Abstract
TAF10 (formerly TAFII30), is a component of TFIID and the TATA box-binding protein (TBP)-free TAF-containing complexes (TFTC/PCAF/STAGA). To investigate the physiological function of TAF10, we disrupted its gene in mice by using a Cre recombinase/LoxP strategy. Interestingly, no TAF10−/− animals were born from intercrosses of TAF10+/− mice, indicating that TAF10 is required for embryogenesis. TAF10−/− embryos developed to the blastocyst stage, implanted, but died shortly after ca. 5.5 days postcoitus. Surprisingly, trophoblast cells from TAF10−/− blastocysts were viable, whereas inner cell mass cells failed to survive, highlighting that TAF10 is not generally required for transcription in all cells. TAF10-deficient cells express normal levels of TBP and TAFs other than TAF10 but contain only partially formed TFIID, are endocycle arrested, and have undetectable levels of transcription. Thus, our results demonstrate that TAF10 is required for TFIID stability, cell cycle progression, and transcription in the early mouse embryo.
Initiation of transcription at eukaryotic class II promoters is a multistep process requiring the coordinated action of many proteins. In general, RNA polymerase II (Pol II) and a host of other factors, including the general transcription factors TFIIA, -B, -D, -E, -F, and -H, work together to form a preinitiation complex (PIC) and allow subsequent transcription. The TATA-box-binding protein (TBP) and 14 evolutionarily conserved TBP-associated factors (TAFs) form TFIID (15). Multiple TFIID complexes exist, indeed nuclear extracts contain at least two subpopulations of TFIID: those that contain TAF10 and those that do not contain TAF10 (39). Electron microscopy has shown that TFIID contains surfaces that could mediate both extensive core promoter and protein-protein interactions (1, 4). Given its early requirement in vitro, as well as its capacity to direct PIC assembly on both TATA-containing and TATA-less promoters, TFIID was proposed to be a central component of the PIC. At present, the role played by TAFs in transcription is not fully understood. Nonetheless, in vitro transcription experiments have suggested that TAFs within TFIID function as coactivators by engaging in direct and selective interactions with transactivators and/or core promoter sequences (2). Furthermore, TAF1 (formerly TAFII250) (39) possesses kinase, histone acetyltransferase (HAT), and ubiquitin conjugating/ligating enzymatic activities (44), suggesting that TAFs can affect transcription at multiple levels.
In yeast TAFs are required for viability, but strains lacking functional TAFs can activate transcription from a variety of inducible genes (38). Moreover, gene expression studies have demonstrated that TAFs have gene-selective effects, with histone fold domain (HFD)-containing TAFs having wide-ranging effects on transcription, whereas the effects of other TAFs appear to be more restricted (18, 23, 24). Taken together with experiments in Drosophila melanogaster and mammalian systems, these results suggest that TAFs, rather than being necessary for all activated transcription, have gene selective effects and therefore play specialized roles at certain subsets of promoters.
Yeast TAF10, along with several other TAFs, is not only an integral component of TFIID but is also found in the SPT-ADA-GCN5 acetylase (SAGA) coactivator complex. Likewise in mammalian cells, TAF10 is shared between TFIID and three closely related multiprotein complexes that we refer to as TFTC-like complexes: the TBP-free TAF-containing complex (TFTC), the p300/CBP-associated factor (PCAF) complex, and the SPT3-TAF9-GCN5-containing complex (STAGA) (25, 30, 45). All three of these complexes contain homologues of the yeast HAT GCN5, as well as a subset of SPT and ADA proteins, the 400-kDa protein TRRAP, and a number of other TAFs (shared TAFs) also found in TFIID. TFTC is structurally similar to TFIID (4) and, although devoid of TBP, is capable of functionally replacing TFIID at both TATA-containing and TATA-less promoters in vitro (45). Due to their HAT subunits, TFTC, PCAF, and STAGA can acetylate histones and nucleosomes (6, 25, 37). Since acetylation of histones within chromatin is positively correlated with transcriptional activity and the establishment of a euchromatic state, it is likely that these complexes, in part, modulate transcription through acetylation of promoter proximal histones.
Analyses of a variety of conditional Saccharomyces cerevisiae TAF10 (scTAF10) mutants have recently been reported (22, 23, 36). Various mutations in scTAF10 induced distinct morphological and cell cycle phenotypes, as well as affected the transcription of different subsets of genes. In Caenorhabditis elegans, RNA interference experiments demonstrated that although TAF10 is required for a large fraction of transcription in the early embryo, it is dispensable for the proper expression of multiple developmental and metazoan specific genes (42). Lack of TAF10 leads to cell cycle arrest and cell death by apoptosis in mouse F9 embryonal carcinoma (EC) cells. Interestingly, the viability of murine F9 EC cells lacking TAF10 can be rescued by retinoic acid (RA) dependent differentiation, showing that TAF10 is essential for undifferentiated F9 cells, whereas it is dispensable for primitive endodermal differentiation and viability (26). Together, these data suggest that TAF10 is required for transcription of a subset of genes essential for specific cell types and differentiation pathways.
Recently, the physiological roles of several cofactors involved in transcription initiation and regulation have been analyzed in mice. Although mouse TFIID has been isolated and partially characterized (46), no disruption of a stoichiometric component of mouse TFIID has been reported so far. However, disruption in mice of the gene encoding TAF4b (formerly TAFII105), a cell-type-specific TAF and substoichiometric component of TFIID, leads to female infertility and altered transcription of a subset genes required for proper folliculogenesis in the ovary (11), suggesting that TAFs may have gene-specific roles in mice. In contrast, disruptions of genes encoding PIC components such the TFIIH subunit MAT1 or the mediator subunits SRB7 and TRAP220 result in embryonic lethality before 9.5 days postcoitus (dpc), suggesting that these factors may be widely needed during development (20, 35, 40).
Disruptions of several genes encoding HATs have also been described in mice. PCAF is dispensable for mouse development, while GCN5 is required for mouse development since null embryos died between 8.5 and 11.5 dpc (47, 48). Embryos null for both GCN5 and PCAF genes showed more severe defects than GCN5 alone and died before 7.5 dpc, indicating that these HATs have overlapping functions during embryogenesis.
Here we show that in mouse F9 TAF10−/− cells the integrity of most TFIID is compromised. Furthermore, we expand the study of TAF10 function from F9 cells to mice and examine the consequences of eliminating a stoichiometric shared TAF, TAF10, on mammalian development, viability, and transcription. By generating a functionally null mutation of the TAF10 gene, we show that it is required for early mouse development and survival of the pluripotent inner cell mass (ICM) but not for survival of mouse trophoblast cells. Distinct from disruptions of SAGA/TFTC-like complex subunits, the molecular and phenotypic results of TAF10 disruption in mice suggest that alterations in the structure of TFIID have important effects on Pol II transcription, cell survival, and early mouse development.
MATERIALS AND METHODS
Immunoprecipitation and Western blot analysis.
F9 EC cells were grown in the abscence or presence of doxicylin, with or without 1 μM RA as previously described (26). Immunoprecipitations were performed overnight at 4°C with 300 μl of the indicated protein fraction, 50 μl of protein G-Sepharose (Pharmacia), and 1 to 10 μg of the indicated antibodies (5, 21). The beads and associated immune complexes were subsequently washed three times with immunoprecipitation buffer (25 mM Tris-HCl [pH 7.9], 10% [vol/vol] glycerol, 0.1% NP-40, 0.5 mM dithiothreitol, 5 mM MgCl2) containing 500 mM KCl, followed by two washes with immunoprecipitation (IP) buffer containing 100 mM KCl. Proteins were boiled in sodium dodecyl sulfate (SDS) sample buffer, separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose membrane, probed with the indicated primary antibodies, and detected by using chemiluminescence according to the manufacturer's instructions.
Targeting of the TAF10 allele.
TAF10(L:LNL) (26) linearized with BamHI and XhoI was purified on a 1% agarose gel and electroporated into SV129 embryonic stem cells as previously described (8). After G418 selection, 118 resistant clones were expanded. Genomic DNA was prepared from each clone and digested with SacI. The digests were analyzed by Southern blotting with a 5′ probe corresponding to an 800-kb EcoRV/KpnI genomic fragment located outside of the sequences present in the targeting vector. Approximately 6% of clones were found to be positive for homologous recombination and were selected for further analysis. Two independent clones were injected into C57BL/6 blastocysts and reimplanted. For excision of the floxed TAF10 exon 2, heterozygous TAF10 mice were interbred with cytomegalovirus (CMV)-Cre mice (9).
Histology and in situ hybridization.
For histology, uteri (i.e., 4.5-dpc embryos) and deciduae (i.e., 5.5-dpc embryos) were split into four parts and fixed in Bouin's fluid for 14 h and then dehydrated and embedded in paraffin. For in situ hybridization, tissues were fixed in 4% paraformaldehyde (PFA)-1× phosphate-buffered saline (PBS) for 12 h at 4°C and embedded in paraffin. Next, 5-μm sections were mounted on SuperFrost Plus slides (O. Kindler & Co.) and stained with hematoxylin and eosin. Digoxigenin (DIG)-labeled RNA in situ hybridizations were based on the protocol of Myat et al. (27) with the following modifications: paraffin sections were cut into 5-μm sections and rehydrated in water, and 120 μl of the heat-inactivated RNA probe (diluted 1/100 in hybridization buffer) was applied to each section. Anti-DIG antibody was applied at 1/2,500 in the blocking solution.
Genotyping of the TAF10 allele.
Genomic DNA was isolated as described previously (10). The various TAF10 alleles were identified by Southern blotting with a 5′ genomic probe (see Fig. 3), and the restriction enzyme SacI.
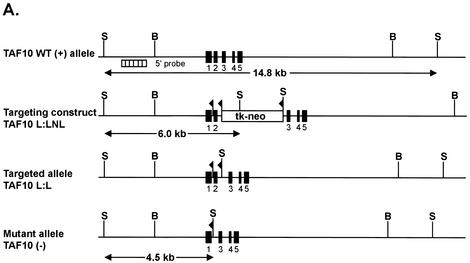
FIG. 3.
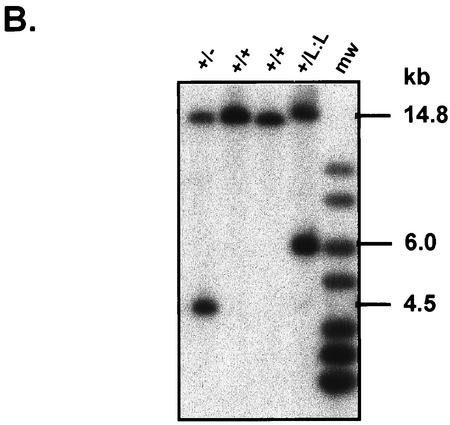
Disruption of the TAF10 gene in mouse. (A) Schematic representation of the mouse TAF10 genomic locus, the targeting construct, and the targeted TAF10 locus and resulting mutated TAF10 locus after Cre-mediated excision of DNA sequences flanked by loxP sites. Exons are represented by filled boxes, and loxP sites are represented by flags. Restriction endonuclease sites: B, BamHI; S, SacI; X, XhoI. The probe used for Southern blot analysis is represented as a hatched box. (B) Genotyping of TAF10 alleles by Southern analysis. Genomic DNA isolated from wild-type (+/+), heterozygote (+/−), and heterozygous targeted (+/L3) TAF10 mice were digested with SacI and Southern blotted. The 14.8-kb band corresponds to the wild-type allele, the 6-kb band corresponds to the targeted allele, and the 4.5-kb band corresponds to the null allele.
Blastocyst culture and genotyping.
Blastocysts recovered at 3.5 dpc were cultured individually on gelatinized culture plates at 37°C at 7% CO2 in blastocyst culture medium containing Dulbecco modified Eagle medium, Glutamax, gentamicin, 10% fetal calf serum, 8 μg of 2-mercaptoethanol/ml, and 1,000 U of leukemia inhibitory factor/ml. Photographs were taken on a Leica DMIRB microscope fitted with a COHU charge-coupled device camera by using coolSNAP software version 1.2 (Roper Scientific, Inc.). For genotyping, blastocysts were recovered by scraping and lysed overnight at 55°C in 1× PCR-PKA buffer (250 mM NaCl, 10 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2, 1% SDS, 100 μg of proteinase K/ml). One-tenth of this mixture was used for nested PCR of the TAF10 genomic locus with oligonucleotides SG12/SE89 and SG12/QX224, resulting in a wild-type band of 756 bp, a L:LNL band of 878 bp, and another band of 405 bp (26).
Immunofluorescence and antibody generation.
Blastocysts (3.5 dpc) were flushed from uteri and cultured for 3 days in blastocyst culture medium. For immunofluorescence, outgrowths were fixed for 15 min in 1% PFA and permeabilized with 0.1% Tween 20-1× PBS prior to immunodetection with anti-TAF4 monoclonal antibody 32TA (34), anti-TBP monoclonal antibody 3G3 (7), anti-YY1 polyclonal antibody (Santa Cruz), anti-TRRAP polyclonal antibody (16), anti-phospho-serine 2-Pol II C-terminal domain (CTD) monoclonal antibody H5 (BabCo), and anti-Pol II CTD monoclonal antibody 7G5 (3). To generate anti-TAF10 and anti-TBN monoclonal antibodies, peptides corresponding to amino acids 88 to 97 of hsTAF10 (EGAISNGVYVLP) and amino acids 151 to 170 of mmTBN (FPDPHTYIKTPTYREPVSDY) were synthesized, coupled to ovalbumin, and used for the immunization of mice. Immunization and monoclonal antibody production have been described (7).
TUNEL test.
TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) tests were performed on 3.5-dpc blastocysts that were fixed for 10 min in 1% PFA, washed in 1× PBS, and dried onto SuperFrost Plus slides (O. Kindler GmbH & Co.). Slides were equilibrated in hybridization buffer, and DIG-labeled nucleotides were incorporated with terminal transferase. DIG-labeled DNA was visualized by using a fluorescent anti-DIG antibody according to the manufacturer's instructions (ApoTag; OnCor Appligene). Images were obtained with a Leica TCS4D fluorescence confocal microscope.
BrdU labeling.
Blastocyst outgrowths grown on coverslips for 2 days were cultured in the presence of 10 μM bromodeoxyuridine (BrdU) for 16 h. For immunodetection cells were washed with PBS and fixed in 70% ethanol at 4°C for 2 h, followed by treatment with 0.1 M HCl-0.5% Triton X-100 for 30 min. Immunolabeling was performed by incubating the cells with 2 μg of anti-BrdU antibody (BMC 9318; Roche)/ml in PBS containing 5% fetal calf serum for 1 h at room temperature, followed by washing and incubation with goat anti-mouse immunoglobulin G-Cy3 secondary antibody (Jackson ImmunoResearch Laboratories, Inc.).
In situ RNA run-on assay.
Blastocyst outgrowths grown on coverslips for 2 days were subjected to in situ run-on assays essentially as described previously (43). Briefly, outgrowths were washed once with PBS and once with PB buffer (100 mM KCl, 50 mM Tris-Cl [pH 7.4], 50 mM MgCl2, 0.5 mM EGTA, 25% glycerol, 5 U of RNasin/ml, and 1 mM phenylmethylsulfonyl fluoride). Cells were subsequently permeabilized with PB buffer supplemented with 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 0.2 mM Br-UTP, and 0.05% Triton X-100. Transcription reactions were carried out in the same buffer as described above without Triton X-100 for 20 min at room temperature. Cells were then washed once in PB buffer and once in PBS containing 0.5% Triton X-100 and 5 U of RNasin/ml and then fixed in 4% PFA at 4°C for 2 h. Immunolabeling was performed by incubation for 1 h with rat anti-BrdU antibody (MAS250; Harlan Sera-Lab), followed by washing and incubation for 1 h with biotinylated anti-rat antisera and then with streptavidin-conjugated Texas red (Gibco-BRL).
RESULTS
Loss of TFIID integrity in cells lacking TAF10.
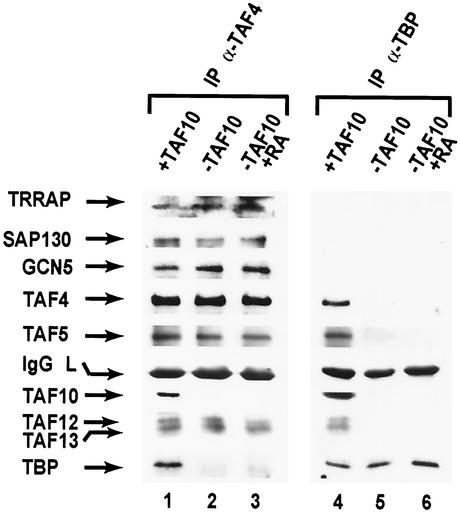
We have reported that disruptions of TAF10 in F9 EC cells do not lead to noticeable changes in the levels of the several other TAFs or TBP, suggesting that the integrity of TFIID and TFTC-like complexes are not seriously affected by lack of TAF10 (26). To further characterize the composition of TAF10 containing complexes in mammalian cells, we used the previously described TAFII30-La/-Lb:R F9 EC cell system, in which expression of TAF10 is under the control of a tetracycline repressor (26). TFIID and TFTC complexes were immunoprecipitated from whole-cell extracts by using monoclonal anti-TBP or monoclonal anti-TAF4 (formerly TAFII135) antibodies. Extracts were isolated from cells expressing TAF10 (plus doxycyclin) or from cells grown in TAF10-depleting conditions (3 days without doxycyclin), with or without RA treatment. Immunocomplexes were washed with 500 mM IP buffer; however, similar results were obtained with 100 mM IP buffer (data not shown). In the presence of TAF10, both antibodies immunoprecipitated TAFs, such as TAF4, TAF5 (formerly TAFII100), TAF12 (formerly TAFII20), and TAF13 (formerly TAFII18), as well as TBP (Fig. 1, lanes 1 and 4). Surprisingly, with extracts from undifferentiated F9 cells depleted of TAF10, immunoprecipitation with anti-TBP antibodies failed to coimmunoprecipitate any of the examined TAFs (Fig. 1, lane 5).
FIG. 1.
Altered TFIID composition in F9 EC cells lacking TAF10. Whole-cell extracts prepared from TAFII30-La/-Lb:R F9 EC cells (26) grown in the presence (+TAF10, lanes 1 and 3) or absence (−TAF10, lanes 2 and 4) of doxycyclin for 3 days. Immunoprecipitations with either anti-TBP (α-TBP) or anti-TAF4 (α-TAF4) antibodies were separated by SDS-polyacrylamide gel electrophoresis, followed by immunoblotting with the indicated antibodies.
Undifferentiated F9 cells depleted of TAF10 undergo apoptosis but can be rescued by RA-dependent differentiation. To analyze TFIID structure in these viable differentiated primitive endodermal cells lacking TAF10, we performed identical immunoprecipitaions with extracts from RA-differentiated F9 cells containing (Fig. 1, lane 3) or depleted of TAF10 (lane 6). Surprisingly, with these RA-differentiated F9 cell extracts depleted of TAF10 immunoprecipitation with anti-TBP antibodies failed to coimmunoprecipitate any of the TAFs examined (Fig. 1, lane 6) (see also Discussion).
In contrast to the anti-TBP antibodies, anti-TAF4 antibodies coimmunoprecipitated TAF4, TAF5, TAF12, and TAF13 from both undifferentiated F9 cells depleted of TAF10 and from RA differentiated F9 cells lacking TAF10, but TBP was nearly undetectable (Fig. 1, lanes 2 and 3). Therefore, under TAF10-depleting conditions most of the TBP dissociates from the other TAFs examined, which remain in a complex. Note that with longer exposure times we were able to detect a small amount of TBP associated with TAFs in these extracts, suggesting that despite the large reduction of TFIID some residual TAF10 lacking TFIID may remain. This agrees with the fact that biochemical characterization of TFIID in human HeLa cells has revealed the existence of TFIID complexes that lack TAF10 (21), although the levels are apparently much lower in mouse F9 EC cells than in human HeLa cells. Taken together, our results strongly suggest that depletion of TAF10 in F9 EC cells alters TFIID structural integrity.
Since TAF4 is shared between TFIID and TFTC, immunoprecipitates with anti-TAF4 antibodies were also examined for the presence of TFTC components TRRAP (6), GCN5 (6), and SAP130 (5). Interestingly, all three proteins were present at comparable levels in the presence or absence of TAF10 (Fig. 1, lanes 1 and 2), suggesting that TFTC devoid of TAF10 remains, at least partially, intact.
Developmental expression patterns of TAF10.
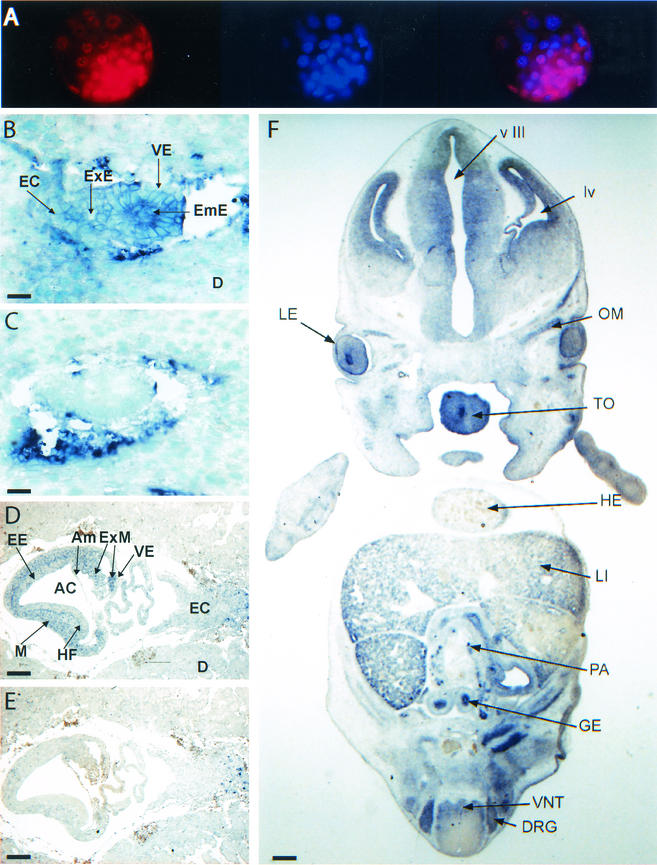
The fact that TAF10 disruption leads to alteration of TFIID, but not TFTC, provides a unique system to examine TFIID function in mammalian cells. Moreover, the selective effects of mutating TAF10 on gene expression and cell survival in mammalian cells raises the possibility that there are other mammalian tissues or cell types in which TAF10 is dispensable. Therefore, to better understand the role of TAF10 in vivo, we examined its expression pattern at specific stages of murine development. TAF10 expression levels were analyzed at the protein level by immunocytochemistry with an anti-TAF10 monoclonal antibody (Fig. 2A) and at the mRNA level by in situ hybridization with a TAF10 antisense probe (Fig. 2B, D, and F) or a sense probe as a control (Fig. 2C and E). At 3.5 dpc, TAF10 protein was uniformly expressed in all cells of the blastocyst, including the ICM and the trophectoderm (Fig. 2A). Likewise at 5.5 dpc, TAF10 mRNA appeared ubiquitously expressed, but high TAF10 mRNA levels were detected in rapidly proliferating embryonic ectoderm cells (Fig. 2, compare panels B to C). At 7.5 dpc, TAF10 mRNA appeared to be expressed at highest levels in the rapidly dividing embryonic and extraembryonic mesoderm (Fig. 2, compare panels D to E). At 12.5 dpc, high TAF10 mRNA levels were seen in the epithelia of the gut, olfactory system, neural retina, and lens, as well as in the liver, kidney, neural tube, pancreas, gonad, muscle, tongue, salivary gland, and head region, notably the dorsal root ganglion lung (Fig. 2F and data not shown), although it was not detected in the heart or the urogenital ridge lung (Fig. 2F and data not shown). Therefore, despite the fact that TAF10 is a component of the general transcription factor TFIID, its mRNA expression level in mouse tissues varies widely throughout development.
FIG. 2.
Expression patterns of TAF10 in wild-type mice embryos. (A) Wild-type blastocysts (3.5 dpc) were fixed and immunostained with a monoclonal TAF10 antibody (left panel). DNA was visualized with DAPI staining (middle panel). A merge of the two images is shown in the right panel. (B to F) Paraffin sections of wild-type embryos isolated at 5.5 dpc (B to C), 7.5 dpc (D and E), or 12.5 dpc (F) were hybridized with a TAF10 specific antisense cRNA probe (B, D, and F) or sense probe (C and E). Tissues positive for TAF10 expression are indicated by arrows AC, amniotic cavity; Am, amnios; D, decidual cells; DRG, dorsal root ganglia; EC, ectoplacental cone; EmE, embryonic ectoderm; ExE, extraembryonic ectoderm, ExM, extraembryonic mesoderm; GE, gut epithelium, HE, heart; HF, head fold; LE, lens epithelium; LI, liver; lv, lateral ventricule; M, mesoderm; OM, ocular muscle; PA, pancreas; TO, tongue; VE, visceral endoderm; VNT, ventral neural tube; v III, third ventricule. Scale bars: B and C, 30 μm; D and E, 110 μm; F, 350 μm.
Generation of mice harboring a null TAF10 allele.
To examine the requirement for TAF10 in mice, its allele was disrupted by using a Cre recombinase/LoxP strategy. Briefly, we introduced into mouse embryonic stem cells a LoxP site and a LoxP-flanked (floxed) neomycin resistance cassette into the introns located upstream and downstream of TAF10 exon 2, respectively, by using the targeting vector pTAF30L:LNL, which was previously used to ablate TAF10 in F9 cells (Fig. 3A) (26). Southern blotting and genomic PCR analyses revealed that the LoxP sites and the floxed cassette were inserted by homologous recombination into one of the TAF10 alleles in seven neomycin-resistant clones (TAF10 L:LNL allele; data not shown). Chimeric males derived from two mutant embryonic stem clones transmitted the TAF10 L:LNL allele through their germ line (TAF10+/L:LNL mice; data not shown). To eliminate the floxed neomycin and exon II sequences, TAF10+/L:LNL mice were bred with CMV-Cre mice, which express the Cre recombinase in the germ cells (9), to generate TAF10+/L:L and TAF10+/− mice (Fig. 3B). TAF10+/− offspring were subsequently bred with C57BL/6 mice, to produce TAF10+/− mice that did not carry the CMV-Cre transgene. TAF10+/− mice developed normally and were indistinguishable from wild-type littermates (data not shown).
Disruption of TAF10 leads to early embryonic lethality.
To analyze the effect of homozygous TAF10 mutation in mice, we genotyped 3-week-old offspring from TAF10+/− intercrosses. Heterozygote and wild-type animals were found at 2 to 1 ratios, but no offspring homozygous for the mutant TAF10 (TAF10−/−) allele were detected (Table 1), indicating that at least one TAF10 allele is required for mouse development.
TABLE 1.
Genotypes of offspring from TAF10+/− intercrosses
| Agea | No. of progeny with genotypeb:
|
No. resorbed | Total no. | ||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | |||
| 3 wk* | 87 | 184 | 0 | 271 | |
| E14.5* | 9 | 18 | 0 | 27 | |
| E8.5* | 10 | 19 | 0 | 3 | 32 |
| E6.5† | 8 | 16 | 0 | 5 | 29 |
| E3.5† | 8 | 15 | 6 | 29 | |
E14.5, embryonic day 14.5, etc.
Genotypes were determined by Southern blotting (*) or PCR (†).
To determine the stage at which TAF10−/− mice arrested development, TAF10+/− intercrosses were staged, and embryos were recovered at 14.5, 8.5, and 6.5 dpc. No TAF10−/− embryos were detected among the 88 analyzed, indicating that the developmental arrest occurred before this time (Table 1). On the other hand, approximately one-quarter of blastocysts recovered at 3.5 dpc (genotyped by PCR) were found to contain TAF10−/− alleles, indicating that TAF10−/− embryos died between days 3.5 and 6.5 dpc. Taken together, these data show that TAF10 is required for early mouse embryonic development and that TAF10−/− embryos suffer a peri-implantation defect.
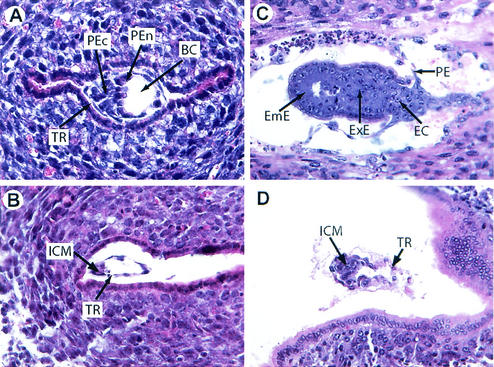
To investigate whether TAF10 mutant blastocysts were able to hatch and implant, decidua from 4.5 and 5.5 dpc mice were recovered and examined at the histological level. Decidua were put into three phenotypic classes: normal, abnormal, and empty (Fig. 4 and data not shown). Of 58 decidua examined, 63% were normal, 25% were abnormal, and 12% were empty. The percentages were similar for both 4.5- and 5.5-dpc deciduae. Note that roughly the same number of empty decidua were observed in intercrosses of TAF10+/+ mice with the same genetic background, indicating that this is the background level of empty decidua in these strains and is not related to TAF10 disruption. Examination of abnormal embryos at 4.5 dpc revealed that they had hatched, begun implantation, and had intact trophectoderm but nevertheless were often developmentally retarded and had a reduced ICM size compared to normal embryos of the same age (Fig. 4B). At 5.5 dpc, developmental defects became more evident. Mutant embryos were often completely disorganized with greatly reduced or absent epiblast, but often some trophectoderm cells remained (Fig. 4D). Although fixation and staining of embryos prevented genotyping, based on the Mendelian ratios it is likely that embryos that appeared normal corresponded to wild-type and heterozygous animals (Fig. 4A and C), while the remaining abnormal decidua corresponded to TAF10−/− embryos. Thus, abnormal decidua most likely represent TAF10−/− embryos that successfully implanted but failed to develop beyond day 5.5 dpc. Taken together, this indicates that TAF10 is a recessive lethal allele whose disruption leads to early embryonic lethality in mice at between 4.5 and 5.5 dpc.
FIG. 4.
Histological analysis of TAF10−/− embryos. Hematoxylin-and-eosin-stained sections of 4.5-dpc (A and B) and 5.5-dpc (C and D) decidua isolated from heterozygous intercrosses. Arrows: BC, blastocoelic cavity; EC, ectoplacental cone; EmE, embryonic ectoderm; ExE, extraembryonic ectoderm; PE, parietal endoderm; PEn, primitive endoderm; PEc, primitive ectoderm; TR, trophectoderm; ICM, inner cell mass.
TAF10 is not required for the survival of trophoblasts.
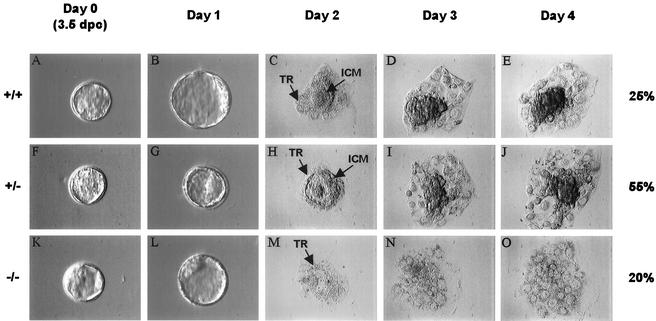
To further characterize the development of TAF10−/− embryos during the implantation process, preimplantation blastocysts were cultured in vitro, and cellular proliferation and differentiation were examined. At the end of the culture period the cells were genotyped. Upon isolation, no discernible differences were evident among individual blastocysts as observed by light microscopy (Fig. 5A, F, and K). All contained recognizable zona pellucida, blastocoelic cavity, ICM, and trophectodermal layer. In culture, trophoblastic cells from TAF10+/+ and TAF10+/− blastocysts attached and spread onto the culture dish surface and supported robust ICM outgrowths after 2 days (Fig. 5C to E and H to J). Trophoblastic cells from TAF10−/− blastocysts also attached and spread onto the culture dish surface but, in contrast to TAF10+/+ and TAF10+/− ICM cells, TAF10−/− ICM cells failed to proliferate and died within 2 days of culture (Fig. 5M to O and data not shown). After 4 days, trophoblastic cells, regardless of the genotype, had a similar morphology (Fig. 5 E, J, and O) and survived in culture for at least 12 days. Therefore, the expansion and survival of the ICM is strictly dependent on TAF10 in vitro. This appears to correlate well with the reduced ICM observed in abnormal 4.5- and 5.5-dpc embryos and suggests that the failure of TAF10−/− embryos in utero is caused by the death of ICM cells. Moreover, these studies identify terminally differentiated murine trophoblast cells as a mammalian cell type that do not require TAF10 for survival.
FIG. 5.
Defective growth of TAF10−/− blastocysts in vitro. Blastocysts (3.5 dpc) were recovered from heterozygous intercrosses and cultured individually for a period of 4 days and genotyped by PCR (n = 28). Representative +/+ (A to E), +/− (F to J), and −/− (K to O) blastocysts are shown. ICM, inner cell mass cells; TR, trophoblastic cells.
TAF10 loss leads to increased apoptosis.
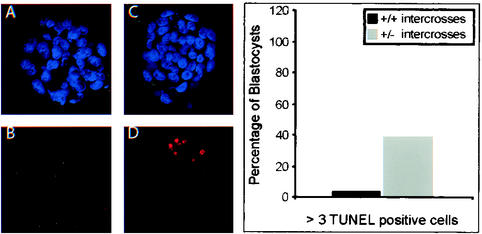
It was previously reported that lack of TAF10 in mouse F9 cells in vitro leads to increased apoptosis (26), suggesting that the growth deficit observed in mutant TAF10−/− ICMs could be a result of increase rate of apoptosis. We therefore compared the number of apoptotic cells present in blastocysts isolated from intercrosses of wild-type mice with those of intercrosses between TAF10 heterozygous mice by using a TUNEL assay to detect chromosomal DNA fragmentation associated with apoptosis. In blastocysts derived from wild-type intercrosses, very few (ca. 4%) contained high numbers of apoptotic cells defined as three to nine TUNEL-positive cells (Fig. 6). In contrast, 40% of the blastocysts derived from +/− intercrosses had high numbers of TUNEL-positive cells. This 10-fold increase most likely represents enhanced apoptosis in TAF10−/− blastocysts, which account for ca. 25% of blastocysts at this stage. TUNEL-positive cells were observed primarily in the ICM of blastocysts, which corresponds with the fact that TAF10−/− ICM cells, but not trophoblast cells, fail to survive in culture. Furthermore, the results suggest that developmental defects occurred as early as 3.5 dpc in TAF10−/− embryos. Therefore, the loss of ICM cells in vitro and in vivo in TAF10−/− embryos is likely due to intrinsically high rates of apoptosis in these cells.
FIG. 6.
Increased ICM cell apoptosis in TAF10 mutant embryos. Blastocysts (3.5 dpc) from wild-type (+/+) (A and B) or heterozygote (+/−) (C and D) intercrosses were subjected to TUNEL analysis. (A and C) nuclear stain DAPI; (B and D) TUNEL. On the right is a bar graph showing the percentages of blastocysts with more than three TUNEL-positive cells.
Gene expression in TAF10−/− trophoblasts.
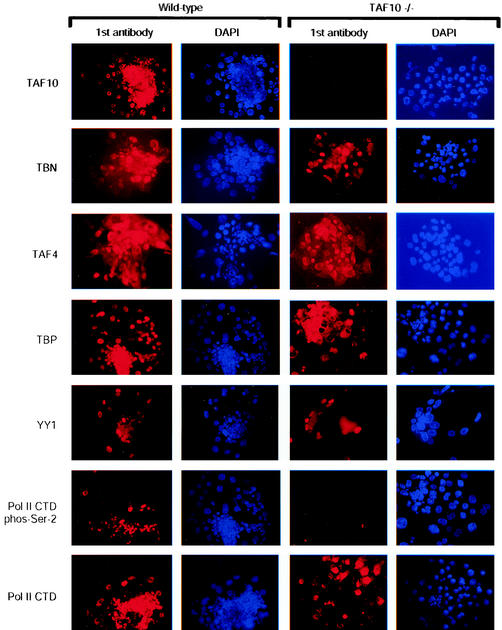
Given that disruption of TAF10−/− in F9 EC cells lead to alterations in TFIID integrity, we examined the effects of TAF10 elimination on protein expression in mouse trophoblast cultures by using immunocytochemistry. As expected, TAF10 protein was detected in wild type, but not mutant, trophoblasts (Fig. 7), confirming the knockout strategy. Similar to our previous findings (26), the levels of expression of TAFs, such as TAF4 and TAF5, appeared to be unaffected in TAF10−/− trophoblasts, even after several days in culture (Fig. 7 and data not shown). Interestingly, the expression levels of Taube nuss (TBN), a mouse homologue of scTAF8 and potential interaction partner of TAF10 (13, 41; unpublished data), also appeared to be unaffected (Fig. 7) in the absence of TAF10. Moreover, the expression levels of TBP and the transcription factor YY1 were not affected in TAF10−/− trophoblasts (Fig. 7).
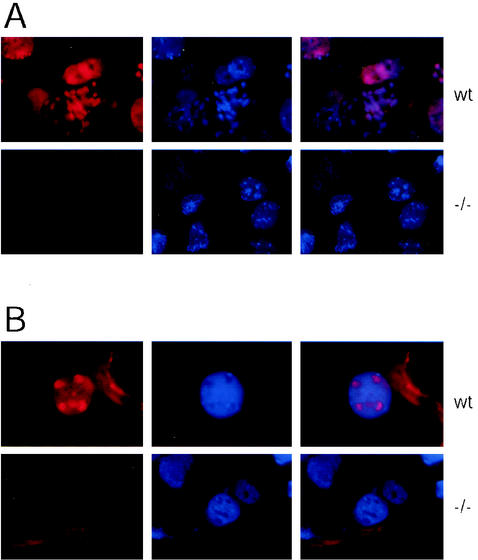
FIG. 7.
Reduced Pol II CTD phosphorylation as a result of TAF10 depletion. Immunocytochemical analysis of the expression of various proteins in TAF10−/− trophoblasts. After 3 days in culture, wild-type or TAF10−/− outgrowths were analyzed by immunofluorescence with the indicated antibodies and then counterstained with DAPI. Magnification, ×200.
RNA Pol II transcription is perturbed by the lack of TAF10.
To examine the transcriptional competence of TAF10−/− trophoblasts, we examined the phosphorylation state of the CTD of the largest subunit of Pol II. Phosphorylation of serine 2 of the CTD has previously been linked with initiation of the zygotic transcription program and elongation of Pol II (28, 31). Despite the apparent lack of effect on gene expression in TAF10−/− trophoblasts, a substantial reduction in the phosphorylation of the CTD was observed when H5 mouse monoclonal antibody that recognizes phospho-serine 2 of the Pol II CTD was used (32) (Fig. 7). We next sought to determine whether the reduction of serine 2 phosphorylation of the Pol II CTD was due to a reduction in the levels of Pol II protein by using a monoclonal antibody that recognizes all RNA Pol II CTD repeats. No reduction in the levels of Pol II CTD were detected in cells lacking TAF10 (Fig. 7), suggesting that the reduction in serine 2 phosphorylation on the CTD of Pol II was indeed the result of reduced phosphorylation. Therefore, despite the fact that the levels of several proteins remain stable, it appears that transcription is perturbed by the loss of TAF10.
Reduction in DNA replication and transcription in TAF10−/− trophoblasts.
To examine whether TAF10−/− trophoblasts are capable of endocycling, we measured BrdU incorporation into replicating DNA in wild-type and TAF10−/− trophoblasts. After 16 h of culture in the presence of BrdU wild-type trophoblasts showed a strong incorporation of label, whereas TAF10−/− trophoblasts showed virtually no BrdU incorporation (Fig. 8A), indicating that TAF10 is required for proper progression through the endocycle in mouse trophoblasts. To examine more directly the transcriptional competence of TAF10−/− trophoblasts, in situ transcription run-on assays were performed on trophoblasts outgrowths. In wild-type trophoblasts, incorporation of Br-UTP into the nascent RNA of permeabilized cells was detected as bright labeling of the entire nucleus, especially the nucleoli (Fig. 8B). In contrast, very little labeling of TAF10−/− trophoblast nuclei was observed, indicating that overall gene transcription is greatly reduced in TAF10 lacking trophoblasts.
FIG. 8.
Reduced DNA replication and transcription in TAF10−/− trophoblasts. (A) DNA synthesis in trophoblast cells. After 16 h of culture in the presence of BrdU, the BrdU incorporation in TAF10−/− (−/−) and wild-type (wt) trophoblasts was visualized by anti-BrdU immunofluorescence and counterstained with DAPI. Magnification, ×630. (B) In situ run-on transcription in trophoblast cells. Blastocyst outgrowths grown on coverslips for 2 days were subjected to in situ run-on assays essentially as described elsewhere (43). Br-UTP incorporation in nascent RNAs in Triton X-100 permeabilized TAF10−/− or wild-type (wt) trophoblasts visualized with an anti-BrdU antibody counterstained with DAPI. Magnification, ×630.
DISCUSSION
TAF10 is required for early mouse development.
The precise role that mammalian TAFs play in Pol II-mediated transcription in vivo is not well understood. Interestingly, we have observed that TAF10 mRNA is ubiquitously expressed during early mouse development, suggesting that it may be widely necessary for cells at these developmental stages. Later in development, TAF10 mRNA expression becomes more variable, with high levels in some rapidly dividing tissues and undetectable levels in tissues such as the heart and urogenital ridge, suggesting that certain cell types either do not need TAF10 or need only trace amounts of TAF10. Whether the observed levels of TAF10 mRNA expression correlate with its protein expression in these tissues is unknown; however, TAF protein levels, including TAF10, have been reported to vary in adult mouse tissues, and TAF10 protein was not detected in heart extracts (33). Taken together, these results suggest that even conserved general TAFs may have selective and variable roles in different cells.
To explore the requirement for TAF10 in mammalian tissues and cell types, we generated a functionally null mutation of the TAF10 gene in mice by homologous recombination and Cre-mediated excision. TAF10−/− embryos developed normally up to the blastocyst stage and implanted but died before the onset of gastrulation. At 3.5 dpc, TAF10−/− blastocysts appeared to be morphologically similar to wild-type blastocysts, although we detected increased numbers of apoptotic cells in the ICMs of mutant TAF10−/−blastocysts. As early as 4.5 dpc, abnormal TAF10 embryos were smaller in size and compromised in epiblast development and, by 5.5 dpc, were severely disorganized with few discernible structures remaining. Therefore, TAF10−/− embryos developed to the blastocyst stage, successfully hatched and implanted, but increased apoptosis in the ICM led to embryo failure soon after implantation, a time when the embryonic epiblast is required to undergo a burst in proliferation in order to further development.
The fact that mutant embryos undergo the processes of cleavage, compaction, ICM and trophectoderm differentiation, blastocoel formation, and implantation can be interpreted in two ways. The first possibility is that, prior to 3.5 dpc, TAF10 is nonessential and its function can be fulfilled by another factor, such as another TAF, during early embryonic development. The second possibility is that maternal stocks of TAF10 persist during implantation and are sufficient for initial embryonic growth and maturation. Examination of blastocysts recovered from heterozygote crosses for the presence of TAF10 by using immunofluorescence clearly revealed blastocysts with reduced TAF10 protein levels; however, we did not detect any blastocysts that were completely devoid of TAF10 at this stage(data not shown). Thus, at 3.5 dpc TAF10 is present in TAF10+/+, TAF10+/−, and TAF10−/− blastocysts, and the survival of homozygous −/− embryos up to 3.5 dpc is probably due to remaining maternally contributed TAF10.
TAF10 is selectively required for cell survival.
Both F9 EC cells and mouse blastocysts exhibit selective cell survival when TAF10 is disrupted. Specifically, TAF10 appears to be required for mitotic undifferentiated F9 EC and ICM cells but is dispensable for terminally differentiated F9 EC and trophoblast cells. Nevertheless, like yeast and murine F9 EC cells, trophoblasts required TAF10 for cell cycle progression since TAF10−/− trophoblasts stopped endocycling. Similar trophoblastic phenotypes have been described for mouse disruptions of MAT1 (35) and TBN (41), suggesting that they affect similar molecular pathways (see also below). On the other hand, disruption of the last three exons of the mouse TRRAP gene leads to a similar and yet distinct trophoblastic phenotype entailing chromosomal missegregations and cell cycle arrest (16). Like TAF10, TRRAP is an integral component of the TFTC-like complexes (6, 25, 30); however, it is also an integral component of at least two other complexes: the TIP60 HAT complex (19) and the p400 complex (12). Therefore, it is possible that the phenotype associated with TRRAP disruption is the result of disruptions in TRRAP-containing complexes other than TFTC-like complexes.
Disruption of TAF10 induces a distinct phenotype from those of HATs.
Disruptions of HATs in several organisms have suggested that, unlike TAF10, they are not generally necessary for survival. For example, the SAGA components GCN5, SPT20, SPT8, SPT7, SPT3, ADA2, and ADA3 are encoded by nonessential genes in yeast. Furthermore, PCAF knockout mice have no phenotype, whereas GCN5 knockout and GCN5/PCAF double-knockout mice die later in life (6.5 to 11.5 dpc) than TAF10−/− mice, demonstrating that these factors are not required for execution of many early mouse developmental programs (47, 48). The fact that double disruptions of PCAF and GCN5 have less-severe effects in mice than TAF10 suggests that the phenotype of TAF10−/− embryos is principally caused by disruption of TFIID function and that TFTC-like complexes become important only at later stages of development.
TAF10 is phenotypically linked to TBN.
Previous studies have indicated that TBN is a HFD-containing protein and shares primary sequence homology with Drosophila transcription factor PRODOS and scTAF8 (13, 17). It has therefore been suggested to associate with HFD-containing TAF10 protein and to be the hitherto-unknown mammalian dimerization partner of TAF10 and the mammalian homologue of scTAF8 (13, 14). Interestingly, TBN and TAF10−/− mice have the same phenotype (41; the present study) in that the lack of either leads to dramatic, but selective, ICM cell death. Together, these findings lend credence to the idea that TBN is the murine homologue of scTAF8 in mouse TFIID and suggest that disruption of any TFIID component could have serious effects on mammalian development.
Alteration of TFIID structure in the absence of TAF10.
F9 EC cells are widely used as a system to model early mouse cell differentiation. Like TAF10−/− trophoblasts, differentiated F9 EC cells are viable in the absence of TAF10 and so these cells provide an excellent model system to study the molecular consequences of TAF10 depletion. Interestingly, immunoprecipitation experiments demonstrated that in the absence of TAF10 the TAFs TAF4, TAF5, TAF12, and TAF13, despite their normal expression, are not tightly associated with TBP and do not form a stable TFIID complex (Fig. 1) in undifferentiated and RA differentiated F9 cells. Whether TFIID TAFs in cells lacking TAF10 form an incomplete TFIID-type complex devoid of TBP and TAF10, or whether they are redistributed into other TAF-containing complexes is unknown. Nevertheless, the fact that the TFTC subunits TRRAP, GCN5, and SAP130 are stably associated with TAF4 suggests that TFTC may remain intact. Therefore, the phenotype observed by the elimination of TAF10 is likely a result of altered TFIID structure and function.
Moreover, immunoprecipitations performed on extracts isolated from RA-differentiated F9 cells suggest that cells lacking TAF10 and thus lacking intact TFIID are able to differentiate into primitive endodermal cells. Since this process requires the transcriptional induction of several genes, for example, the RARβ gene, it seems likely that RA-stimulated transcription can occur without TAF10 and consequently intact TFIID. We have examined the consequences of TAF10 depletion on RA-induced cells by using gene arrays and found that the expression of many genes is unaffected at the mRNA level, whereas a certain percentage of genes is affected either positively or negatively (A. Soldatov, W. S. Mohan II, and L. Tora, unpublished data). These results suggest that, in the case of the unaffected genes, transcription can occur in the absence of intact TFIID but in the presence of either TBP alone, separated TAF core (see Fig. 1), or other factors that can mediate transcription initiation (i.e., TFTC).
Altered Pol II CTD phosphorylation and transcription in TAF10−/− trophoblasts.
The Pol II CTD is composed of 52 repeats in mammals of the consensus heptapeptide sequence YSPTSPS and is the target of various kinases that influence Pol II transcriptional activity (29). A variety of studies have shown that Ser-2 and Ser-5 phosphorylation play major roles in modulating overall Pol II activity. In keeping with the severe effects of eliminating TAF10, we observed radical changes in CTD phosphorylation at Ser-2 of the largest subunit of Pol II. Reduction in Ser-2 phosphorylation in TAF10−/− trophoblasts does not appear to reflect the fact that trophoblast cells in particular are defective for Ser-2 phosphorylation because Ser-2 phosphorylation was observed in trophoblasts derived from wild-type blastocysts (Fig. 7). The fact that Ser-2 phosphorylation is reduced in the absence of TAF10 suggests that directly or indirectly TAF10 is involved in phosphorylation of the CTD. In agreement with our results, a similar reduction in Pol II CTD phosphorylation at Ser-2 was observed in C. elegans when it was depleted of TAF10 (42). We observed, along with the reduced phosphorylation of the Pol II CTD, a large reduction in transcription in TAF10−/− trophoblasts as measured by an in situ run-on assay. Since Fig. 8 demonstrates that transcription is essentially not detectable in TAF10−/− trophoblasts by in situ run-on assays, it would appear that protein expression detected in Fig. 7 is probably not dependent on transcription. Interestingly, the loss of TAF10 leads to a large reduction in both Pol II and Pol I transcription, although the latter is likely the result of secondary effects since TAF10 has not been detected in the nucleolus. The dramatic effects on Pol II transcription after TAF10 disruption appears to agree with the fact that TFIID structure is altered in the absence of TAF10. Even so, we cannot rule out the possibility that some transcription may occur in the absence of TAF10.
Thus, we have for the first time shown that a mammalian TAF is absolutely necessary for establishing gene expression patterns during early mouse development. Furthermore, we have found molecular evidence that the structure of endogenous TFIID is altered in TAF10−/− cells, apparently resulting in abnormal RNA Pol II CTD phosphorylation, revealing a potential molecular mechanism to explain the development defects observed.
Acknowledgments
We are grateful to M. Oulad-Abdelghani, I. Davidson, S. P. Jackson, and M. Vigneron for antibodies; S. Viville, A. Dierich, and the embryonic stem cell facility for advice with blastocyst genotyping and microinjections; J.-M. Bornert, F. Cammas, M. Lemeur, E. Metzger, and the mouse facility for advice and reagents; J. Hergueux for sectioning embryos; F. Ruffenach for oligonucleotide synthesis; M. Boeglin for confocal images; and C. Mendelsohn, B. Bell, and E. vom Baur for critically reading the manuscript.
W.S.M. was supported by fellowships from the Association pour la Recherche sur le Cancer and the Ligue Nationale contre le Cancer. This work was also supported by funds from the Institut National de la Santé et de la Recherche Médicale, the CNRS, the Hôpital Universitaire de Strasbourg, the Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Medicale, the Ligue Nationale contre le Cancer, European Community grants (HPRN-CT-2000-00087 and HPRN-CT-2000-00088), and the Human Frontier Science Program (RG 196/98).
REFERENCES
- 1.Andel, F., III, A. G. Ladurner, C. Inouye, R. Tjian, and E. Nogales. 1999. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science 286:2153-2156. [DOI] [PubMed] [Google Scholar]
- 2.Bell, B., and L. Tora. 1999. Regulation of gene expression by multiple forms of TFIID and other novel TAFII-containing complexes. Exp. Cell Res. 246:11-19. [DOI] [PubMed] [Google Scholar]
- 3.Besse, S., M. Vigneron, E. Pichard, and F. Puvion-Dutilleul. 1995. Synthesis and maturation of viral transcripts in herpes simplex virus type 1 infected HeLa cells: the role of interchromatin granules. Gene Expr. 4:143-161. [PMC free article] [PubMed] [Google Scholar]
- 4.Brand, M., C. Leurent, V. Mallouh, L. Tora, and P. Schultz. 1999. Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science 286:2151-2153. [DOI] [PubMed] [Google Scholar]
- 5.Brand, M., J. G. Moggs, M. Oulad-Abdelghani, F. Lejeune, F. J. Dilworth, J. Stevenin, G. Almouzni, and L. Tora. 2001. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 20:3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brand, M., K. Yamamoto, A. Staub, and L. Tora. 1999. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 274:18285-18289. [DOI] [PubMed] [Google Scholar]
- 7.Brou, C., S. Chaudhary, I. Davidson, Y. Lutz, J. Wu, J. M. Egly, L. Tora, and P. Chambon. 1993. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 12:489-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolle, P., A. Dierich, M. LeMeur, T. Schimmang, B. Schuhbaur, P. Chambon, and D. Duboule. 1993. Disruption of the Hoxd-13 gene induces localized heterochrony leading to mice with neotenic limbs. Cell 75:431-441. [DOI] [PubMed] [Google Scholar]
- 9.Dupe, V., M. Davenne, J. Brocard, P. Dolle, M. Mark, A. Dierich, P. Chambon, and F. M. Rijli. 1997. In vivo functional analysis of the Hoxa-1 3′ retinoic acid response element (3′RARE). Development 124:399-410. [DOI] [PubMed] [Google Scholar]
- 10.Feil, R., J. Brocard, B. Mascrez, M. LeMeur, D. Metzger, and P. Chambon. 1996. Ligand-activated site-specific recombination in mice. Proc. Natl. Acad. Sci. USA 93:10887-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freiman, R. N., S. R. Albright, S. Zheng, W. C. Sha, R. E. Hammer, and R. Tjian. 2001. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science 293:2084-2087. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297-307. [DOI] [PubMed] [Google Scholar]
- 13.Gangloff, Y. G., J. C. Pointud, S. Thuault, L. Carre, C. Romier, S. Muratoglu, M. Brand, L. Tora, J. L. Couderc, and I. Davidson. 2001. The TFIID components human TAFII140 and Drosophila BIP2 (TAFII155) are novel metazoan homologues of yeast TAFII47 containing a histone fold and a PHD finger. Mol. Cell. Biol. 21:5109-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gangloff, Y. G., S. L. Sanders, C. Romier, D. Kirschner, P. A. Weil, L. Tora, and I. Davidson. 2001. Histone folds mediate selective heterodimerization of yeast TAFII25 with TFIID components yTAFII47 and yTAFII65 and with SAGA component ySPT7. Mol. Cell. Biol. 21:1841-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hampsey, M. 1998. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 62:465-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herceg, Z., W. Hulla, D. Gell, C. Cuenin, M. Lleonart, S. Jackson, and Z. Q. Wang. 2001. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat. Genet. 29:206-211. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez-Hernandez, A., and A. Ferrus. 2001. Prodos is a conserved transcriptional regulator that interacts with dTAFII16 in Drosophila melanogaster. Mol. Cell. Biol. 21:614-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 19.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 20.Ito, M., C. X. Yuan, H. J. Okano, R. B. Darnell, and R. G. Roeder. 2000. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell 5:683-693. [DOI] [PubMed] [Google Scholar]
- 21.Jacq, X., C. Brou, Y. Lutz, I. Davidson, P. Chambon, and L. Tora. 1994. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell 79:107-117. [DOI] [PubMed] [Google Scholar]
- 22.Kirchner, J., S. L. Sanders, E. Klebanow, and P. A. Weil. 2001. Molecular genetic dissection of TAF25, an essential yeast gene encoding a subunit shared by TFIID and SAGA multiprotein transcription factors. Mol. Cell. Biol. 21:6668-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirschner, D. B., E. vom Baur, C. Thibault, S. L. Sanders, Y. G. Gangloff, I. Davidson, P. A. Weil, and L. Tora. 2002. Distinct mutations in yeast TAFII25 differentially affect the composition of TFIID and SAGA complexes as well as global gene expression patterns. Mol. Cell. Biol. 22:3178-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, T. I., H. C. Causton, F. C. Holstege, W. C. Shen, N. Hannett, E. G. Jennings, F. Winston, M. R. Green, and R. A. Young. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405:701-704. [DOI] [PubMed] [Google Scholar]
- 25.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metzger, D., E. Scheer, A. Soldatov, and L. Tora. 1999. Mammalian TAFII30 is required for cell cycle progression and specific cellular differentiation programmes. EMBO J. 18:4823-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myat, A., D. Henrique, D. Ish-Horowicz, and J. Lewis. 1996. A chick homologue of Serrate and its relationship with Notch and Delta homologues during central neurogenesis. Dev. Biol. 174:233-247. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 29.Oelgeschlager, T. 2002. Regulation of RNA polymerase II activity by CTD phosphorylation and cell cycle control. J. Cell Physiol. 190:160-169. [DOI] [PubMed] [Google Scholar]
- 30.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schiltz, T. Howard, X. J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 31.Palancade, B., S. Bellier, G. Almouzni, and O. Bensaude. 2001. Incomplete RNA polymerase II phosphorylation in Xenopus laevis early embryos. J. Cell Sci. 114:2483-2489. [DOI] [PubMed] [Google Scholar]
- 32.Patturajan, M., R. J. Schulte, B. M. Sefton, R. Berezney, M. Vincent, O. Bensaude, S. L. Warren, and J. L. Corden. 1998. Growth-related changes in phosphorylation of yeast RNA polymerase II. J. Biol. Chem. 273:4689-4694. [DOI] [PubMed] [Google Scholar]
- 33.Perletti, L., J. C. Dantonel, and I. Davidson. 1999. The TATA-binding potein and its associated factors are differentially expressed in adult mouse tissues. J. Biol. Chem. 274:15301-15304. [DOI] [PubMed] [Google Scholar]
- 34.Perletti, L., E. Kopf, L. Carre, and I. Davidson. 22 March 2001, posting date. Coordinate regulation of RARgamma2, TBP, and TAFII135 by targeted proteolysis during retinoic acid-induced differentiation of F9 embryonal carcinoma cells. BMC Mol. Biol. 2. [Online.] http://www.biomedcentral.com/1471-2199/2/4. [DOI] [PMC free article] [PubMed]
- 35.Rossi, D. J., A. Londesborough, N. Korsisaari, A. Pihlak, E. Lehtonen, M. Henkemeyer, and T. P. Makela. 2001. Inability to enter S phase and defective RNA polymerase II CTD phosphorylation in mice lacking Mat1. EMBO J. 20:2844-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanders, S. L., E. R. Klebanow, and P. A. Weil. 1999. TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J. Biol. Chem. 274:18847-18850. [DOI] [PubMed] [Google Scholar]
- 37.Schiltz, R. L., C. A. Mizzen, A. Vassilev, R. G. Cook, C. D. Allis, and Y. Nakatani. 1999. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 274:1189-1192. [DOI] [PubMed] [Google Scholar]
- 38.Struhl, K. 1997. Selective roles for TATA-binding-protein-associated factors in vivo. Genes Funct. 1:5-9. [DOI] [PubMed] [Google Scholar]
- 39.Tora, L. 2002. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16:673-675. [DOI] [PubMed] [Google Scholar]
- 40.Tudor, M., P. J. Murray, C. Onufryk, R. Jaenisch, and R. A. Young. 1999. Ubiquitous expression and embryonic requirement for RNA polymerase II coactivator subunit Srb7 in mice. Genes Dev. 13:2365-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voss, A. K., T. Thomas, P. Petrou, K. Anastassiadis, H. Scholer, and P. Gruss. 2000. Taube nuss is a novel gene essential for the survival of pluripotent cells of early mouse embryos. Development 127:5449-5461. [DOI] [PubMed] [Google Scholar]
- 42.Walker, A. K., J. H. Rothman, Y. Shi, and T. K. Blackwell. 2001. Distinct requirements for C. elegans TAFIIs in early embryonic transcription. EMBO J. 20:5269-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wansink, D. G., W. Schul, I. van der Kraan, B. van Steensel, R. van Driel, and L. de Jong. 1993. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J. Cell Biol. 122:283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wassarman, D. A., and F. Sauer. 2001. TAFII250: a transcription toolbox. J. Cell Sci. 114:2895-2902. [DOI] [PubMed]
- 45.Wieczorek, E., M. Brand, X. Jacq, and L. Tora. 1998. Function of TAFII-containing complex without TBP in transcription by RNA polymerase II. Nature 393:187-191. [DOI] [PubMed] [Google Scholar]
- 46.Wu, S. Y., M. C. Thomas, S. Y. Hou, V. Likhite, and C. M. Chiang. 1999. Isolation of mouse TFIID and functional characterization of TBP and TFIID in mediating estrogen receptor and chromatin transcription. J. Biol. Chem. 274:23480-23490. [DOI] [PubMed] [Google Scholar]
- 47.Xu, W., D. G. Edmondson, Y. A. Evrard, M. Wakamiya, R. R. Behringer, and S. Y. Roth. 2000. Loss of Gcn512 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26:229-232. [DOI] [PubMed] [Google Scholar]
- 48.Yamauchi, T., J. Yamauchi, T. Kuwata, T. Tamura, T. Yamashita, N. Bae, H. Westphal, K. Ozato, and Y. Nakatani. 2000. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. USA 97:11303-11306. [DOI] [PMC free article] [PubMed] [Google Scholar]