Abstract
A deficit in the Jerky protein in mice causes recurrent seizures reminiscent of temporal lobe epilepsy. Jerky is present in mRNA particles in neurons. We show that the N-terminal 168 amino acids of Jerky are necessary and sufficient for mRNA binding. The binding domain is similar to the two tandemly arranged homeodomain-like helix-turn-helix DNA binding motifs of centromere binding protein B. The putative helix-turn-helix motifs of Jerky can also bind double-stranded DNA and represent a novel mammalian RNA/DNA binding domain. Microarray analysis identified mRNAs encoding proteins involved in ribosome assembly and cellular stress response that specifically bound to the RNA binding domain of Jerky both in vitro and in vivo. These data suggest that epileptogenesis in Jerky-deficient mice most likely involves pathways associated with ribosome biogenesis and neuronal survival and/or apoptosis.
It was previously reported that deletion of the jerky gene in mice causes recurrent limbic seizures (9, 54). The manifestation and course of the seizure abnormality in Jerky-deficient mice are reminiscent of familiar temporal lobe epilepsy, an inherited and relatively benign form of temporal lobe epilepsy (3, 4, 7, 13, 18, 36, 37, 42). A de novo nonconservative mutation to a potential glycosylation site in the human homologue of jerky (JRK/JH8) in an epileptic patient was recently described (29), but a pathogenic role for this mutation in seizures has not been established yet.
Although jerky mRNA is detectable in various organs in the mouse, expression of the Jerky protein is restricted to the central nervous system (26). Jerky is found in both the nuclei and the cytoplasms of neurons. In the cytoplasm, Jerky is present in messenger ribonucleoprotein (mRNP) complexes and is not associated with ribosomes or actively translating mRNAs (26). These data suggest that Jerky may regulate the availability of mRNAs to the translational machinery in neurons. Jerky not only is part of mRNP complexes but also binds mRNAs directly with high affinity.
Besides Jerky, the absence of fragile X protein, another mRNA binding protein, can also cause seizures in human and mouse (8, 15, 31, 32, 55, 56). This protein regulates the polyribosome association of target mRNAs and is believed to control their translation (5, 12, 21, 23, 25, 44). These data suggest that a deficit in mRNA binding proteins may represent a novel disease mechanism of epilepsy.
To learn more about possible mechanisms that cause or increase the vulnerability to epileptic seizures in Jerky-deficient mice, we mapped the RNA binding domain of Jerky and identified mRNA species bound to the protein. Sequence analysis showed that the RNA binding domain of Jerky is similar to the DNA binding domain of centromere binding protein B (CENP-B). Structural studies revealed that the DNA binding domain of CENP-B consists of two helix-turn-helix (HTH) repeats. Jerky interacts with a large number of brain mRNAs, and the highest-affinity mRNA targets of Jerky could be clustered into functional groups.
MATERIALS AND METHODS
Generation of jerky deletion constructs and production of truncated GST-Jerky fusion proteins.
N-terminal deletion mutants of Jerky were produced by PCR using pCMV2-jerky (26) as a template. 5′ primers containing an EcoRI site corresponded to residues 49 to 54, 119 to 124, and 169 to 174, while the 3′ primer corresponded to the final six amino acids of Jerky. For C-terminal deletions, a primer corresponding to the first six amino acids of Jerky was used in conjunction with 3′ primers containing sequences corresponding to residues 113 to 118 and 163 to 168. Internal deletions were produced with 5′ primers corresponding to residues 49 to 54 and 119 to 124 and 3′ primers corresponding to residues 113 to 118 and 163 to 168. The EcoRI sequence on the 5′ primers allowed the cloning of various mutants in frame with the sequence encoding glutathione S-transferase (GST). PCR fragments were first cloned into the TOPO-PCR2.1 vector (Invitrogen, Carlsbad, Calif.) followed by recloning via EcoRI sites into pGEX-6P2 (Amersham, Piscataway, N.J.). The sequences of all mutant plasmids were confirmed by sequencing. Expression of mutant proteins was induced in BL21 Escherichia coli cells by 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). GST-Jerky was purified as described by Liu et al. (26).
RNA and DNA labeling and nucleic acid binding assays.
mRNA (1 μg; Clontech, Palo Alto, Calif.) was 3′ end labeled by using T4 RNA ligase in the presence of [32P]pCp (3,000 Ci/mmol; 10 mCi/ml) at 4°C overnight. For DNA labeling, mouse genomic DNA was first isolated from mouse embryonic stem cells by using DNAzol (Invitrogen). Then DNA was digested with Sau3A followed by labeling with Klenow polymerase in the presence of [α-32P]dATP (3,000 Ci/mmol; 10 mCi/ml) at 37°C for 30 min. In some experiments, mRNA and cut DNA were preselected with GST-Jerky (see below) before labeling. To obtain RNA and DNA probes with identical specific activities, labeled and unlabeled RNA and DNA were mixed. Labeled RNA and DNA (106 cpm) were used to hybridize nitrocellulose blots containing various amounts of purified Jerky, Jerky1-168, and Jerky169-557 as described by Liu et al. (26). Filter binding assays with Jerky proteins were carried out as described by Liu et al. (26).
Selection of mRNAs with Jerky and oligonucleotide microarray analysis.
Mouse brain mRNA (1 μg; Clontech) was incubated with 0.5 μg of GST-Jerky or GST-Jerky169-557 immobilized on agarose beads for 20 min in RNA binding buffer (50 mM LiCl, 10 mM Tris [pH 7.5], 1 mM EDTA) containing 2 μg of yeast tRNA at room temperature. Beads were washed five times with RNA binding buffer for 15 min each. Bound mRNA was then recovered by phenol-chloroform extraction followed by ethanol precipitation in the presence of 1 μl of glycogen (20 mg/ml). As internal controls, four Bacillus subtilis RNAs (dap, phe, thr, and trp) were synthesized from pGIBS-dap, -phe, -thr, and -trp plasmids (ATCC, Manassas, Va.) by using a MEGAscript T3 high-yield transcription kit (Ambion, Austin, Tex.). One microgram of input brain mRNA or Jerky-selected mRNAs derived from 1 μg of brain mRNA was mixed with 2 × 10−3 pmol of each of the four B. subtilis poly(A) RNAs, and cRNA was generated according to the manufacturer's instructions (Affymetrix, Santa Clara, Calif.). Briefly, double-stranded cDNA was synthesized with the SuperScript Choice system (Invitrogen) by using an oligo(dT) primer containing T7 polymerase promoter sequences (Genset, San Diego, Calif.). Next, the double-stranded cDNA was used as a template to synthesize cRNA with a BioArray high-yield RNA transcript-labeling kit (Enzo Diagnostics, Farmingdale, N.Y.) in the presence of biotinylated UTP and CTP. Purified (RNeasy minikit; Qiagen, Valencia, Calif.) and partially hydrolyzed cRNA was used to probe Affymetrix murine genome U74A arrays (MG-U74Av2) in a GeneChip hybridization oven and fluidics station. These experiments were repeated with three independently isolated and prepared pools of selected mRNAs. A high-resolution image of the hybridization pattern was obtained in a Hewlett-Packard GeneArray scanner with Microarray Suite software 5.0 (Affymetrix). Signals corresponding to individual mRNAs were normalized to the signal of the four spiked bacterial RNAs.
In vivo pulldown and Atlas arrays.
Jerky-containing complexes were pulled down from human HEK293 and mouse Neuro-2A cells. Transfection with Jerky-expressing constructs was carried out with Lipofectamine (Invitrogen) and Superfect (Qiagen) for HEK293 and Neuro-2A cells (109), achieving approximately 50 and 3% transfection rates, respectively. Two days later, cells were harvested and washed three times with 10 volumes of phosphate-buffered saline. Cells were then lysed mildly with lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 30 mM EDTA, 0.5% Triton X-100) containing complete protease inhibitor cocktail for 45 min on ice. Nuclei were pelleted at 3,000 × g for 10 min at 4°C. The cytoplasmic supernatant was precleared for 1 h with 250 μl of protein A agarose. After centrifugation, the supernatant was cleared again with 200 μl of anti-Flag M1 agarose beads for 20 min (Sigma, St. Louis, Mo.). Following centrifugation, the supernatant was immunoprecipitated with 200 μl of anti-Flag M2 agarose bead (Sigma) for 3 h. The immunoprecipitated material was recovered by centrifugation and washed twice with 1 ml of wash buffer (50 mM Tris [pH 7.5], 150 mM NaCl) for 15 min at 4°C. Then the material was washed again with the wash buffer containing 50 U of RNase-free DNase I (Roche, Indianapolis, Ind.) in the presence of 200 U of RNasin (Promega, Madison, Wis.). Finally, the immunoprecipitated material was washed with wash buffer containing 200 U of RNasin and pelleted. Ten percent of the pellet was used for protein analysis. To recover the RNA content from the complexes, the rest of the pellet was first incubated with lysis buffer containing 100 μg of proteinase K (Life Technologies, Gaithersburg, Md.) and 200 U of RNasin at 37° for 15 min, followed by phenol-chloroform extraction, and the mRNA was ethanol precipitated. RNA was used in reverse transcription (RT)-PCR experiments (see below).
Human mRNA isolated from HEK293 Jerky immunocomplexes was also used in RT reactions to generate probes for Atlas nylon cDNA arrays (Atlas 1.2; Clontech). A mixture of specific primers corresponding to the Atlas array and provided by Clontech was used for the labeling. Arrays were hybridized in ExpressHyb solution (Clontech) at 68°C overnight and washed four times with a solution of 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 1% sodium dodecyl sulfate (SDS) at 68°C for 30 min each, then with a solution of 0.1× SSC and 0.5% SDS at 68°C for 30 min, and finally with 2× SSC for 5 min at room temperature.
Quantitative RT-PCR assays.
One-step real-time RT-PCR was performed with human and mouse mRNAs isolated from Jerky complexes. Reactions were carried out in an Opticon (MJ Research, Incline Village, Nev.) cycler with SYBR-Green I (Molecular Probes, Eugene, Oreg.) by using Superscript one-step RT-PCR Platinum Taq (Invitrogen) and gene-specific primers. Human gene-specific primer pairs were as follows: P1, 5′-CATCTACTCGGCCCTCATTC-3′ and 5′-AGACCAAAGCCCATGTCATC-3′; S100A, 5′-AGGGAGGCTGGAGATCATTT-3′ and 5′-CAGAGTGCTGAACCCAGGA-3′; GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-CGAGATCCCTCCAAAATCAA-3′ and 5′-TGTGGTCATGAGTCCTTCCA-3′; L27A, 5′-TAATGCTGGTGGTCTGCATC-3′ and 5′-TGCTGAAGAATTTGGCCTTC-3′; S11, 5′-CACCCAAGGAGGCTATTGAG-3′ and 5′-GTGACCTTGAGCACGTTGAA-3′; SOD1 (superoxide dismutase), 5′-GAAGGTGTGGGGAAGCATTA-3′ and 5′-ACCTTTGCCCAAGTCATCTG-3′; actin, 5′-AGACCTGTACGCCAACACAG-3′ and 5′-GCGCAAGTTAGGTTTTGTCA-3′; tubulin, 5′-CCTACTGCATCGACAACGAG-3′ and 5′-TTCTTGGCATCGAACATCTG-3′; GNBR (guanine nucleotide binding protein β1), 5′-TACACCACCAACAAGGTCCA-3′ and 5′CAGTGTGTCCGGTAAACGTG-3′. Mouse gene-specific primer pairs were as follows: P1, 5′-ATCTACTCCGCCCTCATCCT-3′ and 5′-ATGAGGCTCCCAATGTTGAC-3′; GAPDH, 5′-CACTGAGCATCTCCCTCACA-3′ and 5′-GTGGGTGCAGCGAACTTTAT-3′; S11, 5′-ACTGGTAACGTCTCCATCCG-3′ and 5′-CCTGCACTCTCCAACTGTGA-3′; SOD1, 5′-TCTAAGAAACATGGTGGCCC-3′ and 5′-GTTTACTGCGCAATCCCAAT-3′; actin, 5′-GCTCTTTTCCAGCCTTCCTT-3′ and 5′-TGATCCACATCTGCTGGAAG-3′; karyopherin, 5′-CCTGAGGCTTGGAGAACAAG-3′ and 5′-TGCTTCAAAGCTGGAAACCT-3′; GNBR, 5′-GCGGGACACACAGGTTATCT-3′ and 5′-GAAGCATCACAAGCACCAGA-3′. PCR conditions and primers were optimized to produce single DNA products ranging from 100 to 400 bp, as demonstrated by melting curves and visualization in agarose gels. Calculations of threshold cycle and difference were based on the method described in the manual for the ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, Calif.).
RESULTS
The N-terminal 168 residues of Jerky are necessary and sufficient for mRNA binding.
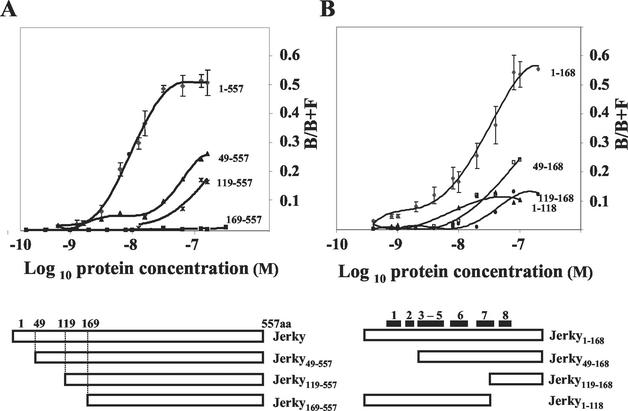
It has been shown that Jerky binds mRNAs with high affinity (26). However, the actual sequence responsible for RNA binding remained elusive, as Jerky contains no known RNA binding domain. Here we used N-terminal deletion mutants to map the RNA binding domain of Jerky. The purity of the various GST-Jerky preparations following affinity purification was assessed on Coomassie blue-stained SDS-polyacrylamide gels, which showed single bands corresponding to the fusion proteins (data not shown). In binding assays, increasing amounts of GST-Jerky was incubated with labeled RNA generated from a pool of mRNAs preselected with Jerky (see Materials and Methods). RNA-protein complexes (bound fraction [B]) were retained on nitrocellulose membranes, while unbound (free [F]) RNA was captured by nylon membranes. As described previously (26), Jerky bound RNA with a low nanomolar affinity (Kd, 8 nM) (Fig. 1A). Removal of the first 48 residues from the N terminus of the 557-amino-acid Jerky resulted in a dramatic decrease in both affinity (Kd) and the amount of the bound fraction of RNA (B/B+F) (Fig. 1A). Nevertheless, Jerky49-557 still bound RNA, as did Jerky119-557, and a complete loss of RNA binding was seen only when 168 residues were removed from the N terminus of Jerky (Jerky169-557) (Fig. 1A). This experiment demonstrated that the first 168 residues in Jerky are necessary for mRNA binding.
FIG. 1.
Identification of the RNA binding domain in Jerky. Filter binding assays were performed with labeled mouse brain mRNA fragments and various Jerky mutants. (A) Mapping of the RNA binding domain in Jerky; (B) dissection of the RNA binding domain of Jerky. The results for full-length Jerky and Jerky1-168 are from four independent experiments, three of them performed in duplicate. Data for the other Jerky mutants are from two independent measurements. Black boxes represent the putative α-helices identified in CENP-B (51).
Figure 1B shows that the 168 N-terminal residues of Jerky are not only necessary but also sufficient for binding. Although the binding affinity of Jerky1-168 was somewhat lower than that of full-length Jerky (20 versus 8 nM), they bound a similar amount of RNA (B/B+F = 0.5 to 0.6). Further dissection of the 1-168 domain again revealed the importance of the first 48 residues, as both the Kd and total binding of Jerky49-168 were reduced compared to those of Jerky1-168 (Fig. 1B). Removal of an additional portion of the N terminus (Jerky119-168) further reduced both the affinity and total binding (Fig. 1B), as was seen with a similar deletion on the full-length protein (Jerky119-557) (Fig. 1A). Since the 119-168 polypeptide still bound mRNA fragments, we tested whether deletion of residues between 119 and 168 has a negative effect on the binding of Jerky1-168. Indeed, Jerky1-118 bound less RNA than Jerky1-168 (Fig. 1B). Taken together, these data demonstrated that all three subfragments (residues 1 to 48, 49 to 118, and 119 to 168) contribute to the high-affinity RNA binding of Jerky1-168.
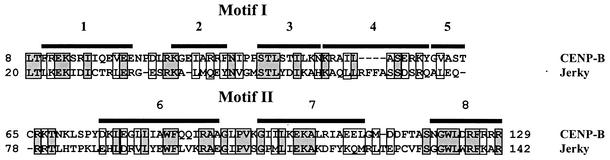
As Fig. 2 shows, there is a high level of similarity between Jerky and CENP-B at their N termini. Since the N terminus of CENP-B is responsible for DNA binding, it was surprising that a homologous region in Jerky serves as an RNA binding domain. Figure 2 also shows the two homeodomain-like HTH structures within the N-terminal 129 residues of CENP-B that are both required for DNA binding (51). The first motif, besides the three canonical helices of homeodomain-like HTHs, contains two additional helices (helices 4 and 5) which are directly adjacent to helix 3. The second domain consists of three helices (helices 6 to 8) where the HTH is formed by helices 7 and 8 (51). There is a significant homology between Jerky and CENP-B within these helices (Fig. 2) suggesting that Jerky has a homeodomain-like HTH structure at its N terminus. Alignment of the two putative HTH sequences in Jerky with deletion mutants showed that the three subfragments (residues 1 to 48, 49 to 118 and 119 to 168) of Jerky1-168, which all required for high-affinity binding, map on helices 1 and 2, helices 3 to 7, and helix 8, respectively (Fig. 1). These data suggest that disrupting any of the two HTH motifs greatly reduces the overall RNA binding of Jerky.
FIG. 2.
Alignment of the mouse Jerky and human CENP-B sequences at the N-terminal regions of the proteins. Identical and similar amino acids are highlighted with shaded and open boxes, respectively. The two HTH structures in CENP-B (51) are labeled as motif I and II. Black boxes indicate the positions of individual helices.
DNA binding of Jerky.
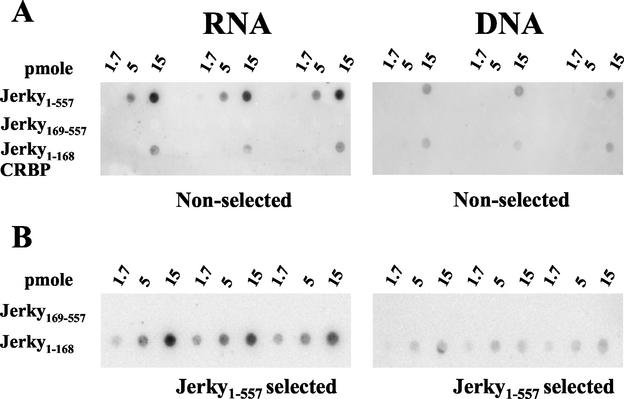
As the Jerky RNA binding domain is homologous to the DNA binding domain of CENP-B, we tested whether Jerky can bind double-stranded DNA (dsDNA). Blots containing triplicate serial dilutions of various Jerky proteins were incubated with radiolabeled RNA and DNA probes with the same specific and total activities. This experiment included full-length Jerky, RNA binding-deficient Jerky (Jerky169-557), the minimal Jerky RNA binding domain (Jerky1-168), and a control protein (cytoplasmic retinoid binding protein [CRBP]) with no RNA binding activity (26). At 15 pmol, both Jerky1-557 and Jerky1-168 interacted with brain mRNAs more strongly than with fragmented mouse DNA (Fig. 3A). At one-third the protein level (5 pmol), only Jerky1-557 incubated with RNA was visible (Fig. 3A). Consistent with previous experiments (Fig. 1), RNA binding of Jerky1-168 was lower than that of Jerky1-557 at equal protein concentrations. Similarly, Jerky1-168 bound less DNA than Jerky1-557. Indeed, DNA binding of Jerky1-168 was barely detectable. Deletion of the N-terminal 168 residues (Jerky169-557) abolished both RNA and DNA binding. To further assess the RNA and DNA binding of Jerky, binding experiments were repeated with Jerky1-557-preselected RNA and DNA species (Fig. 3B). Labeled probes, prepared from preselected RNA and DNA with the same specific and total activities, were incubated with the RNA/DNA binding domain of Jerky (Jerky1-168) and binding-deficient Jerky169-557. Again, RNA binding resulted in a higher signal than DNA binding at identical Jerky1-168 concentrations, and binding was not seen with Jerky169-557. We concluded that Jerky can bind both RNA and DNA, but its binding of mouse mRNAs is stronger than that of fragmented mouse DNA. These data also showed that the same N-terminal 168 residues of Jerky are responsible for both RNA and DNA binding.
FIG. 3.
Jerky binds both RNA and DNA. (A) Interaction of labeled mouse brain mRNA fragments and mouse dsDNA (right panel) with various amounts of GST-Jerky, GST-Jerky169-557, and GST-Jerky1-168 immobilized on nitrocellulose membranes. GST-CRBP was the negative control. (B) Interaction of brain mRNA fragments and mouse dsDNA preselected on GST-Jerky beads with GST-Jerky1-168 and GST-Jerky169-557.
Microarray identification of mRNA species bound to Jerky in vitro.
Next we asked the question of whether Jerky binds specific subsets of mRNAs. Tenenbaum and coworkers (52) recently demonstrated that the mRNA content of mRNP complexes can be determined by cDNA arrays. To identify mRNA targets for Jerky, brain mRNAs were captured by recombinant Jerky and the eluted RNA was used to probe Affymetrix MG-U74Av2 oligonucleotide microarrays. These arrays contain about 6,000 sequences from the Unigene database that have been functionally characterized and approximately 6,000 expressed sequence tag clusters. In parallel, identical MG-U74Av2 arrays were probed with input mRNA not selected by Jerky. To monitor labeling and hybridization, all samples were spiked with predetermined amounts of four different bacterial RNAs. This also allowed the normalization of signals; thus, arrays hybridized with various RNA pools could be directly compared. Microarrays probed with input mRNA showed strong fluorescence consistent with the hybridization of 50.14% of the 12,473 genes present on the array (Fig. 4). When Jerky-selected mRNAs were used to probe arrays, fewer, but still a substantial number (34.0%), of the genes were hybridized. To control for nonspecific binding of mRNAs, a pool of brain mRNAs was also incubated with RNA binding-deficient Jerky (Jerky169-557). Only 104 genes, representing 0.83% of the total, were present on microarrays. Also, the normalized average signal in these arrays was 3.03, a very low value compared to the average signals of 101.4 and 22.88 for arrays hybridized with input and Jerky-associated mRNAs, respectively. These data show that RNA binding-deficient Jerky does not bind a significant number of mRNAs. We concluded that the large majority of mRNAs captured by Jerky1-557 bind to its RNA binding domain.
FIG. 4.
Probing Affymetrix MG-U74Av2 arrays with mouse input brain mRNA and mRNAs selected with the indicated protein.
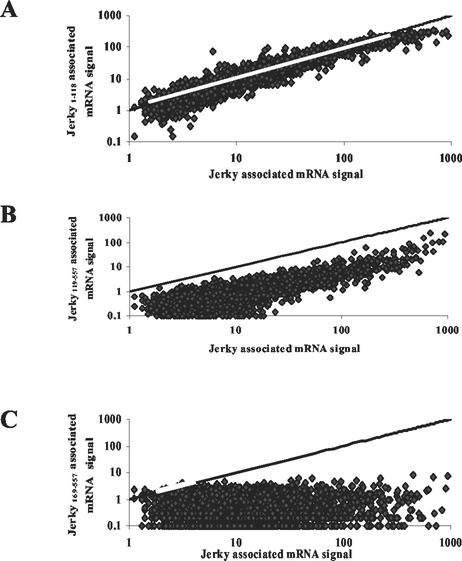
Microarrays were also used to characterize how deletions within the N terminus of Jerky alter the binding of individual mRNA species. For each individual mRNA, signal obtained on arrays probed with Jerky-selected RNA was plotted against the signal measured on arrays probed with mRNA selected with Jerky1-118 and Jerky119-557. As described earlier, these two mutants have reduced binding compared to full-length Jerky (Fig. 1A). Figure 5A shows that Jerky1-118 bound the same species as Jerky1-557, but RNA signals corresponding to this mutant were somewhat lower. Jerky1-118 contains putative helices 1 to 5, which form the first HTH motif, but lacks helix 8, which is required for the second HTH motif (Fig. 1B). Another comparison shows that when this sequence was deleted (Jerky119-557), resulting in a protein with only helix 8 intact, signals corresponding to most mRNA species became too low to be considered positive (signal is generally <1) (Fig. 5B). Nevertheless, species with high signal levels on arrays probed with Jerky-selected RNAs were also bound by this mutant. Finally, as Fig. 5C shows, no individual mRNAs that were bound to RNA binding-deficient Jerky169-557 could be identified by microarray analysis.
FIG. 5.
Microarray analysis of mouse brain mRNAs captured by various Jerky mutants in comparison with mRNAs bound to full-length Jerky. Signal obtained with mRNAs selected with Jerky1-118 (A), Jerky119-557 (B), and Jerky169-557 (C) was plotted against signal corresponding to mRNAs bound by full-length Jerky. The line represents identical signals on the arrays.
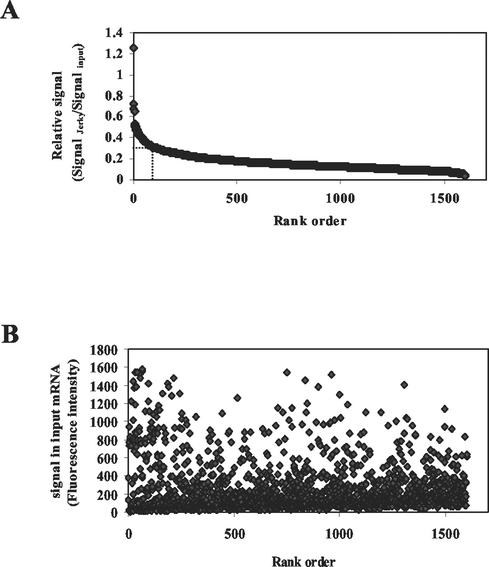
After these initial microarray tests, new sets of brain mRNA samples were prepared and analyzed on three parallel arrays to specify the number and nature of mRNAs that can bind to Jerky. A total of 1,603 genes (12.85%) hybridized with Jerky-selected mRNA in all three arrays. Using triplicate arrays eliminated many genes with low signal levels. Also, tRNA was present during the selection procedure in these experiments to further increase specificity. tRNA does not interfere with overall binding of high-affinity targets but prevents the association of weakly bound mRNAs with Jerky (data not shown). Since signal strength in the selected pool is dependent on both binding and abundance, Jerky-selected mRNA signals were normalized to signals obtained with input RNAs (relative signal) (Fig. 6A). Based on the relative signal, mRNAs were ranked. As Fig. 6A shows, a small portion (114 mRNAs) of the ∼1,600 selected mRNAs had a relative signal of more than 0.3. The large majority of Jerky-bound mRNAs had a low relative signal, 0.05 to 0.2. Importantly, signals measured in the input showed a similar distribution for high-, medium-, and low-ranking mRNAs (Fig. 6B), indicating that top-ranking mRNAs were not biased toward species with high abundance.
FIG. 6.
(A) Distribution of signals normalized to abundance of all mouse brain mRNAs bound to Jerky. Relative signal is displayed as a function of rank in signal strength. The dotted line represents a relative signal of 0.3. A total of 114 mRNAs, including 60 that are functionally characterized, have a relative signal higher than 0.3. (B) Distribution of signal of selected mRNAs in input samples as a function of rank.
Since preferential targets of Jerky presumably have high relative signals, we further analyzed the first 114 highest-ranking targets (relative signal > 0.3), especially the 60 mRNAs which encode proteins with known functions (Table 1). The highest-ranking targets in this group of mRNAs had relative signals of 0.65 to 1.25, clustering around the theoretical maximum of 1.0, indicating that these species are captured very effectively by Jerky. Surprisingly, the majority of the top 60 functionally characterized targets were not neuron or brain specific even when mouse brain mRNA pools were used for selection. Interestingly, mRNAs encoding certain proteins, such as ribosomal proteins (14 mRNAs), cellular-stress-related proteins (7 mRNAs), and cytoskeletal proteins (6 mRNAs), were enriched in the group of preferential targets of Jerky (Table 1).
TABLE 1.
In vitro mRNA targets with a relative signal higher than 0.3 encoding proteins with known functions
| Ranka | Identifier | Gene product | Assignment to functional cluster |
|---|---|---|---|
| 1 | AI840733 | Prostaglandin D2 synthase | |
| 4 | AF110520 | Fas death domain-associated protein | Cellular stress-related/apoptosis associated protein |
| 9 | U29402 | Ribosomal protein, large, P1 | Ribosomal protein |
| 10 | Y12474 | Centrin 3 | |
| 11 | AV329607 | Aldehyde dehydrogenase 2 | |
| 12 | M28729 | α1-Tubulin | Cytoskeletal protein |
| 13 | AV007820 | S100 calcium binding protein A13 | |
| 15 | X52940 | Cytochrome c oxidase, subunit VIIc | Cellular stress-related/apoptosis-associated protein |
| 17 | M32599 | GAPDH | |
| 19 | X05021 | Ribosomal protein L27a | Ribosomal protein |
| 21 | AW045418 | Ribosomal protein L44 | Ribosomal protein |
| 23 | AI851603 | NADH dehydrogenase | |
| 24 | X51703 | Ubiquitin B | |
| 25 | M28727 | α2-Tubulin | Cytoskeletal protein |
| 26 | U93864 | Ribosomal protein S11 | Ribosomal protein |
| 28 | M13441 | α6-Tubulin | Cytoskeletal protein |
| 30 | U11248 | Ribosomal protein S28 | Ribosomal protein |
| 31 | X04591 | Creatine kinase, brain | |
| 33 | M35725 | SOD-1, soluble | Cellular stress-related/apoptosis-associated protein |
| 36 | Y17223 | ATP synthase, H+ transporting | |
| 38 | AA880275 | Metallothionein-I activator | |
| 40 | X80699 | Ribosomal protein L26 | Ribosomal protein |
| 42 | X17320 | Purkinje cell protein 4 | |
| 43 | M29015 | Ribosomal protein L7 | Ribosomal protein |
| 45 | X59382 | Parvalbumin | |
| 47 | U41626 | Split hand/foot deleted gene 1 | |
| 51 | U28917 | Ribosomal protein L13 | Ribosomal protein |
| 53 | X60367 | Retinol binding protein 1, cellular | |
| 54 | V00754 | H3 histone, family 3A | |
| 55 | AF035939 | Mago-nashi homolog | |
| 57 | U59282 | ATP synthase, H+ transporting | |
| 58 | K02928 | Ribosomal protein L30 | Ribosomal protein |
| 60 | AF058955 | Succinate-coenzyme A ligase | |
| 65 | M27844 | Calmodulin 2 | |
| 66 | X52803 | Peptidylprolyl isomerase A | |
| 67 | X53157 | Cytochrome c oxidase, subunit Vb | Cellular stress-related/apoptosis-associated protein |
| 68 | AJ223066 | Fatty acid binding protein 5 | |
| 70 | L10244 | Spermidine/spermine transferase | |
| 73 | AI845819 | Retinoblastoma-binding protein 9 | |
| 75 | X15962 | Ribosomal protein S12 | Ribosomal protein |
| 78 | U62674 | H3 histone, family 2 | |
| 79 | D00466 | Apolipoprotein E | |
| 82 | M12481 | β-Actin, cytoplasmic | Cytoskeletal protein |
| 85 | AI852230 | Mitochondrial ribosomal protein L3 | Ribosomal protein |
| 86 | AW048899 | Ribosomal protein S19 | Ribosomal protein |
| 88 | AF085809 | Synapsin I | |
| 89 | AI851740 | Actin-related protein 2/3 complex | Cytoskeletal protein |
| 91 | AV358770 | Lysophospholipase 1 | |
| 92 | AI852363 | Cytochrome c oxidase, subunit XVII | Cellular stress-related/apoptosis-associated protein |
| 95 | V00835 | Metallothionein 1 | |
| 97 | U08440 | Cytochrome c oxidase, subunit VIa | Cellular stress-related/apoptosis-associated protein |
| 99 | X01756 | Cytochrome c somatic | Cellular stress-related/apoptosis-associated protein |
| 100 | U08354 | Melanocortin 5 receptor | |
| 101 | AF020185 | Dynein, cytoplasmic, light chain 1 | Cytoskeletal protein |
| 102 | X06407 | Translationally regulated transcript | |
| 107 | M93310 | Metallothionein 3 | |
| 109 | AF013715 | Periplakin | |
| 110 | AA666635 | Mitochondrial ribosomal protein L13 | Ribosomal protein |
| 113 | AF043285 | Ribosomal protein S7 | Ribosomal protein |
| 114 | AF100956 | Procollagen, type XI, alpha 2 |
Based on the relative signal on Affymetrix arrays.
Interaction of in vitro-selected target mRNAs with Jerky in vivo.
Selection of mouse mRNAs in the previous experiments was carried out with highly purified recombinant Jerky protein. To determine whether these mRNAs are associated with Jerky in cells, we employed HEK293T cells because they express many of the human homologues of the identified mouse targets and are also amenable to transfection to express Jerky. Jerky was expressed as a Flag-tagged protein to facilitate immunoprecipitation of Jerky-RNA complexes with a monoclonal anti-Flag antibody. Immunoprecipitation with Jerky antibodies was less specific and was not used in these experiments. The presence of Jerky in the immunocomplexes was verified by Western blotting (data not shown). Immunocomplexes were analyzed for the presence of mRNAs by Atlas 1.2 macroarrays.
The Atlas array contained ∼1,000 functionally characterized human genes and a total of 12 genes hybridized with mRNA coimmunoprecipitated with Jerky from HEK293 cells (Table 2). The mouse homologues of three of these mRNAs (α1-tubulin, GAPDH, and SOD1) ranked among the top 60 functionally characterized targets on the Affymetrix arrays (ranked 12, 17, and 33, respectively). The mouse homologue of another Jerky-associated mRNA encoding the small ribosomal protein L13a ranked 420 on the Affymetrix array. The remaining eight genes, which included the small ribosomal protein S9 and others that are involved in cellular stress response and apoptosis, such as heat shock proteins (27 and 90 kDa) and GST isoforms (microsomal and pi), were unique for the Atlas array, and their mouse homologues were not present on the Affymetrix array. However 13 other ribosomal proteins (besides L13a) and 6 other cellular-stress-related proteins (besides SOD) were among the group of preferential targets identified by Affymetrix arrays (Table 1). Also, besides the α1-tubulin mRNA bound to Jerky both in vivo and in vitro, five other mRNAs encoding cytoskeletal proteins were among the in vitro targets (Table 1). Taken together, these data showed that the human homologues of all four Jerky-bound mouse mRNAs that could be tested on Atlas arrays were associated with Jerky in HEK293 cells.
TABLE 2.
Atlas array identification of mRNAs present in Jerky immunocomplexes from transfected HEK293 cells
| GenBank no. | Gene or gene product | Function | Ranka |
|---|---|---|---|
| M13267 | Cytosolic SOD1 | Cellular stress-related protein | 33 |
| X54079 | 27-kDa heat shock protein 1 | Cellular stress-related protein | NA |
| X07270 | 90-kDa heat shock protein A | Cellular stress-related protein | NA |
| L19185 | Peroxiredoxin 2 | Cellular stress-related/apoptosis-associated protein | NA |
| J03746 | Microsomal GST 1 | Cellular stress-related/apoptosis-associated protein | NA |
| X15480 | GST pi | Cellular stress-related/apoptosis-associated protein | NA |
| L22474 | BCL2-associated X protein (BAX beta) | Apoptosis-associated protein | NA |
| X01677 | GAPDH | DNA damage signaling/repair proteins and DNA ligases | 17 |
| U07418 | mutL (E. coli) homolog 1 | DNA damage signaling/repair proteins and DNA ligases | NA |
| X56932 | Ribosomal protein L13a | Ribosomal proteins | 420 |
| U14971 | Ribosomal protein S9 | Ribosomal proteins | NA |
| K00558 | α-Tubulin | Cytoskeletal proteins | 12 |
Rank of the mouse homolog in the Affymetrix array (see Table 1). NA, not applicable (not present on Affymetrix arrays).
Because Atlas arrays contained a limited number of genes, we extended these studies by probing Jerky immunocomplexes pulled down from HEK293 cells for the presence of additional mRNAs by real-time RT-PCR. In these experiments, levels of mRNAs from Jerky and control immunocomplexes were compared to identify mRNAs that specifically associated with Jerky (Table 3). To assess nonspecific association of mRNAs with immunocomplexes, we selected GNBR mRNA, which, despite its high abundance, was not among the Jerky-associated mRNAs on the Affymetrix array. Also, we measured the level of β4-tubulin mRNA, which had a very low relative signal and was among the last of Jerky-selected mRNAs (ranked 1595 of the 1,603 selected mRNAs) (Fig. 6A). As Table 3 shows, there was a <2-fold difference in the level of GNBR and β4-tubulin mRNAs in Jerky and control immunocomplexes, demonstrating that they are not specifically associated with Jerky in cells. In contrast, GAPDH and SOD mRNAs, targets that were among the top 60 targets encoding known proteins and were also shown to be associated with Jerky in HEK293 cells by Atlas arrays, were present at 7- and 10-times-higher levels in Jerky than in control immunocomplexes, respectively. Next we analyzed actin β and three ribosomal protein mRNAs (P1, L27A, and S11) that were among the preferential targets on the Affymetrix array. All of these mRNAs were present in a higher level in Jerky (3.8- to 6.8-fold) than in control immunocomplexes (Table 3). Although S100A mRNA was among the preferential targets, it was not found to be specifically associated with Jerky in HEK293 cells. Combining the Atlas array and RT-PCR results shows that Jerky interacts with most of its in vitro targets (eight of the nine tested) in HEK293 cells. Data also indicate that mouse Jerky can recognize targets regardless of whether they are mouse or human in origin.
TABLE 3.
RT-PCR identification of mRNAs present in Jerky immunocomplexes from transfected HEK293 cells
| Ranka | Gene product | Difference (fold)b |
|---|---|---|
| 9 | P1 | 3.76 |
| 13 | S100 | 1.79 |
| 17 | GAPDH | 7.26 |
| 19 | L27A | 5.57 |
| 26 | S11 | 4.53 |
| 33 | SOD1 | 10.13 |
| 82 | β-Actin | 6.77 |
| 1595 | β4-Tubulin | 1.91 |
| NA | GNBR | 0.83 |
See Table 1. NA, not applicable.
Difference in mRNA levels in Jerky and control immunocomplexes, calculated as 2C(t)vector−C(t)Jerky, where C(t) is the threshold cycle for target application.
Although these data showed that Jerky can recognize its in vitro targets in HEK293 cells, Jerky is a neuronal protein and may form complexes in HEK293 cells that do not exist in neuronal cells. Therefore, we tested Jerky-mRNA association in mouse Neuro-2A cells expressing Flag-tagged mouse Jerky. These mRNAs were mostly chosen from the set tested in the HEK293 pulldown experiments (Table 3) and included five (P1, GAPHD, S11, SOD, and actin) that were among the top-ranking mRNAs selected with Jerky in vitro and two (GNBR and karyopherin) that were not bound to Jerky in vitro (Table 4). To identify mRNAs that specifically interact with the RNA binding domain of Jerky, neuronal cells expressing Flag-Jerky169-557 were used as a control (rather than cells expressing no Jerky) in these experiments. Four (P1, GAPDH, S11, and SOD) of the five mRNAs that were among the top in vitro targets were present at 4.2- to 8.7-times-higher levels in Jerky than in Jerky169-557 immunocomplexes. Interestingly, mRNAs with an 8.2- to 8.7-fold difference in Jerky and Jerky168-557 immunocomplexes (P1, GAPDH, and S11) were on the very top of the target list (rank, 9 to 26), while SOD1, with a 4.2-fold difference, had a rank of 33. Moreover, actin mRNA, whose rank was the lowest (i.e., 82), showed no specific binding to Jerky. The difference in mRNA levels in Jerky/Jerky169-557 immunocomplexes for the controls (GNBR and karyopherin) was within twofold and cannot be regarded as significant. We concluded that Jerky interacts with in vitro-identified target mRNAs in neuronal cells. In fact, there was a good correlation between the in vitro target rank and the specificity of the association in Neuro-2A cells.
TABLE 4.
RT-PCR identification of mRNAs present in Jerky immunocomplexes from transfected Neuro-2A cells
| Ranka | Gene product | Difference (fold)b |
|---|---|---|
| 9 | P1 | 8.663 |
| 17 | GAPDH | 8.663 |
| 26 | S11 | 8.281 |
| 33 | SOD1 | 4.192 |
| 82 | β-Actin | 1.061 |
| NA | GNBR | 2.000 |
| NA | Karyopherin | 1.610 |
See Table 1. NA, not applicable.
Relative difference in mRNA levels in Jerky and Jerky169-557 immunocomplexes, calculated as 2[C(t)vector−C(t)Jerky]/2[C(t)vector−C(t)Jerky169-557], where C(t) is the threshold cycle for target application.
DISCUSSION
The RNA binding domain of Jerky is homologous to HTH motifs found in CENP-B.
Although Jerky binds RNAs with high affinity, none of the well-known RNA binding motifs could be located within its sequence. Well-characterized RNA binding domains include the RNA recognition motif, with a consensus sequence of (L/R)G(F/Y)(G/A)FVX(F/Y) (1, 20, 50), the arginine-rich motif (24), the K homology motif, with a core sequence of VIGXXGXXI (46), and the RGG box, which contains between 6 and 18 copies of closely spaced Arg-Gly-Gly (RGG) repeats (22). Instead of these domains, our mapping revealed that two HTH-like motifs, located at the N-terminal 168 residues, are responsible for the high-affinity RNA binding property of Jerky.
The HTH structure of the Jerky RNA binding domain is inferred from its high sequence similarity to the N terminus of CENP-B. The HTH structure of CENP-B1-56 was first revealed by nuclear magnetic resonance (19). The CENP-B HTH structure could be superimposed on the Fushi tarazu homeodomain and the homeodomain-like structures found in Myb and RAP1 (19). Homeodomains contain three helical regions that are folded into a compact globular structure. Helices 1 and 2 lie parallel to each other and across from the third helix (2). Further, X-ray crystallography studies by Tanaka et al. (51) showed tandemly arranged two-HTH structures in CENP-B1-129. This sequence shows high homology with the N-terminal 141 residues of Jerky. Since deletion analysis showed that the first 168 residues of Jerky are required and sufficient for mRNA binding, these data suggest that a tandem repeat of two HTH structures serves as the RNA binding domain of Jerky. Additional deletion mutants demonstrated that both of these putative HTH structures in Jerky have to be intact for high-affinity RNA binding. Database searches with the putative Jerky HTH motifs identified, in addition to CENP-B, six mammalian genes, indicating that such a structure can be found in a family of proteins (W. Liu and M. Toth, unpublished data). It may be predicted that Jerky-like proteins also bind RNA and that the Jerky family of proteins represents a novel group of RNA binding proteins.
HTH structures, including homeodomains and the homeodomain-like motifs of Myb and RAP1, are primarily known for their dsDNA binding. However, the homeodomain of the Drosophila Bicoid protein has recently been recognized as a functional RNA binding motif (11, 40). Although not entirely composed of α-helices as are homeodomains and homeodomain-like structures, the RNA binding domains of E1AV-TAT and the ribosomal protein L11 can also fold into HTH-like structures (41).
Although Jerky, similar to Bicoid, binds RNA and may assume an HTH structure, there are differences between these two proteins. While Jerky binds a large set of mRNAs, Bicoid interacts with only one known species, caudal mRNA (11, 40). Structurally, Bicoid contains a single homeodomain, while Jerky may harbor two adjacent homeodomain-like HTH motifs. To the best of our knowledge, no mammalian HTH structure with the ability to bind RNA has been identified, and the two tandemly arranged homeodomain-like HTH motifs in Jerky may represent a novel mammalian RNA binding domain.
Jerky binds both RNA and DNA.
CENP-B1-129 binds a specific dsDNA sequence called the B box. There is extensive homology between CENP-B and Jerky at their N termini. When tested with either total fragmented genomic DNA or a Jerky-preselected pool of genomic DNA fragments, Jerky showed an interaction which was less robust than its binding to total or Jerky-preselected mRNAs. Once DNA sequences that specifically bind Jerky are identified, it will be possible to determine if their binding affinities reach a level that is biologically relevant.
Data obtained with deletion mutants clearly showed that the 168 residues that are responsible for RNA binding and form the putative tandem HTH structure are also involved in binding dsDNA. Another HTH sequence which binds both DNA and RNA is the Bicoid homeodomain. Bicoid, by binding to specific DNA sequences, transcriptionally activates zygotic segmentation genes such as Hunchback in Drosophila (6, 10, 14, 48).
How is it possible that certain HTH structures can bind both RNA and dsDNA? The DNA binding of HTH can be easily explained by the accommodation of an α-helix, in particular helix 3, into the major groove of the DNA. It has also been shown that the third helix is crucial in binding of RNA by Bicoid (33). However, the major and minor grooves of RNA do not form a smooth surface to interact with α-helices, and the HTH structure may recognize partially unfolded RNA that is presented at specific stem-loop or bulged structures.
Jerky binds specific sets of mRNAs.
Reviewing the target list obtained by Affymetrix arrays indicated the selection of specific sets of mRNAs by Jerky. In particular, mRNAs encoding proteins involved in ribosome biogenesis, cellular stress response and/or apoptosis, and cytoskeleton organization were enriched in the Jerky-selected mRNA pool. Most of the target mRNAs tested in subsequent pulldown experiments demonstrated an interaction with Jerky and specifically with its RNA binding domain in HEK293 and Neuro-2A cells. Enrichment of Jerky targets in Jerky immunocomplexes from Neuro-2A cells was somewhat higher and correlated better with the rank obtained in the in vitro binding. This may be due to potential differences in the protein composition of Jerky complexes formed in neuronal (Neuro-2A) and epithelial (HEK293) cells. In terms of function, Jerky-mRNA interactions in Neuro-2A cells are more relevant, as Jerky is a predominantly neuronal protein. Species differences could have also accounted for some of the differences. Indeed, in both the in vitro binding and Neuro-2A pulldown assays, complexes were formed between mouse Jerky and mouse mRNAs, while in the HEK293 pulldown experiment, the interaction was between mouse Jerky and human mRNAs.
Importantly, the interaction of Jerky with mRNAs encoding ribosomal proteins and proteins linked to cellular stress response and/or apoptosis has been confirmed in Neuro-2A cells. In contrast, the mRNA for the cytoskeletal protein β-actin did not interact with Jerky in these cells. Determining whether mRNAs that encode cytoskeletal proteins are genuine Jerky targets will require testing of additional members of this group of mRNAs in Neuro-2A cells.
Ribosomal proteins are required for ribosome biogenesis, and mRNAs for 14 and 2 of the total of approximately 70 ribosomal proteins were confirmed to be associated with Jerky in vitro and in vivo (in Neuro-2A cells), respectively. Regulating the expression of ribosomal proteins is challenging because the cell needs equimolar amounts of the individual components of the ribosome and ribosomal proteins. While downregulation of these proteins may interfere with translation, overexpression of ribosomal proteins, due to their highly charged nature, could endanger the cell unless they are assembled into ribosomes. Vertebrates utilize translation as a primary means of regulating ribosomal protein synthesis (28), and Jerky, by controlling mRNA accessibility, could be one of the regulatory factors.
A total of 11 targets of Jerky encode proteins involved in cellular stress response and/or apoptosis. Cells are constantly exposed to environmental, physical, or chemical stresses that can induce either a cellular response which promotes survival or apoptosis, characterized by a sequence of regulated events culminating in cell death. One of the key molecules in cell survival or apoptosis is SOD, which accelerates the formation of H2O2 from O2·− (17, 34, 45). The mRNA encoding SOD was associated with Jerky in vitro as well as in HEK293 and Neuro-2A cells. GST mRNAs, whose encoded proteins are believed to exert a critical role in cellular protection against oxidative stress, were also found to be associated with Jerky in vitro and in HEK293 cells. These proteins detoxify a variety of electrophilic compounds, including oxidized lipid, DNA, and catechol products generated by reactive oxygen species (16). Peroxiredoxin, another Jerky-target encoded protein, has similar functions (39). Two heat shock protein mRNAs were also among those that interacted with Jerky in HEK293 cells. It has been shown that heat shock proteins promote neuronal survival by facilitating both recovery and suppression of apoptosis (43). During apoptosis, cytochrome c enzymes are released from the mitochondria (27, 30, 49), and five mRNAs encoding these proteins were bound to Jerky in vitro. Once in the cytosol, cytochrome c activates caspase 9, whose cleavage is followed by activation of downstream caspases and apoptosis (47). Bax, a Bcl-associated protein, has been shown to promote apoptosis (35), and its mRNA was among the Jerky-associated mRNAs in HEK293 cells. Finally, the mRNA for the Fas death domain-associated protein that is associated with Fas receptor and procaspases to form the “death-inducing signaling complex” (38) was found in Jerky complexes isolated from HEK293 cells. The synthesis of these proteins may be altered in Jerky-deficient cells, which in turn could increase the vulnerability of neurons to physical and chemical stressors.
It seems that association of these functionally related mRNAs with Jerky is not coincidental, because targets that could be analyzed were found in association with Jerky both in vitro and in vivo. Also, functionally similar targets were often found on both the Affymetrix and Atlas arrays. Although these data show that Jerky binds to mRNAs within cells, we do not know whether Jerky regulates these targets. It has been reported that Jerky is present in mRNA particles but is absent from ribosomal complexes (26), which suggests that Jerky may regulate the accessibility of bound mRNAs and their utilization by the translational machinery. However, it is also possible that Jerky controls the transport, localization, and/or stability of these mRNAs.
Epilepsy is a heterogeneous disorder, and mutations in many genes can lead to seizure disorders. Although several single-gene mutations in neuronal channels and/or receptors that cause epilepsies have been identified, seizures can be elicited by mutations in other genes whose protein products are not directly involved in membrane excitability (53). These include genes associated with neuronal migration defects and neurodegeneration. Jerky, as an RNA binding protein, may regulate the expression of numerous genes, and its absence could cause defects in several molecular or cellular pathways. The nature of mRNAs associated with Jerky suggests that the most likely pathways that can be affected are ribosome biogenesis and translation and neuronal survival and apoptosis.
Acknowledgments
This work was supported by National Institute of Health grant R01NS34151 (to M.T.), Cancer Pharmacology training grant T32 CA62948 (W.L.), and National Institute on Drug Abuse grant DA 07274 (J.S.).
REFERENCES
- 1.Bandziulis, R. J., M. S. Swanson, and G. Dreyfuss. 1989. RNA-binding proteins as developmental regulators. Genes Dev. 3:431-437. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee-Basu, S., and A. D. Baxevanis. 2001. Molecular evolution of the homeodomain family of transcription factors. Nucleic Acids Res. 29:3258-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkovic, S. F., M. L. Kennerson, R. A. Howell, I. E. Scheffer, P. A. Hwang, and G. A. Nicholson. 1994. Phenotypic expression of benign familial neonatal convulsions linked to chromosome 20. Arch. Neurol. 51:1125-1128. [DOI] [PubMed] [Google Scholar]
- 4.Berkovic, S. F., A. McIntosh, R. A. Howell, A. Mitchell, L. J. Sheffield, and J. L. Hopper. 1996. Familial temporal lobe epilepsy: a common disorder identified in twins. Ann. Neurol. 40:227-235. [DOI] [PubMed] [Google Scholar]
- 5.Brown, V., P. Jin, S. Ceman, J. C. Darnell, W. T. O'Donnell, S. A. Tenenbaum, X. Jin, Y. Feng, K. D. Wilkinson, J. D. Keene, et al.. 2001. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107:477-487. [DOI] [PubMed] [Google Scholar]
- 6.Burz, D. S., R. Rivera-Pomar, H. Jackle, and S. D. Hanes. 1998. Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J. 17:5998-6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cendes, F., I. Lopes-Cendes, E. Andermann, and F. Andermann. 1998. Familial temporal lobe epilepsy: a clinically heterogeneous syndrome. Neurology 50:554-557. [DOI] [PubMed] [Google Scholar]
- 8.Chen, L., and M. Toth. 2001. Fragile X mice develop sensory hyperreactivity to auditory stimuli. Neuroscience 103:1043-1050. [DOI] [PubMed] [Google Scholar]
- 9.Donovan, G. P., C. Harden, J. Gal, L. Ho, E. Sibille, R. Trifiletti, L. J. Gudas, and M. Toth. 1997. Sensitivity to jerky gene dosage underlies epileptic seizures in mice. J. Neurosci. 17:4562-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Driever, W., and C. Nusslein-Volhard. 1989. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature 337:138-143. [DOI] [PubMed] [Google Scholar]
- 11.Dubnau, J., and G. Struhl. 1996. RNA recognition and translational regulation by a homeodomain protein. Nature 379:694-699. [DOI] [PubMed] [Google Scholar]
- 12.Eberhart, D. E., H. E. Malter, Y. Feng, and S. T. Warren. 1996. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum. Mol. Genet. 5:1083-1091. [DOI] [PubMed] [Google Scholar]
- 13.Gambardella, A., D. Messina, E. Le Piane, R. L. Oliveri, G. Annesi, M. Zappia, E. Andermann, A. Quattrone, and U. Aguglia. 2000. Familial temporal lobe epilepsy autosomal dominant inheritance in a large pedigree from southern Italy. Epilepsy Res. 38:127-132. [DOI] [PubMed] [Google Scholar]
- 14.Gao, Q., and R. Finkelstein. 1998. Targeting gene expression to the head: the Drosophila orthodenticle gene is a direct target of the Bicoid morphogen. Dev. Suppl. 125:4185-4193. [DOI] [PubMed] [Google Scholar]
- 15.Guerrini, R., C. Dravet, A. R. Ferrari, A. Battaglia, M. G. Mattei, P. Salvadori, P. Genton, and P. Pfanner. 1993. The evolution of epilepsy in the most common genetic forms with mental retardation (Down's syndrome and the fragile X syndrome). Pediatr. Med. Chiragu 15(Suppl. 1):19-22. [PubMed] [Google Scholar]
- 16.Hayes, J. D., and R. C. Strange. 2000. Glutathione S-transferase polymorphisms and their biological consequences. Pharmacology 61:154-166. [DOI] [PubMed] [Google Scholar]
- 17.Hyslop, P. A., Z. Zhang, D. V. Pearson, and L. A. Phebus. 1995. Measurement of striatal H2O2 by microdialysis following global forebrain ischemia and reperfusion in the rat: correlation with the cytotoxic potential of H2O2 in vitro. Brain Res. 671:181-186. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda, A., T. Kunieda, S. Miyamoto, H. Fukuyama, and H. Shibasaki. 2000. Autosomal dominant temporal lobe epilepsy in a Japanese family. J. Neurol. Sci. 176:162-165. [DOI] [PubMed] [Google Scholar]
- 19.Iwahara, J., T. Kigawa, K. Kitagawa, H. Masumoto, T. Okazaki, and S. Yokoyama. 1998. A helix-turn-helix structure unit in human centromere protein B (CENP-B). EMBO J. 17:827-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenan, D. J., C. C. Query, and J. D. Keene. 1991. RNA recognition: towards identifying determinants of specificity. Trends Biochem. Sci. 16:214-220. [DOI] [PubMed] [Google Scholar]
- 21.Khandjian, E. W., F. Corbin, S. Woerly, and F. Rousseau. 1996. The fragile X mental retardation protein is associated with ribosomes. Nat. Genet. 12:91-93. [DOI] [PubMed] [Google Scholar]
- 22.Kiledjian, M., and G. Dreyfuss. 1992. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 11:2655-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laggerbauer, B., D. Ostareck, E. M. Keidel, A. Ostareck-Lederer, and U. Fischer. 2001. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum. Mol. Genet. 10:329-338. [DOI] [PubMed] [Google Scholar]
- 24.Lazinski, D., E. Grzadzielska, and A. Das. 1989. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell 59:207-218. [DOI] [PubMed] [Google Scholar]
- 25.Li, Z., Y. Zhang, L. Ku, K. D. Wilkinson, S. T. Warren, and Y. Feng. 2001. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 29:2276-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, W., J. Seto, G. Donovan, and M. Toth. 2002. Jerky, a protein deficient in a mouse epilepsy model, is associated with translationally inactive mRNA in neurons. J. Neurosci. 22:176-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matz, P. G., A. Lewen, and P. H. Chan. 2001. Neuronal, but not microglial, accumulation of extravasated serum proteins after intracerebral hemolysate exposure is accompanied by cytochrome c release and DNA fragmentation. J. Cereb. Blood Flow Metab. 21:921-928. [DOI] [PubMed] [Google Scholar]
- 28.Meyuhas, O., D. Avni, and S. Shama. 1996. Translational control of ribosomal protein mRNAs in eukaryotes, p. 363-388. In J. W. B. Hershey, M. B. Mathews, and N. Sonenberg, (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Moore, T., S. Hecquet, A. McLellann, D. Ville, D. Grid, F. Picard, B. Moulard, P. Asherson, A. J. Makoff, D. McCormick, et al. 2001. Polymorphism analysis of JRK/JH8, the human homologue of mouse jerky, and description of a rare mutation in a case of CAE evolving to JME. Epilepsy Res. 46:157-167. [DOI] [PubMed] [Google Scholar]
- 30.Morita-Fujimura, Y., M. Fujimura, M. Kawase, S. F. Chen, and P. H. Chan. 1999. Release of mitochondrial cytochrome c and DNA fragmentation after cold injury-induced brain trauma in mice: possible role in neuronal apoptosis. Neurosci. Lett. 267:201-205. [DOI] [PubMed] [Google Scholar]
- 31.Musumeci, S. A., P. Bosco, G. Calabrese, C. Bakker, G. B. De Sarro, M. Elia, R. Ferri, and B. A. Oostra. 2000. Audiogenic seizures susceptibility in transgenic mice with fragile X syndrome. Epilepsia 41:19-23. [DOI] [PubMed] [Google Scholar]
- 32.Musumeci, S. A., R. J. Hagerman, R. Ferri, P. Bosco, B. Dalla Bernardina, C. A. Tassinari, G. B. De Sarro, and M. Elia. 1999. Epilepsy and EEG findings in males with fragile X syndrome. Epilepsia 40:1092-1099. [DOI] [PubMed] [Google Scholar]
- 33.Niessing, D., W. Driever, F. Sprenger, H. Taubert, H. Jackle, and R. Rivera-Pomar. 2000. Homeodomain position 54 specifies transcriptional versus translational control by Bicoid. Mol. Cell 5:395-401. [DOI] [PubMed] [Google Scholar]
- 34.Olanow, C. W. 1993. A radical hypothesis for neurodegeneration. Trends Neurosci. 16:439-444. [DOI] [PubMed] [Google Scholar]
- 35.Oltvai, Z. N., C. L. Milliman, and S. J. Korsmeyer. 1993. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 74:609-619. [DOI] [PubMed] [Google Scholar]
- 36.Picard, F., S. Baulac, P. Kahane, E. Hirsch, R. Sebastianelli, P. Thomas, F. Vigevano, P. Genton, R. Guerrini, C. A. Gericke, et al. 2000. Dominant partial epilepsies. A clinical, electrophysiological and genetic study of 19 European families. Brain 123:1247-1262. [DOI] [PubMed] [Google Scholar]
- 37.Poza, J. J., A. Saenz, A. Martinez-Gil, N. Cheron, A. M. Cobo, M. Urtasun, J. F. Marti-Masso, D. Grid, J. S. Beckmann, J. F. Prud'homme, and A. Lopez de Munain. 1999. Autosomal dominant lateral temporal epilepsy: clinical and genetic study of a large Basque pedigree linked to chromosome 10q. Ann. Neurol. 45:182-188. [DOI] [PubMed] [Google Scholar]
- 38.Qiu, J., M. J. Whalen, P. Lowenstein, G. Fiskum, B. Fahy, R. Darwish, B. Aarabi, J. Yuan, and M. A. Moskowitz. 2002. Upregulation of the Fas receptor death-inducing signaling complex after traumatic brain injury in mice and humans. J. Neurosci. 22:3504-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rabilloud, T., M. Heller, F. Gasnier, S. Luche, C. Rey, R. Aebersold, M. Benahmed, P. Louisot, and J. Lunardi. 2002. Proteomics analysis of cellular response to oxidative stress. Evidence for in vivo overoxidation of peroxiredoxins at their active site. J. Biol. Chem. 277:19396-19401. [DOI] [PubMed] [Google Scholar]
- 40.Rivera-Pomar, R., D. Niessing, U. Schmidt-Ott, W. J. Gehring, and H. Jackle. 1996. RNA binding and translational suppression by bicoid. Nature 379:746-749. [DOI] [PubMed] [Google Scholar]
- 41.Rosch, P., and D. Willbold. 1996. Is EIAV Tat protein a homeodomain? Science 272:1672.. [DOI] [PubMed] [Google Scholar]
- 42.Saenz, A., J. Galan, C. Caloustian, F. Lorenzo, C. Marquez, N. Rodriguez, M. D. Jimenez, J. J. Poza, A. M. Cobo, D. Grid, et al. 1999. Autosomal dominant nocturnal frontal lobe epilepsy in a Spanish family with a Ser252Phe mutation in the CHRNA4 gene. Arch. Neurol. 56:1004-1009. [DOI] [PubMed] [Google Scholar]
- 43.Samali, A., and S. Orrenius. 1998. Heat shock proteins: regulators of stress response and apoptosis. Cell Stress Chaperones 3:228-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaeffer, C., B. Bardoni, J. L. Mandel, B. Ehresmann, C. Ehresmann, and H. Moine. 2001. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 20:4803-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simonian, N. A., and J. T. Coyle. 1996. Oxidative stress in neurodegenerative diseases. Annu. Rev. Pharmacol. Toxicol. 36:83-106. [DOI] [PubMed] [Google Scholar]
- 46.Siomi, H., M. C. Siomi, R. L. Nussbaum, and G. Dreyfuss. 1993. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell 74:291-298. [DOI] [PubMed] [Google Scholar]
- 47.Slee, E. A., M. T. Harte, R. M. Kluck, B. B. Wolf, C. A. Casiano, D. D. Newmeyer, H. G. Wang, J. C. Reed, D. W. Nicholson, E. S. Alnemri, et al. 1999. Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J. Cell Biol. 144:281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Struhl, G., K. Struhl, and P. M. Macdonald. 1989. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell 57:1259-1273. [DOI] [PubMed] [Google Scholar]
- 49.Sugawara, T., M. Fujimura, Y. Morita-Fujimura, M. Kawase, and P. H. Chan. 1999. Mitochondrial release of cytochrome c corresponds to the selective vulnerability of hippocampal CA1 neurons in rats after transient global cerebral ischemia. J. Neurosci. 19:RC39. [DOI] [PMC free article] [PubMed]
- 50.Swanson, M. S., T. Y. Nakagawa, K. LeVan, and G. Dreyfuss. 1987. Primary structure of human nuclear ribonucleoprotein particle C proteins: conservation of sequence and domain structures in heterogeneous nuclear RNA, mRNA, and pre-rRNA-binding proteins. Mol. Cell. Biol. 7:1731-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka, Y., O. Nureki, H. Kurumizaka, S. Fukai, S. Kawaguchi, M. Ikuta, J. Iwahara, T. Okazaki, and S. Yokoyama. 2001. Crystal structure of the CENP-B protein-DNA complex: the DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA. EMBO J. 20:6612-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tenenbaum, S. A., C. C. Carson, P. J. Lager, J. D. Keene. 2000. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. USA 97:14085-14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toth, M. 2001. RNA binding proteins and epilepsy. Gene Function Dis. 2:95-98. [Google Scholar]
- 54.Toth, M., J. Grimsby, G. Buzsaki, and G. P. Donovan. 1995. Epileptic seizures caused by inactivation of a novel gene, jerky, related to centromere binding protein-B in transgenic mice. Nat. Genet. 11:71-75. [DOI] [PubMed] [Google Scholar]
- 55.Wisniewski, K. E., J. H. French, S. Fernando, W. T. Brown, E. C. Jenkins, E. Friedman, A. L. Hill, and C. M. Miezejeski. 1985. Fragile X syndrome: associated neurological abnormalities and developmental disabilities. Ann. Neurol. 18:665-669. [DOI] [PubMed] [Google Scholar]
- 56.Wisniewski, K. E., S. M. Segan, C. M. Miezejeski, E. A. Sersen, and R. D. Rudelli. 1991. The Fra(X) syndrome: neurological, electrophysiological, and neuropathological abnormalities. Am. J. Med. Genet. 38:476-480. [DOI] [PubMed] [Google Scholar]