Abstract
The type III transforming growth factor β (TGFβ) receptor (TβRIII) binds both TGFβ and inhibin with high affinity and modulates the association of these ligands with their signaling receptors. However, the significance of TβRIII signaling in vivo is not known. In this study, we have sought to determine the role of TβRIII during development. We identified the predominant expression sites of ΤβRIII mRNA as liver and heart during midgestation and have disrupted the murine TβRIII gene by homologous recombination. Beginning at embryonic day 13.5, mice with mutations in ΤβRIII developed lethal proliferative defects in heart and apoptosis in liver, indicating that TβRIII is required during murine somatic development. To assess the effects of the absence of TβRIII on the function of its ligands, primary fibroblasts were generated from TβRIII-null and wild-type embryos. Our results indicate that TβRIII deficiency differentially affects the activities of TGFβ ligands. Notably, TβRIII-null cells exhibited significantly reduced sensitivity to TGFβ2 in terms of growth inhibition, reporter gene activation, and Smad2 nuclear localization, effects not observed with other ligands. These data indicate that TβRIII is an important modulator of TGFβ2 function in embryonic fibroblasts and that reduced sensitivity to TGFβ2 may underlie aspects of the TβRIII mutant phenotype.
Members of the transforming growth factor β (TGFβ) family are potent regulators of multiple cellular functions, including cell proliferation, differentiation, migration, and death (35, 64). As such, the TGFβs are critical regulators of the growth and morphogenesis of a variety of tissues. Three TGFβ isoforms (TGFβ1 to -3) have been described in mammals and are encoded by distinct genes (36). Although the three ligands have similar biological activities in many in vitro assays, null mutations in the three genes result in mice with distinct phenotypes, suggesting that each ligand has a unique role during murine somatic development (14, 42, 50). In mammalian cells, the diverse actions of the TGFβs are mediated by two distinct type I and type II serine/threonine kinase receptors (TβRI and TβRII, respectively), which are expressed on most cell types and tissues (35). TβRI and TβRII can form a latent receptor complex, and ligand binding is required for the activation of the receptor complex (65). Upon TGFβ binding, the receptors rotate relatively within the complex (65, 66), resulting in phosphorylation and activation of TβRI by the constitutively active and autophosphorylated TβRII (62). The activated TβRI then directly signals to downstream intracellular substrates, e.g., Smads (21, 61).
Many other cell surface receptors have been identified (64). Among them is the type III TGFβ receptor TβRIII, which binds to all three TGFβs (32). In contrast to the type I and II receptors, TβRIII, also known as betaglycan, appears dispensable for TGFβ-mediated signal transduction since most cells that lack functional TβRIII still respond to TGFβ (8). The murine form of TβRIII is an 850-amino-acid proteoglycan with heparin sulfate and chondroitin sulfate glycosaminoglycan (GAG) side chains attached to a 125- to 130-kDa core protein (41). The core protein contains a large extracellular domain, consisting of two putative TGFβ binding sites and two GAG attachment sites as well as a short intracellular tail with no known signaling motif (2, 17, 23, 30, 58). TβRIII is the most abundant TGFβ binding protein on many cell types and binds each of the three TGFβ isoforms with high affinity (8, 38, 53). However, the role played by TβRIII in TGFβ biology remains poorly understood. Membrane-associated TβRIII appears to facilitate TGFβ action by presenting TGFβ to TβRII (31, 32, 58). This function of TβRIII is perhaps most important with regard to TGFβ2. A number of studies have indicated that TGFβ2 has low affinity for TβRII in the absence of TβRIII (17, 32, 51). Overexpression of TβRIII in vitro in cells that normally lack its expression increases the binding of TGFβ to the signaling receptors and in some cases has been shown to augment TGFβ actions, particularly those of TGFβ2 (17, 32, 51). TβRIII also modulates the actions of activin and inhibin, two members of the TGFβ superfamily which functionally antagonize each other (29). Activin and inhibin exist as disulfide-linked dimers that are composed of either α- and β-subunits (inhibin) or two β-subunits (activin). Like TGFβ, activin signals through heterodimeric complexes of type I (ActRI) and type II (ActRII) serine/threonine kinase receptors. Signaling receptors for inhibin have yet to be found, but inhibin is capable of binding ActRII through its β-subunit. Lewis et al. (29) have shown that TβRIII binds inhibin with high affinity and increases its binding to ActRII, thereby antagonizing activin function by preventing ActRI from forming complexes with ActRII. Inhibin also antagonizes the binding of bone morphogenetic proteins to ActRII and bone morphogenetic protein RII, and the presence of TβRIII increases the efficacy of inhibin in these assays (60), suggesting that TβRIII may broadly influence the activities of a number of TGFβ superfamily members.
These data indicate that the membrane-bound form of TβRIII plays a regulatory role in determining cellular sensitivity to inhibin and the three mammalian TGFβ isoforms. However, additional data point to other functions. Notably, the TβRIII extracellular domain can be released from the cell surface as a soluble proteoglycan and is found in the extracellular matrix and serum (27). Soluble TβRIII sequesters TGFβ and inhibits its action in cell cultures, suggesting that the soluble form of TβRIII may have opposing actions to those of its membrane-bound counterpart (3, 31, 56). Adding another level of complexity, a recent study has shown that in certain contexts, the membrane-bound form of TβRIII can also inhibit TGFβ function via TβRIII's GAG side chains (16).
Collectively, these data indicate complex roles for TβRIII in modulating the access of certain TGFβ superfamily members to their signaling receptors. However, the impact of TβRIII activity in vivo is not known. In this study, we have examined the role of TβRIII during murine development. As a first step, we used in situ hybridization histochemistry to study the embryonic expression pattern of TβRIII mRNA in order to compare the sites of TβRIII synthesis with the previously published developmental expression patterns of the TGFβs, inhibins, and activins as well as their signaling receptors. We then disrupted the murine TβRIII gene by homologous recombination in order to study its function. TβRIII−/− embryos are not viable, dying between embryonic day 16.5 (E16.5) and birth with defects in hepatic and cardiovascular development. Using murine embryonic fibroblasts (MEFs) isolated from wild-type and mutant embryos, we found that the cellular sensitivity to TGFβ2, but not to other known TβRIII ligands, was greatly reduced in mutant cells compared to wild-type cells. These data suggest that the TβRIII−/− phenotype may be in part a reflection of disrupted TGFβ2-mediated developmental processes.
MATERIALS AND METHODS
Generation of TβRIII knockout mice.
Genomic clones for the generation of the targeting construct were isolated from a 129/Sv mouse genomic library as previously described (58). A 6.6-kb clone containing putative exon 2 of the murine TβRIII gene was subcloned and analyzed by a combination of restriction enzyme mapping and sequencing. To construct the targeting vector, the genomic clone was digested with AvaI and was inserted into pBluescript II SK (Stratagene) by blunt-end ligation. A 2.55-kb phosphoglycerokinase-neomycin (PGK-Neo) expression cassette was inserted in a sense orientation with respect to the genomic clone into the DraIII site in exon 2. W9.5 embryonic stem (ES) cells (129/Sv) were electroporated with the linearized targeting vector and selected for Neo resistance. Correctly targeted clones were identified by Southern blot analysis and injected into C57BL/6 blastocysts to generate chimeric mice with the targeted allele incorporated into the germ line. Chimeric males were bred to C57BL/6 females, and the genotypes of the progeny produced were determined by Southern blot analysis.
Southern and Northern blot analyses.
Tail and embryo samples were digested with proteinase K, and DNA was precipitated in isopropanol. For Southern blot analysis, DNA was digested with concentrated SpeI and BamHI, separated on a 1% agarose gel, and blotted onto a Hybond-N nylon membrane (Amersham Pharmacia Biotech). A BamHI-HincII restriction fragment of the TβRIII genomic clone, external to the targeting construct, was labeled with [α32P]dCTP by using a random priming kit (Amersham Pharmacia Biotech). Blots were incubated at 65°C for 4 h in a commercially available buffer (Amersham Pharmacia Biotech) containing the radiolabeled probe, washed thoroughly in 1× standard sodium citrate-0.1% sodium dodecyl sulfate, and analyzed with a phosphorimager (Molecular Dynamics). A single integration site was verified by reprobing with a PCR-generated 500-bp cDNA that corresponds to sequences within the PGK-Neo cassette.
Total RNA was isolated from E14.5 to E15.5 embryos by standard methods (10). Poly(A)+-enriched RNA was selected by incubating total RNA fractions with oligo(dT) cellulose according to the manufacturer's instructions (Roche Diagnostics). Poly(A)+ RNA (5 μg) was size fractionated on a 1% formaldehyde gel, transferred to a Hybond N nylon membrane (Amersham Pharmacia Biotech), and hybridized with α-32P-labeled cDNA probes in a solution containing 0.2 M NaH2PO4, 0.3 M Na2HPO4, 0.5 M EDTA, and 7% sodium dodecyl sulfate overnight at 65°C. Radiolabeling of probes, washing of blots, and signal detection were performed as for Southern blotting. A 335-bp EcoRI/XbaI restriction fragment of a rat TβRIII cDNA, corresponding to nucleotides 2087 to 2421 of the published murine TβRIII sequence (41) (cDNA kindly provided by X.-F. Wang, Duke University, Durham, N.C.), was subcloned into pBluescript II KS (Stratagene) and used to detect murine TβRIII transcripts.
Reverse transcriptase PCR (RT-PCR).
RT-PCR was performed by using 1.5 μg of total RNA derived from embryos and random decamer primers of a first-strand synthesis kit (Ambion) according to the manufacturer's instructions. PCR primers included (i) an exon 1 sense primer (5′ ATGGCAGTGACATCCCACCA 3′); (ii) a sense primer originating within the 3′ coding region of the Neo gene (5′ GCGAATGGGCTGACCGCTTC 3′); (iii) an antisense PCR primer against the 5′ end of the Neo gene (5′ GGACAGGTCGGTCTTGACAA 3′); (iv) an antisense primer to the 3′ region of exon 2 (5′ TTTAGGATGTGAACCTCCCTT 3′); and (v) an antisense primer to exon 3 (5′ TCCGAAACCAGGAAGAGTCT 3′). The RT-PCR results were analyzed based on the assessment of product sizes upon ethidium bromide-agarose gel electrophoresis. Each reaction was repeated a minimum of three times with RNA derived from at least two wild-type and TβRIII−/− embryos. Novel products generated from TβRIII−/− RNA were cloned into pCR 2.1 TOPO (Invitrogen) and subjected to automated sequencing.
Embryo collection, histology, and immunocytochemistry.
For histological analysis, embryos from heterozygous matings were obtained from pregnant dams between E12.5 and E18.5, with noon on the day of vaginal plugging designated as E0.5. Embryos, placentas, and yolk sacs were immersion fixed in 4% paraformaldehyde at 4°C. After fixation, embryos were dehydrated through graded alcohols and embedded in paraffin wax prior to sectioning. Prepared tissue sections from selected embryos were stained with hematoxylin and eosin. Immunohistochemistry was performed on tissue sections treated with an antigen retrieval reagent (DAKO Corporation) by using antibodies against the proliferating cell nuclear antigen (DAKO Corporation) and active caspase 3 (R&D Systems) according to the manufacturers' instructions. A minimum of 200 cells per tissue per embryo was counted. A minimum of four littermate pairs (knockouts compared to either heterozygote or wild-type mice) was examined.
Embryos were prepared for wholemount bone and cartilage analysis according to Lufkin et al. (33). Peripheral blood for smears was collected at dissection from neck blood vessels of embryos from E16.5 to E18.5 litters with a 0.56-mm heparinized capillary tube, and smears were stained with May-Grunwald and Giemsa stains. In situ hybridization histochemistry was carried out as described previously (55). cRNA probes were prepared by in vitro transcription by using a riboprobe kit (Promega Corporation) and a combination of 35S-rUTP and 35S-rCTP (1,250 Ci/mmol; NEN Life Science). Antisense and sense probes were derived from the same TβRIII cDNA template that was used in Northern blot analyses (see above).
Primary embryonic tissue culture.
Fibroblasts were isolated from E12.5 embryos following the removal of head and liver structures. Tissue was dispersed through 21-gauge needles and digested with trypsin-vercene prior to plating. Adherent cells were selected and cultured in Dulbecco's modified essential medium (DMEM; Life Technologies, Inc.), 10% fetal calf serum (FCS) (CSL, Melbourne, Australia), 60 μg of penicillin per ml, 100 μg of streptomycin per ml, and 2 mM glutamine at 37°C in the presence of 10% CO2. Passages three to eight were used. For liver cell cultures, livers from E13.5 embryos were dispersed with forceps and incubated for 20 min in 330 μg of collagenase per ml (Sigma) at 37°C. Single-cell suspensions were generated by pipetting, and red blood cells were lysed with ammonium chloride (pH 7.2). Liver cells were plated in BGJb media (Gibco) on 8-well chamber slides (Nunc, Naperville, Ill.) or coverslips in 24-well plates that had been coated with type I collagen (BD Biosciences). Liver cells were cultured for 2 days at 37°C in the presence of 10% CO2 prior to growth factor treatment (0.5 ng of TGFβ1 or TGFβ2 per ml) and/or immunostaining (anti-Smad2; anti-E-cadherin, Transduction Laboratories). Liver cells were fixed in formalin and permeabilized with 0.2% Triton X-phosphate-buffered saline (PBS) prior to processing for immunofluorescence (see below). In total, livers from five litters were cultured on collagen, including 7 knockout, 10 wild-type, and 22 heterozygote embryos. Phase microscopy was used to assess hepatocyte adherence to collagen. For all experiments, cultures derived from littermates were examined in the same assay for comparison purposes.
Binding and affinity cross-linking.
For affinity labeling assays, E14.5 to E15.5 fibroblast cultures were grown to near confluence in 35-mm dishes. The method of Zhu and Sizeland (65) was followed, with 50 pM 125I-TGFβ1 (Amersham Pharmacia Biotech) used per dish. Total protein was assessed in cell lysates with the Bio-Rad protein assay (Bio-Rad Laboratories). Standardized amounts of protein were run on a 4 to 20% Tris-glycine gradient gel (Novex). Gels were stained with Coomassie brilliant blue, dried under vacuum, and analyzed with a Molecular Dynamics phosphorimager.
[3H]thymidine incorporation and basal cell proliferation assays.
Fibroblasts derived from TβRIII+/+ and TβRIII−/− embryos were plated in 96-well plates at concentrations of 2,000 cells/well in DMEM-10% FCS and grown for 24 h. Quadruplicate wells were treated with growth factors (R&D Systems) at the concentrations indicated for 24 h and then incubated with 0.2 μCi of [3H]thymidine/well for an additional 24 h. Cells were lysed with 0.5 M NaOH and harvested by using a Filtermate Harvester (Packard Instrument Co., Meriden, Conn.). The incorporated [3H]thymidine was measured with a Microplate Scintillation Counter (Packard Instrument Co.). Origin 6 was used to fit approximate sigmoidal curves to the data in order to derive 50% inhibitory concentrations (IC50s). Basal proliferation rates of TβRIII+/+ and TβRIII−/− MEFs were examined by culturing 1.0 × 105 cells/well in six-well plates in DMEM-10% FCS. Counts of viable cells were performed after 3 days by using trypan blue staining and a hemacytometer. Counts of live and dead cells were performed on duplicate wells for two pairs of TβRIII+/+ and TβRIII−/− MEF lines.
Transient transfection and luciferase reporter assays.
The pGL3-(CAGA)12-Luc luciferase reporter construct (13) (a generous gift of Aris Moustakas, Ludwig Institute for Cancer Research, Uppsala, Sweden) was used to assess the TGFβ responsiveness of wild-type and knockout MEFs. For transient transfection, MEFs were plated at 6.6 × 105 cells in 92-mm dishes and grown overnight. Cultures were then transfected by using Fugene 6 transfection reagent according to the manufacturer's protocol (Roche Molecular Biochemicals). Cells were cotransfected with 7 μg of pGL3-(CAGA)12-Luc and 0.7 μg of pRL-TK, a control reporter vector in which Renilla luciferase expression is driven by the thymidine kinase promoter (Promega Corporation). The next day, each dish was split into two 24-well plates and allowed to adhere. The cells were starved in DMEM-0.2% FCS for 4 h, and then triplicate wells were treated for 24 h with either TGFβ1 or TGFβ2 in DMEM-0.2% FCS at the indicated concentrations. Total protein lysates were extracted, and firefly luciferase and Renilla luciferase activities were assessed with a dual luciferase reporter assay system (Promega Corporation) in conjunction with a ML3000 Microtiter Plate Luminometer (Dynatech Laboratories, Inc., Chantilly, Va.). The experimental data were normalized to the Renilla luciferase activity/well to control for differences in transfection efficiency and then expressed as the multiple of difference (x-fold) relative to basal conditions.
Immunofluorescence.
MEFs were plated at 1.0 × 104 cells/well in eight-well chamber slides (Nunc) and cultured in 10% FCS-DMEM overnight. Cells were starved for 4 h and treated with the indicated concentrations of TGFβ1 and TGFβ2 in 0.2% bovine serum albumin (BSA)-DMEM for the indicated times. Cells were fixed with −20°C methanol, blocked in 5% skim milk in PBS containing 0.1% Tween 20 (PBS-T), and incubated with the primary antibody (anti-Smad2; Transduction Laboratories) overnight at 4°C. After extensive washing in PBS-T, the cells were incubated with an Alexa 488-conjugated goat anti-mouse immunoglobulin G secondary antibody in 2% BSA-PBS-T (Molecular Probes) for at least 2 h. Cells were washed extensively with PBS-T and water and then air dried. Immunofluorescent images were obtained by using confocal microscopy (Bio-Rad, model MRC-1024). For analyses of Smad2 cellular localization, a minimum of 200 cells per cell line per condition was assessed; two littermate pairs from different litters were examined. For analyses of E-cadherin staining of primary embryonic hepatocyte cultures, three knockouts from different litters were compared to littermate controls.
Flow cytometry.
Fetal livers from E13.5 (4 +/+, 4 +/−, and 1 −/−), E14.5 (6 +/+, 22 +/−, and 10 −/−), and E15.5 (3 +/+, 4 +/−, and 2 −/−) embryos were collected and separated into single-cell suspensions by manipulation with forceps and pipetting. Fluorescence-activated cell sorter (FACS) analyses were performed as described previously (20) with the following modifications for four-color analysis. Cells were washed and resuspended in 2% FCS-2 mM EDTA-PBS before treatment with the rat monoclonal antibody 2.4G2 to block Fc receptors. Cells were then stained with the following monoclonal antibodies: CD45.2 (104), Mac1 (M1/70), Gr-1 (Rb6-8C5), CD71 (C2), Ter119 (Ly-76), CD4 (H129.19), CD8 (53-6.7), B220 (RA3-6B2), Thy1 (30-H12), and rat IgG2bκ isotype control (A95-1). Dead cells were excluded on the basis of propidium iodide (PI) uptake. For four-color analysis, cells were stained with CD45.2-biotin-streptavidin-allophycocyanin in combination with appropriate phycoerythrin- and fluorescein isothiocyanate-labeled monoclonal antibodies. Ten thousand CD45-positive, PI-negative events were acquired. For annexin-V assays, cells were first stained with either CD45 or CD71 before resuspension in annexin-V buffer containing annexin-V-fluorescein isothiocyanate (Becton Dickinson, San Jose, Calif.). PI-negative cells were analyzed. All FACS data were acquired on a FACScalibur and analyzed with CELLQuest-Pro software (Becton Dickinson). All antibodies were obtained from Pharmingen.
Immunoblotting.
For Western blot analyses, MEFs were grown to near confluence in 10% FCS-DMEM, starved for 4 h in a solution containing 0.2% BSA-DMEM, and treated with 1 ng of TGFβ1 or TGFβ2 per ml in 0.2% BSA-DMEM for the indicated times. For most assays, cells and tissues were lysed for 30 min at 4°C in 1% Triton X-100, 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 1 mM dithiothreitol, and complete protease inhibitors (Boehringer Mannheim). Lysates were clarified, and total protein was assessed by using the bicinchoninic acid protein assay (Pierce). Ten embryonic hearts of the same genotype were pooled at each age. To detect phospho-Akt and Akt, livers were lysed in 9 M urea-4% CHAPS, and lysates were clarified by ultracentrifugation at 100,000 rpm (Beckman TL-100 ultracentrifuge). Standardized amounts of protein were run on a 4 to 20% NuPage Tris-bis gradient gel (Invitrogen). Immunoblotting and chemiluminescence detection were performed with antibodies directed against the following proteins, according to manufacturers' protocols: actin, smooth muscle actin (Sigma); myosin heavy chain, Akt, Erk1/2 (Santa Cruz); β1-integrin (Chemicon); phospho-Erk1/2, phospho-SAPK/JNK, p38, phospho-p38, phospho-Akt (NEB/Cell Signaling Technology); phospho-Smad2 (Upstate Biotechnology); and Smad2 (Transduction Laboratories). Densitometry was performed on autoradiograms by using ImageQuant 4.2 software (Molecular Dynamics) to quantify the data.
Statistical analyses.
Statistics were performed on the IC50 data by using a one-way analysis of variance. Differences between wild-type and mutant cells in all other assays were determined by using two-way independent t tests. Differences were considered significant when values of P were < 0.05.
RESULTS
Embryonic liver and heart express TβRIII mRNA.
To aid in understanding the function of TβRIII, we examined its mRNA expression pattern in wild-type embryos by in situ hybridization histochemistry. Specificity of the hybridization signal obtained with antisense riboprobes was verified by hybridization of adjacent sections with a TβRIII sense strand cRNA probe, which showed no hybridization above background (data not shown). TβRIII mRNA was widely expressed throughout gestation, but during midgestation, when developmental processes first appear to be disrupted by TβRIII deficiency (see below), the predominant embryonic sites of TβRIII mRNA expression were heart and liver.
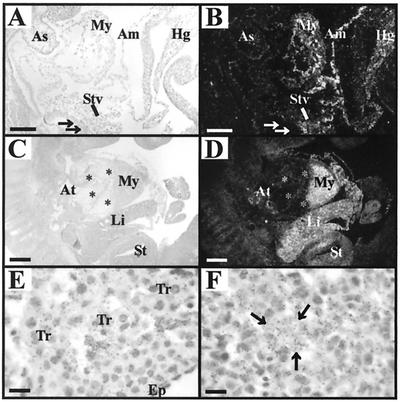
In the mouse, the liver anlage arises from the foregut endoderm at E9.5 when endodermal cells are induced to proliferate and invade the surrounding mesenchyme of the septum transversum in response to a signal from the cardiac mesoderm. Subsequently, the liver bud appears at E10.5 and grows rapidly due to proliferating hepatic and biliary cells and colonizing hematopoietic stem cells (reviewed by Zaret [63]). During the stage of initial hepatic induction at E9.5, TβRIII mRNA was expressed within the septum transversum surrounding the presumptive hepatic endoderm but not within the endoderm itself (Fig. 1A and B). Between E10.5 and E11.5, TβRIII mRNA was expressed at high levels throughout the growing liver bud (Fig. 1C and D) and was localized to most liver cell types, including the immature hepatocytes (Fig. 1F).
FIG. 1.
ΤβRIII mRNA expression within heart and liver during midgestation. Paired light- and dark-field photomicrographs of sections through mouse embryos, processed by in situ hybridization histochemistry by using an antisense cRNA probe for ΤβRIII. (A and B) A sagittal section through an E9.5 embryo shows ΤβRIII mRNA expression localized to the myocardium (My) of the developing ventricles and to the septum transversum (STv), amnion (Am), and hindgut (Hg). No expression is detected in the aortic sac (As) or presumptive hepatic endoderm (arrows). (C and D) A sagittal section through an E11.5 embryo shows high TβRIII mRNA expression in the myocardium of the ventricle (My) and atrium (At) but not in endocardial cushion tissue (*) of the heart. Strong expression is also now evident within the liver (Li) and stomach (St). (E) A light-field image of an E11.5 heart at high magnification shows silver grains localized over the myocytes within the trabeculae (Tr), with little expression within the myocytes immediately beneath the epicardium (Ep). (F) A similar image of E11.5 liver demonstrates strong TβRIII mRNA expression throughout the parenchyma. The arrows point to one of the islands of hepatocytes. Bars, 100 μm (A and B); 200 μm (C and D); 10 μm (E and F).
At E8.5, the earliest age examined, TβRIII mRNA was detected at low levels within cardiomyocytes of the developing ventricle (data not shown), and myocardial expression of TβRIII mRNA increased dramatically by E9.5 to E11.5 (Fig. 1A through D). Within the ventricle, expression was consistently higher in the trabeculating cardiomyocytes in the lumen of the ventricle, with no or very low expression within the subepicardial cardiomyocytes (Fig. 1E). Low, transient expression of ΤβRIII mRNA was also observed within the endothelia of the atrioventricular canal at E8.5 and in isolated endothelial cells adjacent to the cushion tissue at E9.5 to E11.5 (data not shown). However, no ΤβRIII mRNA expression was detected within the mesenchyme of the endocardial cushion tissue or any of its derivatives (Fig. 1C and D).
Targeted disruption of the TβRIII gene causes embryonic lethality.
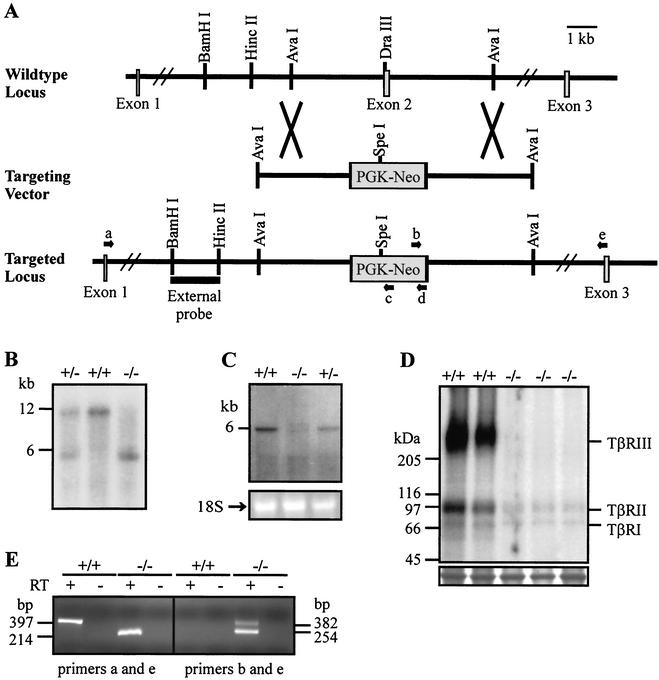
A targeted mutation in the murine TβRIII gene was introduced into ES cells by using a targeting vector in which exon 2 of the published murine TβRIII sequence (41) was disrupted with a PGK-Neo cassette (Fig. 2A). Integration of the targeting vector into the mouse genome by homologous recombination was verified in targeted ES cell clones and offspring by Southern blot analysis using a probe external to the targeted region (Fig. 2A and B). Blastocysts from C57BL/6 females were injected with the targeted ES cells and implanted in pseudopregnant mice to generate chimeric mice with the targeted allele incorporated into the germ line. Chimeric males were bred to C57BL/6 females to generate TβRIII+/− mice. Offspring obtained from TβRIII+/− matings consisted of 33.1% wild-type mice (438/1323), 66.6% heterozygous mice (881/1323), and 0.3% homozygous null mice (4/1323), indicating that, in general, TβRIII−/− mice do not survive to term. The small number of surviving TβRIII−/− mice (three males and one female) were poorly fertile but otherwise apparently healthy. These mice were culled at 12 to 16 months of age and autopsied. No gross pathology was evident in the male mice. The female mouse had a bifurcated spleen, which exhibited slightly reduced white pulp compared to an age-matched heterozygote female (data not shown).
FIG. 2.
Generation of TβRIII−/− embryos. (A) A schematic representation of the targeting strategy. Exon 2 of the murine TβRIII gene was disrupted with a PGK-Neo cassette as detailed in the text. (B) Southern blot analysis of embryonic DNA digested with BamHI and SpeI and hybridized with the 5′ external probe shown in panel A revealed the expected 12-kb fragment from the wild-type allele and 6-kb fragment from the targeted allele. (C) Northern blot analysis of poly(A)+ RNA derived from embryos with a cDNA probe corresponding to the TβRIII 3′ coding sequence revealed the presence of a single 6-kb transcript in wild-type samples and aberrant transcripts in TβRIII−/− samples. The corresponding formaldehyde gel stained with ethidium bromide is shown below the blot, with the band representing 18S rRNA indicated as a loading control. (D) 125I-TGFβ1 affinity cross-linking analysis of ligand binding in embryonic fibroblasts. The TGFβ receptor types are indicated in the right margin, and the corresponding molecular masses in kDa are listed in the left margin. TβRIII−/− fibroblasts showed no binding to TβRIII and reduced binding to the TβRI and TβRII compared to wild-type fibroblasts. A band from the same gel visualized by Coomassie blue staining is shown below the blot as a loading control. (E) Representative ethidium bromide-stained agarose gel visualizing amplified products of RT-PCRs. The orientation and approximate locations of primers a through e are shown in panel A. The arrows are not to scale. Novel products were detected in ΤβRIII−/− samples using primers a and e (left-hand gel) and primers b and e (right-hand gel). No products were obtained by using primer a paired with either primer c or primer d in TβRIII−/− samples.
To confirm that TβRIII−/− mice did not produce functional ΤβRIII, Northern blot analysis of poly(A)+ RNA derived from E14.5 to E15.5 embryos obtained from TβRIII+/− matings was performed using a cDNA probe corresponding to the 3′ coding region of the ΤβRIII gene. A 6-kb transcript was detected in wild-type and heterozygous samples, corresponding to the size of the wild-type ΤβRIII transcript (Fig. 2C). Unexpectedly, two weak bands corresponding to aberrant transcripts were detected in TβRIII−/− samples. To verify that no protein capable of binding TGFβ was being produced from these aberrant transcripts, we undertook 125I-TGFβ1 affinity labeling analysis of fibroblasts derived from E14.5 to E15.5 embryos (Fig. 2D). In wild-type embryonic fibroblasts, 125I-TGFβ1 was cross-linked to proteins of approximately 65 kDa and 85 to 100 kDa, corresponding to TβRI and TβRII, respectively. In addition, a smear of high-molecular-mass proteins (>200 kDa) was detected, representing TβRIII in its differentially glycosylated forms. In contrast, in TβRIII−/− fibroblasts, binding to TβRIII was absent, and binding to TβRI and TβRII was greatly reduced. This latter finding was not unexpected since previous data had indicated that binding to the signaling receptors is facilitated by the presence of the membrane-bound form of TβRIII (32, 58).
To ensure that targeted disruption of exon 2 disrupted the entire coding sequence of the ΤβRIII gene, total RNA from wild-type and ΤβRIII−/− embryos was subjected to RT-PCR analysis to determine the identity of the aberrant transcripts detected by Northern blot analysis (Fig. 2E). Cloning and sequencing of the products from these reactions established that no wild-type TβRIII transcripts were generated in TβRIII−/− embryos (data not shown). Instead, three novel transcripts were produced from the targeted allele by aberrant splicing. The three transcripts included: (i) a transcript generated by splicing from the end of exon 1 directly to exon 3 (nucleotide 300 to nucleotide 484 of the published murine sequence [41]) and (ii) two additional transcripts which originate from within the Neo gene, splice around the 3′ regulatory region of PGK, and join with either nucleotide 484 in exon 3 or nucleotide 335 in exon 2 (Fig. 2E). The first splicing event positions exon 3 and the downstream coding sequence out of frame with respect to exon 1 and, if translated, would produce an aberrant peptide of 97 amino acids. The other two sets of splicing events preserve the stop codon within the Neo-encoding portion of the transcripts and place the downstream TβRIII coding sequence out of frame. We therefore conclude that neither soluble nor membrane-associated forms of TβRIII are expressed in TβRIII−/− embryos.
Liver defects in TβRIII−/− embryos.
To determine the age at which the TβRIII−/− mice died, embryos from TβRIII+/− matings were obtained by Caesarean section delivery at time points between E12.5 and E18.5, and genotypes were determined by Southern blot analysis (Fig. 2B). Analysis of the embryos obtained from timed pregnancies of TβRIII+/− mice revealed few dead TβRIII−/− embryos between E12.5 and E15.5, but thereafter the relative number of dead TβRIII−/− embryos increased sharply (Table 1). At E18.5, knockouts represented only 5% of the living embryos (19 of 371), and live TβRIII−/− embryos between E16.5 and E18.5 were frequently pale, small in size, and moribund (data not shown).
TABLE 1.
Genotypes of embryos derived from heterozygous matings
| Gestational ages | +/+ (dead) | +/− (dead) | −/− (dead) | Resorptions (genotypes unknown) |
|---|---|---|---|---|
| E12.5-E13.5 | 52 (0) | 172 (4) | 56 (1) | 0 |
| E14.5 | 53 (0) | 149 (2) | 50 (2) | 1 |
| E15.5 | 38 (2) | 73 (6) | 30 (3) | 0 |
| E16.5 | 77 (8) | 162 (17) | 55 (17) | 3 |
| E17.5-E18.5 | 120 (11) | 208 (21) | 43 (24) | 17 |
| Total | 340 (21) | 764 (50) | 234 (47) | 21 |
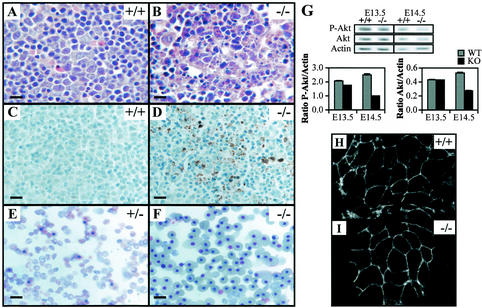
Examination of hematoxylin- and eosin-stained histological sections revealed that E12.5 TβRIII−/− embryos resembled wild-type and heterozygous littermates, with no gross disturbances in organogenesis. From E13.5 to E14.5, knockout embryos began to exhibit foci of apoptotic cell death in the liver (Fig. 3A and B). An increase in intraparenchymal blood was not apparent at this stage, suggesting that the liver cell death was not due to congestion (Fig. 3B). Approximately 75% of TβRIII−/− embryos exhibited some degree of liver pathology, with embryos at E14.5 and older frequently showing severe liver degeneration with hemorrhaging and loss of ultrastructure (data not shown). Immunohistochemistry performed on liver sections by using an antibody against the active form of caspase 3, a key regulatory enzyme in the apoptosis pathway, indicated the presence of numerous apoptotic cells in TβRIII−/− liver, but none in wild-type liver (Fig. 3C and D). To determine whether the apoptosis in TβRIII−/− liver was linked to a loss of pro-survival factors, the antiapoptotic proteins Akt and Bcl-XL were examined by Western blot analyses of embryonic liver. No significant differences were detected in the level of Bcl-XL relative to actin between mutant and wild-type E13.5 and E14.5 liver homogenates (data not shown). In addition, the ratio of phospho-Akt to Akt was not significantly reduced in mutant liver (data not shown). However, both phospho-Akt and Akt were reduced in mutant liver at E14.5 relative to actin (Fig. 3G), suggesting that total Akt may be down-regulated in mutant liver. As an index of the number of proliferating cells in embryonic liver, cells positive for the proliferating cell nuclear antigen (PCNA) were examined. In wild-type livers, on average 64.2% ± 11.4% and 57.0% ± 0.41% of liver cells at E13.5 and E14.5, respectively, expressed PCNA. Similar results were obtained in E13.5 (62.3% ± 7.9%) and E14.5 (53.5% ± 1.8%) knockout liver, suggesting that wild-type and knockout liver did not significantly differ in the proportion of liver cells that were undergoing proliferation.
FIG. 3.
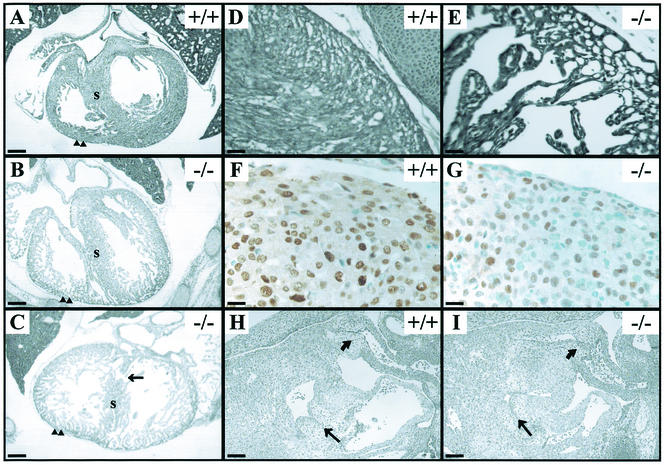
Liver defects in TβRIII−/− embryos. Hematoxylin and eosin staining of wild-type (A) and knockout (B) E14.5 liver sections shows light purple-stained hepatocytes, dark purple-stained hematopoietic precursor cells, and pink RBCs. Note the dramatic loss of parenchyma and disruption of liver architecture in the TβRIII−/− liver (B). Active caspase 3 immunostaining of E14.5 wild-type (C) and knockout (D) liver sections. No staining was noted in wild-type livers, while knockout livers show abundant staining (brown) in areas where cell death was histologically evident. (E and F) Smears of peripheral blood from E16.5 embryos. The TβRIII+/− smear almost entirely comprises nonnucleated RBCs that are indicative of normal definitive erythropoiesis. In contrast, the TβRIII−/− blood smear contains numerous nucleated red blood cells, suggesting a reduction in definitive erythrocyte differentiation. (G) (Top) Western blots of phospho-Akt and Akt in E13.5 and E14.5 liver lysates. Actin is shown as a loading control. (Bottom) Densitometry results for the bands shown above. The ratios of phospho-Akt and Akt to actin are shown. Both phospho-Akt and Akt are reduced in E14.5 mutant liver. Data presented are means ± standard deviations (SD) of triplicate densitometry readings. Data are representative of two littermate pairs at each age. (H and I) E-cadherin immunostaining of E13.5 hepatocytes cultured on collagen. Staining is primarily membranous in both wild-type and knockout cultures, indicating that cell-cell adhesions between hepatocytes are intact in mutant liver. Bars, 10 μm (A through D); 20 μm (E and F).
A cell adhesion defect was reported in Smad2+/−; Smad3+/− double heterozygote liver (59) which involved a loss of E-cadherin membrane staining in mutant liver and a reduction in hepatocyte adhesion to collagen in vitro. Since Smad2 and Smad3 are mediators of TGFβ and activin signaling (35), we investigated whether a similar defect occurred in TβRIII−/− liver. Cultured wild-type and knockout TβRIII liver cells exhibited similar adherence to collagen, remaining fairly small and rounded for 2 days in culture. Immunostaining of cultures using an antibody against E-cadherin revealed that mutant and wild-type hepatocytes showed similar membrane staining (Fig. 3H and I), suggesting that cell-cell adhesion between hepatocytes of mutants was not disrupted. In addition, changes in the levels of β1-integrin and phosphorylated Erk1/2, both notably altered in Smad2+/−; Smad3+/− double heterozygote liver, were not detected in TβRIII mutant liver by Western blot analysis (data not shown). Therefore, the phenotypes of the TβRIII−/− and Smad2+/−; Smad3+/− double heterozygotes appear to differ in several key aspects.
Hematopoiesis in TβRIII−/− embryos.
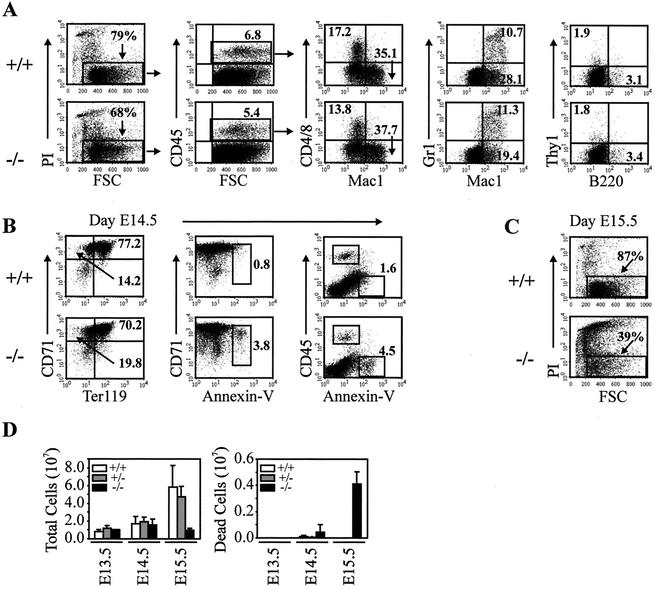
Between E11.5 and E16.5, the liver is the primary hematopoietic organ (25). Examination of red blood cells (RBCs) in smears of embryonic peripheral blood and in E16.5 to E18.5 tissue sections revealed a predominance of nucleated RBCs compared to nonnucleated RBCs in ΤβRIII−/− samples (Fig. 3E and F), indicating that definitive erythropoiesis was disrupted in mutant liver. The relative proportion of nucleated to nonnucleated RBCs varied from embryo to embryo and correlated with the degree of liver cell loss, indicating that the disruption in definitive erythropoiesis may be secondary to the liver pathology. To further investigate liver hematopoiesis in mutant embryos, FACS analyses were performed on E13.5 to E15.5 fetal livers (Fig. 4). For FACS analysis, PI uptake was used to detect dead cells, which were then excluded from further analyses. No significant differences in the percentage of PI-positive E13.5 liver cells were detected between knockout and wild-type embryos (data not shown). However, at E14.5 (Fig. 4A), a significant increase in the percentage of PI-positive fetal liver cells was detected in mutant liver, and by E15.5, mutant livers exhibited dramatic cell death (Fig. 4C). Using trypan blue exclusion as an indication of viability, we also performed manual counts of viable cells in the livers used for FACS analysis. This analysis revealed similar numbers of total cells at E13.5 and E14.5 but greatly reduced numbers of viable cells in E15.5 mutant liver (Fig. 4D). The extensive cell death at E15.5 precluded in-depth analyses of hematopoiesis at this age.
FIG. 4.
Evidence of apoptosis in TβRIII−/− fetal liver with relatively normal hematopoiesis. Single-cell suspensions of fetal liver from wild-type (+/+) and TβRIII knockout animals (−/−) were stained with various antibodies directed against specific lineages of hematopoietic cells. (A) Nucleated, CD45.2-positive hematopoietic cells were gated based on cell size (FSC) and PI exclusion (PI negative) before analysis with lineage-specific antibodies such as Mac1, Gr-1 (monocyte, macrophage, granulocyte), CD4 and CD8 (T cells), or B220 (B cells). Ten thousand CD45-positive/PI-negative events were analyzed. Relatively normal percentages of each population are shown. (B) Viable (PI-negative) fetal liver cells were stained with Ter119 and CD71 (a marker of immature nucleated red blood cells) to investigate fetal liver erythropoiesis. Cell staining with CD71, CD45, and annexin-V illustrate a unique population of CD71-positive, CD45-negative cells that stain with the apoptotic cell marker annexin-V. (C) By E15.5, mutant fetal livers exhibit dramatic cell death as revealed by PI uptake. (D) Counts of fetal liver cells used for FACS analyses. Trypan blue exclusion was used to identify viable cells. A significant reduction in viable cells was only detected in E15.5 knockout liver.
Staining of fetal liver cells from E14.5 wild-type and mutant embryos was conducted using various combinations of antibodies directed against specific lineages of hematopoietic cells (Fig. 4A). The relative proportion of CD45-positive cells did not differ significantly between genotypes, representing between 5 and 8% of total nucleated fetal liver cells (Fig. 4A). The majority of cells (70 to 85%) were CD45 negative or low and Ter119/CD71 positive (Fig. 4B). Viable (PI-negative) CD45.2-positive, myeloid (Mac-1 positive), T-cell (CD4/CD8-positive), granulocyte (Mac-1/Gr-1-positive), and B-cell (B220-positive) populations were all found in similar percentages in both knockout and wild-type fetal livers (Fig. 4A). This suggests that fetal liver hematopoiesis occurs relatively normally in E14.5 mutant embryos. Viable nucleated immature RBCs in fetal liver were also assessed (Fig. 4B) by using Ter119 (a marker of the erythroid lineage) and CD71 (a putative marker of immature nucleated RBCs). Staining E14.5 liver cells with CD71 and Ter119 indicated similar levels of total viable immature RBCs in mutant and wild-type liver (Fig. 4B). Staining with CD71, CD45, and annexin-V identified a unique population of CD71-positive, CD45-negative cells which stain with the apoptotic cell marker annexin-V (Fig. 4B). These data may indicate that a population of immature RBCs within mutant liver is apoptotic, which may account for the reduction in liver-derived mature RBCs observed in mutant blood smears (Fig. 3E and F). However, the degree to which CD71 binds to hepatocytes and other liver cells has not yet been determined, and staining with both Ter119 and annexin-V did not work in combination. Thus, it is not clear if TβRIII has a direct role in erythropoiesis or whether loss of fetal liver support is the cause of defective definitive erythropoiesis.
Heart defects in TβRIII−/− embryos.
Approximately 50% of embryos between E14.5 and E18.5 TβRIII−/− also exhibited defects of varying severity in the development of the myocardial wall of the heart ventricles (Fig. 5A through C). In these embryos, the ventricular wall failed to thicken in the region of the compact zone, resulting in a significantly thinner myocardial wall (Fig. 5A through C). The failure of the ventricular wall to thicken was associated with a thin, poorly cohesive muscular ventricular septum, which in severely affected embryos was accompanied by a ventricular septal defect (Fig. 5C). Trabeculation occurred, but the trabeculae were frequently reduced in mass, and myocyte morphology was abnormal (Fig. 5D and E). No significant differences in the amount of cardiac myosin heavy-chain and smooth-muscle actin were detected by Western blot analyses of mutant and wild-type E13.5 and E14.5 heart homogenates (data not shown). However, the percentage of proliferating myocytes within the ventricular wall, as indexed by PCNA immunostaining (Fig. 5F and G), was significantly reduced in E14.5 knockouts (49.1% ± 10.0%) compared to littermate controls (75.1% ± 5.67%). Antiactive caspase 3 immunostaining revealed little staining within the heart wall and trabeculae in both knockouts and littermate controls (data not shown), suggesting that apoptosis did not contribute to the thinning of the ventricular wall in mutants. The nonmuscular structures in the TβRIII−/− embryos have not yet been studied in detail but appeared grossly normal (Fig. 5H and I). However, a slight delay in the fusion of the endocardial cushion tissue with the muscular interventricular septum was detected in some E14.5 TβRIII−/− embryos (data not shown).
FIG. 5.
Heart defects in TβRIII−/− embryos. (A through C) Transverse sections through E16.5 hearts, near the level of the inlet valves, show thin, poorly compacted myocardial walls (arrowheads) and poorly formed septa in the TβRIII−/− hearts. (C) A severely affected heart, displaying a ventricular septal defect (arrow). (D and E) High-powered views of the compact zone in E18.5 hearts show an extreme example of compact zone thinness in the TβRIII−/− section. (F and G) PCNA immunostaining (brown) in E14.5 wild-type and knockout heart walls. Note the increase in the number of myocytes that do not express PCNA (blue cells) in TβRIII−/− heart. (H and I) Sagittal sections through E12.5 hearts show grossly normal atrioventricular (arrow) and outflow tract (curved arrow) endocardial cushion tissues in both wild-type and TβRIII−/− sections. Bars, 100 μm (A through C, H and I); 20 μm (D and E); 10 μm (F and G).
Liver and heart defects in TβRIII−/− mutants have lethal systemic effects.
The deleterious effects of the liver and heart defects on erythropoiesis and cardiac function are most likely the cause of death of the TβRIII−/− embryos during late gestation. Mutant embryos appeared systemically affected, with an arrest in development after E14.5 evident in many organs, thereby precluding analysis of the specific effects of TβRIII deficiency in these organs. Unfortunately, this was also true of the skeletal system, which was of particular interest since the phenotype of the TGFβ2-null mice includes a number of bone defects (50), and TβRIII has been suggested to be necessary for high-affinity binding of TGFβ2 to the signaling receptors (32, 51). An analysis of the skeletons of E18.5 TβRIII−/− embryos (n = 6) and their littermates (n = 120) using wholemount preparations stained with alcian blue and alizarin red S to detect cartilage and bone, respectively, revealed that 50% of ΤβRIII−/− embryos exhibited a generalized reduction in the size and ossification of the craniofacial, axial, and appendicular skeletons (data not shown). However, a small number of TβRIII+/+ and TβRIII+/− embryos that resembled the ΤβRIII-deficient embryos, in that they exhibited pallor and poor health during late gestation, showed a similar hypoplasia of the skeleton (data not shown). We were therefore unable to discern whether the reduction in bone size and ossification observed in ΤβRIII−/− embryos was a specific effect of TβRIII gene disruption. However, ΤβRIII-deficient embryos clearly lack some of the specific skeletal defects observed in TGFβ2 mutants, i.e., missing deltoid tuberosity and third trochanter, sternum malformations, and rib fusions (50).
Reduced sensitivity to TGFβ2 in TβRIII−/− MEFs.
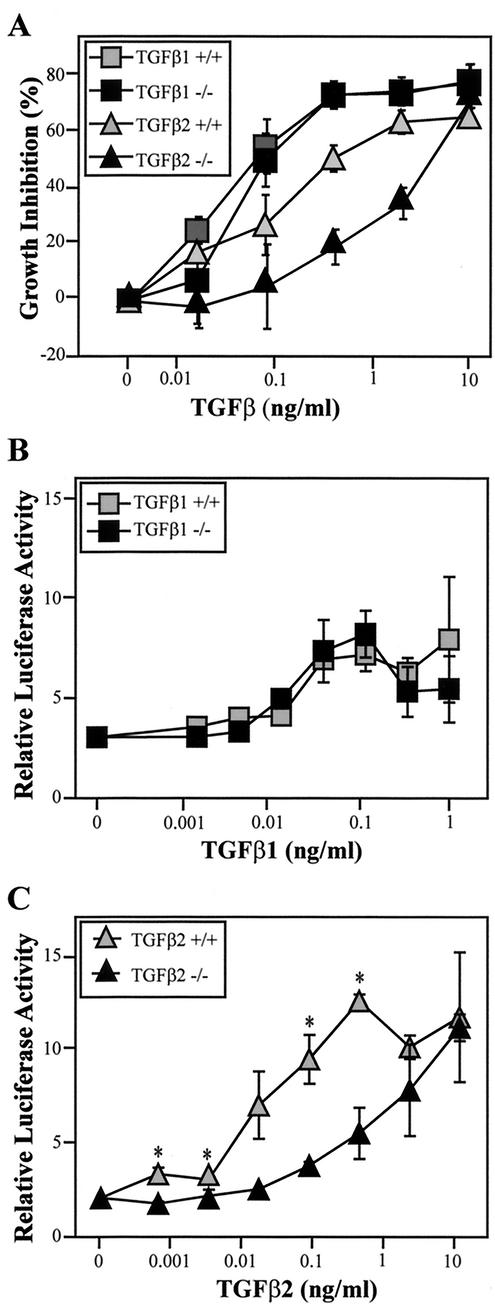
To study the cellular function of TβRIII in terms of the activities of the known TβRIII ligands, fibroblasts were derived from mutant and wild-type embryos. To assess the effect of TβRIII deficiency on TGFβ-mediated growth inhibition, MEFs were cultured in the presence of TGFβ1 or TGFβ2, and [3H]thymidine incorporation was measured as an index of cellular proliferation. As shown in Fig. 6, TGFβ1 and TGFβ2 inhibited [3H]thymidine incorporation in wild-type and mutant MEFs in a dose-dependent fashion. However, in TβRIII−/− cells, significantly higher doses of TGFβ2 were needed to induce the inhibition of [3H]thymidine incorporation (Fig. 6A). IC50s calculated from the TGFβ dose-response data indicate that, on average, TGFβ2 was 10-fold less potent on TβRIII−/− MEFs than on wild-type MEFs (Table 2). It should be noted that these values are only approximate because the upper asymptote of the dose-response curve for TGFβ2 in TβRIII−/− cells was generally not achieved.
FIG. 6.
Reduced responsiveness to TGFβ2 in TβRIII−/− embryonic fibroblasts. (A) MEFs derived from E12.5 littermates were as-sayed for TGFβ-mediated growth inhibition by measurement of [3H]thymidine incorporation. A representative graph of one littermate pair of cell lines shows the percentage of growth inhibition (relative to untreated controls for each cell line) induced by either TGFβ1 or TGFβ2 in the indicated concentrations. Quadruplicate wells were examined for each growth factor concentration, and means ± SD are shown. (B and C) MEFs were transiently transfected with pGL3-(CAGA)12-Luc. Sensitivity to TGFβ1 (B) was not significantly altered by TβRIII gene deletion but TGFβ2-induced gene expression (C) was significantly reduced (* indicates P < 0.05). Data presented are means ± SD from a representative littermate pair and have been corrected to an internal Renilla luciferase control. Four independent assays were conducted, each using different littermate pairs.
TABLE 2.
Mean IC50s (ng/ml) from growth inhibition assays of genotypes
| Growth factor | +/+ | +/− | −/− |
|---|---|---|---|
| TGFβ1 | 0.191 ± 0.047 (n = 8) | 0.183 ± 0.036 (n = 8) | 0.194 ± 0.044 (n = 15) |
| TGFβ2a | 0.340 ± 0.081 (n = 7) | 0.830 ± 0.172 (n = 8) | 3.13 ± 0.424 (n = 15) |
Significantly different by analysis of variance; P = 1.95 × 10−5.
In contrast to TGFβ2, TGFβ1-induced growth inhibition was not significantly affected by the loss of TβRIII (Fig. 6A, Table 2). Mutant MEFs derived from the same litters reproducibly exhibited slightly more or less sensitivity to TGFβ1 than their wild-type counterparts, but the average IC50s for TGFβ1 in mutant and wild-type MEFs were nearly identical (Table 2). Inhibin, in doses as high as 100 ng/ml, produced little change in [3H]thymidine incorporation in MEFs under the culture conditions used for these assays (data not shown). Similarly, while high doses of activin (100 ng/ml or greater) induced inhibition of [3H]thymidine incorporation in both wild-type and mutant cells, inhibin did not clearly antagonize this effect in either cell type (data not shown). Cell number and viability counts over 3 days verified that wild-type and TβRIII−/− cells did not significantly differ in their basal proliferation rates in DMEM-10% FCS (data not shown).
After TGFβ binds to its receptors, activated TβRI phosphorylates Smad2 and Smad3, which then forms a complex with Smad4 and translocates to the nucleus to propagate TGFβ signaling (35). We chose pGL3-(CAGA)12-Luc, a TGFβ-responsive reporter construct in which luciferase gene transcription is under the control of a Smad3/Smad4 binding element (13) to characterize the effect of TβRIII deficiency on TGFβ-induced reporter gene activation. MEFs were transiently transfected with pGL3-(CAGA)12-Luc and then treated with either TGFβ1 or TGFβ2 over a 1,000-fold concentration range. Activation of the reporter construct occurred over a similar range of TGFβ1 concentrations in wild-type and mutant MEFs (Fig. 6B), but luciferase expression was greatly reduced in TGFβ2-treated TβRIII−/− MEFs relative to wild-type cells (Fig. 6C). These differences in TGFβ1 and TGFβ2 sensitivity were consistent across all four littermate pairs examined.
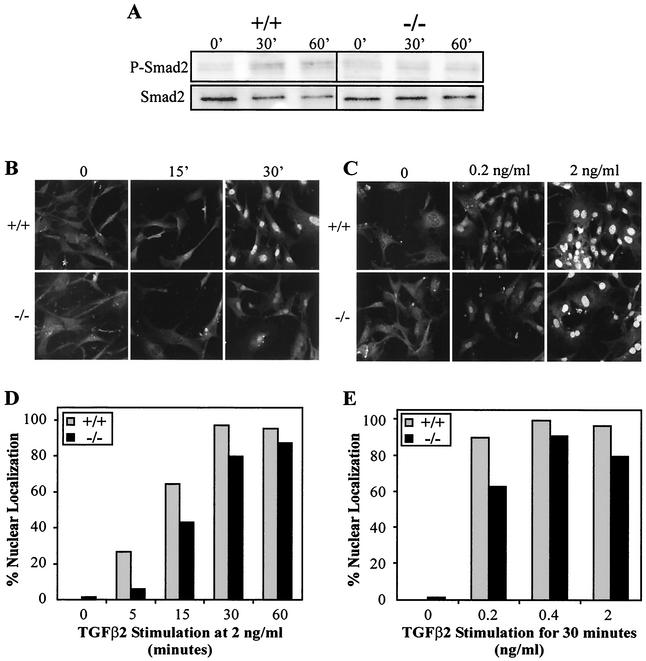
We then directly assessed Smad2 activation in wild-type and mutant cells. Following TGFβ stimulation, the level of Smad2 phosphorylation was measured by Western blot analysis of total cell lysates by using an antibody against the phosphorylated form of Smad2. Peak Smad2 phosphorylation occurred within 30 min of TGFβ1 or TGFβ2 treatment in wild-type MEFs (Fig. 7A). A slight reduction in the level of Smad2 phosphorylation was consistently detected in mutant MEFs following TGFβ2 treatment (Fig. 7A) but not following TGFβ1 treatment (data not shown). To achieve a more quantitative assessment of Smad2 activation, we examined the subcellular localization of Smad2 after TGFβ treatment in individual cells by using confocal microscopy. The cellular sensitivity to TGFβ2 was assessed by varying the amount of time to which the cells were exposed to TGFβ2 (Fig. 7B through D) or titrating the dose of TGFβ2 used (Fig. 7C through E). Examination of the cells under confocal microscopy revealed that both the intensity of anti-Smad2 nuclear staining (Fig. 7B and C) and the number of cells showing nuclear localization (Fig. 7D and E) were reduced in TβRIII−/− MEFs compared to wild-type cells. Unexpectedly, TGFβ1-treated TβRIII−/− MEFs also showed a reduction in Smad2 nuclear localization relative to wild-type cells, although the reduction was less reproducible across the two littermate pairs examined than that for TGFβ2 (data not shown). Attempts to use this same methodology to assess Smad2 activation in hepatocyte culture were unsuccessful due to high levels of constitutive Smad2 nuclear localization under the culture conditions used (data not shown), which was perhaps due to autocrine TGFβ/activin activity. Other potential TGFβ signal transduction molecules were examined in Western blots of lysates of wild-type and mutant cells exposed to TGFβ, but no significant differences between genotypes in the phosphorylation levels of several molecules (Erk1/2, p38, and SAPK/JNK) were detected by this method (data not shown).
FIG. 7.
Reduced Smad2 activation in TβRIII−/− embryonic fibroblasts. (A) Western blots of phospho-Smad2 and Smad2 in MEFs following treatment with 1 ng of TGFβ2 per ml for the times indicated. A slight reduction in TGFβ2-induced Smad2 phosphorylation was consistently detected in TβRIII−/− MEFs. (B through E) TβRIII−/− MEFs exhibit reduced Smad2 nuclear localization. MEFs were cultivated in the presence of TGFβ2 and then processed through immunohistochemistry using an anti-Smad2 antibody. Confocal microscopy revealed that TβRIII−/− MEFs exhibit reduced sensitivity to TGFβ2 in terms of Smad2 nuclear localization. (B and C) Confocal images. (D and E) Percentage of cells showing nuclear localization (data are from a representative litter).
DISCUSSION
TβRIII binds both inhibin and TGFβ with high affinity in vitro, but physiological roles for TβRIII in particular cell or tissue types in mouse are unknown. In this study, we demonstrate that heart muscle and liver are the major sites of TβRIII mRNA synthesis during midgestation. Consistent with the high TβRIII mRNA expression in fetal heart and liver, TβRIII−/− embryos display defects in the development of these tissues, indicating essential roles for TβRIII in cardiovascular and hepatic organogenesis. Based on in vitro data, TβRIII has been proposed to have complex positive and negative regulatory roles in the function of its ligands (16, 17, 23, 29, 30, 58). Experiments in which TβRIII was overexpressed in cells that lack the endogenous receptor indicate that TβRIII increases the binding of its ligands to their respective type II receptors and increases ligand efficacy in biological assays, particularly in regard to TGFβ2 (32). In accordance with these previous studies, our experiments performed with TβRIII-null MEFs indicate that mutant MEFs are significantly less sensitive to TGFβ2 than wild-type cells in terms of cellular proliferation, reporter gene transcription, and Smad2 activation, thus supporting the view that TβRIII modulates endogenous TGFβ2 responses. Based on our present data, however, it is unclear what role TβRIII has in regulating the function of its other ligands, i.e., TGFβ1 and inhibin, which elicited nearly identical responses in mutant and wild-type MEFs. The lack of measurable differences in TGFβ1 or inhibin activity in our assays may simply point to more subtle defects in TGFβ1 or inhibin signaling in the mutant MEFs.
Reduced sensitivity to TGFβ may underlie the TβRIII−/− heart phenotype.
During late gestation, the subepicardial myocytes proliferate to generate the outer wall of the heart and the muscular interventricular septum, both of which are poorly formed in TβRIII−/− embryos. At E14.5, TβRIII mutants exhibited a significant reduction in the proportion of ventricular myocytes expressing PCNA, suggesting that reduced myocyte proliferation may be the cause of the muscular heart phenotype. In accordance with a role for TβRIII in heart muscle formation, TβRI, TβRII, and TβRIII have been localized to cardiomyocytes during murine somatic development (34), indicating that embryonic cardiomyocytes are target cells for TGFβ. Indeed, TGFβ has been shown to regulate cardiomyocyte differentiation and proliferation in vitro (24, 39, 54). The predominant isoform produced in heart muscle is TGFβ2, which has been localized to cardiomyocytes during gestation, starting from the time when these cells first appear in the murine embryo (15). In addition, TGFβ2−/− mice display severe cardiac malformations (4, 50). Notably, within heart muscle, TGFβ2-null embryos show spongy ventricular myocardial walls and reduced compact zone formation (4), features shared by TβRIII−/− mutants. A large body of data indicates that TβRIII is required for the high affinity binding and signaling of TGFβ2 (32), suggesting that reduced sensitivity of cardiomyocytes to TGFβ2 may underlie the heart muscle phenotype in TβRIII−/− embryos. In addition, TGFβ1 is expressed within the endocardium of the developing mouse heart (1), and TGFβ1−/− pups of homozygous-null mothers show severe ventricular defects (28), indicating that TGFβ1 function in heart may also be affected by TβRIII deficiency.
Previous studies have supported a role for TβRIII in the formation of the endocardial cushion tissue, a mesenchymal tissue that arises from an epithelial-mesenchymal transformation of endothelial cells in the outflow tract and atrioventricular canal (6, 7). In explanted chick atrioventricular cushion tissue, a blocking antibody to TβRIII significantly reduced the number of mesenchymal cells formed in the explants in response to induction by cushion myocardium (7). However, the initial formation of cushion tissue in the TβRIII−/− hearts did not seem to be impaired, suggesting that TβRIII does not play a requisite role in epithelial-mesenchymal transformation within the murine atrioventricular canal. Nevertheless, because a slight lag in the fusion of the endocardial cushion tissue with the muscular interventricular septum was detected in TβRIII−/− heart, we cannot yet formally rule out subtle defects in the development or remodeling of the atrioventricular endocardial cushion tissue.
TβRIII is required in fetal liver development.
The expression of TβRIII mRNA in liver from E10.5 to E16.5 is in good agreement with the localization of the TGFβs and TGFβ receptor mRNAs and proteins within the liver during the course of murine development (15, 34, 40, 52), suggesting that TβRIII plays a role in TGFβ-mediated events during liver development. TGFβ is a major regulator of hepatocyte biology in fetal and adult liver, negatively regulating cell proliferation and survival and stimulating extracellular matrix elaboration (11, 47-49). Of particular interest, TGFβ induces apoptosis in fetal rat hepatocytes by a mechanism involving caspase 3 activation (9, 18, 22). This mechanism of cell death can be blocked by phosphatidylinositol 3-kinase/Akt pathway activation, which inhibits TGFβ-induced caspase 3 activation independent of Smad pathway involvement (9, 18). Interestingly, TGFβ-treated fetal rat hepatocytes that have undergone an epithelial-mesenchymal transformation are resistant to TGFβ-induced apoptosis and express higher levels of phospho-Akt than fetal hepatocytes (57). These studies suggest that reduced Akt activity may increase the susceptibility of embryonic hepatocytes to apoptosis. A similar mechanism may underlie the apoptosis in TβRIII−/− embryonic liver, as reduced levels of Akt and phospho-Akt were detected in liver at the time of the onset of the liver pathology. However, the mechanism by which the absence of TβRIII leads to a reduction in Akt levels is not yet clear. The potential for reciprocal feedback regulations between TGFβ and Akt is suggested by a recent report that demonstrates that active Akt can down-regulate TGFβ2 mRNA (46).
In contrast to TGFβ, available evidence suggests that inhibin is not an important regulator of fetal liver development, although inhibin is apparently required after birth to prevent activin-mediated hepatocyte necrosis (37). Transcripts for the inhibin α-subunit, ActRII, and ActRIIB were not detected in fetal rodent liver (19, 43). In addition, inhibin-null mice exhibit no embryonic liver phenotype, dying in adulthood of gonadal tumors and a cancer cachexia-like syndrome (37). Based on these data, disruption of inhibin functioning is unlikely to solely account for the TβRIII−/− liver phenotype.
Despite the overlap in expression patterns between TβRIII mRNA and other components of the TGFβ system, the TβRIII−/− liver phenotype is also not similar to any of the phenotypes produced by TGFβ-null mutations (14, 26, 42, 50). This suggests that TβRIII deficiency does not disrupt activated TGFβ receptor signaling in fetal liver, at least not in terms of any single TGFβ isoform. However, a hypoplastic liver phenotype was observed in Smad2; Smad3 double heterozygous embryos, which was not observed in either the Smad2- or Smad3-null homozygotes (59), suggesting that these two Smads are required in concert to regulate liver outgrowth. Since Smad2 and Smad3 are major signal transducers for both activin and TGFβ (35), these data suggest a possible role for TGFβ/activin in liver development. It is possible that TβRIII deficiency may disrupt cooperative regulatory processes among TGFβ superfamily members or between TGFβ and other growth factors in embryonic mouse liver. However, it should be noted that the phenotype observed in the double heterozygotes appears to differ from the TβRIII−/− phenotype. Notably, the Smad2+/−; Smad3+/− phenotype appears to be the result of defects in liver outgrowth and cell-cell and cell-substrate adhesions within liver (59). In contrast, in TβRIII−/− mutants, the liver phenotype appears after initial liver outgrowth, and no cell adhesion defect was detected.
ΤβRIII is not required for all TGFβ2-mediated developmental processes.
Although the late-gestation lethality caused by TβRIII gene deletion confounds a comparison of the TβRIII mutants with the TGFβ2 mutants (and the other ligand knockouts), certain aspects of the TGFβ2 knockout phenotype, i.e., particular bone defects (14), were not evident in TβRIII mutants. Thus, despite the large body of in vitro data that indicates that TβRIII is essential for TGFβ2 function, our data indicate that there may be exceptions in vivo. Recent studies point to possible mechanisms by which TGFβ2 signaling could occur in the absence of TβRIII. Notably, a splice variant of the murine TβRII (termed TβRIIB), which does not require TβRIII to bind TGFβ2 with high affinity, was found on bone cells (45), indicating that TβRIII may be dispensable for some TGFβ2-mediated processes in bone. Additionally, in some contexts, coexpression of TβRI with TβRII (44) or treating cells with high levels of TGFβ (12) was sufficient to overcome the low affinity of TGFβ2 for TβRII. Therefore, the ability of TβRIII to facilitate TGFβ2 binding may be nonessential in certain developing organs due to the availability of alternative TGFβ2 signaling complexes or adequate levels of ligand to meet threshold binding to the conventional signaling receptors.
In conclusion, our cell culture data and the ventricle phenotype of the TβRIII mutant suggest that TβRIII may be required for optimal TGFβ2 function during development. However, the TβRIII and ligand knockout phenotypes are different in many respects, suggesting that TβRIII is dispensable for facilitating certain TGFβ-mediated and inhibin-mediated developmental processes and may have actions independent of these ligands. Proper regulation of TGFβ activity is critical for the normal development and maintenance of most tissues, and dysregulation of this system is implicated in many pathological conditions (5). The TβRIII knockout mice should continue to prove useful in further defining the physiological roles of TβRIII in modulating TGFβ and inhibin cellular responsiveness.
Acknowledgments
This work was supported in part by Public Health Service fellowship F32 CA90034 (K.L.S.) from the National Cancer Institute and by project grants from the Australian National Health & Medical Research Council (164815 to K.L.S. and 164812 to H.-J.Z.).
We thank A. W. Burgess for critical reading and support, Helen Abud for helpful advice on PCNA and caspase immunohistochemistry, X. F. Wang for the gift of the rat TβRIII cDNA, and A. Moustakas for pGL3-(CAGA)12-Luc reporter. We also thank the Ludwig Institute animal facility staff for animal husbandry, Janna Stickland for help with photography, and Valerie Feakes for histological sectioning.
REFERENCES
- 1.Akhurst, R. J., S. A. Lehnert, A. Faissner, and E. Duffie. 1990. TGF beta in murine morphogenetic processes: the early embryo and cardiogenesis. Development 108:645-656. [DOI] [PubMed] [Google Scholar]
- 2.Andres, J. L., D. DeFalcis, M. Noda, and J. Massague. 1992. Binding of two growth factor families to separate domains of the proteoglycan betaglycan. J. Biol. Chem. 267:5927-5930. [PubMed] [Google Scholar]
- 3.Bandyopadhyay, A., Y. Zhu, M. L. Cibull, L. Bao, C. Chen, and L. Sun. 1999. A soluble transforming growth factor beta type III receptor suppresses tumorigenicity and metastasis of human breast cancer MDA-MB-231 cells. Cancer Res. 59:5041-5046. [PubMed] [Google Scholar]
- 4.Bartram, U., D. G. Molin, L. J. Wisse, A. Mohamad, L. P. Sanford, T. Doetschman, C. P. Speer, R. E. Poelmann, and A. C. Gittenberger-de Groot. 2001. Double-outlet right ventricle and overriding tricuspid valve reflect disturbances of looping, myocardialization, endocardial cushion differentiation, and apoptosis in TGF-beta(2)-knockout mice. Circulation 103:2745-2752. [DOI] [PubMed] [Google Scholar]
- 5.Border, W. A., and E. Ruoslahti. 1992. Transforming growth factor-beta in disease: the dark side of tissue repair. J. Clin. Investig. 90:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer, A. S., I. I. Ayerinskas, E. B. Vincent, L. A. McKinney, D. L. Weeks, and R. B. Runyan. 1999. TGFbeta2 and TGFbeta3 have separate and sequential activities during epithelial-mesenchymal cell transformation in the embryonic heart. Dev. Biol. 208:530-545. [DOI] [PubMed] [Google Scholar]
- 7.Brown, C. B., A. S. Boyer, R. B. Runyan, and J. V. Barnett. 1999. Requirement of type III TGF-beta receptor for endocardial cell transformation in the heart. Science 283:2080-2082. [DOI] [PubMed] [Google Scholar]
- 8.Cheifetz, S., H. Hernandez, M. Laiho, P. ten Dijke, K. K. Iwata, and J. Massague. 1990. Distinct transforming growth factor-beta (TGF-beta) receptor subsets as determinants of cellular responsiveness to three TGF-beta isoforms. J. Biol. Chem. 265:20533-20538. [PubMed] [Google Scholar]
- 9.Chen, R. H., Y. H. Su, R. L. Chuang, and T. Y. Chang. 1998. Suppression of transforming growth factor-beta-induced apoptosis through a phosphatidylinositol 3-kinase/Akt-dependent pathway. Oncogene 17:1959-1968. [DOI] [PubMed] [Google Scholar]
- 10.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 11.Clouthier, D. E., S. A. Comerford, and R. E. Hammer. 1997. Hepatic fibrosis, glomerulosclerosis, and a lipodystrophy-like syndrome in PEPCK-TGF-beta1 transgenic mice. J. Clin. Investig. 100:2697-2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, X., S. Bellis, Z. Yan, and E. Friedman. 1999. Differential responsiveness to autocrine and exogenous transforming growth factor (TGF) beta1 in cells with nonfunctional TGF-beta receptor type III. Cell Growth Differ. 10:11-18. [PubMed] [Google Scholar]
- 13.Dennler, S., S. Itoh, D. Vivien, P. ten Dijke, S. Huet, and J. M. Gauthier. 1998. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17:3091-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickson, M. C., J. S. Martin, F. M. Cousins, A. B. Kulkarni, S. Karlsson, and R. J. Akhurst. 1995. Defective haematopoiesis and vasculogenesis in transforming growth factor-beta 1 knock out mice. Development 121:1845-1854. [DOI] [PubMed] [Google Scholar]
- 15.Dickson, M. C., H. G. Slager, E. Duffie, C. L. Mummery, and R. J. Akhurst. 1993. RNA and protein localisations of TGF beta 2 in the early mouse embryo suggest an involvement in cardiac development. Development 117:625-639. [DOI] [PubMed] [Google Scholar]
- 16.Eickelberg, O., M. Centrella, M. Reiss, M. Kashgarian, and R. G. Wells. 2002. Betaglycan inhibits TGF-beta signaling by preventing type I-type II receptor complex formation. Glycosaminoglycan modifications alter betaglycan function. J. Biol. Chem. 277:823-829. [DOI] [PubMed] [Google Scholar]
- 17.Esparza-Lopez, J., J. L. Montiel, M. M. Vilchis-Landeros, T. Okadome, K. Miyazono, and F. Lopez-Casillas. 2001. Ligand binding and functional properties of betaglycan, a coreceptor of the transforming growth factor-beta superfamily. Specialized binding regions for transforming growth factor-beta and inhibin A. J. Biol. Chem. 276:14588-14596. [DOI] [PubMed] [Google Scholar]
- 18.Fabregat, I., B. Herrera, M. Fernandez, A. M. Alvarez, A. Sanchez, C. Roncero, J. J. Ventura, A. M. Valverde, and M. Benito. 2000. Epidermal growth factor impairs the cytochrome C/caspase-3 apoptotic pathway induced by transforming growth factor beta in rat fetal hepatocytes via a phosphoinositide 3-kinase-dependent pathway. Hepatology 32:528-535. [DOI] [PubMed] [Google Scholar]
- 19.Feijen, A., M. J. Goumans, and A. J. Eijnden-van Raaij. 1994. Expression of activin subunits, activin receptors and follistatin in postimplantation mouse embryos suggests specific developmental functions for different activins. Development 120:3621-3637. [DOI] [PubMed] [Google Scholar]
- 20.Harder, K. W., L. M. Parsons, J. Armes, N. Evans, N. Kountouri, R. Clark, C. Quilici, D. Grail, G. S. Hodgson, A. R. Dunn, and M. L. Hibbs. 2001. Gain- and loss-of-function Lyn mutant mice define a critical inhibitory role for Lyn in the myeloid lineage. Immunity 15:603-615. [DOI] [PubMed] [Google Scholar]
- 21.Heldin, C. H., K. Miyazono, and P. ten Dijke. 1997. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature 390:465-471. [DOI] [PubMed] [Google Scholar]
- 22.Herrera, B., M. Fernandez, A. M. Alvarez, C. Roncero, M. Benito, J. Gil, and I. Fabregat. 2001. Activation of caspases occurs downstream from radical oxygen species production, Bcl-xL down-regulation, and early cytochrome C release in apoptosis induced by transforming growth factor beta in rat fetal hepatocytes. Hepatology 34:548-556. [DOI] [PubMed] [Google Scholar]
- 23.Kaname, S., and E. Ruoslahti. 1996. Betaglycan has multiple binding sites for transforming growth factor-beta 1. Biochem. J. 315(Pt 3):815-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kardami, E. 1990. Stimulation and inhibition of cardiac myocyte proliferation in vitro. Mol. Cell. Biochem. 92:129-135. [DOI] [PubMed] [Google Scholar]
- 25.Kaufman, M. H. 1992. The atlas of mouse development. Academic Press, London, United Kingdom.
- 26.Kulkarni, A. B., C. G. Huh, D. Becker, A. Geiser, M. Lyght, K. C. Flanders, A. B. Roberts, M. B. Sporn, J. M. Ward, and S. Karlsson. 1993. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc. Natl. Acad. Sci. USA 90:770-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamarre, J., J. Vasudevan, and S. L. Gonias. 1994. Plasmin cleaves betaglycan and releases a 60 kDa transforming growth factor-beta complex from the cell surface. Biochem. J. 302(Pt 1):199-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letterio, J. J., A. G. Geiser, A. B. Kulkarni, N. S. Roche, M. B. Sporn, and A. B. Roberts. 1994. Maternal rescue of transforming growth factor-beta 1 null mice. Science 264:1936-1938. [DOI] [PubMed] [Google Scholar]
- 29.Lewis, K. A., P. C. Gray, A. L. Blount, L. A. MacConell, E. Wiater, L. M. Bilezikjian, and W. Vale. 2000. Betaglycan binds inhibin and can mediate functional antagonism of activin signalling. Nature 404:411-414. [DOI] [PubMed] [Google Scholar]
- 30.Lopez-Casillas, F., S. Cheifetz, J. Doody, J. L. Andres, W. S. Lane, and J. Massague. 1991. Structure and expression of the membrane proteoglycan betaglycan, a component of the TGF-beta receptor system. Cell 67:785-795. [DOI] [PubMed] [Google Scholar]
- 31.Lopez-Casillas, F., H. M. Payne, J. L. Andres, and J. Massague. 1994. Betaglycan can act as a dual modulator of TGF-beta access to signaling receptors: mapping of ligand binding and GAG attachment sites. J. Cell Biol. 124:557-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez-Casillas, F., J. L. Wrana, and J. Massague. 1993. Betaglycan presents ligand to the TGF beta signaling receptor. Cell 73:1435-1444. [DOI] [PubMed] [Google Scholar]
- 33.Lufkin, T., M. Mark, C. P. Hart, P. Dolle, M. LeMeur, and P. Chambon. 1992. Homeotic transformation of the occipital bones of the skull by ectopic expression of a homeobox gene. Nature 359:835-841. [DOI] [PubMed] [Google Scholar]
- 34.Mariano, J. M., L. M. Montuenga, M. A. Prentice, F. Cuttitta, and S. B. Jakowlew. 1998. Concurrent and distinct transcription and translation of transforming growth factor-beta type I and type II receptors in rodent embryogenesis. Int. J. Dev. Biol. 42:1125-1136. [PubMed] [Google Scholar]
- 35.Massague, J. 1998. TGF-beta signal transduction. Annu. Rev. Biochem. 67:753-791. [DOI] [PubMed] [Google Scholar]
- 36.Massague, J., and Y. G. Chen. 2000. Controlling TGF-beta signaling. Genes Dev. 14:627-644. [PubMed] [Google Scholar]
- 37.Matzuk, M. M., M. J. Finegold, J. P. Mather, L. Krummen, H. Lu, and A. Bradley. 1994. Development of cancer cachexia-like syndrome and adrenal tumors in inhibin-deficient mice. Proc. Natl. Acad. Sci. USA 91:8817-8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell, E. J., L. Fitz-Gibbon, and M. D. O'Connor-McCourt. 1992. Subtypes of betaglycan and of type I and type II transforming growth factor-beta (TGF-beta) receptors with different affinities for TGF-beta 1 and TGF-beta 2 are exhibited by human placental trophoblast cells. J. Cell. Physiol. 150:334-343. [DOI] [PubMed] [Google Scholar]
- 39.Parker, T. G., S. E. Packer, and M. D. Schneider. 1990. Peptide growth factors can provoke “fetal” contractile protein gene expression in rat cardiac myocytes. J. Clin. Investig. 85:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelton, R. W., M. E. Dickinson, H. L. Moses, and B. L. Hogan. 1990. In situ hybridization analysis of TGF beta 3 RNA expression during mouse development: comparative studies with TGF beta 1 and beta 2. Development 110:609-620. [DOI] [PubMed] [Google Scholar]
- 41.Ponce-Castaneda, M. V., J. Esparza-Lopez, M. M. Vilchis-Landeros, V. Mendoza, and F. Lopez-Casillas. 1998. Murine betaglycan primary structure, expression and glycosaminoglycan attachment sites. Biochim. Biophys. Acta 1384:189-196. [DOI] [PubMed] [Google Scholar]
- 42.Proetzel, G., S. A. Pawlowski, M. V. Wiles, M. Yin, G. P. Boivin, P. N. Howles, J. Ding, M. W. Ferguson, and T. Doetschman. 1995. Transforming growth factor-beta 3 is required for secondary palate fusion. Nat. Genet. 11:409-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts, V. J., and S. L. Barth. 1994. Expression of messenger ribonucleic acids encoding the inhibin/activin system during mid- and late-gestation rat embryogenesis. Endocrinology 134:914-923. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez, C., F. Chen, R. A. Weinberg, and H. F. Lodish. 1995. Cooperative binding of transforming growth factor (TGF)-beta 2 to the types I and II TGF-beta receptors. J. Biol. Chem. 270:15919-15922. [DOI] [PubMed] [Google Scholar]
- 45.Rotzer, D., M. Roth, M. Lutz, D. Lindemann, W. Sebald, and P. Knaus. 2001. Type III TGF-beta receptor-independent signalling of TGF-beta2 via TbetaRII-B, an alternatively spliced TGF-beta type II receptor. EMBO J. 20:480-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samatar, A. A., L. Wang, A. Mirza, S. Koseoglu, S. Liu, and C. C. Kumar. 2002. Transforming growth factor-beta 2 is a transcriptional target for Akt/protein kinase B via forkhead transcription factor. J. Biol. Chem. 277:28118-28126. [DOI] [PubMed] [Google Scholar]
- 47.Sanchez, A., A. M. Alvarez, M. Benito, and I. Fabregat. 1995. Transforming growth factor beta modulates growth and differentiation of fetal hepatocytes in primary culture. J. Cell. Physiol. 165:398-405. [DOI] [PubMed] [Google Scholar]
- 48.Sanchez, A., A. M. Alvarez, M. Benito, and I. Fabregat. 1996. Apoptosis induced by transforming growth factor-beta in fetal hepatocyte primary cultures: involvement of reactive oxygen intermediates. J. Biol. Chem. 271:7416-7422. [DOI] [PubMed] [Google Scholar]
- 49.Sanderson, N., V. Factor, P. Nagy, J. Kopp, P. Kondaiah, L. Wakefield, A. B. Roberts, M. B. Sporn, and S. S. Thorgeirsson. 1995. Hepatic expression of mature transforming growth factor beta 1 in transgenic mice results in multiple tissue lesions. Proc. Natl. Acad. Sci. USA 92:2572-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanford, L. P., I. Ormsby, A. C. Gittenberger-de Groot, H. Sariola, R. Friedman, G. P. Boivin, E. L. Cardell, and T. Doetschman. 1997. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development 124:2659-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sankar, S., N. Mahooti-Brooks, M. Centrella, T. L. McCarthy, and J. A. Madri. 1995. Expression of transforming growth factor type III receptor in vascular endothelial cells increases their responsiveness to transforming growth factor beta 2. J. Biol. Chem. 270:13567-13572. [DOI] [PubMed] [Google Scholar]
- 52.Schmid, P., D. Cox, G. Bilbe, R. Maier, and G. K. McMaster. 1991. Differential expression of TGF beta 1, beta 2 and beta 3 genes during mouse embryogenesis. Development 111:117-130. [DOI] [PubMed] [Google Scholar]
- 53.Segarini, P. R., D. M. Rosen, and S. M. Seyedin. 1989. Binding of transforming growth factor-beta to cell surface proteins varies with cell type. Mol. Endocrinol. 3:261-272. [DOI] [PubMed] [Google Scholar]
- 54.Slager, H. G., W. Van Inzen, E. Freund, A. J. Eijnden-van Raaij, and C. L. Mummery. 1993. Transforming growth factor-beta in the early mouse embryo: implications for the regulation of muscle formation and implantation. Dev. Genet. 14:212-224. [DOI] [PubMed] [Google Scholar]
- 55.Stacker, S. A., K. Stenvers, C. Caesar, A. Vitali, T. Domagala, E. Nice, S. Roufail, R. J. Simpson, R. Moritz, T. Karpanen, K. Alitalo, and M. G. Achen. 1999. Biosynthesis of vascular endothelial growth factor-D involves proteolytic processing which generates non-covalent homodimers. J. Biol. Chem. 274:32127-32136. [DOI] [PubMed] [Google Scholar]
- 56.Sun, L., and C. Chen. 1997. Expression of transforming growth factor beta type III receptor suppresses tumorigenicity of human breast cancer MDA-MB-231 cells. J. Biol. Chem. 272:25367-25372. [DOI] [PubMed] [Google Scholar]
- 57.Valdes, F., A. M. Alvarez, A. Locascio, S. Vega, B. Herrera, M. Fernandez, M. Benito, M. A. Nieto, and I. Fabregat. 2002. The epithelial mesenchymal transition confers resistance to the apoptotic effects of transforming growth factor β in fetal rat hepatocytes. Mol. Cancer Res. 1:68-78. [PubMed] [Google Scholar]
- 58.Wang, X. F., H. Y. Lin, E. Ng-Eaton, J. Downward, H. F. Lodish, and R. A. Weinberg. 1991. Expression cloning and characterization of the TGF-beta type III receptor. Cell 67:797-805. [DOI] [PubMed] [Google Scholar]
- 59.Weinstein, M., S. P. Monga, Y. Liu, S. G. Brodie, Y. Tang, C. Li, L. Mishra, and C. X. Deng. 2001. Smad proteins and hepatocyte growth factor control parallel regulatory pathways that converge on beta1-integrin to promote normal liver development. Mol. Cell. Biol. 21:5122-5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiater, E., and W. Vale. 2003. Inhibin is an antagonist of bone morphogenetic protein signaling. J. Biol. Chem. 278:7934-7941. [DOI] [PubMed] [Google Scholar]
- 61.Wrana, J. L. 2000. Regulation of Smad activity. Cell 100:189-192. [DOI] [PubMed] [Google Scholar]
- 62.Wrana, J. L., L. Attisano, R. Wieser, F. Ventura, and J. Massague. 1994. Mechanism of activation of the TGF-beta receptor. Nature 370:341-347. [DOI] [PubMed] [Google Scholar]
- 63.Zaret, K. S. 2002. Regulatory phases of early liver development: paradigms of organogenesis. Nat. Rev. Genet. 3:499-512. [DOI] [PubMed] [Google Scholar]
- 64.Zhu, H. J., and A. W. Burgess. 2001. Regulation of transforming growth factor-beta signaling. Mol. Cell Biol. Res. Commun. 4:321-330. [DOI] [PubMed] [Google Scholar]
- 65.Zhu, H. J., and A. M. Sizeland. 1999. A pivotal role for the transmembrane domain in transforming growth factor-beta receptor activation. J. Biol. Chem. 274:11773-11781. [DOI] [PubMed] [Google Scholar]
- 66.Zhu, H. J., and A. M. Sizeland. 1999. Extracellular domain of the transforming growth factor-beta receptor negatively regulates ligand-independent receptor activation. J. Biol. Chem. 274:29220-29227. [DOI] [PubMed] [Google Scholar]