host immunity, and the potential usefulness of the trp region as a novel molecular epidemiological marker are discussed.
Methods
C. trachomatis and HeLa cell propagation.
C. trachomatis serovars were propagated in HeLa 229 cells and EBs were purified as described previously (27). HeLa cells were maintained at 37°C with 5% CO2 in DMEM (Gibco Invitrogen, Grand Island, New York, USA) supplemented with 10% fetal bovine serum (Hyclone, Logan, Utah, USA), 2 mM L-glutamine (Gibco Invitrogen), 0.01 M HEPES (Gibco Invitrogen), 1 mM sodium pyruvate (Gibco Invitrogen), 0.055 mM β-mercaptoethanol (Gibco Invitrogen), and 10 μg/ml gentamicin. This medium is referred to as complete DMEM-10.
Clinical samples.
Ocular chlamydial samples or isolates were obtained from conjunctival swabs of male and female village residents in trachoma-endemic areas of Kongwa, Rombo Tanzania, and Gambia as described previously (28, 29). One trachoma sample was obtained from China. For genital chlamydial isolates/samples, cervical, urethral, and anorectal swabs and urine specimens were collected as previously described (30–32). Male and female patients were enrolled at geographically diverse clinical sites that included both low- and high-prevalence populations. The clinical sites were Nairobi, Kenya; Winnipeg, Manitoba, Canada; Ottawa, Ontario, Canada; Calgary, Alberta, Canada; Indianapolis, Indiana, USA; San Francisco, California, USA; Baltimore, Maryland, USA; New Orleans, Louisiana, USA; Birmingham, Alabama, USA; and Seattle, Washington, USA. The sites included sexually transmitted disease (STD) clinics, obstetrics and gynecology clinics, family planning clinics, adolescent clinics, and emergency rooms. The majority of the patients were seen at STD or family planning clinics.
Indole rescue of clinical C. trachomatis isolates.
Tryptophan-starved HeLa 229 monolayers in 96-well plates were infected with reference serovars D and B and clinical isolates at an moi of 0.1. Infected cells were fed with DMEM without L-tryptophan (University of California at San Francisco, Cell Culture Facility, San Francisco, California, USA) supplemented with 10% dialyzed fetal bovine serum (Hyclone), 2 mM L-glutamine (Gibco Invitrogen), 0.01 M HEPES (Gibco Invitrogen), 1 mM sodium pyruvate (Gibco Invitrogen), 0.055 mM β-mercaptoethanol (Gibco Invitrogen), and 10 μg/ml gentamicin. This medium is referred to as DMEM-10(-Trp). The medium was replaced at 20–24 hours after infection with complete DMEM-10, DMEM-10(-Trp), or DMEM-10(-Trp) plus 10 μM indole. Monolayers were fixed in methanol at 48 hours after infection and stained for inclusions using antichlamydial LPS and FITC goat anti-mouse Ig antiserum. The monolayers were examined for inclusions by fluorescence microscopy.
C. trachomatis growth assays.
Monolayers of HeLa cells in 6-well plates were inoculated with C. trachomatis EBs at an moi of 3–5. The infected wells were incubated with complete DMEM-10 as a control or with complete DMEM-10 further supplemented with 5 ng/ml recombinant human IFN-γ with or without the addition of 50 μM indole or 1 mg/ml tryptophan. Following incubation for 48–72 hours at 37°C, the culture supernatant was collected and the HeLa cells were lysed with cold distilled H2O. Aliquots of the combined HeLa cell lysates and culture medium were used to infect fresh HeLa cell monolayers for enumeration of recoverable inclusion forming units (IFUs) as previously described (33). Briefly, monolayers were fixed in methanol 48 hours after infection. Chlamydiae were detected by reacting fixed cells with an anti-LPS mAb followed by staining with a FITC goat anti-mouse Ig antiserum.
Sequencing of MOMP and tryptophan synthase from clinical samples.
A 100-μl aliquot of each clinical sample or culture isolate was added to lysis buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2, 0.022% gelatin, 0.5% Nonidet P-40, 0.5% Tween-20) and digested with proteinase K at a final concentration of 100 μg/ml at 55°C for 2 hours. The samples were then boiled for 10 minutes to inactivate the proteinase K. The primers used for PCR and sequencing are listed in Table 1. To determine the serovar of each clinical isolate, nested PCR was performed using primers designed to amplify the ompA gene. Five microliters of DNA (approximately 50 ng) from each sample were amplified in a 100-μl reaction mixture containing 0.5 μM primers, 0.2 mM dNTPs, 1.5 mM MgCl2, 1 × Taq reaction buffer, and 1.25 U of Taq DNA polymerase (Invitrogen, Carlsbad, California, USA). The first round of PCR used primers CT1 and CT5 to generate an 1142-bp product. The cycling program was 5 minutes at 95°C followed by 35 cycles of 30 seconds at 95°C, 30 seconds at 60°C, and 1.5 minutes at 72°C, with a final extension of 10 minutes at 72°C. For the second round of the nested PCR, 1 μl of the first-round PCR product was amplified as described above using primers VD1 and VD4 to generate an 879-bp product. The cycling program for the second round of PCR was 5 minutes at 95°C followed by 40 cycles of 30 seconds at 94°C, 30 seconds at 60°C, and 1 minute at 72°C, with a final extension of 10 minutes at 72°C. The nested PCR product was purified using the QIAprep spin miniprep kit (Qiagen) and sequenced using all four primers.
Table 1.
Primer sequences for PCR, nucleotide sequencing, and RT-PCR
For determination of the trpA and trpB gene sequences, PCR was performed on samples using the primers listed in Table 1. DNA (50 ng) was used in PCR reactions (Invitrogen) with primers that amplified the entire trpRBA operon (Trp.1 and Trp.2 for trpRBA). For sequencing of trpB, nested PCR was performed using primers 5′ TrpB-L and 3′ TrpB-L for the first round of PCR, followed by 5′ TrpB-M and 3′ TrpB-M for the second round of PCR. The nested PCR product was sequenced using primer 5′ TrpB-seq. For sequencing of the mutational “hotspot” around nucleotide 531 of trpA, nested PCR was performed using primers 5′ TrpA-L and 3′ TrpA-L for the first round of PCR, followed by 5′ TrpA-M and 3′ TrpA-M for the second round of PCR. The nested PCR product was purified as described above and sequenced using primer 5′ TrpA-seq. A second nested PCR was performed to sequence the region around nucleotide 118 of trpA using primers 5′ TrpA-L#2 and 3′TrpA-L#2 for the first round of PCR, followed by 5′ TrpA-M#2 and 3′ TrpA-M#2 for the second round of PCR. The cycling program was 5 minutes at 95°C, followed by 35 cycles of 30 seconds at 94°C, 30 seconds at 60°C, and 1 minute at 72°C, with a final extension of 10 minutes at 72°C. The nested PCR product was purified and sequenced using primer 5′ TrpA-#2-seq. All PCR products were sequenced by the DNA Core Facility (Health Canada, Winnipeg, Manitoba, Canada), using an ABI PRISM 377 DNA Sequencer.
RT-PCR analysis of C. trachomatis transcript expression.
HeLa cell monolayers in T175 flasks were infected with C. trachomatis EBs at an moi of 3–5 IFUs/cell. The infected flasks were incubated with complete DMEM-10 as a control or with complete DMEM-10 further supplemented with 5 ng/ml recombinant human IFN-γ with or without the addition of 50 μM indole or 1 mg/ml tryptophan. The cells were incubated for 24–36 hours at 37°C, and total RNA was prepared from the cells using Trizol reagent according to the manufacturer’s instructions (Invitrogen). One microgram of total RNA was treated with DNase I (Invitrogen), reverse transcribed using random hexamer primers and Thermoscript reverse transcriptase (Invitrogen), and treated with RNase H (Invitrogen). The nucleotide sequences of the primers used are listed in Table 1. For quantitative PCR, amplifications were carried out according to the manufacturer’s instructions using the LightCycler and SYBR Green I as the fluorophore (Roche Diagnostics, Laval, Quebec, Canada). Briefly, 2 μl of cDNA was amplified in a 20-μl reaction mixture containing 0.5 μM primers, 5 mM MgCl2, and 2 μl of FastStart DNA Master SYBR Green I mix. The cycling program was 95°C for 10 minutes followed by 40 cycles of 5 seconds at 95°C, 10 seconds at 60°C, and 15 seconds at 72°C. Melting curves determined for all products indicated that primer dimers were not amplified in any of the reactions. Transcript levels were quantified using the LightCycler Data Analysis software from standard curves generated by amplification of known quantities of plasmid DNA containing the amplicon of interest. Results were normalized against the copy number of 16S rRNA transcripts in each cDNA preparation and are presented as transcript copy number per microgram of total RNA.
Western blot analyses.
HeLa cell monolayers in T175 flasks were infected with C. trachomatis EBs at an moi of 3–5 IFUs/cell. The infected flasks were incubated with complete DMEM-10 as a control or with complete DMEM-10 further supplemented with 5 ng/ml recombinant human IFN-γ with or without the addition of 50 μM indole or 1 mg/ml tryptophan. After incubation at 37°C for 30–48 hours, the cells were harvested by gentle scraping and were sonicated briefly. The sonicated cells were then centrifuged at 500 g for 10 minutes to pellet host cell debris. The supernatant was then ultracentrifuged for 1 hour at 16,000 g over a 30% MD-76 (Mallinckrodt) cushion. The pellet of semipurified EBs, reticulate bodies (RBs), and aberrant forms was collected, resuspended in 200 μl of 2× Laemmli electrophoresis sample buffer, and heated at 95°C for 10 minutes. The proteins were resolved on a 12% acrylamide gel and electrophoretically transferred to a nitrocellulose membrane. TrpB and TrpA were detected using polyclonal antibodies raised against the corresponding C. trachomatis L2 recombinant proteins (16), followed by enhanced chemiluminescence (Amersham Biosciences, Baie d’Urfe, Quebec, Canada). As a positive control, purified recombinant TrpB and TrpA were included on each gel. Duplicate blots were probed with monoclonal antibody to the chlamydial Hsp-60 to ensure equal loading of all samples.
Results
IFN-γ inhibition of C. trachomatis infectivity and rescue of infectivity by indole.
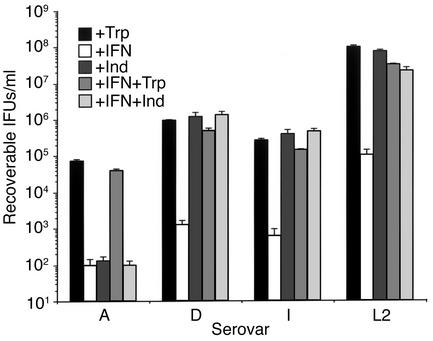
We have previously shown that the infectivity of all human C. trachomatis serovars was inhibited when grown in tryptophan-deficient medium and, importantly, that the addition of exogenous indole to the growth medium rescued the infectivity of genital (D–K, L1–L3) but not ocular (A–C, Ba) serovars (16). This indole “rescuable” phenotype was unambiguously associated with a functional tryptophan synthase present in genital but not ocular serovars. We expanded on these findings here to include experiments that more closely mimicked the immunobiological conditions that exist in vivo. This was accomplished by treating infected HeLa cells with IFN-γ to deprive chlamydiae of tryptophan and then elucidating whether infectivity could be rescued from treated cultures by the addition of exogenous indole. We chose to use serovars A, D, I, and L2 for these experiments because they represent the four trpA genotypes defined in our previous work (16): an ocular serovar with a truncated trpA (A) and genital serovars with a full-length trpA but possessing changes in trpA that result in TrpA amino acid polymorphisms CQ, YQ, and YE (D, I, and L2, respectively) (Figure 1a). The infectivity of all serovars was markedly reduced in IFN-γ–treated HeLa cell cultures (Figure 2), a finding consistent with other reports (34). Infectivity of both ocular and genital serovars was reversed by the addition of exogenous tryptophan, an expected finding because rescue did not depend on any of the serovars possessing a functional tryptophan synthase. Relevant to this study was the finding that the infectivity of the genital serovars, but not the ocular serovar, was rescued from the effect of IFN-γ by the addition of exogenous indole. Thus, these findings corroborate and expand on our earlier work but, most importantly, illustrate the potential relevance of indole rescue of infectivity to natural infection; specifically evasion of host immunity.
Figure 2.
Effect of indole or tryptophan on the growth of C. trachomatis reference serovars A, D, I, and L2 cultured in the presence of IFN-γ. HeLa cell monolayers were infected with EBs at an moi of 3–5 IFUs/cell. For serovar L2 but not A, D, or I, HeLa cell monolayers were pretreated with IFN-γ (5 ng/ml) for 24 hours before infection. Infected HeLa cells were cultured in the presence of complete DMEM-10 (+Trp), complete DMEM-10 plus 5 ng/ml IFN-γ (+IFN), complete DMEM-10 plus IFN-γ and 100 μM indole (IFN+Ind), and complete DMEM-10 plus IFN-γ and supplemented with 1 g/l tryptophan (IFN+Trp). After 48 hours for serovar L2 and 72 hours for serovars A, D and I, infected cells and culture supernatants were collected and used to infect a new HeLa cell monolayer for enumeration of recoverable IFUs. Data are presented as IFUs (log10) and represent the means ± SD of triplicate determinations.
Sequence analysis of trp genes from C. trachomatis clinical isolates.
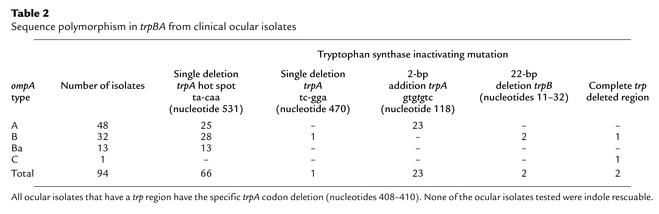
Sequence analysis of the trpRBA operon genes of the 14 C. trachomatis reference serovars showed that these serovars possessed a mutational hot spot at nucleotides position 531 of the trpA gene; the exception was serovar B, in which the entire trpRBA operon was deleted from the chromosome (Figure 1a) (16). It was imperative to extend the findings obtained with C. trachomatis reference serovars to more recently isolated naturally occurring strains because of the potential importance these mutations might have in the pathogenesis of infection and as novel epidemiological markers. We therefore sequenced the trpRBA region from a total of 94 trachoma isolates and 214 genital isolates obtained from distinct geographical areas within Africa, China, Canada, and the USA. A summary of the clinical isolates by disease, ompA, and trpA genotype together with the trpA genotype from each isolate is shown in Tables 2 and 3 and schematically in Figure 1b. The results showed that there was an absolute correlation between ocular and genital isolates and the presence of specific mutations in the trpA genes. Remarkably, every ocular isolate displayed mutations that resulted in an interrupted trpA or trpB open reading frame or a complete loss of both genes due to large deletions (Table 2 and Figure 1b). As striking, we found no such mutations in trpA for any of the genital isolates (Table 3 and Figure 1b). Six different types of inactivating mutations were identified among the ocular isolates.
Table 2.
Sequence polymorphism in trpBA from clinical ocular isolates
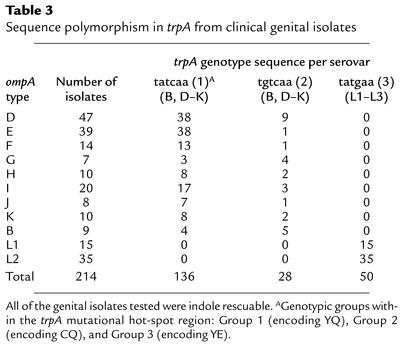
Table 3.
Sequence polymorphism in trpA from clinical genital isolates
The vast majority (65 of 66) of Gambian isolates had the same mutation (single-nucleotide deletion at position 531) previously described for trachoma reference serovars A, Ba, and C (16, 35). The bulk of Tanzanian isolates, and a single Gambian isolate, were found to have a 2-bp (TG) insertion at nucleotide 118 in trpA. Two Tanzanian isolates had a 22-bp deletion (nucleotides 11–32) in trpB, and one had a single-base deletion (nucleotides 470) in trpA. Finally, one Tanzanian isolate that ompA typed as serovar B and one isolate from China, ompA type serovar C, had the entire trp region deleted, much like the mutation in reference serovar B (16, 35), although the limits of the deleted regions were different (data not shown). Of particular interest, we observed a three-base deletion (nucleotides 408–410) in trpA for all ocular isolates that had trp genes, a finding we previously reported for reference ocular strains (16). This deletion was not present in the trpA of any genital strain and occurred irrespective of the geographical location or ompA genotype of the ocular isolate. Thus, the trpA genotypic marker might prove useful for epidemiological typing of ocular and genital strains.
Conversely, the trpA genes of all genital serovars encoded intact open reading frames giving rise to a functional tryptophan synthase, regardless of ompA genotype or the geographic location of the isolate (Table 3 and Figure1b). Furthermore, each isolate typed as one of the three distinct trpA genotypes that we had previously found with the reference serovars based on changes surrounding the mutational hot spot at nucleotides 531 (16). These changes resulted in a region of polymorphism in TrpA which we refer to as Group 1 (YQ), Group 2 (CQ), and Group 3 (YE); the Group 3 genotype was found exclusively in LGV isolates. In contrast, Group 1 and 2 polymorphisms occurred independently of ompA (D–K) genotype, and there was a predominance of Group 1 over Group 2 sequences (136 out of 170) among genital isolates.
trpRBA gene expression in clinical ocular and genital C. trachomatis isolates.
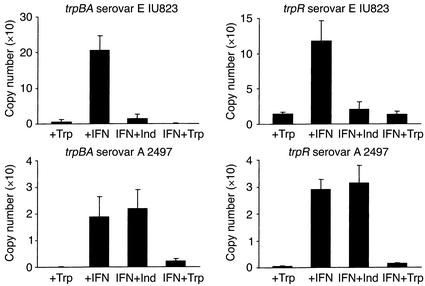
To determine if the trp genes were expressed and regulated by tryptophan availability in the clinical isolates during a HeLa cell infection, representative ocular (A-2497) and genital (E-IU823) serovars were chosen for more in-depth analysis. For real-time quantitative RT-PCR analysis, total RNA was isolated from midcycle (36-hour) infected HeLa cell cultures grown in the presence of complete DMEM-10, complete DMEM-10 plus IFN-γ, complete DMEM-10 plus IFN-γ and indole, and complete DMEM-10 plus IFN-γ and excess tryptophan. As shown in Figure 3, trpR and trpBA expression is detectable in HeLa cells infected with either the ocular or the genital isolate. In general, the level of expression is lower in cells infected with the ocular isolate, a result that could be caused by Rho-dependent termination due to the premature stop codon in trpA of this isolate. In both the ocular and the genital isolates, trpR and trpBA expression are dramatically upregulated by the presence of IFN-γ, suggesting that there is induction of expression under tryptophan-limiting conditions. Addition of excess exogenous tryptophan to IFN-γ–treated cultures repressed the expression of trpR and trpBA in both serovars. In contrast, indole reversed the IFN-γ–induced increase in trp gene expression in the genital but not the ocular isolate. This result suggests that, unlike the genital isolate, the ocular isolate is incapable of converting indole into tryptophan.
Figure 3.
RT-PCR quantitation of trpBA and trpR mRNA isolated from HeLa cell monolayers infected with C. trachomatis ocular (serovar A 2497) and genital (serovars E IU823) clinical isolates cultured in the presence of IFN-γ. Culture and chlamydiae infection conditions were as described in the legend to Figure 2. Infected HeLa cells were cultured in the presence of complete DMEM-10 (+Trp), complete DMEM-10 plus 5 ng/ml IFN-γ (+IFN), complete DMEM-10 plus IFN-γ and 100 μM indole (IFN+Ind), and complete DMEM-10 plus IFN-γ and supplemented with 1 g/l tryptophan (IFN+Trp). RNA was isolated 36 hours after infection. The RNA was reverse transcribed using random hexamer primers, and the cDNA was used for quantitative PCR amplifications with primers specific for trpBA or trpR. Quantitative PCR reactions were carried out using the LightCycler and SYBR Green I as the fluorophore. See Methods for details.
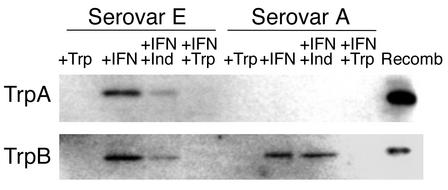
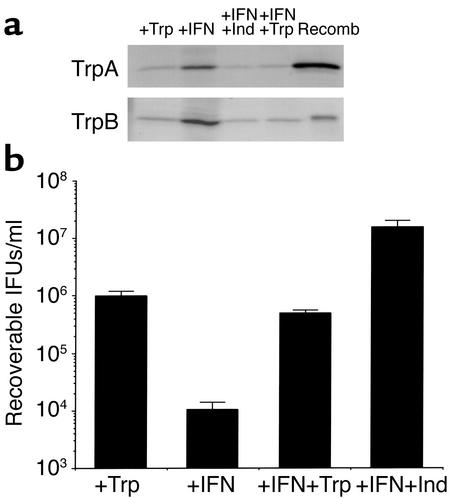
Polyclonal antibodies to TrpA and TrpB were used for Western blot analysis to assess whether the encoded messenger RNA was translated into protein products (Figure 4). For the genital serovar E strain, protein bands of the same electrophoretic mobility as purified recombinant TrpA and TrpB were detected in semipurified chlamydiae prepared from IFN-γ–treated cultures, which corresponds well with the increased level of transcript detected under these culture conditions. The amounts of immunoreactive TrpA and TrpB are substantially reduced when indole is added and are below the level of detection in control cultures or cultures treated with IFN-γ in the presence of excess tryptophan. Different results were observed for the ocular serovar A clinical isolate in that no TrpA protein band was detected under any of the conditions tested. This is not surprising since the two-base insertion mutation in trpA in this isolate occurs at bases 118–119. This frame shift mutation gives rise to a new stop codon (nucleotides 136–138) with a resulting truncated protein product of 4.7 kDa, a size not detectable under our experimental conditions. An anti-TrpB immunoreactive band, of the correct mobility, was detected in IFN-γ–treated cultures and IFN-γ–treated cultures plus indole but not in control or IFN-γ–treated cultures plus excess tryptophan. These results correlate well with the RT-PCR data and suggest that, even in the ocular serovars in which tryptophan synthase is inactive, trp gene expression is regulated by tryptophan availability.
Figure 4.
Western blot analysis of TrpA and TrpB expression in HeLa cell monolayers infected with C. trachomatis ocular clinical isolate serovar A 2497 or genital clinical isolate serovar E IU823 cultured in the presence of IFN-γ. Infected HeLa cells were cultured in the presence of complete DMEM-10 (+Trp), complete DMEM-10 plus 5 ng/ml IFN-γ (+IFN), complete DMEM-10 plus IFN-γ and 100 μM indole (IFN+Ind), and complete DMEM-10 plus IFN-γ and supplemented with 1 g/l tryptophan (IFN+Trp). Infected cells were harvested 48 hours after infection and sonicated, and chlamydial particles were semipurified over a 30% MD-76 cushion. The resulting pellet was lysed in Laemmli sample buffer, and proteins were separated by SDS-PAGE (12%) and then transferred to nitrocellulose. Proteins were detected using polyclonal antibodies raised against recombinant L2 TrpB or TrpA. The respective recombinant proteins (Recomb) were included as positive controls.
Effect of various culture conditions on the yield of infectious chlamydial EBs.
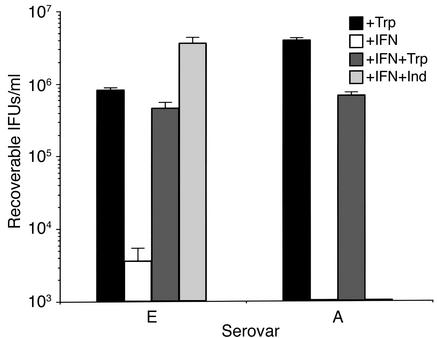
To determine if indole could rescue the clinical serovar A and E isolates from the effects of IFN-γ in vivo, we calculated the level of recoverable IFUs from infected HeLa cell cultures treated with IFN-γ. As shown in Figure 5, both serovars were sensitive to IFN-γ, and the inhibition was reversed by the addition of excess exogenous tryptophan. Addition of indole to the culture medium reversed the growth inhibitory effect of IFN-γ on the genital serovar E but not the ocular serovar A. These findings are in keeping with the expression data and suggest that the genital, but not the ocular, isolate produces a functional tryptophan synthase.
Figure 5.
Effect of indole or tryptophan on the growth of C. trachomatis ocular (serovar A 2497) and genital (serovar E IU823) clinical isolates cultured in the presence of IFN-γ. Culture and chlamydiae infection conditions were as described in the legend to Figure 2. Infected HeLa cells were cultured in the presence of complete DMEM-10 (+Trp), complete DMEM-10 plus 5 ng/ml IFN-γ (+IFN), complete DMEM-10 plus IFN-γ and 100 μM indole (IFN+Ind), and complete DMEM-10 plus IFN-γ and supplemented with 1 g/l tryptophan (IFN+Trp). After 72 hours, cultures were harvested and recoverable IFUs were enumerated. Data are presented as IFUs (log10) and represent the means ± SD of triplicate determinations.
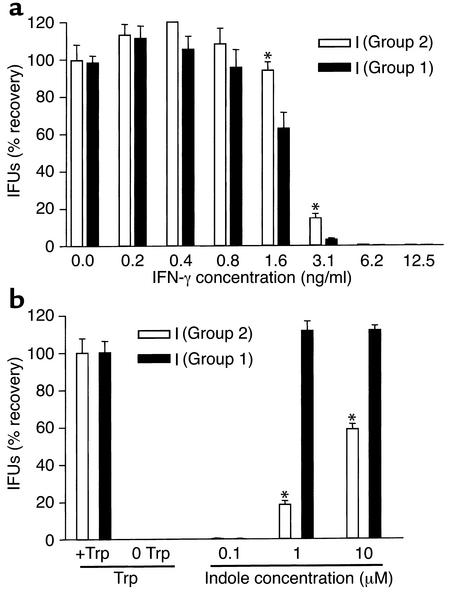
Comparison of the IFN-γ sensitivity and indole rescuability of clinical serovar I isolates with a YQ or CQ genotype.
The amino acid changes in TrpA of the Group 1, 2, and 3 genotypes occur in loop 6, a region of the protein important for substrate binding and subunit-subunit interaction (17, 18). After sequencing the trp region of all reference serovars (16), we noticed that there was an association between the genital serovar trpA genotype and the IFN-γ sensitivity as reported by Morrison (34). The Group 1 serovars (I, J, G, and F) were most sensitive to IFN-γ followed by Group 2 serovars (D, K, E), with the Group 3 (LGV) serovars being the most resistant. To determine if the Group 1 and 2 genotypes were associated with IFN-γ sensitivity and/or indole rescuability, we chose to compare these properties in a pair of clinical serovar I isolates, one Group 1 and the other Group 2. The results shown in Figure 6a indicate that the Group 1 serovar I isolate was more sensitive to IFN-γ and could be rescued with lower concentrations of indole than could its counterpart Group 2 serovar I isolate (Figure 6b).
Figure 6.
IFN-γ sensitivity and indole rescue of paired I serovariants as a function of the Group 1/2 polymorphism in trpA. (a) Chlamydial isolates were grown in the presence of a range of IFN-γ concentrations in complete DMEM-10 in the absence of cyclohexamide on HeLa monolayers at an moi of 0.1. After 48 hours, cultures were harvested and recoverable IFUs were enumerated. Graphed values represent the percent recovery from each concentration of IFN-γ based on the 0 IFN-γ well of each isolate equaling 100% recovery. (b) Chlamydial isolates were grown in complete DMEM-10 or DMEM-10(-Trp) on HeLa monolayers at an moi of 0.5. Tryptophan minus wells were rescued 24 hours after infection with three different indole concentrations. After a further 48 hours, cultures were harvested and recoverable IFUs were enumerated. Graphed values represent the percent recovery from each indole concentration based on the plus tryptophan well of each serovar equaling 100% recovery. The differences in recovery were significant (P < 0.05) at the 1 and 10 μM indole concentrations.
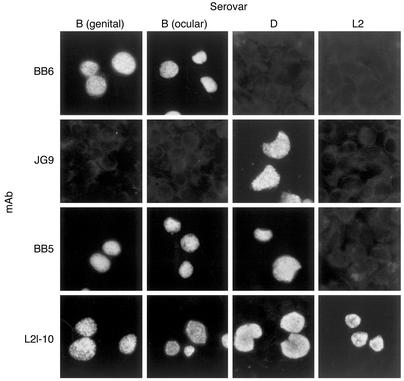
Characterization of C. trachomatis genital serovar B.
An interesting group of clinical isolates among the samples we tested were the genital samples ompA sequenced that typed as serovar B. Although serovar B is most commonly associated with ocular trachoma (2, 4, 5), it has also been isolated from patients with urogenital disease (6–8, 36). Since these genital Bs represent an unusual example of a crossover serovar, we decided to characterize them in more detail. To confirm the serotype specificity of the genital B clinical isolates, we tested a representative isolate by immunofluorescence antibody (IFA) against mAbs specific to OmpA that discriminate among C. trachomatis serovars based on differences in OmpA antigenicity. HeLa cells infected with a B genital isolate (B clinical), or the reference laboratory serovars B, D, or L2 were tested by IFA using the following mAbs specific to OmpA: mAb BB6 (serovar B specific), mAb JG9 (serovar D specific), mAb BB5 (B/D specific), and mAb L2I-10 (C. trachomatis species specific, all serovars). As shown in Figure 7, the serovar B–specific mAb BB6 was immunoreactive with both the B reference strain and the B genital isolate. In contrast, the serovar D–specific mAb JG9 was not reactive with either the B reference serovar or genital B isolate. Interestingly, the B clinical isolate was reactive with mAb BB5 (B/D specific), clearly demonstrating antigenic relatedness between OmpA of the B and D serovars. As expected, mAb L2-10, a species-specific antibody, reacted with all the strains. To confirm the serotyping, we sequenced the complete ompA gene from several of these genital B isolates from distinct geographic locations. All the ompA sequences were identical (data not shown). Furthermore, the ompA sequence of these isolates was identical to that of the genital Bb isolate initially reported by Frost et al. (36) and the genital isolate B/IU1226 subsequently reported by Stothard et al. (7). Interestingly, these isolates differ from serovar D and H genotypes by one base in conserved segment 1 but are identical to serovar B/Ba throughout the rest of the gene (7). We also sequenced the trpA and trpB genes from these clinical B isolates. In contrast to what we found with the ocular B serovars, all of the genital B isolates showed intact tryptophan synthase genes similar to those found with all the classical genital serovars (D–K). As with the other genital isolates, we found both the Group 1 and 2 genotypes in the trpA hot-spot region.
Figure 7.
Indirect fluorescent antibody staining of C. trachomatis–infected HeLa cells with mAbs specific to ompA. Note that the B genital isolate reacts with the same specificity as the ocular B laboratory reference strain. Strains tested are shown across the top of the figure; mAbs used to stain infected cells are shown on the left hand side of the figure. HeLa cells were infected with an moi of 0.2. Monolayers were fixed in methanol at 36–48 hours after infection. Chlamydiae were detected by reacting fixed cells with individual mAbs followed by staining with an FITC goat anti-mouse Ig antiserum. The specificity of the mAbs are as follows: BB6 (serovar B), JG9 (serovar D), BB5 (serovars B and D), and L2I-10 (all C. trachomatis serovars, A-L3).
To determine if these isolates expressed TrpA and TrpB and whether their expression was regulated, we carried out Western blot analysis on semipurified chlamydiae particles prepared from IFN-γ–treated and untreated infected HeLa cells. The results shown in Figure 8a indicate that immunoreactive material with the same electrophoretic mobility as full-length recombinant TrpA and Trp B was detected. The expression of both TrpA and TrpB was upregulated by IFN-γ, and the induction was repressed by the addition of excess exogenous tryptophan or indole. Finally, just as we had seen with the genital serovar E, genital serovar B growth, as assessed by quantitation of recoverable IFUs, was inhibited by IFN-γ, and the inhibition was reversed by the addition of exogenous tryptophan or indole (Figure 8b).
Figure 8.
(a) Western blot analysis of TrpA and TrpB expression in HeLa cell monolayers infected with a C. trachomatis clinical B isolate. Cell culture conditions, harvesting, chlamydial protein sample preparation, and Western blot procedure were as described in the legend to Figure 4. (b) Effect of indole or tryptophan on the growth of C. trachomatis genital serovar B clinical isolate cultured in the presence of IFN-γ. Cell culture conditions, harvesting at 72 hours after infection, and enumeration of recoverable infectious chlamydial EBs were as described in the legend to Figure 2. Data are presented as IFUs (log10) and represent the means ± SD of triplicate determinations.
Discussion
Interestingly, only the genes required for the last stages of the tryptophan biosynthetic pathway (trpRBA) are contained in the C. trachomatis PZ (15, 16, 20), explaining chlamydial tryptophan auxotrophy but leaving no obvious connection for retaining the ability to both regulate and make tryptophan synthase. The possible role of these genes as virulence factors becomes clearer when they are considered in the context of the key role of IFN-γ in host defenses against chlamydial infection. In vivo, IFN-γ is a key cytokine in the development of antichlamydial protective immunity (12, 14, 23, 37). Although its effector function has yet to be precisely determined in vivo, the antichlamydial activity of IFN-γ in chlamydia-infected epithelial cells is well understood. Binding of IFN-γ to its receptor activates the L-tryptophan decyclizing enzyme IDO, which degrades tryptophan to N-formyl kynurenine (38). Deprivation of L-tryptophan inhibits chlamydial replication and prevents transmission of infection by blocking the differentiation of metabolically active RBs to infectious EBs (21, 39). Thus, the ability to synthesize tryptophan within an IFN-γ–rich environment would be an important virulence determinant and a parasitic strategy for evading the host defense.
We recently described the genomic sequences of the tryptophan operon region (trpRBA) from all 14 human C. trachomatis serovars (16) because of its possible importance in the pathogenesis of chlamydial infection. We found a striking correlation among ocular serovars and genital serovars with respect to genetic mutations within the trpA gene. Ocular serovars (A, Ba, C) all had a frame-shift mutation in trpA resulting in the synthesis of a truncated protein. In contrast, all genital serovars (D–K) encoded a full-length TrpA. Further, these mutations resulted in measurable biological differences in the TrpA function between ocular and genital serovars; the tryptophan synthase from ocular serovars was enzymatically inactive, whereas the synthase from genital serovars was active when indole was provided as a substrate.
Because these original findings were made with reference serovars that had been passaged many times in vitro, we felt it necessary to verify the results using recent clinical specimens that represented diverse epidemiological and geographical populations. Here, we show that all C. trachomatis specimens sequenced, regardless of geographical origin, adhered to the paradigm established using laboratory reference strains — namely, that ocular serovars possess a dysfunctional tryptophan synthase, whereas genital serovars possess a functional tryptophan synthase. Perhaps the strongest evidence in support of this paradigm was the finding that the rare genital isolates of serovar B, a serovariant usually associated with ocular disease (2, 4), had intact trp genes that encoded a functional TrpBA complex, a characteristic associated exclusively with genital serovars. From an epidemiological perspective, this makes the trp locus useful for differentiating ocular and genital infections with serovar B isolates.
Sequencing of the trpA region from ocular clinical isolates uncovered four new inactivating mutations in addition to the complete trp region deletion and trpA frame-shift mutation (nucleotide 531) originally described in the ocular reference strains (16, 35). Interestingly, the chlamydial isolate(s) infecting a particular geographic region appear to be clonal with regard to the trp locus. For example, 65 out of 66 isolates from Gambia, regardless of ompA serotype, had a frame-shift mutation at base 531 of trpA, the same mutation originally described with reference serovars A, Ba, and C (16, 35). Similarly, all isolates from Tanzania had the same inactivating mutation, a two-base insertion at nucleotide 118 of trpA. These data suggest that the chlamydiae strains infecting these populations are clonal and that the trp inactivating mutation predates ompA variation. The clonality of these populations may reflect the limited opportunities for interchange of the strains that occur between the remote trachoma-endemic villages where the samples were collected. Determination of the trp genotype may prove useful for molecular epidemiological studies aimed at identifying chlamydial reservoirs and evaluating transmission patterns.
In free living organisms, de novo tryptophan biosynthesis requires the sequential action of seven gene products, encoded by trpA through trpG, by a repressor encoded by trpR (40, 41). Intriguingly, none of the chlamydial genomes sequenced to date contains the full complement of genes required to synthesize tryptophan — that is, none of the genes in C. pneumoniae (15, 42), one (trpF) in C. trachomatis MoPn (15), four (trpFRBA) in C. trachomatis serovar D (20) and six (trpA, B, C, D, F, R) in C. psittaci GPIC (http://www.tigr.org). Since mammalian cells are auxotrophic for tryptophan, these findings imply that to be able to synthesize tryptophan, chlamydiae must acquire the required biosynthetic intermediates from the environment. Previously, we showed that indole could rescue the growth of genital but not ocular reference serovars of chlamydiae, when cultured in medium lacking tryptophan (16). Here, we show that the addition of exogenous indole can reverse the effects of tryptophan starvation induced in a more physiologically relevant manner, namely the addition of IFN-γ. Together, these findings support the previously expressed view that, as with Toxoplasma gondii (43, 44), the inhibitory effect of IFN-γ on chlamydiae is through IDO-induced tryptophan degradation (25, 26, 45). Taken together, our findings generate the fascinating hypothesis that genital strains have evolved to utilize indole for the synthesis of tryptophan as an immunoavoidance strategy, thereby explaining the retention of the trpRBA operon.
Two obvious questions arise from our observations. First, what selective pressure exists on genital but not ocular serovars of chlamydiae to retain a functional TrpBA complex? Second, what is the in vivo source of indole that is needed as a precursor for tryptophan biosynthesis by genital isolates? Our studies indicate that there is strong selective pressure for genital but not ocular serovars to retain the ability to synthesize tryptophan. Because they lack the other genes of the tryptophan operon needed for de novo indole biosynthesis (trpE, G, D, C), and because under normal physiological conditions indole is not present in mammalian cells, genital strains probably acquire indole from their environment. We suggest that the most likely source of exogenous indole is the other microorganisms known to colonize the female genital tract. Interestingly, the predominant organism present in the normal vagina, lactobacillus, does not produce indole. However, many other organisms present in the vaginal microflora, including Peptostreptococcus species, Fusobacterium species, Bacteriodes species, and aerobic gram-negative rods such as E. coli, do produce indole (46–48). Interestingly, many of these microbes, especially the anaerobes, are present in much higher concentrations in women with bacterial vaginosis, whereas lactobacilli are reduced or absent in these women (49–52). It is known that coinfection with the organisms that cause bacterial vaginosis and chlamydiae is common. Moreover, these coinfections significantly increase the risk of complications such as pelvic inflammatory disease (53–55). Thus, it is possible that genital strains evolved the capability to synthesize tryptophan from indole produced by other vaginal microbes and that this host microbe-microbe interplay is essential in the ability of these strains to avoid IFN-γ–mediated host defense mechanisms and establish persistent infection, particularly in the female host. Our studies indicate that even low concentrations of indole (0.1–1.0 μM) allow recovery of EBs in the presence of IFN-γ. This highly efficient escape mechanism from the inhibitory effect of IFN-γ could result in the development of infectious EBs that would maintain the pathogen’s ability to persist in the presence of an active immune response and to be transmitted from person to person. The ability to establish such a unique polymicrobial host interaction may explain why chronic infection and associated harmful sequelae are more common in women than men, in whom the sparse urethral flora would most likely generate little indole. It would also establish women as a key reservoir of infection, an important epidemiological niche since the pathogen does not have a known nonhuman infection reservoir (2).
We described three genotypes in our previous work using genital reference strains that result from changes in a trpA mutational hot spot (16). Here, we have defined three genotypic groups within this region: Group 1 (encoding YQ), Group 2 (encoding CQ), and Group 3 (encoding YE). All LGV clinical isolates are Group 3 genotypes. In contrast, all other clinical genital serovars (D–K) may be Group 1 or 2. The fact that no other amino acid changes were found suggests that the spectrum of mutations allowed may be limited, presumably by the need for a functional enzyme. Given that these amino acids lie in TrpA loop 6, a region identified in the tryptophan synthase crystal structure as being important for subunit-subunit interactions between TrpB and TrpA, metabolite channeling, and substrate binding (17, 18, 56) it is quite possible that the mutations have an effect on enzyme catalytic activity or efficiency. This hypothesis can be tested by direct enzyme kinetic characterization using purified chlamydial recombinant TrpA and TrpB. There are two lines of experimental evidence that link these mutational changes with function. First, the mutations do correlate, albeit not perfectly, with the resistance of the reference serovars to the in vitro inhibitory effect of IFN-γ. For example, all three LGV serovars (Group 3) are significantly more resistant to IFN-γ than the other serovars, followed by serovars with intermediate sensitivity (Group 2) and those with the greatest sensitivity (Group 1) to IFN-γ, respectively (34). Second, our results using clinical pairs of Group 1 and Group 2 genotypes of the same serovariant (serovar I) showed that the in vitro growth of the Group 1 clinical isolate was more susceptible to IFN-γ than the Group 2 isolate. Moreover, the infectivity of the Group 1 variant was more readily rescued with lower amounts of exogenous indole than the Group 2 trpA genotype. Clearly, definitive conclusions await experimental findings using more serovar pairs with different hot-spot sequences. Whether these genotypic differences in trpA, unlike those reported for ompA (7), will reflect differences in susceptibility to host defenses, persistence, or transmission awaits future molecular epidemiological investigations.
We have presented what we believe to be a reasonable hypothesis of why genital strains have maintained a functional tryptophan synthase. However, we are left with explaining the more difficult observation of why trachoma serovariants have evolved an equally powerful negative selection to mutate the synthase.
Not unlike chlamydial genital infection, chlamydial ocular infections also persist, and persistence has been suggested as a mechanism that leads to the late damaging inflammatory and scarring stages of the disease (2, 10, 11). Genital serovars do infect the conjunctivae of infants during their passage from the birth canal. However, these infections produce a self-limiting neonatal conjunctivitis that does not progress to more chronic eye disease characteristic of trachoma (2, 4). This suggests that these strains are highly susceptible to the inhibitory effects of IFN-γ in the environment of the eye, perhaps because the eye represents a more sterile habitat lacking other microflora that could provide a source of exogenous indole. In the late stages of trachoma, there are reports that coinfection might contribute to disease transmission (2, 57, 58).
In contrast to the flora of the female genital tract, however, the ocular microflora are more limited in diversity (57, 58), and of those associated with trachoma only H. influenza is an established producer of indole (59). Perhaps there are alternate sites of infection in the eye than the conjunctival epithelium, and the trachoma serovars have adapted a strategy of interacting with these different cellular environments that allows for both productive and persistent infection. The conjunctival epithelium is an immunocompetent tissue in which infection induces IFN-γ, and analysis of both systemic and local cytokine profiles among subgroups of individuals differing in trachoma disease status implicate an IL-12 Th-1 type of immunity in resistance to the progression of chronic scarring inflammatory disease (60, 61). Thus, trachoma conjunctival infection of adolescents would mimic in many aspects neonatal conjunctivitis caused by genital serovars in that it is would be self-limiting and effectively cleared from this tissue by the host immune response (2, 4).
Why would it be an advantage for these serovars to have a dysfunctional (non–indole rescuable) synthase? Perhaps secondary infection targets exist in immunoprivileged areas of the eye that are sites in which chlamydiae establish persistent growth. If this were the case, then the ocular serovars might require a strategy that would allow for their growth and survival in multiple host cells of the eye. Clearly, there would be little need to maintain a functional synthase if (a) there were not environmental sources of exogenous indole (conjunctivae) and (b) secondary infection sites were devoid of lymphocytes and IFN-γ, as is the situation in immunoprivileged tissues of the eye (62, 63). In fact, being “rescuable” in a persistent immunocompromised environment could be a disadvantage, since it would compromise the ability of the pathogen to sustain a persistent infection. The possibility that the ocular serovars might reside in an immunoprivileged environment is indirectly supported by the limited number of serovariants (A, B, and C) associated with trachoma, as compared with the numerous (D, E, F, G, H, I, J, K, L1, L2, L3) genital serovariants. This indicates that the ocular strains have been exposed to less immune selection than the genital strains.
Although we are at present unsure of the precise molecular mechanisms involved, the evidence is irrefutable that there is a strict correlation among ocular and genital strains in their ability to maintain a functional tryptophan synthase. We believe that this strong evolutionary selection is critical to the pathogen’s ability to both infect and persist in different mucosal tissues of the human host. Our results indicate that the ability to synthesize tryptophan is a biological characteristic that has been retained or lost in order to allow chlamydiae to adapt to different environments within a human host. Moreover, our findings showing that polymorphisms in tryptophan synthase reflect functional changes in the organism’s sensitivity to IFN-γ justify future molecular epidemiological studies focused on defining whether these specific mutations correlate with differing clinical manifestations of chlamydial genital infection. Our findings also support the development of in vivo animal models to directly test the hypothesis that polymicrobial host interactions of the female genital tract are important determiners of chlamydial pathogenesis. Clearly, our interpretations are limited by our current incomplete knowledge of the biological differences between ocular and genital serovars. To address this, we are sequencing the genome of serovar A so that a comparative genomic analysis can be done with the published serovar D genome. It will be of interest to see what other if any genetic differences exist between ocular and genital strains.
Acknowledgments
This work was supported by research grants from the Canadian Institutes of Health Research (GR-13301 to G. McClarty) and the Health Canada Office of the Chief Scientist Fund (to G. McClarty) and a postgraduate scholarship from the Natural Sciences and Engineering Research Council of Canada (to H. Wood). We would like to thank Pam Small for helpful discussions and Hua Su for help with the figures.
Footnotes
See the related Commentary beginning on page 1647.Harlan D. Caldwell and Grant McClarty contributed equally to this work.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: tryptophan synthase (trpBA); major outer membrane protein (MOMP); plasticity zone (PZ); trpBA operon (trpRBA); indole glycerol 3-phosphate (IGP); cellular immune response (CMI); indoleamine 2,3-dioxygenase (IDO); elementary body (EB); sexually transmitted disease (STD); inclusion forming unit (IFU); reticulate body (RB); immunofluorescence antibody (IFA).
References
- 1.Moulder JW. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schachter, J. 1999. Infection and disease epidemiology. American Society for Microbiology Press. Washington, DC, USA. 139–170.
- 3.Stephens, R.S. 1999. Genomic autobiographies of Chlamydiae. American Society for Microbiology Press. Washington, DC, USA. 9–27.
- 4.An BB, Adamis AP. Chlamydial ocular diseases. Int. Ophthalmol. Clin. 1998;38:221–230. doi: 10.1097/00004397-199803810-00017. [DOI] [PubMed] [Google Scholar]
- 5.Mabey D, Bailey R. Eradication of trachoma worldwide. Br. J. Ophthalmol. 1999;83:1261–1263. doi: 10.1136/bjo.83.11.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang SP, Grayston JT. Immunologic relationship between genital TRIC, lymphogranuloma venereum, and related organisms in a new microtiter indirect immunofluorescence test. Am. J. Ophthalmol. 1970;70:367–374. doi: 10.1016/0002-9394(70)90096-6. [DOI] [PubMed] [Google Scholar]
- 7.Stothard DR, Boguslawski G, Jones RB. Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect. Immun. 1998;66:3618–3625. doi: 10.1128/iai.66.8.3618-3625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikehata M, Numazaki K, Chiba S. Analysis of Chlamydia trachomatis serovars in endocervical specimens derived from pregnant Japanese women. FEMS Immunol. Med. Microbiol. 2000;27:35–41. doi: 10.1111/j.1574-695X.2000.tb01409.x. [DOI] [PubMed] [Google Scholar]
- 9.Fraiz J, Jones RB. Chlamydial infections. Annu. Rev. Med. 1988;39:357–370. doi: 10.1146/annurev.me.39.020188.002041. [DOI] [PubMed] [Google Scholar]
- 10.Schachter J. Pathogenesis of chlamydial infections. Pathol. Immunopathol. Res. 1989;8:206–220. doi: 10.1159/000157149. [DOI] [PubMed] [Google Scholar]
- 11.Smith A, et al. OmpA genotypic evidence for persistent ocular Chlamydia trachomatis infection in Tanzanian village women. Ophthalmic Epidemiol. 2001;8:127–135. doi: 10.1076/opep.8.2.127.4164. [DOI] [PubMed] [Google Scholar]
- 12.Brunham, R.C. 1999. Human immunity. American Society for Microbiology Press. Washington, DC, USA. 211–238.
- 13.Ward, M.E. 1999. Mechanisms of disease. American Society for Microbiology Press. Washington, DC, USA. 171–210.
- 14.Ward ME. The immunobiology and immunopathology of chlamydial infections. APMIS. 1995;103:769–796. doi: 10.1111/j.1699-0463.1995.tb01436.x. [DOI] [PubMed] [Google Scholar]
- 15.Read TD, et al. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fehlner-Gardiner C, et al. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 2002;277:26893–26903. doi: 10.1074/jbc.M203937200. [DOI] [PubMed] [Google Scholar]
- 17.Miles EW. Tryptophan synthase. Structure, function, and protein engineering. Subcell. Biochem. 1995;24:207–254. [PubMed] [Google Scholar]
- 18.Miles EW. Tryptophan synthase: a multienzyme complex with an intramolecular tunnel. Chem. Rec. 2001;1:140–151. doi: 10.1002/tcr.4. [DOI] [PubMed] [Google Scholar]
- 19.Miles EW. Structural basis for catalysis by tryptophan synthase. Adv. Enzymol. Relat. Areas Mol. Biol. 1991;64:93–172. doi: 10.1002/9780470123102.ch3. [DOI] [PubMed] [Google Scholar]
- 20.Stephens RS, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 21.Beatty WL, Morrison RP, Byrne GI. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol. Rev. 1994;58:686–699. doi: 10.1128/mr.58.4.686-699.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loomis WP, Starnbach MN. T cell responses to Chlamydia trachomatis. Curr. Opin. Microbiol. 2002;5:87–91. doi: 10.1016/s1369-5274(02)00291-6. [DOI] [PubMed] [Google Scholar]
- 23.Morrison RP, Caldwell HD. Immunity to murine chlamydial genital infection. Infect. Immun. 2002;70:2741–2751. doi: 10.1128/IAI.70.6.2741-2751.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim SK, DeMars R. Epitope clusters in the major outer membrane protein of Chlamydia trachomatis. Curr. Opin. Immunol. 2001;13:429–436. doi: 10.1016/s0952-7915(00)00237-5. [DOI] [PubMed] [Google Scholar]
- 25.Beatty WL, Belanger TA, Desai AA, Morrison RP, Byrne GI. Tryptophan depletion as a mechanism of gamma interferon-mediated chlamydial persistence. Infect. Immun. 1994;62:3705–3711. doi: 10.1128/iai.62.9.3705-3711.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beatty WL, Morrison RP, Byrne GI. Reactivation of persistent Chlamydia trachomatis infection in cell culture. Infect. Immun. 1995;63:199–205. doi: 10.1128/iai.63.1.199-205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tipples G, McClarty G. Isolation and initial characterization of a series of Chlamydia trachomatis isolates selected for hydroxyurea resistance by a stepwise procedure. J. Bacteriol. 1991;173:4932–4940. doi: 10.1128/jb.173.16.4932-4940.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh YH, Bobo LD, Quinn TC, West SK. Determinants of trachoma endemicity using Chlamydia trachomatis ompA DNA sequencing. Microbes Infect. 2001;3:447–458. doi: 10.1016/s1286-4579(01)01400-9. [DOI] [PubMed] [Google Scholar]
- 29.Conway DJ, et al. Scarring trachoma is associated with polymorphism in the tumor necrosis factor alpha (TNF-alpha) gene promoter and with elevated TNF-alpha levels in tear fluid. Infect. Immun. 1997;65:1003–1006. doi: 10.1128/iai.65.3.1003-1006.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brunham RC, et al. The epidemiology of Chlamydia trachomatis within a sexually transmitted diseases core group. J. Infect. Dis. 1996;173:950–956. doi: 10.1093/infdis/173.4.950. [DOI] [PubMed] [Google Scholar]
- 31.Geisler WM, Whittington WL, Suchland RJ, Stamm WE. Epidemiology of anorectal chlamydial and gonococcal infections among men having sex with men in Seattle: utilizing serovar and auxotype strain typing. Sex. Transm. Dis. 2002;29:189–195. doi: 10.1097/00007435-200204000-00001. [DOI] [PubMed] [Google Scholar]
- 32.Van Der Pol B, et al. Multicenter evaluation of the BDProbeTec ET System for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urine specimens, female endocervical swabs, and male urethral swabs. J. Clin. Microbiol. 2001;39:1008–1016. doi: 10.1128/JCM.39.3.1008-1016.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caldwell HD, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrison RP. Differential sensitivities of Chlamydia trachomatis strains to inhibitory effects of gamma interferon. Infect. Immun. 2000;68:6038–6040. doi: 10.1128/iai.68.10.6038-6040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw AC, Christiansen G, Roepstorff P, Birkelund S. Genetic differences in the Chlamydia trachomatis tryptophan synthase alpha-subunit can explain variations in serovar pathogenesis. Microbes Infect. 2000;2:581–592. doi: 10.1016/s1286-4579(00)00368-3. [DOI] [PubMed] [Google Scholar]
- 36.Frost EH, Deslandes S, Gendron D, Bourgaux-Ramoisy D, Bourgaux P. Variation outside variable segments of the major outer membrane protein distinguishes trachoma from urogenital isolates of the same serovar of Chlamydia trachomatis. Genitourin. Med. 1995;71:18–23. doi: 10.1136/sti.71.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perry LL, Feilzer K, Caldwell HD. Immunity to Chlamydia trachomatis is mediated by T helper 1 cells through IFN-gamma-dependent and -independent pathways. J. Immunol. 1997;158:3344–3352. [PubMed] [Google Scholar]
- 38.Taylor MW, Feng GS. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 1991;5:2516–2522. [PubMed] [Google Scholar]
- 39.Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-gamma. Annu. Rev. Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 40.Crawford IP. Evolution of a biosynthetic pathway: the tryptophan paradigm. Annu. Rev. Microbiol. 1989;43:567–600. doi: 10.1146/annurev.mi.43.100189.003031. [DOI] [PubMed] [Google Scholar]
- 41.Crawford IP, Stauffer GV. Regulation of tryptophan biosynthesis. Annu. Rev. Biochem. 1980;49:163–195. doi: 10.1146/annurev.bi.49.070180.001115. [DOI] [PubMed] [Google Scholar]
- 42.Kalman S, et al. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 1999;21:385–389. doi: 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- 43.Pfefferkorn ER. Interferon gamma blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade tryptophan. Proc. Natl. Acad. Sci. U. S. A. 1984;81:908–912. doi: 10.1073/pnas.81.3.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai W, Pan H, Kwok O, Dubey JP. Human indoleamine 2,3-dioxygenase inhibits Toxoplasma gondii growth in fibroblast cells. J. Interferon Res. 1994;14:313–317. doi: 10.1089/jir.1994.14.313. [DOI] [PubMed] [Google Scholar]
- 45.Beatty WL, Morrison RP, Byrne GI. Immunoelectron-microscopic quantitation of differential levels of chlamydial proteins in a cell culture model of persistent Chlamydia trachomatis infection. Infect. Immun. 1994;62:4059–4062. doi: 10.1128/iai.62.9.4059-4062.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibbs RS. Microbiology of the female genital tract. Am. J. Obstet. Gynecol. 1987;156:491–495. doi: 10.1016/0002-9378(87)90318-8. [DOI] [PubMed] [Google Scholar]
- 47.Larsen B, Monif GR. Understanding the bacterial flora of the female genital tract. Clin. Infect. Dis. 2001;32:e69–e77. doi: 10.1086/318710. [DOI] [PubMed] [Google Scholar]
- 48.Balows, A. 1991. The prokaryotes. Springer-Verlag. New York, New York, USA. 1886–1887.
- 49.Morris M, Nicoll A, Simms I, Wilson J, Catchpole M. Bacterial vaginosis: a public health review. BJOG. 2001;108:439–450. doi: 10.1111/j.1471-0528.2001.00124.x. [DOI] [PubMed] [Google Scholar]
- 50.Hillier SL. Diagnostic microbiology of bacterial vaginosis. Am. J. Obstet. Gynecol. 1993;169:455–459. doi: 10.1016/0002-9378(93)90340-o. [DOI] [PubMed] [Google Scholar]
- 51.Hill GB. The microbiology of bacterial vaginosis. Am. J. Obstet. Gynecol. 1993;169:450–454. doi: 10.1016/0002-9378(93)90339-k. [DOI] [PubMed] [Google Scholar]
- 52.Hellberg D, Nilsson S, Mardh PA. The diagnosis of bacterial vaginosis and vaginal flora changes. Arch. Gynecol. Obstet. 2001;265:11–15. doi: 10.1007/s004040000109. [DOI] [PubMed] [Google Scholar]
- 53.Jossens MO, Schachter J, Sweet RL. Risk factors associated with pelvic inflammatory disease of differing microbial etiologies. Obstet. Gynecol. 1994;83:989–997. doi: 10.1097/00006250-199406000-00019. [DOI] [PubMed] [Google Scholar]
- 54.Paavonen J, et al. Microbiological and histopathological findings in acute pelvic inflammatory disease. Br. J. Obstet. Gynaecol. 1987;94:454–460. doi: 10.1111/j.1471-0528.1987.tb03125.x. [DOI] [PubMed] [Google Scholar]
- 55.Wiesenfeld HC, et al. Lower genital tract infection and endometritis: insight into subclinical pelvic inflammatory disease. Obstet. Gynecol. 2002;100:456–463. doi: 10.1016/s0029-7844(02)02118-x. [DOI] [PubMed] [Google Scholar]
- 56.Weyand M, Schlichting I. Crystal structure of wild-type tryptophan synthase complexed with the natural substrate indole-3-glycerol phosphate. Biochemistry. 1999;38:16469–16480. doi: 10.1021/bi9920533. [DOI] [PubMed] [Google Scholar]
- 57.Chern KC, et al. Alterations in the conjunctival bacterial flora following a single dose of azithromycin in a trachoma endemic area. Br. J. Ophthalmol. 1999;83:1332–1335. doi: 10.1136/bjo.83.12.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peterson J, Treadway G. Impact of community-based azithromycin treatment of trachoma on carriage and resistance of Streptococcus pneumoniae. Clin. Infect. Dis. 1998;26:248–249. doi: 10.1086/517045. [DOI] [PubMed] [Google Scholar]
- 59.Alrawi AM, et al. Biotypes and serotypes of Haemophilus influenzae ocular isolates. Br. J. Ophthalmol. 2002;86:276–277. doi: 10.1136/bjo.86.3.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bobo L, et al. Evidence for a predominant proinflammatory conjunctival cytokine response in individuals with trachoma. Infect. Immun. 1996;64:3273–3279. doi: 10.1128/iai.64.8.3273-3279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holland MJ, et al. T helper type-1 (Th1)/Th2 profiles of peripheral blood mononuclear cells (PBMC); responses to antigens of Chlamydia trachomatis in subjects with severe trachomatous scarring. Clin. Exp. Immunol. 1996;105:429–435. doi: 10.1046/j.1365-2249.1996.d01-792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geiger K, Sarvetnick N. Local production of IFN-gamma abrogates the intraocular immune privilege in transgenic mice and prevents the induction of ACAID. J. Immunol. 1994;153:5239–5246. [PubMed] [Google Scholar]
- 63.Geiger KD, Sarvetnick NE. Ocular production of interferon-gamma and lack of major histocompatibility complex molecules induce immunological changes in the intraocular environment. Ger. J. Ophthalmol. 1996;5:309–314. [PubMed] [Google Scholar]