Abstract
The AP-1 transcription factor plays an essential role in cell proliferation and tumorigenesis. It was previously shown that the fra-1 gene product is upregulated by various oncogenes and is involved in the in vitro and in vivo transformation of thyroid cells. Here we show that the ras oncogene-dependent accumulation of Fra-1 is mediated by a positive feedback mechanism which requires both transcriptional autoregulation and posttranslational stabilization of the protein. The oncogene-dependent transcriptional activation involves the cooperation between both Raf-dependent and Raf-independent pathways and is mediated by an AP-1 site within the fra-1 first intron, which becomes stably occupied by a transcriptionally active Fra-1-containing complex in ras-transformed cells. The posttranslational stabilization results in a drastic increase in the Fra-1 half-life in ras-transformed cells and is totally dependent on the activity of the MEK/ERK phosphorylation pathway. The analysis of the Fra-1 transactivation potential shows that the protein is able to stimulate a heterologous promoter in a ras-dependent manner, but the transactivating activity requires the recruitment of a heterodimeric partner. These data show that the alteration of multiple regulatory mechanisms is required for the constitutive activation of Fra-1 as a nuclear target of ras transformation.
During the course of oncogenesis, the establishment of the transformed and tumorigenic phenotype requires the activity of multiple nuclear targets of transformation, including the activator protein 1 (AP-1) transcription factor, which mediates a wide range of the effects of activated oncogenes on cell growth, apoptosis, and differentiation.
The AP-1 complex is formed by homo- and heterodimerization between Jun (c-Jun, JunB, and JunD), Fos (c-Fos, FosB, Fra-1, and Fra-2), and several ATF family members, all belonging to the bZIP protein superfamily, which is characterized by the basic DNA-interacting region and the leucine zipper dimerization domain (50). Given the combinatorial diversity resulting in a large number of distinct dimers along with the multiple control levels of expression and posttranslational modification of each component, the regulation of AP-1 activity is very complex, allowing the performance of highly disparate biological functions by a single transcription factor complex (reviewed in references 5, 25, 27, 32, and 51).
In normal cells, AP-1 is involved in the control of cell proliferation and, depending on the cellular context, can have both proapoptotic and antiapoptotic functions in response to a wide range of environmental stimuli, such as mitogens, stress-inducing agents, inflammatory cytokines, etc. (50).
Recently, important information was provided on the role of Jun proteins in the control of cell cycle, showing that both the positive effect of c-Jun and the negative control by JunB are mediated by their dual effects on the expression of cyclin D1 (6, 62) and the p16 and p21 cdk inhibitors (41, 47), while the role of JunD appears to be cell context dependent and at least in part mediated by the p19Arf tumor suppressor (60). Importantly, the effect of c-Jun on the p21 promoter is mediated by p53 (47) and is essential for cell cycle reentry during mammalian UV response (52).
Besides cyclins and cell cycle inhibitors, AP-1-regulated genes include several growth factors (granulocyte-macrophage colony-stimulating factor, keratinocyte growth factor, heparin-binding epidermal growth factor, vascular endothelial growth factor D, and FL1) and components of the apoptotic pathway (FasL, Fas, and Bcl3; reviewed in references 50, 58, and 59). In addition, another important group of AP-1 targets is represented by the genes encoding proteins involved in cell migration. These include various extracellular matrix components (fibronectin, SPARC) and degrading enzymes (collagenase and stromelysin matrix metalloproteases, urokinase plasminogen activator) along with their cognate inhibitors (TIMP and PAI-1), altogether implicated in cancer cell invasion (reviewed in references 58 and 59).
The relationship between AP-1 and tumorigenesis was suggested by the response to tumor promoters and the increased AP-1 activity in transformed cell lines. The components most frequently upregulated by oncogenic transformation are represented by c-Jun, JunB, Fra-1, and Fra-2, while the levels of c-Fos and FosB are generally not affected. The oncogenic role of AP-1 has been established by multiple approaches, including the overexpression of individual components, the functional inhibition by dominant-negative derivatives and antisense expression vectors, and the use of knockout and transgenic mice (27, 32, 58, 59).
In vitro, a dominant-negative c-Jun derivative lacking the amino-terminal carboxy-terminal region is able to antagonize the AP-1 transactivation and ras-mediated transformation in mouse fibroblasts (12, 30), while in a transgenic mouse model the epidermis-specific expression of the dominant-negative c-Jun is able to suppress tumor promoter-induced papillomas (63). The role of ras-dependent c-Jun amino-terminal phosphorylation (53) has been established by use of cell lines harboring the mutant allele of c-Jun with the JNK phosphoacceptor serines changed to alanine (JunAA). In vivo, the development of ras-inducible skin tumors is impaired in mouse strains expressing the nonphosphorylatable derivative of c-Jun (9).
Important information on the role of c-fos and junB has been provided by knockout mouse analysis, showing that the c-fos gene product is essential for malignant progression in the animal model exhibiting keratinocyte-specific expression of the v-H-ras oncogene (46), while the junB antioncogenic role is strongly supported by the analysis of knockout mice developing chronic myeloid leukemia as a consequence of the lack of JunB expression in the myeloid cell lineage (40).
Although genetic evidence based on knockout mice is not yet available, the information on the role of Fra-1 in neoplastic transformation has been obtained mainly by ectopic overexpression in various cell lines, including rat embryo fibroblasts (10), NIH 3T3 cells (31), and a mouse mammary adenocarcinoma cell line, in which the stable or inducible accumulation of the protein, alone or in combination with c-Jun, was able to recapitulate several aspects of the transformed phenotype (29).
The role of the highly related Fra-2 has been addressed in detail with v-src-transformed chicken embryo fibroblasts (34, 35), for which the mitogen-activated protein (MAP) kinase-dependent phosphorylation of the protein has been characterized. The corresponding phosphoacceptor sites have been proposed to be essential for the ERK-dependent transactivation of Fra-1 (64). Because of the very similar expression as delayed early genes, Fra-1 and Fra-2 have been largely considered functionally equivalent. However, the use of the recently described tethered AP-1 pseudoheterodimers has allowed the ascribing of a specific cyclin A-dependent role to the c-Jun/Fra-2 but not the c-Jun/Fra-1 and c-Jun/c-Fos heterodimers in the control of cell growth of mouse fibroblasts (7). On the other hand, with regard to ras-mediated transformation, Fra-1 but not Fra-2 has been identified among the transcriptional targets most dramatically induced by the activated oncogene in a systematic screening of ras transformation targets (65).
Most studies on AP-1 in neoplastic transformation have addressed the mechanisms triggered by the ras oncogene resulting from mutational activation of the ras protooncogene, which encodes a small GTP-binding protein playing a central role in mitogenic signaling pathways. Ras activity is mediated by multiple downstream pathways, including the Raf/MEK/ERK phosphorylation cascade, essential for transducing the mitogenic signal from growth factor receptors to nuclear targets; the phosphatidylinositol 3 (PI3)-kinase/AKT pathway, mainly involved in cell survival signaling; the Cdc42/Rac/Rho pathway, implicated in cytoskeletal reorganization; and the JNK and p38 pathways, mediating stress responses and apoptosis signaling (13, 20). The cooperation between multiple Ras-dependent pathways is essential for cell cycle progression (23). Although most of the ras oncogene-dependent modifications are usually attributed to the Raf/MEK-mediated pathway, the Rho/Rac pathway is also required for ras transformation, and a constitutively active form of Rac can act as a weak oncogene in mouse fibroblasts (43).
Mutational activation of Ras plays a major role in human tumorigenesis and occurs at high frequency in several kinds of epithelial cancer. The study of Ras mutations in both benign and malignant follicular tumors indicates that Ki-ras is the most frequently altered gene in thyroid cancer and that its activation is an early event in thyroid tumorigenesis (54). In vitro, follicular rat thyroid cells are fully transformed by the ras oncogene, which results in growth factor independence, morphological transformation, and tumorigenicity in nude mice; the transformed phenotype is associated with the decrease or loss of differentiation (21). Thus, the rat thyroid cell lines, representing a model system for studying the epithelial tumorigenesis, have allowed the dissection of the mechanisms of ras-dependent extinction of thyroid-specific gene expression, mediated by the inactivation of tissue-specific transcription factors, responsible for the loss of expression of thyroid-specific genes (14, 16, 22, 33).
It was previously shown that the main compositional change of AP-1 in ras-transformed thyroid cells is represented by the drastic increase of Fra-1 and JunB and that Fra-1 accumulation is essential for the establishment of the fully transformed phenotype (57). In vivo, the increase of AP-1 activity represents a general event in thyroid transformation (8), and Fra-1 gene induction occurs at high frequency in human thyroid carcinoma cell lines and tissues (15).
In this study we have dissected the mechanisms regulating Fra-1 accumulation in ras-transformed cells and used Ras effector mutants and constitutive activators to investigate the signal transduction pathways involved in Fra-1 accumulation. We show that the Raf/MEK/ERK cascade is necessary but insufficient for full oncogenic induction of Fra-1 and that ERK-dependent stabilization is required for the robust accumulation of the protein in ras-transformed cells. Interestingly, the transformation-dependent transcriptional induction is mediated by positive autoregulation of fra-1 by its own gene product, which is involved in the ras-dependent occupancy and transactivation of the AP-1 regulatory element in the first intron of the gene. We have further dissected the Fra-1-dependent transcriptional activation in response to the oncogenic Ras and have shown that the Fra-1-mediated transactivation totally depends upon recruitment of the heterodimeric partner.
MATERIALS AND METHODS
Cell cultures, DNA constructs, and transfection assays.
The rat thyroid follicular FRTL-5 cell line and the FRTL-5KRas-transformed derivative have been described previously (21). Both cell lines were maintained in Coon's modified Ham's F-12 medium (Euro Clone) as previously described (2). For transfection assays, approximately 2 × 106 cells were electroporated with the indicated amounts of reporter constructs and/or expression vectors, keeping the total amount of DNA at 20 to 25 μg in 0.5 ml of complete medium at 250 V and 960 μF, by using a Gene Pulser apparatus (Bio-Rad) and were plated in fresh medium for the indicated time periods. For stable clones, FRTL-5 and FRTL-5KRas cell lines were transfected with the DNA vector encoding the selectable marker, alone or in combination with the linearized fra-1/β-globin wild type or the fra-1/β-globin-Δ construct. After selection in G418 (800 μg/ml; Calbiochem), pools of stably transfected cell clones were derived from a similar number (∼80 to 100) of G418-resistant colonies. The comparable copy number of stably integrated constructs in each pool of G418-resistant transfectants was verified by Southern blot hybridization with a rabbit β-globin probe detecting an internal 3.8-kb EcoRI fragment (data not shown). The pcDNA3 Ras (V12), (V12/S35), (V12/C40), and (V12/G37) constructs were kindly provided by Julian Downward. Both the wild-type and the mutated version of the fra-1/β-globin vectors and the vectors coding Gal4/Fra-1, Gal4/Fra-1-ΔZip, Gal4/c-Fos, and Gal4/c-Fos-insZip were obtained from Bergers et al. (10). The trans-reporter plasmid FrLuc (Stratagene) contains the luciferase reporter gene cloned downstream of a basic promoter element (TATA box) and was joined to five tandem repeats of the 17-bp GAL4 binding element. For measuring luciferase activity cells were collected 24 h after transfection and were processed by using the Dual-Luciferase Reporter Assay system (Promega) as indicated by the manufacturer in a Lumat LB 9501 luminometer (Berthold).
RNA extraction and Northern analysis.
Total RNA was extracted with a one-step purification kit (TRI-REAGENT; Molecular Research Center, Inc.) following the procedure indicated by the manufacturer. For Northern blots, 20 to 30 μg of total RNA was separated, transferred onto Immobilon-Ny+ membranes (Millipore), and UV cross-linked by using a Stratalinker apparatus. The EcoRI/EcoRI 1.5-kb rat fra-1 cDNA fragment (17) was labeled with [α-32P]dCTP by using a random oligonucleotide primer (Ready-To-Go; Pharmacia). Hybridizations were carried out overnight (O/N) at 42°C in a solution containing 50% formamide and 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
RT-PCR analysis.
Usually, 2 μg of DNase-treated total RNA was reverse transcribed by using random hexanucleotide primers (12.5 μg/μl), deoxynucleoside triphosphate (1.25 mM), dithiothreitol (10 mM), RNase OUT (40 U) (Promega), and Moloney murine leukemia virus reverse transcriptase (RT; 40 U) (Life Technologies). Approximately 200 ng of cDNA was amplified in a reaction mixture containing 0.2 mM deoxynucleoside triphosphates, 1.5 mM MgCl2, a 0.4 μM concentration of each primer, and 1 U of Taq DNA polymerase (Life Technologies). A 76-bp cDNA fragment from the rabbit β-globin cDNA was amplified by using the oligonucleotide primers 5′-CTGCACGTGGATCCTGAGAACTTCA-3′ (forward) and 5′-GCCAAAATGATGAGACAGCACAACA-3′ (reverse) (for details see above). As an internal control a 370-bp cDNA fragment of the rat hypoxanthine phosphoribosyltransferase (HPRT) gene was coamplified with the primers 5′-CCTGCTGGATTACATTAAAGCACTG-3′ (forward) and 5′-CCTGAAGTACTCATTATAGTCAAGG-3′ (reverse). The expression of the chloramphenicol acetyltransferase (CAT) reporter gene was utilized to normalize for the transfection efficiency by coamplifying a 346-bp cDNA fragment with the primers 5′-GGGCGAAGAAGTTGTCCATA-3′ (forward) and 5′-CCGTAAAGAAAAATAAGCAC-3′ (reverse). To ensure no contamination of RNA samples with DNA, the β-globin primers were generated encompassing an intronic portion of the gene, and negative controls were obtained by performing the PCRs on samples that were not reverse transcribed but that were otherwise identically processed.
Immunoblotting analysis.
Whole-cell lysates were prepared directly in sample buffer or in lysis buffer (20 mM Tris [pH 7.9], 120 mM KCl, 5 mM MgCl2, 0.2% NP-40, 5 mM EDTA, 10% glycerol, 0.5 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml, 1 mM sodium vanadate, 50 mM NaF) and normalized for equal protein concentration by the Bradford assay (Bio-Rad). Cell extracts were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) and were transferred onto Immobilon-P membranes (Millipore). After being blocked with 5% nonfat dry milk in Tris-buffered saline-0.2% Tween 20, membranes were incubated with the indicated primary antibodies as instructed by the manufacturer. Bound antibodies were detected by the appropriate horseradish peroxidase-conjugated secondary antibodies (Sigma), and immunocomplexes were revealed by enhanced chemiluminescence (ECLplus; Amersham International). Image acquisition and quantification of chemiluminescence signals was performed with the Gel-Doc apparatus and the QuantityOne software package (Bio-Rad).
Pulse-chase analysis.
Usually, 18 h after plating or transfection cells were starved for 3 h in 5% dialyzed fetal bovine serum (FBS) in minimum essential medium lacking methionine and cysteine (Sigma). Cells were labeled with 35S (Pro-Mix; 100 μC in 1 ml; Amersham) for 1 h and then were washed twice with 1× phosphate-buffered saline and switched in fresh F-12 appropriate medium for the time periods indicated in the figure legends. Whole-cell lysates were prepared by collecting cells in 200 μl of lysis buffer (50 mM Tris [pH 7.9], 0.5% SDS, 70 mM β-mercaptoethanol) and boiling the lysates at 95°C for 5 min. The concentration of SDS was adjusted to 0.1% by adding 800 μl of radioimmunoprecipitation assay buffer without SDS (10 mM Tris [pH 7.9], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate). Five hundred to 800 μg of total cell extracts was immunoprecipitated with 2 μg of anti-Fra-1 overnight. Immunocomplexes were recovered on protein A-Sepharose beads and were washed three times in 0.1% SDS radioimmunoprecipitation assay buffer. Proteins were resuspended in Laemmli sample buffer supplemented with 5% β-mercaptoethanol and resolved by SDS-10% PAGE. The gel was fixed, treated with the fluorographic reagent Amplify (Amersham), and subjected to autoradiography.
Protein kinase assay.
For the myelin basic protein (MBP) kinase assay, FRTL-5 cells were cotransfected with different Ras constructs and ERK2-hemagglutinin (HA), allowed to adhere in complete F-12 medium, and then switched to F-12 medium containing 0.5% FBS for 18 h. Cells were then lysed in ERK lysis buffer (10 mM Tris [pH 7.4], 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 10% glycerol, protease and phosphatase inhibitors). After preclearing, 2 to 300 μg of total cell extracts was immunoprecipitated with 2 μg of the 12CA5 monoclonal anti-HA antibody (Boehringer). Immunocomplexes recovered on protein A-Sepharose beads were washed three times in lysis buffer and were equilibrated in kinase buffer (50 mM HEPES-Na [pH 7.4], 10 mM MgCl2, 5 mM MnCl2) and were incubated successively for 15 min at 30°C in kinase buffer containing 1 mM dithiothreitol, 1 μM ATP, and 0.5 μl of [32P]ATP (3,000 Ci/mmol) in the presence of 10 μg of MBP. The kinase reaction was stopped by adding 4× Laemmli sample buffer. Samples were subjected to SDS-15% PAGE and autoradiography.
Nuclear extracts and EMSA.
Nuclear extract preparation and electrophoretic mobility shift assay (EMSA) were performed as described previously (57). The oligonucleotide used as the probe was fra-1 TRE (5′-TGAGGCTGAGTCACCCTGGG-3′). For the antibody supershift analysis the binding reaction products were incubated with 1 μg of each antibody at 4°C for 3 h and were resolved by 6% PAGE. For in vitro dephosphorylation, nuclear extracts were processed without phosphatase inhibitors and were incubated for 2 h at 37°C alone or with 20 U of calf intestine phosphatase (CIP; Boehringer).
In vitro binding assay. (i) Preparation of fra-1 TRE DNA affinity beads.
The biotinylated fra-1 tetradecanoyl phorbol acetate-responsive element (TRE) oligonucleotide 5′-GAGTCACTGAGGCTGAGTCACCCTGGGTGC-3′, containing a wild-type TRE sequence (in bold), was derived from nucleotides 1844 to 1873 of the first intron of the fra-1 gene. In this paper we refer to this sequence as the fra-1 TRE DNA. To disrupt the AP-1 binding site we generated a mutated version of this oligonucleotide by substituting two nucleotides in the palindromic TRE (TC) and in the flanking region (CT). This sequence is referred as mut fra-1 TRE DNA (5′-GAGTCACTGAGGCTGAGCTACCCTGGGTGC-3′). Both the wild-type and the mutated versions and their inverse complementary oligonucleotides were custom synthesized by MWG-Biotech AG. Binding of annealed, double-stranded biotinylated fra-1 TRE oligonucleotide to streptavidin beads (Pierce) was performed according to the manufacturer's instructions.
(ii) fra-1 TRE DNA affinity chromatography.
fra-1 TRE DNA affinity chromatography was performed as follows. As little as 25 μg of input nuclear extract proteins and 5 μg of DNA on beads were used (1:5 ratio of DNA to input protein). AP-1-associated proteins were isolated by single-step batchwise DNA affinity chromatography, incubating nuclear extracts with fra-1 TRE DNA affinity beads in the presence of 6 μg of poly(dI-dC), in binding buffer (25 mM HEPES [pH 7.9], 40 mM KCl, 2 mM spermidine, 0.1% NP-40, 20% glycerol added with protease and phosphatase inhibitors) O/N at 4°C on a rotary shaker. For competition assays, nuclear extracts were preincubated with a 25-fold molar excess of fra-1 TRE or mut fra-1 TRE double-stranded oligonucleotide for 20 min at 4°C before adding the DNA beads. To recover the unbound proteins the beads were centrifuged and the supernatant was trichloroacetic acid precipitated. Beads were then washed twice with binding buffer and were boiled in SDS sample buffer to elute the bound material to be subjected to on SDS-PAGE. The Fra-1 protein was detected by immunoblotting as described above.
Chromatin immunoprecipitation (ChIp).
Approximately 108 cells were washed with phosphate-buffered saline and were fixed in the presence of 1% formaldehyde for 10 min at 37°C. After being cross-linked, the chromatin was isolated and then was subjected to sonication essentially as described previously (39). Chromatin fragments containing the protein of interest were purified by immunoprecipitation by using antibodies anti-Fra-1 and anti-acetylated histone H3 (Upstate). Sonicated material was incubated O/N with the various antibodies at 4°C, and immunocomplexes were isolated following incubation with protein A-Sepharose at room temperature for 2 h. The recovered DNA was analyzed by radioactive PCR with primers (fra-1-Int) located in the first intron and flanking the defined intronic fra-1 TRE (10) site: (+178) 5′-GCTGTTCGTGTGTCGCTGTA-3′ (forward) and (+752) 5′-ATGGGGGGCTGAGTGTCCTG −3′ (reverse). Oligonucleotides for HPRT were used as controls: (+1546) 5′-TGCTGCGTCCCTTTTGATTT-3′ (forward) and (+2084) 5′-GGCTTTCCCGCTTTACCATT-3′ (reverse). To ensure that PCR was in the linear range, serial dilutions of DNA were amplified for a maximum of 30 cycles. The products were resolved by native gel electrophoresis (6% polyacrylamide in 1× Tris-borate-EDTA) and were detected by autoradiography of dried gels.
RESULTS
Roles of different Ras downstream effectors in the induction of Fra-1.
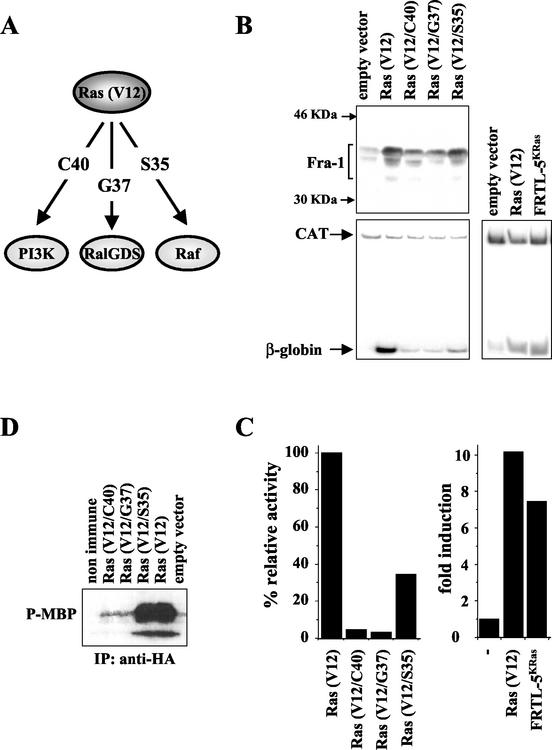
We have previously shown that fra-1 plays a major role as a target of the activated ras oncogene, which dramatically upregulates the Fra-1 mRNA and protein in the in vitro-transformed FRTL-5 rat thyroid cell line. To understand the roles of the individual Ras-dependent signaling pathways in the oncogene-dependent induction of Fra-1, we took advantage of the previously described Ras effector loop mutants (19, 45), characterized by their ability to individually activate each of the major Ras downstream effectors PI3-kinase, RalGDS, and Raf (Fig. 1A). The Ras effector mutants in a V12 oncogenic background were coexpressed with a reporter construct containing a portion of the fra-1 gene (spanning from −710 to 2741) fused to the rabbit β-globin gene, along with the cytomegalovirus-CAT reporter plasmid, as an internal control. Ras-dependent accumulation of the endogenous Fra-1 and activation of the fra-1/β-globin minigene were analyzed in parallel by transient transfection of the activated ras in FRTL-5 cells (immunoblotting and RT-PCR in Fig. 1B). The results showed that both the accumulation of the endogenous Fra-1 (upper panel) and the activity of the transfected promoter (lower panel) were strongly increased by the Ras (V12). In addition, we found the fra-1/β-globin reporter exhibited a similar activity when coexpressed with the oncogenic Ras in FRTL-5 cells or when transfected in the Ki-ras-transformed FRTL-5KRas cell line.
FIG. 1.
Roles of different Ras downstream effectors in the induction of Fra-1. (A) Scheme of the activity of the Ras (V12) effector mutants. The C40, G37, and S35 effector loop mutations enable the Ras (V12) oncoprotein to activate selectively the PI3-kinase (C40)-, the RalGDS (G37)-, or the Raf (S35)-dependent cascade. (B) Immunoblotting and RT-PCR analysis of the activity of Ras (V12) effector mutants on the expression of fra-1 in thyroid cells. The fra-1/β-globin reporter construct (10 μg) was cotransfected with the empty vector (pCDNA3) or the indicated Ras expression vector (5 μg) in FRTL-5 cells. As a control of activity in the transformed cell line, the reporter construct was transfected in FRTL-5Kras cells. The total DNA was kept to 20 μg, and 3 μg of the pCMV-CAT reporter was cotransfected as an internal control for transfection efficiency. After 36 h cell extracts or total RNA was prepared. For immunoblotting (upper panel), 50 μg of cell extracts was processed as described in Materials and Methods and was probed with anti-Fra-1 antibody (Santa Cruz Biotechnology, Inc.). As a control for equal loading, the blotted proteins were stained with Red-Ponceau (not shown). For the RT-PCR (lower panel), total RNA was reverse transcribed and the 76-bp β-globin transcript was coamplified with the 346-bp CAT mRNA in the presence of [α32-P]dCTP and was analyzed by 5% PAGE. (C) Diagram of PhosphorImager quantitation (ImageQuant software) of the RT-PCR data. The relative activity of the Ras effector double mutants is expressed as a percentage of the activity of the Ras (V12) construct which resulted in the maximal stimulation of the fra-1/β-globin reporter. In the right-hand diagram the results are shown as fold induction of the reporter gene relative to its activity in FRTL-5 cells. These experiments were repeated three times with similar results. (D) In vitro MBP phosphorylation assay of ERK activation by Ras (V12) effector mutants. The Ras expression constructs or the empty vector were cotransfected in FRTL-5 cells along with the vector encoding the epitope-tagged ERK2 (pcDNA3-ERK2-HA). After 24 h cells were collected and equal amounts of cell lysates were immunoprecipitated (IP) with α-HA antibody or nonimmune serum and were subjected to in vitro phosphorylation reaction as described in Materials and Methods. Reaction products were analyzed by SDS-10% PAGE.
The comparison of the three Ras effector mutants showed that Fra-1 induction was severely affected both with the V12/C40 and with the V12/G37 construct, while a significant activation was still retained with the Ras (V12/S35) derivative. PhosphorImager quantitation and normalization with the internal control revealed less than 5% residual activity relative to Ras (V12) for the V12/C40 and V12/G37 mutants, compared to 35% residual activity detected with the V12/S35 Ras derivative (Fig. 1C). Stimulation of MAP kinase activity by Ras effector mutants in FRTL-5 cells was recorded by in vitro phosphorylation of MBP (Fig. 1D). In agreement with the results reported for other cell types (61), mutants V12/C40 and V12/G37 failed to increase the MAP kinase activity above background levels while the Ras (V12/S35) activated the ERK pathway to the same extent as the oncogenic Ras (V12). Therefore, these data point to an essential role played by the ERK pathway in the Ras-dependent induction of Fra-1 expression.
ERK activity is necessary for both mRNA induction and posttranslational stabilization of Fra-1 but not for its DNA binding activity.
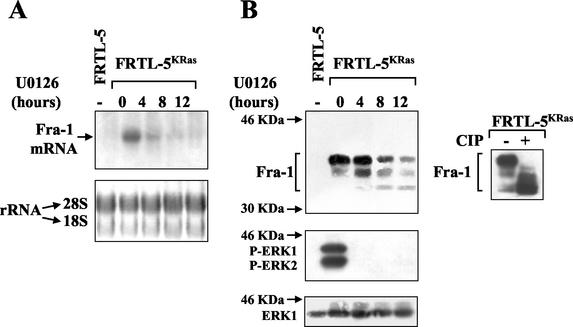
To further investigate the role of the extracellular signal-regulated kinase pathway in the regulation of Fra-1, we analyzed the effect of the MEK/ERK-specific chemical inhibitor (U0126) on Fra-1 protein and mRNA levels in the FRTL-5KRas cell line. Northern blot hybridization showed a drastic reduction of fra-1 expression, which was almost completely suppressed within 4 h from the onset of ERK inhibition (Fig. 2A).
FIG. 2.
Effect of chemical inhibition of the MEK/ERK pathway on Fra-1 expression. FRTL-5KRas cells were treated with 10 μM U0126 (Promega) for 4, 8, or 12 h. Total RNA or whole-cell extracts were prepared from untreated FRTL-5 (lane 1) and FRTL-5KRas (lane 2) cells or from FRTL-5KRas cells treated with U0126 (lanes 3 to 5). (A) For Northern blotting, 30 μg of total RNA was analyzed by hybridization to the radiolabeled rat fra-1 cDNA probe as indicated in Materials and Methods. The ethidium bromide staining of rRNAs (28S and 18S) was utilized as a control for RNA loading. (B) For immunoblotting analysis, 50-μg samples of cell extracts were processed as described in Materials and Methods. The same membrane was first incubated with the α-Fra-1 antibody and subsequently was stripped and reprobed with α-P-ERK1/2 (New England Biolabs) as a control for the inhibitory activity of the drug. Finally, the blot was incubated with α-ERK1 antibody (New England Biolabs) as a control for equal protein loading. For the in vitro dephosphorylation reaction (B, right-hand panel) the cell extracts from untreated FRTL-5KRas cells were incubated for 2 h at 37°C with or without 20 U of CIP prior to SDS-10% PAGE and immunoblotting. Northern blotting and immunoblotting data were confirmed by multiple experiments, and similar results were obtained by using the PD 98059 MEK inhibitor.
Immunoblotting results showed that the inhibition of ERK activity resulted in both quantitative and qualitative changes in the Fra-1 protein (Fig. 2B). During the 12-h time course the amount of Fra-1 declined rather slowly, and the decrease of the 40- to 44-kDa polypeptides was associated with the appearance of a faster-migrating species of about 38 kDa, likely representing a hypophosphorylated (or nonphosphorylated) form of Fra-1. The role of phosphorylation in determining the migration of the proteins was strongly supported by the comigration of the products of in vitro alkaline phosphatase treatment with the fastest-migrating species resulting from the inhibition of the MEK/ERK pathway. As a control of the inhibition of MAP kinase, the effect of the drug was monitored by the use of phosphospecific antibodies recognizing the catalytically active forms of ERK-1 and ERK-2, showing that the phosphorylation of both kinases was completely suppressed within the first time point (4 h) from drug addition (Fig. 2B).
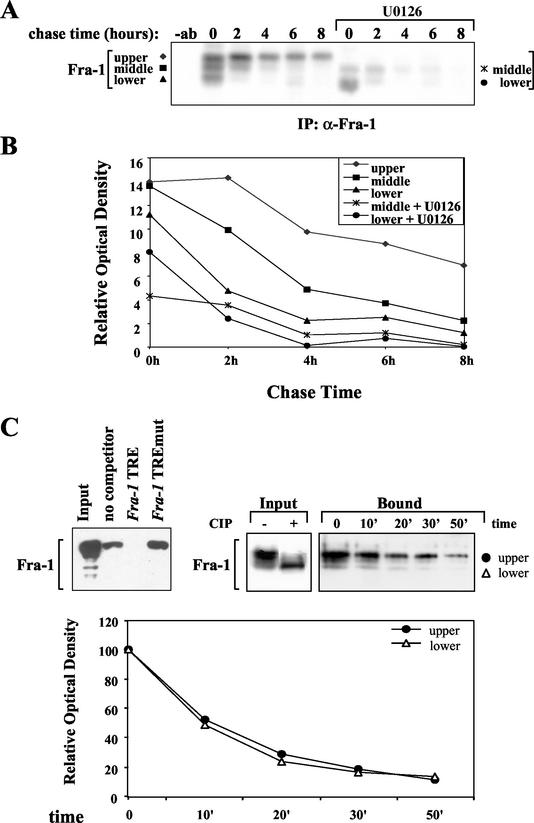
The very abundant expression of Fra-1, along with its extensive posttranslational modification in FRTL-5KRas cells, suggested that its ras-dependent accumulation might be sustained by a posttranslational mechanism in addition to its transcriptional induction. To test whether the ERK-dependent modification was indeed involved in the control of Fra-1 stability, we determined the half-life of the protein by pulse-chase labeling in the absence or presence of the MEK inhibitor in the FRTL-5KRas cells. We found that in the untreated samples the immunoprecipitated pulse-labeled protein was represented by three major electrophoretic species with apparent molecular sizes ranging from 38 to 44 kDa. Time course analysis revealed quite different half-lives for the three isoforms, showing a high stability for the slower species compared to the rapid disappearance of the faster-migrating species and an intermediate stability of the middle isoform, thus suggesting a positive correlation between Fra-1 phosphorylation and stability (Fig. 3A). In the cells treated with the MEK inhibitor, the slower Fra-1 isoform was totally suppressed and the major species was represented by the faster isoform exhibiting the same short half-life of the corresponding isoform detected in the untreated cells. Interestingly, the half-life of the intermediate isoform was not affected by the U0126 treatment, suggesting the existence of Fra-1 modifications independent of ERK activity. Densitometric quantitation confirmed our observations, showing a dramatic difference in half-life between the phosphorylated isoform (about 8 h) and the nonphosphorylated species (less than 2 h), while the half-life of the ERK-independent isoform was about 3.5 h (Fig. 3B). Therefore, the ERK-dependent phosphorylation is essential for the stabilization of Fra-1, although the detection of another phosphorylated isoform, insensitive to MEK inhibition, suggests that at least another kinase activity, independent of MEK pathway, is involved in Fra-1 phosphorylation.
FIG. 3.
Effect of chemical inhibition of the MEK/ERK pathway on Fra-1 stability and DNA binding activity. (A) Pulse-chase analysis of the Fra-1 half-life in FRTL-5KRas cells treated with the MEK inhibitor. Cells pretreated for 30 min with 10 μM U0126 were subjected to pulse-chase labeling, as described in Materials and Methods, alone or in the presence of U0126. Control cells were treated with the vehicle (dimethyl sulfoxide). Cells were collected at the indicated time points. Whole-cell extracts were immunoprecipitated with the α-Fra-1 antibody and were analyzed by SDS-PAGE. −ab, without antibody. (B) Diagram of densitometric quantitation of the results (QuantityOne software) showing the kinetics of decay of distinct Fra-1 electrophoretic isoforms in untreated cells (upper, middle, and lower bands) compared to that of U0126-treated cells (middle and lower bands) expressed as the relative optical density of the autoradiographic image. Similar results were obtained in four different pulse-chase experiments. (C) Immunoblotting analysis of Fra-1 following DNA affinity chromatography of nuclear extracts subjected to in vitro dephosphorylation. The upper left-most panel shows 25 μg of nuclear extract from FRTL-5KRas cells incubated with 5 μg of fra-1 TRE DNA-agarose beads in the absence of competitor oligonucleotide (no competitor) or after preincubation with a 25-fold molar excess of fra-1 TRE or mut fra-1 TRE competitor oligonucleotides. The eluted TRE-bound material was subjected to SDS-10% PAGE along with the same amount of untreated proteins (input). The two upper right panels show 25 μg of nuclear extract subjected to in vitro dephosphorylation (+CIP) and compared to the untreated control (−CIP). The lower panel is on off-rate analysis of an equivalent untreated sample (25 μg) subjected to DNA affinity chromatography, as described in Materials and Methods. A 25-fold molar excess of competitor fra-1 TRE oligonucleotide was added, and the DNA-agarose-bound complex was allowed to dissociate at room temperature during the indicated time course prior to elution and SDS-PAGE. The diagram was obtained by quantitation of the chemiluminescence signal by use of the Gel-Doc image acquisition apparatus and QuantityOne software (Bio-Rad). The data, representing the average of two independent experiments, were normalized by attributing the 100% value to the first time point for each of the two electrophoretic isoforms (upper and lower). IP, immunoprecipitation.
It has been previously reported that in vitro phosphorylation by MAP kinase affects DNA binding activity of both Fra-1 and Fra-2 in mouse fibroblasts (24). To test whether the in vivo modification might play a role in the DNA binding activity of Fra-1 in the FRTL-5KRas cells, we analyzed the effect of the in vitro dephosphorylation of the protein on its binding to a consensus TRE. To this end we performed DNA affinity chromatography by linking a biotinylated oligonucleotide containing the high-affinity AP-1 binding element of the fra-1 first intron (fra-1 TRE; see below) to streptavidin-agarose beads. Following pull-down the bound and unbound fractions were analyzed by immunoblotting. Competition with wild-type and mutated oligonucleotides ensured the specificity of binding to the streptavidin-bound TRE (Fig. 3C, left panel).
The pattern of Fra-1 electrophoretic isoforms eluted from the solid support (bound) was identical to that of the input (Fig. 3C, lane input −CIP) of the in vitro binding reaction, and the relative migration of the faster TRE-bound isoform was the same as that of the faster species generated by in vitro dephosphorylation (Fig. 3C, lane input + CIP), thus suggesting that phosphorylation is not essential for the binding of Fra-1 to DNA (Fig. 3C). In order to rule out possible quantitative differences of affinity between isoforms, we analyzed the dissociation rate of the faster (lower) compared to that of the slower (upper) isoform in the presence of an excess of free competitor oligonucleotide. The results, quantified and represented in the diagram of Fig. 3C, show the same dissociation rate for the two electrophoretic isoforms, further supporting the evidence that the ERK-dependent phosphorylation does not affect the DNA binding activity of the Fra-1-containing AP-1 complexes expressed in FRTL-5KRas cells. In summary, the constitutive activation of the ERK pathway is involved in the control of both synthesis (Fig. 2) and stability (Fig. 3) of Fra-1, but it is not essential for its DNA binding activity (Fig. 3) in ras-transformed thyroid cells.
The ras-dependent induction of fra-1 is mediated by an AP-1 element within its first intron.
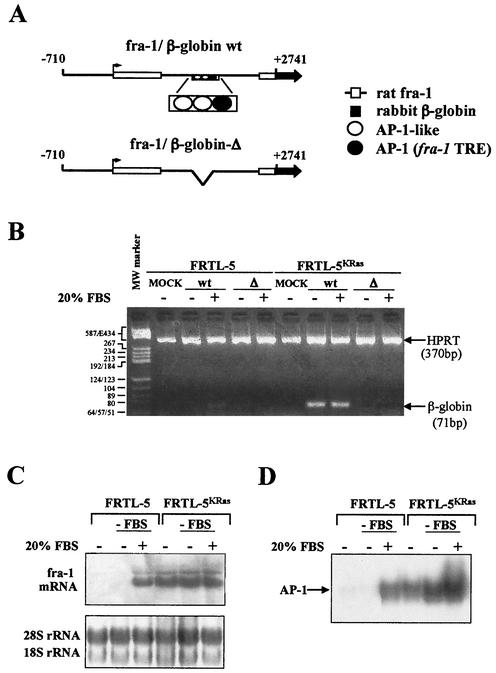
The data depicted in Fig. 1 showed that the fra-1/β-globin fusion construct utilized as a reporter along with the Ras (V12) effector mutants is functional as a target of the oncogenic Ras. Such a construct (Fig. 4A), encompassing the rat fra-1 promoter region and including the first exon, the first intron, and part of the second exon (−710 to +2741) and fused to the rabbit β-globin gene (exons 2 and 3), has previously been utilized to identify regulatory sequences within the fra-1 first intron (10). It was shown that one AP-1 and two AP-1-like elements play an essential role in the activation of the fra-1 promoter by c-Fos and other AP-1 components. To test the role played by these regulatory sites in the ras-dependent induction of the gene, we isolated pools of stably transfected cell clones expressing the fra-1/β-globin gene fusion and containing either the wild-type sequence or a small deletion encompassing the AP-1 elements. Southern hybridization revealed that a similar copy number of fra-1/β-globin genes was integrated in the pools of stably transfected cells (data not shown). The results showed that the fra-1/β-globin transcript, undetectable in FRTL-5 cells, was strongly accumulated in the FRTL-5KRas cell line. In addition, serum stimulation of FRTL-5 cells resulted in a smaller induction of the chimeric transcript (Fig. 4B). The serum-dependent expression of the fra-1 promoter in the normal cells and its oncogene-dependent induction in FRTL-5KRas cells paralleled the expression of the endogenous fra-1 mRNA (Fig. 4C), indicating that the reporter construct contained sequences necessary and sufficient for serum- and ras-dependent induction. The deletion of the AP-1 sites was associated with the complete disappearance of the fra-1/β-globin transcript, both in the Ki-ras-transformed cell line and in serum-stimulated normal cells. Therefore, the AP-1 elements localized in the first intron are essential for the full oncogene-dependent induction of the fra-1 promoter.
FIG. 4.
Characterization of the Ras-responsive element of the fra-1 gene. (A) Schematic representation of the fra-1/β-globin reporter constructs. The −710 to +2741 region of the rat fra-1 gene was fused to a portion of the rabbit β-globin gene (exons 2 and 3, black arrow) to generate a stable chimeric transcript. The fra-1-derived sequence includes the (−710) promoter region, the first exon, the first intron, and part of the second exon (white boxes). The first intron contains two AP-1-like sites (empty circles) and an AP-1 consensus element (fra-1 TRE, black circle) deleted in the mutated version (fra-1/β-globin-Δ). For stable clones, FRTL-5 and FRTL-5KRas cell lines were transfected with the DNA vector encoding the selectable marker (pCMV-Neo), alone (MOCK clones) or in combination with the linearized fra-1/β-globin wild type (wt) or the fra-1/β-globin-Δ construct. After selection in G418 (800 μg/ml; Calbiochem), pools of stably transfected cell clones were derived from a similar number (∼80 to 100) of G418-resistant colonies. The comparable copy number of stably integrated constructs in each pool of G418-resistant transfectants was verified by Southern blot hybridization with a rabbit β-globin probe detecting an internal 3.8-kb EcoRI fragment (data not shown). (B) RT-PCR analysis of the basal and serum-inducible expression of the chimeric transcript in the FRTL-5- and FRTL-5KRas-derived pools of transfected cell clones. Cells were maintained in normal growth conditions before stimulation with 20% fetal calf serum for 3 h. Total RNA was prepared from MOCK cell clones (lanes 1 and 6) and from cell clones expressing the wild type (lanes 2 to 3 and 7 to 8) or the deletion-containing (lanes 4 to 5 and 9 to 10) chimeric minigene. For RT-PCR, 2 μg of DNase-treated RNA was reverse transcribed as described in Materials and Methods and the 76-bp β-globin cDNA fragment was amplified together with the 370-bp HPRT cDNA as an internal control. PCR products were resolved by 4% agarose gel electrophoresis. (C and D) Serum induction of fra-1 and mRNA and protein binding to fra-1 TRE. FRTL-5 and FRTL-5KRas cells were maintained in complete medium or were switched into a medium containing 0.5% FBS for 48 h, and then 20% FBS was added 3 h prior to harvest. Total RNA or nuclear proteins were extracted from cycling (lanes 1 and 4), serum-arrested (lanes 2 and 5), and serum-stimulated (lanes 3 and 6) cells. (C) For Northern blot analysis, 30 μg of total RNA/sample was hybridized to the radiolabeled rat fra-1 cDNA probe, as indicated in Materials and Methods. Equal loading was verified by ethidium bromide staining of rRNAs (bottom panel). (D) EMSA of the serum-induced binding to the fra-1 TRE. Nuclear proteins were incubated with the 5′-end-labeled fra-1 TRE oligonucleotide before 5% polyacrylamide gel retardation. The arrows indicate the oligoncleotide/AP-1 complex. All results were confirmed in at least three independent experiments.
It has been shown that, among the three AP-1-like sequences clustered within a 40-bp region of the fra-1 first intron, only the high-affinity AP-1 binding site is able to bind in vitro the Fos/Jun heterodimer, and it is essential for the c-Fos-responsive activity (10). Thus, we investigated the interaction of this element with the AP-1 complex in normal and ras-transformed cells. Gel retardation revealed a very high constitutive and weakly serum-inducible level of nuclear protein binding to the fra-1 TRE in the Ki-ras-transformed cell line compared to the very low but strongly serum-inducible binding level of the FRTL-5 cell line (Fig. 4D).
Fra-1 mediates its MEK-dependent positive autoregulation by interacting in vitro and in vivo with the fra-1 TRE.
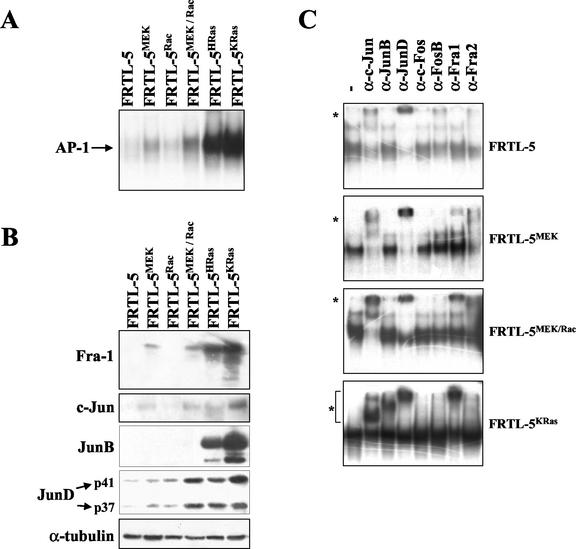
The results depicted in Fig. 1 and 2 indicate that the activity of the MEK-dependent pathway is necessary for Fra-1 accumulation but is insufficient for full induction of the fra-1 promoter. To understand the role of the MEK pathway and its possible cooperation with other Ras-dependent pathways in regulating the interaction of AP-1 with its essential target site in the fra-1 promoter, we investigated protein binding to the fra-1 TRE in cell clones stably expressing the constitutively active derivative of MEK alone or in combination with a mutationally activated Rac isoform. For this purpose we utilized the previously described FRTL-5-derived cell clones (16) expressing constitutive MEK-1 (MEKΔN3/S218E/S222D), Rac-1 (Rac-1/V12), or both (MEK/Rac). The results showed that the amount of fra-1 TRE-bound complex was significantly increased in the MEK-expressing cell clone, was unaffected in the FRTL-5Rac cell clone, and was further augmented in the FRTL-5MEK/Rac cell clone with respect to the FRTL-5 parental cell line. However, the binding to the fra-1 TRE in the FRTL-5KRas- and FRTL-5HRas-transformed cells was much stronger compared to that of cell clones expressing both constitutive activators (Fig. 5A).
FIG. 5.
In vitro analysis of the fra-1 TRE binding complex in the cell clones expressing the MEK and Rac constitutive derivatives. (A) EMSA of AP-1 binding to the fra-1 TRE oligonucleotide in the FRTL-5-derived cell clones expressing the constitutively active form of MEK and/or Rac (FRTL-5MEK, FRTL-5Rac, and FRTL-5MEK/Rac). Nuclear proteins (3 μg) were incubated with the labeled fra-1 TRE oligonucleotide before PAGE. (B) Immunoblotting analysis of Fra-1 and Jun proteins in the normal (FRTL-5), Ha-ras- or Ki-ras-transformed (FRTL-5Hras and FRTL-5KRas), and stably transfected cell lines (FRTL-5MEK, FRTL-5Rac, and FRTL-5MEK/Rac). Nuclear proteins were separated by SDS-PAGE (20 μg/lane) and were transferred to a polyvinylidene difluoride membrane. Western blots were sequentially incubated with anti-Fra-1, anti-c-Jun, anti-JunB, and anti-JunD followed by anti-α-tubulin antibodies as a control for equal loading. The arrows indicate the major isoforms of Fra-1 (38 kDa), c-Jun (39 kDa), and JunB (38 kDa) and the two isoforms of JunD (41 and 37 kDa). (C) Antibody supershift analysis of the complex bound to the fra-1 TRE oligonucleotide. After protein binding to the labeled oligonucleotide, nuclear extracts were incubated for 3 h with the indicated antibodies before gel retardation. Different autoradiographic exposures were chosen for different panels (FRTL-5, 24 h; FRTL-5MEK and FRTL-5 MEK/Rac, 16 h; FRTL-5Kras, 8 h) to allow the optimal visualization of supershifted complexes. The arrows indicate the AP-1/oligonucleotide complex, while the asterisks refer to the supershifted ternary complexes. In vitro binding and supershift assays were repeated at least twice with comparable results.
To investigate the correlation between the observed TRE binding activity and the level of Fra-1 expression, we analyzed by immunoblotting the FRTL-5-derived cell clones and found that Fra-1, essentially undetectable in the exponentially growing FRTL-5 cells, was accumulated in the cell clone expressing the constitutively active MEK but not in the cell clones expressing the constitutive Rac (V12) derivative (Fig. 5B). In the cell clone stably expressing both of the constitutive activators (FRTL-5MEK/Rac) we found a further increased level of Fra-1 with respect to the FRTL-5MEK cell line. The fully transformed lines, however, exhibited much higher Fra-1 accumulation than the FRTL-5MEK, FRTL-5Rac, and FRTL-5MEK/Rac cell clones, with higher Fra1 overexpression in the Ki-ras-transformed cells than in the Ha-ras-transformed cells (Fig. 5B).
We also analyzed the expression of the three Jun family members in the FRTL-5MEK, FRTL-5Rac, and FRTL-5MEK/Rac cell clones (Fig. 5B). The level of both c-Jun and JunD was increased in the presence of both the constitutively active MEK and Rac, with a slightly higher accumulation in the cell clone expressing both activators. However, JunB was undetectable in each of the stably transfected cell clones, thus suggesting that alternative or additional pathways are required for its expression. Therefore, in agreement with the data of Fig. 1, constitutive activation of the MEK pathway is quantitatively insufficient to reproduce the strong increase of Fra-1 elicited by the ras oncogene. In addition, the level of the Jun family partners indicates that the fra-1 gene product accumulated in the MEK and MEK/Rac cell clones is likely to be present in the form of heterodimeric complexes with c-Jun or JunD.
We therefore investigated the composition of the complex in the cell clones exhibiting increased binding to the fra-1 first intron by gel supershift analysis with antibodies recognizing individual members of the Jun and Fos protein families. The data showed that Fra-1 was absent from the complex bound to the fra-1 TRE in the parental cell line, while it gave rise to a weak retarded complex in the FRTL-5MEK clone, an intermediate supershift in FRTL-5MEK/Rac, and a strong supershifted complex in FRTL-5KRas (Fig. 5C). The data with the Jun-specific antibodies revealed that, while c-Jun and JunD were both present in the four cell lines and exhibited an increased supershift in the fully transformed cells and in the MEK and MEK/Rac cell clones, JunB was detectable only in the FRTL-5KRas cells, as predicted from the results shown in Fig. 5B. On the basis of the in vitro gel retardation data, it can be predicted that three different Fra-1-containing heterodimers (Fra-1/c-Jun, Fra-1/JunB, and Fra-1/JunD) can compete for the binding to the c-Fos- and ras-oncogene-responsive element of the fra-1 promoter in the Ki-ras-transformed cells.
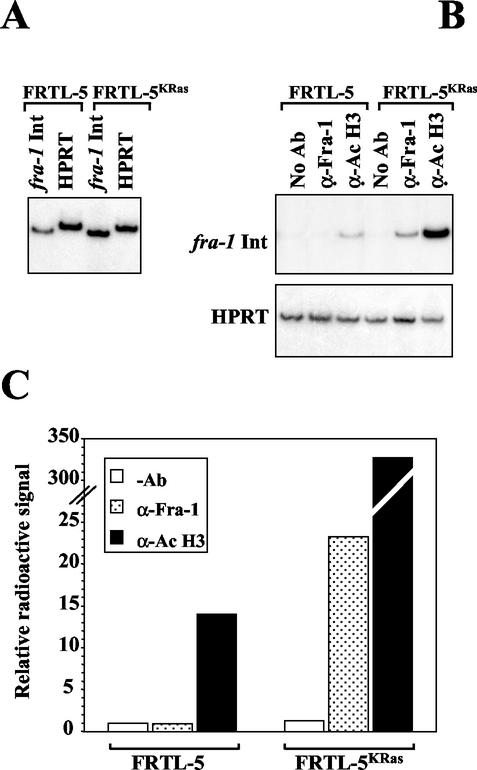
To verify the interaction of Fra-1 with its genomic regulatory sequence within the native chromatin environment, we performed ChIp of in vivo cross-linked extracts from FRTL-5 and FRTL-5KRas cells followed by PCR with primers encompassing the fra-1 TRE region. As a positive control for the amplification, we first checked by PCR the naked DNA extracted from FRTL-5 and FRTL-5KRas cells (Fig. 6A). The results of the ChIp with the Fra-1-specific antibody showed almost no amplification product with the extract from FRTL-5 cells, while a considerable amount of amplified fragment could be detected with the FRTL-5KRas chromatin immunoprecipitate (Fig. 6B). To investigate the correlation with the status of chromatin activation within the analyzed region, we also performed ChIp with the antibody recognizing the acetylated isoform of histone H3 (α-AcH3). Unlike Fra-1, some acetylated histone H3 was detected in the parental cell line, but the relative intensity of the amplification products revealed a dramatic increase of the acetylated histone H3 bound to the fra-1 intronic region in the FRTL-5KRas compared to that of the FRTL-5 cell line. Densitometric scanning revealed a very similar fold increase (about 25-fold) for the acetylated histone and Fra-1 (compare the ratio between the α-AcH3 black bars of FRTL-5Kras versus those of FRTL-5 to the ratio between the α-Fra-1 dotted bars of FRTL-5Kras versus those of FRTL-5 in Fig. 6B). Hence, the fra-1 gene product is bound in vivo to its transcriptionally active genomic region in ras-transformed cells, and its binding is associated with the increased acetylation of histone H3 within the surrounding chromatin region.
FIG. 6.
In vivo occupancy of the fra-1 TRE in normal and ras-transformed cells. (A) Radioactive PCR on naked DNA from normal and transformed cells as a control for the amplification products. Chromosomal DNA extracted from FRTL-5 and FRTL-5KRas was analyzed by radioactive PCR. The fra-1-Int primers amplified a 594-bp DNA region containing the intronic fra-1 TRE, while the primers for the HPRT gene gave rise to a 558-bp PCR product. (B) ChIp of the +178- to +772-nucleotide region encompassing the fra-1 TRE. Following in vivo formaldehyde cross-linking, chromatin extracted from FRTL-5 and FRTL-5KRas cells was immunoprecipitated with anti-Fra-1 or anti-acetylated histone H3 antibodies (α-AcH3). Following reversal of cross-linking, the purified DNA fragments were amplified by radioactive PCR with the fra-1-Int and HPRT primers as a control for equal input. The products were resolved by native gel electrophoresis and were detected by autoradiography. (C) The radioactive signal of the amplification products was quantified by PhosphorImager (with ImageQuant software), normalized for the HPRT internal control and expressed as relative to the no-antibody (−Ab)/FRTL-5 control sample. The reproducibility of these results was confirmed by repeating the ChIp experiments two times independently.
The ras-inducible transactivating function of Fra-1 requires the recruitment of heterodimeric partners.
We then asked whether the function of Fra-1 was solely to allow the stable DNA binding of the Jun family partners required for recruitment of transcriptional coactivators or if Fra-1 itself might contribute to the transcriptional activation mediated by the AP-1 complex in ras-transformed cells. We postulated that a latent Fra-1 transactivation domain might be activated by the ERK-mediated phosphorylation triggered by the oncogenic Ras. Therefore, we analyzed the Ras-responsive activity of GAL4/Fra-1 chimeras by coexpression with RasVal12 in FRTL-5 cells. As controls, we adopted the GAL4/c-Fos fusion, along with the GAL4/Fra-1-ΔZip and GAL4/c-Fos-insZip derivatives, carrying mutations that inactivate the dimerization function of the proteins.
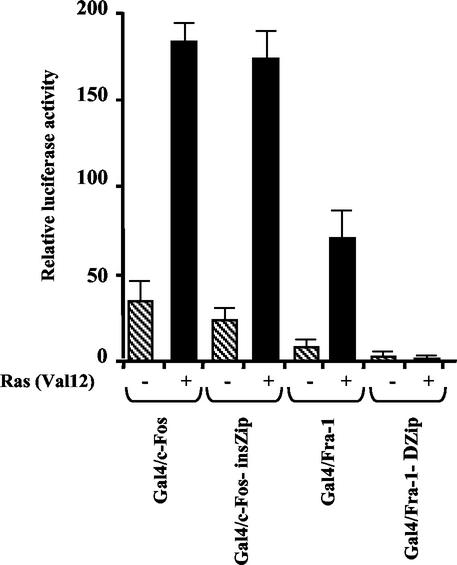
The results (Fig. 7) showed that the GAL4/Fra-1 fusion could efficiently activate the target promoter, although it did so (about fourfold) less efficiently than the c-Fos-containing chimera, and the activity of both fusion proteins was significantly increased (more than fivefold) by coexpression with the Ras (Val12) oncoprotein. Similar results were obtained by cotransfection with the vector expressing the constitutively active MEK derivative (MEK-EE) (data not shown).
FIG. 7.
Analysis of the Fra-1-dependent transactivation in response to the ras oncogene and transactivation activity of Gal4/Fra-1 fusion proteins. The reporter plasmid FrLuc (5 μg) was coexpressed with the vectors (10 μg) expressing the Gal4/Fra-1, GAL4/Fra-1-ΔZip, GAL4/c-Fos, or GAL4/c-Fos-insZip chimeric protein, along with the pCDNA3-Ras (V12) expression vector (5 μg) or the pCDNA3 empty vector. As an internal control, a vector encoding Renilla Luciferase (0.5 μg) was cotransfected. Thirty-six hours after transfection cells were harvested and assayed for both firefly and Renilla luciferase activities (Dual-Luciferase Reporter Assay system) to allow for normalizing of the FrLuc reporter activity for variations of transfection efficiency. The diagram shows the relative luciferase activity in the absence (hatched boxes) or presence (black boxes) of pCDNA3-Ras (V12). The results represent the average of three independent experiments, with the error bars indicating the standard errors.
However, an important difference between the two fusion proteins emerged from the functional assay of the respective dimerization-defective derivatives: while GAL4/c-Fos-insZip exhibited a significant basal level and a Ras-dependent fold induction even higher than that of GAL4/c-Fos, GAL4/Fra-1-ΔZip displayed only a background basal activity which was unaffected by the coexpressed ras oncogene. These findings indicate that although both GAL4/c-Fos and GAL4/Fra-1 are capable of transducing the effects of Ras-dependent signaling on the heterologous promoter, the action of Fra-1 requires the leucine zipper-mediated recruitment of a transcriptionally active partner, thus supporting the notion that Fra-1 does not harbor an autonomous transactivation domain.
DISCUSSION
We have studied the effect of the ras oncogene on the AP-1 component Fra-1 in rat thyroid cells and found that the high constitutive activity of Fra-1 results from the conjunction of multiple mechanisms, including transcriptional induction, posttranslational stabilization, and recruitment of transcriptionally active partners (summarized in Fig. 8). We have shown that such control mechanisms largely depend on the activity of the MEK/ERK phosphorylation cascade, in agreement with the essential role played by the Raf-dependent pathway in neoplastic transformation (56), and the requirement of ERK-mediated signaling for the induction of Fra-1 synthesis (18, 55). However, in the rat thyroid epithelial cell system the cooperation with other pathways, in addition to the MEK- and Rac-dependent signaling, appears necessary for optimal signaling from the constitutive Ras oncoprotein to the fra-1 promoter. In addition, we have found that ERK-dependent phosphorylation plays an essential role in the stabilization of the protein in ras-transformed cells while it is not essential for Fra-1 DNA binding activity.
FIG. 8.
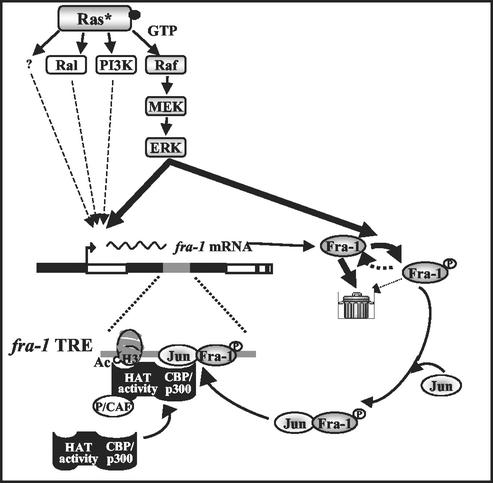
Model of multistep positive autoregulation of Fra-1 in ras-transformed cells. The thickness of the arrows originating from the constitutively GTP-bound oncogene (Ras*) indicates the relative contribution of different Ras-dependent pathways in the activation of the fra-1 promoter. Following ERK-dependent phosphorylation, Fra-1 is stabilized in transformed cells and interacts with Jun family partners. The Jun/Fra-1 heterodimer can activate fra-1 gene transcription by binding the autoregulatory site in its first intron (fra-1 TRE) and recruiting a transcriptional coactivator (CBP/p300). White boxes, fra-1 exons; thick black line, fra-1 5′-flanking and first intron; grey line, fra-1 TRE; trash bin, degradation apparatus.
It has been previously shown that, during the mitogenic response of mouse fibroblasts, the delayed kinetics of the serum-dependent induction of fra-1 requires the presence of intact c-fos and c-jun gene products (48) acting through regulatory sequences within the fra-1 first intron. In this respect, fra-1 was depicted as a unique member of the fos gene family under direct control of AP-1 activity, although a weaker AP-1-dependent induction also has been described for fra-2 (10). Here we show that the previously characterized intronic regulatory element is essential for the ras-dependent induction of the fra-1 promoter, that Fra-1 is the predominant component of the AP-1 complex interacting in vitro and in vivo with the fra-1 intronic site, and that the activation of the surrounding chromatin region is reflected by the increased histone H3 acetylation. Therefore, in response to the activated ras oncogene, Fra-1 becomes implicated in its serum-independent positive autoregulation, which is likely to play a major role in maintaining a constitutively high level of AP-1 activity in transformed cells.
The identification of Fra-1 as part of an in vivo transcriptionally active complex raised the question of its role as a bona fide transcriptional activator. The original report showing that Fra-1 lacks an autonomous transactivation domain (10) is in apparent contradiction with a recent study that supports the evidence of its phosphorylation-dependent transactivating activity (64). Our results, showing that the activity of the GAL4 fusion protein is strongly inducible by Ras, apparently support the hypothesis of a phosphorylation-dependent transactivation domain within the Fra-1 molecule. However, unlike c-Fos, the requirement for Fra-1 of an intact leucine zipper suggests that the Ras-dependent transactivation may depend on the recruitment of a heterodimeric partner. We propose that the major role of Fra-1 is to allow the DNA binding and transcriptional activation mediated by its Jun partners and that the ERK-dependent phosphorylation is not needed for transactivation, although it cannot be ruled out that an inducible transactivation domain, modulated by phosphorylation of the essential Thr-231 residue (64), might act in cooperation with functional domains of the heterodimeric partner. Interestingly, a recent study showing the discrepancy between the transactivation potential and the ability to induce cellular morphological alterations by Fos family members led to the suggestion that Fra-1 biological activity might reflect its specific role within the molecular context of natural enhancers driving the expression of genes selectively upregulated by Fra-1 rather than c-Fos and Fra-2, such as the uPA (urokinase plasminogen activator) gene (3).
Our analysis shows multiple roles for the ERK-dependent regulation of Fra-1, including the MAP kinase-dependent stabilization of the protein in ras-transformed cells. The role of MAP kinase in controlling the half-life of Fra-1 in response to serum mitogens has been described previously in a detailed study on the regulation of Fra-1 and Fra-2 phosphorylation during the cell cycle of murine fibroblasts (24). We have found that ras transformation results in the serum-independent Fra-1 stabilization, which likely supports the constitutively high levels of the protein in transformed cells. It remains to be investigated whether the expression of the protein is totally deregulated or is still subjected to cell cycle-dependent variations in the transformed cell lines.
The relationship between phosphorylation and protein stabilization has been analyzed in detail for the two prototype AP-1 components, c-Fos and c-Jun. The first evidence on the role of MAP kinases in c-Fos stabilization came from the study of the v-mos-dependent c-Fos stabilization (38), which is likely unrelated to neoplastic transformation, since mos is not involved in human tumorigenesis. Recently it has been shown that the duration of ERK signaling allows c-Fos to discriminate between sustained (mitogenic) and transient (nonmitogenic) stimuli, by a complex mechanism involving multiple sequential phosphorylation steps, required for full protein stabilization. Interestingly, it has been proposed that an identical putative DEF (ERK docking) domain, identified both in Fra-1 and Fra-2 (but not in FosB), might be implicated in similar signaling mechanisms (36). Accordingly, the cluster of serine and threonine residues previously identified as MAP kinase phosphorylation sites in the Fra-2 carboxy-terminal region is adjacent to the putative DEF domain (34) and includes the conserved Thr-231 residue of Fra-1 which, although not yet biochemically identified as a phosphoacceptor site, was found to be essential for ERK-dependent transactivation in response to tetradecanoyl phorbol acetate (64). An additional level of complexity, implying a possible stimulus-specific regulation of Fra-1 phosphorylation, has been suggested from the recent analysis of Fra-1 modification in response to insulin, which showed that the activation of ERK1/ERK2 resulted in serine (but not threonine) phosphorylation of the carboxy-terminal region of Fra-1 (26).
The essential role of c-Jun amino-terminal phosphorylation in ras-mediated transformation (53) and in protection from ubiquitin-dependent degradation (37) has been well established. In our experimental system we have found that the increased half-life is a major determinant of ras-dependent c-Jun accumulation, and we analyzed the mechanism of the stabilization in the transformed thyroid cell lines (unpublished data). In summary, at least for c-Jun and Fra-1, the Ras- and phosphorylation-dependent stabilization appears to be an important regulatory mechanism which might favor the accumulation of c-Jun/Fra-1 heterodimers during tumorigenesis.
Since the induction of Fra-1 and Fra-2 cannot take place in the absence of sustained MEK/ERK activation (55), both proteins would be synthesized and immediately converted into the phosphorylated stable isoforms. Therefore, it can be speculated that another important regulatory step, potentially altered during oncogenesis, would be the dephosphorylation of Fra-1 and/or Fra-2, following their ERK-dependent synthesis and modification, to allow their turnover and interchange with other Fos family members within the AP-1 complex during the cell cycle.
It has been proposed that, both for Fra-1 and Fra-2 (but not c-Fos and FosB), MAP kinase phosphorylation might also regulate DNA binding activity (24). In our cell system we have shown that phosphorylation is not essential for the DNA binding of Fra-1. To explain the apparent discrepancy, it can be observed that the previous study relied on the in vitro phosphorylation of recombinant proteins, while we have analyzed the in vivo phosphorylated Fra-1. Alternatively, the effect on DNA binding might be specific for c-Jun/Fra-1 (analyzed in the previous study) but not for the JunB/Fra-1 and JunD/Fra-1 heterodimers, which represent a significant component of the AP-1 complex in ras-transformed thyroid cells.
Besides the common ERK-dependent control on their synthesis and stability, important differences exist in the ras-dependent regulation of Fra-1 and Fra-2. In the thyroid cell system we have found a dramatic induction of the Fra-1 transcript compared to small variations of the Fra-2 mRNA (57). Importantly, in rat fibroblasts a genome-wide survey of ras transformation targets allowed the identification of Fra-1 as the most upregulated transcription factor (over 100-fold), pinpointing Fra-1 as the major AP-1 component in ras-transformed fibroblasts, while Fra-2 was not detected.
The study of the role of Fra-1 in the cell cycle will take great advantage of the utilization of fra-1 knockout cell lines. The availability of primary fibroblasts derived from mouse embryos lacking individual AP-1 components allows the establishment of important relationships between AP-1 and cell cycle regulators, showing the dual effects of c-Jun, both as a positive regulator of cyclin D1 (6, 62) and a negative regulator of the p21 cdk cyclin inhibitor (47), and JunB, as a negative regulator of cyclin D1 (6) and a positive regulator of the p16 cell cycle inhibitor (41). The analysis of c-fos−/− and fosB−/− mouse embryo fibroblasts revealed redundancy among these fos family members, showing that while the lack of individual components had no effect on cell cycle control, the double knockout exhibited a proliferation defect, at least partially consequent to insufficient induction of cyclin D1 promoter (11). Since the in vitro analysis did not show any cell-autonomous proliferation defect in fra-1 null fibroblasts (49) and because of a mitogen-dependent regulation very similar to that of fra-2 (24, 28), it will be important to explore both the potential functional overlap between the fra-1 and fra-2 gene products and the possibility that the two Fos family members might play nonredundant roles in cell types different from fibroblasts.
A major function of Fra-1 might be unrelated to normal cell cycle control. In light of the evidence suggesting a role for Fra-1 in tumor progression in a variety of naturally occurring neoplasms and experimental models, including breast (29, 42), lung (44), epidermis (64), and thyroid (15), the accumulation of Fra-1 and the activation of target genes in tumor cells would require multiple genetic lesions. In this respect it can be noticed that the above-mentioned survey of ras transformation targets was based on differential screening between a preneoplastic cell line and the malignant ras-transformed derivative (65). Likewise, the analysis of the Fra-1 ability to induce morphological transformation and activation of multiple transcriptional targets was based on the ectopic overexpression of the protein in a mouse mammary adenocarcinoma revertant cell line, selected for the complete loss of metastatic potential, compared to that of the parental malignant cell line (3, 29). Interestingly, the Fra-1-upregulated gene products (mts1, uPA, uPAR, and PAI-1) have been strongly implicated in metastasis, cell migration, and angiogenesis (1, 4), thus indicating the crucial role played by Fra-1 in tumor progression.
Acknowledgments
We thank Julian Downward for providing the plasmids expressing the Ras effector mutants and Meinrad Busslinger for fra-1/β-globin reporter constructs and the Gal4/Fra-1 fusion derivatives. We are grateful to Maurizio D'Esposito and Maria R. Matarazzo for help with ChIp and to Rita Vito for technical assistance. We thank Roberto Di Lauro for the generous gift of the FRTL-5-derived cell clones. We also thank Alfredo Fusco and Massimo Santoro for cell lines and helpful discussions.
This work was supported by grants from Associazione Italiana per la Ricerca sul Cancro, from Regione Campania (L.R. 41/94), and from MURST-CNR Program Legge 488/92 (cluster 02).
REFERENCES
- 1.Ambartsumian, N., J. Klingelhofer, M. Grigorian, C. Christensen, M. Kriajevska, E. Tulchinsky, G. Georgiev, V. Berezin, E. Bock, J. Rygaard, R. Cao, Y. Cao, and E. Lukanidin. 2001. The metastasis-associated Mts1(S100A4) protein could act as an angiogenic factor. Oncogene 20:4685-4695. [DOI] [PubMed] [Google Scholar]
- 2.Ambesi-Impiombato, F. S., L. A. Parks, and H. G. Coon. 1980. Culture of hormone-dependent functional epithelial cells from rat thyroids. Proc. Natl. Acad. Sci. USA 77:3455-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen, H., S. Mahmood, V. Tkach, M. Cohn, O. Kustikova, M. Grigorian, V. Berezin, E. Bock, E. Lukanidin, and E. Tulchinsky. 2002. The ability of Fos family members to produce phenotypic changes in epithelioid cells is not directly linked to their transactivation potentials. Oncogene 21:4843-4848. [DOI] [PubMed] [Google Scholar]
- 4.Andreasen, P. A., R. Egelund, and H. H. Petersen. 2000. The plasminogen activation system in tumor growth, invasion, and metastasis. Cell Mol. Life Sci. 57:25-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angel, P., A. Szabowski, and M. Schorpp-Kistner. 2001. Function and regulation of AP-1 subunits in skin physiology and pathology. Oncogene 20:2413-2423. [DOI] [PubMed] [Google Scholar]
- 6.Bakiri, L., D. Lallemand, E. Bossy-Wetzel, and M. Yaniv. 2000. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J. 19:2056-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakiri, L., K. Matsuo, M. Wisniewska, E. F. Wagner, and M. Yaniv. 2002. Promoter specificity and biological activity of tethered AP-1 dimers. Mol. Cell. Biol. 22:4952-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Battista, S., F. de Nigris, M. Fedele, G. Chiappetta, S. Scala, D. Vallone, G. M. Pierantoni, T. Mega, M. Santoro, G. Viglietto, P. Verde, and A. Fusco. 1998. Increase in AP-1 activity is a general event in thyroid cell transformation in vitro and in vivo. Oncogene 17:377-385. [DOI] [PubMed] [Google Scholar]
- 9.Behrens, A., W. Jochum, M. Sibilia, and E. F. Wagner. 2000. Oncogenic transformation by ras and fos is mediated by c-Jun N-terminal phosphorylation. Oncogene 19:2657-2663. [DOI] [PubMed] [Google Scholar]
- 10.Bergers, G., P. Graninger, S. Braselmann, C. Wrighton, and M. Busslinger. 1995. Transcriptional activation of the fra-1 gene by AP-1 is mediated by regulatory sequences in the first intron. Mol. Cell. Biol. 15:3748-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, J. R., E. Nigh, R. J. Lee, H. Ye, M. A. Thompson, F. Saudou, R. G. Pestell, and M. E. Greenberg. 1998. Fos family members induce cell cycle entry by activating cyclin D1. Mol. Cell. Biol. 18:5609-5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown, P. H., R. Alani, L. H. Preis, E. Szabo, and M. J. Birrer. 1993. Suppression of oncogene-induced transformation by a deletion mutant of c-jun. Oncogene 8:877-886. [PubMed] [Google Scholar]
- 13.Campbell, S. L., R. Khosravi-Far, K. L. Rossman, G. J. Clark, and C. J. Der. 1998. Increasing complexity of Ras signaling. Oncogene 17:1395-1413. [DOI] [PubMed] [Google Scholar]
- 14.Casamassimi, A., M. G. Miano, A. Porcellini, G. De Vita, F. de Nigris, M. Zannini, R. Di Lauro, T. Russo, V. E. Avvedimento, and A. Fusco. 1998. p53 genes mutated in the DNA binding site or at a specific COOH-terminal site exert divergent effects on thyroid cell growth and differentiation. Cancer Res. 58:2888-2894. [PubMed] [Google Scholar]
- 15.Chiappetta, G., G. Tallini, M. C. De Biasio, F. Pentimalli, F. de Nigris, S. Losito, M. Fedele, S. Battista, P. Verde, M. Santoro, and A. Fusco. 2000. FRA-1 expression in hyperplastic and neoplastic thyroid diseases. Clin. Cancer Res. 6:4300-4306. [PubMed] [Google Scholar]
- 16.Cobellis, G., C. Missero, and R. Di Lauro. 1998. Concomitant activation of MEK-1 and Rac-1 increases the proliferative potential of thyroid epithelial cells, without affecting their differentiation. Oncogene 17:2047-2057. [DOI] [PubMed] [Google Scholar]
- 17.Cohen, D. R., and T. Curran. 1988. fra-1: a serum-inducible, cellular immediate-early gene that encodes a fos-related antigen. Mol. Cell. Biol. 8:2063-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cook, S. J., N. Aziz, and M. McMahon. 1999. The repertoire of fos and jun proteins expressed during the G1 phase of the cell cycle is determined by the duration of mitogen-activated protein kinase activation. Mol. Cell. Biol. 19:330-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Downward, J. 1996. Control of ras activation. Cancer Surv. 27:87-100. [PubMed] [Google Scholar]
- 20.Downward, J. 1998. Ras signalling and apoptosis. Curr. Opin. Genet. Dev. 8:49-54. [DOI] [PubMed] [Google Scholar]
- 21.Fusco, A., M. T. Berlingieri, P. P. Di Fiore, G. Portella, M. Grieco, and G. Vecchio. 1987. One- and two-step transformations of rat thyroid epithelial cells by retroviral oncogenes. Mol. Cell. Biol. 7:3365-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallo, A., A. Feliciello, A. Varrone, R. Cerillo, M. E. Gottesman, and V. E. Avvedimento. 1995. Ki-ras oncogene interferes with the expression of cyclic AMP-dependent promoters. Cell Growth Differ. 6:91-95. [PubMed] [Google Scholar]
- 23.Gille, H., and J. Downward. 1999. Multiple ras effector pathways contribute to G1 cell cycle progression. J. Biol. Chem. 274:22033-22040. [DOI] [PubMed] [Google Scholar]
- 24.Gruda, M. C., K. Kovary, R. Metz, and R. Bravo. 1994. Regulation of Fra-1 and Fra-2 phosphorylation differs during the cell cycle of fibroblasts and phosphorylation in vitro by MAP kinase affects DNA binding activity. Oncogene 9:2537-2547. [PubMed] [Google Scholar]
- 25.Herdegen, T., and V. Waetzig. 2001. AP-1 proteins in the adult brain: facts and fiction about effectors of neuroprotection and neurodegeneration. Oncogene 20:2424-2437. [DOI] [PubMed] [Google Scholar]
- 26.Hurd, T. W., A. A. Culbert, K. J. Webster, and J. M. Tavare. 2002. Dual role for mitogen-activated protein kinase (Erk) in insulin-dependent regulation of Fra-1 (fos-related antigen-1) transcription and phosphorylation. Biochem. J. 368:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jochum, W., E. Passegue, and E. F. Wagner. 2001. AP-1 in mouse development and tumorigenesis. Oncogene 20:2401-2412. [DOI] [PubMed] [Google Scholar]
- 28.Kovary, K., and R. Bravo. 1992. Existence of different Fos/Jun complexes during the G0-to-G1 transition and during exponential growth in mouse fibroblasts: differential role of Fos proteins. Mol. Cell. Biol. 12:5015-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kustikova, O., D. Kramerov, M. Grigorian, V. Berezin, E. Bock, E. Lukanidin, and E. Tulchinsky. 1998. Fra-1 induces morphological transformation and increases in vitro invasiveness and motility of epithelioid adenocarcinoma cells. Mol. Cell. Biol. 18:7095-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lloyd, A., N. Yancheva, and B. Wasylyk. 1991. Transformation suppressor activity of a Jun transcription factor lacking its activation domain. Nature 352:635-638. [DOI] [PubMed] [Google Scholar]
- 31.Mechta, F., D. Lallemand, C. M. Pfarr, and M. Yaniv. 1997. Transformation by ras modifies AP1 composition and activity. Oncogene 14:837-847. [DOI] [PubMed] [Google Scholar]
- 32.Mechta-Grigoriou, F., D. Gerald, and M. Yaniv. 2001. The mammalian Jun proteins: redundancy and specificity. Oncogene 20:2378-2389. [DOI] [PubMed] [Google Scholar]
- 33.Missero, C., M. T. Pirro, and R. Di Lauro. 2000. Multiple ras downstream pathways mediate functional repression of the homeobox gene product TTF-1. Mol. Cell. Biol. 20:2783-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murakami, M., M. H. Sonobe, M. Ui, Y. Kabuyama, H. Watanabe, T. Wada, H. Handa, and H. Iba. 1997. Phosphorylation and high level expression of Fra-2 in v-src transformed cells: a pathway of activation of endogenous AP-1. Oncogene 14:2435-2444. [DOI] [PubMed] [Google Scholar]
- 35.Murakami, M., M. Ui, and H. Iba. 1999. Fra-2-positive autoregulatory loop triggered by mitogen-activated protein kinase (MAPK) and Fra-2 phosphorylation sites by MAPK. Cell Growth Differ. 10:333-342. [PubMed] [Google Scholar]
- 36.Murphy, L. O., S. Smith, R. H. Chen, D. C. Fingar, and J. Blenis. 2002. Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol. 4:556-564. [DOI] [PubMed] [Google Scholar]
- 37.Musti, A. M., M. Treier, and D. Bohmann. 1997. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science 275:400-402. [DOI] [PubMed] [Google Scholar]
- 38.Okazaki, K., and N. Sagata. 1995. The Mos/MAP kinase pathway stabilizes c-Fos by phosphorylation and augments its transforming activity in NIH 3T3 cells. EMBO J. 14:5048-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orlando, V., H. Strutt, and R. Paro. 1997. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11:205-214. [DOI] [PubMed] [Google Scholar]
- 40.Passegue, E., W. Jochum, M. Schorpp-Kistner, U. Mohle-Steinlein, and E. F. Wagner. 2001. Chronic myeloid leukemia with increased granulocyte progenitors in mice lacking junB expression in the myeloid lineage. Cell 104:21-32. [DOI] [PubMed] [Google Scholar]
- 41.Passegue, E., and E. F. Wagner. 2000. JunB suppresses cell proliferation by transcriptional activation of p16(INK4a) expression. EMBO J. 19:2969-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Philips, A., C. Teyssier, F. Galtier, C. Rivier-Covas, J. M. Rey, H. Rochefort, and D. Chalbos. 1998. FRA-1 expression level modulates regulation of activator protein-1 activity by estradiol in breast cancer cells. Mol. Endocrinol. 12:973-985. [DOI] [PubMed] [Google Scholar]
- 43.Qiu, R. G., J. Chen, D. Kirn, F. McCormick, and M. Symons. 1995. An essential role for Rac in Ras transformation. Nature 374:457-459. [DOI] [PubMed] [Google Scholar]
- 44.Risse-Hackl, G., J. Adamkiewicz, A. Wimmel, and M. Schuermann. 1998. Transition from SCLC to NSCLC phenotype is accompanied by an increased TRE-binding activity and recruitment of specific AP-1 proteins. Oncogene 16:3057-3068. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez-Viciana, P., P. H. Warne, A. Khwaja, B. M. Marte, D. Pappin, P. Das, M. D. Waterfield, A. Ridley, and J. Downward. 1997. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell 89:457-467. [DOI] [PubMed] [Google Scholar]
- 46.Saez, E., S. E. Rutberg, E. Mueller, H. Oppenheim, J. Smoluk, S. H. Yuspa, and B. M. Spiegelman. 1995. c-fos is required for malignant progression of skin tumors. Cell 82:721-732. [DOI] [PubMed] [Google Scholar]
- 47.Schreiber, M., A. Kolbus, F. Piu, A. Szabowski, U. Mohle-Steinlein, J. Tian, M. Karin, P. Angel, and E. F. Wagner. 1999. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 13:607-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schreiber, M., C. Poirier, A. Franchi, R. Kurzbauer, J. L. Guenet, G. F. Carle, and E. F. Wagner. 1997. Structure and chromosomal assignment of the mouse fra-1 gene, and its exclusion as a candidate gene for oc (osteosclerosis). Oncogene 15:1171-1178. [DOI] [PubMed] [Google Scholar]
- 49.Schreiber, M., Z. Q. Wang, W. Jochum, I. Fetka, C. Elliott, and E. F. Wagner. 2000. Placental vascularisation requires the AP-1 component fra1. Development 127:4937-4948. [DOI] [PubMed] [Google Scholar]
- 50.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131-E136. [DOI] [PubMed] [Google Scholar]
- 51.Shaulian, E., and M. Karin. 2001. AP-1 in cell proliferation and survival. Oncogene 20:2390-2400. [DOI] [PubMed] [Google Scholar]
- 52.Shaulian, E., M. Schreiber, F. Piu, M. Beeche, E. F. Wagner, and M. Karin. 2000. The mammalian UV response: c-Jun induction is required for exit from p53-imposed growth arrest. Cell 103:897-907. [DOI] [PubMed] [Google Scholar]
- 53.Smeal, T., B. Binetruy, D. A. Mercola, M. Birrer, and M. Karin. 1991. Oncogenic and transcriptional cooperation with Ha-Ras requires phosphorylation of c-Jun on serines 63 and 73. Nature 354:494-496. [DOI] [PubMed] [Google Scholar]
- 54.Suarez, H. G. 1998. Genetic alterations in human epithelial thyroid tumours. Clin. Endocrinol. (Oxford) 48:531-546. [DOI] [PubMed] [Google Scholar]
- 55.Treinies, I., H. F. Paterson, S. Hooper, R. Wilson, and C. J. Marshall. 1999. Activated MEK stimulates expression of AP-1 components independently of phosphatidylinositol 3-kinase (PI3-kinase) but requires a PI3-kinase signal to stimulate DNA synthesis. Mol. Cell. Biol. 19:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Troppmair, J., J. T. Bruder, H. Munoz, P. A. Lloyd, J. Kyriakis, P. Banerjee, J. Avruch, and U. R. Rapp. 1994. Mitogen-activated protein kinase/extracellular signal-regulated protein kinase activation by oncogenes, serum, and 12-O-tetradecanoylphorbol-13-acetate requires Raf and is necessary for transformation. J. Biol. Chem. 269:7030-7035. [PubMed] [Google Scholar]
- 57.Vallone, D., S. Battista, G. M. Pierantoni, M. Fedele, L. Casalino, M. Santoro, G. Viglietto, A. Fusco, and P. Verde. 1997. Neoplastic transformation of rat thyroid cells requires the junB and fra-1 gene induction which is dependent on the HMGI-C gene product. EMBO J. 16:5310-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Dam, H., and M. Castellazzi. 2001. Distinct roles of Jun : Fos and Jun : ATF dimers in oncogenesis. Oncogene 20:2453-2464. [DOI] [PubMed] [Google Scholar]
- 59.Vogt, P. K. 2001. Jun, the oncoprotein. Oncogene 20:2365-2377. [DOI] [PubMed] [Google Scholar]
- 60.Weitzman, J. B., L. Fiette, K. Matsuo, and M. Yaniv. 2000. JunD protects cells from p53-dependent senescence and apoptosis. Mol. Cell 6:1109-1119. [DOI] [PubMed] [Google Scholar]
- 61.White, M. A., C. Nicolette, A. Minden, A. Polverino, L. Van Aelst, M. Karin, and M. H. Wigler. 1995. Multiple Ras functions can contribute to mammalian cell transformation. Cell 80:533-541. [DOI] [PubMed] [Google Scholar]
- 62.Wisdom, R., R. S. Johnson, and C. Moore. 1999. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 18:188-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Young, M. R., J. J. Li, M. Rincon, R. A. Flavell, B. K. Sathyanarayana, R. Hunziker, and N. Colburn. 1999. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc. Natl. Acad. Sci. USA 96:9827-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young, M. R., R. Nair, N. Bucheimer, P. Tulsian, N. Brown, C. Chapp, T. C. Hsu, and N. H. Colburn. 2002. Transactivation of Fra-1 and consequent activation of AP-1 occur extracellular signal-regulated kinase dependently. Mol. Cell. Biol. 22:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zuber, J., O. I. Tchernitsa, B. Hinzmann, A. C. Schmitz, M. Grips, M. Hellriegel, C. Sers, A. Rosenthal, and R. Schafer. 2000. A genome-wide survey of RAS transformation targets. Nat. Genet. 24:144-152. [DOI] [PubMed] [Google Scholar]