Abstract
DNA methylation in the promoter of certain genes is associated with transcriptional silencing. Methylation affects gene expression directly by interfering with transcription factor binding and/or indirectly by recruiting histone deacetylases through methyl-DNA-binding proteins. In this study, we demonstrate that the human lung cancer cell line H719 lacks p53-dependent and -independent p21Cip1 expression. p53 response to treatment with gamma irradiation or etoposide is lost due to a mutation at codon 242 of p53 (C→W). Treatment with depsipeptide, an inhibitor of histone deacetylase, was unable to induce p53-independent p21Cip1 expression because the promoter of p21Cip1 in these cells is hypermethylated. By analyzing luciferase activity of transfected p21Cip1 promoter vectors, we demonstrate that depsipeptide functions on Sp1-binding sites to induce p21Cip1 expression. We hypothesize that hypermethylation may interfere with Sp1/Sp3 binding. By using an electrophoretic mobility shift assay, we show that, although methylation within the consensus Sp1-binding site did not reduce Sp1/Sp3 binding, methylation outside of the consensus Sp1 element induced a significant decrease in Sp1/Sp3 binding. Depsipeptide induced p21Cip1 expression was reconstituted when cells were pretreated with 5-aza-2′-deoxycytidine. Our data suggest, for the first time, that hypermethylation around the consensus Sp1-binding sites may directly reduce Sp1/Sp3 binding, therefore leading to a reduced p21Cip1 expression in response to depsipeptide treatment.
Although a strong correlation between promoter methylation and gene silencing has been extensively demonstrated (5, 24, 35), the molecular mechanisms of this methylation-modulated gene inactivation remains unclear. Two hypotheses have been proposed to explain transcriptional inactivation from promoter methylation. One of them is based on the finding that methyl-CpG-binding proteins (MBPs), such as MeCP2, specifically bind to symmetrically methylated DNA through a methyl-CpG-binding domain (11, 41). MBPs then recruit transcriptional repressors such as Sin3, NuRD, and histone deacetylases (HDACs) through its transcriptional-repression domain (25, 32, 54). Since Sin3 and HDACs are known transcriptional repressors (2, 50), methylated DNA may repress gene expression indirectly through MeCP2 and other MBPs. In addition, deacetylation of histones results in a net increase in positively charged lysines and arginines at the N-terminal tail of the histones (18, 21), thus inducing a tighter noncovalent linkage between the positively charged histones and the negatively charged DNA (3, 47). Consequently, transcription factors have difficulty accessing their DNA-binding sites (4, 29, 47), with a reduction or silencing of gene transcription. This hypothesis, based on the interaction between DNA methylation and histone acetylation status, has been extensively supported by accumulated experimental evidence (7, 16, 37, 40). For example, trichostatin A (TSA), an inhibitor of HDAC, induces a robust reexpression of silenced genes when used with minimal doses of the demethylating agent, 5-aza-2′-deoxycytidine (5-aza-CdR), although TSA or 5-aza-CdR alone do not lead to gene reexpression (7). Our previous data also show a link between histone acetylation status and DNA methylation, such that 5-aza-CdR significantly enhances acetylation of histones H3 and H4 induced by a HDAC inhibitor, depsipeptide. Related to this, depsipeptide-induced apoptosis is dramatically increased in cells pretreated with 5-aza-CdR (56). In addition, p19INK4D expression is greatly enhanced when human lung cancer cells are treated with depsipeptide and 5-aza-CdR together compared to treatment with each agent alone (55). These studies support the notion that methylation and histone acetylation work cooperatively to influence gene expression and other biological processes.
Another explanation for methylation-induced gene repression emphasizes a direct influence of methylation on the binding of transcription factors (6, 38, 49). Since many mammalian transcription factors have CG-rich binding sites in their DNA recognition element, methylation occurring at the CG of the specific binding element may sterically interfere with binding of transcription factors to DNA, thus inhibiting transcription (5). For instance, a HpaII site (CCGG) is located in an element to which the transcription factor AP-2 binds. Methylation at this HpaII site (CCmGG) within the AP-2-binding site inhibits AP-2 binding and suppresses AP-2-regulated gene transcription in C6-glioma and CV-1 cells (10). This direct effect of methylation on binding of transcription factors is also observed in other genes including cyclic AMP (cAMP)/cAMP-responsive element (CRE, TGACGTCA) (23) and the retinoblastoma binding factor 1 recognition sequence (AGCTGCCGCGGGCGGAAGT) (33). However, reports regarding the effect of methylation at the Sp1-binding site on Sp1 binding are conflicting. Several researchers have shown that methylation at Sp1-binding sites has no influence on Sp1 binding and gene expression (20, 33). In contrast, others have reported that methylated CpG dinucleotides variably interfere with Sp1 binding by 50 to 95% depending on the configuration of methylated cytosines within the consensus Sp1-binding element (9, 28). Together, which mechanism is predominant in the methylation-induced gene repression may be dependent on cell type, transcription factor, or received stimuli.
The present study focuses on whether methylation influences p21Cip1 expression in human lung cancer cell lines in response to p53-dependent and -independent stimuli. p21Cip1 was selected as the target gene since it plays an important role in controlling the cell cycle (42) and has previously been shown to be induced by depsipeptide treatment (36). In addition, since p21Cip1 is rarely mutated or deleted in tumors and cancer cell lines (17, 44), it is an ideal candidate for studying the role of methylation in gene expression in cancer cells. Results from the present study show a causal relation between methylation of the p21Cip1 promoter and p21Cip1 silencing in some cell lines of human lung cancer. We also demonstrate that methylation at CG sites outside of the consensus Sp1-binding site may directly reduce the ability of Sp1/Sp3 to bind its DNA recognition element.
MATERIALS AND METHODS
Cell culture and chemical treatments.
The human lung cancer cell lines H23, H69, H82, H125, H211, H290, H360, H513, H719, H792, H841, H1781, H1299, H1977, H2052, and A549 (obtained from the American Type Culture Collection) were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum and penicillin-streptomycin in an incubator (37°C, 5% CO2). Cells were plated onto 100-cm2 dishes 18 to 24 h prior to experiments. 5-Aza-CdR was freshly added into cells every 24 h as described previously (12). Depsipeptide (kindly provided by Kenneth Chan, School of Pharmacy, The Ohio State University) was added to cells for 6 h at various concentrations. Sodium butyrate (Sigma, St. Louis, Mo.) was added to cells at 5 mM for 24 h.
Irradiation treatment.
Fresh medium was added to cells 1 h prior to irradiation. Cells were then gamma irradiated at 1.02 Gy/min. Plates containing cells were kept on ice for the entire irradiation period. The irradiated cells were then incubated at 37°C in 5% CO2 until harvest.
Flow cytometry assay.
After various treatments, cells were trypsinized and washed with cold phosphate-buffered saline (PBS) once. Cells were then fixed with 70% ethanol and stored at −20°C overnight. Propidium iodide (10 μg/ml; Sigma) was added to cells for staining. A Becton-Coulter (Miami, Fla.) Elite flow cytometer was used to analyze cellular DNA content.
Immunoblotting.
Protein expression was detected by Western immunoblotting as previously described with minor modifications (55, 56). Briefly, cells were harvested with a scraper and then washed with cold PBS once. Cells were then lysed in lysis buffer (50 mM Tris-HCl, 250 mM NaCl, 5 mM EDTA, 50 mM NaF, 0.15% Igepal CA-630, 1.5 mM phenylmethylsulfonyl fluoride [PMSF]). Equal amounts of proteins (100 to 150 μg) were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 9 to 15% polyacrylamide gels. Proteins were then transferred onto a nitrocellulose membrane. Nonspecific protein binding to the membrane was blocked with blocking buffer (5% nonfat milk, 200 mM NaCl, 50 mM Tris, 0.05% Tween 20). The blocked membrane was then incubated with primary antibodies at 4°C overnight with rocking. After the membrane was washed six times with TBS-T (20 mM Tris, 500 mM NaCl, 0.1% Tween 20) for 5 min each time, the membrane was incubated with secondary antibody at 4°C for 1 h. The detection of specific protein binding was performed with a chemilumenescence kit (Amersham Pharmacia Biotech, Uppsala, Sweden). The antibodies and concentrations used were 1 μg of anti-p21Cip1 (Santa Cruz Biotechnology, Santa Cruz, Calif.)/ml, 0.5 μg of anti-p53 (DO-1 [Oncogene Research Products])/ml, and 0.3 μg of α-tubulin (Oncogene Research Products)/ml.
Northern blotting.
Total RNA was extracted with TRIzol reagent (Gibco-BRL, Gaithersburg, Md.). Full-length p21Cip1 cDNA was cut from pGEX-p21 (a kind gift of Yue Xiong, University of North Carolina) and labeled as a probe. Northern blot analysis was performed by using standard procedures. Briefly, 20 μg of total RNA was size separated by using an agarose-formaldehyde gel (1%) and transferred to nylon membrane (Hybond-N; Amersham). Hybridization with random-primed 32P-labeled probes was performed at 42°C overnight. Filters were washed twice for 5 min each time with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at room temperature, twice for 5 min each time with 0.2 SSC-0.1% SDS at room temperature, and finally twice for 15 min each time at 42°C in 0.1 SSC-0.1% SDS and twice for 15 min each time at 68°C. The washed membrane was exposed to X-ray film at −80°C for 4 to 7 days or scanned with a phosphorimager.
Methylation-specific PCR and methylation detection.
DNA was extracted and then treated with bisulfite as previously described with minor modifications (19). Briefly, genomic DNA (1 μg) in a volume of 50 μl was denatured by NaOH (final concentration, 0.275 M) for 10 min at 42°C. The denatured DNA was then treated with 10 μl of 10 mM hydroquinone and 520 μl of 3 M sodium bisulfite at 50°C overnight. The bisulfite-modified DNA was purified with a Qiaquick gel extraction kit (Qiagen) according to the manufacturer's instruction. The DNA was then precipitated with sodium acetate (final concentration, 0.45 M) and isopropanol. DNA was eluted with distilled H2O and used for PCR. The primers and PCR conditions for p21Cip1 were follows: forward primer, 5′-GGG AGG AGG GAA GTG TTT TT-]3′; and reverse primer, 5′-ACA ACT ACT CAC ACC TCA ACT-3′. The conditions were as follows: 95°C for 10 min; 35 cycles of 96°C for 30 s, 52°C for 20 s, and 72°C for 20 s; and then 72°C for 7 min at the end. The PCR products were purified with a purification kit (Qiaquick Spin) and then incubated with HhaI at 37°C for 2 h or with TaqI at 65°C for 2 h. Digested DNA was then size fractionated via PAGE to detect the methylation status (52).
Bisulfite sequencing.
DNA was treated with bisulfite and purified for PCR as described above. The PCR products were gel extracted (Qiagen) and ligated into a plasmid vector, pCR2.1-TOPO, by using the TA cloning system (Invitrogen, Carlsbad, Calif.). Plasmid-transformed bacteria TOP10 F′ was cultured overnight, and the plasmid DNA was isolated (Qiagen). At least 20 separate clones were chosen for sequence analysis.
Detection of p53 mutation.
The detection of p53 mutation was performed by reverse transcription-PCR (RT-PCR) and sequencing analysis. The primers were 5′-GAC ACT TTG CGT TCG GGC T-3′ (forward primer) and 5′-CGG GAC AAA GCA AAT GGA AGT-3′ (reverse primer). PCR conditions were as follows: 95°C for 2 min and 28 cycles of 95°C for 30 s, 58°C for 105 s, 72°C for 105 s, and 72°C for 4 min. The PCR product was gel purified and then sequenced.
Transient transfection and measurement of relative luciferase activity.
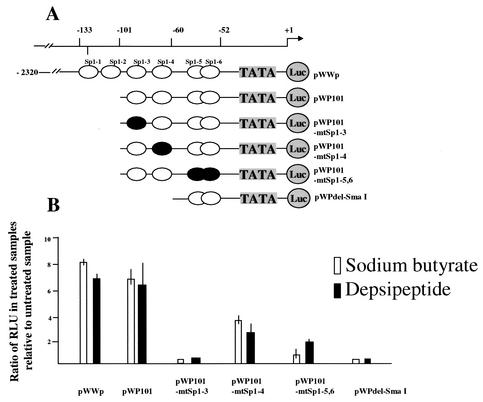
The vectors used for transfection experiments included pWWP-Luc, pWP101, pWPdel-SmaI, and three mutated Sp1 vectors: pWP101mtSp1-3, pWP101mtSp1-4, and pWP101mtSp1-5,6. The human wild-type p21Cip1 promoter luciferase fusion plasmid, pWWP-Luc, was made from a 2.4-kb genomic fragment of p21Cip1 promoter containing the transcriptional start site subcloned into the luciferase reporter vector, pGL3Basic. pWP101 contains four Sp1 sites termed Sp1-3, Sp1-4, and Sp1-5,6 that are located between −101 and −61 of the p21Cip1 promoter relative to the transcriptional start site and removed ca. 1,100 bp from the 5′ end of the p21Cip1 promoter. pWPdel-SmaI contains only Sp1-5,6 binding sites. These vectors, including pWP101mtSp1-3 (mutated at the Sp1-3 site compared to pWP101), pWP101mtSp1-4 (mutated at the Sp1-4 site compared to pWP101), and pWP101mtSp1-5,6 (mutated at the Sp1-5,6 sites compared to pWP101), were kindly provided by T. Sakai (Department of Preventive Medicine, Kyoto Prefectural University of Medicine, Kyoto, Japan). H719 cells transfected with the indicated p21Cip1-luciferase vectors were treated with 0.05 μM depsipeptide for 6 h or 5 mM sodium butyrate for 24 h and then harvested to analyze the luciferase activity. The luciferase activity was normalized for the amount of protein in cell lysate. All of the luciferase experiments were carried out at least twice in triplicate.
Extraction of nuclear proteins.
Nuclear protein was extracted as described previously with modifications (13). Briefly, 107 H719 cells were scraped into a 1.5-ml tube and centrifuged at room temperature for 5 min at 1,000 rpm. The cell pellet was washed with 1 to 2 ml of cold PBS and then centrifuged at 1,500 rpm for 20 to 30 s at 4°C. The resulting pellet was incubated in buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], 0.5 mM PMSF; protease inhibitor cocktail [Roche Diagnostics, Mannheim, Germany]) and then incubated on ice for 15 min. The cells were centrifuged at 1,500 rpm for 5 min at 4°C, and the resulting pellet was resuspended in buffer A and homogenized with a glass homogenizer (Kontes Glass Co., Vineland, N.J.). Checked under microscope with trypan blue, >90% free nuclei were confirmed. After centrifugation at 1,000 rpm at 4°C, the supernatant was discarded, and the pellet was suspended in 1/2 volume of buffer B (20 mM HEPES, 0.2 mM EDTA, 1.5 mM MgCl2, 0.02 M KCl, 25% glycerol, 0.5 mM DTT, 0.5 mM PMSF; protease inhibitors). The suspension was then gently resuspended in 1/2 volume of buffer C (20 mM HEPES, 0.2 mM EDTA, 1.5 mM MgCl2, 1.2 M KCl, 25% glycerol, 0.5 mM DTT, 0.5 mM PMSF; protease inhibitors), incubated at 4°C for 30 min with rotation, and then centrifuged at 4°C at 14,000 rpm for 30 min. The nuclear protein was then dialyzed three times against dialysis buffer (20 mM HEPES, 0.2 mM EDTA, 0.1 M KCl, 20% glycerol, 0.5 mM DTT, 0.5 mM PMSF; protease inhibitors) for 2 h each time. Finally, the concentration of nuclear protein was determined and saved at −80°C for experiments.
EMSA.
Electrophoretic mobility shift assay (EMSA) experiments were performed as described previously with minor modifications (22). Oligonucleotides and methylated oligonucleotides were purchased from Qiagen Operon Technologies, Inc. (Alameda, Calif.). These included the wild-type consensus Sp1-3 element of p21Cip1 promoter (CGA GCG CGG GTC CCG CCT CCT), methylated Sp1-3 element (CGA GCG CGG GTC CCmG CCT CCT), and two mutated Sp1-3 elements (AGC GCG GGT CTT GCC TCC TTG and CGA GCG CGG GTC TCT CCT CCT). The consensus Sp1-3-binding site is underlined, and the mutated nucleotides are in boldface. Two pairs of methylated oligonucleotides, in which the methylated cytosines are not located within the consensus Sp-1-binding site, were also designed for the EMSA experiments. The first of these pairs includes CmGA GCG CGG GTC CCG CCT CCT and CmGA GCmG CmGG GTC CCG CCT CCT. In addition, another pair of oligonucleotides, in which methylated cytosines are located both in and outside of the consensus binding site (CmGA GCmG CmGG GTC CCmG CCT CCT), was used in the experiments. All listed oligonucleotides included antiparallel partner oligonucleotides. In the case of the methylated oligonucleotides, the antisense oligonucleotides pair was methylated at the corresponding cytosine as the sense strand. These oligonucleotides were end labeled with [γ-32P]ATP (3,000 mCi/mmol; NEN Life Science Products) after pair annealing as the EMSA probes. Then, 10 μg of nuclear proteins in 20 μl of reaction solution [4% Ficoll [wt/vol], 25 mM HEPES, 1 mM EDTA, 0.5 mM DTT, 50 mM KCl, 0.375 mg of bovine serum albumin/ml, 0.15 μg of poly(dI-dC)/ml)] were incubated on ice for 30 min, followed by the addition of 250,000 cpm of probe for another 30 min at 4°C. For supershift experiments, Sp1 or Sp3 antibodies (Santa Cruz) were added into the reaction solution 30 min prior to the addition of the probes. The DNA-protein complex was then size fractionated by using a 5% polyacrylamide gel and subjected to autoradiography.
RESULTS
p21Cip1 expression in H719 cells is suppressed after both p53-dependent and -independent stimuli.
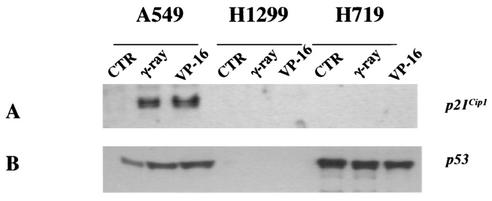
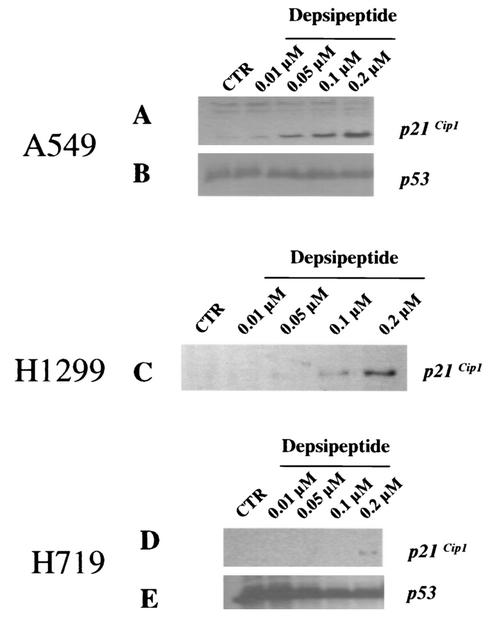
To investigate p21Cip1 expression in human lung cancer cell lines, A549 (wild-type p53), H1299 (p53 null), and H719 (p53 status unknown) were used in the present study. Cells were gamma irradiated at 8 Gy or treated with 50 μg of etoposide (VP-16)/ml for 24 h and then harvested to determine p21Cip1 expression 24 h after treatment. As shown in Fig. 1A, increased p21Cip1 expression upon treatment with gamma irradiation or VP-16 was observed only in A549 cells and not in H1299 and H719 cells. The p21Cip1 expression upon gamma irradiation or VP-16 treatments in A549 cells was correlated with wild-type p53 increase (Fig. 1B). Since there is no endogenous p53 in H1299 cells, a lack of p21Cip1 expression was expected (Fig. 1A). Expression of p53, however, was not changed in H719 cells after treatment with either VP-16 or gamma irradiation, implying that H719 cells lack DNA damage-induced p53 response (Fig. 1B). Since depsipeptide induces a p53-independent increase in p21Cip1 expression (36), we further investigated p53-independent p21Cip1 expression by treating cells with depsipeptide (0.0125 to 0.2 μM) for 6 h. Depsipeptide induced a dose-dependent p21Cip1 expression in A549 cells and H1299 cells but not in H719 cells (Fig. 2A, C, and D). Depsipeptide-induced p21Cip1 expression in H719 cells was observed only after high doses (Fig. 2D). One of the potential causes of the low-level p21Cip1 expression seen in H719 cells after treatment with 0.2 μM depsipeptide is heterogeneity of promoter methylation (see below). By analyzing cell cycle changes in gamma irradiation- or depsipeptide-treated cells, a p21Cip1-modulated G1 or G2 arrest was observed in A549 or H1299 cells but not in H719 cells (data not shown). These data showed that p21Cip1 expression in H719 cells is suppressed after both p53-dependent and p53-independent stimuli.
FIG. 1.
Expression of p21Cip1 and p53 in human lung cancer cells in response to gamma irradiation or etoposide treatment. Cells were exposed to gamma irradiation (8 Gy) or VP-16 treatment (50 μg/ml for 24 h) and then incubated at 37°C for 24 h. Protein was harvested, and Western immunoblot was performed to detect the expression of p21Cip1 (A) and p53 (B).
FIG. 2.
Expression of p21Cip1 and p53 in human lung cancer cells after depsipeptide treatment. Cells were treated with depsipeptide (0.0125 to 0.2 μM for 6 h), washed with cold PBS, and incubated at 37°C for 24 h. Protein was harvested, and Western immunoblot analysis was performed to detect the expression of p21Cip1 (A, C, and D) or p53 (B and E) in response to depsipeptide treatment. H1299 cells have no endogenous p53.
CpG island of p21Cip1 promoter of H719 cells is hypermethylated.
Since p21Cip1 expression in H719 cells is very low at baseline (Fig. 1 to 2) and no detectable increase in p21Cip1 expression was observed after DNA-damaging treatments or low-dose depsipeptide (Fig. 1 and 2), we considered two possibilities: (i) p53 is mutated and/or (ii) the p21Cip1 promoter is mutated or methylated in this cell line. To test these possibilities, total RNA from asynchronously growing H719 cells was extracted, and RT-PCR and sequencing of the p53 cDNA were performed. We found a missense mutation at codon 242 of the p53 gene, in which a cysteine is changed to a tryptophan (TGC to TGG; data not shown). This result is consistent with the lack of p53-dependent p21Cip1 response after H719 cells were treated with gamma irradiation or VP-16 (Fig. 1A). We also subjected the p21Cip1 promoter to sequence analysis and found no mutations or deletions (data not shown).
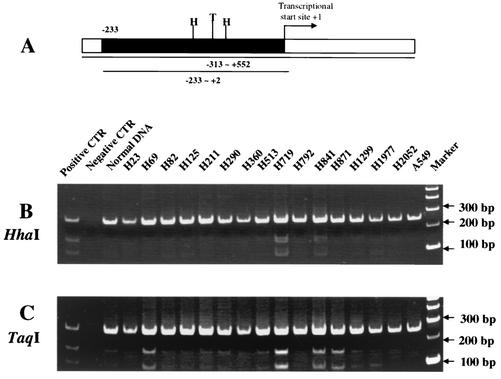
Subsequently, to determine the methylation status of the p21Cip1 promoter in human lung cancer cell lines, primers were designed to amplify a fragment within the CpG island of the p21Cip1 promoter (Fig. 3A). DNA from 16 human lung cancer cell lines, including H719, A549, and H1299, was treated with bisulfite, and then PCR was performed with oligonucleotide primers that flanked two HhaI sites and one TaqI site. The PCR products were purified and then incubated with the enzymes HhaI or TaqI. In this assay, bisulfite modification results in a change from C to T when the C is unmethylated in the context of a CG dinucleotide; however, when the cytosine is methylated, bisulfite modification leaves the methylated cytosine intact. Therefore, after bisulfite modification and PCR amplification, methylated CG dinucleotides will be intact, and restriction digestion with a CG containing restriction endonuclease can occur. In contrast, unmethylated CG dinucleotides are altered to TG, and CG-containing restriction enzymes, such as TaqI or HhaI, are unable to digest the fragment. Human lung cancer cell lines H719 and H841 showed a methylated pattern in the CpG island when cut with both enzymes, i.e., HhaI (GCGC) (Fig. 3B) and TaqI (TCGA) (Fig. 3C), whereas H69 and H871 showed a methylated pattern only when cut with TaqI (TCGA) (Fig. 3C). In addition, a slight band upon TaqI incubation was observed in the DNA from normal human blood and H82, H290, H513, H1299, and H1977 cells (Fig. 3C), indicating that a small part of DNA from these cell lines is methylated at the TaqI site.
FIG. 3.
Methylation status of p21Cip1 promoter in human lung cancer cells. (A) Schematic diagram of the p21Cip1 promoter. The CpG island of p21Cip1 promoter (positions −313 to +552 relative to the transcription start site; National Center for Biotechnology Information [NCBI] no. U24170) was determined by computer program (WebGene [http://www.itba.mi.cnr.it/webgene/]). Primers were designed to amplify a fragment spanning positions −233 to +2 (235 bp, black box). Within this fragment there is one TaqI recognition site (TCGA, presented as T in the figure) and two HhaI recognition sites (GCGC, presented as H in the figure). (B and C) DNA from 16 human lung cancer cell lines and normal human blood cells was treated with bisulfite and then PCR amplified. The PCR products were purified and digested with the CG-containing enzymes HhaI (B) or TaqI (C). Digested samples were size separated by 8% PAGE. DNA not treated with bisulfite served as a negative control. DNA treated with Sss methylase was the positive control. Digested fragments correspond to methylated DNA.
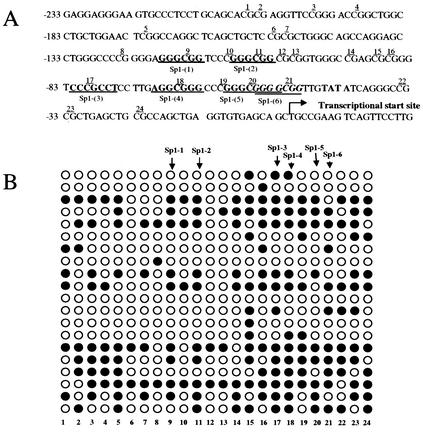
To further identify the methylation pattern within the CpG island of the p21Cip1 promoter in H719 cells, PCR products were subcloned into the vector pCR2.1, and 20 separate subclones were sequenced. By analyzing the sequence of the human p21Cip1 promoter, we noted that there are six Sp1-binding sites in the tested CpG island (Fig. 4A). Figure 4B shows the methylation status for all CGs in the tested CpG island (24 CGs) between the −233 to +1 positions relative to the transcriptional start site of p21Cip1 promoter. Overall, 40% of CGs were methylated in this fragment (20 clones, 192 of 480 of CGs were methylated) (Fig. 4B). About 44% of CGs of Sp1-binding sites and their boundaries are methylated (from the 9th CG to the 21st CG) (Fig. 4B). The highest proportion of methylated CGs (53%) was found in the region from the 14th to the 24th CG that spans the 3rd to 6th Sp1-binding sites (Fig. 4B). No methylated CGs in A549 cells and very few methylated CGs (< 1%) in H1299 cells were found by the bisulfite sequencing in the tested CpG island of p21Cip1 promoter (data not shown). As mentioned above, the heterogeneity of promoter methylation observed in H719 cells might explain why there was any p21Cip1 expression even after high-level exposure to depsipeptide (Fig. 2D).
FIG. 4.
Bisulfite sequencing of CpG island in the p21Cip1 promoter (A) The CpG island of p21Cip1 promoter (NCBI no. U24170) was analyzed. This sequence spans 250 bp between positions −233 to +17 relative to the transcription start site, including 24 CGs upstream of the transcriptional start site. There are six CG-containing Sp1-binding sites within this sequence that are indicated as underlined and emboldened, corresponding to the 9th, 11th, 17th, 18th, 20th, and 21st CGs within this island. (B) DNA from H719 cells was treated with bisulfite, and the p21Cip1 promoter was PCR amplified. The PCR product was ligated into pCR2.1-TOPO by using the TA cloning system. Twenty subclones were picked and sequenced. Symbols: ○, unmethylated cytosines; •, methylated cytosines.
Decreased Sp1/Sp3 binding is responsible for the reduced p21Cip1 expression upon depsipeptide treatment.
To demonstrate whether depsipeptide induces p21Cip1 expression through Sp1-binding sites, luciferase vectors containing different mutated Sp1-binding fragments were transiently transfected into H719 cells to detect relative luciferase activity after depsipeptide treatment. As illustrated in Fig. 5A, H719 cells were transfected with a wild-type p21Cip1 promoter-luciferase fusion plasmid, pWWP-Luc, or with pWP101, pWPdel-SmaI, pWP101mtSp1-3, pWP101mtSp1-4, or pWP101mtSp1-5,6, followed by treatment with 0.05 μM depsipeptide for 6 h. The cells were then harvested at 24 h after treatment to measure the relative luciferase activity. As a positive control, the cells transfected with the various vectors were also treated with 5 mM sodium butyrate for 24 h to detect the relative luciferase activity (31). As shown in Fig. 5B, the relative luciferase activity in wild-type pWWP-Luc-transfected cells was increased sevenfold in depsipeptide-treated cells compared to nontreated cells. This depsipeptide-induced increase in luciferase activity is almost identical to that for sodium butyrate-treated cells (8.5-fold, Fig. 5B). We observed, in assays with different mutated Sp1 luciferase vectors, that Sp1-3 and Sp1-4 sites were most responsive to depsipeptide treatments: the relative luciferase activities were decreased 2.5- to 100-fold in the mutant vectors compared to the wild-type pWWP-Luc-transfected cells (Fig. 5B). The Sp1-3-binding site appears to be most important in depsipeptide-induced p21Cip1 expression (Fig. 5B).
FIG. 5.
Analysis of relative luciferase activity in cells treated with depsipeptide or sodium butyrate. (A) p21Cip1 promoter constructs used for the luciferase transfection assay. The human wild-type p21Cip1 promoter luciferase fusion plasmid, pWWP-Luc, contains all six Sp1 sites and the transcription start site (2.4 kb). pWP101 contains four Sp1 sites termed Sp1-3 to Sp1-6. pWPdel-SmaI only contains the Sp1-5 and Sp1-6 sites. Three mutated Sp1 vectors are pWP101mtSp1-3 (mutated at the Sp1-3 site), pWP101mtSp1-4 (mutated at the Sp1-4 site), and pWP101mtSp1-5,6 (mutated at the Sp1-5 and Sp1-6 sites). •, mutated Sp1-binding sites. (B) At 24 h after transfection, H719 cells were treated with 0.05 μM depsipeptide for 6 h or 5 mM sodium butyrate for 24 h. The cells were harvested for analysis of luciferase activity at 24 h after the treatments. The luciferase activity of each sample was normalized for the amount of protein in the cell lysate. The experiments were carried out at least two times in triplicate. The luciferase activity of the untreated cells that was transfected with wild-type pWWP-Luc vector served as a control. The relative luciferase activity for each sample was then compared to the control, and the ratios were calculated.
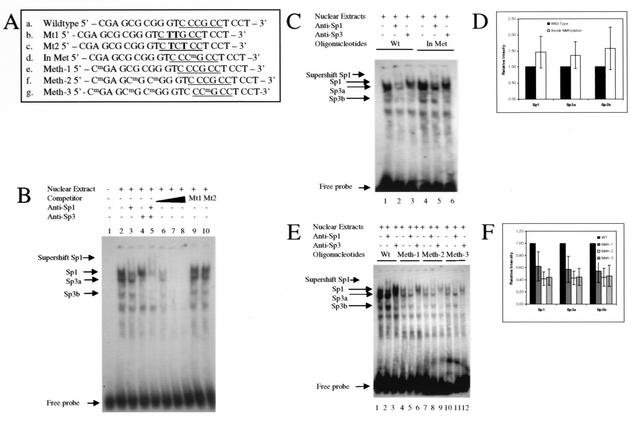
Subsequently, EMSA was performed to investigate whether methylation of the Sp1 site influences binding of Sp1 or Sp3 to their recognition elements. A consensus Sp1-3 fragment that contains Sp1/Sp3 recognition binding site (CCCGCC) was selected to test Sp1/Sp3 binding. In addition, two mutated Sp1-3 fragments (as illustrated in Fig. 6A) were used as controls. These experiments showed that the Sp1-3 fragment was able to bind H719 nuclear extracts (Fig. 6B). Incubation of H719 nuclear extracts with wild-type Sp1-3 probe (Fig. 6B, lane 2) revealed a pattern of shifted bands representing Sp1- and Sp3-binding activity. The Sp1 and Sp3 specific binding was confirmed by decreased intensity of the Sp1 and Sp3 bands, as well as by a supershift seen when Sp1 or Sp3 antibodies were added to the incubation mixture (Fig. 6B, lane 3 to 5). In addition, an unlabeled consensus Sp1-3 element (oligonucleotide a) competed effectively with labeled probe (Fig. 6B, lane 6 to 8). However, fragments with mutated Sp1 sites were unable to compete with the consensus Sp1-3 element (Fig. 6B, lanes 9 and 10). Unexpectedly, however, methylation of the CG within the Sp1-3 element did not lead to a decrease in the binding of Sp1 or Sp3 (Fig. 6C). In Fig. 6D, the intensities of the Sp1 and Sp3 bands were determined for the wild-type and internally methylated oligonucleotides (from Fig. 6C, lanes 1 and 4), showing no decrease in intensity. In order to investigate whether methylation outside of the consensus Sp1-3 element influences the binding of Sp1/Sp3, three methylated fragments with methylated CGs located outside of the consensus Sp1-binding site were used for EMSA experiments (oligonucleotides e, f, and g in Fig. 6A). As shown in Fig. 6E, the binding of Sp1/Sp3 to the recognition binding site was significantly decreased when methylation occurred at CpG dinucleotides adjacent to the consensus Sp1-3-binding sites compared to nonmethylated fragments. The density of methylation at sites outside of the Sp1/Sp3 site did not appear to have a major impact on binding (Fig. 6F, compare Meth-1 to Meth-2, corresponding to oligonucleotides e and f in Fig. 6A). As shown in Fig. 6E and F, the decrease in Sp1/Sp3 binding is approximately equivalent when three CGs outside of the consensus Sp1-binding site are methylated (Fig. 6E, lanes 7 to 9) compared to one methylated CG (Fig. 6E, lanes 4 to 6). The results represented in Fig. 6D and F were obtained from four replicate EMSA experiments and represent the relative intensity of binding without antibody supershift (i.e., lanes 1 and 4 from Fig. 6C, and lanes 1, 4, 7, and 10 from Fig. 6E). When methylation occurred both within and outside of the consensus Sp1-3 site, the Sp1/Sp3 binding is also significantly decreased (Meth-3, oligonucleotide g; Fig. 6E, lanes 10 to 12) but without additional decrease.
FIG. 6.
Analysis of binding of Sp1/Sp3 to the recognition Sp1-3-binding sites assayed by EMSA. (A) Sequence of oligonucleotides used for EMSA experiments (positions −93 to −72 relative to transcription start site of the p21Cip1 promoter). Sequences: a, wild-type Sp1-3-binding site of p21Cip1 promoter (Sp1-binding site is underlined); b and c, mutated Sp1-3-binding sites, with the mutated nucleotides in boldface; d, methylated Sp1-3-binding site within the Sp1-3-binding site (the methylated C corresponds to CG site 17 in Fig. 4); e and f, methylated cytosines located upstream of the consensus Sp1-3-binding site (the methylated C's correspond to CG sites 14, 15, and 16 in Fig. 4); g, methylated cytosines located both in and upstream of the Sp1-3-binding site. (B) EMSA experiments show specific binding for Sp1 and Sp3 (from H719 nuclear extracts) to the Sp1-3 recognition binding site. Lane 1, radiolabeled probe in the absence of nuclear extract; lane 2, Sp1/Sp3 binding is depicted by arrows on left of figures; lane 3, anti-Sp1 antibody is added to extracts; lane 4, anti-Sp3 supershift; lane 5, anti-Sp1 and anti-Sp3 supershift; lanes 6 to 8, excess unlabeled wild-type competitor (lane 6 [1:10], lane 7 [1:50], and lane 8 [1:100]) competes for binding with the labeled element; lanes 9 and 10, unlabeled mutated Sp1-3 oligonucleotide (1:100) could not compete with the labeled Sp1 probe. (C) Comparison of nuclear extract binding to the consensus wild-type and methylated Sp1-3-binding sites (oligonucleotides a [wild type] and d [In Met], respectively). Lanes 1 to 3 show binding reactions with labeled wild-type Sp1-3 as a probe, and lanes 4 to 6 show binding reactions with labeled methylated Sp1-3 as a probe. Lanes 1 and 4, no antibody added; lanes 2 and 5, anti-Sp1 antibody; lanes 3 and 6, anti-Sp3 antibody. (D) Graphic representation of the the relative binding affinities of nuclear extracts to the oligonucleotides in panel C (lanes 1 and 4). The relative intensity of binding was determined in four replicate experiments. (E) Comparison of nuclear extract binding to the consensus Sp1-3-binding element with wild-type and nonconsensus methylated oligonucleotides (see panel A, oligonucleotides e to g). Lanes 1 to 3, wild-type Sp1-3 element; lanes 4 to 6, oligonucleotide e (Meth-1); lanes 7 to 9, oligonucleotide f (Meth-2); lanes 10 to 12, oligonucleotide g (Meth-3); lanes 1, 4, 7, and 10, no antibody; lanes 2, 5, 8, and 11, anti-Sp1 antibody; lanes 3, 6, 9, and 12, anti-Sp3 antibody. (F) Graphic representation of the relative binding affinities of nuclear extracts to the oligonucleotides in panel E (lanes 1, 4, 7, and 10). The relative intensities of oligonucleotide binding were determined in four replicate experiments.
Depsipeptide-induced p21Cip1 expression is reconstituted in H719 cells after treatment with DNA-demethylating agent.
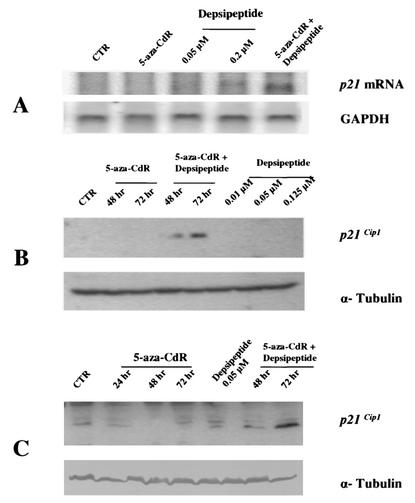
To confirm that methylation is the dominant mechanism for the reduced p21Cip1 expression in response to depsipeptide treatment in vivo, H719 cells were treated with 5-aza-CdR to observe whether the depsipeptide-induced p21Cip1 expression is recovered. A representative Northern blot shown in Fig. 7A demonstrates that 5-aza-CdR at 1 μM alone does not induce an increase in p21Cip1 expression, and high doses of depsipeptide induced only minimal expression of p21Cip1 mRNA. However, p21Cip1 mRNA was increased upon depsipeptide treatment after cells were pretreated with 5-aza-CdR for 48 to 72 h (Fig. 7A). Consistent with this, p21Cip1 protein expression was also dramatically increased when cells were treated with depsipeptide after demethylating treatment, as demonstrated by Western blotting (Fig. 7B). We analyzed the methylation status of p21Cip1 promoter in H719 cells after treatment with 5-aza-CdR for 48 or 72 h by bisulfite sequencing and confirmed that the previously methylated promoter region was now unmethylated (data not shown). Finally, we show similar results in another human lung cancer cell line (H841) that is similarly hypermethylated in the p21Cip1 promoter. Depsipeptide-induced p21Cip1 expression was much increased in the cells pretreated with 5-aza-CdR (Fig. 7C).
FIG. 7.
Reconstitution of p21Cip1 expression in mRNA and protein after depsipeptide treatment when cells are pretreated with DNA-demethylating agent. (A) H719 cells were treated with 1 μM 5-aza-CdR for 48 h alone or in combination with 0.05 μM depsipeptide for 6 h at 42 to 48 h. H719 cells were also treated with depsipeptide alone (at 0.05 or 0.2 μM) for 6 h. RNA from these treated cells was then extracted, and Northern blot analysis was performed by using a p21Cip1 cDNA probe. The membrane was stripped and then probed with GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as a loading control. (B) The p21Cip1 changes were also observed by Western immunoblotting in cells that were treated with 5-aza-CdR alone (1 μM) or depsipeptide alone (0.05 to 0.125 μM) or in combination (5-aza-CdR for 48 to 72 h at 1 μM plus depsipeptide at 0.05 μM for 6 h of treatment at 42 to 48 h or 66 to 72 h). Alpha-tubulin was used as a loading control. (C) H841 cells were treated with 5-aza-CdR (1 μM, 24 to 72 h), depsipeptide (0.05 μM, 6 h), or both to detect p21Cip1 expression.
DISCUSSION
Other investigators have shown that the inhibition of DNA methyltransferase (DNMT) (14, 30, 34, 43, 53) leads to a significant increase in p21Cip1 expression, whereas the CpG island of p21Cip1 promoter was totally unmethylated in the tested cell lines. This increased p21Cip1 in response to DNA-demethylating treatments suggests an alternative mechanism of gene reexpression after DNMT inhibition unrelated to DNA methylation. In contrast, a causal relation between DNA methylation and p21Cip1 silencing has been confirmed in tumors (8), cancer cell lines (1), and patients (39). Consistent with these reports, our study indicates that hypermethylation in the CpG island of the p21Cip1 promoter is associated with reduced p21Cip1 expression in response to depsipeptide treatments in human lung cancer cell line, H719 (Fig. 2D, 3B and C, and 4B). In support of this, depsipeptide-induced p21Cip1 expression was reconstituted in both H719 and H841 cells after treatment with 5-aza-CdR (Fig. 7).
As has been described with other HDAC inhibitors, such as TSA (46, 51) or sodium butyrate (31), we now have shown that depsipeptide can induce p21Cip1 expression through Sp1-binding sites (Fig. 5B). This led us to investigate whether methylation occurring within the Sp1-binding sites directly influences Sp1/Sp3 binding. Unexpectedly, however, methylation at the CG dinucleotide within the consensus Sp1-3-binding site did not interfere with Sp1/Sp3 binding in our study (Fig. 6C and D). In fact, several earlier reports showed that methylation of cytosine within the consensus Sp1-binding sites is not associated with decreased Sp1 binding and gene repression. Holler et al. (20) performed EMSA experiments to compare Sp1-binding ability by using methylated or unmethylated consensus Sp1-binding element and found that neither binding in vitro nor transcription in vivo and in vitro are affected by methylation status in HeLa nuclear extracts. Ohtani-Fujita et al. (33) also investigated the influence of methylation in the binding sites of several transcription factors in the RB promoter and found that methylation in the ATF-like site and the RBF-1 site was significantly correlated with a decrease in their binding but not in the Sp1 site. However, methylated cytosines at a different configuration in the consensus Sp1-binding site may induce reduced Sp1 binding and activity. As indicated by Clark et al. (9), methylation of the central CpG dinucleotides on either or both strands had no effect on the efficiency of Sp1 binding, whereas methylation of the outer cytosine of mCpmCpG on Sp1-binding sites inhibited binding almost completely. In addition, Mancini et al. (28) used the EMSA to compare the Sp1-binding ability in the neurofibromatosis gene (NF-1) with variably methylated oligonucleotides. These authors found that methylation occurring at central CpG dinucleotide did not impede Sp1 binding but that Sp1 binding was significantly decreased when the first two cytosines of the Sp1 recognition sequence presented on the antisense strand were methylated.
It is intriguing in the present study that methylation at sites outside of the consensus Sp1-3 element significantly reduced Sp1/Sp3 binding in H719 nuclear extracts (Fig. 6E and F). There did not appear to be a major difference in Sp1/Sp3 binding at this site with respect to methylation of CG sites 14 alone or 14 to 16 together, with ca. 40 to 60% of binding seen whether one or all three sites were methylated upstream of the third Sp1/Sp3 binding element in the p21Cip1 promoter. We note that in the 20 clones evaluated for CG methylation by bisulfite sequencing (Fig. 4B), 40% of site 14, 60% of site 15, and 55% of site 16 CGs were methylated (cumulative of 52%). Whether in vitro EMSA binding would be equally affected by methylation of the closest CG site alone (i.e., site 16) is not clear at this time. The exact mechanism of the methylation-involved reduction in Sp1/Sp3 binding is not clear. However, a relation between mutation or deletion outside of consensus DNA-binding sites and decreased affinity of binding of transcription factors has been reported in several studies (26, 45). For example, wild-type rat androgen receptor interacts with an androgen response element (ARE) as a dimer (26). When mutants outside of the ARE DNA-binding domain were incubated with nuclear extracts, however, the affinity of androgen binding to the ARE was decreased 2.6-fold compared to that of the wild-type sequence as determined by the EMSA methodology. When different mutated elements outside of the ARE for EMSA experiments were examined, it was found that a single nucleotide mutation between two ARE-half sites could abolish the full-length receptor's ability to form a stable complex with DNA. In addition, the Epstein-Barr virus EBNA2 protein is a transcriptional activator and its responsive enhancer in the viral latency C promoter binds two cellular factors: CBF1 and CBF2. To investigate the effect of mutation on CBF2 binding, several oligonucleotides including mutated sequences within or outside the conserved core sequence were designed for determining the affinity of CBF2 to the binding site (15). Surprisingly, mutation outside of the binding site had the greatest effect on CBF2 binding compared with mutation in the core binding sites. Sun et al. (48) also reported that the transcriptional activity of vascular smooth muscle alpha-actin gene is dependent on its 30-bp polypurine-polypyrimidine tract. When elements upstream of the binding site (GGAATG) of its transcriptional enhancer factor 1 (TEF-1) were mutated, TEF-1 binding was greatly impaired (48).
Recently, the possibility that methylation at the boundaries of Sp1 sites affects Sp1 binding has been suggested (27). The 5′-flanking region of cyclin D1 in rat leukemia cell lines was found to be methylated around two continuous Sp1-binding sites in the cyclin D promoter. Methylation that was not within but adjacent to the two Sp1 sites in this promoter significantly influenced cyclin D expression as induced by Sp1 activity. That study indirectly showed that a critical role of methylation at boundaries of Sp1 may interfere with Sp1 binding. Our study, for the first time, shows directly that methylation at CGs outside of consensus Sp1 site reduces Sp1/Sp3 binding when assayed by EMSA experiments (Fig. 6E and 6F). Two additional lines of evidence reported here support the finding that methylation adjacent to transcription factor-binding sites (Sp1/Sp3 in this case) can affect binding and activity. The data in Fig. 4B show that the CG dinucleotides around the Sp1-3 site (from 14th CG to 19th CG) are methylated ca. 56% of the time in asynchronously growing cells. The strongest support of this methylation suppressing p21Cip1 comes from the ability of the DNA methyltransferase inhibitor, 5-aza-CdR to reconstitute depsipeptide responsiveness to p21Cip1 expression (Fig. 7). The data presented in Fig. 7 support the notion that DNA methylation is dominant to histone acetylation status with respect to gene transcription in that it is only after removal of the cytosine methylation that p21Cip1 expression is affected by HDAC inhibitor treatment.
Finally, there are likely to be multiple mechanisms leading to the expression or lack of expression of p21Cip1 after treatment with depsipeptide. First, since depsipeptide is an HDAC inhibitor, it is conceivable that the synergistic effect of depsipeptide and 5-aza-CdR on recovering p21Cip1 expression may result from cooperative inhibition of histone deacetylation and DNA methylation linked through the DNA methyl-binding proteins. The ability of HDAC inhibitors such as depsipeptide to affect transcriptional activity is clearly dependent on the program of DNA methylation at a particular gene promoter, as we have shown here. The results presented here uniquely demonstrate that methylation outside of a transcription factors cognate recognition sequence can affect DNA-binding activity (and presumably functional activity) of the transcription factor and that inhibition of this methylation is correlated with reexpression of the relevant gene. We plan to investigate whether the synergistic effect of HDAC and DNMT inhibitors on p21Cip1 expression is mediated through MBPs. Second, depsipeptide may function on other transcription factors that bind upstream of Sp1-binding sites and through them interact with Sp1 or Sp3 to induce p21Cip1 expression. This hypothesis also needs to be further determined in the future. Nevertheless, in the present study, hypermethylated CpG dinucleotides outside of the consensus Sp1-binding sites significantly inhibits Sp1/Sp3 binding. Therefore, we have shown that hypermethylation-induced inhibition of Sp1/Sp3 binding is one of mechanisms by which p21Cip1 expression in H719 cells was suppressed in response to depsipeptide treatment.
Acknowledgments
We thank Y. Xiong, XH. Pei, T. Sakai, and KK Chan for their materials that are used in this study. We also thank Z. Hu, Y. Zhang, and J. Xiao for technical help. We also express appreciation for helpful discussions with Gustavo Leone.
This research was supported by grant P30 CA16058 from the National Cancer Institute, Bethesda, Md., and by a Translational Research Grant from the V Foundation.
REFERENCES
- 1.Allan, L. A., T. Duhig, M. Read, and M. Fried. 2000. The p21WAF1/CIP1 promoter is methylated in Rat-1 cells: stable restoration of p53-dependent p21WAF1/CIP1 expression after transfection of a genomic clone containing the p21WAF1/CIP1 gene. Mol. Cell. Biol. 20:1291-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alland, L., R. Muhle, H. Hou, Jr., J. Potes, L. Chin, N. Schreiber-Agus, and R. A. DePinho. 1997. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387:49-55. [DOI] [PubMed] [Google Scholar]
- 3.Baylin, S. B., M. Esteller, M. R. Rountree, K. E. Bachman, K. Schuebel, and J. G. Herman. 2001. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 10:687-692. [DOI] [PubMed] [Google Scholar]
- 4.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 5.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 6.Bird, A. P., and A. P. Wolffe. 1999. Methylation-induced repression: belts, braces, and chromatin. Cell 99:451-454. [DOI] [PubMed] [Google Scholar]
- 7.Cameron, E. E., K. E. Bachman, S. Myohanen, J. G. Herman, and S. B. Baylin. 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 21:103-107. [DOI] [PubMed] [Google Scholar]
- 8.Chen, B., L. He, V. H. Savell, J. J. Jenkins, and D. M. Parham. 2000. Inhibition of the interferon-gamma/signal transducers and activators of transcription (STAT) pathway by hypermethylation at a STAT-binding site in the p21WAF1 promoter region. Cancer Res. 60:3290-3298. [PubMed] [Google Scholar]
- 9.Clark, S. J., J. Harrison, and P. L. Molloy. 1997. Sp1 binding is inhibited by (m)Cp(m)CpG methylation. Gene 195:67-71. [DOI] [PubMed] [Google Scholar]
- 10.Comb, M., and H. M. Goodman. 1990. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 18:3975-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross, S. H., J. A. Charlton, X. Nan, and A. P. Bird. 1994. Purification of CpG islands using a methylated DNA binding column. Nat. Genet. 6:236-244. [DOI] [PubMed] [Google Scholar]
- 12.Dai, Z., R. R. Lakshmanan, W. G. Zhu, D. J. Smiraglia, L. J. Rush, M. C. Fruhwald, R. M. Brena, B. Li, F. A. Wright, P. Ross, G. A. Otterson, and C. Plass. 2001. Global methylation profiling of lung cancer identifies novel methylated genes. Neoplasia 3:314-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fournel, M., P. Sapieha, N. Beaulieu, J. M. Besterman, and A. R. MacLeod. 1999. Down-regulation of human DNA-(cytosine-5) methyltransferase induces cell cycle regulators p16ink4A and p21WAF/Cip1 by distinct mechanisms. J. Biol. Chem. 274:24250-24256. [DOI] [PubMed] [Google Scholar]
- 15.Fuentes-Panana, E. M., and P. D. Ling. 1998. Characterization of the CBF2 binding site within the Epstein-Barr virus latency C promoter and its role in modulating EBNA2-mediated transactivation. J. Virol. 72:693-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuks, F., W. A. Burgers, A. Brehm, L. Hughes-Davies, and T. Kouzarides. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 24:88-91. [DOI] [PubMed] [Google Scholar]
- 17.Gartel, A. L., and A. L. Tyner. 1999. Transcriptional regulation of the p21WAF1/CIP1 gene. Exp. Cell Res. 246:280-289. [DOI] [PubMed] [Google Scholar]
- 18.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 19.Herman, J. G., J. R. Graff, S. Myohanen, B. D. Nelkin, and S. B. Baylin. 1996. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 93:9821-9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holler, M., G. Westin, J. Jiricny, and W. Schaffner. 1988. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 2:1127-1135. [DOI] [PubMed] [Google Scholar]
- 21.Hong, L., G. P. Schroth, H. R. Matthews, P. Yau, and E. M. Bradbury. 1993. Studies of the DNA binding properties of histone H4 amino terminus: thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J. Biol. Chem. 268:305-314. [PubMed] [Google Scholar]
- 22.Hu, Z. Z., L. Zhuang, J. Meng, and M. L. Dufau. 1998. Transcriptional regulation of the generic promoter III of the rat prolactin receptor gene by C/EBPβ and Sp1. J. Biol. Chem. 273:26225-26235. [DOI] [PubMed] [Google Scholar]
- 23.Iguchi-Ariga, S. M., and W. Schaffner. 1989. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 3:612-619. [DOI] [PubMed] [Google Scholar]
- 24.Jones, P. A., and D. Takai. 2001. The role of DNA methylation in mammalian epigenetics. Science 293:1068-1070. [DOI] [PubMed] [Google Scholar]
- 25.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCp2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 26.Kallio, P. J., J. J. Palvimo, M. Mehto, and O. A. Janne. 1994. Analysis of androgen receptor-DNA interactions with receptor proteins produced in insect cells. J. Biol. Chem. 269:11514-11522. [PubMed] [Google Scholar]
- 27.Kitazawa, S., R. Kitazawa, and S. Maeda. 1999. Transcriptional regulation of rat cyclin D1 gene by CpG methylation status in promoter region. J. Biol. Chem. 274:28787-28793. [DOI] [PubMed] [Google Scholar]
- 28.Mancini, D. N., S. M. Singh, T. K. Archer, and D. I. Rodenhiser. 1999. Site-specific DNA methylation in the neurofibromatosis (NF1) promoter interferes with binding of CREB and SP1 transcription factors. Oncogene 18:4108-4119. [DOI] [PubMed] [Google Scholar]
- 29.Marks, P., R. A. Rifkind, V. M. Richon, R. Breslow, T. Miller, and W. K. Kelly. 2001. Histone deacetylases and cancer: causes and therapies. Nat. Rev. Cancer 1:194-202. [DOI] [PubMed] [Google Scholar]
- 30.Milutinovic, S., J. D. Knox, and M. Szyf. 2000. DNA methyltransferase inhibition induces the transcription of the tumor suppressor p21WAF1/CIP1/sdi1. J. Biol. Chem. 275:6353-6359. [DOI] [PubMed] [Google Scholar]
- 31.Nakano, K., T. Mizuno, Y. Sowa, T. Orita, T. Yoshino, Y. Okuyama, T. Fujita, N. Ohtani-Fujita, Y. Matsukawa, T. Tokino, H. Yamagishi, T. Oka, H. Nomura, and T. Sakai. 1997. Butyrate activates the WAF1/Cip1 gene promoter through Sp1 sites in a p53-negative human colon cancer cell line. J. Biol. Chem. 272:22199-22206. [DOI] [PubMed] [Google Scholar]
- 32.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-389. [DOI] [PubMed] [Google Scholar]
- 33.Ohtani-Fujita, N., T. Fujita, A. Aoike, N. E. Osifchin, P. D. Robbins, and T. Sakai. 1993. CpG methylation inactivates the promoter activity of the human retinoblastoma tumor-suppressor gene. Oncogene 8:1063-1067. [PubMed] [Google Scholar]
- 34.Periyasamy, S., S. Ammanamanchi, M. P. Tillekeratne, and M. G. Brattain. 2000. Repression of transforming growth factor-beta receptor type I promoter expression by Sp1 deficiency. Oncogene 19:4660-4667. [DOI] [PubMed] [Google Scholar]
- 35.Plass, C., and P. D. Soloway. 2002. DNA methylation, imprinting, and cancer. Eur. J. Hum. Genet. 10:6-16. [DOI] [PubMed] [Google Scholar]
- 36.Rajgolikar, G., K. K. Chan, and H. C. Wang. 1998. Effects of a novel antitumor depsipeptide, FR901228, on human breast cancer cells. Breast Cancer Res. Treat. 51:29-38. [DOI] [PubMed] [Google Scholar]
- 37.Robertson, K. D., S. Ait-Si-Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1, and HDAC1, and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338-342. [DOI] [PubMed] [Google Scholar]
- 38.Robertson, K. D., and P. A. Jones. 2000. DNA methylation: past, present, and future directions. Carcinogenesis 21:461-467. [DOI] [PubMed] [Google Scholar]
- 39.Roman-Gomez, J., J. A. Castillejo, A. Jimenez, M. G. Gonzalez, F. Moreno, C. Rodriguez Mdel, M. Barrios, J. Maldonado, and A. Torres. 2002. 5′ CpG island hypermethylation is associated with transcriptional silencing of the p21CIP1/WAF1/SDI1 gene and confers poor prognosis in acute lymphoblastic leukemia. Blood 99:2291-2296. [DOI] [PubMed] [Google Scholar]
- 40.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 41.Rountree, M. R., K. E. Bachman, J. G. Herman, and S. B. Baylin. 2001. DNA methylation, chromatin inheritance, and cancer. Oncogene 20:3156-3165. [DOI] [PubMed] [Google Scholar]
- 42.Sherr, C. J. 2000. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 60:3689-3695. [PubMed] [Google Scholar]
- 43.Shin, J. Y., H. S. Kim, J. Park, J. B. Park, and J. Y. Lee. 2000. Mechanism for inactivation of the KIP family cyclin-dependent kinase inhibitor genes in gastric cancer cells. Cancer Res. 60:262-265. [PubMed] [Google Scholar]
- 44.Shiohara, M., K. Koike, A. Komiyama, and H. P. Koeffler. 1997. p21WAF1 mutations and human malignancies. Leuk. Lymphoma 26:35-41. [DOI] [PubMed] [Google Scholar]
- 45.Shuman, S., and J. Turner. 1993. Site-specific interaction of vaccinia virus topoisomerase I with base and sugar moieties in duplex DNA. J. Biol. Chem. 268:18943-18950. [PubMed] [Google Scholar]
- 46.Sowa, Y., T. Orita, S. Minamikawa-Hiranabe, T. Mizuno, H. Nomura, and T. Sakai. 1999. Sp3, but not Sp1, mediates the transcriptional activation of the p21/WAF1/Cip1 gene promoter by histone deacetylase inhibitor. Cancer Res. 59:4266-4270. [PubMed] [Google Scholar]
- 47.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 48.Sun, S., E. S. Stoflet, J. G. Cogan, A. R. Strauch, and M. J. Getz. 1995. Negative regulation of the vascular smooth muscle alpha-actin gene in fibroblasts and myoblasts: disruption of enhancer function by sequence-specific single-stranded-DNA-binding proteins. Mol. Cell. Biol. 15:2429-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tate, P. H., and A. P. Bird. 1993. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 3:226-231. [DOI] [PubMed] [Google Scholar]
- 50.Wade, P. A., P. L. Jones, D. Vermaak, G. J. Veenstra, A. Imhof, T. Sera, C. Tse, H. Ge, Y. B. Shi, J. C. Hansen, and A. P. Wolffe. 1998. Histone deacetylase directs the dominant silencing of transcription in chromatin: association with MeCP2 and the Mi-2 chromodomain SWI/SNF ATPase. Cold Spring Harbor Symp. Quant. Biol. 63:435-445. [DOI] [PubMed] [Google Scholar]
- 51.Xiao, H., T. Hasegawa, and K. Isobe. 2000. p300 collaborates with Sp1 and Sp3 in p21waf1/cip1 promoter activation induced by histone deacetylase inhibitor. J. Biol. Chem. 275:1371-1376. [DOI] [PubMed] [Google Scholar]
- 52.Xiong, Z., and P. W. Laird. 1997. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 25:2532-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young, J. I., and J. R. Smith. 2001. DNA methyltransferase inhibition in normal human fibroblasts induces a p21-dependent cell cycle withdrawal. J. Biol. Chem. 276:19610-19616. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Y., H. H. Ng, H. Erdjument-Bromage, P. Tempst, A. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu, W. G., Z. Dai, H. Ding, K. Srinivasan, J. Hall, W. Duan, M. A. Villalona-Calero, C. Plass, and G. A. Otterson. 2001. Increased expression of unmethylated CDKN2D by 5-aza-2′-deoxycytidine in human lung cancer cells. Oncogene 20:7787-7796. [DOI] [PubMed] [Google Scholar]
- 56.Zhu, W. G., R. R. Lakshmanan, M. D. Beal, and G. A. Otterson. 2001. DNA methyltransferase inhibition enhances apoptosis induced by histone deacetylase inhibitors. Cancer Res. 61:1327-1333. [PubMed] [Google Scholar]