Abstract
The Eleven Lysine-rich Leukemia (ELL) gene undergoes translocation and fuses in frame to the Multiple Lineage Leukemia (MLL) gene in a substantial proportion of patients suffering from acute forms of leukemia. Molecular mechanisms of cellular transformation by the MLL-ELL fusion are not well understood. Although both MLL-ELL and wild-type ELL can reduce functional activity of p53 tumor suppressor, our data reveal that MLL-ELL is a much more efficient inhibitor of p53 than is wild-type ELL. We also demonstrate for the first time that ELL extreme C terminus [ELL(eCT)] is required for the recruitment of p53 into MLL-ELL nuclear foci and is both necessary and sufficient for the MLL-ELL inhibition of p53-mediated induction of p21 and apoptosis. Finally, our results demonstrate that MLL-ELL requires the presence of intact ELL(eCT) in order to disrupt p53 interactions with p300/CBP coactivator and thus significantly reduce p53 acetylation in vivo. Since ELL(eCT) has recently been shown to be both necessary and sufficient for MLL-ELL-mediated transformation of normal blood progenitors, our data correlate ELL(eCT) contribution to MLL-ELL transformative effects with its ability to functionally inhibit p53.
Chromosomal rearrangements are frequently detected in leukemic cells and are considered cytogenetic traits of leukemic disease. In a subset of patients suffering from both de novo and chemotherapy-related acute myeloid and lymphoblastic leukemias (AML and ALL, respectively), chromosomal translocation t(11;19)(q23;p13.1) (28) results in the generation of MLL-ELL chimeric protein in which almost an entire open reading frame of ELL, a general transcription elongation factor (23), is fused to the first 1,400 amino acid (aa) residues of MLL, a protein that normally positively regulates Hox gene expression. ELL is one of multiple fusion partners of MLL in leukemia-associated translocations (for a review, see reference 1).
The critical contribution of the MLL-ELL fusion protein to the pathogenesis of acute leukemia has been vividly demonstrated in transgenic animal models. Transplantation of blood progenitor cells transduced with MLL-ELL into sublethally irradiated normal mice results in morphological and clinical disease manifestations closely resembling those observed in patients suffering from leukemia (16). It has also been demonstrated that ELL extreme C terminus [ELL(eCT); aa 508 to 621] is required and sufficient for malignant transformation of bone marrow cells by MLL-ELL and the onset of leukemia in animals (19). However, the precise mechanisms of leukemogenesis by the MLL-ELL fusion remain elusive. It has been hypothesized that MLL, which contains DNA-binding domain (DBD), gains strong transactivation function once fused to ELL or other partner proteins, and the resulting constitutively active leukemic protein abnormally regulates expression of genes involved in blood progenitor cell differentiation (1). However, recent evidence suggests that acquisition of high transcriptional activity by MLL cannot completely account for tumorigenic effects of leukemic MLL fusions (25).
Recently, both MLL-ELL and ELL, but not wild-type MLL, have been shown to inhibit p53 transcriptional activity (20, 24). Even though the mechanism of this inhibitory effect had not been defined, it was speculated that repression of p53 activity by MLL-ELL may contribute to the transformation of blood progenitor cells and development of leukemia. p53, which is considered one of the most significant tumor suppressor genes discovered to date, mediates cell cycle arrest and/or apoptosis in response to various forms of cytotoxic insult (17). p53 acts as a transcription factor to induce various gene expression, including that of the cyclin-dependent kinase inhibitor p21 and a number of apoptotic regulators such as Bax, p53AIP1, PUMA, NOXA, PIG3, and others (for a review, see reference 2). The transcriptional activity of p53 has been recently shown to be absolutely required for its antitumor effects (12). Interestingly, although mutations or loss of p53 are found in a vast majority of human malignancies (11), very few p53 genetic alterations have been detected in patients diagnosed with AML (6, 7, 26, 27), suggesting that p53 is functionally, rather than genetically, inactivated in this disease.
These findings prompted us to further investigate the MLL-ELL/p53 interactions and to gain insight into the mechanism of p53 repression by this leukemic protein. Our results show that the MLL-ELL fusion is substantially more potent in suppressing p53 function than wild-type ELL. For the first time, we demonstrate that MLL-ELL and p53 colocalize in the nucleus, and this colocalization completely depends on the intact ELL(eCT) and transactivation domain of p53 [p53(TAD)]. We further show that these regions within ELL and p53 are also required for in vivo binding between the two proteins, as well as the inhibitory effect on p53. In addition, our data demonstrate that through binding to p53(TAD), MLL-ELL and ELL disrupt p53 interactions with its important transcriptional coactivator p300/CBP in vivo, thus identifying it as one of the mechanisms of p53 suppression by the leukemic fusion protein.
(Part of this work was presented at the 2002 Annual Meeting of American Association for Cancer Research in San Francisco, Calif.)
MATERIALS AND METHODS
Plasmid construction.
The p53 and p73 expression plasmids have been described earlier (9). p53/p73 chimeric proteins were generated by using two-step PCR as previously described (9). p536KR mutant was obtained by substituting six lysine residues in the p53 C terminus (Lys 370, 372, 373, 381, 382, and 386) with arginines (10). Previously constructed pMSCV-MLL-ELL (4) was digested with EcoRI and EcoRV, and the resulting MLL-ELLΔaa340-621 was ligated into pcDNA3.0 (Invitrogen) and named MLL-ELLΔCT. The remaining part of ELL (aa 341 to 621) was PCR amplified and cloned into EcoRV site of pcDNA3.0MLL-ELLΔCT, resulting in the generation of full-length MLL-ELL expression construct. p53 and ELL deletion mutants were created by PCR with specific primers and cloned into either Flag- or GFP-tagged vector. ELL(508-621) was cloned into pCMV/myc/Nuc (Invitrogen). PCR-amplified p53, ELL or p300 sequences were also cloned in frame into the pcDNA3.0-Gal4 DBD (aa 1 to 147) and pcDNA3.0-FlagVP16 transactivating domain (TAD; aa 413 to 491) (Y. Shi, Harvard Medical School). Construct identity was verified by digestion with appropriate restriction enzymes, DNA sequencing (Harvard Cancer Center Core Facility), and Western blot analysis.
Cell culture, transient transfections, and luciferase assays.
Human small cell lung carcinoma H1299 cells, human kidney 293T cells, human osteosarcoma U2OS cells, human breast carcinoma MCF-7 cells (American Type Culture Collection), or p53−/− mouse embryonic fibroblasts (C. Maki, Harvard School of Public Health) were maintained in Dulbecco modified Eagle medium (Mediatech) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 10 U of penicillin/ml, and 10 μg of streptomycin/ml at 37°C in 5% CO2 humidified atmosphere. Human colon carcinoma HCT116 cell line (B. Vogelstein, Johns Hopkins University) was maintained as described above except that McCoy's media was used. Human leukemia U937 cells were grown in RPMI medium with supplements as indicated for other cell lines. Cells were transfected with various constructs by using either the calcium phosphate precipitation method (for luciferase assays) as previously described (30) or Lipofectamine (Invitrogen) reagent (for Western blot, apoptosis analysis, and immunostaining) according to the manufacturer's instructions. Lipofectamine method was always used if transfection included MLL-ELL or its mutants.
For luciferase activity measurements, H1299 cells were cotransfected in 35-mm dishes with various constructs and with either PG13-Luc or with pGL3-G5SV (Y. Shi), along with pRL-TK plasmid (Promega) to control for transfection efficiency. Empty vector was used to standardize for total DNA amount. Firefly (PG13, pGL3-G5SV) and Renilla (pRL-TK) luciferase was measured 24 to 36 h posttransfection by using the Promega dual luciferase system according to the manufacturer's instructions. Firefly activity was standardized to Renilla expression.
Immunostaining.
293T or U2OS cells were grown in 60-mm tissue culture dishes and transfected with various constructs as described above. At 24 to 36 h posttransfection, cells were washed with cold phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde solution (Sigma). After three washes with PBS, cells were permeabilized with ice-cold 0.2% Triton X-100, blocked with 0.5% bovine serum albumin (Sigma) for 30 min, and incubated with anti-ELL antibody (1:300). After three washes with PBS, the specimens were incubated with Texas red mouse immunoglobulin G (Santa Cruz Biotechnology) that contained 20 mg of DAPI (4′,6′-diamidino-2-phenylindole) stain/ml. After being mounted in Fluoromount-G (Southern Biotechnology Associates) containing 2.5 mg of n-propyl gallate (Sigma)/ml, the cells were examined by using a Nikon Eclipse E600 fluorescent microscope at ×100 magnification and photographed.
Preparation of whole-cell lysates and Western blot analysis.
Cells were transfected in 60-mm plates with various amounts of plasmid DNA and harvested at 24 h posttransfection. Cells were lysed in 100 μl of lysis buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1% Triton X-100, 150 mM NaCl, 1 mM dithiothreitol, 10% glycerol, 0.2 mM phenylmethylsulfonyl fluoride, and protease inhibitors) by incubation on ice for 30 min, and the extracts were centrifuged at 13,000 rpm for 15 min to remove cell debris. Protein concentrations were determined by using Bio-Rad protein assay (Bio-Rad). After the addition of 5× loading buffer, the samples were incubated at 95°C for 5 min and then resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were transferred onto nitrocellulose membranes (Schleicher & Schuell) and probed with anti-Flag (M5; Sigma), anti-p53 (Ab6; Calbiochem), anti-p21 (Ab3; Calbiochem), anti-acetyl p53 (Lys 373 and Lys 382; Upstate Biotechnology), anti-GFP (Clontech), anti-β-actin (AC5; Sigma), anti-Gal4 (Santa Cruz Biotechnology), anti-HA (Boehringer Mannheim), anti-p300 (Upstate Biotechnology), anti-Myc (Dieter Wolf, Harvard School of Public Health), or anti-ELL (A. Shilatifard) antibodies. Proteins were visualized with an enhanced chemiluminescence detection system (Perkin-Elmer Life Sciences).
Apoptosis analysis.
H1299 cells were transfected with p53 or empty vector and with various MLL-ELL or ELL constructs as indicated in the figures. Us9-GFP vector (13) was included as a transfection efficiency indicator. Cells were harvested by trypsinization 36 h posttransfection and after being washed in PBS fixed in 70% ice-cold ethanol for 24 to 72 h. After three PBS washes, the cells were stained with propidium iodide solution (50 μg/ml) containing 100 μg of RNase A/ml for 30 min at 37°C, and the DNA content of ca. 10,000 green fluorescent protein (GFP)-positive cells from each transfection was analyzed by using Beckman-Coulter Elite flow cytometer (Kresge Center for Environmental Health Sciences, Harvard School of Public Health).
RESULTS
The extreme C terminus of ELL is both necessary and sufficient to inhibit p53 transcriptional activity.
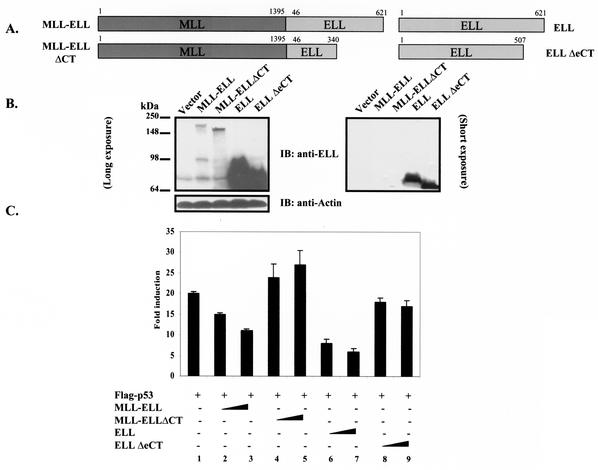
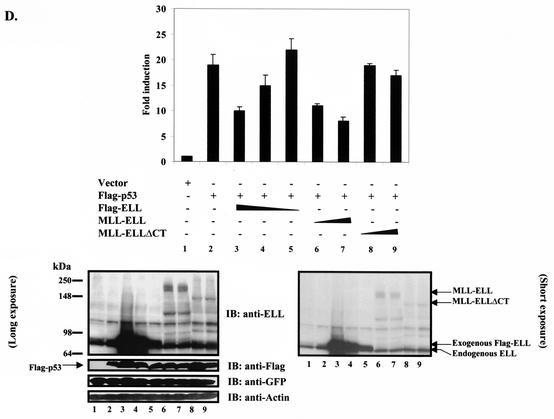
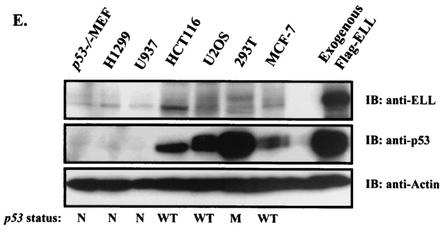
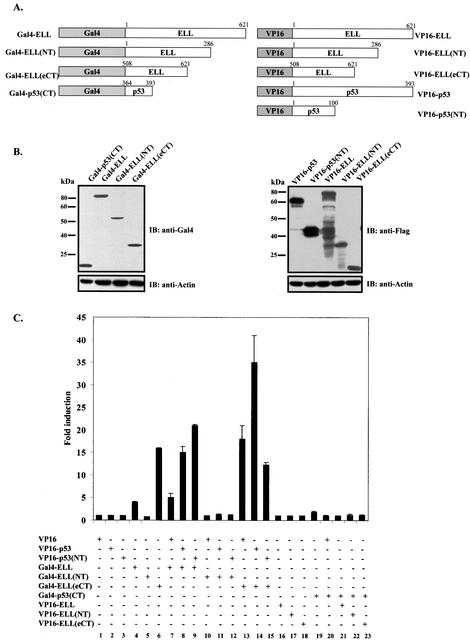
It has been recently shown that the ELL C terminus is both necessary and sufficient for immortalization of normal bone marrow cells by MLL-ELL and the development of leukemia-like disease in animals (4, 19). We were therefore interested to determine if the same domain of MLL-ELL is required for the inhibition of p53 functional activity. Having generated full-length MLL-ELL and MLL-ELLΔCT, as well as full-length ELL and ELLΔeCT expression plasmids (Fig. 1A), we confirmed protein expression of these constructs by Western blot utilizing monoclonal anti-ELL antibody. When transfected into 293T cells, ELL and its C-terminally truncated mutant were robustly expressed, whereas MLL-ELL and MLL-ELLΔCT protein levels were markedly lower, similar to previously reported results (Fig. 1B) (4, 15). Next, we examined the effect of MLL-ELL and ELL expression on p53 transcriptional activity in p53-null H1299 cells by using the luciferase reporter containing 13 repeats of p53 consensus binding site in its promoter region (PG13). Transfection of equal amounts of either MLL-ELL or ELL expression plasmids caused a dose-dependent downregulation in p53 transactivating function as previously reported by others (20, 24); however, the C-terminally truncated mutants of both MLL-ELL and ELL had no inhibitory effect on p53 (Fig. 1C). These data demonstrate that ELL(eCT) is required for the inhibition of p53 transcriptional activity. Because dramatic differences in MLL-ELL and ELL protein levels were observed when equal amounts of plasmids were transfected, we reduced the dose of transfected ELL while slightly increasing the quantity of MLL-ELL in order to evaluate their inhibitory effect on p53 at similar levels of protein expression. MLL-ELLΔCT was included as a negative control. As shown in Fig. 1D, ELL completely lost its capacity to suppress p53 transcriptional activity even before its protein levels reached those of MLL-ELL (compare lane 5 to lanes 6 and 7). Interestingly, levels of exogenous ELL at which it was ineffective in inhibiting p53 strongly resembled those of endogenous protein in H1299 cells (Fig. 1D, lane 5 versus lane 1). These results demonstrate that MLL-ELL leukemic fusion is a much more potent inhibitor of p53 transactivation than wild-type ELL and suggest that endogenous ELL might not be suppressing p53 function. To address this further, we screened a number of cell lines by using anti-ELL antibody to determine the level of endogenous ELL expression. Our data revealed that ELL is generally expressed at low levels compared to transiently transfected plasmid, whereas the amounts of endogenous p53 varies widely depending on its genetic status in the cell (i.e., p53-null, mutant, or wild-type) (Fig. 1E). These results suggest that endogenous ELL might not contribute significantly to p53 inhibition in the absence of MLL-ELL translocation.
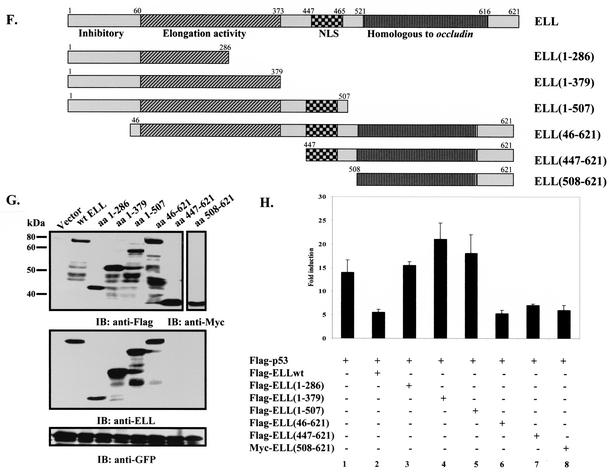
FIG. 1.
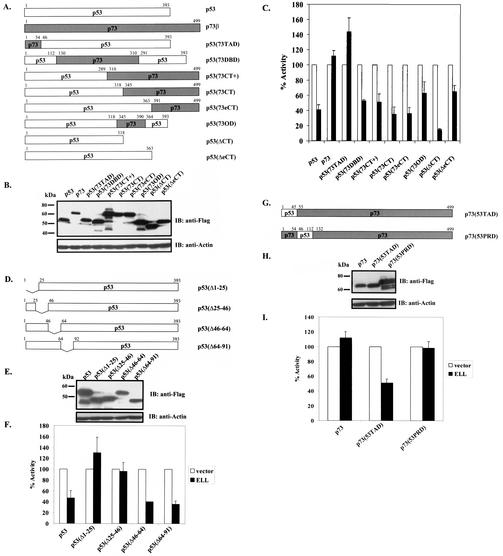
ELL(eCT) is necessary and sufficient to inhibit p53 transcriptional activity. (A) Full-length or truncated mutants of MLL-ELL and ELL. (B) The indicated constructs were transfected into 293T cells (5 μg of each cDNA), and cell lysates were prepared 24 h after transfection. Western blot analysis was carried out with the indicated antibodies. Actin served as loading control. Examples of short and long film exposures are shown. (C) Flag-p53 expression vector (100 ng) was cotransfected into H1299 cells with PG13 luciferase reporter (0.25 μg) in the presence or absence of various MLL-ELL and ELL constructs (1 and 2 μg for both) by using the Lipofectamine method. pRL-TK was used as a transfection efficiency control. Both Firefly (PG13) and Renilla (pRL-TK) luciferase activity was measured 36 h posttransfection by using the Promega dual luciferase system. The relative luciferase activity was calculated as a ratio of the Firefly activity to the Renilla activity and is expressed as the fold induction over the vector control (mean ± the standard deviation [SD] of three independent experiments, each done in duplicate). (D) H1299 cells were Lipofectamine transfected as indicated with Flag-p53 (100 ng), Flag-ELL (1 μg, 100 ng, and 10 ng), and MLL-ELL and MLL-ELLΔCT (2 and 4 μg for both). PG13 and pRL-TK plasmids were also included in the transfection mixture. The cells were lysed 36 h posttransfection, and the luciferase activity was determined as described in panel A (the mean ± the SD of three separate experiments, each done in duplicate, is shown in the upper panel). Proteins in the remainder of cell lysates were resolved by SDS-PAGE, transferred onto nitrocellulose membranes, and analyzed by Western blot with the indicated antibodies (lower panel). Sample numbers on the luciferase activity chart correspond to those on the Western blot. (E) Cell lysate of 293T cells transfected with Flag-ELL (1 μg) or cell lysates from untransfected cell lines as shown were probed with anti-ELL and anti-p53 antibody. Actin levels were examined to ensure equal loading. N, null; WT, wild type; M, mutant. (F) Schematic depiction of ELL functional domains and ELL truncated mutants. (G) cDNA of ELL or its truncated constructs (5 μg) was transfected into 293T cells. pEGFP (0.5 μg) was included as a transfection efficiency control. Cell lysates were prepared 24 h later and analyzed with the indicated antibody. (H) Plasmids (1 μg) as shown in panel D were cotransfected with Flag-p53 (1 μg), along with PG13-Luc and pRL-TK, into H1299 cells by calcium phosphate precipitation. Luciferase activity, which was recorded 36 h posttransfection, is expressed as the fold induction over vector alone and is shown for three independent transfection experiments, each done in duplicate (mean ± the SD).
We went on to exploit the overexpressed ELL inhibitory effect on p53 to further evaluate the contribution of each ELL domain to p53 suppression and to investigate whether ELL(eCT) might be sufficient to inhibit p53 functional activity. To accomplish this, a number of ELL C- and N-terminal deletion mutants were generated and cloned into Flag- or Myc-tagged vectors (Fig. 1F). Since both wild-type ELL and MLL-ELL leukemic proteins localize exclusively in the nucleus (4), and ELL(508-621) would have lost its natural nuclear localization signal (NLS), this ELL sequence was cloned into a vector containing artificial NLS. Comparable protein expression of all ELL mutants was verified by monoclonal anti-Flag, anti-Myc, or anti-ELL antibodies (Fig. 1G). Upon cotransfection with p53 and p53-driven luciferase reporter into H1299 cells, both wild-type ELL and ELL(46-621), which is present in the MLL-ELL fusion, efficiently inhibited p53 transcriptional activity (Fig. 1H, columns 2 and 6 versus column 1). On the contrary, ELL mutants that lacked both the NLS and the C terminus [ELL(1-286) and ELL(1-379)], as well as an ELL construct that only lacked the extreme C terminus [ELL(1-507)], failed to suppress p53 activity (Fig. 1H, columns 3, 4, and 5 versus column 2). ELL(447-621) that included the NLS and the extreme C terminus was as efficient in p53 inhibition as wild-type ELL (Fig. 1H, column 7 versus 2). Even more remarkable was the observed strong inhibitory effect of ELL(508-621) on p53, revealing that this region of ELL is both necessary and sufficient for downregulation of p53 functional activity (Fig. 1H, column 8 versus column 1). These data indicate that ELL(eCT) is solely responsible for the inhibitory effect on p53.
The transactivation domain is necessary and sufficient to render p53 sensitive to ELL inhibitory effect.
In order to elucidate potential mechanisms of MLL-ELL inhibitory effect on p53, we next sought to identify the p53 domain that is required for repression by the leukemic protein. In our studies, we utilized full-length ELL and p53 chimeric proteins in which specific p53 domains have been replaced with corresponding portions of its close structural homologue p73 that is capable of transactivating p53-responsive promoters (9). Since ELL in vitro binding sites have been previously mapped to the p53 C terminus, we also generated p53 mutants that lacked either the entire C terminus [p53(ΔCT)] or just its extreme portion [p53(ΔeCT)] (Fig. 2A). Comparable protein expression of all constructs is shown in Fig. 2B. When cotransfected with either p53 or p73 and p53-driven luciferase reporter in H1299 cells, ELL significantly suppressed p53 transcriptional activity but had no effect on p73 transactivation function (Fig. 2C). Lack of ELL inhibitory effect on p73 therefore further justified the use of p53/p73 chimeras in our experiments. Chimeric proteins that contained p53 backbone and various p73 portions, either from the C terminus [p53(73eCT), p53(73CT), p53(73CT+), and p53(73OD)] or the DBD [p53(73DBD)], were as sensitive to the inhibition by ELL as was wild-type p53. A similar inhibitory effect was observed when ELL was coexpressed with p53(ΔCT) or p53(ΔeCT). We therefore concluded that neither p53 CT nor DBD is functionally required for the suppression of p53 activity by ELL. In contrast, the transactivation domain [p53(73TAD)] swap chimera was completely resistant to the inhibition by ELL (Fig. 2C), indicating that p53(TAD) is necessary for the negative regulatory effect of ELL.
FIG. 2.
p53 (aa 1 to 45) is necessary and sufficient to render p53 sensitive to the inhibition by ELL. (A) p53/p73 chimeras were generated by using two-step PCR. p53 deletion mutants were PCR amplified with specific primers. (B) Protein expression of all constructs was verified by a anti-Flag Western blot of 293T-cell lysates transfected with 5 μg of each cDNA. Actin served as an equal loading control. (C) Chimeric or deletion mutants (1 μg) were cotransfected into p53−/− H1299 cells by calcium phosphate precipitation with either Flag-ELL (1 μg) (▪) or with empty vector (□) and with PG13 and pRL-TK plasmids. The relative luciferase activity measured 36 h posttransfection was set to 100% for every construct in the presence of vector alone. All transfections were repeated at least three times, in duplicate experiments (mean ± the SD). (D) Systematic 25-aa deletions in p53(NT) were generated by using standard PCR. (E) Protein expression of p53 deletions was confirmed by a Western blot with anti-Flag antibody. (F) The transcriptional activity of p53 deletion constructs in the presence of vector alone or Flag-ELL was assayed 36 h posttransfection as described in the text. The relative luciferase activity in the presence of vector was set at 100%. All transfections were repeated at least three times, with each done in duplicate (mean ± the SD). (G and H) Protein expression of the indicated p73/p53 chimeric constructs (G) was confirmed by Western blot with anti-Flag (H), and their transcriptional activity was measured by cotransfecting in the presence of either vector alone or Flag-ELL in H1299 cells. (I) The mean ± the SD of three separate transfections, each done in duplicate, is shown.
We confirmed this finding by utilizing systematic p53 N-terminal deletion mutants (Fig. 2D), comparable protein expression of which was verified by Western blot (Fig. 2E). Upon cotransfection into H1299 cells with ELL and p53-driven luciferase reporter, mutants that lacked aa 1 to 45 of p53 were refractory to ELL inhibition (Fig. 2F). To further substantiate the requirement of p53 aa 1 to 45 for ELL-mediated inhibition, we tested whether p73 would gain sensitivity to ELL when aa 1 to 45 of p53 were swapped into it. Strikingly, the introduction of aa 1 to 45 of p53 into p73 (Fig. 2G) rendered p73 sensitive to the inhibition by ELL, whereas the introduction of aa 46 to 112 of p53 had no such effect (Fig. 2I). Once again, equal protein expression level of chimeric constructs is shown in Fig. 2H. These data clearly show that p53TAD (aa 1 to 45) is both necessary and sufficient for ELL repression of p53 function. These results were reproduced in p53−/− mouse embryonic fibroblasts, eliminating the possibility of cell type-specific effects (data not shown).
Colocalization of p53 with MLL-ELL in the nucleus depends on the presence of intact p53(TAD) and ELL(eCT).
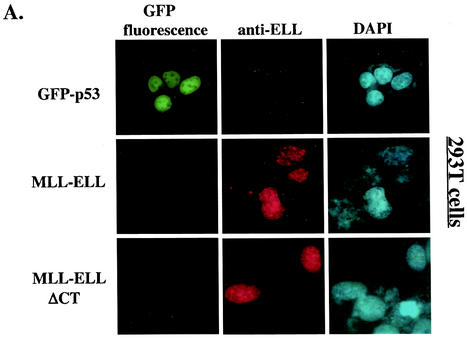
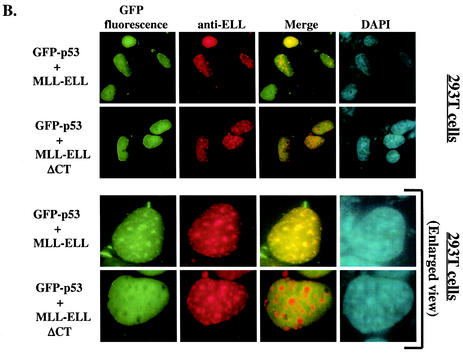
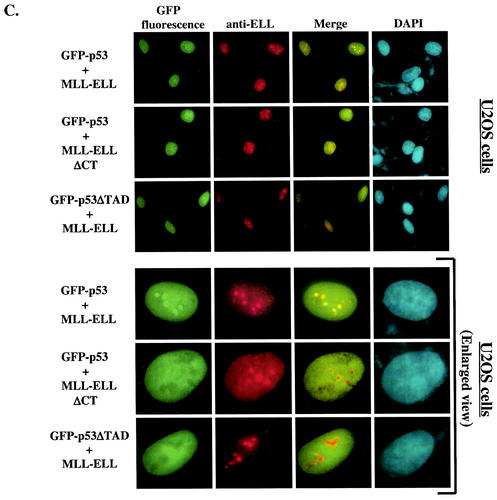
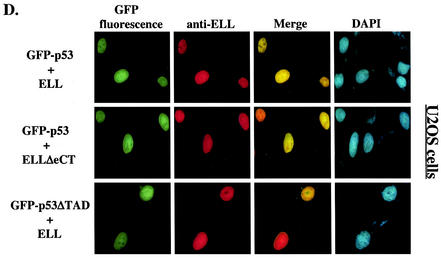
The results presented above implicated ELL(eCT) and p53(TAD) in the inhibitory effect of MLL-ELL and ELL on p53 functional activity. We then sought to determine whether MLL-ELL and p53 can colocalize in the cell and if such colocalization is mediated by the same regions in the two molecules that we identified as necessary for the inhibitory effect. To address this question, 293T cells were transfected with GFP-p53 in the presence or absence of MLL-ELL or MLL-ELLΔCT expression and subsequently stained with anti-ELL antibodies. When transfected alone, GFP-p53 displayed typical diffused nuclear distribution (Fig. 3A). In contrast, MLL-ELL and its C-terminally truncated mutant, while also exclusively localizing to the nucleus, formed distinct nuclear foci as noted previously (3, 4). Remarkably, upon cotransfection with MLL-ELL, but not with MLL-ELLΔCT, p53 appeared to be recruited into MLL-ELL foci and displayed strong colocalization with full-length leukemic fusion protein (Fig. 3B). Similar results were obtained in U2OS cells (Fig. 3C), thus ruling out cell type-specific effects. We then investigated whether p53TAD is required for colocalization of MLL-ELL and p53. As shown in Fig. 3C, deletion of p53TAD resulted in the loss of colocalization between the two proteins and diffuse p53 staining in the nucleus. Our immunostaining analysis therefore demonstrates that MLL-ELL expression alters normal p53 subnuclear distribution and that colocalization of p53 with leukemic fusion protein requires both TAD of p53 and eCT of ELL.
FIG. 3.
Nuclear colocalization of MLL-ELL with p53 requires ELL(eCT) and p53(TAD). 293T (A and B) or U2OS (C and D) cells were cotransfected with 4 μg of the indicated constructs and after 24 h were stained with anti-ELL antibody as described in Materials and Methods. Cells were visualized by using a fluorescence microscope, and representative fields were photographed. The images show GFP-p53 (green); MLL-ELL, ELL, or their truncated mutants (red); and nuclei (DAPI; blue). Merge images were produced by using SPOT advanced software. An enlarged view of representative cells coexpressing various constructs is also shown.
Finally, to confirm that the lack of inhibitory effect of ELLΔeCT on p53 was not due to abnormal subcellular localization of this mutant, we examined its colocalization with full-length p53. A similar experiment was performed with p53ΔTAD and full-length ELL. Both ELLΔeCT and p53ΔTAD displayed diffuse nuclear staining, a characteristic similar to that of their wild-type counterparts (Fig. 3D), thus excluding altered subcellular distribution as the reason for the mutants' lack of inhibitory function.
In vivo binding between p53 and ELL involves p53(NT) and ELL(eCT).
Even though the C terminus of p53 (aa 346 to 393) has previously been shown to be required for ELL binding to p53 in vitro (24), the results of our functional assays, as well as the pattern of colocalization of p53 with leukemic proteins, indicated that p53(TAD) located in the N terminus of p53 [p53(NT)] is functionally required for the inhibition by ELL. To resolve this apparent inconsistency, we used the mammalian two-hybrid system (5, 22) to analyze the in vivo binding between p53 and ELL. As depicted in Fig. 4A, various portions of ELL and p53 were fused in frame to either Gal4DBD or VP16TAD. Comparable protein expression was verified by Western blot and is presented in Fig. 4B. When cotransfected into H1299 cells, along with the luciferase reporter under the control of Gal4-responsive elements (pGL3-G5SV), neither of the VP16 constructs alone nor Gal4-ELL(NT) alone exhibited any activity above vector control (Fig. 4C, columns 1, 2, 3, and 5), whereas both full-length ELL and its C terminus fused to Gal4 [Gal4-ELL and Gal4-ELL(eCT), respectively] possessed various degrees of basal transcriptional activity (Fig. 4C, columns 4 and 6). Cotransfection of Gal4-ELL with VP16-p53 or VP16-p53(NT) caused further increases in luciferase expression (Fig. 4C, columns 8 and 9 versus column 4), indicating in vivo binding between these fusion proteins. Interestingly, cointroduction of Gal4-ELL(eCT) with VP16-p53 also resulted in a substantial increase of luciferase activity (Fig. 4C, column 14 versus column 6), indicating their in vivo association. In contrast, cotransfection of Gal4-p53(CT) with various ELL VP16 fusion constructs (Fig. 4C, columns 19 to 23) or coexpression of Gal4-ELL(NT) with p53 VP16 fusions (Fig. 4C, columns 11 and 12 versus column 5) did not lead to any appreciable increases in luciferase expression, pointing to the lack of in vivo binding between ELL(NT) and p53 or p53(CT) and ELL. We also did not observe direct association between Gal4-ELL(eCT) and VP16-p53(NT) (Fig. 4C, column 6 versus column 15), indicating perhaps that correct conformational folding of full-length p53 might be required for the efficient binding of ELL(eCT).
FIG. 4.
In vivo binding between p53 and ELL is mediated by ELL(eCT) and p53(NT). (A) Various segments of p53 and ELL were fused in frame to either Gal4DBD or VP16TAD. (B) Protein expression of all constructs was confirmed by Western blot analysis of 293T-cell lysates with the indicated antibody. (C) Gal4 fusions of ELL were cotransfected into H1299 cells with VP16 constructs of p53 along with pGL3-G5SV and pRL-TK, and luciferase expression was measured as previously described 36 h posttransfection. In a separate experiment, Gal4-p53(CT) was cotransfected into H1299 cells with VP16 ELL fusions as shown. Solid bars represent the fold induction of luciferase activity compared to the vector control (mean ± the SD of three separate transfections, each done in duplicate).
These results, in concert with the data obtained from functional assays and immunostaining, firmly establish p53(TAD) and ELL(eCT) as the regions responsible for in vivo interaction between p53 and MLL-ELL or ELL.
MLL-ELL and ELL disrupt p53 interactions with p300 coactivator in vivo.
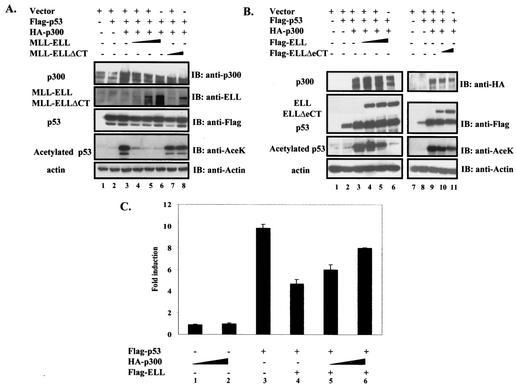
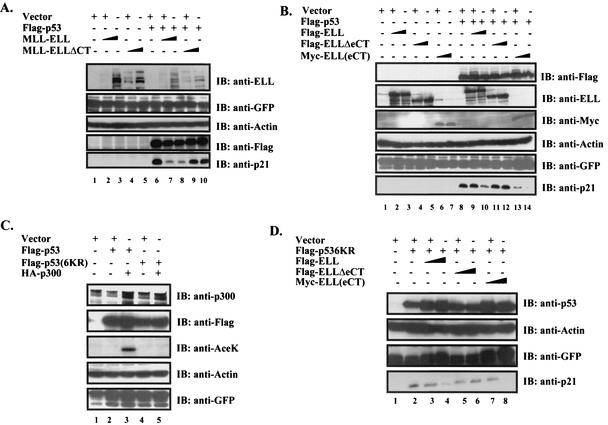
p53 recruits transcriptional coactivators such as p300/CBP to function at its full transactivation potential (18). Since p300 binds to p53(TAD) (8), the same region that we identified as necessary and sufficient for ELL inhibitory effect, we hypothesized that MLL-ELL and ELL might be disrupting p53 interactions with its coactivator at the N terminus. Since p300 is believed to achieve most of its positive effects on p53 by acetylating several lysine residues located in the extreme C terminus of p53 (18), we investigated whether MLL-ELL and ELL can interfere with p300-mediated acetylation of p53 in vivo. As shown in Fig. 5A, cotransfection of p53 with p300 resulted in a dramatic increase in the acetylated form of p53, as detected by p53-specific anti-AceK antibody (fourth panel, lane 3 versus lane 2). Simultaneous expression of full-length MLL-ELL, but not its C-terminal deletion mutant, led to an almost complete abrogation of p300-mediated p53 acetylation in a dose-dependent manner (Fig. 5A, fourth panel, lanes 4, 5, and 6 versus lanes 7 and 8). A similar inhibitory effect on p53 acetylation was observed when ELL, but not ELLΔeCT, was used (Fig. 5B, lanes 4, 5, and 6 versus lanes 10 and 11).
FIG. 5.
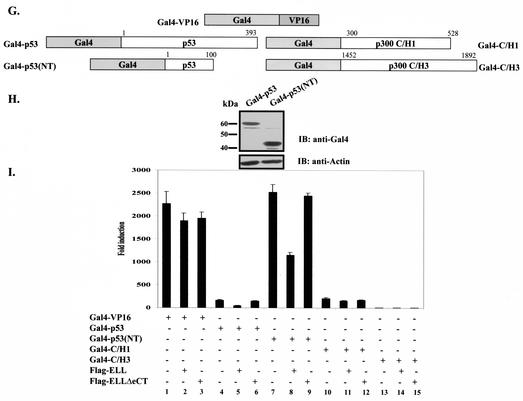
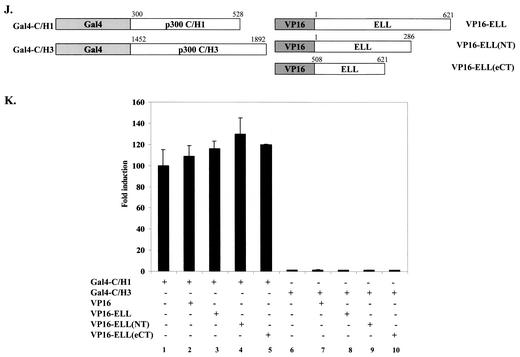
MLL-ELL and ELL disrupt p53 interactions with p300 transcriptional coactivator. (A) H1299 cells were transfected with the following constructs as indicated: Flag-p53 (0.3 μg), MLL-ELL (2, 5, and 10 μg), MLL-ELLΔCT (2 and 5 μg), and HA-p300 (1 μg). pEGFP (0.5 μg) served as a transfection efficiency control. Empty vector was used to correct for total DNA amount. Cell lysates were prepared 24 h posttransfection and Western blot analysis was carried out with the indicated antibody. (B) A similar experiment was carried out with 1, 2.5, and 5 μg of ELL and with 1 and 2.5 μg of ELLΔeCT. (C) Flag-p53 plasmid (25 ng) was cotransfected into H1299 cells with PG13 luciferase reporter and with or without Flag-ELL (250 ng) and HA-p300 (2 and 4 μg) as indicated. pRL-TK was included to control for transfection efficiency. Values shown are the fold induction in luciferase expression over the vector control (mean ± the SD of three separate experiments). (D) Gal4 and VP16 fusions of p300 and p53. (E) Protein expression of constructs depicted in panel D was analyzed by transfection of 293T cells, followed by Western blot with anti-Gal4 as described. (F) The indicated plasmids, as well as pGL3-G5SV and pRL-TK plasmids, were cotransfected into H1299 cells in the presence or absence of 2 μg of Flag-ELL or Flag-ELLΔeCT. The luciferase activity measured 36 h posttransfection is expressed as the fold induction over the vector control (mean ± the SD of three independent experiments). (G and H) The indicated Gal4 fusions (G), the protein levels of which were examined by Western blot (H), were transfected into H1299 cells with or without Flag-ELL or Flag-ELLΔeCT and luciferase reporters (pGL3-G5SV and pRL-TK). (I) The luciferase expression was measured as described 36 h after transfection and is shown as the fold induction compared with vector control in three separate transfection experiments (mean ± the SD). (J) The indicated p300 Gal4 and ELL VP16 constructs were cotransfected with pGL3-G5SV and pRL-TK into H1299 cells. (K) The luciferase activity was assessed 36 h posttransfection (the fold induction over the vector control is shown as the mean ± the SD of three separate experiments, each done in duplicate).
Since both MLL-ELL and ELL significantly interfered with p53 acetylation by p300, we hypothesized that p53 might be rescued from the inhibition by leukemic proteins if p300 is overexpressed. To test this possibility, H1299 cells were cotransfected with p53 and ELL, either in the presence or in the absence of increasing amounts of p300. Our data show that p53 transcriptional activity is efficiently inhibited by ELL (Fig. 5C, column 3 versus column 4) and that this inhibitory effect can indeed be alleviated, albeit partially, by overexpression of p300 (Fig. 5C, column 4 versus columns 5 and 6). These data show that disruption of p53 acetylation is one of the mechanisms of MLL-ELL and ELL inhibitory effect on p53.
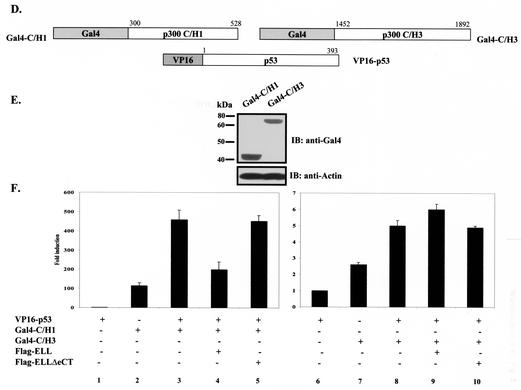
The C/H1 domain of p300 (aa 300 to 582) has been shown to bind to p53 in vivo and to be both necessary and sufficient to cause an increase in p53 transcriptional activity (29, 31). On the other hand, the C/H3 domain, which is also capable of binding to p53, is not required for transcriptional coactivator function of p300 (29). We therefore, investigated whether ELL can specifically disrupt p53 interactions with either of these two p300 domains in vivo by again utilizing the mammalian two-hybrid system. p300 sequences were fused to Gal4DBD as shown in Fig. 5D, and protein expression was confirmed by Western blot (Fig. 5E). Cotransfection of Gal4-C/H1 or Gal4-C/H3, both of which possessed endogenous transactivation activity, with VP16-p53 resulted in a further increase in Gal4 transactivation that is indicative of in vivo binding between full-length p53 and C/H1 or C/H3 domains of p300 (Fig. 5F, column 2 versus column 3 and column 7 versus column 8). This increase was completely abrogated in VP16-p53/Gal4-C/H1 but not in VP16-p53/Gal4-C/H3 cotransfection experiment when wild-type ELL, but not ELLΔeCT, was present (Fig. 5F, columns 4 and 5 versus column 3 and columns 9 and 10 versus column 8).
To confirm that ELL does not act as a general inhibitor of Gal4-VP16-mediated transcription and exerts its effects on p53 and not p300, Gal4-p53 and Gal4-p53(NT) were generated and, along with other Gal4 fusions (Fig. 5G), used to address this question. Equal protein expression of p53 Gal4 fusions is shown in Fig. 5H. As previously reported by others (20), ELL had no effect on Gal4-VP16 transcriptional activity (Fig. 5I, columns 1 to 3). The expression of ELL also did not alter the endogenous transcriptional activity of either Gal4-C/H1 or Gal4-C/H3 (Fig. 5I, columns 10 to 15) while substantially suppressing the activity of Gal4-p53 and Gal4-p53(NT) in the same assay (Fig. 5I, columns 4 to 9). Consistently, ELLΔeCT had no inhibitory effect. These data indicate that ELL specifically interferes with p53/p300 C/H1 domain binding in vivo and confirm the ELL inhibitory effect on the p53 N terminus rather than on the p300 C/H1 domain.
To provide further direct evidence that ELL elicits its effects by binding to p53 and not p300, we conducted in vivo binding experiments with Gal4/VP16 constructs of p300 and ELL (Fig. 5J). Cotransfection of Gal4-C/H1 or Gal4-C/H3 with VP16 constructs of various parts of ELL did not cause any increase in luciferase activity, indicating a lack of direct in vivo interactions between p300 C/H1 or C/H3 domains and ELL (Fig. 5K).
Collectively, our data demonstrate that the disruption of p53/p300 association and the subsequent decrease in acetylated p53 is an important component of MLL-ELL and ELL inhibitory effect.
The extreme C terminus of ELL is necessary and sufficient for MLL-ELL inhibition of p53-mediated induction of endogenous p21 and apoptosis.
Cellular transformation by an oncoprotein such as MLL-ELL depends in part on its ability to successfully override p53 antiproliferative effects. Since ELL(eCT) has been shown to mediate malignant transformation of normal blood progenitors by MLL-ELL and our data have implicated this region in the inhibition of p53 transactivation in luciferase assays, we sought to determine whether ELL(eCT) might be necessary and sufficient to suppress p53-mediated induction of endogenous p21 and apoptosis. To address this question, H1299 cells were transfected with various MLL-ELL and ELL constructs in the presence or absence of exogenous p53 expression, and cell lysates were analyzed for p21 expression at 24 h posttransfection. As shown in Fig. 6A (fifth panel, lanes 1 to 5) and B (sixth panel, lanes 1 to 7), endogenous p21 was extremely low in the absence of p53 and the expression of leukemic proteins did not have any effect on the basal p21 protein level. Once p53 was introduced, a marked increase in p21 expression was recorded in cells transfected with either vector alone, MLL-ELLΔCT (Fig. 6A, fifth panel, lanes 6, 9, and 10), or ELLΔeCT (Fig. 6B, sixth panel, lanes 8, 11, and 12). In sharp contrast, p53-mediated p21 induction was severely diminished in cells expressing either full-length MLL-ELL or ELL (Fig. 6A, fifth panel, lanes 7 and 8, and B, sixth panel, lanes 9 and 10, respectively). Coexpression of ELL(eCT) also resulted in substantially reduced p21 level (Fig. 6B, sixth panel, lanes 13 and 14), even more so than when full-length ELL was used, demonstrating that the extreme C terminus of ELL is both necessary and sufficient to suppress p21 upregulation mediated by p53.
FIG. 6.
ELL(eCT) is necessary and sufficient to suppress both p53- and p53(6KR)-mediated induction of p21. (A) H1299 cells were transfected with the following constructs using Lipofectamine: Flag-p53 (1 μg) or MLL-ELL or MLL-ELLΔCT (5 or 10 μg of each cDNA). pEGFP (0.5 μg) was included as a transfection efficiency control. (B) A similar experiment was conducted with 2 and 5 μg of Flag-ELL, Flag-ELLΔeCT, and Myc-ELL(eCT). Cell lysates prepared 24 h posttransfection were probed with the indicated antibody. (C) H1299 cells transfected with Flag-p53 (0.3 μg), Flag-p53(6KR) (0.3 μg), and HA-p300 (1 μg) were harvested 24 h posttransfection, and cell lysates were analyzed by Western blot with the indicated antibody. (D) H1299 cells were transfected as shown with p53(6KR) (1 μg) and ELL constructs (2 and 5 μg), and after 24 h the cell lysates were analyzed with the indicated antibody.
Since we have previously shown that leukemic proteins can disrupt p53 acetylation by p300, we next set out to investigate whether abrogation of p53 acetylation is an exclusive mechanism by which MLL-ELL and ELL inhibit p53. We utilized p53 mutant in which all C-terminal lysines, two of which are subject to acetylation by p300 (Lys 373 and 382), have been mutated to arginines [p53(6KR)]. As expected, when p300 was coexpressed, anti-AceK antibody directed against acetylated p53 Lys 372 and Lys 382 failed to detect p53(6KR), whereas acetylated forms of wild-type p53 were readily recognized (Fig. 6C, third panel, lane 4 vesus 3). Nonetheless, both ELL and ELL(eCT) had a discernible inhibitory effect on p53(6KR)-mediated induction of p21 (Fig. 6D, fourth panel, lanes 3, 4, 7, and 8 versus lane 2), indicating that disruption of p300-mediated acetylation of p53 C terminus is not the only mechanism of p53 inhibition by leukemic proteins.
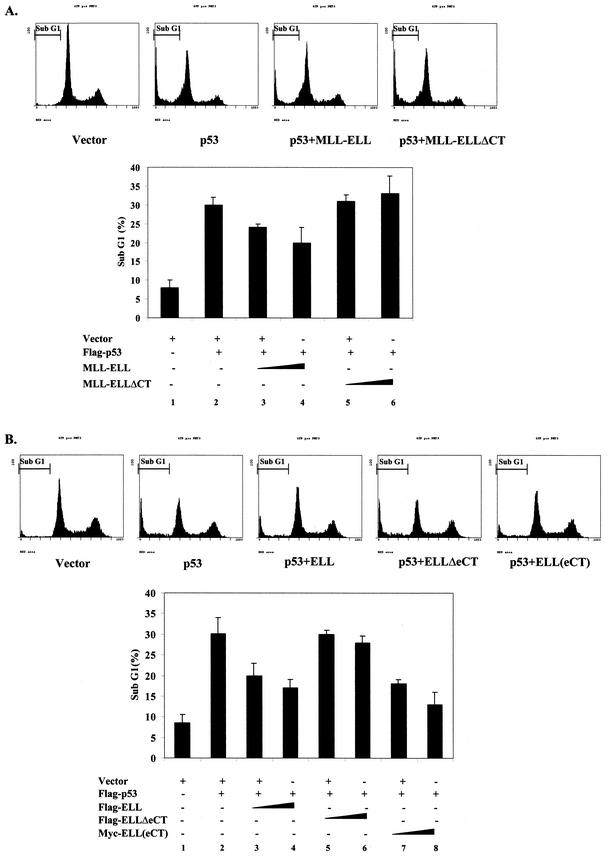
Finally, we examined the effect MLL-ELL and ELL on p53-driven apoptosis. H1299 cells were transfected with p53 in the presence or absence of MLL-ELL or ELL expression plasmids, as well as with membrane-anchored GFP construct that served as an indicator of transfection. After propidium iodide staining at 36 h posttransfection, positively transfected cells were analyzed for their DNA content by using flow cytometry. The number of cells in the sub-G1 phase of the cell cycle, which represents apoptotic population, was very low in cells transfected with vector only (Fig. 7A, column 1, and B, column 1). Expression of neither leukemic protein had any significant effect on the baseline apoptosis in H1299 cells (data not shown). Exogenous p53 caused a >3-fold increase in apoptosis in cells coexpressing either vector alone, MLL-ELL(ΔCT), or ELL(ΔeCT) (Fig. 7A, columns 2, 5, and 6, and B, columns 2, 5, and 6). However, cells cotransfected with either full-length MLL-ELL (Fig. 7A, columns 3 and 4) or ELL or ELL(eCT) (Fig. 7B, columns 3 and 4 and columns 7 and 8, respectively) displayed dose-dependent resistance to p53 apoptosis. Once again, ELL(eCT) was more effective than full-length ELL in inhibiting the apoptotic function of p53. These data demonstrate that MLL-ELL and ELL can suppress p53-mediated apoptosis and that ELL(eCT) is both necessary and sufficient for this effect.
FIG. 7.
ELL(eCT) is necessary and sufficient to inhibit p53-induced apoptosis. (A) Flag-p53 (1 μg), MLL-ELL or MLL-ELLΔCT (5 and 10 μg each), and Us9-GFP (0.5 μg) were transfected into H1299 cells with Lipofectamine. After 36 h of incubation, cells were fixed in 70% ethanol for 24 to 72 h and stained with propidium iodide solution. Approximately 10,000 GFP-positive cells from each sample were then analyzed for their DNA content by using flow cytometry. The percentage of cells in sub-G1 phase is shown as the mean ± the SD of three independent experiments. Representative histograms from each transfection are also shown. (B) An experiment identical to that in panel A was conducted with Flag-ELL, Flag-ELLΔeCT, and Myc-ELL(eCT) (5 and 10 μg each).
DISCUSSION
It is believed that leukemic fusion proteins that result from chromosomal translocations directly contribute to clinical progression and the outcome of leukemia. MLL-ELL translocation has been associated with particularly poor patient response to conventional chemotherapy and diminished survival (21). Since p53 is a major mediator of apoptosis and cell cycle arrest, we sought to further elucidate the complex molecular interactions between MLL-ELL and p53 tumor suppressor.
In our experiments, we were able to achieve only low levels of MLL-ELL expression, which is consistent with a number of previous reports that showed barely detectable MLL-ELL protein in both stable and transient transfections (4, 15). Whether low MLL-ELL levels are due to the general toxicity of this fusion protein or to its inherent instability is not entirely clear. However, normal blood progenitors expressing low amounts of MLL-ELL readily undergo malignant transformation and, upon transplantation into mice, cause leukemic disease (4). We show here that MLL-ELL, even at low levels, is a powerful inhibitor of p53 functional activity, which could be contributing to its overall pro-oncogenic function. On the other hand, a much higher expression of ELL, far above its physiological level, was needed to inhibit p53, which suggests that under normal conditions ELL might not be acting as a suppressor of p53 function. Nevertheless, overexpression of ELL has been shown to transform Rat1 fibroblasts, thus placing it in the category of potential oncogenes (14). Therefore, further evaluation of ELL status in human malignancies is warranted.
In our study, overexpression of ELL and its truncated mutants served as a useful tool to examine in detail the contribution of each ELL domain to suppression of p53. We found that ELL elongation activity is completely dispensable to its ability to inhibit p53 but that intact ELL(eCT) is required for in vivo binding and inhibition of p53 transactivation. These results tightly correlate with and provide a molecular basis for the previously reported requirement of ELL(eCT) for transformation of normal bone marrow cells by MLL-ELL (4, 19). Interestingly, we found ELL(eCT) to be more effective than full-length ELL in suppressing p53-mediated induction of p21 and apoptosis, suggesting that ELL might undergo conformational changes upon fusion to MLL, resulting in an enhanced interaction of ELL(eCT) with p53.
Conforming to the observed functional requirement of ELL(eCT) to p53 inhibition by the leukemic proteins, the same region of ELL was also necessary for the colocalization of MLL-ELL with p53. It is well established that MLL-ELL fusion follows the wild-type MLL pattern of localization and forms distinct nuclear foci, a phenomenon that we observed in our study, while ELL displays diffuse nuclear staining (3, 24). As our results show, p53 is actively recruited into MLL-ELL nuclear foci. However, foci formation per se, as was seen with MLL-ELLΔCT, does not affect normal p53 subnuclear localization. It is plausible that altered distribution of p53 in the presence of MLL-ELL is one of the mechanisms by which the leukemic protein inhibits p53 function.
To further address the mechanism of MLL-ELL inhibitory effect on p53, we sought to identify the p53 domain that renders it sensitive to the inhibition by MLL-ELL. Our results implicate p53(TAD) as the region that is both necessary and sufficient for p53 association with MLL-ELL and ELL. Our experimental approach was different from that previously used by other investigators. Instead of utilizing GST pull-down assays and in vitro-translated proteins, we assessed functional contribution of each p53 domain to the inhibitory effect of ELL by studying transcriptional activity of p53/p73 chimeras in the presence or absence of ELL in vivo. We then supplemented our functional data with in vivo binding experiments and immunostaining, all of which convincingly demonstrate that p53(TAD) is both necessary and sufficient to render p53 sensitive to the inhibition by MLL-ELL and ELL.
Indeed, the association of leukemic proteins with p53(TAD) can better explain their inhibitory effect on p53-driven transcription. The N terminus of p53, and TAD in particular, is the site of multiple interactions with both basal transcriptional machinery and important coactivators, such as p300/CBP, all of which are required for p53 transactivation function (17). This region is also subject to negative control by inhibitors of p53 function. For example, MDM2, a major negative regulator of p53, binds to p53(NT) and interferes with the recruitment of p300, resulting in the inhibition of p53 transcriptional activity (29). Another repressor of p53 function, Rad23, has been shown to compete with p53 for binding to p300 C/H1 domain, also leading to significant downregulation of p53 transactivation function (31). We show here that ELL acts directly on p53 to interfere with p300 recruitment, without any detectable association with p300 itself. Our data also show that abrogation of p300 acetylation is not the only mechanism by which MLL-ELL and ELL negatively regulate p53, since ELL inhibitory effect was only partially rescued by overexpression of p300 and p53(6KR) mutant was susceptible to the inhibition by ELL.
However, positive effects of p300 on p53 might not be limited to the acetylation of two C-terminal lysines and could include recruitment of additional coactivators to p53, as well as acetylation of other lysine residues that are not recognized by the antibody used in our study. Abrogation of these auxiliary stimulatory effects of p300 might explain the ELL inhibitory effect on p53(6KR). Apart from interference with p53/p300 interactions, MLL-ELL and ELL might also be preventing basal transcriptional machinery from binding to p53(TAD) and/or impinging on phosphorylation of several critical residues in this region of p53.
In conclusion, our data reveal a crucial role for ELL(eCT) and p53(TAD) in the inhibition of p53 activity by the leukemic protein MLL-ELL. Specifically disrupting the p53/MLL-ELL complex in cells that contain the t(11;19)(q23;p13.1) translocation could prove to be an attractive therapeutic option in treating patients suffering from leukemia.
Acknowledgments
We thank Vanessa Lopez-Pajares and Aida Kuri for excellent technical assistance, Amy Imrich for help with flow cytometry analysis, Y. Shi (Harvard Medical School) for Gal4DBD and VP16TAD expression plasmids, and Dieter Wolf (Harvard School of Public Health) for anti-Myc antibodies.
D.W. is supported by a training grant from the National Institute of Environmental Health Sciences (T32ES07155) and by Harvard Merit fellowship. This work was supported by NIH (RO1CA85679-01) and American Cancer Society (RSG GMC-104886) grants to Z.-M.Y.
REFERENCES
- 1.Ayton, P. M., and M. L. Cleary. 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20:5695-5707. [DOI] [PubMed] [Google Scholar]
- 2.Bates, S., and K. H. Vousden. 1999. Mechanisms of p53-mediated apoptosis. Cell Mol. Life Sci. 55:28-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler, L. H., R. Slany, X. Cui, M. L. Cleary, and D. Y. Mason. 1997. The HRX proto-oncogene product is widely expressed in human tissues and localizes to nuclear structures. Blood 89:3361-3370. [PubMed] [Google Scholar]
- 4.DiMartino, J. F., T. Miller, P. M. Ayton, T. Landewe, J. L. Hess, M. L. Cleary, and A. Shilatifard. 2000. A carboxy-terminal domain of ELL is required and sufficient for immortalization of myeloid progenitors by MLL-ELL. Blood 96:3887-3893. [PubMed] [Google Scholar]
- 5.Downes, M., L. J. Burke, P. J. Bailey, and G. E. Muscat. 1996. Two receptor interaction domains in the corepressor, N-CoR/RIP13, are required for an efficient interaction with Rev-erbA alpha and RVR: physical association is dependent on the E region of the orphan receptors. Nucleic Acids Res. 24:4379-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleckenstein, D. S., C. C. Uphoff, H. G. Drexler, and H. Quentmeier. 2002. Detection of p53 gene mutations by single strand conformational polymorphism (SSCP) in human acute myeloid leukemia-derived cell lines. Leukoc. Res. 26:207-214. [DOI] [PubMed] [Google Scholar]
- 7.Graf, J., B. Merk, U. Maurer, E. Muller, and L. Bergmann. 2001. Identification of novel polymorphisms in intron 7 of the human p53 gene in acute myeloid leukemia and healthy donors. Leukoc. Lymphoma 41:655-658. [DOI] [PubMed] [Google Scholar]
- 8.Grossman, S. R. 2001. p300/CBP/p53 interaction and regulation of the p53 response. Eur. J. Biochem. 268:2773-2778. [DOI] [PubMed] [Google Scholar]
- 9.Gu, J., D. Chen, J. Rosenblum, R. M. Rubin, and Z. M. Yuan. 2000. Identification of a sequence element from p53 that signals for Mdm2-targeted degradation. Mol. Cell. Biol. 20:1243-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu, J., L. Nie, D. Wiederschain, and Z. M. Yuan. 2001. Identification of p53 sequence elements that are required for MDM2-mediated nuclear export. Mol. Cell. Biol. 21:8533-8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollstein, M., D. Sidransky, B. Vogelstein, and C. C. Harris. 1991. p53 mutations in human cancers. Science 253:49-53. [DOI] [PubMed] [Google Scholar]
- 12.Jimenez, G. S., M. Nister, J. M. Stommel, M. Beeche, E. A. Barcarse, X. Q. Zhang, S. O'Gorman, and G. M. Wahl. 2000. A transactivation-deficient mouse model provides insights into Trp53 regulation and function. Nat. Genet. 26:37-43. [DOI] [PubMed] [Google Scholar]
- 13.Kalejta, R. F., A. D. Brideau, B. W. Banfield, and A. J. Beavis. 1999. An integral membrane green fluorescent protein marker, Us9-GFP, is quantitatively retained in cells during propidium iodide-based cell cycle analysis by flow cytometry. Exp. Cell Res. 248:322-328. [DOI] [PubMed] [Google Scholar]
- 14.Kanda, Y., K. Mitani, M. Kurokawa, T. Yamagata, Y. Yazaki, and H. Hirai. 1998. Overexpression of the MEN/ELL protein, an RNA polymerase II elongation factor, results in transformation of Rat1 cells with dependence on the lysine-rich region. J. Biol. Chem. 273:5248-5252. [DOI] [PubMed] [Google Scholar]
- 15.Kanda, Y., K. Mitani, T. Tanaka, K. Tanaka, S. Ogawa, Y. Yazaki, and H. Hirai. 1997. Subcellular localization of the MEN, MLL/MEN and truncated MLL proteins expressed in leukemic cells carrying the t(11;19)(q23;p13.1) translocation. Int. J. Hematol. 66:189-195. [DOI] [PubMed] [Google Scholar]
- 16.Lavau, C., R. T. Luo, C. Du, and M. J. Thirman. 2000. Retrovirus-mediated gene transfer of MLL-ELL transforms primary myeloid progenitors and causes acute myeloid leukemias in mice. Proc. Natl. Acad. Sci. USA 97:10984-10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 18.Lill, N. L., S. R. Grossman, D. Ginsberg, J. DeCaprio, and D. M. Livingston. 1997. Binding and modulation of p53 by p300/CBP coactivators. Nature 387:823-827. [DOI] [PubMed] [Google Scholar]
- 19.Luo, R. T., C. Lavau, C. Du, F. Simone, P. E. Polak, S. Kawamata, and M. J. Thirman. 2001. The elongation domain of ELL is dispensable but its ELL-associated factor 1 interaction domain is essential for MLL-ELL-induced leukemogenesis. Mol. Cell. Biol. 21:5678-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maki, K., K. Mitani, T. Yamagata, M. Kurokawa, Y. Kanda, Y. Yazaki, and H. Hirai. 1999. Transcriptional inhibition of p53 by the MLL/MEN chimeric protein found in myeloid leukemia. Blood 93:3216-3224. [PubMed] [Google Scholar]
- 21.Moorman, A. V., A. Hagemeijer, C. Charrin, H. Rieder, and L. M. Secker-Walker. 1998. The translocations, t(11;19)(q23;p13.1) and t(11;19)(q23;p13.3): a cytogenetic and clinical profile of 53 patients. Leukemia 12:805-810. [DOI] [PubMed] [Google Scholar]
- 22.Seol, W., M. Chung, and D. D. Moore. 1997. Novel receptor interaction and repression domains in the orphan receptor SHP. Mol. Cell. Biol. 17:7126-7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shilatifard, A., W. S. Lane, K. W. Jackson, R. C. Conaway, and J. W. Conaway. 1996. An RNA polymerase II elongation factor encoded by the human ELL gene. Science 271:1873-1876. [DOI] [PubMed] [Google Scholar]
- 24.Shinobu, N., T. Maeda, T. Aso, T. Ito, T. Kondo, K. Koike, and M. Hatakeyama. 1999. Physical interaction and functional antagonism between the RNA polymerase II elongation factor ELL and p53. J. Biol. Chem. 274:17003-17010. [DOI] [PubMed] [Google Scholar]
- 25.So, C. W., and M. L. Cleary. 2002. MLL-AFX requires the transcriptional effector domains of AFX to transform myeloid progenitors and transdominantly interfere with forkhead protein function. Mol. Cell. Biol. 22:6542-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stirewalt, D. L., and J. P. Radich. 2000. Malignancy: tumor suppressor gene aberrations in acute myelogenous leukemia. Hematology 5:15-25. [DOI] [PubMed] [Google Scholar]
- 27.Sweetser, D. A., C. S. Chen, A. A. Blomberg, D. A. Flowers, P. C. Galipeau, M. T. Barrett, N. A. Heerema, J. Buckley, W. G. Woods, I. D. Bernstein, and B. J. Reid. 2001. Loss of heterozygosity in childhood de novo acute myelogenous leukemia. Blood 98:1188-1194. [DOI] [PubMed] [Google Scholar]
- 28.Thirman, M. J., D. A. Levitan, H. Kobayashi, M. C. Simon, and J. D. Rowley. 1994. Cloning of ELL, a gene that fuses to MLL in a t(11;19)(q23;p13.1) in acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 91:12110-12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wadgaonkar, R., and T. Collins. 1999. Murine double minute (MDM2) blocks p53-coactivator interaction, a new mechanism for inhibition of p53-dependent gene expression. J. Biol. Chem. 274:13760-13767. [DOI] [PubMed] [Google Scholar]
- 30.Yuan, Z. M., H. Shioya, T. Ishiko, X. Sun, J. Gu, Y. Y. Huang, H. Lu, S. Kharbanda, R. Weichselbaum, and D. Kufe. 1999. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature 399:814-817. [DOI] [PubMed] [Google Scholar]
- 31.Zhu, Q., G. Wani, M. A. Wani, and A. A. Wani. 2001. Human homologue of yeast Rad23 protein A interacts with p300/cyclic AMP-responsive element binding (CREB)-binding protein to downregulate transcriptional activity of p53. Cancer Res. 61:64-70. [PubMed] [Google Scholar]