Abstract
Calcium induces transcriptional activation of the fos promoter by activation of the cyclic AMP response element (CRE)-binding protein (CREB), and in some cells its effect is enhanced synergistically by cyclic GMP (cGMP) through an unknown mechanism. We observed calcium-cGMP synergism in neuronal and osteogenic cells which express type II cGMP-dependent protein kinase (G-kinase); the effect on the fos promoter was mediated by the CRE and proportional to G-kinase activity. Dominant negative transcription factors showed involvement of CREB- and C/EBP-related proteins but not of AP-1. Expression of C/EBP-β but not C/EBP-α or -δ enhanced the effects of calcium and cGMP on a CRE-dependent reporter gene. The transactivation potential of full-length CREB fused to the DNA-binding domain of Gal4 was increased synergistically by calcium and cGMP, and overexpression of C/EBP-β enhanced the effect, while a dominant negative C/EBP inhibited it. With a mammalian two-hybrid system, coimmunoprecipitation experiments, and in vitro binding studies, we demonstrated that C/EBP-β and CREB interacted directly; this interaction involved the C terminus of C/EBP-β but occurred independently of CREB's leucine zipper domain. CREB Ser133 phosphorylation was stimulated by calcium but not by cGMP; in cGMP-treated cells, 32PO4 incorporation into C/EBP-β was decreased and C/EBP-β/CRE complexes were increased, suggesting regulation of C/EBP-β functions by G-kinase-dependent dephosphorylation. C/EBP-β and CREB associated with the fos promoter in intact cells, and the amount of promoter-associated C/EBP-β was increased by calcium and cGMP. We conclude that calcium and cGMP transcriptional synergism requires cooperation of CREB and C/EBP-β, with calcium and cGMP modulating the phosphorylation states of CREB and C/EBP-β, respectively.
The c-fos proto-oncogene product is a transcription factor of the AP-1 family that is involved in cell proliferation, differentiation, and survival (34, 63). It is thought to couple short-term signals elicited by cell surface stimuli to long-term changes in cellular phenotype by regulating specific target genes (32). The cooperation of four cis-acting elements within the c-fos promoter, i.e., the sis-inducible element, the serum response element (SRE), the fos AP-1 site, and the cyclic AMP (cAMP) response element (CRE), is required for tissue- and stimulus-specific regulation (32, 59).
Increased intracellular calcium stimulates c-fos transcription in many cell types, including neuronal cells in response to synaptic activity (32, 64, 76). Calcium induction of c-fos can be mediated by calcium/calmodulin-dependent protein kinases (Cam-kinases) and/or the Ras/Raf/MEK/extracellular signal-regulated kinase (ERK)/RSK pathway; both pathways lead to phosphorylation of the CRE binding protein (CREB) at Ser133 (19, 64, 76). In addition, calcium can increase the intracellular cAMP concentration leading to CREB phosphorylation via activation of cAMP-dependent protein kinase (A-kinase) and/or the Rap1/B-Raf/MEK/ERK pathway (19, 76). Calcium targets both the CRE and the SRE in the c-fos promoter, with cooperation between both elements (64). CREB phosphorylation on Ser133 appears to be necessary but not sufficient for calcium induction of c-fos through the CRE, and Cam-kinase-mediated phosphorylation of Ser142/143 in CREB and/or phosphorylation of Ser301 in the CREB-binding protein CBP may also be important (12, 30, 36, 64, 76).
An increase in the intracellular cGMP concentration also induces c-fos mRNA, e.g., during activation of soluble and membrane-bound guanylate cyclases by nitric oxide (NO) and natriuretic peptides, respectively (15, 17, 25, 51, 55, 68). We have shown that the effect of NO/cGMP on the fos promoter is mediated by cGMP-dependent protein kinases (G-kinases) but that the soluble type I and the membrane-bound type II G-kinases regulate transcription by different mechanisms (21, 22, 29). In some cell types, including C6 glioma cells, G-kinase I activation causes nuclear translocation of the enzyme which can directly phosphorylate CREB (20, 23). However, extranuclear G-kinase II transcriptionally activates the fos promoter in a cell type-specific manner without inducing CREB Ser133 phosphorylation (21; T. Gudi and R. B. Pilz, unpublished observation). When the calcium and NO/cGMP signaling pathways are activated together, they synergistically induce high levels of c-fos mRNA expression in cells of neuronal origin, even in cells which show little or no c-fos mRNA induction by NO/cGMP alone (5, 37, 53). The mechanism of the synergism is unclear, although the involvement of A-kinase and mitogen-activated protein kinases (MAP kinases) has been suggested (37, 53). We found that G-kinase is required for synergistic activation of the fos promoter by calcium and NO/cGMP and that G-kinase II is more effective than G-kinase I (21).
G-kinase II is the predominant G-kinase isoform in bone, small intestinal mucosa, and most areas of the central nervous system, while G-kinase I is most abundant in vascular and visceral smooth muscle cells and in the cerebellum (16, 71). G-kinase II is required for normal growth and differentiation of bone-forming cells (48, 54), and c-fos is a key regulator of normal bone development and remodeling (34, 50), but the effect of cGMP/G-kinase on c-fos expression in osteogenic cells has not been studied. In the central nervous system, NO/cGMP signaling may modulate synaptic plasticity and neuronal survival (3, 39, 41, 65), but G-kinase functions are not well defined, since mice lacking G-kinase I and II have no obvious neurological deficits (35, 54). CREB mediates both activity-dependent synaptic plasticity and trophic-factor-dependent neuronal survival, with synaptic activity leading to calcium influx and activation of CREB and its downstream target c-fos (12, 44, 76).
We now show that synergistic transcriptional regulation of the fos promoter by calcium and cGMP in osteoblast-derived cells as well as in G-kinase II-expressing glioma cells requires the cooperation of CREB and C/EBP-β at the CRE.
MATERIALS AND METHODS
Plasmids.
The expression vector encoding G-kinase II (pRC/CMV-GKII) was from S. Lohmann (33). The control vectors pRSV-Luc and pRSV-βGal were described before (22), and the reporter pCRE-Luc (containing four tandem canonical CREs), pSRE-Luc, containing four copies of the fos SRE including the C/EBP-β binding site (46), and pAP1-Luc (containing four canonical AP-1 binding sites) were from Stratagene. Expression vectors for A-CREB, A-C/EBP, and A-Fos were from C. Vinson (2, 52). M. Montmini provided the reporter pGAL4-Luc (containing five GAL4 binding sites) and the expression vectors pGal4-CREB(full-length) and pGal4-CREB(Δbzip), which encode the DNA-binding domain of the yeast transcription factor Gal4 (amino acids 1 to 147) fused to amino acids 1 to 354 or 1 to 284 of CREB, respectively (14). pGal4-ATF-1(full-length) and Δbzip were from N. C. Jones and R. A. Maurer, respectively (28, 67). Mammalian and bacterial expression vectors encoding epitope (EE)-tagged or glutathione S-transferase (GST)-tagged CREB sequences were constructed by placing amino acids 1 to 354, 1 to 284, 1 to 199, or 1 to 99 of CREB in frame downstream of a leader sequence encoding EEEEYMPME (EE tag, in pcDNA3) or downstream of GST (in pGEX-KG), respectively. Expression vectors encoding C/EBP-α, -β, and -δ were from P. F. Johnson (77); vectors encoding p20 and p20-VP16 were from L. Sealy (27).
Cell culture and transfections.
Rat UMR106 osteosarcoma cells and COS7 cells were from the American Type Culture Collection, rat C6 glioma cells were from M. Ellisman, and murine MC3T3-E1 osteoblast cells were from E. J. Murray. All cells were routinely cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and maintained for <16 passages. Cells were transfected with Lipofectamine Plus (Life Sciences) as previously described (21). After transient transfection, cells were transferred to DMEM supplemented with 0.1% FBS plus 0.1% bovine serum albumin (BSA), and 24 h later, 0.3 μM A23187 (calcium ionophore, Calbiochem) or 250 μM 8-chlorophenylthio-cGMP (CPT-cGMP; BioLog) or both agents were added for 8 h prior to harvesting. To stably transfect C6 cells with G-kinase II, cells were plated at 10 to 1,000 cells per 10-cm dish 24 h after transfection, and 24 h later 2 mg of G418 per ml was added in full growth medium; after 10 to 14 days, single colonies were picked, expanded, and screened for G-kinase expression by activity assay (see below). COS7 cells were transfected by electroporation with a Bio-Rad Gene Pulser at 0.27 kV, 550 μF, and a pulse length of 18 ms with 10 μg of DNA and 4 × 106 cells.
Subcellular fractionation and G-kinase activity assay.
For subcellular fractionation, cells were harvested in late log phase and extracted at 4°C by Dounce homogenization in lysis buffer containing 10 mM Na-HEPES (pH 7.0), 2 mM EDTA, 1 mM MgCl2, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, and 10 μg of aprotinin per ml. After centrifugation for 5 min at 500 × g to remove nuclei and debris, the homogenates were centrifuged for 30 min at 37,000 × g to yield a supernatant designated the cytosol. The pellet was washed once in lysis buffer to yield membranes, which were resuspended in lysis buffer containing 500 mM NaCl and 1% Triton X-100.
G-kinase assays were performed as described before (21) with 100 μM [γ-32P]ATP, 0.3 mM Kemptide, and 0.3 μM protein kinase inhibitor (PKI) by measuring the difference in Kemptide phosphorylation observed in the presence and absence of 3 μM cGMP. To measure G-kinase II activity in whole-cell lysates, cells were extracted in the presence of 1% Triton and 500 mM NaCl (72).
Western blots and antibodies.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed as described previously (20, 23), with polyclonal rabbit antibodies specific for G-kinase I (StressGen, KAP-PK005) or G-kinase II (provided by S. Lohmann) (43) at a dilution of 1:1,000. Antibodies specific for CREB and CREB phosphorylated on Ser133 were from Cell Signaling Technology; all other antibodies, including additional anti-CREB antibodies, were from Santa Cruz Biotechnology. They were used according to the manufacturers' protocols, and Western blots were developed with horseradish peroxidase-coupled secondary antibodies and enhanced chemiluminescence as described before (20, 23).
Northern blots.
Total cytoplasmic RNA was prepared and analyzed by denaturing agarose gel electrophoresis; Northern blots were prepared and hybridized to a 32PO4-labeled rat c-fos cDNA probe as described previously (55). Blots were reprobed by hybridization to a cDNA probe encoding glyceraldehyde-3-phosphate dehydrogenase.
Reporter gene assays.
The activities of luciferase and β-galactosidase were measured with luminescence-based assays as described previously (20, 22).
Electrophoretic mobility shift assays.
Nuclear extracts were prepared, incubated with 5′-end-labeled double-stranded oligodeoxynucleotide probes, and analyzed by nondenaturing PAGE and autoradiography as described previously (55). Oligodeoxynucleotide probes encoding canonical CRE, C/EBP, and SP-1 recognition sites were from Promega Life Sciences. For supershift assays, nuclear extracts were preincubated for 20 min at 4°C with specific antibodies or control rabbit immunoglobulin G (IgG) as described before (55).
C/EBP-β phosphorylation studies.
C6 cells were transfected with expression vectors encoding G-kinase II and/or C/EBP-β; 24 h posttransfection, cells were transferred to phosphate-free DMEM and labeled for 4 h with 32PO4 (100 μCi/six-well dish). After that, some cells were treated with 0.3 μM A23187 or 250 μM CPT-cGMP or both agents for the indicated time. Whole-cell lysates were subjected to immunoprecipitation with a C/EBP-β-specific rabbit antibody or control rabbit immunoglobulin G as described below, and immunoprecipitates were analyzed by SDS-PAGE, electroblotting, and autoradiography. The amount of C/EBP-β present in the immunoprecipitates was determined by blotting with a mouse anti-C/EBP-β antibody.
Protein interaction studies.
COS7 cells were electroporated as described above with a total of 10 μg of DNA, including expression vectors encoding the indicated EE-CREB constructs and/or C/EBP-β or the appropriate empty vector. Cells were plated on 10-cm dishes and, 72 h after transfection, lysed in 10 mM Tris-HCl (pH 7.5)-1 mM EDTA-150 mM NaCl-10 mM β-glycerol phosphate-0.25% Nonidet P-40 with a protease inhibitor cocktail (Calbiochem) and subjected to a 10-s burst of sonication. After centrifugation at 12,000 × g for 10 min, cell extracts were incubated for1 h with a mouse monoclonal anti-EE epitope antibody (20) or a rabbit polyclonal antibody specific for C/EBP-β. Protein G-agarose beads were added for an additional hour, collected by centrifugation, and washed three times with lysis buffer. Immunoprecipitates were analyzed by SDS-PAGE and Western blotting with antibodies of a different species than the immunoprecipitating antibody. In vitro GST pulldown assays were performed as described previously (7) with GST-tagged CREB sequences and His- and Myc-tagged C/EBP-β (p35 or p20) purified from bacteria.
Chromatin immunoprecipitation.
About 107 UMR106 cells were fixed in situ with 1% formaldehyde for 5 min at room temperature, washed, and scraped into 1 ml of lysis buffer containing 10 mM Tris-HCl (pH 8.0), 85 mM KCl, 0.5% Nonidet P-40, and a protease inhibitor cocktail. Nuclei were spun through a cushion of 12.5% glycerol in lysis buffer and resuspended and sonicated in a chromatin precipitation buffer as described previously (30). After centrifugation at 30,000 × g for 15 min, the supernatant was preabsorbed with 25 μl of protein A-agarose which had been blocked with 1 mg of BSA and 1 mg of sheared salmon sperm DNA per ml. The supernatant was shaken gently for 16 h at 4°C with 5 μg of either control rabbit immunoglobulin G or antibodies specific for C/EBP-β or CREB; after a 10-min centrifugation at 11,000 × g, 25 μl of blocked protein A-agarose was added for 2 more hours of incubation. Immunoprecipitates were washed, eluted, and heated as described previously (6). Eluates were digested with proteinase K and purified with a Qiagen PCR purification kit. Semiquantitative PCR was performed as described previously (30) with various amounts of template and 32PO4-end-labeled primers encoding rat fos promoter sequences flanking the CRE (−235 to −2 relative to the transcription start site).
Data presentation.
All results presented in bar graphs represent the means ± standard deviations of at least three independent experiments performed in duplicate. Autoradiographs and Western blots demonstrate a representative experiment which was performed at least three times with similar results.
RESULTS
G-kinase II expression in UMR106 and C6 cells.
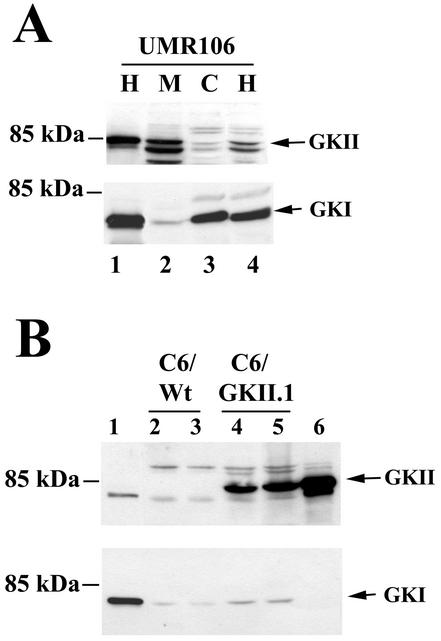
Although G-kinase II protein is easily detected in small intestine and brain, we have not been able to detect the enzyme in established cell lines of intestinal or neuronal origin (16, 21, 35, 72). We found significant amounts of type II (as well as type I) G-kinase protein in the rat osteosarcoma cell line UMR106 and in the murine osteoblast-derived cell line MC3T3-E1 (Fig. 1A shows UMR106 cells). Type II G-kinase was almost exclusively associated with membranes, whereas type I G-kinase was predominantly cytosolic, as reported for other cell types (71, 72).
FIG. 1.
Expression of G-kinases I and II in UMR106 and C6 cells. (A) UMR106 cells were transfected with expression vectors encoding either G-kinase II (GKII, lane 1, upper panel) or G-kinase I (GKI, lane 1, lower panel) or left untransfected (lanes 2 to 4). Postnuclear homogenates (H), membranes (M), and cytosol (C) were prepared as described in Materials and Methods, and proteins were resolved by SDS-PAGE; Western blots were developed with antibodies specific for G-kinase II (upper panel) or G-kinase I (lower panel). The faster-migrating bands in the upper panel likely represent degradation products of G-kinase II, as they were decreased when denaturing SDS sample buffer was added immediately after cell lysis, as shown in lane 1. (B) C6 cells were transiently transfected with expression vectors encoding G-kinase I (lane 1) or G-kinase II (lane 6) or left untransfected (C6/Wt, lanes 2 and 3); C6 cells stably transfected with G-kinase II are shown for comparison (C6/GKII.1, lanes 4 and 5). Whole-cell homogenates were analyzed by Western blotting as described above.
C6 glioma cells lack G-kinase II and are largely deficient in G-kinase I; they provide the opportunity to study G-kinase II functions and demonstrate the specificity of G-kinase I and II antibodies in cells transfected with the corresponding expression vectors (Fig. 1B). Although the G-kinase II antibody cross-reacted to a small degree with G-kinase I (shown in Fig. 1B, lane 1), the latter enzyme was clearly distinguished by its faster migration, and the G-kinase I antibody did not significantly cross-react with G-kinase II (shown in Fig. 1B, lane 6).
G-kinase activity in membrane preparations from untransfected UMR106 cells was 91 ± 21 pmol/min/mg of protein and represented mostly G-kinase II activity; this specific activity is <1/20 of that reported for G-kinase II in membranes of the intestinal epithelium (72), and we noted a decline in activity with increasing cell passage. G-kinase activity in the UMR106 cytosol was 142 ± 36 pmol/min/mg of protein and represented mostly G-kinase I activity. G-kinase activity measured in stably transfected C6/GKII.1 cells was 296 ± 50 pmol/min/mg of protein in whole-cell lysates, compared with a G-kinase activity of about 20 pmol/min/mg of protein in untransfected C6 cells.
The specific activity of G-kinase II in transiently transfected C6 and UMR106 cells was less than one-fourth of that reported for the intestinal epithelium; although the specific activity of G-kinase II in brain and developing bone is unknown, immunohistochemical studies of G-kinase II in mouse embryos (day 19 postcoitum) demonstrate signal intensities in specific areas of forebrain and developing bone comparable to those in the intestinal epithelium (21, 72; T. Gudi and R. B. Pilz, unpublished observation). Thus, G-kinase activity in the transfected C6 and UMR106 cells should be in a physiologically relevant range.
Induction of c-fos mRNA by calcium and cGMP in UMR106 and C6 cells.
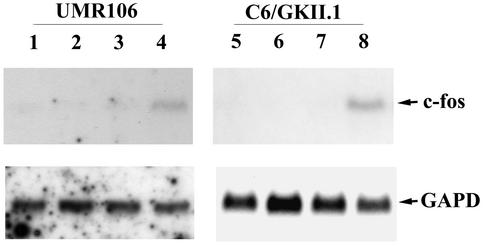
In osteoblasts, c-fos mRNA induction by mechanical strain is thought to be mediated by increased intracellular calcium (9, 50); involvement of the NO/cGMP pathway is suggested by the finding that NO synthase inhibition can partially prevent c-fos induction by mechanical strain (8). We used the calcium ionophore A23187 to increase intracellular calcium and the membrane-permeating cGMP analog CPT-cGMP to activate G-kinase and examined the effect of these agents on c-fos mRNA expression in UMR106 osteosarcoma and MC3T3-E1 osteoblast cells. Cells were treated for 30 min with either agent alone or a combination of both, and total cytoplasmic RNA was analyzed by Northern blotting. Neither agent alone increased c-fos mRNA levels detectably, but together they increased c-fos levels significantly above background (Fig. 2 shows results for UMR106 cells on the left; similar results were obtained with MC3T3-E1 cells).
FIG. 2.
Calcium and cGMP synergistically induce c-fos mRNA in UMR106 cells and G-kinase II-expressing C6 cells. Untransfected UMR106 cells (lanes 1 to 4) and stably transfected C6 cells expressing G-kinase II (C6/GKII.1, lanes 5 to 8) were cultured in DMEM containing 0.1% FBS and 0.1% BSA for 24 h; cells were treated for 30 min with buffer (lanes 1 and 5), calcium ionophore A23187 (0.3 μM, lanes 2 and 6), CPT-cGMP (250 μM, lanes 3 and 7), or both A23187 and CPT-cGMP (lanes 4 and 8). Northern blots were prepared with 30 μg (UMR106) or 20 μg (C6/GKII) of total cytoplasmic RNA per lane and hybridized to a c-fos cDNA probe as described in Materials and Methods (upper panels). Blots were reprobed with a cDNA probe encoding glyceraldehyde-3-phosphate dehydrogenase to demonstrate equal loading (GAPD, lower panels).
In certain cells of neuronal and glial origin, increased intracellular calcium and cGMP induce c-fos mRNA synergistically, while cGMP alone is sufficient to induce c-fos in some cells (5, 25, 53, 76). When we treated G-kinase II-expressing C6 cells with either calcium ionophore A23187 or CPT-cGMP, we did not detect c-fos expression, but when we combined the two agents, c-fos mRNA was induced synergistically (Fig. 2, right panel, shows results for C6/GKII.1 cells). No c-fos signal was observed when G-kinase-deficient C6 cells were treated with A23187 and CPT-cGMP (not shown).
Synergistic activation of CRE- but not SRE-driven reporter genes by calcium and cGMP.
We (21) and others (37, 53) have shown synergistic transcriptional activation of the full-length fos promoter by calcium and cGMP in cells of neuronal or glial origin, with G-kinase II mediating this effect more effectively than G-kinase I (21). By stepwise deletion of the fos promoter sequences, we confirmed that the isolated CRE responded to calcium and cGMP, as shown previously (53).
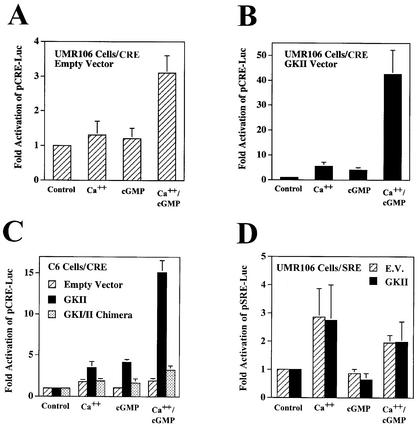
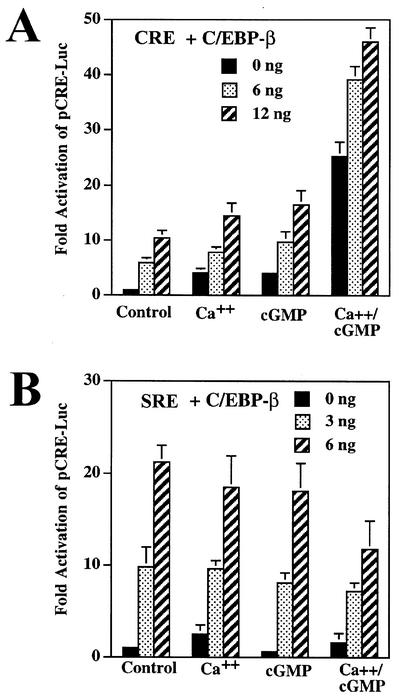
In UMR106 osteosarcoma cells transfected with a CRE-driven luciferase reporter gene (pCRE-Luc), we observed a threefold increase in luciferase activity when the cells were treated simultaneously with calcium ionophores A23187 and CPT-cGMP, but there was no significant effect of either agent alone (Fig. 3A). Since G-kinase activity was low in UMR106 cells and represented a mixture of G-kinases I and II, we examined the effect of overexpressing G-kinase II to levels about fivefold higher than that for endogenous G-kinase II (as shown in Fig. 1A); we found that CRE-driven luciferase activity was increased modestly by calcium or cGMP alone, but the combination of these agents resulted in a dramatic synergistic response (Fig. 3B).
FIG. 3.
Synergistic activation of CRE- but not SRE-driven reporter genes by calcium and cGMP. UMR106 (A, B, and D) and C6 cells (C) were transiently transfected with the reporter plasmid pCRE-Luc (A to C) or pSRE-Luc (D) and with the control plasmid pRSV-βGal; cells were cotransfected with either empty vector (A, C, and D), G-kinase II expression vector (B to D), or a membrane-targeted G-kinase I construct (G-kinase I/II chimera; C) as described in Materials and Methods. Cells were maintained in low-serum medium for 24 h before they were treated for 8 h with 0.3 μM calcium ionophore A23187 (Ca++), 250 μM CPT-cGMP (cGMP), or both, as indicated. Luciferase and β-galactosidase activities were measured as described in Materials and Methods, and the ratio of luciferase to β-galactosidase activity of untreated cells was assigned a value of 1. Cotransfection of G-kinase II or the G-kinase I/II chimera had no significant effect in untreated cells.
Similar results were obtained in C6 cells cotransfected with pCRE-Luc and G-kinase II and in stably transfected C6/GKII.1 cells (Fig. 3C). In C6 cells cotransfected with pCRE-Luc and empty vector, there was only a modest increase in luciferase activity with calcium but no effect of cGMP at all (Fig. 3C). The C6 cells used in the present studies were selected for low NO production, but increasing the intracellular calcium concentration can increase NO synthase activity, explaining some of calcium's stimulation of pCRE-Luc in G-kinase II-expressing cells (21). A G-kinase I/II chimera containing G-kinase I fused to the membrane-targeting domain of G-kinase II failed to mediate the synergistic effects of calcium and cGMP on pCRE-Luc (Fig. 3C); this construct produced G-kinase activity similar to that of the wild-type G-kinase I or II vector but failed to mediate cGMP transactivation of the fos promoter (21). Taken together, these results indicate a synergistic interaction between calcium and cGMP on a CRE-driven promoter, with the magnitude of the synergism depending on the amount of G-kinase II present in the cells.
Since calcium has been reported to activate the SRE in the c-fos promoter, we transfected C6 and UMR106 cells with an SRE-driven luciferase reporter in the absence and presence of G-kinase II to examine the effect of calcium and cGMP on this promoter element. The calcium ionophore increased luciferase expression two- to threefold in the absence or presence of G-kinase, but the cGMP analog had no effect on the SRE by itself or in combination with the ionophore (Fig. 3D shows UMR106 cells, with similar results obtained in C6 cells).
Synergistic activation of the CRE by calcium and cGMP is prevented by dominant negative CREB and C/EBP constructs but not by a dominant negative Fos.
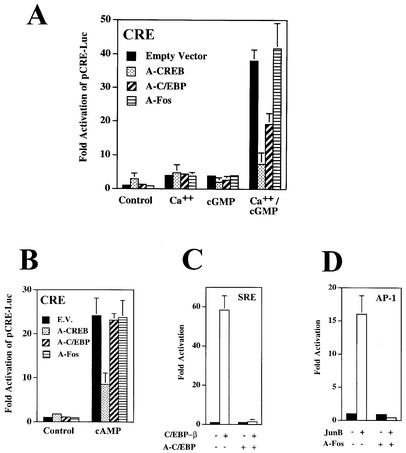
Transcriptional regulators which bind to the CRE sequence (TGAGCTCA) include members of the CREB/CREM/ATF family as well as members of the AP-1 and C/EBP families (61, 62, 74, 78). To examine which transactivating factors might be involved in synergistic activation of the CRE by calcium and cGMP, we employed the dominant negative constructs A-CREB, A-C/EBP, and A-Fos; they contain the leucine zipper domains of CREB, C/EBP, and Fos, respectively, with an N-terminal acidic extension stabilizing dimerization with endogenous transcription factors and effectively prevent DNA binding with extremely high specificity (2, 52).
Cotransfection of A-CREB with pCRE-Luc and G-kinase II into UMR106 cells (Fig. 4A) slightly increased luciferase expression from the reporter in untreated cells, probably reflecting the removal of endogenous, unphosphorylated CREB or CREM-related proteins from the CRE, which may inhibit transcription (18, 44, 73). A-CREB had little effect on reporter gene expression in calcium ionophore- or cGMP-treated cells, probably because the separate effects of calcium and cGMP were very modest and similar to the effect of A-CREB by itself. However, A-CREB completely prevented the synergistic effect of calcium and cGMP. A-C/EBP had no effect on basal luciferase activity and little effect on calcium- or cGMP-induced luciferase expression from pCRE-Luc (Fig. 4A); however, it reduced by >50% the synergistic stimulation of CRE-dependent transcription by calcium and cGMP. In contrast, A-Fos had no effect on the calcium and cGMP synergism (Fig. 4A). Results similar to those shown in Fig. 4A were obtained in C6 cells. The specificity and effectiveness of the dominant negative constructs were demonstrated: cAMP-induced transcription from pCRE-Luc was inhibited only by A-CREB, not by A-C/EBP or A-Fos (Fig. 4B); A-C/EBP prevented stimulation of an SRE-driven reporter by C/EBP-β (Fig. 4C); and A-Fos prevented JunB-mediated stimulation of an AP-1 response element-driven reporter gene (Fig. 4D). We conclude that synergistic activation of CRE-dependent transcription by calcium and cGMP required transcription factors of the CREB and C/EBP families but appeared to be independent of the AP-1 (Fos/Jun) family.
FIG. 4.
Synergistic activation of CRE-dependent transcription by calcium and cGMP is prevented by dominant negative CREB and C/EBP constructs but not by a dominant negative Fos. (A and B) UMR106 cells were transfected with pCRE-Luc, pRSV-βGal, or G-kinase II vector as described in the legend to Fig. 3; some cells were cotransfected with 40 ng of expression vector encoding the dominant negative A-CREB, A-C/EBP, A-Fos, or empty vector. Cells were treated for 8 h with 0.3 μM A23187 (Ca++), 250 μM CPT-cGMP (cGMP), or both (A), or cells were treated with 100 μM CPT-cAMP (B), as indicated. The ratio of luciferase to β-galactosidase activity was normalized as described in the legend to Fig. 3. (C) Cells were transfected with pSRE-Luc, pRSV-βGal, and G-kinase II and cotransfected with empty vector (solid bars) or C/EBP-β (20 ng; open bars); some cells also received vector encoding A-C/EBP (40 ng) as indicated. (D) Cells were transfected with pAP1-Luc, pRSV-βGal, and G-kinase II and cotransfected with empty vector (solid bars) or JunB (20 ng; open bars); some cells also received vector encoding A-Fos (40 ng) as indicated.
C/EBP-β enhances the effect of calcium and cGMP on a CRE-dependent reporter gene.
Since the dominant negative A-C/EBP inhibited synergistic activation of the CRE-driven reporter gene by calcium and cGMP, we examined the effect of overexpressing C/EBP-β, a member of the C/EBP family expressed in cells of neuronal and osteogenic origin (24, 81). When we cotransfected increasing amounts of an expression vector encoding C/EBP-β with pCRE-Luc and G-kinase II into UMR106 or C6 cells, we found a dose-dependent increase in luciferase expression (Fig. 5A shows UMR106 cells, with similar results seen in G-kinase II-expressing C6 cells). C/EBP-β enhanced the separate effects of calcium and cGMP as well as their synergistic effect. In contrast, the stimulatory effect of C/EBP-β on an SRE-driven reporter gene was not enhanced by calcium and/or cGMP but was slightly inhibited by the drugs (Fig. 5B). These results suggest that calcium and cGMP specifically modulated the transactivation potential of C/EBP-β on a CRE-containing promoter but not on an SRE-containing promoter. Since C/EBP-β can bind to both DNA elements (46, 60, 73), calcium and cGMP may modulate cooperation of C/EBP-β with a protein that differentially binds to the CRE but not the SRE. When we tested C/EBP-α and C/EBP-δ in this system, we found that neither C/EBP isoform significantly enhanced the effects of calcium and cGMP on the CRE-containing promoter (data not shown).
FIG. 5.
C/EBP-β enhances the effects of calcium and cGMP on a CRE-dependent reporter. (A) UMR106 cells were transfected with pCRE-Luc, pRSV-βGal, and G-kinase II as described in the legend to Fig. 3; cells additionally received either empty vector (0 ng) or expression vector encoding C/EBP-β (6 ng and 12 ng). (B) Cells were transfected as in panel A except that pSRE-Luc was substituted for pCRE-luc and only 3 or 6 ng of C/EBP-β was cotransfected. Luciferase/β-galactosidase activity ratios were normalized as described in the legend to Fig. 3.
Effect of calcium and cGMP on CRE-binding proteins and expression of CREB- and C/EBP-related proteins.
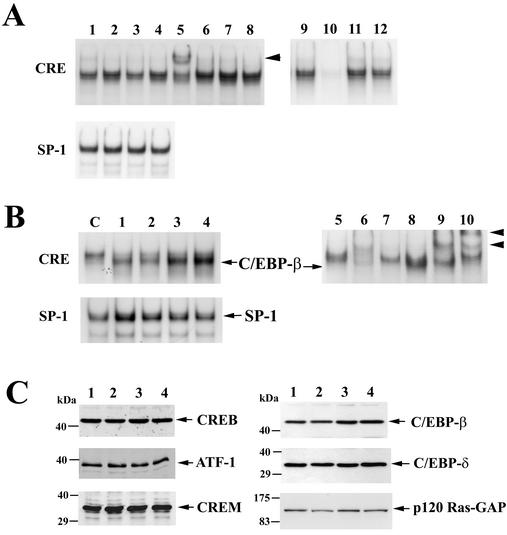
To determine whether calcium and/or cGMP altered the binding of nuclear proteins to the CRE, we performed electrophoretic mobility shift assays with a radioactively labeled oligodeoxynucleotide probe encoding a single CRE. When this probe was incubated with nuclear extracts from UMR106 or C6/GKII.1 cells, several protein-DNA complexes were specifically competed away with excess unlabeled CRE oligodeoxynucleotide but not with unrelated oligodeoxynucleotides (Fig. 6A shows UMR106 cells; competition with unlabeled CRE oligodeoxynucleotide is shown in lane 10). Treating cells with the calcium ionophore A23187 slightly increased the intensity of several protein-DNA complexes (Fig. 6A, lane 2), but treatment with CPT-cGMP by itself had no discernible effect (lane 3), and the combination of A23187 and CPT-cGMP did not produce changes in addition to the effect of A23187 alone (lane 4).
FIG. 6.
Effect of calcium and cGMP on CRE-binding proteins and expression of CREB- and C/EBP-related proteins. (A) UMR106 cells were left untreated (lanes 1 and 9 to 12) or treated for 4 h with 0.3 μM A23187 (lane 2), 250 μM CPT-cGMP (lane 3), or both drugs (lanes 4 to 8). Nuclear extracts were prepared, and electrophoretic mobility shift assays were performed with a radioactively labeled oligodeoxynucleotide probe encoding a CRE consensus sequence (upper panel) or an SP-1 consensus sequence (lower panel) as described in Materials and Methods. Some nuclear extracts were incubated with antibodies specific for CREB (lane 5), C/EBP-α (lane 6), C/EBP-β (lane 7), or control IgG (lane 8) prior to incubation with the CRE probe; the arrowhead indicates a supershifted complex obtained with anti-CREB antibody. To demonstrate competition of sequence-specific binding proteins, unlabeled CRE oligodeoxynucleotide (lane 10), SP-1 oligodeoxynucleotide (lane 11), or C/EBP consensus oligodeoxynucleotide (lane 12) was added at a 50-fold excess. (B) C6 cells were cotransfected with G-kinase II and either empty vector (lanes C and 5 to 7) or a vector encoding C/EBP-β (lanes 1 to 4 and 8 to 10); the cells in lanes 1 to 4 were treated as described for lanes 1 to 4 of panel A, and the cells in lane C and lanes 5 to 10 were treated with both A23187 and CPT-cGMP. Electrophoretic mobility shift assays were performed with the CRE probe (upper panel) or the SP-1 probe (lower panel) as described for panel A. Migration of a protein-DNA complex containing the transfected C/EBP-β is indicated in the upper panel. Some nuclear extracts were incubated with buffer (lanes 5 and 8) or with antibodies specific for CREB (lanes 6 and 9) or C/EBP-β (lanes 7 and 10) prior to incubation with the CRE probe. (C) UMR106 cells in lanes 1 to 4 were treated as described for lanes 1 to 4 of panel A. Equal amounts of nuclear extract protein were analyzed by SDS-PAGE and Western blotting with antibodies specific for CREB, ATF-1, CREM, C/EBP-β, C/EBP-δ, and p120 Ras-GTPase activating protein (GAP), serving as a control for contaminating cytoplasmic proteins.
To identify some of the proteins binding to the CRE, we incubated nuclear extracts from A23187- plus CPT-cGMP-treated UMR106 cells with antibodies specific for CREB- and C/EBP-related proteins. Several of the protein-DNA complexes were supershifted (i.e., retarded in their mobility) when extracts were incubated with antibodies specific for CREB (Fig. 6A, lane 5), CREM, or ATF-1 (not shown). Incubation with antibodies specific for C/EBP-α (Fig. 6A, lane 6), C/EBP-β (lane 7), or C/EBP-δ (not shown) produced no definitive supershift.
To determine whether C/EBP-β bound to the CRE probe, we transfected C6 cells with expression vectors encoding C/EBP-β and G-kinase II and treated the cells with A23187, CPT-cGMP, or both agents (Fig. 6B, lanes 1 to 4; lane C shows cells transfected only with G-kinase II). Transfection of C/EBP-β produced a complex that migrated faster than the main protein-DNA complexes formed in cells transfected only with G-kinase II. Treating cells with CPT-cGMP or A23187 plus CPT-cGMP significantly enhanced this C/EBP-β-containing complex (Fig. 6B, lanes 3 and 4; equal expression of transfected C/EBP-β was demonstrated by Western blotting, not shown). The protein-DNA complex present in cells transfected only with G-kinase II (lanes C and 5) contained proteins that could be supershifted by addition of CREB-specific (lane 6) but not C/EBP-β-specific (lane 7) antibodies. The faster-migrating complex present in G-kinase II- plus C/EBP-β-transfected cells (lanes 4 and 8) was supershifted by a C/EBP-β-specific antibody (lane 10). Nuclear extract quality and loading were examined with a probe for the SP-1 transcription factor (Fig. 6A and B, lower panels).
CREB, ATF-1, CREM, C/EBP-β, and C/EBP-δ were easily detectable in nuclear extracts from UMR106 and C6/GKII.1 cells by Western blotting (Fig. 6C shows UMR106 cells, with similar results obtained in C6/GKII.1 cells). None of the transcription factor levels were affected significantly when cells were treated for 4 h with A23187, CPT-cGMP, or both drugs. Treating cells with CPT-cGMP very modestly increased the amount of C/EBP-β in nuclear extracts (1.3- ± 0.2-fold, result of scanning three independent experiments), but this increase is of unknown significance. There was no detectable increase in total C/EBP-β protein in whole-cell extracts and no consistent change in the amount of a cytosolic protein (p120 Ras-GAP in Fig. 6C) contaminating nuclear extracts in cGMP-treated cells. We conclude that C/EBP-β can bind to the CRE but that it constitutes only a minor fraction of multiple CRE-binding proteins in UMR106 and C6 cells; increased DNA binding of C/EBP-β to the CRE probe was detectable in C/EBP-β-overexpressing cells treated with CPT-cGMP.
Dephosphorylation of C/EBP-β in cGMP-treated cells.
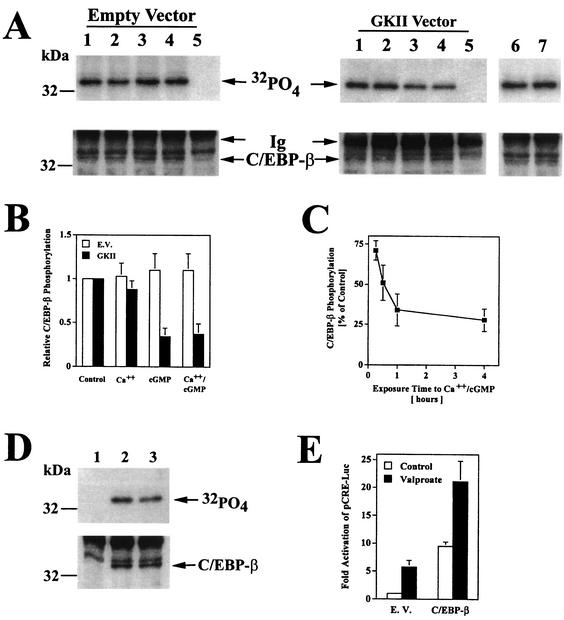
Since cGMP treatment appeared to modulate the amount of nuclear C/EBP-β-CRE complexes, we examined the effect of cGMP on the phosphorylation state of C/EBP-β. C6 cells transfected with C/EBP-β and either empty vector or the G-kinase II expression vector were incubated with 32PO4 and treated with the calcium ionophore, CPT-cGMP, or both drugs for 1 h (Fig. 7A). The amount of 32PO4 incorporation into C/EBP-β immunoprecipitates was large in untreated cells and was not significantly affected by the calcium ionophore. However, CPT-cGMP treatment decreased 32PO4 incorporation into C/EBP-β in G-kinase II-expressing cells in the absence or presence of A23187, while the same treatment had no effect in G-kinase-deficient cells (the amount of C/EBP-β present in immunoprecipitates is shown in the lower panel of Fig. 7A). Figure 7B summarizes the results of three independent experiments. In C6 cells expressing the G-kinase I/II chimera, there was no effect of calcium or cGMP on C/EBP-β phosphorylation, consistent with the lack of cGMP transcriptional effects in cells expressing this construct (Fig. 7A, lanes 6 and 7; Fig. 3C shows transcriptional effects). In G-kinase II-expressing cells, decreased C/EBP-β phosphorylation was detectable as early as 15 min after addition of calcium or cGMP, was maximal by 1 h, and persisted for >4 h (Fig. 7C), correlating with increased DNA binding of C/EBP-β in calcium- and cGMP-treated cells (not shown).
FIG. 7.
Decreased C/EBP-β phosphorylation in cGMP-treated cells. (A) C6 cells were transfected with an expression vector encoding C/EBP-β and either empty vector (left panel, lanes 1 to 5), G-kinase II (right panel, lanes 1 to 5) or a G-kinase I/II chimera (right panel, lanes 6 and 7); cells were incubated with 32PO4 for 4 h and treated for 1 h with buffer (lanes 1, 5, and 6), 0.3 μM A23187 (lane 2), 250 μM CPT-cGMP (lane 3), or both agents (lanes 4 and 7) as described in Materials and Methods. Cell lysates were subjected to immunoprecipitation with a rabbit anti-C/EBP-β antibody (lanes 1 to 4, 6, and 7) or control rabbit IgG (lane 5). Immunoprecipitates were analyzed by SDS-PAGE, electroblotting, and autoradiography (upper panel) and by blotting with a murine anti-C/EBP-β antibody (lower panel). The immunoglobulin heavy chain band is labeled Ig. (B) 32PO4 incorporation into C/EBP-β was quantitated by scanning densitometry of autoradiographs of three independent experiments performed as described for panel A; cells were transfected with C/EBP-β plus either empty vector (open bars) or G-kinase II (solid bars). (C) Cells were transfected with C/EBP-β and G-kinase II and labeled for 4 h in 32PO4-containing medium as described for panel A. Half of the cultures were treated with 0.3 μM A23187 and 250 μM CPT-cGMP for the indicated times, and 32PO4 incorporation into C/EBP-β was quantitated as described for panel B. Data are expressed as percentages of the level in untreated controls. (D) Cells transfected with C/EBP-β were labeled with 32PO4 for 4 h and either left untreated (lanes 1 and 2) or treated for 1 h with 5 mM sodium valproate (lane 3); cell lysates were processed as described for panel A (lane 1, control IgG; lanes 2 and 3, anti-C/EBP-β antibody). (E) Cells were transfected with pCRE-Luc, pRSV-βGal, and either empty vector (E.V.) or 12 ng of C/EBP-β vector; cultures were either left untreated (open bars) or treated with 5 mM sodium valproate for 8 h (solid bars). Luciferase activity was normalized to β-galactosidase activity as described for Fig. 3.
These results suggest that cGMP activation of G-kinase II either inhibited a C/EBP-β kinase or activated a C/EBP-β phosphatase. Glycogen synthase-3 (GSK-3) is a possible candidate, because it is active in unstimulated cells and phosphorylates C/EBP-β in vitro (57). Incubating C6 cells with the GSK-3 inhibitor sodium valproate (79) decreased 32PO4 incorporation into C/EBP-β (Fig. 7D) and increased transactivation of the CRE-dependent reporter gene by C/EBP-β (Fig. 7E), supporting a role for GSK-3 in the regulation of C/EBP-β activity. Similar results were obtained when GSK-3 was inhibited by lithium chloride (data not shown) (79).
Effect of calcium and cGMP on CREB phosphorylation.
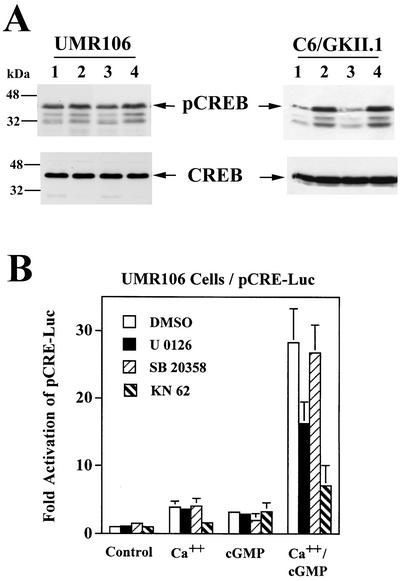
Since synergistic activation of CRE-dependent transcription by calcium and cGMP required CREB binding to the CRE (Fig. 4A), and since transcriptional activation by CREB requires Ser133 phosphorylation, we studied the effect of calcium and cGMP on CREB phosphorylation with a phospho-Ser133-specific antibody (44, 64). In UMR106 cells, basal CREB Ser133 phosphorylation was high, and calcium only modestly increased Ser133 phosphorylation (about 1.5-fold in multiple experiments), while cGMP had no detectable effect (Fig. 8A, left panel). In C6/GKII cells, calcium ionophore treatment increased CREB Ser133 phosphorylation about threefold, but again, CPT-cGMP had no effect on CREB phosphorylation and did not enhance the calcium-induced CREB phosphorylation (Fig. 8A, right panel). Since the duration of CREB phosphorylation may determine the degree of transcriptional activation of CRE-dependent promoters (64, 76), we also examined the effect of cGMP on the kinetics of calcium-induced CREB phosphorylation in C6/GKII.1 cells. Calcium-induced CREB phosphorylation peaked at 1 h and declined thereafter to levels still about twofold above control levels at 8 h; adding CPT-cGMP to the cells did not alter the level of calcium-induced CREB phosphorylation at any time point (not shown). These results suggest that cGMP does not enhance the transcriptional effects of calcium by enhancing the level of CREB phosphorylation, either through activation of additional CREB kinases or through inhibition of phosphatases.
FIG. 8.
Effect of calcium and cGMP on CREB phosphorylation and effect of Cam-kinase and MAP kinase inhibitors on calcium- and cGMP-stimulated transcription. (A) UMR106 cells (left panel) and C6/GKII.1 cells (right panel) were treated for 1 h with 0.3 μM A23187 (lane 2), 250 μM CPT-cGMP (lane 3), or both (lane 4). Equal amounts of cell extracts were analyzed by SDS-PAGE and Western blotting with an antibody specific for Ser133-phosphorylated CREB (pCREB, upper panel) and an antibody that recognizes CREB irrespective of its phosphorylation status (CREB, lower panel). The phospho-CREB antibody cross-reacts with phosphorylated ATF-1 and probably CREM (64). (B) UMR106 cells were transfected with pCRE-Luc, pRSV-βGal, and G-kinase II and treated with A23187 (Ca++), CPT-cGMP (cGMP), or both, as described in the legend to Fig. 3. At 24 h before harvesting, some cells were treated with 0.1% dimethyl sulfoxide (vehicle; DMSO), 10 μM U0126, 10 μM SB20358, or 10 μM KN62. Reporter gene activities were normalized as described for Fig. 3.
Calcium can induce CREB phosphorylation through activation of several pathways, involving Cam-kinases, activation of Raf/MEK/ERK, p38/MAPKAP-K2/3, or calcium-sensitive adenylate cyclase/A-kinase (64). To examine whether signaling by Cam-kinases, MEK/ERK, p38, or A-kinase contributed to the synergistic activation of CRE-dependent transcription by calcium and cGMP, we treated cells transfected with pCRE-Luc and G-kinase II vectors with the pharmacologic inhibitors KN62, U0126, and SB20358 or cotransfected cells with the specific A-kinase inhibitor PKI. The Cam-kinase inhibitor KN62 significantly reduced reporter gene activation by calcium and prevented the synergism between calcium and cGMP almost completely (Fig. 8B; data for UMR106 cells are shown, but similar results were obtained with C6 cells). KN62 also inhibited calcium-induced CREB Ser133 phosphorylation in both cell types (not shown). In UMR106 cells, the MEK inhibitor U0126 reduced the synergistic effect of calcium and cGMP by 42%, and cotransfection of PKI reduced the effect by 28%; both of these agents were without effect in C6 cells, and the p38 inhibitor SB20358 had no significant effect in either cell type (Fig. 8B, PKI data not shown). We conclude that Cam-kinase signaling is required for calcium-induced CREB phosphorylation and for regulation of CRE-dependent transcription by calcium and cGMP. MEK/ERK signaling appears to contribute to calcium's effects in UMR106 but not in C6 cells.
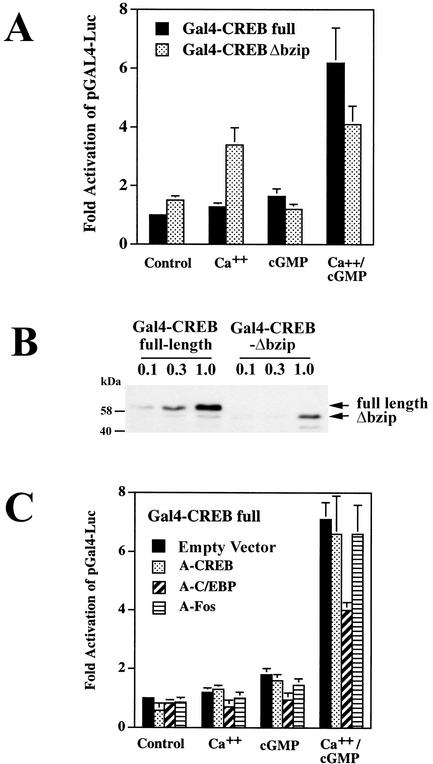
Calcium and cGMP synergistically increase the transactivation potential of full-length Gal4-CREB but not of Gal4-CREB-Δbzip or Gal4-ATF-1.
Although CREB phosphorylation on Ser133 is necessary for CREB activation, it is not sufficient, and other factors, including CREB association with transcriptional coactivators, determine CREB's transactivation potential (12, 44, 64). We examined the effect of calcium and cGMP on the transactivation potential of CREB with chimeric Gal4-CREB constructs encoding the DNA-binding domain of the yeast transcription factor Gal4 fused to either full-length CREB (pGal4-CREB) or amino acids 1 to 232 of CREB (pGal4-CREBΔbzip, missing the leucine zipper/dimerization and basic/DNA binding domains of CREB) (14). When similar amounts of vector encoding full-length Gal4-CREB or Gal4-CREBΔbzip were transfected, Gal4-CREBΔbzip produced three- to fourfold-higher basal reporter gene activity than full-length Gal4-CREB, although the protein levels expressed from pGal4-CREBΔbzip were only about one-third those expressed by pGal4-CREB (Fig. 9A shows Gal4-dependent reporter gene expression in the presence of 5 ng of pGal4-CREBΔbzip and 15 ng of pGal4-CREB to allow comparison on the same scale; Fig. 9B shows a Western blot developed with a Gal4-specific antibody). Thus, the truncated protein appears to be a more efficient transactivator than the full-length protein, possibly because the C-terminal deletion causes a general change in protein conformation or because full-length CREB can dimerize with inhibitory CREB-related proteins (64).
FIG. 9.
Calcium and cGMP synergistically increase the transactivation potential of full-length Gal4-CREB but not of Gal4-CREB-Δbzip; effect of A-C/EBP. (A) UMR106 cells were transfected with the reporter plasmid pGAL4-Luc, pRSV-βGal, and G-kinase II; cells were cotransfected with a vector encoding either full-length Gal4-CREB or Gal4-CREB-Δbzip. Cells were treated with A23187 (Ca++) and/or CPT-cGMP (cGMP), and reporter gene activities were measured as described in the legend to Fig. 3. The luciferase/β-galactosidase activity ratio of untreated cells transfected with full-length Gal4-CREB was assigned a value of 1. We transfected different amounts of vector encoding full-length Gal4-CREB (15 ng) or Gal4-CREB-Δbzip (5 ng) to produce similar reporter gene activities in untreated cells. When the same amount of each vector was transfected, the activity of Gal4-CREB-Δbzip was 2.8- ± 0.4-fold higher than the activity of full-length Gal4-CREB (not shown). (B) In parallel experiments, cells were transfected with 0.1 μg, 0.3 μg, or 1 μg of full-length Gal4-CREB or Gal4-CREB-Δbzip, as indicated, to examine the expression levels of both constructs by Western blotting with an antibody specific for the Gal4 DNA-binding domain. (C) UMR106 cells were transfected with full-length Gal4-CREB, pGAL4-Luc, pRSV-βGal, and G-kinase II as described for panel A; cells were cotransfected with either empty vector or 40 ng of expression vector encoding A-CREB, A-C/EBP, or A-Fos. Cells were treated, and luciferase/β-galactosidase activity ratios were determined as described for panel A.
As has been reported for cortical neurons (36), we found that increasing the intracellular calcium concentration enhanced the transactivation potential of Gal4-CREBΔbzip more efficiently than that of full-length Gal4-CREB (Fig. 9A shows UMR106 cells cotransfected with G-kinase II, with similar results obtained in C6 cells). Treating cells with CPT-cGMP did not significantly affect the activity of Gal4-CREBΔbzip and had only a small effect on the activity of full-length Gal4-CREB. Treating cells with both the calcium ionophore and CPT-cGMP synergistically increased the activity of full-length Gal4-CREB but not of Gal4-CREBΔbzip.
To determine whether the effects of calcium and cGMP were specific for full-length CREB, we examined the CREB-related protein ATF-1. As observed with the Gal4-CREB constructs, the basal activity of full-length Gal4-ATF1 was lower than the activity of truncated Gal4-ATF1Δbzip, and Gal4-ATF1Δbzip was more responsive to calcium than the full-length construct. Neither ATF-1 construct was activated by CPT-cGMP, and there was no enhancement of the calcium effect by CPT-cGMP (data not shown).
C/EBP-β modulates the transactivation potential of Gal4-CREB.
Since the differential effect of calcium and cGMP on full-length Gal4-CREB and Gal4-CREBΔbzip could involve heterodimerization of full-length Gal4-CREB with other leucine zipper proteins, we examined the effect of A-CREB, A-C/EBP, and A-Fos on Gal4-CREB activity in UMR106 cells (Fig. 9C). Cotransfection of A-CREB or A-Fos had no significant effect on the regulation of Gal4-CREB by calcium and cGMP, indicating that binding of A-CREB to the leucine zipper domain of Gal4-CREB was not inhibitory and binding of A-CREB and A-Fos to the leucine zipper domains of endogenous CREB- and AP1-related proteins was without effect. Gal4-CREB binds DNA and forms homodimers via the DNA binding and dimerization domains provided by Gal4; therefore, cotransfection of A-CREB would be expected to interfere only with binding of accessory proteins to the CREB leucine zipper domain. Interestingly, A-C/EBP inhibited the separate and combined effects of calcium and cGMP on the transcriptional activity of full-length Gal4-CREB (Fig. 9C), suggesting that a C/EBP-related protein binds to and regulates the activity of Gal4-CREB in response to calcium and cGMP and that this interaction involves the leucine zipper domain of the C/EBP protein independent of the leucine zipper domain of CREB.
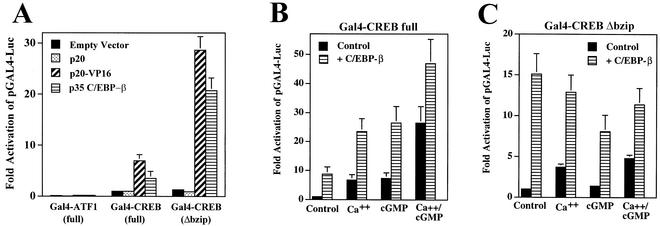
Since overexpression of C/EBP-β enhanced the synergistic stimulation of CRE-dependent transcription by calcium and cGMP while C/EBP-α and -δ were without effect (Fig. 5A and data not shown), we examined the effect of C/EBP-β on Gal4-CREB activity. Transfection of an expression vector encoding C/EBP-β together with the reporter pGAL4-Luc did not affect luciferase activity, indicating that the reporter does not contain cryptic C/EBP-β binding sites (data not shown). Cotransfection of C/EBP-β with Gal4-CREB in UMR106 cells significantly enhanced the transactivation potential of Gal4-CREB and Gal4-CREBΔbzip, suggesting an interaction between C/EBP-β and CREB that is independent of the CREB leucine zipper domain (Fig. 10A). Cotransfection of p20, a truncated version of C/EBP-β containing the leucine zipper and DNA-binding domain but lacking a typical transactivation domain (38), had no effect (Fig. 10A). However, cotransfection of p20 fused to the transactivation domain of the viral protein VP16 (p20-VP16) strongly enhanced Gal4-CREB-dependent activation of the reporter gene (Fig. 10A). These results suggest that C/EBP-β interacts with CREB through its C terminus but that enhancement of CREB's transactivation potential by C/EBP-β requires the presence of a transactivation domain attached to p20. In contrast, cotransfection of p20-VP16 or C/EBP-β (p35) with Gal4-ATF-1 only minimally affected the transactivation potential of ATF-1, indicating that C/EBP-β interacted specifically with CREB and not with the Gal4 DNA-binding domain.
FIG. 10.
C/EBP-β enhances the transactivation potential of Gal4-CREB and Gal4-CREB-Δbzip. (A) UMR106 cells were transfected with pGAL4-Luc, pRSV-βGal, and Gal4-ATF-1 (100 ng), Gal4-CREB (15 ng), or Gal4-CREB-Δbzip (5 ng) as indicated; cells were cotransfected with either empty vector, 100 ng of expression vector encoding p20, 100 ng of p20 fused to the activation domain of VP16, or 12 ng of full-length C/EBP-β (p35). Different amounts of vector encoding p20, p20-VP16, and p35 were used because Western blotting with an antibody specific for the C terminus of C/EBP-β showed that the p20 and p20-VP16 vectors expressed about eightfold-lower protein levels than the same amount of p35 C/EBP-β vector (not shown). The luciferase/β-galactosidase activity ratio of untreated cells transfected with full-length Gal4-CREB was assigned a value of 1. (B and C) C6 cells were transfected with pGAL4-Luc, pRSV-βGal, G-kinase II, and either full-length Gal4-CREB (15 ng, panel B) or pGal4-CREBΔbzip (5 ng, panel C); cells were cotransfected with either empty vector (control) or 12 ng of expression vector encoding C/EBP-β. Cells were treated with A23187 (Ca++) and/or CPT-cGMP (cGMP), and luciferase/β-galactosidase activity ratios were determined as described in the legend to Fig. 3.
C/EBP-β (p35) potentiated the effects of calcium and cGMP on full-length Gal4-CREB in G-kinase II-expressing C6 cells, with the additive rather than synergistic effect of calcium and cGMP likely due to the presence of saturating concentrations of C/EBP-β (Fig. 10B). While C/EBP-β also enhanced the transactivation potential of Gal4-CREBΔbzip, neither calcium nor cGMP nor the combination of the two further stimulated reporter gene expression (Fig. 10C). These results suggest that C/EBP-β can interact with CREB outside of the CREB leucine zipper domain to enhance the transactivation potential of CREB, but the conformation of the truncated CREBΔbzip does not appear to allow modulation of transcriptional activity by calcium and cGMP.
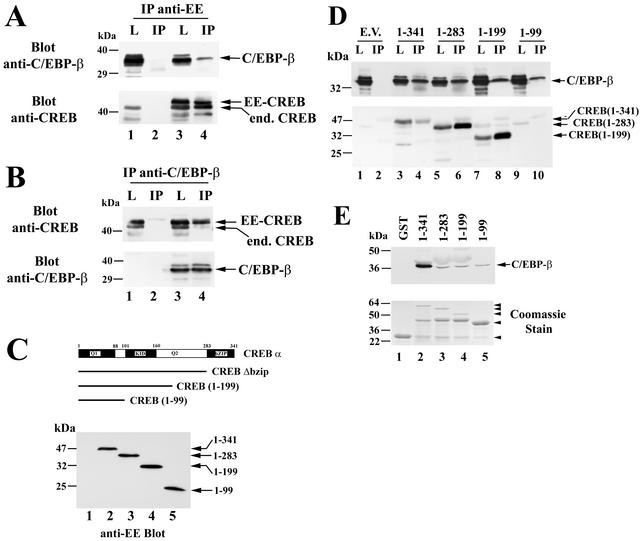
C/EBP-β associates with CREB in vitro and in intact cells.
To determine whether the functional interaction between CREB and C/EBP-β involved direct protein-protein interaction, we performed coimmunoprecipitation experiments. Unfortunately, the efficiency of immunoprecipitation and the levels of endogenous C/EBP-β in C6 and UMR106 cells were too low for detection of C/EBP-β in anti-CREB immunoprecipitates. Therefore, we transfected COS7 cells with expression vectors encoding C/EBP-β and either empty vector or a vector encoding EE epitope-tagged CREB and subjected cell lysates to immunoprecipitation with the anti-EE epitope antibody. Western blotting with a C/EBP-β-specific antibody demonstrated the presence of C/EBP-β in anti-EE immunoprecipitates of cells expressing both C/EBP-β and EE-CREB but not in precipitates of cells expressing C/EBP-β alone (Fig. 11A, upper panel). Reprobing the blot with a CREB-specific antibody showed endogenous CREB in cells transfected with C/EBP-β only (Fig. 11A, lower panel, lysate in lane 1), while lysates and anti-EE immunoprecipitates from cells cotransfected with EE-CREB showed both endogenous (faster-migrating) and EE epitope-tagged (slower-migrating) CREB (Fig. 11A, lower panel, lanes 3 and 4). These results indicate that both C/EBP-β and endogenous CREB coimmunoprecipitated with the EE epitope-tagged CREB.
FIG. 11.
CREB and C/EBP-β association in vitro and in intact cells. (A) COS7 cells were electroporated with C/EBP-β vector (lanes 1 to 4) and either empty vector (lanes 1 and 2) or vector encoding EE epitope-tagged CREB (lanes 3 and 4). Cell lysates were subjected to immunoprecipitation with an anti-EE epitope antibody and washed immunoprecipitates (IP) or 5% of the input lysates (L) were analyzed by SDS-PAGE and Western blotting. Blots were developed with antibodies specific for C/EBP-β (upper panel) and reprobed with anti-CREB antibody (lower panel). Note the presence of endogenous CREB in cell lysates (lanes 1 and 3) as well as in immunoprecipitates containing EE-CREB, which heterodimerizes with endogenous CREB (lane 4); the lowest band most likely represents a proteolytic breakdown product, as it varied in intensity in different experiments. (B) COS7 cells were electroporated with EE-CREB (lanes 1 to 4) and either empty vector (lanes 1and 2) or vector encoding C/EBP-β (lanes 3 and 4). Immunoprecipitates (IP) were obtained with a C/EBP-β-specific antibody and analyzed side by side with 5% of input lysates (L) by Western blotting with antibodies specific for CREB (upper panel) and C/EBP-β (lower panel). Note that endogenous C/EBP-β in COS cells migrates with an apparent molecular mass of 42 to 45 kDa and is only visualized on longer exposures (not shown). (C) COS7 cells were cotransfected with C/EBP-β and either empty vector (lane 1) or vectors encoding EE epitope-tagged full-length CREB (lane 2), CREBΔbzip (lane 3), CREB truncated at amino acid 199 (lane 4), or CREB truncated at amino acid 99 (lane 5). Western blots were developed with anti-EE antibody. (D) Cell lysates of the COS7 cells described for panel C were subjected to immunoprecipitation with anti-EE antibody as described for panel A, and the immunoprecipitates (IP) as well as 5% of cell lysate input (L) were analyzed by Western blotting with an anti-C/EBP-β-specific antibody (upper panel) or a CREB-specific antibody (lower panel). Note that the anti-CREB antibody does not recognize the construct encoding amino acids 1 to 99 (lanes 9 and 10), but expression of the protein was demonstrated on the anti-EE blot shown in panel C. (E) GST (lane 1) or GST fusion proteins encoding full-length CREB or the indicated C-terminal CREB truncations (lanes 2 to 5) were bound to glutathione-agarose beads and incubated with purified Myc epitope-tagged C/EBP-β as described in Materials and Methods. Washed beads were analyzed by Western blotting with anti-Myc antibody to detect C/EBP-β (upper panel). An aliquot of the GST proteins was analyzed by SDS-PAGE and Coomassie staining (lower panel). The arrowheads indicate the migration of full-length proteins; in lanes 2 to 4, an ≈45-kDa proteolytic breakdown product is present.
The reciprocal experiment was performed to confirm the C/EBP-β-CREB interaction. COS7 cells were transfected with EE-CREB and either empty vector or a vector encoding p35 C/EBP-β, and cell lysates were subjected to immunoprecipitation with a C/EBP-β-specific antibody. COS7 cells contain small amounts of C/EBP-β migrating with an apparent molecular mass of 42 to 45 kDa (27) that were visualized only on longer exposures (not shown). Anti-C/EBP-β immunoprecipitates from cells transfected only with EE-CREB contained significant amounts of neither C/EBP-β nor CREB (Fig. 11B, lane 2), but immunoprecipitates from cells transfected with C/EBP-β and EE-CREB contained both endogenous and EE epitope-tagged CREB besides C/EBP-β (Fig. 11B, lane 4). We noted that the association of C/EBP-β and CREB was sensitive to the salt concentration, as it was lost in buffer containing 350 mM NaCl, suggesting the involvement of charged residues.
To map the interaction between CREB and C/EBP-β further, we expressed EE epitope-tagged C-terminal truncations of CREB comprising amino acids 1 to 283 (CREBΔbzip), amino acids 1 to 199 (Q1 and KID domain), or amino acids 1 to 99 (Q1 domain) in COS7 cells (Fig. 11C). Cells were transfected with p35 C/EBP-β and either empty vector or the EE-tagged CREB constructs. C/EBP-β was found in anti-EE immunoprecipitates from cells transfected with each of the EE-CREB constructs (Fig. 11D, upper panel), although the amount of C/EBP-β relative to the amount of full-length CREB appeared greater than the amount of C/EBP-β associated with the other CREB constructs (an anti-CREB blot is shown in the lower panel of Fig. 11D; note that the shortest construct was not recognized by the anti-CREB antibody, but its expression is shown on the anti-EE blot in Fig. 11C). These results suggest that the Q1 domain of CREB is sufficient to mediate interaction with CEBP-β, although the CREB-C/EBP-β interaction may involve multiple domains, including the leucine zipper.
To determine whether C/EBP-β and CREB can interact directly in vitro, we performed a glutathione-agarose pulldown assay after incubating bacterially expressed GST-tagged CREB constructs with Myc epitope-tagged p35 C/EBP-β (Fig. 11E; similar results were obtained with p20, not shown). No C/EBP-β was found bound to beads coated with GST only (lane 1), but C/EBP-β was reproducibly recovered from beads coated with truncated GST-CREB constructs containing as few as the first 99 amino acids (lane 5). Significantly more C/EBP-β was bound to full-length GST-CREB than to the truncated constructs, suggesting that at high protein concentrations in vitro, heterodimerization via the leucine zipper domain was more efficient than binding of C/EBP-β to N-terminal CREB sequences. We conclude that C/EBP-β can directly bind to the N-terminal Q1 domain of CREB in addition to binding to the leucine zipper domain.
Association of CREB and C/EBP-β with the fos promoter in intact cells.
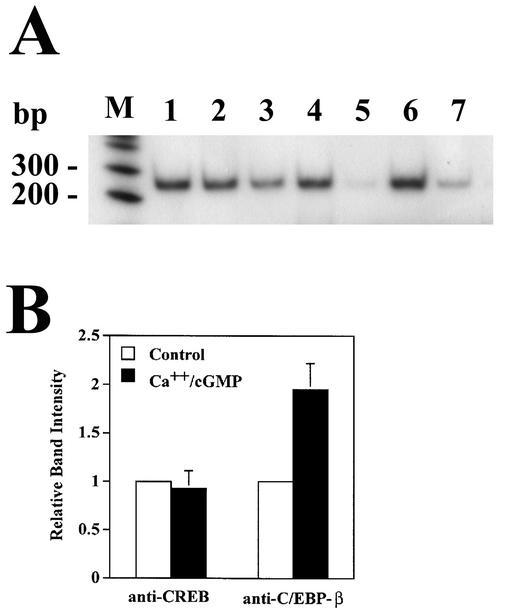
Since we were unable to demonstrate the association of endogenous CREB and C/EBP-β, we asked whether we could detect endogenous CREB and C/EBP-β bound to the fos promoter in UMR106 cells with a chromatin immunoprecipitation assay. Cell lysates containing chromatin fragments were subjected to immunoprecipitation with antibodies specific for CREB, C/EBP-β, and control IgG. PCR amplification with fos promoter-specific primers flanking the fos promoter CRE produced a strong signal with DNA isolated from both anti-CREB and anti-C/EBP-β immunoprecipitates but not from control IgG precipitates (Fig. 12A). PCR amplification was semiquantitative, as the amount of amplified product was proportional to the amount of input DNA (Fig. 12A, lanes 6 and 7).
FIG. 12.
Association of CREB and C/EBP-β with the fos promoter in intact cells. (A) UMR106 cells were cultured for 4 h in the absence (lanes 1, 3, and 5 to 7) or presence (lanes 2 and 4) of 0.3 μM A23187 and 250 μM CPT-cGMP. After a brief treatment with 1% formaldehyde to cross-link proteins and DNA, cells were lysed, and chromatin was sheared as described in Materials and Methods. These cell lysates were subjected to immunoprecipitation with antibodies specific for CREB (lanes 1 and 2), C/EBP-β (lanes 3 and 4), or control IgG (lane 5). DNA was isolated from the immunoprecipitates and amplified with radioactively labeled primers flanking the CRE of the fos promoter; amplification products were analyzed by nondenaturing SDS-PAGE and autoradiography. Lanes 6 and 7 show amplification products obtained with a fraction of the cell lysate, corresponding to 10−5 (lane 6) and 3 × 10−6 (lane 7) of the input. (B) Densitometric analysis of autoradiographs from three independent experiments performed as described for panel A.
The amounts of fos promoter DNA amplified from anti-CREB immunoprecipitates of untreated and calcium ionophore- plus cGMP-treated cells were the same, consistent with constitutive DNA binding of CREB (64). However, anti-C/EBP-β immunoprecipitates from calcium ionophore- and cGMP-treated cells contained twofold more fos promoter DNA than those from untreated cells, suggesting that more C/EBP-β was bound to the fos promoter in cells treated with both agents (Fig. 12B summarizes the results of three independent experiments). A positive DNA amplification signal in these C/EBP-β immunoprecipitates may represent C/EBP-β binding to DNA-bound CREB or C/EBP-β directly binding to the fos promoter; since chromatin fragments of >1 kb were included, we cannot distinguish between C/EBP-β bound to the fos CRE, SRE, or AP-1 site.
DISCUSSION
Synergistic cooperation between CREB and C/EBP-related proteins occurs at promoters containing binding sites for both proteins; for example, cAMP induction of the phosphoenolpyruvate carboxykinase promoter requires simultaneous binding of CREB and C/EBP-α or -β to separate sites (60, 78). Cooperation between CREB and C/EBP at this promoter requires a specific distance between binding sites, the kinase-inducible domain (KID) of CREB, and a cAMP-inducible domain in the N terminus of C/EBP-α or -β (78). Regulation of the human immunodeficiency virus type 1 long terminal repeat or the human pre-interleukin-1β promoter by cAMP appears to involve heterodimer formation between the leucine zippers of CREB and C/EBP-β, with binding to hybrid sites composed of half CREB and half C/EBP recognition sites (13, 61, 70). Since C/EBP-β, as a homo- or heterodimer with CREB-related proteins, may also bind typical CRE sequences (60, 73, 74, 78) (Fig. 6B), it is possible that cooperation between CREB and C/EBP-β bound to tandem CRE sites in the pCRE-Luc reporter contributes to the synergistic effect of calcium and cGMP.
We observed a modest increase in C/EBP-β-CRE complexes in cGMP-treated cells overexpressing C/EBP-β, but endogenous C/EBP-β constituted only a minor component of CRE binding proteins. As reported for hippocampal neurons and HeLa cells (45, 81), we found C/EBP-β immunofluorescence to be largely nuclear in UMR106 and G-kinase II-expressing C6 cells, without a significant increase in cGMP-treated cells (T. Gudi and R. B. Pilz, unpublished observation). In contrast, in PC-12 pheochromocytoma cells and human colon cancer cells, C/EBP-β is mostly cytoplasmic, and A-kinase activation increases phosphorylation, nuclear translocation, and DNA binding of C/EBP-β (10, 45). In pituitary cells, constitutively active Cam-kinase II increases the transcriptional activity of wild-type C/EBP-β but not mutant C/EBP-β (Ala276), and treatment with the Cam-kinase inhibitor KN62 reduces, while calcium ionophore modestly increases, 32PO4 incorporation into C/EBP-β (75).
We found high basal 32PO4 incorporation into C/EBP-β and no increase in calcium-ionophore-treated C6 cells; however, cGMP significantly reduced C/EBP-β phosphorylation in a G-kinase II-dependent fashion, suggesting inhibition of a C/EBP-β kinase or activation of a phosphatase. We suggest GSK-3 as a possible candidate, because it phosphorylates C/EBP-β in vitro, and in vivo GSK-3 appears to negatively regulate C/EBP-β DNA binding and transcriptional activity (57). The GSK-3 inhibitors valproate and lithium mimicked the effects of cGMP on C/EBP-β phosphorylation and CEBP-β-dependent transactivation of the pCRE-Luc reporter, and preliminary results suggest that activation of G-kinase II by cGMP increases phosphorylation of GSK-3β on Ser9 (Y. Chen and R. B. Pilz, unpublished observation); phosphorylation of Ser9 is known to inhibit GSK-3 activity (79).
Growth hormone stimulation of appropriate cells causes C/EBP-β dephosphorylation and increased DNA binding through inhibition of GSK-3; however, the responsible phosphorylation site(s) has not been mapped (56, 57). As observed in growth hormone-stimulated cells, cGMP may induce complex changes in C/EBP-β phosphorylation, resulting in a net decrease in 32PO4 incorporation (56). Inhibition of GSK-3 signaling by NO/cGMP has been suggested by authors studying the neuroprotective effect of NO/cGMP (11). Moreover, modulation of C/EBP-β activity by cGMP and calcium has been observed in hypothalamic cells, in which treatment with NO/cGMP and a calcium ionophore suppresses gonadotropin-releasing hormone gene expression; this effect requires binding of C/EBP-β and Oct-1 to the cGMP/calcium-responsive repressor element, but the mechanism of C/EBP-β regulation by cGMP and calcium was not elucidated (4).
A model for C/EBP-β function suggests that the protein assumes a folded conformation in which the DNA-binding and transactivation domains are masked by interaction with a centrally located regulatory domain; to function efficiently, C/EBP-β must undergo an activation step which may be phosphorylation or interaction with other transcription factors. Indeed, phosphorylation of C/EBP-β can regulate its subcellular localization, DNA binding, transactivation potential, and interaction with other proteins (10, 27, 45, 69, 75). C/EBP-β interacts with several transcription factors, including serum response factor, Runx2, NF-κB, c-Myb, and CREB-binding protein (CBP)/p300, but to our knowledge C/EBP-β binding to CREB, other than leucine zipper-mediated heterodimerization, has not been reported previously (24, 27, 47, 66). The interaction of C/EBP-β with CBP occurs at a site distinct from that of the CREB-CBP interaction (47), suggesting that C/EBP-β binding to CREB could serve to recruit additional CBP into the complex.
Although we found that C/EBP-β can interact with CREB independently of CREB's bzip domain, the C/EBP-β-CREBΔbzip complex was no longer activated by calcium and cGMP, suggesting that the bzip domain of CREB may be required for a conformational change and/or for the recruitment of another, yet to be identified cofactor(s). Recently, Kornhauser et al. (36) reported that mutation of Ser142 to alanine in full-length Gal4-CREB led to an enhancement of transcription, whereas the same mutation in Gal4-CREBΔzip inhibited transcriptional activity. Since Ser142 phosphorylation is induced by calcium (36), it could be involved in the synergistic activation of full-length Gal4-CREB by calcium and cGMP, and the different effects of Ser142 phosphorylation in full-length and truncated Gal4-CREB might explain the lack of calcium-cGMP synergy in the context of Gal4-CREBΔbzip.
Peunova and Enikolopov (53) suggested that synergistic regulation of c-fos mRNA expression by calcium and NO/cGMP involved A-kinase, but they used a recombinant peptide inhibitor of A-kinase which likely also inhibited G-kinase. Lee et al. (37) concluded that ERK activation was required for the synergistic effect of calcium and cGMP on c-fos mRNA. Although calcium increased ERK-1/2 activity in C6 and UMR106 cells and this increase was inhibited by the MEK inhibitor U0126 (data not shown), we found that U0126 had no effect on calcium and cGMP regulation of CRE-dependent transcription in C6 cells and only modestly decreased the synergistic effect in UMR106 cells. We found that the synergistic effect of calcium and cGMP required the activity of a Cam-kinase and G-kinase, with Cam-kinase being necessary for CREB Ser133 phosphorylation and G-kinase being necessary for modulating C/EBP-β phosphorylation. We cannot exclude the possibility that calcium may also influence C/EBP-β activity.
The physiological significance of the calcium and NO/cGMP synergy on the fos promoter is most obvious in the central nervous system and in developing bone (16, 48, 71). Multiple studies have implicated NO/cGMP signaling in long-term potentiation and depression in hippocampal and cerebellar neurons, respectively (3, 39, 41, 65). The synergistic induction of c-fos by calcium and NO/cGMP may be important for modulation of synaptic plasticity, and NO synthase inhibitors diminish different forms of learning associated with increased c-fos expression in the hippocampus and cerebral cortex in rats (41, 49, 53, 58). Mice deficient in endothelial NO synthase, C-type natriuretic peptide, or G-kinase II demonstrate severe defects in bone development, suggesting that the NO/cGMP/G-kinase signaling pathway plays a central role in growth and differentiation of osteogenic cells, and c-fos is a key regulator of normal bone development (1, 34, 48, 54). NO synthase inhibitors diminish the induction of c-fos and decrease bone formation after mechanical strain in vivo, while low concentrations of NO/cGMP promote osteoblast proliferation, new bone formation, and osteoblast-specific gene expression in vitro (8, 26, 42, 80). Dominant negative inhibitors of Fos and CREB disrupt proliferation and maturation of chondrocytes, and expression of a dominant negative CREB in chondrocytes of transgenic mice results in dwarfism similar to that in G-kinase II−/− mice (31, 40). Our finding that cGMP/G-kinase II in cooperation with calcium regulates c-fos expression in cells of osteogenic and neuronal origin, requiring direct interaction and cooperation between CREB and C/EBP-β, provides a physiologically important new link between NO/cGMP signaling and gene expression.
Acknowledgments
We are grateful to M. Ellisman, P. F. Johnson, N. C. Jones, S. Lohmann, R. A. Maurer, M. Montmini, E. J. Murray, L. Sealy, and C. Vinson for providing cell lines and plasmids.
This work was supported by Public Health Service grants 1R01 GM-55586 (to R.B.P.) and 1R01 CA-90932 (to G.R.B.).
REFERENCES
- 1.Aguirre, J., L. Buttery, M. O'Shaughnessy, F. Afzal, I. F. de Marticorena, M. Hukkanen, P. Huang, I. Maclntyre, and J. Polak. 2001. Endothelial nitric oxide synthase gene-deficient mice demonstrate marked retardation in postnatal bone formation, reduced bone volume, and defects in osteoblast maturation and activity. Am. J. Pathol. 158:247-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, S., M. Olive, S. Aggarwal, D. Krylov, D. D. Ginty, and C. Vinson. 1998. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol. Cell. Biol. 18:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arancio, O., I. Antonova, S. Gambaryan, S. M. Lohmann, J. S. Wood, D. S. Lawrence, and R. D. Hawkins. 2001. Presynaptic role of cGMP-dependent protein kinase during long-lasting potentiation. J. Neurosci. 21:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belsham, D. D., and P. L. Mellon. 2000. Transcription factors Oct-1 and C/EBPβ (CCAAT/enhancer-binding protein β) are involved in the glutamate/nitric oxide/cyclic-guanosine 5′-monophosphate-mediated repression of gonadotropin-releasing hormone gene expression. Mol. Endocrinol. 14:212-228. [DOI] [PubMed] [Google Scholar]
- 5.Belsham, D. D., W. C. Wetsel, and P. L. Mellon. 1996. NMDA and nitric oxide act through the cGMP signal transduction pathway to repress hypothalamic gonadotropin-releasing hormone gene expression. EMBO J. 15:538-547. [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, K. E., J. Wells, J. Gutman, S. M. Bartley, and P. J. Farnham. 1998. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc. Natl. Acad. Sci. USA 95:13887-13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casteel, D. E., S. Zhuang, T. Gudi, J. Tang, M. Vuica, S. Desiderio, and R. B. Pilz. 2002. cGMP-dependent protein kinase I β physically and functionally interacts with the transcriptional regulator TFII-I. J. Biol. Chem. 277:32003-32014. [DOI] [PubMed] [Google Scholar]
- 8.Chambers, T. J., J. W. Chow, S. W. Fox, C. J. Jagger, and J. M. Lean. 1998. The role of prostaglandins and nitric oxide in the response of bone to mechanical stimulation, p. 295-298. In H. Sinzinger (ed.), Recent advances in prostaglandin, thromboxine and leukotriene research. Plenum Press, New York, N.Y.
- 9.Chen, N. X., K. D. Ryder, F. M. Pavalko, C. H. Turner, D. B. Burr, J. Qiu, and R. L. Duncan. 2000. Ca2+ regulates fluid shear-induced cytoskeletal reorganization and gene expression in osteoblasts. Am. J. Physiol. 278:C989-C997. [DOI] [PubMed] [Google Scholar]
- 10.Chinery, R., J. A. Brockman, D. T. Dransfield, and R. J. Coffey. 1997. Antioxidant-induced nuclear translocation of CCAAT/enhancer-binding protein β. J. Biol. Chem. 272:30356-30361. [DOI] [PubMed] [Google Scholar]
- 11.Ciani, E., M. Virgili, and A. Contestabile. 2002. Akt pathway mediates a cGMP-dependent survival role of nitric oxide in cerebellar granule neurons. J. Neurochem. 81:218-228. [DOI] [PubMed] [Google Scholar]
- 12.Deisseroth, K., and R. W. Tsien. 2002. Dynamic multiphosphorylation passwords for activity-dependent gene expression. Neuron 34:179-182. [DOI] [PubMed] [Google Scholar]
- 13.Dumais, N., S. Bounou, M. Olivier, and M. J. Tremblay. 2002. Prostaglandin E2-mediated activation of HIV-1 long terminal repeat transcription in human T cells necessitates CCAAT/enhancer binding protein (C/EBP) binding sites in addition to cooperative interactions between C/EBPβ and cyclic adenosine 5′-monophosphate response element binding protein. J. Immunol. 168:274-282. [DOI] [PubMed] [Google Scholar]
- 14.Dwarki, V. J., M. Montminy, and I. M. Verma. 1990. Both the basic region and the ‘leucine zipper’ domain of the cyclic AMP response element binding (CREB) protein are essential for transcriptional activation. EMBO J. 9:225-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eigenthaler, M., S. M. Lohmann, U. Walter, and R. B. Pilz. 1998. Signal transduction by cGMP-dependent protein kinases and their emerging roles in the regulation of cell adhesion and gene expression. Rev. Physiol. Biochem. Pharmacol. 135:174-209. [DOI] [PubMed] [Google Scholar]
- 16.El-Husseini, A. E. D., C. Bladen, and S. R. Vincent. 1995. Molecular characterization of a type II cyclic GMP-dependent protein kinase expressed in the rat brain. J. Neurochem. 64:2814-2817. [DOI] [PubMed] [Google Scholar]
- 17.Felley-Bosco, E., S. Ambs, C. J. Lowenstein, L. K. Keefer, and C. C. Harris. 1994. Constitutive expression of inducible nitric oxide synthase in human bronchial epithelial cells induces c-fos and stimulates the cGMP pathway. Am. J. Respir. Cell Mol. Biol. 11:159-164. [DOI] [PubMed] [Google Scholar]
- 18.Foulkes, N. S., B. M. Laoide, F. Schlotter, and P. Sassone-Corsi. 1991. Transcriptional antagonist cAMP-responsive element modulator (CREM) down-regulates c-fos cAMP-induced expression. Proc. Natl. Acad. Sci. USA 88:5448-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grewal, S. S., R. D. York, and P. J. S. Stork. 1999. Extracellular-signal-regulated kinase signalling in neurons. Curr. Opin. Neurobiol. 9:544-553. [DOI] [PubMed] [Google Scholar]
- 20.Gudi, T., D. E. Casteel, C. Vinson, G. R. Boss, and R. B. Pilz. 2000. NO activation of fos promoter elements requires nuclear translocation of G-kinase I and CREB phosphorylation but is independent of MAP kinase activation. Oncogene 19:6324-6333. [DOI] [PubMed] [Google Scholar]
- 21.Gudi, T., G. K. P. Hong, A. B. Vaandrager, S. M. Lohmann, and R. B. Pilz. 1999. Nitric oxide and cGMP regulate gene expression in neuronal and glial cells by activating type II cGMP-dependent protein kinase. FASEB J. 13:2143-2152. [PubMed] [Google Scholar]
- 22.Gudi, T., I. Huvar, M. Meinecke, S. M. Lohmann, G. R. Boss, and R. B. Pilz. 1996. Regulation of gene expression by cGMP-dependent protein kinase. J. Biol. Chem. 271:4597-4600. [DOI] [PubMed] [Google Scholar]
- 23.Gudi, T., S. M. Lohmann, and R. B. Pilz. 1997. Regulation of gene expression by cyclic GMP-dependent protein kinase requires nuclear translocation of the kinase: identification of a nuclear localization signal. Mol. Cell. Biol. 17:5244-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutierrez, S., A. Javed, D. K. Tennant, M. van Rees, M. Montecino, G. S. Stein, J. L. Stein, and J. B. Lian. 2002. CCAAT/enhancer-binding proteins (C/EBP) β and δ activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J. Biol. Chem. 277:1316-1323. [DOI] [PubMed] [Google Scholar]
- 25.Haby, C., F. Lisovoski, D. Aunis, and J. Zwiller. 1994. Stimulation of the cyclic GMP pathway by NO induces expression of the immediate early genes c-fos and junB in PC12 cells. J. Neurochem. 62:496-501. [DOI] [PubMed] [Google Scholar]
- 26.Hagiwara, H., A. Inoue, A. Yamaguchi, S. Yokose, M. Furuya, S. Tanaka, and S. Hirose. 1996. cGMP produced in response to ANP and CNP regulates proliferation and differentiation of osteoblastic cells. Am. J. Physiol. 270:C1311-C1318. [DOI] [PubMed] [Google Scholar]
- 27.Hanlon, M., and L. Sealy. 1999. Ras regulates the association of serum response factor and CCAAT/enhancer-binding protein β. J. Biol. Chem. 274:14224-14228. [DOI] [PubMed] [Google Scholar]
- 28.Hurst, H. C., N. F. Totty, and N. C. Jones. 1991. Identification and functional characterization of the cellular activating transcription factor 43 (ATF-43) protein. Nucleic Acids Res. 19:4601-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Idriss, S. D., T. Gudi, D. E. Casteel, V. G. Kharitonov, R. B. Pilz, and G. R. Boss. 1999. Nitric oxide regulation of gene transcription via soluble guanylate cyclase and type I cGMP-dependent protein kinase. J. Biol. Chem. 274:9489-9493. [DOI] [PubMed] [Google Scholar]
- 30.Impey, S., A. L. Fong, Y. Wang, J. R. Cardinaux, D. M. Fass, K. Obrietan, G. A. Wayman, D. R. Storm, T. R. Soderling, and R. H. Goodman. 2002. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron 34:235-244. [DOI] [PubMed] [Google Scholar]
- 31.Ionescu, A. M., E. M. Schwarz, C. Vinson, J. E. Puraz, R. Rosier, P. R. Reynolds, and R. J. O'Keefe. 2001. PTHrP modulates chondrocyte differentiation through AP-1 and CREB signaling. J. Biol. Chem. 276:11639-11647. [DOI] [PubMed] [Google Scholar]
- 32.Janknecht, R., M. A. Cahill, and A. Nordheim. 1995. Signal integration at the c-fos promoter. Carcinogenesis 16:443-450. [DOI] [PubMed] [Google Scholar]
- 33.Jarchau, T., C. Haeusler, T. Markert, D. Poehler, J. Vandekerckhove, H. R. De Jonge, S. M. Lohmann, and U. Walter. 1994. Cloning, expression, and in situ localization of rat intestinal cGMP-dependent protein kinase II. Proc. Natl. Acad. Sci. USA 91:9426-9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jochum, W., E. Passegué, and E. F. Wagner. 2001. AP-1 in mouse development and tumorigenesis. Oncogene 20:2401-2412. [DOI] [PubMed] [Google Scholar]
- 35.Kleppisch, T., A. Pfeifer, P. Klatt, P. Ruth, A. Montkowshi, R. Fassler, and F. Hofmann. 1999. Long-term potentiation in the hippocampal CA1 region of mice lacking cGMP-dependent kinases is normal and susceptible to inhibition of nitric oxide synthase. Neuroscience 19:48-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kornhauser, J. M., C. W. Cowan, A. J. Shaywitz, R. E. Dolmetsch, E. C. Griffith, L. S. Hu, C. Haddad, Z. Xia, and M. E. Greenberg. 2002. CREB transcriptional activity in neurons is regulated by multiple, calcium-specific phosphorylation events. Neuron 34:221-233. [DOI] [PubMed] [Google Scholar]
- 37.Lee, S. A., J. K. Park, E. K. Kang, H. R. Bae, K. W. Bae, and H. T. Park. 2000. Calmodulin-dependent activation of p38 and p42/44 mitogen-activated protein kinases contributes to c-fos expression by calcium in PC12 cells: modulation by nitric oxide. Mol. Brain Res. 75:16-24. [DOI] [PubMed] [Google Scholar]
- 38.Lekstrom-Himes, J., and K. Xanthopoulos. 1998. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 273:28545-28548. [DOI] [PubMed] [Google Scholar]
- 39.Lev-Ram, V., T. Jiang, J. Wood, D. S. Lawrence, and R. Y. Tsien. 1997. Synergies and coincidence requirements between NO, cGMP, and Ca2+ in the induction of cerebellar long-term depression. Neuron 18:1025-1038. [DOI] [PubMed] [Google Scholar]
- 40.Long, F., E. Schipani, H. Asahara, H. Kronenberg, and M. Montminy. 2001. The CREB family of activators is required for endochondral bone development. Development 128:541-550. [DOI] [PubMed] [Google Scholar]
- 41.Lu, Y.-F., E. R. Kandel, and R. D. Hawkins. 1999. Nitric oxide signaling contributes to late-phase LTP and CREB phosphorylation in the hippocampus. J. Neurosci. 19:10250-10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mancini, L., N. Moradi-Bidhendi, L. Becherini, V. Martineti, and I. MacIntyre. 2000. The biphasic effects of nitric oxide in primary rat osteoblasts are cGMP dependent. Biochem. Biophys. Res. Commun. 274:477-481. [DOI] [PubMed] [Google Scholar]
- 43.Markert, T., A. B. Vaandrager, S. Gambaryan, D. Poehler, C. Haeusler, U. Walter, H. R. De Jonge, T. Jarchau, and S. M. Lohmann. 1995. Endogenous expression of type II cGMP-dependent protein kinase mRNA and protein in rat intestine. J. Clin. Investig. 96:822-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayr, B., and M. Montminy. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell. Biol. 2:599-609. [DOI] [PubMed] [Google Scholar]
- 45.Metz, R., and E. Ziff. 1991. cAMP stimulates the C/EBP-related transcription factor rNFIL-6 to trans-locate to the nucleus and induce c-fos transcription. Genes Dev. 5:1754-1766. [DOI] [PubMed] [Google Scholar]
- 46.Metz, R., and E. Ziff. 1992. The helix-loop-helix protein rE12 and the C/EBP-related factor rNFIL-6 bind to neighboring sites within the c-fos serum response element. Oncogene 6:2165-2177. [PubMed] [Google Scholar]
- 47.Mink, S., B. Haenig, and K.-H. Klempnauer. 1997. Interaction and functional collaboration of p300 and C/EBPβ. Mol. Cell. Biol. 17:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyazawa, T., Y. Ogawa, H. Chusho, A. Yasoda, N. Tamura, Y. Komatsu, A. Pfeifer, F. Hofmann, and K. Nakao. 2002. Cyclic GMP-dependent protein kinase II plays a critical role in C-type natriuretic peptide-mediated endochondral ossification. Endocrinology 143:3604-3610. [DOI] [PubMed] [Google Scholar]
- 49.Nakazawa, K., L. Karachot, Y. Nakabeppu, and T. Yamamori. 1993. The conjunctive stimuli that cause long-term desensitization also predominantly induce c-Fos and Jun-B cerebellar Purkinje cells. Neuroreport 4:1275-1278. [DOI] [PubMed] [Google Scholar]
- 50.Nomura, S., and T. Takano-Yamamoto. 2000. Molecular events caused by mechanical stress in bone. Matrix Biol. 19:91-96. [DOI] [PubMed] [Google Scholar]
- 51.Ohki, K., K. Yoshida, M. Hagiwara, T. Harada, M. Takamura, T. Ohashi, H. Matsuda, and J. Imaki. 1995. Nitric oxide induces c-fos gene expression via cyclic AMP element binding protein (CREB) phosphorylation in rat retinal pigment epithelium. Brain Res. 696:140-144. [DOI] [PubMed] [Google Scholar]
- 52.Olive, M., D. Krylov, D. R. Echlin, K. Gardner, E. Taparowsky, and C. Vinson. 1997. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J. Biol. Chem. 272:18586-18594. [DOI] [PubMed] [Google Scholar]
- 53.Peunova, N., and G. Enikolopov. 1993. Amplification of calcium-induced gene transcription by nitric oxide in neuronal cells. Nature 364:450-453. [DOI] [PubMed] [Google Scholar]
- 54.Pfeifer, A., A. Aszòdi, U. Seidler, P. Ruth, F. Hofmann, and R. Fässler. 1996. Intestinal secretory defects and dwarfism in mice lacking cGMP-dependent protein kinase II. Science 274:2082-2086. [DOI] [PubMed] [Google Scholar]
- 55.Pilz, R. B., M. Suhasini, S. Idriss, J. L. Meinkoth, and G. R. Boss. 1995. Nitric oxide and cGMP analogs activate transcription from AP-1-responsive promoters in mammalian cells. FASEB J. 9:552-558. [DOI] [PubMed] [Google Scholar]
- 56.Piwien-Pilipuk, G., O. MacDougald, and J. Schwartz. 2002. Dual regulation of phosphorylation and dephosphorylation of C/EBPβ modulate its transcriptional activation and DNA binding in response to growth hormone. J. Biol. Chem. 277:44557-44565. [DOI] [PubMed] [Google Scholar]
- 57.Piwien-Pilipuk, G., D. Van Mater, S. E. Ross, O. A. MacDougald, and J. Schwartz. 2001. Growth hormone regulates phosphorylation and function of CCAAT/enhancer- binding protein β by modulating Akt and glycogen synthase kinase-3. J. Biol. Chem. 276:19664-19671. [DOI] [PubMed] [Google Scholar]
- 58.Qiang, M., J. Xie, H. Wang, and J. Qiao. 1999. Effect of nitric oxide synthesis inhibition on c-fos expression in hippocampus and cerebral cortex following two forms of learning in rats. Behav. Pharmacol. 10:215-222. [DOI] [PubMed] [Google Scholar]
- 59.Robertson, L. M., T. K. Kerppola, M. Vendrell, D. Luk, R. J. Smeyne, C. Bocchiaro, J. I. Morgan, and T. Curran. 1995. Regulation of c-fos expression in transgenic mice requires multiple interdependent transcription control elements. Neuron 14:241-252. [DOI] [PubMed] [Google Scholar]
- 60.Roesler, W. J., J. G. Graham, R. Kolen, D. J. Klemms, and P. J. McFie. 1995. The cAMP response element binding protein synergizes with other transcription factors to mediate cAMP responsiveness. J. Biol. Chem. 270:8225-8232. [DOI] [PubMed] [Google Scholar]
- 61.Ross, H. L., M. R. Nonnemacher, T. H. Hogan, S. J. Quiterio, A. Henderson, J. J. McAllister, F. C. Krebs, and B. Wigdahl. 2001. Interaction between CCAAT/enhancer binding protein and cyclic AMP response element binding protein 1 regulates human immunodeficiency virus type 1 transcription in cells of the monocyte/macrophage lineage. J. Virol. 75:1842-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ryseck, R.-P., and R. Bravo. 1991. c-Jun, JunB, and JunD differ in their binding affinities to AP-1 and CRE consensus sequences: effect of FOS proteins. Oncogene 6:533-542. [PubMed] [Google Scholar]
- 63.Shaulian, E., and M. Karin. 2001. AP-1 in cell proliferation and survival. Oncogene 20:2390-2400. [DOI] [PubMed] [Google Scholar]
- 64.Shaywitz, A. J., and M. E. Greenberg. 1999. CRE(B) a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 68:821-861. [DOI] [PubMed] [Google Scholar]
- 65.Son, H., R. D. Hawkins, K. Martin, M. Kiebler, P. L. Huang, M. C. Fishman, and E. R. Kandel. 1996. Long-term potentiation is reduced in mice that are doubly mutant in endothelial and neuronal nitric oxide synthase. Cell 87:1015-1023. [DOI] [PubMed] [Google Scholar]
- 66.Stein, B., and M. X. Yang. 1995. Repression of the interleukin-6 promoter by estrogen receptor is mediated by NF-κB and C/EBPβ. Mol. Cell. Biol. 15:4971-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun, P., L. Lou, and R. A. Maurer. 1996. Regulation of activating transcription factor-1 and the cAMP response element-binding protein by Ca2+/calmodulin-dependent protein kinases type I, II, and IV. J. Biol. Chem. 271:3066-3073. [DOI] [PubMed] [Google Scholar]
- 68.Thiriet, N., L. Esteve, D. Aunis, and J. Zwiller. 1997. Immediate early gene induction by natriuretic peptides in PC12 phaeochromocytoma and C6 glioma cells. Mol. Neurosci. 8:399-402. [DOI] [PubMed] [Google Scholar]
- 69.Trautwein, C., C. Caelles, P. Van der Geer, T. Hunter, M. Karin, and M. Chojkier. 1993. Transactivation by NF-IL6/LAP is enhanced by phosphorylation of its activation domain. Nature 364:544-547. [DOI] [PubMed] [Google Scholar]
- 70.Tsukada, J., K. Saito, W. R. Waterman, A. C. Webb, and P. E. Auron. 1994. Transcription factors NF-IL6 and CREB recognize a common essential site in the human prointerleukin 1β gene. Mol. Cell. Biol. 14:7285-7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vaandrager, A. B., and H. R. De Jonge. 1996. Signalling by cGMP-dependent protein kinases. Mol. Cell. Biochem. 157:23-30. [DOI] [PubMed] [Google Scholar]
- 72.Vaandrager, A. B., M. Edixhoven, A. G. M. Bot, M. A. Kroos, T. Jarchau, S. Lohmann, H.-G. Genieser, and H. R. De Jonge. 1997. Endogenous type II cGMP-dependent protein kinase exists as a dimer in membranes and can be functionally distinguished from the type I isoforms. J. Biol. Chem. 272:11816-11823. [DOI] [PubMed] [Google Scholar]
- 73.Vallejo, M., M. E. Gosse, W. Beckman, and J. F. Habener. 1995. Impaired cyclic AMP-dependent phosphorylation renders CREB a repressor of C/EBP-induced transcription of the somatostatin gene in an insulinoma cell line. Mol. Cell. Biol. 15:415-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vallejo, M., D. Ron, C. P. Miller, and J. F. Habener. 1993. C/ATF, a member of the activation transcription factor family of DNA-binding proteins, dimerizes with CAAT/enhancer-binding proteins and directs their binding to cAMP response elements. Proc. Natl. Acad. Sci. USA 90:4679-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wegner, M., Z. Cao, and M. G. Rosenfeld. 1992. Calcium-regulated phosphorylation within the leucine zipper of C/EBPβ. Science 256:370-373. [DOI] [PubMed] [Google Scholar]
- 76.West, A. E., W. G. Chen, M. B. Dalva, R. E. Dolmetsch, J. M. Kornhauser, A. J. Shaywitz, M. A. Takasu, X. Tao, and M. E. Greenberg. 2001. Calcium regulation of neuronal gene expression. Proc. Natl. Acad. Sci. USA 98:11024-11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Williams, S. C., C. A. Cantwell, and P. F. Johnson. 1991. A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev. 5:1553-1567. [DOI] [PubMed] [Google Scholar]
- 78.Wilson, H. L., and W. J. Roesler. 2002. CCAAT/enhancer binding proteins: do they possess intrinsic cAMP-inducible activity? Mol. Cell. Endocrinol. 188:15-20. [DOI] [PubMed] [Google Scholar]
- 79.Woodgett, J. R. 18September2001, posting date. Judging a protein by more than its name: GSK-3. Sci. STKE 100:RE12. [Online.] [DOI] [PubMed]
- 80.Yasoda, A., Y. Ogawa, M. Suda, N. Tamura, K. Mori, Y. Sakuma, H. Chusho, K. Shiota, K. Tanaka, and K. Nakao. 1998. Natriuretic peptide regulation of endochondral ossification. Evidence for possible roles of the C-type natriuretic peptide/guanylyl cyclase-B pathway. J. Biol. Chem. 273:11695-11700. [DOI] [PubMed] [Google Scholar]
- 81.Yukawa, K., T. Tanaka, S. Tsuji, and S. Akira. 1998. Expressions of CCAAT/enhancer-binding proteins β and δ and their activities are intensified by cAMP signaling as well as Ca2+/calmodulin kinase activation in hippocampal neurons. J. Biol. Chem. 273:31345-31351. [DOI] [PubMed] [Google Scholar]