Abstract
Regulation of gene transcription by nuclear receptors involves association with numerous coregulators. Receptor-interacting protein 140 (RIP140) is a corepressor that negatively regulates the ligand-induced activity of several nuclear receptors, including the glucocorticoid receptor (GR). In the present study, we have characterized the role of the intranuclear localization of RIP140 in its corepressor activity. In the absence of ligand-activated GR, RIP140 is localized in small nuclear foci targeted by a 40-amino-acid-long sequence. Although the focus-targeting domain overlaps with a binding sequence for the corepressor CtBP (C-terminal binding protein), interaction with CtBP is not involved in the localization. RIP140 foci do not correspond to PML bodies but partly colocalize with domains harboring the corepressor SMRT. Upon ligand binding, GR and RIP140 are redistributed to large nuclear domains distinct from the RIP140 foci. The redistribution requires regions of RIP140 with corepressor activity, as well as the DNA-binding domain of GR. Furthermore, we show that full RIP140 corepressor activity is contributed both by C-terminal receptor-binding LXXLL motifs and interaction with the CtBP corepressor. In conclusion, our results suggest that the corepressor function of RIP140 is multifaceted and involves binding to nuclear receptors, as well as additional functions mediated by the formation and intranuclear relocalization of a repressive protein complex.
Nuclear hormone receptors are ligand-regulated transcription factors that may either activate or repress gene transcription. Transcriptional activation by nuclear receptors is dependent on the recruitment of numerous coregulator proteins to the receptors (for a review, see reference 18). These encompass ATP-dependent chromatin-remodeling proteins, proteins with histone acetyltransferase activity, and protein complexes involved in recruitment of the basal transcriptional machinery, including RNA polymerase II. Ligand-induced gene activation by nuclear receptors has been suggested to constitute a switch from a repressed state that is maintained by receptor interaction with corepressor proteins such as SMRT and NCoR, recruiting histone deacetylases (HDACs), to an activated state induced by coactivator proteins such as p160/SRC-1 proteins that have histone acetylase activity or recruit proteins that do. The C-terminal ligand-binding domain (LBD) of the receptor is the main docking site for the coregulators (1). Many coregulators have specific LXXLL motifs (NR boxes) that are involved in the interaction with the receptor LBDs (7, 13, 24).
Receptor-interacting protein 140 (RIP140), also called nuclear receptor-interacting protein 1 (Nrip1), is a unique coregulator of nuclear receptors (3). It acts mainly as a corepressor reducing gene transcription but, in contrast to other corepressors that interact with unliganded or antagonist-activated receptors, RIP140 interacts with ligand-activated receptors. RIP140 has a restricted number of target proteins that only includes some nuclear receptors and the aryl hydrocarbon receptor (12, 14, 15, 25, 32). Genetic knockout studies with mice showed that the RIP140 protein has important physiological functions, especially in female fertility (31). RIP140 has been suggested to repress receptor activity by competing with binding of coactivators to the receptor (25). However, RIP140 also has an intrinsic repression activity possibly mediated through interaction with HDAC proteins (29, 30) and/or the corepressor CtBP (C-terminal binding protein) (26), which represses gene transcription by an unknown mechanism (5).
We have previously shown that the glucocorticoid receptor (GR), a member of the nuclear receptor family, interacts with RIP140 and that RIP140 acts as a corepressor on GR-regulated genes (21, 32, 35). In the absence of glucocorticoid ligand, GR is mainly located in the cytoplasm in a multiprotein complex including heat shock proteins. Upon ligand binding, the receptor translocates to the nucleus and can bind specific DNA response elements and function as a transcriptional activator. However, GR also mediates glucocorticoid responses by mechanisms not involving DNA binding and gene activation (for a review, see reference 16).
Recently, the role of intranuclear compartmentalization in gene regulation has become appreciated. The cell nucleus contains several different identified areas or domains associated with distinct functions or defined by the colocalization of specific proteins (for a review, see reference 2). These include the nucleolus, the splicing factor speckles, PML bodies, and domains enriched in HDAC activity. The aims of the present study were to characterize the intranuclear localization of RIP140 and determine the role of this localization in its corepressor activity on GR.
MATERIALS AND METHODS
Plasmids.
Green fluorescent protein (GFP)-fused RIP140 was expressed from plasmid pEGFP-RIP140 as previously described (35). GFP-fused RIP/1-281 (amino acids [aa] 1 to 281) and RIP/431-1158 (aa 431 to 1158) were made by subcloning fragments from corresponding pBK-CMV expression plasmids into pEGFP-C2 (Clontech) (25). GFP-fused RIP/1-472 (aa 1 to 472), RIP/431-745 (aa 431 to 745), and RIP/747-1158 (aa 747 to 1158) were generated by subcloning of corresponding PCR fragments into pEGFP-C2. Plasmids expressing GFP-GR (aa 1 to 777), the GFP-GR DNA-binding domain (DBD; aa 418 to 503), GFP-GR DBD-LBD (aa 418 to 777), and GFP-GR LBD (aa 485 to 777) were made by subcloning fragments from plasmids pEG202-GR, pEG202-GR DBD, pEG202-GR DBD-LBD, and pEG202-GR LBD into pEGFP-C2 (27). pSG5-HA, for expression of hemagglutinin (HA)-tagged proteins, was made by insertion of a double-stranded oligonucleotide encoding the influenza virus HA epitope into plasmid pSG5 (Stratagene). pSG5-HA-RIP140, expressing HA-tagged RIP140, has been described previously (25). Plasmids for expression of HA-tagged RIP140 domains were made by subcloning the corresponding fragments from the pEGFP-C2 plasmids. Plasmid pFLAG-CMV-6b-GR, for expression of FLAG-tagged GR, was made by transfer of a fragment from pEG202-GR into pFLAG-CMV-6b (Sigma). Plasmids expressing GAL4 DBD-fused RIP140 and RIP140 domains were made by subcloning the corresponding fragments from pEGFP-C2 plasmids into plasmid pM (Clontech). The expression plasmid for FLAG-tagged CtBP was a kind gift from Catharina Svensson, Uppsala University. Expression plasmid pCMX-SMRT was a kind gift from Ronald Evans, The Salk Institute. Expression plasmid pCMV4-hGR and reporter plasmids p19-tk-luc, pGAL4-luc, and pCMV-βgal were described previously (27, 28). Point mutations and deletions in RIP140 were introduced by using the QuikChange XL site-directed mutagenesis kit (Stratagene).
Confocal microscopy.
COS-7, HeLa, or A549 cells were plated on coverslips in six-well plates and, on the following day, transfected with expression plasmids by using the Fugene 6 transfection reagent (Roche). One hundred nanograms of the expression plasmid for GFP fusion proteins and 200 ng of the expression plasmid for FLAG or HA fusion proteins were used. On the next day, fresh medium was added and the cells were maintained in the presence of 100 nM dexamethasone (Dex) or ethanol vehicle for 3 h. Thereafter, the cells were fixed with 3% paraformaldehyde in 5% sucrose-phosphate-buffered saline (PBS), rinsed with PBS, and permeabilized with PBS containing 0.1% Tween 20 (PBS-T). Cells were then blocked with 5% goat serum (Jackson ImmunoResearch) in PBS-T and incubated with a primary antibody diluted in PBS-T for 1 h at room temperature. Anti-FLAG M5 monoclonal antibody (Sigma), anti-HA.11 monoclonal antibody (BabCO), anti-SMRT monoclonal antibody (MA1-843; Affinity Bioreagents), anti-PML polyclonal antibody (H-238; Santa Cruz Biotechnology), and anti-GR polyclonal antibody (E-20; Santa Cruz Biotechnology) were used at a dilution of 1:200. After being washed, the cells were again blocked with 5% goat serum in PBS-T and incubated with appropriate secondary antibodies conjugated with tetramethyl rhodamine isothiocyanate (Jackson ImmunoResearch) diluted 1:200 in PBS-T for 1 h at room temperature. Cells were washed and fixed to slides with Immu-Mount (Shandon). Subcellular images were determined with a TCS SP Multiband Confocal Imaging System (Leica). Fluorescence images from more than 100 cotransfected cells were studied for each experiment, and the images shown are representative of the majority of the cells studied, as indicated in each figure legend.
Reporter gene assays.
COS-7 or A549 cells were plated in 24-well plates and, on the following day, transfected with plasmids by using the Fugene 6 transfection reagent (Roche). The plasmid amounts used were as follows: 100 ng of p19-tk-luc, 100 ng of pGAL4-luc, 5 ng of pCMV-βgal, 2.5 ng of pCMV4-hGR, 10 ng of pSG5-HA-RIP140, and 100 ng of pM-RIP140. The plasmid amount was kept constant in each transfection by addition of the corresponding empty plasmid. On the next day, fresh medium containing 1 μM Dex or ethanol as a vehicle was added and after 24 h, the cells were harvested. Luciferase and β-galactosidase assays were performed with a Gen-Glow-1000 kit (Bioorbit) and a Galacto-Light plus kit (Tropix), respectively, in an Anthos Lucy 3 luminometer (Anthos Labtec-Instruments).
Coimmunoprecipitation.
Rat hepatoma HTC cells were incubated in the presence or absence of 1 μM triamcinolone acetonide (TA) for 2 h. The cells were harvested in PBS, and the cell pellet was snap-frozen in liquid nitrogen and resuspended in extraction buffer (50 mM phosphate buffer [pH 7.6], 0.4 M KCl, 10% glycerol, 1 mM EDTA, Complete protease inhibitors [Roche]). The buffer for non-TA-treated cells contained 10 mM Na2MoO4 to stabilize the nonactivated GR, and that for TA-treated cells contained 1 μM TA. The cells were homogenized and incubated on ice for 1 h, and the cell extract was centrifuged (50,000 rpm [Beckman TL-100 ultracentrifuge], 30 min, 4°C). The supernatant was diluted 1:8 with EPG buffer (20 mM phosphate buffer [pH 7.4], 10% glycerol, 1 mM EDTA, Complete protease inhibitors [Roche]) and incubated with normal mouse immunoglobulin G-Sepharose for 2 h at 4°C and thereafter with GR-specific mouse monoclonal antibody 250-Sepharose or, as a negative control, with normal mouse immunoglobulin G-Sepharose overnight at 4°C. The Sepharose pellet was washed with EPG buffer containing 50 mM NaCl three times, and bound proteins were eluted by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and analyzed by SDS-PAGE and Western blotting with anti-RIP140 polyclonal antibody (H-300; Santa Cruz Biotechnology) and anti-GR monoclonal antibody (250; Ann-Charlotte Wikström, Karolinska Institutet).
GST pulldown experiments.
The GST-GR LBD was expressed in Escherichia coli strain BL21(DE3)pLysS, and extract was prepared as previously described (35). [35S]methionine-labeled proteins were synthesized in vitro with the TnT-coupled reticulocyte lysate system (Promega). GST fusion protein (5 μg) bound to 30 μl of glutathione beads (Sigma) was incubated with 10 μl of in vitro-translated protein in 200 μl of pulldown buffer (20 mM HEPES-KOH [pH 7.9], 10% glycerol, 100 mM KCl, 5 mM MgCl2, 0.2 mM EDTA, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 1 mg of bovine serum albumin per ml, 0.01% Igepal CA-630) overnight at 4°C. The beads were recovered by centrifugation and washed six times with pulldown buffer without bovine serum albumin, and bound proteins were eluted with SDS-PAGE sample buffer, analyzed by SDS-PAGE, and visualized by autoradiography. In experiments with the GST-GR LBD, the fusion protein bound to glutathione beads was incubated with 1 μM Dex for 2 h before being mixed with translated proteins.
Western blotting.
Whole-cell extracts were prepared from COS-7, HeLa, A549, and HTC cells. The cell pellet was washed in PBS and resuspended in extraction buffer (10 mM Tris-HCl [pH 7.8], 150 mM NaCl, 1 mM EDTA, 0.1% Igepal CA-630, Complete protease inhibitors [Roche]), incubated for 15 min on ice, and homogenized. The supernatant after centrifugation (13,000 rpm [Heraeus Biofuge Fresco], 10 min) was analyzed by SDS-PAGE and Western blotting with anti-RIP140 polyclonal antibody (H-300; Santa Cruz Biotechnology) or anti-GR polyclonal antibody (E-20; Santa Cruz Biotechnology).
RESULTS
RIP140 is localized in small, discrete foci in the nucleus.
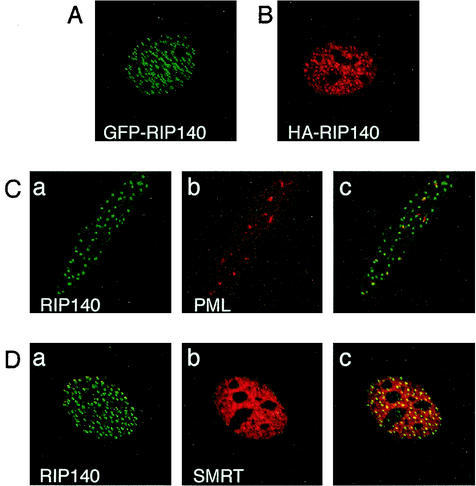
To analyze the intracellular localization of RIP140, we transfected COS-7 cells with a plasmid expressing GFP-tagged RIP140. GFP-RIP140 was localized in the nucleus in discrete, small, bright foci that often appeared as doublets (Fig. 1A). The same localization was also observed for HA-tagged RIP140 (Fig. 1B). Furthermore, GFP-RIP140 expressed in HeLa cells also localizes in discrete foci in the nucleus (Fig. 1C, part a). A similar localization has previously been described for the endogenous RIP140 in COS-1 cells, showing that the localization in nuclear foci is not due to overexpression of the protein (3).
FIG. 1.
RIP140 is localized in discrete foci in the nucleus. (A) COS-7 cells were transfected with a GFP-RIP140 expression plasmid. (B) COS-7 cells were transfected with an HA-RIP140 expression plasmid. (C) HeLa cells were transfected with a GFP-RIP140 expression plasmid. The localization of endogenous PML protein in GFP-RIP140-transfected cells was detected with a PML-specific antibody. Part a shows the localization of GFP-RIP140 in green, and part b shows the localization of PML protein in red in the same cell. Part c shows the merged image, where colocalization of the proteins is in yellow. (D) COS-7 cells were cotransfected with expression plasmids for GFP-RIP140 and SMRT. The localization of SMRT protein in GFP-RIP140-transfected cells was detected with a SMRT-specific antibody. Part a shows the localization of GFP-RIP140 in green, and part b shows the localization of SMRT protein in red in the same cell. Part c shows the merged image, where colocalization of the proteins is in yellow. Protein localization was visualized by confocal microscopy. The images shown in panels A to D are representative of 85, 85, 100, and 40% of the cells studied, respectively.
To further analyze the nature of the RIP140 foci in the nucleus, the colocalization of RIP140 with previously described nuclear domains was analyzed. PML nuclear bodies are characterized by the presence of the PML protein and have been implicated in regulation of transcription (34). However, GFP-RIP140 did not colocalize with PML bodies identified by using an antibody against the endogenous PML protein in HeLa cells (Fig. 1C, parts a to c). The corepressors SMRT and Hr colocalize with HDACs in specific nuclear domains termed MAD bodies (6, 17). The possible localization of GFP-RIP140 in MAD bodies in COS-7 cells was investigated by using cotransfected SMRT identified by a SMRT-specific antibody. SMRT was expressed in a diffuse pattern in the nucleus, with some localization in large domains. Some of the RIP140 foci colocalized with SMRT-containing domains, indicating that some of the RIP140 foci are related to the MAD bodies (Fig. 1D, parts a to c). However, the MAD bodies identified by SMRT were larger than the RIP140 foci. Collectively, these data show that RIP140 is localized in discrete punctate bodies in the nucleus that are distinct from other nuclear domains.
A domain of 40 aa is necessary for targeting of RIP140 into small nuclear foci.
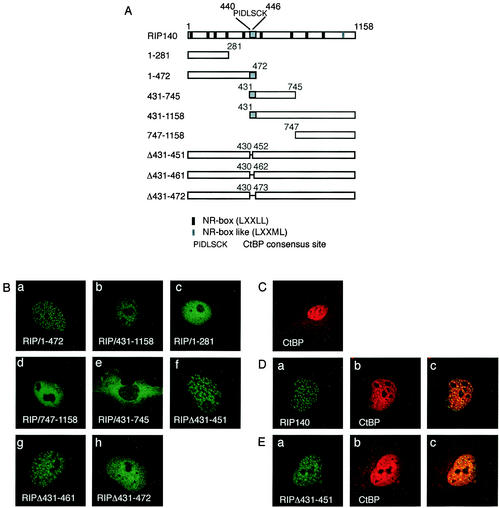
To investigate which domain of RIP140 was responsible for localization of the protein into nuclear foci, we made GFP fusions of several RIP140 domains (Fig. 2A). Analyses of the intracellular localization of overlapping fragments encompassing the N terminus (RIP/1-472) and the middle and C-terminal domains (RIP/431-1158) showed that both localized in small foci in the nucleus (Fig. 2B, parts a and b). In contrast, shorter fragments of the N terminus (RIP/1-281) or the C terminus (RIP/747-1158) were evenly localized in the nucleus (Fig. 2B, parts c and d). These results suggested that the region encompassing aa 431 to 472 present in both RIP/1-472 and RIP/431-1158 is responsible for the localization in the punctate nuclear bodies. To further prove the role of the region in targeting RIP140 to the nuclear dots, deletions of aa 431 to 451, 431 to 461, and 431 to 472 in full-length GFP-RIP140 were performed. RIPΔ431-451 and RIPΔ431-461 were still localized in small foci (Fig. 2B, parts f and g). However, RIPΔ431-472 was not localized in small RIP140 foci but rather evenly distributed (Fig. 2B, part h). This result showed that the domain encompassing aa 431 to 472 is necessary for the localization of RIP140 in the small nuclear dots and, further, that aa 462 to 472 play the major role. RIP/431-745, however, containing the dot-targeting domain, was evenly distributed in the cytoplasm (Fig. 2B, part e), indicating that factors other than the dot-targeting domain are also necessary for the localization of RIP140 to nuclear foci. To investigate whether forced nuclear localization of RIP/431-745 would result in focus formation, RIP/431-745 was expressed fused to the GAL4 DBD containing a nuclear localization signal (NLS) or fused to GFP coupled to an NLS. However, both constructs were still localized in the cytoplasm (data not shown), suggesting that RIP/431-745 contains a nuclear export sequence or a cytoplasmic retention signal that overcomes the NLS. The CRM1 protein is involved in the nuclear export of many proteins that have a prototypic nuclear export sequence (9). However, nuclear export by CRM1 is not responsible for the cytoplasmic localization of RIP/431-745 since the CRM1 inhibitor leptomycin B did not change the localization of the protein (data not shown).
FIG. 2.
The domain targeting RIP140 to small nuclear foci is mapped to aa 431 to 472. (A) Schematic representation of the RIP140 domains and deletion mutant forms used. NR boxes (LXXLL motifs), the CtBP-binding motif (PIDLSCK), and the dot-targeting domain (in gray) are indicated. (B) COS-7 cells were transfected with expression plasmids for GFP-fused RIP140 domains or deletion proteins. Part a, GFP-RIP140 aa 1 to 472; part b, GFP-RIP140 aa 431 to 1158; part c, GFP-RIP140 aa 1 to 281; part d, GFP-RIP140 aa 747 to 1158; part e, GFP-RIP140 aa 431 to 745; part f, GFP-RIP140 with a deletion of aa 431 to 451; part g, GFP-RIP140 with a deletion of aa 431 to 461; part h, GFP-RIP140 with a deletion of aa 431 to 472. (C) COS-7 cells were transfected with an expression plasmid for FLAG-CtBP. The localization of FLAG-CtBP was visualized with a FLAG-specific antibody. (D) COS-7 cells were cotransfected with expression plasmids for GFP-RIP140 and FLAG-CtBP. Part a shows the localization of GFP-RIP140 in green, and part b shows the localization of the FLAG-CtBP protein in red in the same cell. Part c shows the merged image, where colocalization of the proteins is in yellow. (E) COS-7 cells were cotransfected with expression plasmids for GFP-RIP140 with a deletion of aa 431 to 451 (RIPΔ431-451) and FLAG-CtBP. Part a shows the localization of RIPΔ431-451 in green, and part b shows the localization of the FLAG-CtBP protein in red in the same cell. Part c shows the merged image, where colocalization of the proteins is in yellow. The images shown in panels B (parts a to h), C, D, and E are representative of 70, 85, 90, 90, 90, 60, 60, 80, 100, 40, and 100%, respectively, of the cells studied.
Interestingly, a consensus binding motif (PIDLSCK; aa 440 to 446) for the corepressor protein CtBP has been identified within the domain targeting RIP140 to nuclear foci (26), presenting the possibility that CtBP interaction recruits RIP140 to the foci. To investigate the intracellular localization of CtBP, FLAG-tagged CtBP was expressed in COS-7 cells. FLAG-CtBP was mainly localized in the nucleus, but some cytoplasmic expression was also observed (Fig. 2C). FLAG-CtBP had an even distribution in the nucleus, distinct from the localization in small foci observed for RIP140, making it unlikely that the typical RIP140 localization is due to CtBP recruiting RIP140 to the foci. However, cotransfection of GFP-RIP140 and FLAG-CtBP resulted in redistribution of CtBP to RIP140-like foci (Fig. 2D, part b) and colocalization of the two proteins in the foci, confirming the interaction between the two proteins. In contrast, GFP-RIPΔ431-451 containing a deletion of the CtBP consensus binding site did not redistribute CtBP to the small foci (Fig. 2E, part b), indicating that the deletion mutant form did not interact with CtBP. Since RIPΔ431-451 was still localized in small foci, this result shows that interaction with CtBP is not necessary for RIP140 localization in small foci.
In conclusion, these results indicate that a 40-aa-long segment encompassing aa 431 to 472 is necessary for the targeting of RIP140 to the typical nuclear dots and that although the CtBP corepressor can bind within this segment, CtBP interaction is not involved in RIP140 targeting to the foci.
CtBP binding and the dot-targeting domain are dispensable for the intrinsic repression activity of RIP140.
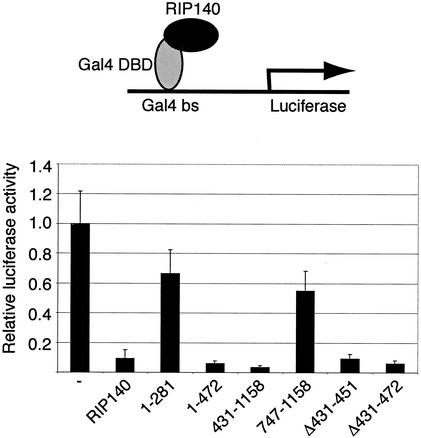
RIP140 has an intrinsic repression activity that can be mediated by the formation of a repression complex involving proteins such as HDAC and CtBP (14, 26). The intrinsic repression activity of RIP140 can be analyzed by GAL4 DBD-fused RIP140-induced repression of a reporter gene regulated by GAL4-binding sites. In order to map the intrinsic repression activity of RIP140, we used GAL4 DBD-fused RIP140 domains (Fig. 3). Interestingly, both the N-terminal domain RIP/1-472 and the middle and C-terminal domains contained in RIP/431-1158 repressed the reporter gene activity to the same extent as the wild-type protein (Fig. 3). In contrast, the shorter fragments, RIP/1-281 and RIP/747-1158, did not repress well. Since both RIP/1-472 and RIP/431-1158 contain the CtBP-interacting domain and the domain targeting RIP140 to the nuclear small foci, we asked whether these domains are necessary for the observed intrinsic repression activity. The RIP140 deletion mutant form RIPΔ431-451, which does not interact with CtBP, could repress the GAL4 reporter gene to the same extent as the wild-type protein (Fig. 3). The mutant form RIP140Δ431-472, which does not localize in nuclear foci, still retained strong repression activity (Fig. 3). This suggests that the intrinsic repression activity of RIP140 can be mediated by both N- and C-terminal domains and that neither the CtBP-binding domain nor the dot-targeting domain is necessary for intrinsic repression.
FIG. 3.
Mapping of the intrinsic repression activity. COS-7 cells were transfected with plasmids expressing the GAL4 DBD, GAL4 DBD-fused RIP140, or RIP140 domain, as well as reporter plasmids pGal4-luc, containing a luciferase reporter gene regulated by GAL4-binding sites (Gal4 bs), and pCMV-βgal, containing a constitutively expressed β-galactosidase reporter gene. Relative luciferase activity was calculated by relating the measured luciferase reporter gene activity to the constitutive β-galactosidase reporter gene activity. The values were related to the activity obtained for the control GAL4 DBD protein. The mean and standard deviation (n = 6) of a representative experiment are shown.
Both CtBP binding and C-terminal NR boxes contribute to the corepressor activity of RIP140.
To analyze whether endogenous GR and RIP140 can interact in cells, we performed coimmunoprecipitation experiments with the rat hepatoma HTC cell line. Precipitation of proteins from cell extracts with an anti-GR antibody coupled to Sepharose showed that endogenous RIP140 protein coprecipitated with endogenous GR (Fig. 4A, lane 2). A slightly enhanced interaction between GR and RIP140 was observed in cells treated with the glucocorticoid ligand TA (Fig. 4A, lanes 2 and 4). The RIP140 protein was not precipitated by a negative control antibody-Sepharose, showing the specificity of the coimmunoprecipitation (Fig. 4A, lane 5).
FIG. 4.
Interaction between endogenous GR and RIP140. (A) RIP140 was coimmunoprecipitated with GR from HTC cells with anti-GR antibody-Sepharose (anti-GR) in the absence (−) or presence (+) of the glucocorticoid ligand TA. Coimmunoprecipitation with normal mouse antibody-Sepharose (control ab) was used as a negative control. Proteins in the cell extract used for the precipitation (input) and precipitated proteins (ppt) were analyzed by Western blotting with anti-RIP140 and anti-GR antibodies. IP, immunoprecipitate. (B) In vitro-translated RIP140 or RIP140 domains were incubated with GST or the GST-GR LBD bound to glutathione beads. The interacting proteins were visualized by SDS-PAGE and autoradiography. The values on the left are molecular sizes in kilodaltons.
To map the interaction site of GR on RIP140, GST pulldown experiments were performed with the GST-fused GR LBD and in vitro-translated fragments of RIP140 (Fig. 4B). Interestingly, all fragments of RIP140, except the middle domain (RIP/431-745), interacted with the GR LBD, indicating that RIP140 can make several contacts with GR.
RIP140 can act as a corepressor for GR and repress GR activation of a glucocorticoid response element (GRE)-regulated reporter gene (21, 32, 35). To determine whether RIP140 also can corepress the activity of endogenous GR in cells, we used the human lung carcinoma A549 cell line, which contains endogenous GR protein. The cells were transfected with a GRE-regulated reporter gene and increasing amounts of a RIP140-expressing plasmid. Endogenous GR showed strong ligand-induced activation of the reporter gene in the absence of the RIP140 protein, whereas increasing amounts of the RIP140 protein resulted in repression of the endogenous GR activity (Fig. 5A).
FIG. 5.
Corepressor activity of RIP140 is dependent on the C-terminal NR boxes and CtBP binding. (A) A549 cells containing endogenous GR protein were transfected with reporter plasmid p19-tk-luc, containing a GRE-regulated luciferase reporter gene, and increasing amounts of a plasmid expressing RIP140. The cells were maintained in the presence or absence of 1 μM Dex. (B and C) COS-7 cells were transfected with a plasmid expressing GR in the presence or absence of a plasmid expressing RIP140 or RIP140 domains or deletions, as well as reporter plasmids p19-tk-luc, containing a GRE-regulated luciferase reporter gene, and pCMV-βgal, containing a constitutively expressed β-galactosidase reporter gene. The cells were maintained in the presence or absence of 1 μM Dex. The values were related to the activity obtained with GR treated with Dex in the absence of the RIP140 protein. (D) COS-7 cells were transfected with a plasmid expressing GR in the presence or absence of a plasmid expressing the C-terminal domain of RIP140 (aa 747 to 1158) or proteins containing mutations of the indicated NR boxes in the context of RIP/747-1158, as well as reporter plasmids p19-tk-luc, containing a GRE-regulated luciferase reporter gene, and pCMV-βgal, containing a constitutively expressed β-galactosidase reporter gene. The mean and standard deviation (n = 6) of a representative experiment are shown.
To gain insight into the mechanism of the corepressor activity, the effects of different RIP140 fragments on GR transactivation were studied with a GRE-regulated reporter gene. The corepressor activity of RIP140 was mainly contained within the C-terminal domain (RIP/747-1158), which strongly repressed GR activity (Fig. 5B). However, the N-terminal domain RIP/1-472, which includes the focus-targeting region and the CtBP-binding site, could also slightly corepress GR activity (Fig. 5B). RIP/431-1158, containing the C-terminal domain, the focus-targeting region, and the CtBP-binding site, was the best corepressor (Fig. 5B). However, although RIP/1-281 could interact with GR, it did not have any corepressor activity (Fig. 5B). RIP/431-745, which did not bind the GR, also did not corepress (Fig. 5B). To further analyze the role of the dot-targeting domain and the CtBP-binding site for the corepressor activity, the deletion mutant forms RIPΔ431-451 and RIPΔ431-472 were used. Interestingly, deletion of the CtBP-binding site (RIPΔ431-451) reduced the corepressor activity to the same level as that of the C-terminal fragment (RIP/747-1158) (Fig. 5C). Further deletion of the dot-targeting domain (RIPΔ431-472) did not have any major additional effect (Fig. 5C).
To analyze the mechanism of the corepressor function of the C-terminal domain (RIP/747-1158), the role of the NR boxes was addressed. RIP140 has nine NR boxes (LXXLL motifs), as well as one NR box-like motif (LXXML) (25). Two of the NR boxes and the NR box-like motif are present within the C-terminal domain that strongly represses GR activity. Mutation of the last two leucine residues to alanine in individual NR boxes or combinations of sequences was performed in the context of RIP/747-1158, and the corepressor activity of the mutated forms was tested (Fig. 5D). Mutation of all three NR boxes (RIPΔNR8,9,10) dramatically reduced corepressor activity (Fig. 5D). NR box 8 seemed to be the most important NR box within the C-terminal domain for the corepressor activity since mutation of NR box 8, individually (RIPΔNR8) or in combination (RIPΔNR8,9 and RIPΔNR8,10), had the most dramatic effect on the activity (Fig. 5D). Mutation of the NR boxes also reduced binding affinity for GR, as determined by GST pulldown assays with the GST-GR LBD and in vitro-translated RIP140 mutant forms (data not shown). Collectively, these data suggest that the corepressor activity of RIP140 involves two distinct mechanisms. First, corepression is mediated by the C-terminal domain (RIP/747-1158) and the importance of the NR boxes implies that competition for binding to the GR is involved. Second, although the C-terminal domain contains the main corepressor activity, CtBP interaction contributes slightly to the corepressor effect. The binding of CtBP seems to be the only mechanism for corepression in addition to the role of the C terminus, since deletion of the CtBP-binding domain reduces corepression activity to the level of the C-terminal domain (RIP/747-1158).
Corepression correlates with an intranuclear redistribution of GR and RIP140.
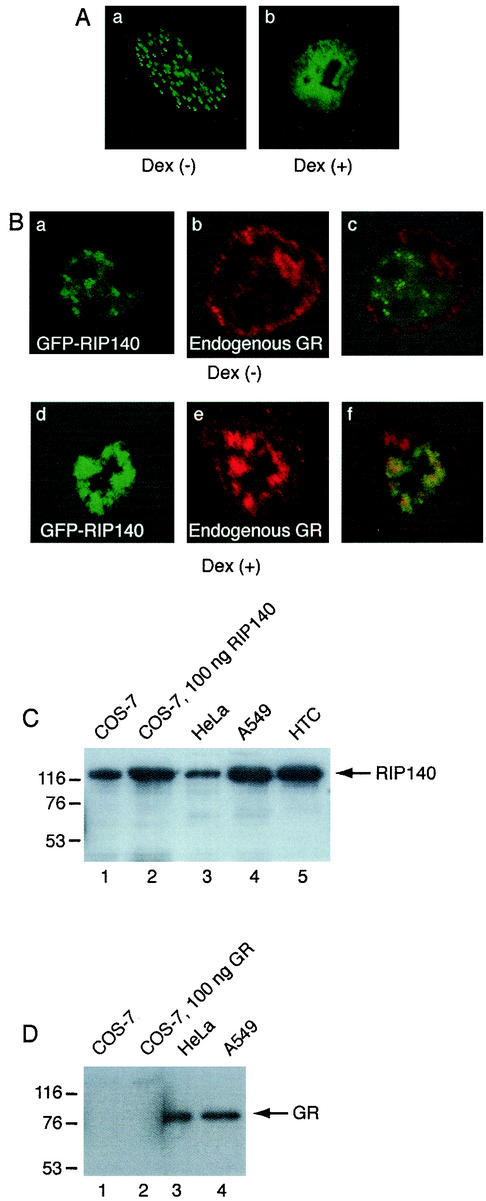
To analyze the intracellular localization of GFP-RIP140 under corepressing conditions in the A549 cell line containing endogenous GR, confocal microscopy studies were performed. GFP-RIP140 was localized in the typical intranuclear foci in the A549 cell line (Fig. 6A, part a). However, upon treatment with the glucocorticoid ligand Dex, GFP-RIP140 was redistributed to a more diffuse pattern (Fig. 6A, part b), indicating that endogenous ligand-activated GR can redistribute RIP140 from the small foci. To analyze the colocalization of RIP140 and endogenous GR in A549 cells, endogenous GR protein was detected with a GR-specific antibody. In the absence of ligand, endogenous GR was mainly localized in the cytoplasm and did not colocalize with GFP-RIP140 (Fig. 6B, parts a to c). However, upon ligand treatment, endogenous GR was partly colocalized with GFP-RIP140 in diffuse larger structures in the nucleus (Fig. 6B, parts d to f).
FIG. 6.
Intranuclear redistribution of RIP140 by ligand-activated endogenous GR. (A) A549 cells containing endogenous GR were transfected with a plasmid expressing GFP-RIP140. The cells were treated with (+) or without (−) 100 nM Dex for 3 h before visualization of the RIP140 protein by confocal microscopy. The images shown in parts a and b are representative of 80 and 73%, respectively, of the cells studied. (B) A549 cells were transfected with a plasmid expressing GFP-RIP140. The cells were treated with (+) or without (−) 100 nM Dex for 3 h. Endogenous GR was detected with a GR-specific antibody. Parts a and d show the localization of GFP-RIP140, parts b and e show the localization of endogenous GR, and parts c and f show the merged images. (C and D) The expression levels of endogenous RIP140 and GR in the COS-7, HeLa, A549, and HTC cell lines were compared to the levels of the transfected RIP140 and GR proteins by Western blotting. The values on the left are molecular sizes in kilodaltons.
To study the intranuclear redistribution of RIP140 and GR in more detail, we used transfected COS-7 cells. Western blotting experiments showed that transfection of COS-7 cells with the amount of RIP140 or GR expression plasmid used for confocal microscopy experiments (100 ng) did not result in overexpression of the proteins but rather gave expression levels similar to or lower than those of endogenous proteins in the HeLa, A549, and HTC cell lines (Fig. 6C and D). We therefore assume that studies with transfected proteins are representative of endogenous proteins. GR was expressed in COS-7 cells tagged with either GFP or FLAG. In the absence of glucocorticoid, GR was mainly localized in the cytoplasm but some nuclear staining was also observed (data not shown). Upon ligand treatment, GR translocated into the nucleus. In the nucleus, GR had an even distribution (Fig. 7A and 8A, part a). The same localization pattern was observed for both GFP-GR and FLAG-GR and resembled the localization of endogenous GR in A549 cells. When GFP-RIP140 and FLAG-GR were coexpressed in COS-7 cells in the absence of ligand, RIP140 was localized in the typical small foci and GR was mainly in the cytoplasm, as when they were individually expressed (data not shown). However, upon Dex treatment for 3 h, the proteins colocalized in diffuse, large domains in the nucleus (Fig. 7B, parts a to c). Thus, the localization of the coexpressed proteins was distinct from that of the individually expressed proteins; i.e., the localization of GFP-RIP140 was changed from the localization in small foci and the localization of FLAG-GR was changed from an even distribution. The same redistribution of coexpressed proteins in the presence of Dex was also seen for HA-RIP140 and GFP-GR, indicating that tagging of the proteins did not contribute to the effect (Fig. 8B, parts a to c). The redistribution confirmed the results obtained for endogenous GR and GFP-RIP140 in A549 cells (Fig. 6B). In contrast, coexpression of GR with other cofactors did not change the GR localization (data not shown). Dex treatment did not have any effect of RIP140 localization in the absence of GR (data not shown).
FIG. 7.
Corepression correlates with intranuclear redistribution of GR and RIP140. (A) COS-7 cells were transfected with a plasmid expressing FLAG-GR. The cells were treated with 100 nM Dex for 3 h before visualization of the expressed protein with a FLAG-specific antibody. (B to G) COS-7 cells were transfected with plasmids expressing GFP-RIP140 or GFP-RIP140 domains and FLAG-GR, and the cells were treated with 100 nM Dex for 3 h. Part a shows the localization of GFP-RIP140 in green, and part b shows the localization of FLAG-GR protein in red in the same cell. Part c shows the merged image, where colocalization of the proteins is in yellow. The images shown in panels A to G are representative of 100, 70, 60, 70, 60, 100, and 100%, respectively, of the cells studied.
FIG. 8.
GR DBD is needed for redistribution of RIP140. (A) COS-7 cells were transfected with plasmids expressing GFP-GR (part a), GFP-GR DBD-LBD (part b), GFP-GR LBD (part c), and GFP-GR DBD (part d). The cells were treated with 100 nM Dex for 3 h. (B to E) COS-7 cells were cotransfected with plasmids expressing HA-RIP140 and GFP-GR or GFP-GR domains, and the cells were treated with 100 nM Dex for 3 h. HA-RIP140 was visualized with an HA-specific antibody. Part a shows the localization of HA-RIP140 in red, and part b shows the localization of the GFP-GR protein in green in the same cell. Part c shows the merged image, where colocalization of proteins is in yellow. The images shown in panels A to E are representative of 100, 70, 70, 100, and 100%, respectively, of the cells studied.
To determine which domains of RIP140 can redistribute GR into the diffuse, large domains, GFP-RIP140 domains were coexpressed with FLAG-GR (Fig. 7). All of the fragments that could corepress GR activity were also shown to redistribute GR within the nucleus (RIP/1-472, RIP/431-1158, and RIP/747-1158) (Fig. 7C, D, and E, parts a to c). In contrast, RIP/1-281, which did not function as a corepressor for GR, did not redistribute GR either, although the protein could bind to GR, as shown by the GST pulldown experiment (Fig. 7F, parts a to c). Cytoplasmically localized RIP/431-745, which does not interact with GR, also did not redistribute GR (Fig. 7G, parts a to c). The fact that RIP/431-745 was still localized in the cytoplasm when coexpressed with GR in the presence of ligand and not cotransported with GR into the nucleus confirmed that the two proteins did not interact. To evaluate the role of CtBP binding and the dot-targeting domain of RIP140 in the redistribution, we analyzed the RIP140 deletion mutant forms. Both RIPΔ431-451 and RIPΔ431-472 redistributed GR (data not shown), indicating that these domains are not required for redistribution.
To determine which domains of GR are involved in the redistribution of RIP140, GFP-GR domains were coexpressed with HA-RIP140 (Fig. 8). The GR DBD-LBD redistributed RIP140 into the large, diffuse domains in the presence of ligand in the same way as the full-length GR (Fig. 8C, parts a to c). Interestingly, the GR LBD did not redistribute RIP140 but rather colocalized with RIP140 in small foci similar to typical RIP140 foci (Fig. 8D, parts a to c), suggesting that the DBD is necessary for redistribution of RIP140. RIP140 interacts with the LBDs of nuclear receptors and, accordingly, the GFP-GR DBD and HA-RIP140 did not colocalize in cells (Fig. 8E, parts a to c). All of the GR domains had an even distribution in the nucleus in the presence of Dex when expressed alone (Fig. 8A). In conclusion, these studies imply that the formation of a repressing complex between GR and RIP140 correlates with the relocalization of both proteins within the nucleus to specific large nuclear domains. The redistribution requires a fragment of RIP140 containing corepressor activity, as well as the GR DBD.
DISCUSSION
The activity of transcription factors and transcriptional coregulators can be modified at many levels, including posttranscriptional regulation, such as phosphorylation and acetylation. An additional important regulatory mechanism involves protein targeting to distinct intracellular sites. Regulation of nuclear translocation has previously been implicated in the regulation of transcription factor activity (2). This mechanism is also, to some extent, used for regulation of GR activity. However, the specific intranuclear localization of nuclear proteins may also be of major importance for the functional activity of those proteins. In this report, we describe a role for the regulation of the intranuclear localization of the corepressor RIP140 in its repression of GR-regulated genes.
We have observed a redistribution of the intranuclear localization of RIP140 correlating with corepression of GR activity. In the absence of ligand-activated GR, RIP140 is localized in specific small foci in the nucleus. However, upon ligand treatment, RIP140 is redistributed to large nuclear domains, where it is colocalized with GR. We have identified a sequence element that is necessary for localization of RIP140 in the specific small foci in the nucleus. The sequence necessary for targeting of RIP140 to the foci has been mapped to a 40-aa-long segment, and the C-terminal third of the segment plays the major role. In addition to the focus-targeting domain, a region for nuclear localization is also needed. Subnuclear targeting domains in nuclear proteins, including proteins localized in the nucleolus and splicing speckles, have been identified previously. For example, the domain targeting the protein phosphatase 1 regulator NIPP1 to nuclear domains called speckles interacts with a splicing factor that is also localized in the speckles, suggesting that the subnuclear targeting domain can act by protein-protein interaction (8). However, although the focus-targeting domain in RIP140 overlaps a CtBP-binding site, CtBP binding is not needed for localization in nuclear foci. It will be interesting to determine the functional mechanism for the targeting domain and identify the putative proteins or nuclear structures involved in the localization of RIP140. Localization in nuclear foci and the targeting domain has been implicated in the function of some transcription-regulatory proteins. The coactivator TAZ and the transcription factor RunX2 both contain domains involved in subnuclear targeting, and the domains are also needed for transcriptional activation (10, 33). The sequence elements needed for localization of RIP140 in the small foci do not seem to be involved in the corepressor activity on GR transcription, and the functional role of the localization of RIP140 in the typical small foci still has to be determined.
In contrast, the colocalization of ligand-activated GR with RIP140 in the large nuclear domains correlates with the corepressing activity of RIP140. The colocalization with RIP140 in the nuclear domain is not specific for GR, since we have also observed the colocalization of RIP140 with ERβ (data not shown); colocalization with the nuclear receptor TR2 and the aryl hydrocarbon receptor in nuclear structures has previously been reported (12, 14). However, the functional significance of the nuclear domains has not previously been addressed. Our data imply that the localization in the large nuclear domains could be involved in the RIP140 corepression mechanism. Several transcriptional repressors are localized in subnuclear domains, including Sall1, RGS12TS-S, and Hr (4, 11, 17). The functional significance of this pattern of localization is still unclear. The intranuclear domains could be sites for assembly of multiprotein complexes needed for the repression effect. Alternatively, it may represent a mechanism by which to inactivate transcription factors by transferring them to domains that are not accessible for active transcription. In analogy, sequestering of proteins in the nucleolus or PML bodies has been shown to be a mechanism for protein regulation (2). Covalent modification of proteins with SUMO and subsequent localization in PML nuclear bodies have been implicated as one mechanism for transcriptional repression. For example, the activity of the transcription factor LEF1 is repressed by localization in PML bodies (19). RIP140 contains several consensus sumoylation sites, and it remains to be determined whether it can be modified by sumoylation. However, RIP140 is not localized in typical PML bodies, but rather, the RIP140 foci appear to constitute distinct subnuclear domains. The mechanism for the redistribution of GR and RIP140 could be related to a conformational change in both proteins upon receptor-corepressor interaction, possibly modifying protein-protein interactions important for the subnuclear localization. Since the DBD of GR is needed for the redistribution, it is possible that the complex formed between RIP140 and GR is modulated by the DBD. Alternatively, the repressing nuclear domain containing GR/RIP140 could be involved in chromatin interaction through the DNA-binding activity of the GR DBD. However, since a nuclear-matrix-targeting signal has been mapped to the GR DBD (23), it is also possible that GR and RIP140 are redistributed to the nuclear matrix. The fact that the GR LBD, which cannot redistribute to the repressing nuclear domain because of the lack of a DBD, is colocalized with RIP140 in RIP140 foci implies that the mechanism of the repressive effect of RIP140 on GR first involves colocalization of the two proteins in RIP140 foci, whereupon the proteins redistribute to the larger repressing domains.
RIP140 corepression of GR-regulated genes involves at least two repression functions, one in the N-terminal part of RIP140 and the other in the C-terminal part. The two functions are distinct and mediated by different mechanisms. The N-terminal repression function is correlated with CtBP interaction since disruption of CtBP binding reduces repression to the same extent as that obtained with the isolated C-terminal domain. CtBP can act as a corepressor for a number of transcription factors involved in growth and differentiation, including E1A, Knirps, and BKLF (reviewed in reference 5). The mechanism of the transcriptional repression by CtBP is not clear, but it may be related to recruitment of HDAC proteins or the Polycomb (PcG) complex (20, 22). Further deletion of both CtBP-binding and focus-targeting domains does not further disrupt repression. The C-terminal repression function is at least partly dependent on the integrity of the receptor-binding sequences, the NR boxes. Especially NR box 8 is important for the repression. This finding supports the idea that RIP140 represses nuclear receptors by competing with coactivators for binding to the receptor LBD. However, since the repression activity was not completely abolished by mutating the C-terminal NR boxes, either additional receptor interaction sequences or additional repressor functions, such as interaction with HDAC proteins, are involved (26).
In conclusion, we have mapped the domain targeting RIP140 to discrete intranuclear foci to a 40-aa segment. Although a CtBP-binding site is present within the N-terminal part of this sequence, we show that CtBP interaction is not involved in RIP140 localization, but rather the C-terminal part of the sequence is important for the focus targeting. Furthermore, we show that RIP140 repression of GR-regulated genes involves both CtBP binding and C-terminal NR box motifs. The corepressor function also involves redistribution of RIP140 and GR.
Acknowledgments
We thank Ronald Evans, The Salk Institute, and Catharina Svensson, Uppsala University, for kind gifts of plasmids; Ann-Charlotte Wikström, Karolinska Institutet, for the kind gift of GR antibody; and Marika Rönnholm, Karolinska Institutet, for technical assistance in coimmunoprecipitation experiments.
This work was supported by grants from the Emil and Wera Cornell Foundation and the Tore Nilson Foundation.
REFERENCES
- 1.Bourguet, W., P. Germain, and H. Gronemeyer. 2000. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol. Sci. 21:381-388. [DOI] [PubMed] [Google Scholar]
- 2.Carmo-Fonseca, M. 2002. The contribution of nuclear compartmentalization to gene regulation. Cell 108:513-521. [DOI] [PubMed] [Google Scholar]
- 3.Cavailles, V., S. Dauvois, F. L'Horset, G. Lopez, S. Hoare, P. J. Kushner, and M. G. Parker. 1995. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 14:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee, T. K., and R. A. Fisher. 2002. RGS12TS-S localizes at nuclear matrix-associated subnuclear structures and represses transcription: structural requirements for subnuclear targeting and transcriptional repression. Mol. Cell. Biol. 22:4334-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinnadurai, G. 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 9:213-224. [DOI] [PubMed] [Google Scholar]
- 6.Downes, M., P. Ordentlich, H. Y. Kao, J. G. Alvarez, and R. M. Evans. 2000. Identification of a nuclear domain with deacetylase activity. Proc. Natl. Acad. Sci. USA 97:10330-10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 8.Jagiello, I., A. Van Eynde, V. Vulsteke, M. Beullens, A. Boudrez, S. Keppens, W. Stalmans, and M. Bollen. 2000. Nuclear and subnuclear targeting sequences of the protein phosphatase-1 regulator NIPP1. J. Cell Sci. 113:3761-3768. [DOI] [PubMed] [Google Scholar]
- 9.Jans, D. A., C. Y. Xiao, and M. H. Lam. 2000. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays 22:532-544. [DOI] [PubMed] [Google Scholar]
- 10.Kanai, F., P. A. Marignani, D. Sarbassova, R. Yagi, R. A. Hall, M. Donowitz, A. Hisaminato, T. Fujiwara, Y. Ito, L. C. Cantley, and M. B. Yaffe. 2000. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 19:6778-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiefer, S. M., B. W. McDill, J. Yang, and M. Rauchman. 2002. Murine Sall1 represses transcription by recruiting a histone deacetylase complex. J. Biol. Chem. 277:14869-14876. [DOI] [PubMed] [Google Scholar]
- 12.Kumar, M. B., R. W. Tarpey, and G. H. Perdew. 1999. Differential recruitment of coactivator RIP140 by Ah and estrogen receptors. Absence of a role for LXXLL motifs. J. Biol. Chem. 274:22155-22164. [DOI] [PubMed] [Google Scholar]
- 13.Le Douarin, B., A. L. Nielsen, J. M. Garnier, H. Ichinose, F. Jeanmougin, R. Losson, and P. Chambon. 1996. A possible involvement of TIF1α and TIF1β in the epigenetic control of transcription by nuclear receptors. EMBO J. 15:6701-6715. [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, C. H., C. Chinpaisal, and L. N. Wei. 1998. Cloning and characterization of mouse RIP140, a corepressor for nuclear orphan receptor TR2. Mol. Cell. Biol. 18:6745-6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, C. H., and L. N. Wei. 1999. Characterization of receptor-interacting protein 140 in retinoid receptor activities. J. Biol. Chem. 274:31320-31326. [DOI] [PubMed] [Google Scholar]
- 16.Newton, R. 2000. Molecular mechanisms of glucocorticoid action: what is important? Thorax 55:603-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potter, G. B., G. M. Beaudoin III, C. L. DeRenzo, J. M. Zarach, S. H. Chen, and C. C. Thompson. 2001. The hairless gene mutated in congenital hair loss disorders encodes a novel nuclear receptor corepressor. Genes Dev. 15:2687-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenfeld, M. G., and C. K. Glass. 2001. Coregulator codes of transcriptional regulation by nuclear receptors. J. Biol. Chem. 276:36865-36868. [DOI] [PubMed] [Google Scholar]
- 19.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sewalt, R. G., M. J. Gunster, J. van der Vlag, D. P. Satijn, and A. P. Otte. 1999. C-terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol. Cell. Biol. 19:777-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramaniam, N., E. Treuter, and S. Okret. 1999. Receptor interacting protein RIP140 inhibits both positive and negative gene regulation by glucocorticoids. J. Biol. Chem. 274:18121-18127. [DOI] [PubMed] [Google Scholar]
- 22.Sundqvist, A., K. Sollerbrant, and C. Svensson. 1998. The carboxy-terminal region of adenovirus E1A activates transcription through targeting of a C-terminal binding protein-histone deacetylase complex. FEBS Lett. 429:183-188. [DOI] [PubMed] [Google Scholar]
- 23.Tang, Y., R. H. Getzenberg, B. N. Vietmeier, M. R. Stallcup, M. Eggert, R. Renkawitz, and D. B. DeFranco. 1998. The DNA-binding and tau2 transactivation domains of the rat glucocorticoid receptor constitute a nuclear matrix-targeting signal. Mol. Endocrinol. 12:1420-1431. [DOI] [PubMed] [Google Scholar]
- 24.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 25.Treuter, E., T. Albrektsen, L. Johansson, J. Leers, and J.-Å. Gustafsson. 1998. A regulatory role for RIP140 in nuclear receptor activation. Mol. Endocrinol. 12:864-881. [DOI] [PubMed] [Google Scholar]
- 26.Vo, N., C. Fjeld, and R. H. Goodman. 2001. Acetylation of nuclear hormone receptor-interacting protein RIP140 regulates binding of the transcriptional corepressor CtBP. Mol. Cell. Biol. 21:6181-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakui, H., A. P. Wright, J.-Å Gustafsson, and J. Zilliacus. 1997. Interaction of the ligand-activated glucocorticoid receptor with the 14-3-3 eta protein. J. Biol. Chem. 272:8153-8156. [DOI] [PubMed] [Google Scholar]
- 28.Wärnmark, A., T. Almlöf, J. Leers, J.-Å. Gustafsson, and E. Treuter. 2001. Differential recruitment of the mammalian mediator subunit TRAP220 by estrogen receptors ERα and ERβ. J. Biol. Chem. 276:23397-23404. [DOI] [PubMed] [Google Scholar]
- 29.Wei, L. N., M. Farooqui, and X. Hu. 2001. Ligand-dependent formation of retinoid receptors, receptor-interacting protein 140 (RIP140), and histone deacetylase complex is mediated by a novel receptor-interacting motif of RIP140. J. Biol. Chem. 276:16107-16112. [DOI] [PubMed] [Google Scholar]
- 30.Wei, L. N., X. Hu, D. Chandra, E. Seto, and M. Farooqui. 2000. Receptor-interacting protein 140 directly recruits histone deacetylases for gene silencing. J. Biol. Chem. 275:40782-40787. [DOI] [PubMed] [Google Scholar]
- 31.White, R., G. Leonardsson, I. Rosewell, M. A. Jacobs, S. Milligan, and M. Parker. 2000. The nuclear receptor co-repressor nrip1 (RIP140) is essential for female fertility. Nat. Med. 6:1368-1374. [DOI] [PubMed] [Google Scholar]
- 32.Windahl, S. H., E. Treuter, J. Ford, J. Zilliacus, J.-Å. Gustafsson, and I. J. McEwan. 1999. The nuclear-receptor interacting protein (RIP) 140 binds to the human glucocorticoid receptor and modulates hormone-dependent transactivation. J. Steroid Biochem. Mol. Biol. 71:93-102. [DOI] [PubMed] [Google Scholar]
- 33.Zaidi, S. K., A. Javed, J. Y. Choi, A. J. van Wijnen, J. L. Stein, J. B. Lian, and G. S. Stein. 2001. A specific targeting signal directs Runx2/Cbfa1 to subnuclear domains and contributes to transactivation of the osteocalcin gene. J. Cell Sci. 114:3093-3102. [DOI] [PubMed] [Google Scholar]
- 34.Zhong, S., P. Salomoni, and P. P. Pandolfi. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2:E85-E90. [DOI] [PubMed] [Google Scholar]
- 35.Zilliacus, J., E. Holter, H. Wakui, H. Tazawa, E. Treuter, and J.-Å. Gustafsson. 2001. Regulation of glucocorticoid receptor activity by 14-3-3-dependent intracellular relocalization of the corepressor RIP140. Mol. Endocrinol. 15:501-511. [DOI] [PubMed] [Google Scholar]