Abstract
The mammalian Bin1/Amphiphysin II gene encodes an assortment of alternatively spliced adapter proteins that exhibit markedly divergent expression and subcellular localization profiles. Bin1 proteins have been implicated in a variety of different cellular processes, including endocytosis, actin cytoskeletal organization, transcription, and stress responses. To gain insight into the physiological functions of the Bin1 gene, we have disrupted it by homologous recombination in the mouse. Bin1 loss had no discernible impact on either endocytosis or phagocytosis in mouse embryo-derived fibroblasts and macrophages, respectively. Similarly, actin cytoskeletal organization, proliferation, and apoptosis in embryo fibroblasts were all unaffected by Bin1 loss. In vivo, however, Bin1 loss resulted in perinatal lethality. Bin1 has been reported to affect muscle cell differentiation and T-tubule formation. No striking histological abnormalities were evident in skeletal muscle of Bin1 null embryos, but severe ventricular cardiomyopathy was observed in these embryos. Ultrastructurally, myofibrils in ventricular cardiomyocytes of Bin1 null embryos were severely disorganized. These results define a developmentally critical role for the Bin1 gene in cardiac muscle development.
Bin1 is a conserved member of the BAR family of genes that have been implicated in diverse cellular processes. All BAR gene products include a unique signature domain termed the BAR domain (49), which is evolutionarily conserved among a wide array of eukaryotic organisms (46, 47, 49). This domain has high α-helix-forming potential and can form both homo- and heterotypic interactions (20, 44, 54, 62) as well as bind acidic phospholipids (18, 56), but it remains functionally ill-defined. Four mammalian BAR protein-encoding genes have been reported in the literature, namely, Amphiphysin I, Bin1 (also known as Amphiphysin II), Bin2, and Bin3 (20, 32, 47, 49, 55). The products of these genes appear to be comprised solely of protein-protein interaction domains, suggesting that BAR proteins define a unique class of adapter proteins.
BAR proteins have been implicated in endocytosis, actin organization, programmed cell death, stress responses, and transcriptional control. The first mammalian BAR protein to be discovered, Amphiphysin I (AmphI), was identified in an immunoscreen for proteins associated with the plasma membranes of synaptic neurons (32). The AmphI-encoded gene product is comprised of an N-terminal BAR domain, a C-terminal SH3 domain, and a midsection that contains binding sites for the endocytic vesicle components clathrin and AP-2 (35, 49). Expression of AmphI is largely restricted to neuronal cells, where it forms interactions with synaptic vesicle components (11, 35, 36, 53). Ectopic overexpression of either the SH3 domain or the AP-2- and clathrin-binding region of AmphI impairs clathrin-coated vesicle endocytosis (50, 54, 63). These observations have been interpreted as evidence that AmphI functions in the control of clathrin-dependent synaptic vesicle endocytosis.
The mammalian Bin1 gene was first identified in a two-hybrid screen for polypeptides that bind to the N-terminal Myc box 1 (MB1) portion of the c-Myc oncoprotein (49). Bin1 is similar to AmphI in overall structure, with an N-terminal BAR domain and a C-terminal SH3 domain (49). However, the Bin1 gene is more complex than the AmphI gene, encoding at least seven different splice variants that differ widely in subcellular localization, tissue distribution, and ascribed functions (7, 43, 58, 60).
Different Bin1 isoforms are distinguished herein by indicating in parentheses alternatively spliced exons that are either missing or added relative to Bin1(+10), the first isoform to be identified which includes the alternately spliced exon 10. The (+10) designation is appended to this form to distinguish it from generic references to the Bin1 gene product. Two splice isoforms, Bin1(−10) and Bin1(−10 −13), are expressed ubiquitously. Bin1(−10) is a nucleocytosolic isoform that can complex with c-Myc, act as a transcriptional corepressor, and suppress the growth of oncogenically transformed cells and tumor cell lines (17, 49). Bin1(−10 −13), which was identified initially in a two-hybrid screen for c-Abl-interacting proteins (26), lacks a critical portion of the c-Myc binding domain. Coexpression of Bin1(−10 −13) with c-Abl induces a transformed morphology in NIH 3T3 cells that is associated with disruption of actin cytoskeletal organization. Five other splice isoforms [Bin1(+10) and at least four Bin1(−10 + 12) isoforms] exhibit tissue-specific expression. Expression of Bin1(+10), the isoform originally identified in the Myc two-hybrid screen, is restricted primarily to skeletal muscle, and this isoform has been implicated in mouse myoblast differentiation (31, 34, 61). Further supporting a role in muscle cells, Bin1(+10) has been localized to T tubules (7), and recent studies involving disruption of the likely Bin1 ortholog in Drosophila suggest that it functions in T-tubule organization (45). Differential splicing of exons 12A to D produces at least four Bin1 isoforms [the Bin1(−10 +12) isoforms that invariably contain exon 12A] which are expressed predominantly in brain (60). These isoforms most closely resemble AmphI both structurally and functionally and are often collectively referred to as Amphiphysin II (AmphII) (7, 30, 43, 62). The Bin1(−10 +12)/AmphII isoforms localize to the cytosol, where they can heterodimerize with AmphI (62) and associate with other components of clathrin-coated synaptic vesicles (40, 42). Like the AmphI SH3 domain, the Bin1/AmphII SH3 domain can, when overexpressed, impede clathrin-coated vesicle endocytosis, which has been interpreted as evidence that Bin1/AmphII has a role in this process (51).
Genetic studies in budding and fission yeast reinforce the theme from mammalian studies that BAR adapter proteins do not have a single, readily classifiable function. Two BAR adapter genes, analogous to mammalian Bin1 and Bin3, are conserved in both the budding yeast Saccharomyces cerevisiae (RVS167 and RVS161) and the fission yeast Saccharomyces pombe (hob1+ and hob3+). The rvs167 and rvs161 mutants were identified in a screen for mutations that cause reduced viability upon starvation (rvs phenotype) (3, 10). Other common phenotypes of rvs mutants include endocytosis defects, a depolarized actin cytoskeleton, and sensitivity to high salt and amino acid analog concentrations (3, 39, 52). rvs161 mutants additionally exhibit a defect in cell fusion during mating (6). The likely Bin1 homolog Rvs167p interacts physically and genetically with actin (1, 5) and may link the actin cytoskeleton to stress responses mediated by Pho85, a cell cycle-dependent kinase related to cdk5 that phosphorylates Rvs167p (8, 23, 29).
The fission yeast genes hob1+ and hob3+ (homolog of Bin1 and Bin3, respectively) (47, 48) share significant homology with their budding yeast counterparts; however, disruption of these genes affects distinct cellular functions. hob1Δ and hob3Δ mutants do not exhibit reduced viability upon nutrient starvation, increased osmolar sensitivity, or defective endocytosis. Instead, hob1Δ mutants exhibit defects in cell cycle arrest (48), whereas hob3Δ mutants exhibit defects in F-actin organization and cytokinesis (47). The mammalian Bin1 and Bin3 genes complement these respective defects (47, 48), arguing for evolutionary conservation of function in these processes.
While genetic investigations in yeast suggest that BAR proteins can be linked to endocytosis, actin cytoskeletal organization, and stress responses, these investigations do not identify a single cellular process in which BAR proteins are consistently required. To investigate the physiological role of the mammalian Bin1 gene, we have evaluated the consequences of homozygous Bin1 disruption in the mouse germ line. The results of this study provide an important genetic context in which to interpret the previous molecular and biochemical data relating to Bin1 function.
MATERIALS AND METHODS
Murine Bin1 gene disruption.
Construction of the Bin1 targeting vector, disruption of the gene in embryonic stem cells by homologous recombination, and generation of mice with germ line transmission of the null allele were conducted at the inGenious targeting laboratory (Stony Brook, N.Y.). The template used for construction of the targeting vector was a BAC clone of ∼100 kb encompassing the entire Bin1 gene within which the exons and introns have been mapped (34). The two arms of the targeting vector were constructed by PCR amplification and ligated into restriction enzyme sites flanking the neomycin resistance gene (Fig. 1). The 10-kb long arm was generated with the two primers 5′-GCCTATTACAGTGGGCCTTG-3′, located within intron 1 at a position 4.5 kb upstream of exon 2, and 5′-GCCAGATAGGTCCGAAGATCCTTC-3′, located within exon 3. The 1.0-kb short arm was generated with the two primers 5′-GGAGCTTGTGGAAGGAGCTGGAG-3′, located within intron 5 at a position 40 bp downstream of exon 5, and 5′-AGCTTCCGCCCCCGCTTGGCAATG-3′, located within exon 6.
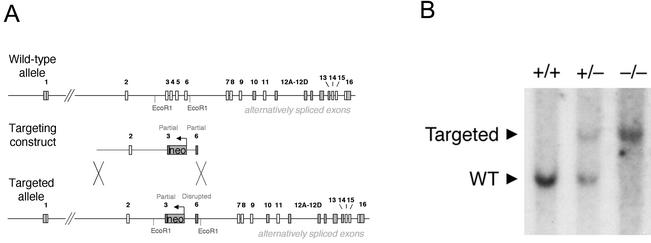
FIG. 1.
Disruption of the murine Bin1 gene by homologous recombination. (A) Bin1 targeting schematic. Organization of the wild-type Bin1 genomic locus has been determined previously (34). The targeting vector is comprised of the neomycin resistance gene flanked by a PCR-generated 10-kb long arm and 1-kb short arm. The targeted allele, produced by homologous recombination, has part of exon 3 and all of exons 4 and 5 replaced by the neomycin resistance gene transcribed in the opposing orientation relative to the Bin1 gene. Also shown are approximate locations of the EcoRI sites used to discriminate between the wild-type and recombinant alleles. (B) Southern blot analysis. Genomic DNA from Bin1+/+, Bin1+/−, and Bin1−/− primary MEF lines, previously genotyped by PCR, was digested with EcoRI. The sizes of the fragments detected by hybridization with a Bin1 cDNA probe are consistent with the pattern expected for the endogenous and targeted alleles (34). WT, wild type.
Ten micrograms of the targeting vector was linearized with NotI and used to transfect IT2 embryonic stem cells by electroporation. Following G418 selection, surviving colonies were expanded and analyzed by PCR to identify clones that had undergone homologous recombination. The primers used for this analysis were 5′-TGCGAGGCCAGAGGCCACTTGTGTAGC-3′, located within the neo gene cassette, and 5′-ATTTTGGCTTCATCCTTCTTTTTGG-3′, located within the middle of exon 6. A 1.2-kb PCR fragment was generated from clones that scored positive. Correctly targeted embryonic stem cell lines were microinjected into C57BL/6J host blastocysts, and chimeric mice were identified that exhibited germ line transmission of the disrupted Bin1 gene. Mice were interbred and maintained on a mixed genetic background of C57BL/6J and 129/SvJ.
Cell culture.
Mouse embryo fibroblasts (MEFs) were obtained from 13.5-day-postcoitus (dpc) embryos and routinely passaged on a 3T3-equivalent schedule (57) in Dulbecco's modified Eagle medium (DMEM) containing 10% calf serum (Colorado Serum). To establish these cultures, embryos were eviscerated, and the torso was minced and digested for approximately 45 min at 37°C in 6 ml of 0.05% trypsin with occasional vortexing. Six milliliters of DMEM + 10% calf serum was then added to neutralize the trypsin. Large chunks of tissue were allowed to settle out, and the supernatant was transferred to another tube. The cells were then spun at 1,000 rpm for 5 min (Sorvall RT6000), and the cell pellet was resuspended in 10 ml DMEM-10% calf serum. Cells from a single embryo were plated on a single 10-cm-diameter tissue culture dish.
Immortalized macrophage cultures were produced by infection of early-passage MEF cultures with retrovirus expressing c-Myc. This resulted in the outgrowth of clusters of rounded, refractile cells which became the predominant cell type following several rounds of passage. These cells were determined to be macrophages by virtue of Mac-1 antigen positivity (fluorescein isothiocyanate [FITC]-conjugated, anti-Mac-1 antibody [Pharmingen]), NO production in response to lipopolysaccharide (27), and phagocytic capability (described below). L-cell conditioned medium was prepared as described previously (19) as a source of colony-stimulating factor 1 to support macrophage growth.
Cortical neuron cultures were prepared from 18.5-dpc embryos as described previously (2). Cells were plated at 200,000 cells/well in 12-well poly-d-lysine-coated plates and grown in B27-neurobasal plating medium supplemented with 10% fetal calf serum. Following 3 weeks in culture, cells were fixed and prepared for electron microscopy.
Molecular biology assays.
Standard PCR-based analysis of genomic DNA was performed to distinguish between Bin1 wild-type and null alleles for routine genotyping of mice. Three primers were employed, namely, Bin1e3F1 (5′-GAGGGTACCCGGCTGCAG-3′), a forward primer within the retained portion of Bin1 exon 3; Bin1e4R1 (5′-CTTGTTTGCTTCATCCCTGCC-3′), a reverse primer within the targeted Bin1 exon 4; and NeoR3 (5′-CCTGCCCATTCGACCACC-3′), a reverse primer within the integrated neomycin resistance gene. Platinum Taq (Life Technologies) was used as recommended by the manufacturer to catalyze reactions run as follows: denaturation at 94°C for 5 min (1 cycle); 94°C for 20s, 65°C for 1 min, and 72°C for 1 min (35 cycles); and extension at 72°C for 5 min (1 cycle). Bin1-null and wild-type products of ∼1.2 and ∼0.3 kb, respectively, were resolved on 1.0% Tris-borate-EDTA-agarose gels and visualized by ethidium bromide staining.
Western blotting procedures have been described for the detection of Bin1 protein with antibodies 99D (49) and 2F11 (14). The antibodies anti-amphiphysin (Transduction Laboratories) and Actin (I-19) (Santa Cruz Biochemicals), which recognizes a broad range of actin isoforms, were used as recommended by the manufacturers.
Cell-based assays.
To prepare retroviruses, plasmids were first transiently transfected with Lipofectamine 2000 (Invitrogen) into Phoenix cells (a 293 cell-derived retroviral packaging line [41]) plated the previous day at 2 × 106 cells per 10-cm-diameter dish. High-titer retrovirus stocks were prepared for use as described previously (38, 41). Briefly, at the end of the day following the transfection, 5 ml of new media was added to the plates. The following morning, the supernatant was collected, combined with 1 ml of fetal calf serum, passed through a 0.5-μm-pore-size filter, and dispensed into two single-use 3-ml aliquots which were stored frozen at −80°C. To infect a 10-cm-diameter plate of cells, 3 ml of virus was rapidly thawed in a 37°C water bath, added to cells together with 4 μg of polybrene/ml, and incubated at 37°C with intermittent rocking for 3 h.
Fluorescent staining of actin filaments with FITC-conjugated phalloidin (Molecular Probes) was performed as recommended by the manufacturer. Endocytic uptake of FITC-conjugated transferrin (Molecular Probes) by MEF lines was assessed as previously described for transfected COS cells (20). Published methodology (21) was also followed to assess phagocytic uptake of FITC-conjugated zymosan (Molecular Probes).
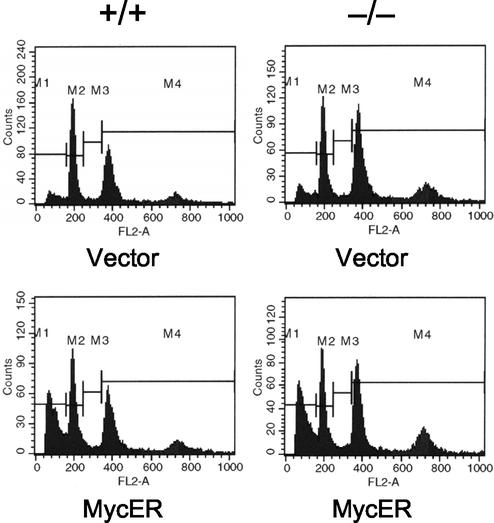
Apoptosis triggered by c-Myc in response to serum deprivation was assessed in embryo fibroblast populations by standard flow cytometric detection of propidium iodide-stained cells exhibiting sub-G1 DNA content. Early-passage MEFs (second passage following establishment of cultures) were plated at 5 × 105 cells per 10-cm-diameter dish and infected with the conditional c-Myc-expressing retrovirus pBabepuro-MycER (a gift of G. Evan [33]) or empty vector the following day. Stably infected cells were selected 48 h later in medium containing 2 μg of puromycin/ml. Three days after selection, 5 × 105 cells were seeded into medium containing 0.1 μM 4-hydroxytamoxifin (4HT). The next day, the cells were washed and incubated in serum-free medium plus 4HT. Additional control populations were treated similarly, except that cells were either maintained in 10% serum or not exposed to 4HT. Cells were harvested 24 and 48 h later and stained with propidium iodide for flow cytometry analysis.
In vivo studies.
Histopathology was performed on embryos that were decapitated, skinned, fixed in 10% buffered formalin overnight, and stored in 70% ethanol prior to mounting in paraffin blocks. For histological evaluation of tissues, sections were stained with hematoxalin and eosin (H&E). Immunohistochemical staining with the anti-Bin1 monoclonal antibody 2F11 at a concentration of 2 μg/ml was performed with the M.O.M. immunodetection kit (Vector Laboratories) as recommended by the manufacturer. Staining with rabbit polyclonal antibody to Ki67 antigen (Novocastra Laboratories) was performed with the Vectastain ABC kit peroxidase rabbit immunoglobulin G (IgG) (Vector Laboratories) as recommended by the manufacturers. To prepare tissue biopsy samples for electron microscopic examination, heart and skeletal muscle samples were obtained from 18.5-dpc embryos with a dissecting microscope. The tissue biopsy samples were fixed in 2.5% paraformaldehyde and processed for embedding and sectioning. The electron micrographs shown were captured at either ×15,000 or ×50,000 magnification.
RESULTS
Disruption of the murine Bin1 gene results in perinatal lethality.
The BAR domain is critical to the antitransforming and proapoptotic activities that have been ascribed to Bin1 (16, 17). We have disrupted the endogenous murine Bin1 gene by replacing exons which encode the most highly conserved central region of the BAR domain with a neomycin resistance gene cassette (34) (Fig. 1A). Integration of the targeting vector is predicted to result in expression of a small, inactive peptide of ∼70 amino acids encoded by exons 1 and 2 and a portion of exon 3. The targeting vector also disrupts the open reading frame in exon 6, so that if exon 3 is bypassed by alternative splicing of exon 2, the translated product will terminate in exon 6.
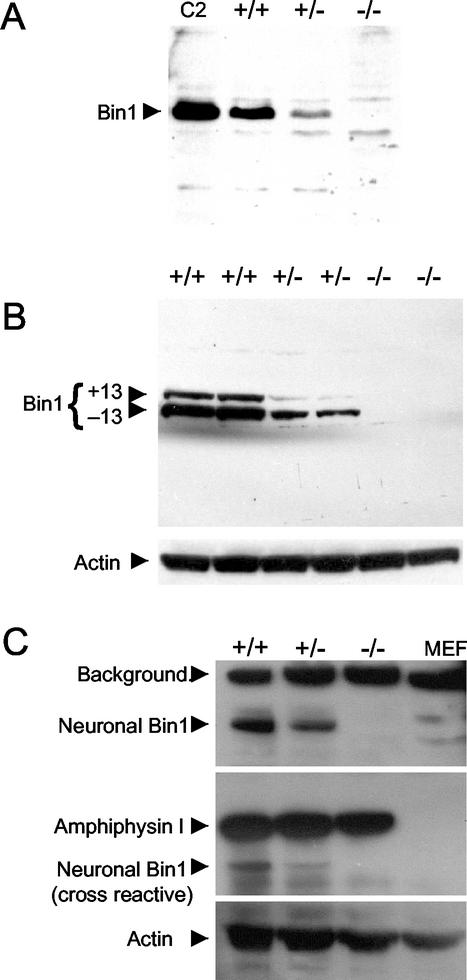
Southern blot analysis of genomic DNA from late-stage embryos confirmed that the integrated targeting vector produces a restriction fragment of the predicted size, consistent with homologous recombination into the genome (Fig. 1B). Northern blot analysis, which demonstrated the absence of detectable Bin1 transcript in Bin1−/− cells, has been reported elsewhere (13). Loss of Bin1 protein expression was confirmed by Western blot analysis of MEF cell lysates with two different Bin1-specific monoclonal antibodies. Antibody 99D recognizes an epitope within the C-terminal Myc binding domain encoded by exon 13 (49), and antibody 2F11 recognizes an epitope within the N-terminal BAR domain encoded by exons 7 and 8 (14). Bin1 protein was undetectable in null cell lysates and was reduced in heterozygous cell lysates relative to wild-type levels (Fig. 2A and B). Disruption of the AmphI gene in mice has recently been reported to result in a dramatic reduction in expression of the neuronal Bin1(−10 +12)/AmphII isoforms, suggesting that the levels of AmphI and neuronal Bin1(−10 +12)/AmphII protein may be coordinately regulated in the brain (12). However, Western blot analysis revealed that Bin1−/− brain tissue has normal levels of AmphI protein, indicating that there is no reciprocal impact of deleting the Bin1 gene on the level of AmphI expressed in the brain (Fig. 2C).
FIG. 2.
Western blot analyses of protein expression following Bin1 gene disruption. (A) Detection of Bin1 protein with anti-Bin1 monoclonal antibody 99D (epitope in alternately spliced exon 13) (59). Extracts were prepared from Bin1+/+, Bin1+/−, and Bin1−/− primary MEF lines, which had been previously evaluated by Southern blot analysis (Fig. 1B). Lane C2, C2C12 myoblast extract (positive control). (B) Detection of Bin1 protein with anti-Bin1 monoclonal antibody 2F11 (epitope in exons 7 to 8) (14). Extracts were prepared from two independent primary MEF lines for each genotype. The blot was stripped and reprobed for actin to evaluate protein loading. (C) Bin1 deletion does not affect AmphI protein expression in mouse brain. Detection of Bin1 protein by using monoclonal antibody 2F11 (top panel), AmphI protein (middle panel), and actin protein loading control (bottom panel). Extracts were prepared from brain tissue of genotyped 18.5-dpc mouse embryos. Extract prepared from Bin1+/+ MEFs was also included as a control.
Nullizygous Bin1 embryos were found to develop to term with no apparent deviation from a normal Mendelian ratio. Genotypic evaluation of 223 embryos from crosses between Bin1 heterozygotes was conducted at 18.5 dpc just prior to parturition. Of these 223 embryos, 50 were Bin1−/−, 117 were Bin1+/−, and 56 were Bin1+/+. Nullizygous Bin1 pups did not, however, survive to weaning age. Of 182 pups born to heterozygous parents, 0 were Bin1−/−, 117 were Bin1+/−, and 65 were Bin1+/+ at 3 to 4 weeks postpartum. To determine more precisely when the onset of mortality occurs, 50 neonates from Bin1 heterozygous parents were closely monitored from birth. Seventeen pups died within the first 24 h, and all were Bin1−/−. The remaining pups were either Bin1+/− or Bin1+/+. Five of the Bin1−/− pups were born either dead or moribund, but the rest were indistinguishable from their littermates at birth. During the next several hours, however, the remaining Bin1−/− pups failed to nurse and grew progressively weaker before expiring.
Anticipated defects in cellular processes are lacking in primary Bin1 null cells.
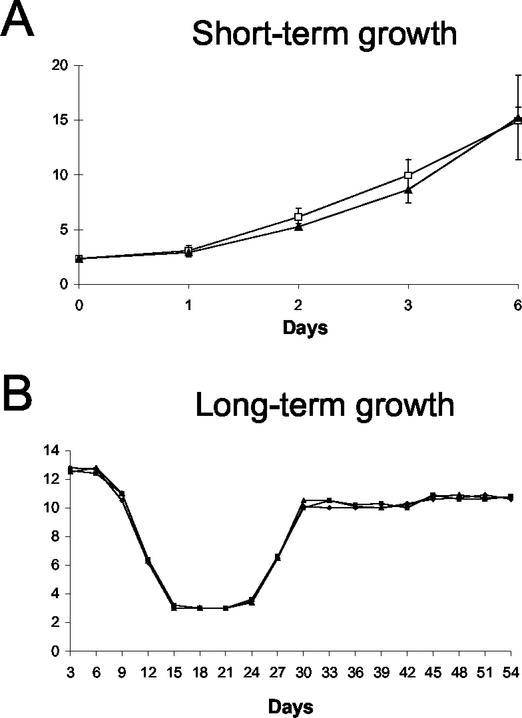
Primary cultures of MEFs were established to evaluate how the loss of Bin1 gene expression affects normal cellular physiology. No significant impact was observed on the short-term growth rate of early-passage MEFs lacking Bin1 expression (Fig. 3A). To evaluate the effect of Bin1 loss on long-term growth, MEF cultures were immortalized by passage on a 3T3 schedule. During establishment, there was no discernible effect of Bin1 loss on proliferation, and Bin1−/− MEFs entered and exited crisis concurrently with Bin1+/+ and Bin1+/− MEFs (Fig. 3B).
FIG. 3.
In vitro proliferation of Bin1 null MEFs. (A) Short-term growth assay. Early passage Bin1+/+ and Bin1−/− MEFs were seeded at an initial plating density of 2.0 × 104 cells per 10-cm2 dish. Viable cell numbers were determined in triplicate by trypan blue exclusion at the times indicated. (B) Long-term growth assay. Early passage Bin1+/+, Bin1+/−, and Bin1−/− MEFs were seeded at 3 × 105 cells per 6-cm2 dish in duplicate and carried thereafter on a 3T3 schedule. Every 3 days, cells were counted and reseeded at to a cell density to 3 × 105 cells per 6-cm2 dish. Cell counts over 18 passages are graphed.
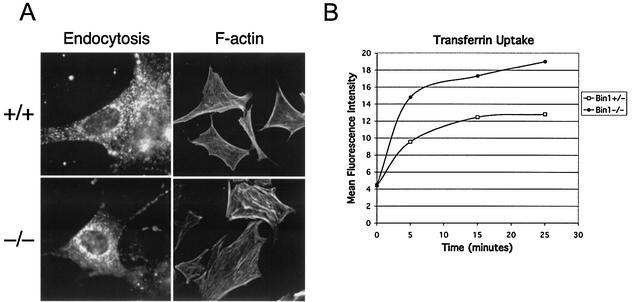
Bin1−/− MEFs clearly retained endocytic function, as assessed by fluorescent microscopic examination of cells incubated with FITC-labeled transferrin (Fig. 4A). A series of fluorescence-activated cell sorter profiles of both Bin1+/− and Bin1−/− MEFs incubated with FITC-labeled transferrin over time were obtained in order to quantitatively assess the impact of Bin1 loss on endocytosis. No demonstrable impairment of transferrin uptake was observed in the Bin1 null MEFs, which actually exhibited somewhat higher levels of transferrin accumulation than did their wild-type counterparts in this particular assay (Fig. 4B). The organization of filamentous actin stress fibers in these cells likewise appeared to be unaffected by Bin1 loss (Fig. 4A). Finally, Bin1 loss had no apparent effect on serum withdrawal-induced apoptosis mediated by ectopic expression of c-Myc in early-passage MEFs (Fig. 5). In these cells, c-Myc does not cause neoplastic transformation, which in several cell systems has been found to be a prerequisite for Bin1 to be proapoptotic (13-15). In summary, there was no demonstrable impact of Bin1 loss on endocytosis, actin cytoskeletal organization, or serum deprival-induced apoptosis triggered by c-Myc in nontransformed, primary MEFs.
FIG. 4.
Endocytic uptake and actin cytoskeletal organization of Bin1 null MEFs. (A) Microscopic evaluation of actively proliferating, early-passage primary MEFs. The first column shows accumulated, intercellular FITC-conjugated transferrin internalized by receptor-mediated endocytosis. The second column shows F-actin organization as revealed by FITC-conjugated phalloidin staining. (B) Flow cytometry assessment of transferrin uptake by early-passage primary MEFs over time. Total mean fluorescence intensity for 10,000 cells was determined for each sample, and the results are presented graphically.
FIG. 5.
Sensitivity of Bin1 null MEFs to c-Myc-mediated apoptosis in response to serum withdrawal. Flow cytometry profiles of propidium iodide-stained, early-passage MEF populations transduced with either MycER-expressing or empty control retroviruses and subsequently deprived of serum for 48 h. All cultures were additionally treated with 4-hydroxytamoxifen, necessary to induce activity of the conditional MycER protein, 24 h prior to serum withdrawal. The proportions of cells with sub-G1 DNA content were 31% of MycER-expressing Bin1+/+ MEFs (bottom left) and 30% of MycER-expressing Bin1−/− MEFs (bottom right).
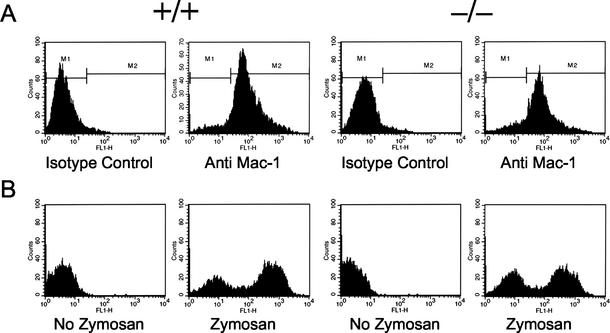
Phagocytosis by macrophages has been reported to require the Bin1(−10 −13) splice isoform (referred to as Amphiphysin IIm), based on the use of a dominant negative strategy to inhibit Bin1 (21). However, as shown in Fig. 6, macrophages lacking Bin1 were quantitatively as efficient at phagocytic internalization of FITC-labeled zymosan as their wild type counterparts, demonstrating that Bin1 is dispensable for the specialized endocytic process of phagocytosis.
FIG. 6.
Phagocytic uptake by Bin1 null macrophages. Flow cytometry results from a representative set of c-Myc immortalized macrophages; identical results were obtained with primary macrophages. (A) Mac-1 staining. Cells were stained with FITC-conjugated antibody recognizing the macrophage-specific surface antigen Mac-1. More than 90% of the cells in each preparation stained positive. (B) Phagocytosis assay. Fluorescence-activated cell sorter profiles of Bin1+/+ and Bin1−/− macrophages exposed to FITC-conjugated zymosan, which is internalized through a phagocytosis. Both Bin1+/+ and Bin1−/− populations exhibit biphasic peaks of brightly and dimly fluorescing cells. The brightly fluorescent peak represents 55% of the Bin1+/+ population and 48% of the Bin1−/− population.
Bin1 null embryos develop ventricular cardiomyopathy.
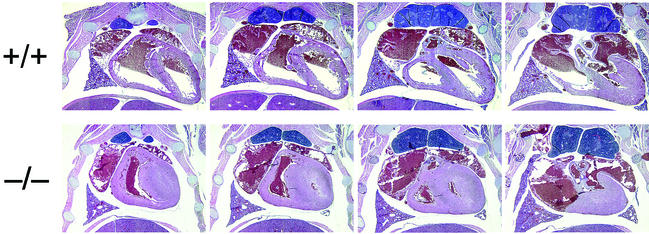
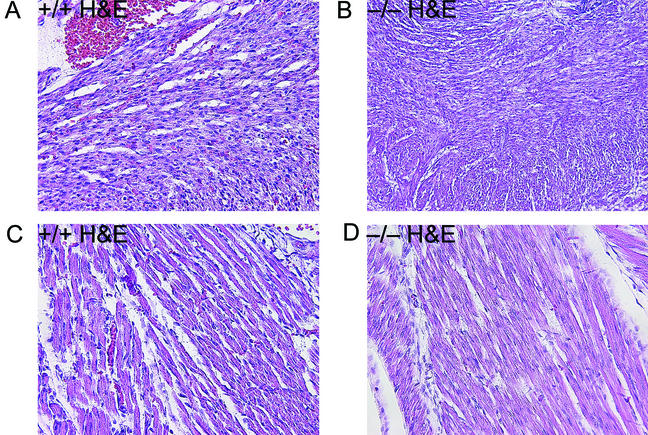
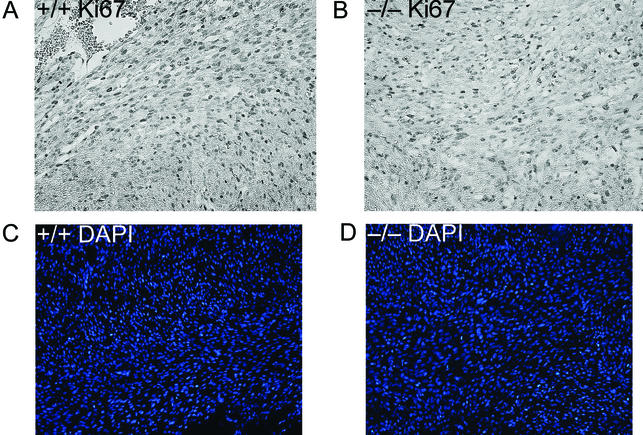
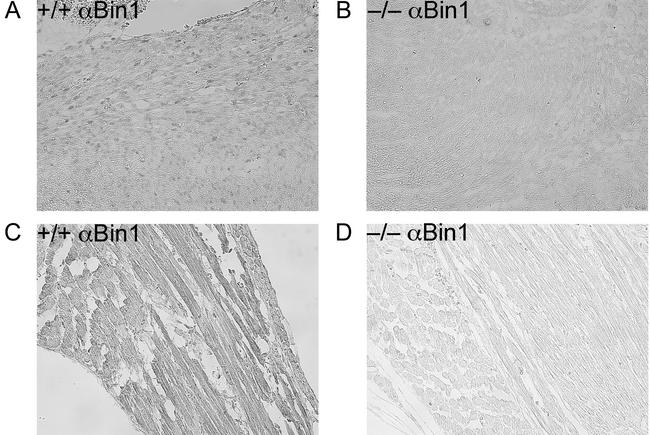
Sagittal and transverse whole-mount serial sections from several litters of late-stage embryos and neonates were examined histologically for evidence of developmental defects. No gross abnormalities were identified in organs lacking Bin1, with the notable exception of the heart. As shown in Fig. 7, the ventricular wall of the Bin1 null heart is markedly increased in thickness, resulting in occlusion of both ventricular chambers. In contrast, Bin1 null skeletal muscle appeared to be developmentally normal throughout the whole-mount sections (data not shown). Also, unlike Bin1 null muscle fibers (Fig. 8C and D), Bin1 null cardiomyocytes appeared to be more densely packed than those of the wild type, with indications of haphazardly distributed myofibers (Fig. 8A and B). The ventricular myocardium lacking Bin1 did not, however, exhibit a significant increase in the frequency of nuclei staining positive for the nuclear proliferation antigen Ki67 (Fig. 9A and B) or a significant increase in density of cell nuclei (Fig. 9C and D). The lack of evidence for hyperplastic growth is consistent with the idea that hypertrophic growth, possibly in response to disarrayed fibers as occurs in human familial hypertrophic cardiomyopathy (4), is the basis for the observed cardiac phenotype in Bin1 null mice. Immunostaining revealed a predominantly nuclear pattern of Bin1 localization in the ventricular cardiomyocytes of wild-type animals (Fig. 10A and B). In differentiated skeletal muscle, a cytosolic pattern of Bin1 staining was observed, which, in accord with previous in vitro and in vivo observations (7, 34, 61), appears to be concentrated along myofibers (Fig. 10C,D).
FIG. 7.
Embryonic cardiomyopathy is associated with Bin1 loss. Serial transverse sections through the thoracic cavities of a pair of Bin1+/+ and Bin1−/− littermates at 1 day postpartum are shown. Along the top row (labeled +/+) and arranged in a ventral-to-dorsal orientation (from left to right) are sections from a Bin1 homozygous wild-type neonate. Along the bottom row (labeled −/−) are comparable sections from a Bin1 homozygous knockout littermate. Sections were stained with H&E and captured at ×20 magnification.
FIG. 8.
Cardiac and skeletal muscle histology. Photomicrographs of H&E-stained whole-embryo sections obtained from 18.5-dpc littermates (magnification, ×200). (A and B) Cardiac muscle from a Bin1+/+ (A) and Bin1−/− (B) animal. (C and D) Skeletal muscle from a Bin1+/+ (C) and Bin1−/− (D) animal.
FIG. 9.
Assessment of cardiomyocyte proliferation and density. Photomicrographs of cardiac muscle from transverse whole-embryo sections obtained from 18.5-dpc littermates (magnification, ×200). (A and B) Staining with antibody to the nuclear proliferation marker Ki67 in a Bin1+/+ (A) and Bin1−/− (B) animal. (C and D) DAPI (4′,6′-diamidino-2-phenylindole) staining of nuclei from a Bin1+/+ (C) and Bin1−/− (D) animal.
FIG. 10.
Immunohistochemical detection of Bin1 in cardiac and skeletal muscle. Photomicrographs of transverse whole-embryo sections obtained from 18.5-dpc littermates and stained with anti-Bin1 monoclonal antibody 2F11 (magnification, ×200). (A and B) Cardiac muscle from a Bin1+/+ (A) and Bin1−/− (B) animal. (C and D) Skeletal muscle from a Bin1+/+ (C) and Bin1−/− (D) animal.
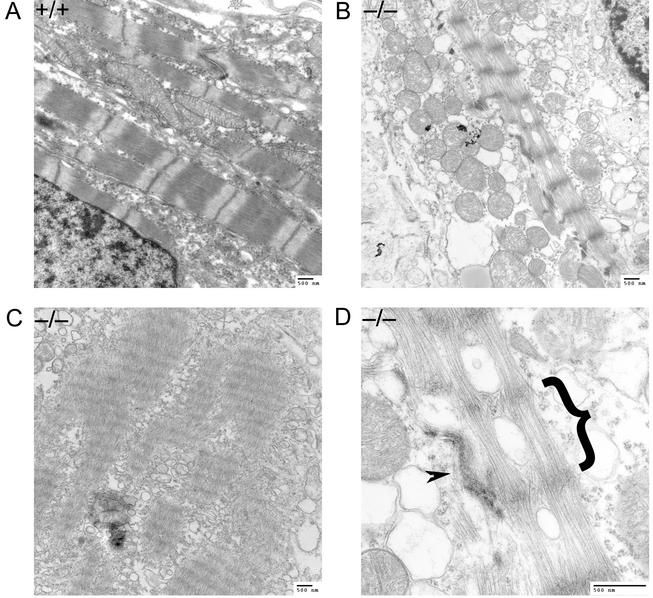
Results of recent studies in Drosophila (45) and in the mouse myoblast cell line C2C12 (28) suggest that Bin1 may be involved in the formation of subcellular structures within muscle fibers, especially of the myofibril-associated T-tubule system. However, as shown in Fig. 11, more severe defects than would be anticipated from these studies are apparent in electron micrographs of myofibrils from Bin1−/− ventricular cardiomyocytes. Compared with myofibrils from a wild-type animal, the myofibrils from null animals are loosely packed and disorganized. The sarcomere units are also clearly defective, with diffuse Z lines and no apparent A bands present. The impact of Bin1 loss on skeletal muscle ultrastructure was less dramatic, and a more thorough electron microscopic analysis of Bin1 null skeletal muscle will be necessary to substantiate the presence of significant defects in this tissue. Indications of loose myofibril packing and mislocalization of T-tubule triads to Z lines were, however, observed (data not shown).
FIG. 11.
Ventricular cardiomyocytes of Bin1 null embryos exhibit aberrant myofibril organization. (A) Electron micrograph showing a typical transverse section of cardiac myofibrils from a wild-type embryo. (B) Cardiac myofibrils from a Bin1−/− embryo. (C) Cardiac myofibrils from a second Bin1−/− embryo. (D) Higher-magnification image of the Bin1−/− embryo cardiac myofibril from panel B. A single aberrant sarcomere from presumptive Z line to Z line is indicated by the bracket. An adjacent intercalated disk, a structure unique to cardiac muscle, is designated with an arrowhead.
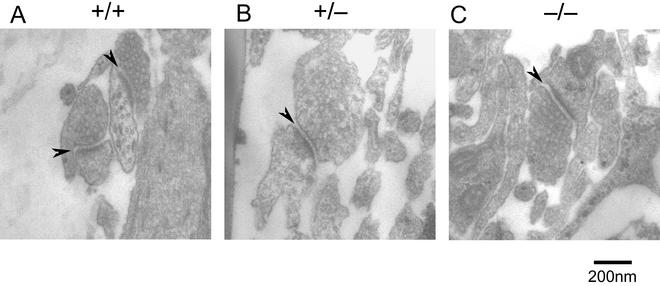
The restricted expression of specific splice isoforms of Bin1 in neuronal cells suggests that Bin1 may have a specialized role(s) in this tissue. No apparent histological defects in brain architecture were observed in either late-stage embryos or neonates lacking Bin1 (data not shown). Bin1(−10 +12)/AmphII has been shown to associate with several components of clathrin-coated vesicles. Mice harboring null alleles of one of these components, the polyphosphoinositide phosphatase Synaptojanin 1, accumulate clathrin-coated vesicles adjacent to the synaptic junction that can be visualized by electron microscopy (9). To examine whether Bin1 loss might cause a similar defect, cortical neuron cultures were prepared from 18.5-dpc embryos and grown for 3 weeks in vitro to permit the maturation of synaptic contacts. Electron microscopy revealed no evidence of increased numbers of electron-dense, clathrin-coated vesicles within synapses formed by Bin1−/− neurons (Fig. 12). Synaptojanin 1 has been proposed to play a role in the regulation of synaptic vesicle uncoating; thus the failure of Bin1 loss to phenocopy Syntaptojanin 1 loss argues that that there is not an essential requirement for Bin1 function in this process.
FIG. 12.
Synaptic junctions formed by Bin1 null neurons in vitro. Electron micrographs of cortical neuron cultures showing typical synaptic profiles from Bin1+/+ (A), Bin1+/− (B), and Bin1−/− (C) cells. Synaptic junctions (indicated by arrows) are recognizable as characteristic electron-dense thickenings at the interface between cell membranes. In all cases, the presynaptic element is tightly packed with vesicles that are morphologically indistinguishable.
DISCUSSION
In this study, we examined the physiological consequences of Bin1 gene deletion in the mouse. Bin1 loss resulted in perinatal lethality, indicating that it has a key role in mouse development. Several independent studies have demonstrated that inhibition of Bin1 can impair the in vitro differentiation of C2C12 mouse myoblasts (28, 31, 61), suggesting a role for Bin1 in muscle differentiation. While Bin1−/− embryos and neonates did not exhibit overt histological changes in skeletal muscle, development of severe ventricular cardiomyopathy was observed in the heart. As with most forms of cardiomyopathy in humans, the observed cardiac phenotype in Bin1 null mice appears to reflect hypertrophy and disarray of ventricular cardiomyocytes rather than hyperplasia. Further work is needed to determine the precise developmental progression of the cardiomyopathy in Bin1 null mice and the possible equivalency to human forms of the disease.
Molecular genetic studies have identified mutations in genes that encode sarcomeric proteins as the basis for familial hypertrophic cardiomyopathies (4). Bin1 previously has been determined to localize within the vicinity of T tubules in skeletal muscle (7), and results obtained from both Drosophila and in C2C12 myoblasts have suggested that Bin1 loss may result in malformation of the T-tubule system (28, 45). The effects of Bin1 loss on myofibril structure in ventricular cardiomyocytes, however, were much more pronounced than would have been expected from these previous studies. The severity of the impact of Bin1 loss suggests the hypothesis that loss and/or disruption of normal sarcomeric structures may be the underlying molecular basis for the cardiomyopathy that develops in Bin1 null mice. Interestingly, the impact of Bin1 loss on skeletal muscle myofibril ultrastructure did not appear to be as pronounced. This correlates with different patterns of Bin1 subcellular localization that were observed in skeletal muscle and cardiac muscle cells. Bin1 staining was predominantly cytoplasmic in skeletal muscle cells, where Bin1 previously has been reported to concentrate along muscle fibers (7), whereas in heart muscle cells, Bin1 staining was predominantly nuclear. This suggests that Bin1, through interaction with nuclear proteins, may have an indirect role in the formation of cardiac myofibrils and that this role may be of greater consequence to myofibril architecture than direct effects that Bin1 may have through interaction with muscle fiber-associated components. Further work will be needed to elucidate the molecular role that Bin1 plays in the formation of myofibrils and associated structures in both cardiac and skeletal muscle.
The lack of any apparent effects of Bin1 loss on endocytotic processes was somewhat unexpected. The notion that Bin1 is important for endocytic function in mammalian cells is based predominantly on evidence that (i) particular Bin1 isoforms associate with the endocytic machinery in neuronal cells and macrophages (7, 21, 30, 43, 62), and that (ii) putative Bin1 dominant negative mutants can impair endocytic and phagocytotic processes (21, 51). It has also been demonstrated that expression of either full-length AmphI or Bin1(−10 +12)/AmphII alone can inhibit endocytosis in COS cells, while coexpression of their two genes rescues this inhibition (62). This result has been interpreted as indicating that the heterodimer of AmphI and Bin1(−10 +12)/AmphII is required for endocytic function and that expression of either gene product alone may have a dominant negative effect.
The simplest interpretation of our observations is that Bin1 lacks a necessary functional role in generalized endocytosis or in phagocytosis. If this is the case, the effects documented in dominant negative studies may represent an artifact of overexpressing protein-binding domains that disrupt other critical interactions. Our findings are supported by recent work in Drosophila Amphiphysin knockouts, which confirm the idea that BAR adapter proteins are not essential components of the endocytic machinery. In two independent studies, disruption of the sole BAR domain-containing gene identified in Drosophila (likely the Bin1 homolog) did not detectably affect synaptic vesicle endocytosis (45, 64). Similarly, recent work in fission yeast likewise demonstrated that disruption of hob1+, the Bin1 homolog in S. pombe, also does not affect endocytosis (48). Direct evaluation in cultured neuronal cells is still necessary to rigorously evaluate whether there is a specialized requirement for Bin1 in mammalian synaptic vesicle endocytosis. However, a recent report by DiPaolo et al. (12) may be germane to this question. In this report, AmphI knockout mice were also found to exhibit dramatically reduced expression of the brain-specific, amphiphysin-like Bin1(−10 +12)/AmphII isoforms (12). Nevertheless, despite the virtual absence of both gene products in the central nervous system, basal synaptic vesicle endocytic uptake was not significantly different than in wild-type synapses, although endocytic uptake after high K+ stimulation was about 60 to 70% of stimulated wild-type levels. This suggests that the Bin1(−10 +12)/AmphII knockout mouse may also retain synaptic vesicle endocytic function but that the response to certain types of regulatory signal transduction events may be impaired.
In budding yeast, BAR proteins have been physically and genetically linked to actin organization. However, this does not appear to hold true for the Bin1 homolog in fission yeast (48), and consistent with these findings, we did not observe any gross impact of Bin1 deletion on actin cytoskeletal organization in mouse fibroblasts. c-Myc triggers apoptosis in response to serum deprival-induced stress, somewhat reminiscent of the rvs stress response phenotype of budding yeast, and Bin1 can promote apoptosis in a c-Myc-dependent manner in transformed cells and tumor cells, although not in nontransformed cells (15, 16). In primary MEFs with constitutively overexpressed c-Myc, we did not observe any effect of Bin1 deletion on apoptosis elicited by serum withdrawal. This suggests that the consequences of Bin1 expression may be different under pathophysiologic and physiologic conditions, consistent with a hypothesized role for Bin1 in stress signaling processes suggested by fission yeast studies (45). Further investigation is needed to assess the connections between Bin1, the actin cytoskeleton, and stress responses, including c-Myc-dependent apoptosis, in other murine cell types, including in tumor-derived and oncogenically transformed cells.
Certain Bin1 splice isoforms have been reported to interact with Myc gene family members both physically and functionally (17). In vivo data implicate Myc gene family members in cardioventricular development. The ectopic expression of a c-Myc transgene in the hearts of developing embryos caused increased cardiac mass primarily as a consequence of cardiomyocyte hyperplasia (24, 25). Perhaps even more germane are reports that mouse embryos that carry one null and one hypomorphic allele of N-Myc develop an abnormally thin ventricular wall resulting from apparent hypoplasia of the compact subepicardial layer of the myocardium (37). Thus it is clear that the level of Myc activity is critical for proper cardioventricular organogenesis. Although the evidence so far suggests that Bin1 loss induces hypertrophy rather than hyperplasia, given that Bin1 is predominantly localized to the nucleus of ventricular cardiomyocytes and can functionally interact with both c-Myc and N-Myc proteins (17, 22), it will be interesting to determine whether Bin1 loss in ventricular cardiomyocytes directly affects the control of Myc activity in a tissue-restricted manner in the heart.
Acknowledgments
We thank Diane Sharp for performing flow cytometry analysis and Elsa Aglow for histology. We gratefully acknowledge support from Robert B. Stein during the course of the work at the former DuPont Pharmaceuticals Company.
This work was supported in part by a grant from the U.S. Army Breast Cancer Research Program (G.C.P.).
REFERENCES
- 1.Balguerie, A., P. Sivadon, M. Bonneu, and M. Aigle. 1999. Rvs167p, the budding yeast homolog of amphiphysin, colocalizes with actin patches. J. Cell Sci. 112(Pt. 15):2529-2537. [DOI] [PubMed] [Google Scholar]
- 2.Banker, G. A., and K. Goslin. 1991. Culturing nerve cells. MIT Press, Cambridge, Mass.
- 3.Bauer, F., M. Urdaci, M. Aigle, and M. Crouzet. 1993. Alteration of a yeast SH3 protein leads to conditional viability with defects in cytoskeletal and budding patterns. Mol. Cell. Biol. 13:5070-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonne, G., L. Carrier, P. Richard, B. Hainque, and K. Schwartz. 1998. Familial hypertrophic cardiomyopathy: from mutations to functional defects. Circ. Res. 83:580-593. [DOI] [PubMed] [Google Scholar]
- 5.Breton, A. M., and M. Aigle. 1998. Genetic and functional relationship between Rvsp, myosin and actin in Saccharomyces cerevisiae. Curr. Genet. 34:280-286. [DOI] [PubMed] [Google Scholar]
- 6.Brizzio, V., A. E. Gammie, and M. D. Rose. 1998. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. J. Cell Biol. 141:567-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler, M. H., C. David, G.-C. Ochoa, Z. Freyberg, L. Daniell, D. Grabs, O. Cremona, and P. De Camilli. 1997. Amphiphysin II (SH3P9; BIN1), a member of the amphiphysin/RVS family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of Ravier in brain and around T tubules in skeletal muscle. J. Cell Biol. 137:1355-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colwill, K., D. Field, L. Moore, J. Friesen, and B. Andrews. 1999. In vivo analysis of the domains of yeast Rvs167p suggests Rvs167p function is mediated through multiple protein interactions. Genetics 152:881-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cremona, O., G. Di Paolo, M. R. Wenk, A. Luthi, W. T. Kim, K. Takei, L. Daniell, Y. Nemoto, S. B. Shears, R. A. Flavell, D. A. McCormick, and P. De Camilli. 1999. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell 99:179-188. [DOI] [PubMed] [Google Scholar]
- 10.Crouzet, M., M. Urdaci, L. Dulau, and M. Aigle. 1991. Yeast mutant affected for viability upon nutrient starvation: characterization and cloning of the RVS161 gene. Yeast 7:727-743. [DOI] [PubMed] [Google Scholar]
- 11.David, C., P. S. McPherson, O. Mundigl, and P. de Camilli. 1996. A role of amphiphysin in synaptic vesicle endocytosis suggested by its binding to dynamin in nerve terminals. Proc. Natl. Acad. Sci. USA 93:331-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Paolo, G., S. Sankaranarayanan, M. R. Wenk, L. Daniell, E. Perucco, B. J. Caldarone, R. Flavell, M. R. Picciotto, T. A. Ryan, O. Cremona, and P. De Camilli. 2002. Decreased synaptic vesicle recycling efficiency and cognitive deficits in amphiphysin 1 knockout mice. Neuron 33:789-804. [DOI] [PubMed] [Google Scholar]
- 13.DuHadaway, J. B., W. Du, P. S. Donover, J. Baker, A. Liu, D. M. Sharp, A. J. Muller, and G. C. Prendergast. Transformation selective apoptosis triggered by farnesyltransferase inhibitors requires Bin1. Oncogene, in press. [DOI] [PubMed]
- 14.DuHadaway, J. B., F. J. Lynch, S. Brisbay, C. Bueso-Ramos, P. Troncoso, T. McDonnell, and G. C. Prendergast. 2003. Immunohistochemical analysis of Bin1/Amphiphysin II in human tissues: diverse sites of nuclear expression and losses in prostate cancer. J. Cell. Biochem. 88:635-642. [DOI] [PubMed] [Google Scholar]
- 15.DuHadaway, J. B., D. Sakamuro, D. L. Ewert, and G. C. Prendergast. 2001. Bin1 mediates apoptosis by c-Myc in transformed primary cells. Cancer Res. 61:3151-3156. [PubMed] [Google Scholar]
- 16.Elliott, K., K. Ge, W. Du, and G. C. Prendergast. 2000. The c-Myc-interacting adaptor protein Bin1 activates a caspase-independent cell death program. Oncogene 19:4669-4684. [DOI] [PubMed] [Google Scholar]
- 17.Elliott, K., D. Sakamuro, A. Basu, W. Du, W. Wunner, P. Staller, S. Gaubatz, H. Zhang, E. Prochownik, M. Eilers, and G. C. Prendergast. 1999. Bin1 functionally interacts with Myc and inhibits cell proliferation via multiple mechanisms. Oncogene 18:3564-3573. [DOI] [PubMed] [Google Scholar]
- 18.Farsad, K., N. Ringstad, K. Takei, S. R. Floyd, K. Rose, and P. De Camilli. 2001. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J. Cell Biol. 155:193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallily, R., and N. Savion. 1983. Cultivation, proliferation and characterization of thymic macrophages. Immunology 50:139-148. [PMC free article] [PubMed] [Google Scholar]
- 20.Ge, K., and G. C. Prendergast. 2000. Bin2, a functionally nonredundant member of the BAR adaptor gene family. Genomics 67:210-220. [DOI] [PubMed] [Google Scholar]
- 21.Gold, E. S., N. S. Morrissette, D. M. Underhill, J. Guo, M. Bassetti, and A. Aderem. 2000. Amphiphysin IIm, a novel amphiphysin II isoform, is required for macrophage phagocytosis. Immunity 12:285-292. [DOI] [PubMed] [Google Scholar]
- 22.Hogarty, M. D., X. Liu, P. M. Thompson, P. S. White, E. P. Sulman, J. M. Maris, and G. M. Brodeur. 2000. BIN1 inhibits colony formation and induces apoptosis in neuroblastoma cell lines with MYCN amplification. Med. Pediatr. Oncol. 35:559-562. [DOI] [PubMed] [Google Scholar]
- 23.Huang, D., G. Patrick, J. Moffat, L. H. Tsai, and B. Andrews. 1999. Mammalian Cdk5 is a functional homologue of the budding yeast Pho85 cyclin-dependent protein kinase. Proc. Natl. Acad. Sci. USA 96:14445-14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson, T., M. F. Allard, C. M. Sreenan, L. K. Doss, S. P. Bishop, and J. L. Swain. 1990. The c-myc proto-oncogene regulates cardiac development in transgenic mice. Mol. Cell. Biol. 10:3709-3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson, T., M. F. Allard, C. M. Sreenan, L. K. Doss, S. P. Bishop, and J. L. Swain. 1991. Transgenic animals as a tool for studying the effect of the c-myc proto-oncogene on cardiac development. Mol. Cell. Biochem. 104:15-19. [DOI] [PubMed] [Google Scholar]
- 26.Kadlec, L., and A. M. Pendergast. 1997. The amphiphysin-like protein 1 (ALP1) interacts functionally with the cABL tyrosine kinase and may play a role in cytoskeletal regulation. Proc. Natl. Acad. Sci. USA 94:12390-12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le, Q., R. Daniel, S. W. Chung, A. D. Kang, T. K. Eisenstein, B. M. Sultzer, H. Simpkins, and P. M. Wong. 1998. Involvement of C-Abl tyrosine kinase in lipopolysaccharide-induced macrophage activation. J. Immunol. 160:3330-3336. [PubMed] [Google Scholar]
- 28.Lee, E., M. Marcucci, L. Daniell, M. Pypaert, O. A. Weisz, G. C. Ochoa, K. Farsad, M. R. Wenk, and P. De Camilli. 2002. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science 297:1193-1196. [DOI] [PubMed] [Google Scholar]
- 29.Lee, J., K. Colwill, V. Aneliunas, C. Tennyson, L. Moore, Y. Ho, and B. Andrews. 1998. Interaction of yeast Rvs167 and Pho85 cyclin-dependent kinase complexes may link the cell cycle to the actin cytoskeleton. Curr. Biol. 8:1310-1321. [DOI] [PubMed] [Google Scholar]
- 30.Leprince, C., F. Romero, D. Cussac, B. Vayssiere, R. Berger, A. Tavitian, and J. H. Camonis. 1997. A new member of the amphiphysin family connecting endocytosis and signal transduction pathways. J. Biol. Chem. 272:15101-15105. [DOI] [PubMed] [Google Scholar]
- 31.Li, F. Q., A. Coonrod, and M. Horwitz. 2000. Selection of a dominant negative retinoblastoma protein (RB) inhibiting satellite myoblast differentiation implies an indirect interaction between MyoD and RB. Mol. Cell. Biol. 20:5129-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lichte, B., R. Veh, H. Meyer, and M. Kilimann. 1992. Amphiphysin, a novel protein associated with synaptic vesicles. EMBO J. 11:2521-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Littlewood, T. D., D. C. Hancock, P. S. Danielian, M. G. Parker, and G. I. Evan. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23:1686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao, N. C., E. Steingrimsson, J. DuHadaway, W. Wasserman, J. C. Ruiz, N. G. Copeland, N. A. Jenkins, and G. C. Prendergast. 1999. The murine Bin1 gene functions early in myogenesis and defines a new region of synteny between mouse chromosome 18 and human chromosome 2. Genomics 56:51-58. [DOI] [PubMed] [Google Scholar]
- 35.McMahon, H. T., P. Wigge, and C. Smith. 1997. Clathrin interacts specifically with amphiphysin and is displaced by dynamin. FEBS Lett. 413:319-322. [DOI] [PubMed] [Google Scholar]
- 36.McPherson, P. S., E. P. Garcia, V. I. Slepnev, C. David, X. Zhang, D. Grabs, W. S. Sossin, R. Bauerfeind, Y. Nemoto, and P. De Camilli. 1996. A presynaptic inositol-5-phosphatase. Nature 379:353-357. [DOI] [PubMed] [Google Scholar]
- 37.Moens, C. B., B. R. Stanton, L. F. Parada, and J. Rossant. 1993. Defects in heart and lung development in compound heterozygotes for two different targeted mutations at the N-myc locus. Development 119:485-499. [DOI] [PubMed] [Google Scholar]
- 38.Muller, A. J., J. C. Young, A. M. Pendergast, M. Pondel, N. R. Landau, D. R. Littman, and O. N. Witte. 1991. BCR first exon sequences specifically activate the BCR/ABL tyrosine kinase oncogene of Philadelphia chromosome-positive human leukemias. Mol. Cell. Biol. 11:1785-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munn, A. L., B. J. Stevenson, M. I. Geli, and H. Riezman. 1995. end5, end6, and end7: mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell 6:1721-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Owen, D. J., P. Wigge, Y. Vallis, J. D. Moore, P. R. Evans, and H. T. McMahon. 1998. Crystal structure of the amphiphysin-2 SH3 domain and its role in the prevention of dynamin ring formation. EMBO J. 17:5273-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramjaun, A. R., and P. S. McPherson. 1998. Multiple amphiphysin II splice variants display differential clathrin binding: identification of two distinct clathrin-binding sites. J. Neurochem. 70:2369-2376. [DOI] [PubMed] [Google Scholar]
- 43.Ramjaun, A. R., K. D. Micheva, I. Bouchelet, and P. S. McPherson. 1997. Identification and characterization of a nerve terminal-enriched amphiphysin isoform. J. Biol. Chem. 272:16700-16706. [DOI] [PubMed] [Google Scholar]
- 44.Ramjaun, A. R., J. Philie, E. de Heuvel, and P. S. McPherson. 1999. The N terminus of amphiphysin II mediates dimerization and plasma membrane targeting. J. Biol. Chem. 274:19785-19791. [DOI] [PubMed] [Google Scholar]
- 45.Razzaq, A., I. M. Robinson, H. T. McMahon, J. N. Skepper, Y. Su, A. C. Zelhof, A. P. Jackson, N. J. Gay, and C. J. O'Kane. 2001. Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Genes Dev. 15:2967-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Razzaq, A., Y. Su, J. E. Mehren, K. Mizuguchi, A. P. Jackson, N. J. Gay, and C. J. O'Kane. 2000. Characterisation of the gene for Drosophila amphiphysin. Gene 241:167-174. [DOI] [PubMed] [Google Scholar]
- 47.Routhier, E. L., T. C. Burn, I. Abbaszade, M. Summers, C. F. Albright, and G. C. Prendergast. 2001. Human BIN3 complements the F-actin localization defects caused by loss of Hob3p, the fission yeast homolog of Rvs161p. J. Biol. Chem. 276:21670-21677. [DOI] [PubMed] [Google Scholar]
- 48.Routhier, E. L., P. S. Donover, and G. C. Prendergast. 2003. hob1+, the fission yeast homolog of Bin1, is dispensable for endocytosis or actin organization, but required for the response to starvation or genotoxic stress. Oncogene 22:637-648. [DOI] [PubMed] [Google Scholar]
- 49.Sakamuro, D., K. J. Elliott, R. Wechsler-Reya, and G. C. Prendergast. 1996. BIN1 is a novel MYC-interacting protein with features of a tumour suppressor. Nat. Genet. 14:69-77. [DOI] [PubMed] [Google Scholar]
- 50.Shupliakov, O., P. Low, D. Grabs, H. Gad, H. Chen, C. David, K. Takei, P. De Camilli, and L. Brodin. 1997. Synaptic vesicle endocytosis impaired by disruption of dynamin-SH3 domain interactions. Science 276:259-263. [DOI] [PubMed] [Google Scholar]
- 51.Simpson, F., N. K. Hussain, B. Qualmann, R. B. Kelly, B. K. Kay, P. S. McPherson, and S. L. Schmid. 1999. SH3-domain-containing proteins function at distinct steps in clathrin-coated vesicle formation. Nat. Cell. Biol. 1:119-124. [DOI] [PubMed] [Google Scholar]
- 52.Sivadon, P., F. Bauer, M. Aigle, and M. Crouzet. 1995. Actin cytoskeleton and budding pattern are altered in the yeast rvs161 mutant: the Rvs161 protein shares common domains with the brain protein amphiphysin. Mol. Gen. Genet. 246:485-495. [DOI] [PubMed] [Google Scholar]
- 53.Slepnev, V. I., G. C. Ochoa, M. H. Butler, and P. De Camilli. 2000. Tandem arrangement of the clathrin and AP-2 binding domains in amphiphysin 1 and disruption of clathrin coat function by amphiphysin fragments comprising these sites. J. Biol. Chem. 275:17583-17589. [DOI] [PubMed] [Google Scholar]
- 54.Slepnev, V. I., G. C. Ochoa, M. H. Butler, D. Grabs, and P. D. Camilli. 1998. Role of phosphorylation in regulation of the assembly of endocytic coat complexes. Science 281:821-824. [DOI] [PubMed] [Google Scholar]
- 55.Sparks, A. B., N. G. Hoffman, S. J. McConnell, D. M. Fowlkes, and B. K. Kay. 1996. Cloning of ligand targets: systematic isolation of SH3 domain-containing proteins. Nat. Biotech. 14:741-744. [DOI] [PubMed] [Google Scholar]
- 56.Takei, K., V. I. Slepnev, V. Haucke, and P. De Camilli. 1999. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat. Cell Biol. 1:33-39. [DOI] [PubMed] [Google Scholar]
- 57.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsutsui, K., Y. Maeda, K. Tsutsui, S. Seki, and A. Tokunaga. 1997. cDNA cloning of a novel amphiphysin isoform and tissue-specific expression of its multiple splice variants. Biochem. Biophys. Res. Commun. 236:178-183. [DOI] [PubMed] [Google Scholar]
- 59.Wechsler-Reya, R., K. Elliott, M. Herlyn, and G. C. Prendergast. 1997. The putative tumor suppressor BIN1 is a short-lived nuclear phosphoprotein, the localization of which is altered in malignant cells. Cancer Res. 57:3258-3263. [PubMed] [Google Scholar]
- 60.Wechsler-Reya, R., D. Sakamuro, J. Zhang, J. Duhadaway, and G. C. Prendergast. 1997. Structural analysis of the human BIN1 gene. Evidence for tissue-specific transcriptional regulation and alternate RNA splicing. J. Biol. Chem. 272:31453-31458. [DOI] [PubMed] [Google Scholar]
- 61.Wechsler-Reya, R. J., K. J. Elliott, and G. C. Prendergast. 1998. A role for the putative tumor suppressor Bin1 in muscle cell differentiation. Mol. Cell. Biol. 18:566-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wigge, P., K. Kohler, Y. Vallis, C. A. Doyle, D. Owen, S. P. Hunt, and H. T. McMahon. 1997. Amphiphysin heterodimers: potential role in clathrin-mediated endocytosis. Mol. Biol. Cell 8:2003-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wigge, P., Y. Vallis, and H. T. McMahon. 1997. Inhibition of receptor-mediated endocytosis by the amphiphysin SH3 domain. Curr. Biol. 7:554-560. [DOI] [PubMed] [Google Scholar]
- 64.Zelhof, A. C., H. Bao, R. W. Hardy, A. Razzaq, B. Zhang, and C. Q. Doe. 2001. Drosophila amphiphysin is implicated in protein localization and membrane morphogenesis but not in synaptic vesicle endocytosis. Development 128:5005-5015. [DOI] [PubMed] [Google Scholar]