Abstract
A series of efficiently spliced pre-mRNA substrates containing single 4-thiouridine residues were used to monitor RNA-protein interactions involving the branch site-3′ splice site-3′ exon region during yeast pre-mRNA splicing through cross-linking analysis. Prior to the assembly of the prespliceosome, Mud2p and the branch point bridging protein cross-link to a portion of this region in an ATP-independent fashion. Assembly of the prespliceosome leads to extensive cross-linking of the U2-associated protein Hsh155p to this region. Following the first step of splicing and in a manner independent of Prp16p, the U5 small nuclear ribonucleoprotein particle-associated protein Prp8p also associates extensively with the branch site-3′ splice site-3′ exon region. The subsequent cross-linking of Prp16p to the lariat intermediate is restricted to the 3′ splice site and the adjacent 3′ exon sequence. Using modified substrates to either mutationally or chemically block the second step, we found that the association of Prp22p with the lariat intermediate represents an authentic transient intermediate and appears to be restricted to the last eight intron nucleotides. Completion of the second step leads to the cross-linking of an unidentified ∼80-kDa protein near the branch site sequence, suggesting a potential role for this protein in a later step in intron metabolism. Taken together, these data provide a detailed portrayal of the dynamic associations of proteins with the branch site-3′ splice site region during spliceosome assembly and catalysis.
The locations of introns within nascent yeast pre-mRNAs is specified largely by just three short conserved sequences located at and near the 5′ and 3′ ends of the intron (6, 47). These sequences are recognized during the assembly of the spliceosome, a massive macromolecular complex consisting of the U1, U2, U4/U6, and U5 small nuclear ribonucleoprotein particles (snRNPs) and at least 60 associated proteins (31, 57). The spliceosome not only must precisely recognize intron boundary sequences to avoid the introduction of catastrophic frameshift errors in coding regions but also must be able to accommodate significant variations in intron size, sequence, and organization. A full understanding of how accurate splice site recognition is achieved will require detailed knowledge of the dynamic contacts between the conserved sequences that define intron boundaries and the components of the spliceosome.
Nuclear pre-mRNA splicing proceeds via two sequential transesterification reactions. In the first catalytic step, the 2′ hydroxyl group of the last adenosine residue in the conserved branch site sequence (UACUAAC) attacks the phosphodiester bond at the 5′ splice site to yield free 5′ exon and branched lariat intermediates. The branch site and 5′ splice site sequences are initially recognized by binding of the branch point bridging protein (BBP) (5) and the U1 snRNA (45), respectively. These sequences are then recognized a second time through interactions involving the U2 and U6 snRNAs prior to the first catalytic step (22, 23). In the second step, the 3′ hydroxyl of the 5′ exon intermediate attacks the phosphodiester bond at the 3′ splice site to produce ligated exons and release the excised intron lariat. Genomic analysis of intron sequences in the yeast Saccharomyces cerevisiae has shown that the conserved YAG↓ sequence that defines the 3′ splice site is located between 9 and 143 nucleotides (nt) downstream of the branch site sequence (47).
The branch site-3′ splice site region is initially recognized through binding of the BBP and Mud2p, the yeast ortholog of the mammalian splicing factor U2AF65 (1, 5). Selection of the 3′ splice site for the second step is tightly coupled to the prior recognition of the branch site sequence, as the first AG downstream of the pre-mRNA branch site sequence is generally chosen, with an optimal spacing of 18 to 22 nt (10, 26, 37). Genetic analyses in yeasts have provided strong evidence that non-Watson-Crick interactions with the conserved ↓GUA at the 5′ splice site and the highly conserved GAG sequence in the U6 snRNA function in binding of the 3′ splice site YAG↓ for the second catalytic step (14, 36). Other genetic analyses have also suggested an important role for intron sequences adjacent to the 3′ splice site in 3′ splice site selection (37).
Six yeast proteins, Prp8p, Prp16p, Prp17p, Slu7p, Prp18p, and Prp22p, have been shown to be involved in the second catalytic step (42, 53). Four of these proteins, Prp8p, Prp16p, Prp22p, and Slu7p, have each been reported to cross-link at or near the 3′ splice site (30, 48, 49, 52). U2 snRNA and ATP-dependent cross-linking of the 3′ splice site to an additional, unidentified ∼122-kDa protein has also been reported, but the role of this protein, if any, in 3′ splice site selection is unknown (30). The relevance of the cross-linking of Prp22p to the 3′ splice site during normal splicing is also unclear, as strong cross-linking of Prp22p has been reported to occur with mutant substrates unable to complete the second step (30). A central role for Prp8p has been suggested from the isolation of prp8 mutants that affect the fidelity of 3′ splice site recognition (13, 15, 50, 51). ATP hydrolysis by Prp16p, as part of a complex involving Prp17p, Slu7p, Prp18p, and Prp22p, facilitates a conformational rearrangement that involves the 3′ splice site and that leads to its protection from oligonucleotide-directed RNase H cleavage. This conformational rearrangement appears to occur in two stages, an initial ATP-dependent stage involving Prp16p and Prp17p and a subsequent ATP-independent stage involving Slu7p, Prp18p, and Prp22p (42, 53). Previous results suggested that the protection of the 3′ splice site from RNase H cleavage is most likely due to the Prp16-dependent interaction of Prp22p with the 3′ splice site (30). In a series of elegant experiments, Schwer and colleagues (5a, 43, 58) showed that the in vitro requirements for Slu7p, Prp18p, and Prp22p in the second step can be alleviated if actin-based pre-mRNA substrates containing short intervals between the branch site and the 3′ splice site are used. These results suggested that Slu7p, Prp18p, and Prp22p may act to form a bridge between or stabilize the interactions of factors bound to the branch site and the 3′ splice site (6, 43, 58; however, see reference 12). ATP hydrolysis by Prp16p has also been linked to the use of a discard pathway for aberrant lariat intermediates in which Prp16p acts as a clock to time the rate of a spontaneous conformational change linked to the identity of the branch site nucleotide (7, 8).
To better understand the spatial organization and dynamics of protein-RNA interactions involved in branch site-3′ splice site selection in S. cerevisiae, we have developed a series of closely related, efficiently spliced cross-linking substrates. Using these substrates, we found that upon prespliceosome formation, the yeast ortholog of the mammalian U2-associated splicing factor SAP155 (19, 38) becomes associated with a large region encompassing the branch site, 3′ splice site, and 3′ exon sequences. Following the first catalytic step of splicing, the U5 snRNP-associated protein Prp8p associates extensively with the 3′ splice site region in a Prp16p-independent manner. Despite the previous implication of Prp16p in the recognition of the branch site nucleotide (7, 16), cross-linking of Prp16p to sequences within or near the branch site is not observed. Instead, cross-linking of Prp16p is restricted to the last four intron nucleotides together with at least 13 nt of the adjacent 3′ exon. Inhibition of the second catalytic step using 3′ splice site mutant substrates results in the accumulation of cross-links to Prp22p in the last eight intron nucleotides. Identical strong cross-linking of Prp22p to wild-type 3′ splice sites can be observed when the second step is blocked by phosphorothioate substitution of the nonbridging Rp oxygen at the 3′ splice site. These results demonstrate that the association of Prp22p with the 3′ end of the intron represents an authentic transient intermediate that normally exists prior to the second catalytic step. Finally, we provide evidence that an unidentified ∼80-kDa cross-linked species interacts specifically with intron sequences downstream of the branch site following exon ligation. The distinct spatial and temporal associations of these and other proteins with the branch site-3′ splice site region provide a detailed portrayal of the overall organization of this region during the initial stages of intron recognition, spliceosome assembly, and catalysis.
MATERIALS AND METHODS
Synthetic oligodeoxyribonucleotides were obtained from either Integrated DNA Technologies, Coralville, Iowa, or Cybersyn, Aston, Pa., and were either cartridge purified by the manufacturer or purified by denaturing polyacrylamide gel electrophoresis (PAGE). To create the plasmid template for PCRs used in these experiments, the fragment containing the wild-type yeast actin intron under the control of an SP6 promoter (24) was cloned by PCR into the PstI-EcoRI sites of pUC18. Then, the sequence encompassing the branch site-3′ splice site and the first 13 nt of the 3′ exon were altered to create pACM13 (actin-modified pre-mRNA with a 13-nt space between the branch site and the 3′ splice site; the parent sequence is shown in Fig. 1). The Δ6 mutation was also incorporated into pACM13 to eliminate a cryptic branch site located immediately upstream of the normal branch site (55).
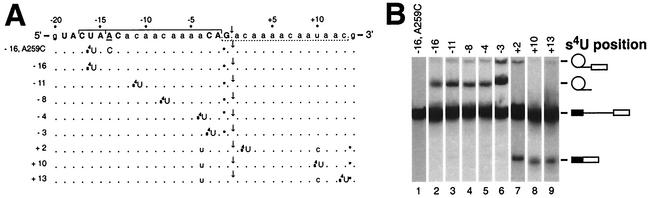
FIG. 1.
Photochemical cross-linking substrates used in this study. (A) Sequences of the branch site-3′ splice site-3′ exon region in ACM13 cross-linking substrates. The top line shows the sequence of plasmid pACM13, which served as the parent for each cross-linking substrate. The conserved branch site and 3′ splice site sequences are shown in boldface, uppercase letters, and the branch site nucleotide is underlined. The brackets above and below this sequence represent the boundaries of the s4U-containing RNase T1 fragments and/or phosphorothioate-containing oligonucleotides ligated together to make individual substrates. The sequence of each cross-linking substrate is shown below with notation of any sequence changes from the parent sequence; a dot indicates bases identical to those in the parent sequence, s4U indicates the site of an s4U substitution, an asterisk indicates the position of the single 32P label, and a downward-pointing arrow indicates the position of 3′ splice site cleavage. (B) Representative splicing assays for each s4U-containing pre-mRNA substrate. A portion of a sample from the 20-min time point was removed prior to cross-linking, and splicing products were analyzed on a 5% (29:1) denaturing polyacrylamide gel. The migration positions are from top to bottom of intron lariat intermediates, intron lariat products, pre-mRNA, and spliced exons at the right.
PCR with Vent DNA polymerase (New England Biolabs) was used to create templates for in vitro transcription and was performed according to the manufacturer's guidelines. A large number of oligodeoxyribonucleotides were used to generate these templates, as bridging oligonucleotides for in vitro RNA ligation, and for the in vitro transcription of short RNAs containing single 4-thiouridine (s4U) residues. A complete list of the synthetic oligodeoxyribonucleotides used in this study is available upon request. In vitro transcription reactions were performed as previously described (30). The ACM13(−11) transcript (s4U at position −11) lacking a 5′ splice site was created by PCR to position an SP6 promoter so that the transcript was initiated at the second conserved G of the normal actin 5′ splice site sequence. Short RNAs containing chiral phosphorothioate substitutions were made by chemical synthesis (Dharmacon, Inc.) followed by separation of Rp and Sp phosphorothioate stereoisomers by reverse-phase high-pressure liquid chromatography (46). These RNAs were incorporated into ACM13 pre-mRNA substrates together with in vitro-transcribed s4U-containing RNAs by four-way in vitro RNA ligations.
In vitro RNA ligations (33) were performed either as previously described (30) or by a modified procedure (27). Typically, in this modified procedure, 45 pmol of unlabeled 5′ RNA (∼348 nt) was combined with 45 pmol of s4U-containing oligonucleotide RNA (∼13 nt), 15 pmol of 5′ 32P-labeled 3′ RNA (∼174 nt), and 15 pmol of bridging oligonucleotide (∼43 nt) in 9.0 μl of double-distilled water, heated to 60°C for 1 min, and incubated on ice. Next, 2.5 μl of 5× DNA dilution buffer followed by 12.5 μl of 2× DNA ligation buffer (Rapid DNA ligation kit; Roche) were added and gently mixed. Subsequently, 1.0 μl of RNasin (40 U/μl; Promega) and 2.5 μl of T4 DNA ligase (5 U/μl; Roche) were added and gently mixed before incubation of the reaction mixture for 1 h at room temperature (final volume, 27.5 μl). Following incubation, 22.0 μl of 2× urea loading buffer (0.8 volume; 2× urea loading buffer contains 0.22 g urea, 194 μl of double-distilled water, 2 μl of 10× Tris-borate-EDTA, 4 μl of 2% [wt/vol] bromophenol blue, and xylene cyanol) was added to the reaction mixture before heating to 65°C for 2 min and purification of the full-length ligation product (535 nt) on 5% (29:1) denaturing polyacrylamide gels.
Whole-cell yeast splicing extracts were prepared as previously described (24) from strain EJ101 (24), EJ101-Hsh155-13Myc (38), EJ101-Mud2-13Myc (this study), or EJ101-BBP-13Myc (this study). The epitope-tagged yeast strains EJ101-Mud2-13Myc and EJ101-BBP-13Myc were constructed as previously described (25). The presence of the 13-Myc tag on the desired protein in each strain was confirmed by both PCR analysis and Western blotting. Oligonucleotide-directed RNase H depletion of specific snRNAs in yeast splicing extracts, splicing reactions, UV cross-linking, and immunoprecipitation reactions were performed as previously described (30). Splicing extracts immunodepleted of Prp16p were prepared as previously described (3) by using affinity-purified Prp16 antibodies (a gift from B. Schwer). Cross-linked proteins were uniformly analyzed on 30-cm sodium dodecyl sulfate (SDS)-7% (29:1) polyacrylamide gels run at 250 V for ∼7 h until the bromophenol blue was ∼1 cm from the bottom. Following electrophoresis, the gels were stained with 2% Coomassie blue-45% methanol-10% acetic acid for 50 min, destained with 45% methanol-10% acetic acid for 50 min, and dried to allow for the visualization of molecular mass standards (Benchmark Protein Ladder; Gibco). These gels allowed for the routine resolution of proteins from ∼40 kDa to greater than 220 kDa and, importantly, for the reproducible resolution of cross-linked Prp16p (native molecular mass, ∼122 kDa) from cross-linked Prp22p (native molecular mass, ∼130 kDa). When higher-percentage SDS-polyacrylamide gels were used to examine possible lower-molecular-mass cross-linked species, only a very few species with apparent molecular masses of less than 40 kDa were observed. The cross-linking of proteins with molecular masses of less than ∼40 kDa was often variable and usually resulted in bands that were diffuse in appearance (data not shown). We attributed the diffuse nature of these lower-molecular-mass bands to partial digestion of the 32P-labeled, cross-linked RNA fragments by an endogenous nuclease(s) in the extracts.
RESULTS
Design and splicing of pre-mRNA substrates for photochemical cross-linking.
To precisely map sites of close association of proteins with sequences in the branch site-3′ splice site region and adjacent 3′ exon sequences, site-specific incorporation of the uridine analog s4U was used. s4U is a short-range (2- to 3-Å) cross-linking reagent that can be selectively photoactivated by using long-wavelength (320-nm) UV light and that efficiently forms cross-links to both proteins and RNAs (17). To facilitate the unique incorporation of s4U, a derivative of the yeast actin pre-mRNA (ACM13) was designed to lack any RNase T1 cleavage sites (G residues) between the branch site and the 3′ splice site as well as any RNase T1 cleavage sites within the first 13 nt of the 3′ exon (Fig. 1A). Site-specific incorporation of s4U at positions −16 to +13 relative to the 3′ splice site was achieved by the in vitro transcription of either of two short RNAs containing only a single uridine residue, depending on whether the s4U residue was located upstream or downstream of the 3′ splice site (Fig. 1A; see Materials and Methods). These short s4U-containing RNAs were then used in either three- or four-way in vitro RNA ligation reactions to create site specifically modified, full-length pre-mRNA substrates. In the ACM13 substrates, most of the intron and exon sequences are identical to those in the efficiently spliced yeast Δ6 actin pre-mRNA (56; see below). During RNA ligations, single 32P labels (∼7,000 Ci/mmol) were also selectively incorporated into each substrate so that, following cross-linking, the 32P label (Fig. 1A) would be retained on the same RNase T1 fragment as the s4U residue.
As shown in Fig. 1B, each of the s4U-containing cross-linking substrates used in this study is efficiently spliced in vitro, even though the 13-base interval between the branch site and the 3′ splice site in the ACM13 substrates is less than the previously determined optimal distance of 18 to 22 bases (26). For substrates containing s4U upstream of the 3′ splice site (positions −16, −11, −8, −4, and −3; Fig. 1B, lanes 1 to 6), only intron-containing RNA species are visible, as the single 32P label is located between intron positions −1 and −2. Similarly, for substrates containing s4U downstream of the 3′ splice site (positions +2, +10, and +13; Fig. 1B, lanes 7 to 9), only 3′ exon-containing RNA species are visible, as the single 32P label is located in the 3′ exon between positions +13 and +14. Reverse transcription-PCR analysis of mRNAs produced in reactions with each of these substrates confirmed that only the 3′ splice site designated in Fig. 1A was utilized (data not shown). Correct 3′ splice site use was also confirmed by strong inhibition of the second step upon mutation of the consensus 3′ splice site sequence in several of the substrates from YAG↓ to ACG↓ (e.g., see Fig. 7). As expected (55), the ACM13 substrate containing an A→C substitution of the branch site nucleotide (A259C) was completely blocked for the first step of splicing (Fig. 1B, lane 1).
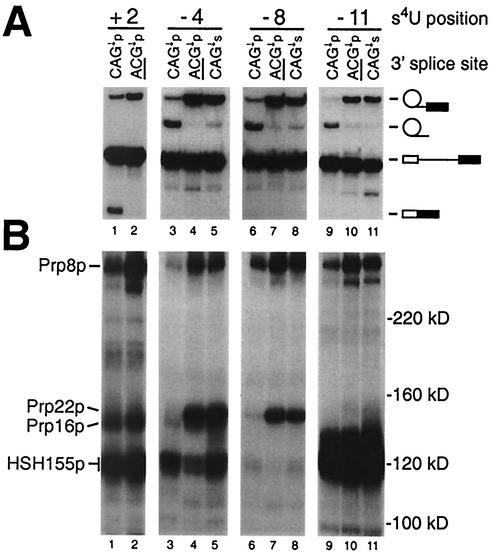
FIG. 7.
The interaction of Prp22p is limited to the 3′ terminus of the intron and represents a normal transient association that exists prior to the second step. Shown are analyses of splicing activity (A) and cross-linked proteins (B) with the ACM13(+2) substrate (lanes 1 and 2), the ACM13(−4) substrate (lanes 1 to 3), the ACM13(−8) substrate (lanes 4 to 6), and the ACM13(−11) substrate (lanes 7 to 9) containing a wild-type 3′ splice site (CAG↓p; lanes 1, 3, 6, and 9), a mutant 3′ splice site (ACG↓p; lanes 2, 4, 7, and 10), or an Rp phosphorothioate substitution of the nonbridging oxygen atom at the 3′ splice site (CAG↓s; lanes 5, 8, and 11). All reactions represent analyses of cross-linked proteins following 20 min of incubation in the presence of ATP.
Analysis of factors associated with intron sequences between the branch site and the 3′ splice site.
To map the extent of the interactions of both previously identified as well as unidentified proteins with intron sequences between the branch site and the 3′ splice site, substrates containing s4U at positions −3, −4, −8, and −11 were incubated with whole-cell yeast splicing extracts in the absence or presence of ATP for various lengths of time, exposed to long-wavelength UV light to induce the formation of cross-links, and treated with RNase T1; the resulting total 32P-labeled, cross-linked proteins were analyzed by SDS-PAGE. Cross-linked species specifically associated with spliceosome formation were identified on the basis of their ATP dependence and time of appearance relative to that of splicing products.
Previous studies demonstrated the ATP-independent interaction of BBP and Mud2p, the yeast ortholog of U2AF65, with the branch site region (2, 5). To facilitate the identification of potential cross-links to either BBP or Mud2p, splicing extracts were prepared from strains constructed to solely express C-terminal 13-Myc-tagged versions of these proteins. Any cross-links to these as well as other 13-Myc-tagged proteins (e.g., Hsh155p-13Myc) were expected to significantly retard their migration during SDS-PAGE, as the 13-Myc tag should increase the apparent molecular mass of each cross-linked species by approximately 13 kDa (see below).
With each of the ACM13 pre-mRNA substrates shown in Fig. 1A, cross-linking to between 9 and 14 ATP-independent species with apparent molecular masses of >50 kDa is observed (Fig. 2; see also Fig. 4 and 5). To enrich for early complexes in spliceosome assembly, such as commitment complexes and prespliceosomes, oligonucleotide-directed RNase H cleavage was also used to deplete the extracts of their endogenous U1, U2, or U6 snRNAs. The use of these approaches to identify proteins that cross-link within the intron at position −3, immediately adjacent to the 3′ splice site, is shown in Fig. 2A. In untagged (EJ101) extracts in both the presence and the absence of ATP (Fig. 2A, lanes 1 to 8) as well as in extracts depleted of their U1, U2, or U6 snRNAs (lanes 9 to 11), cross-linking to an ∼62-kDa species is observed. In the U1-depleted Mud2p-13Myc extract (Fig. 2A, lane 12), this 62-kDa band is replaced by a new, ∼94-kDa cross-linked species, demonstrating that the 62-kDa band corresponds to Mud2p. After 10 min of incubation of EJ101 extracts lacking added ATP (Fig. 2A, lane 2) or depleted of U2 snRNA (lane 10), significantly enhanced cross-linking of Mud2p but none of the other ATP-independent species is observed. In the U1-depleted BBP-13Myc extract, weak cross-linking to an ∼102-kDa species representing 13-Myc-tagged BPP is observed. However, there is no discernible disappearance of any of the lower-molecular-mass cross-linked species that would represent cross-linking to untagged (native) BBP (molecular mass, ≈53 kDa; see Discussion).
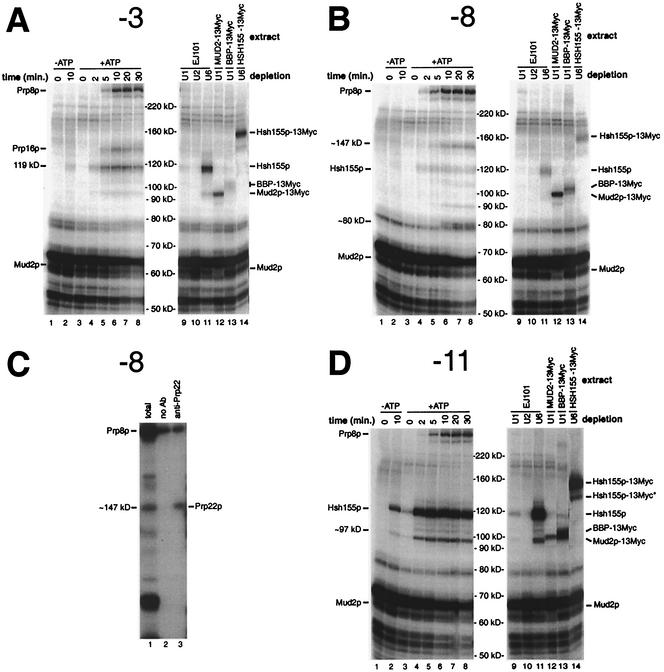
FIG. 2.
Analysis of total cross-linked proteins associated with the intron branch site-3′ splice site region. (A, B, and D) Analyses of proteins cross-linked to the ACM13(−3), ACM13(−8), and ACM13(−11) substrates, respectively. Lanes 1 and 2 show splicing reaction mixtures incubated in the absence of ATP for the indicated times prior to cross-linking; lanes 3 to 8 show splicing reaction mixtures incubated in the presence of ATP for the indicated times prior to cross-linking; and lanes 9 to 14 show cross-linked proteins from 10-min splicing reactions (+ATP) performed with the indicated extracts, which had been pretreated with oligonucleotides complementary to the U1, U2, or U6 snRNAs to effect the RNase H-mediated cleavage of these RNAs. Molecular masses shown correspond to the migration of Coomassie blue-stained molecular mass standards. (C) Identification of the ∼147-kDa cross-linked species at position −8 as Prp22p by immunoprecipitation analysis. All reaction mixtures correspond to samples cross-linked following 20 min of incubation in the presence of ATP. Lane 1 shows total cross-linked proteins; lane 2 shows cross-linked proteins bound to protein A-agarose beads only; and lane 3 shows cross-linked proteins bound to anti-Prp22p-protein A-agarose bead conjugates. Ab, antibody.
FIG. 4.
Analysis of total cross-linked proteins associated with the intron branch site. (A and B) Proteins cross-linked to the wild-type and A→C branch nucleotide mutant (A259C) versions of the ACM13(−16) substrate, respectively. In panel A, lanes 1 and 2 show splicing reaction mixtures incubated in the absence of ATP for the indicated times prior to cross-linking, and lanes 3 to 8 show splicing reaction mixtures incubated in the presence of ATP for the indicated times prior to cross-linking. Lanes 9 to 14 in panel A and lanes 1 to 6 in panel B show 10-min splicing reactions (+ATP) performed with the indicated extracts, which had been pretreated with oligonucleotides complementary to the U1, U2, or U6 snRNAs to effect the RNase H-mediated cleavage of these RNAs.
FIG. 5.
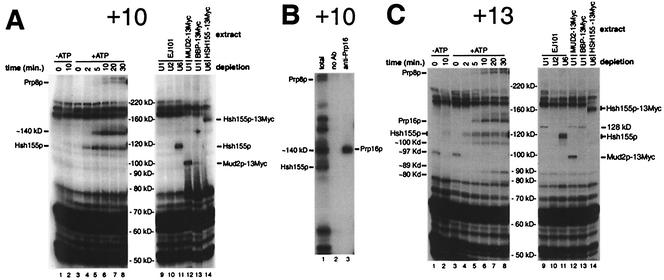
Analysis of total cross-linked proteins associated with the 3′ exon. (A and C) Analysis of proteins cross-linked to the ACM13(+10) and ACM13(+13) substrates, respectively. Lanes 1 and 2 show splicing reaction mixtures incubated in the absence of ATP for the indicated times prior to cross-linking; lanes 3 to 8 show splicing reaction mixtures incubated in the presence of ATP for the indicated times prior to cross-linking; and lanes 9 to 14 correspond to 10-min splicing reactions (+ATP) performed with the indicated extracts, which had been pretreated with oligonucleotides complementary to the U1, U2, or U6 snRNAs to effect RNase H-mediated cleavage of these RNAs. (B) Identification of the ∼136-kDa cross-linked species at position +10 as Prp16p by immunoprecipitation analysis. All reaction mixtures correspond to samples cross-linked following 20 min of incubation in the presence of ATP. Lane 1 shows total cross-linked proteins; lane 2 shows cross-linked proteins bound to protein A-agarose beads only; and lane 3 shows cross-linked proteins bound to anti-Prp16p-protein A-agarose bead conjugates.
Weak cross-linking to an ∼119-kDa species is seen after 10 min of incubation in the absence of added ATP with the ACM13(−3) substrate (Fig. 2A, lane 2) (30). The extracts in our experiments were not depleted of endogenous ATP and permit the formation of a small amount of prespliceosomes, the first ATP-dependent complexes in spliceosome assembly (21) (data not shown). Consistent with the ∼119-kDa cross-linked species being associated with prespliceosome formation, this species is the first cross-linked species observed in reaction mixtures supplemented with ATP. Cross-links to this ∼119-kDa species in the presence of added ATP are visible as early as ∼2 min (Fig. 2A, lanes 3 to 8), and this cross-linking requires intact U1 and U2 but not U6 RNA (lanes 9 to 11). Cross-linking of proteins of known sizes to a 14- to 19-nt RNA fragment results in a shift in the apparent molecular mass of between ∼2 and ∼17 kDa (unpublished data), suggesting the actual molecular mass of the un-cross-linked form of the 119-kDa species is between ∼102 and 117 kDa. Only one yeast protein involved in prespliceosome formation, Hsh155p (predicted molecular mass, ≅110 kDa), the yeast ortholog of the mammalian U2 snRNP-associated splicing factor SAP155, met these criteria (19, 38, 56). In U6-depleted extracts prepared from Hsh155p-13Myc, the addition of the 13-Myc tag to Hsh155p results in a shift in the apparent molecular mass of the ∼119-kDa species to ∼160 kDa (Fig. 2A, lane 14) and demonstrates that the ∼119-kDa cross-linked species corresponds to Hsh155p. Western blot analysis of Hsh155p-13Myc as well as both BBP-13Myc and Mud2p-13Myc in extracts showed only the expected increase in the molecular mass due to the addition of the 13-Myc tag (data not shown). The reason for the unexpectedly large shift (∼40 kDa versus ∼13 kDa) in the apparent molecular masses of the cross-linked 13-Myc-tagged forms of these proteins is not known.
Consistent with the lack of any detectable Mud2p in prespliceosomes (41), the increase in cross-linking of Hsh155p at position −3 occurs concurrently with a decrease in the cross-linking of Mud2p (Fig. 2A, lanes 3 to 8). As previously reported (30), following the appearance of cross-links to Hsh155p, cross-links to both Prp16p and Prp8p appear at ∼5 min (Fig. 2A, lanes 5 to 8). As expected, the appearance of cross-links to both Prp8p and Prp16p is dependent on intact U6 snRNA (Fig. 2A, lanes 9 to 11). With the ACM13(−4) substrate (Fig. 1A), identical patterns of cross-links to Mud2p, BBP-13Myc, Prp16p, and Prp8p are observed (data not shown). Strong cross-linking to both ∼117- and ∼119-kDa species at position −4 is also observed. In Hsh155p-13Myc extracts, both of these bands are replaced by higher-molecular-mass species, suggesting that they likely represent cross-linking to two different positions on the native protein. Weak cross-linking to an ∼147-kDa species at position −4, representing Prp22p, is also visible after prolonged incubation in the presence of ATP (see below).
Analyses of proteins that cross-link at either position −8 (Fig. 2B and C) or position −11 (Fig. 2D) show several significant similarities to as well as differences from the cross-linking observed at intron positions −3 and −4. Early ATP-independent cross-linking of Mud2p is visible at both positions −8 and −11 (Fig. 2B and D, lanes 1, 2, and 12), and this cross-linking appears to be enhanced in either ATP- or U2-depleted EJ101 extracts (lanes 2 and 10). In contrast to the weak cross-linking of BBP-13Myc seen at position −8 (Fig. 2B, lane 13), extremely strong cross-linking of BBP-13Myc is seen at position −11 (Fig. 2D, lane 13). However, despite the strong cross-linking of BBP-13Myc, no decrease in the intensity of any lower-molecular-mass cross-linked species corresponding to untagged BBP is apparent, strongly suggesting that this cross-linking involves the 13-Myc tag and not BBP itself.
Extremely strong cross-linking of Hsh155p is also seen 3 nt downstream of the branch site adenosine at position −11 (Fig. 2D, lanes 3 to 8, 11, and 16) and mirrors the previously observed strong cross-linking of SAP155 at the equivalent position in a mammalian pre-mRNA (19). The strong cross-linking of Hsh155p is not observed when an ACM13(−11) substrate lacking an intact 5′ splice site sequence is used (data not shown), indicating that U1 snRNP binding is necessary for the stable association of Hsh155p. Accompanying the appearance of strong cross-links to Hsh155p at position −11, cross-links to a U1- and U2-dependent, ∼97-kDa species are observed (Fig. 2D, lanes 3 to 8 and 11). In Hsh155p-13Myc extracts, this ∼97-kDa band appears to shift concomitantly with the ∼119-kDa band representing Hsh155p (Fig. 2D, lane 14, Hsh155p-13Myc*). Because this ∼97-kDa band is smaller than that predicted for un-cross-linked Hsh155p (110 kDa), it may represent either a degraded or an alternative form of Hsh155p. Similar lower-molecular-mass forms of Hsh155p are also faintly visible at several other positions (Fig. 2A; see also Fig. 5A and C; also, data not shown).
No cross-linking of Prp16p is observed at either position −8 or position −11 (Fig. 2B and D, lanes 3 to 8). Instead, cross-linking to an ∼147-kDa species at position −8 is seen at later times in the presence of ATP (≥10 min; Fig. 2B, lanes 3 to 8). The cross-linking of this band to the “wild-type” ACM13(−8) substrate parallels the appearance of the much stronger cross-links to Prp22p at positions −3 and −4 previously observed with 3′ splice site mutant substrates (30). As shown in Fig. 2C, the ∼147-kDa species is efficiently immunoprecipitated by Prp22p antisera, demonstrating that Prp22p is associated with intron sequences as far upstream as position −8 (see below).
At positions −3, −4, and −11, cross-linking of Prp8p is not apparent until approximately 5 min (Fig. 2A and D, lanes 3 to 8; also, data not shown). At position −8, however, cross-linking of Prp8p is visible as early as 2 min (Fig. 2B, lanes 3 to 8). The early cross-linking of Prp8p is dependent on intact U6 snRNA (Fig. 2B, lane 12) and coincides with the first appearance of lariat intermediates (data not shown). These data are in agreement with previous data showing that the cross-linking of Prp8p near the 3′ splice site does not occur until after the first step (48, 52) as well as experiments described below that demonstrate that the association of Prp8p with the 3′ splice site is independent of Prp16p (see Fig. 6).
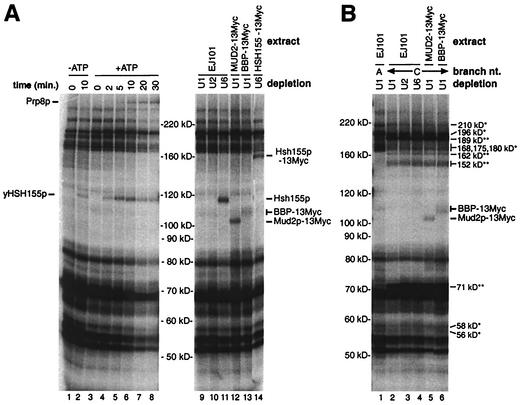
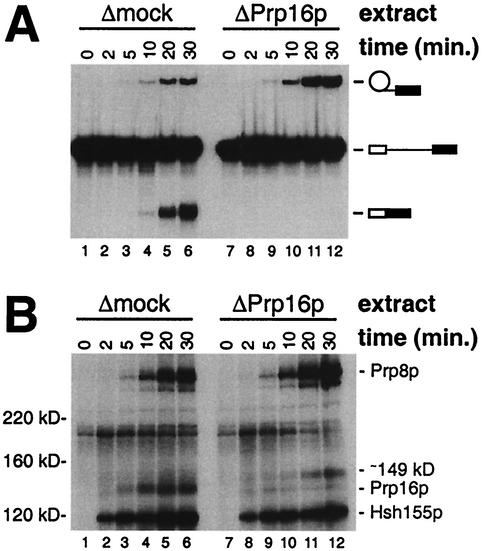
FIG. 6.
Comparison of splicing activity and cross-linking of proteins at position +2 of the 3′ exon in mock-depleted (Δmock) and Prp16p-immunodepleted (ΔPrp16p) splicing extracts. (A) Analysis of splicing products from reactions performed with the Δmock extract (lanes 1 to 6) or the ΔPrp16p extract (lanes 7 to 12) and the ACM13(+2) substrate in the presence of ATP for the indicated times. (B) Analysis of total cross-linked proteins in samples from the splicing reaction mixtures shown in panel A following incubation for the indicated times.
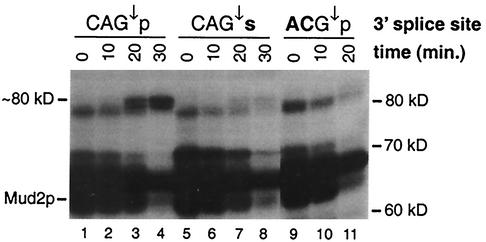
The association of a novel ∼80-kDa species with position −8 occurs only after completion of the second step.
At position −8, ATP-dependent cross-links to a unique ∼80-kDa species appear to lag slightly behind those to Prp22p (Fig. 2B, lanes 6 to 8). To determine whether the association of this 80-kDa species occurs prior to or following completion of the second step, cross-linking to pre-mRNA substrates having s4U at position −8 and either a “wild-type” 3′ splice site (CAG↓p), a mutant 3′ splice site (ACG↓p) (“CA” changed to “AC”), or an Rp phosphorothioate substitution (CAG↓s) at the 3′ splice site was examined. Sulfur substitution of the Rp (nonbridging) phosphoryl oxygen atom at the site of 3′ splice site cleavage blocks the second step (28, 32). In a high-resolution SDS-PAGE analysis, the ∼80-kDa species appears as a doublet that cross-links at late times (≥10 min) to the wild-type ACM13(−8) substrate (Fig. 3, lanes 1 to 4). In contrast, no strong accumulation of cross-links to the ∼80-kDa doublet is seen with either the phosphorothioate substitution substrate (Fig. 3, lanes 5 to 8) or the mutant 3′ splice site substrate (lanes 7, 8, and 11). These results demonstrate that the association of the ∼80-kDa doublet with the intron sequences downstream of the branch site occurs following completion of the second catalytic step. Very small amounts of the ∼80-kDa species are seen at late times with the phosphorothioate substitution substrate (Fig. 3, lanes 5 to 8) but not with the mutant 3′ splice site substrate (lanes 9 to 11). This small amount of cross-linking most likely arises from adventitious oxygen contamination that permits a small fraction of the phosphorothioate substrate to proceed through the second step (see Fig. 7A, lanes 5, 8, and 11).
FIG. 3.
Evidence that the association of an ∼80-kDa protein with the intron occurs only after successful completion of the second step. Lanes 1 to 4, proteins cross-linked to the ACM13(−8) substrate containing a wild-type 3′ splice site (CAG↓p) following incubation for the indicated times in the presence of ATP; lanes 5 to 8, proteins cross-linked to the ACM13(−8) substrate containing an Rp phosphorothioate substitution of the nonbridging oxygen atom at the 3′ splice site (CAG↓s); lanes 9 to 11, proteins cross-linked to the ACM13(−8) substrate containing a mutant 3′ splice site (ACG↓p). Boldface letters indicate differences from the wild-type 3′ splice site sequence.
The primary sequence of the highly conserved branch site influences the association of a large number of factors.
To examine proteins associated with the highly conserved yeast branch site sequence (UACUAAC) during spliceosome assembly, the second uridine (position −16) of this sequence was substituted with s4U in both “wild-type” and A259C mutant versions of the ACM13 substrate (Fig. 1A). In the absence of added ATP, the wild-type ACM13(−16) substrate shows significant cross-linking to a large number of novel proteins with apparent molecular masses of >160 kDa as well as weak cross-linking to the 119-kDa band representing Hsh155p (Fig. 4A, lanes 1 and 2). Incubation for 10 min of extracts in the absence of ATP (Fig. 4A, lane 2) or of extracts depleted of either U1 or U2 snRNA (lanes 9 and 10) produces no increase in the cross-linking of these ATP-independent species and suggests they are not components of commitment complexes. Although no cross-linking to untagged Mud2p or BBP is apparent, weak cross-linking to the 13-Myc-tagged versions of these proteins is seen (Fig. 4, lanes 12 and 13). Within 2 to 5 min of incubation in the presence of ATP, weak U2-dependent cross-linking to Hsh155 is observed, followed by weak U6-dependent cross-linking to Prp8p (Fig. 4A, lanes 2 to 10 and 14). One possible explanation for the apparent lack of ATP-dependent cross-linked species is that upon U2 snRNP binding, the branch site sequence becomes much more highly constrained due to base pairing to the U2 snRNA than the adjacent downstream sequences and is therefore much more restricted in its ability to form cross-links (17).
It was previously established that mutation of the branch site nucleotide severely attenuates BBP binding to the branch site sequence in a purified two-component system (5) and prevents prespliceosome formation in yeast splicing extracts (11). To examine the effects of an A→C mutation of the branch site nucleotide (A259C), cross-linking of proteins to ACM13(−16) substrates that were either wild type or contained the A259C branch site mutation was analyzed (Fig. 4B). Most notably, under conditions that permit the assembly of prespliceosomes, cross-linking of Hsh155p to the A259C mutant substrate is eliminated (Fig. 4B, lane 4). In contrast, the weak cross-linking of both Mud2p-13Myc and BBP-13Myc appears to be only moderately reduced by the A259C mutation (Fig. 4B, lanes 5 and 6). Surprisingly, the cross-linking of a large number of other unidentified species is also significantly affected by the A259C mutation. Compared to the results obtained with the wild-type substrate (Fig. 4B, lane 1), the cross-linking of at least five species with apparent molecular masses of ∼168, 175, 180, 196, and 210 kDa is significantly reduced by the A259C mutation (single asterisks in lane 2). Conversely, the cross-linking to the A259C mutant substrate of several species with apparent molecular masses of ∼71, ∼152, ∼162, and ∼189 kDa appears to be significantly enhanced (double asterisks in lane 2 of Fig. 4B). Whereas it is possible that members of the first group of proteins are either directly or indirectly involved in recognition of the branch site sequence, the increased cross-linking of the second group of proteins is likely to be a direct consequence of the lack of proper recognition of the mutant branch site sequence.
Unexpected complexity in the association of proteins with exon sequences adjacent to the 3′ splice site.
To determine the arrangement of proteins associated with nonconserved 3′ exon sequences, cross-linking with substrates with s4U located at positions +2, +10, and +13 relative to the 3′ splice site was analyzed. At all positions of the 3′ exon examined, the apparent molecular mass of each cross-linked species was slightly reduced relative to that observed when s4U was located in the upstream intron sequences (data not shown). This decrease was caused by the smaller size of the cross-linked 32P-labeled RNase T1 fragments in the 3′ exon (14 nt) than in the intron (19 nt) (Fig. 1A).
Cross-linking analysis with the ACM13(+10) and ACM13(+13)substrates is shown in Fig. 5. With these substrates, ATP-dependent cross-linking to either an ∼117-kDa species or an ∼120-kDa doublet is observed at early times (Fig. 5A and C, lanes 1 to 8). Incubation of these substrates with Hsh155p-13Myc extracts results in a large shift in the mobility of these bands (Fig. 5A and C, lane 14), demonstrating that they represent cross-linking to Hsh155p (38). With these substrates, very weak cross-linking of an ∼110-kDa species can also be observed; this species shifts when Hsh155p-13Myc extracts are used (Fig. 5A and C, lane 14), suggesting that it may represent the same degraded or alternative form of Hsh155p that is seen with s4U at positions −3 and −4. At later times (≥5 min), cross-links to an ∼140-kDa species are visible at both positions +10 and +13. Because both the apparent size and timing of this cross-linked species resemble those observed for the cross-linking of Prp16p at position −3 (Fig. 2A) (30), immunoprecipitation analysis was performed. The ∼140-kDa species cross-linked at position +10 is efficiently immunoprecipitated by anti-Prp16 sera (Fig. 5B, lane 3). At later times (≥10 min), weak cross-linking to a very-high-molecular-mass doublet (≫220 kDa) is observed with both the ACM13(+10) and the ACM13(+13) substrates, but only the lower band of this doublet comigrates with Prp8p cross-linked within the intron (data not shown). Although Prp8p was previously reported to cross-link to at least 13 nt of the 3′ exon (48), the identity or relationship of the apparently higher-molecular-mass species to Prp8p has not been investigated further. Cross-links to Hsh155p, Prp16p, and Prp8p at position +2 identical to those at position +10 are observed (30) (Fig. 6; also, data not shown). Interestingly, although cross-linking to Mud2p-13Myc is observed at positions +2, +10, and +13 (Fig. 5A and C, lane 12; also, data not shown), no decrease in cross-linking to any species corresponding to native Mud2p (∼60 kDa) at positions +10 and +13 is apparent (e.g., lane 11); these results suggest that cross-linking at these positions within the 3′ exon may involve the 13-Myc tag and not Mud2p itself.
Cross-linking to several novel species with unique properties is observed 13 nt downstream of the 3′ splice site (Fig. 5C). Distinct cross-linking of an unidentified ∼97-kDa species is seen in the zero-time-point reactions (Fig. 5C, lanes 1 and 3) but not at later times with the ACM13(+13) substrate (lanes 2 and 4 to 8). Although the identity of this ∼97-kDa species is unknown, one possible explanation for its complete disappearance at later times is that it is displaced by a factor(s) that associates with the 3′ exon in an ATP-independent manner during the initial stages of splicing complex formation. In a similar fashion, an unidentified ∼128-kDa cross-linked species at position +13 is observed only in U1-depleted extracts (Fig. 5C, lanes 9, 12, and 13) and not in either nondepleted extracts (lanes 1 to 8) or U2- or U6-depleted extracts (lanes 10, 11, and 14). The unique behavior of this ∼128-kDa species suggests the possibility that the association of this protein with the 3′ exon is affected directly or indirectly by binding of the U1 snRNP. Weak cross-linking to both ∼80- and ∼89-kDa species is also observed at very late times (Fig. 5C, lanes 6 to 8) and suggests the possibility that these species associate with the 3′ exon after the completion of splicing (40). Experiments to determine the exact timing of their association with the 3′ exon as well as their identities are in progress.
The association of Ppr8p with the 3′ splice site region is independent of Prp16p.
Analysis of the total cross-linked proteins at intron position −8 (Fig. 2B) suggested that the association of Prp8p with the 3′ splice site region occurs earlier than that of Prp16p. To determine whether the association of Prp8p with the 3′ splice site region requires Prp16p, a time course analysis of cross-linking in mock-depleted (Δmock) and Prp16p-immunodepleted (ΔPrp16p) extracts was performed. The ACM13(+2) substrate was chosen for this analysis because it cross-links readily to Prp16p and Prp8p (30) and facilitates the easy assessment of both catalytic steps due to the single 32P label within the 3′ exon (Fig. 1). As expected from earlier studies showing that Prp16p is required specifically for the second catalytic step in vitro (44), a splicing time course analysis of the ΔPrp16p extract shows the generation of only the lariat-3′ exon intermediate (Fig. 6A, lanes 7 to 12), while both the lariat-3′ exon intermediate and the mature mRNA product are produced in the Δmock extract (lanes 1 to 6). An analysis of proteins that cross-link to position +2 in samples from these reactions is shown in Fig. 6B. As expected, no difference in the timing or intensity of cross-linking to the U2-associated protein Hsh155p is observed between the Δmock (Fig. 6B, lanes 1 to 6) and ΔPrp16p (lanes 7 to 12) extracts. Importantly, while very little, if any, cross-linking to Prp16p is apparent in the ΔPrp16p extract compared to the Δmock extract, the timing and intensity of the initial cross-linking to Prp8p are almost indistinguishable in the two extracts (Fig. 6B, lanes 1 to 6 and lanes 7 to 12). These results clearly show that following the first step of splicing, Prp16p is not required for the subsequent association of Prp8p with the 3′ splice site region. Curiously, in the reactions performed with the ΔPrp16p extract, weak cross-links to a novel ∼149-kDa species accumulate at late times (Fig. 6B, lanes 7 to 12). Although the apparent molecular mass of this protein is similar to that observed for cross-linked Prp22p (Fig. 2 and 7), in other experiments with a 3′ splice site mutant version of the ACM13(+2) substrate to enhance the possible cross-linking of Prp22, no cross-linking to Prp22p is observed (30) (Fig. 7B).
The association of Prp22p with the 3′ terminus of the intron prior to the second step represents an authentic transient intermediate in splicing.
Previous experiments with substrates containing a short interval between the branch site and the 3′ splice site (13 nt) revealed that mutational inactivation of the 3′ splice site results in the dramatic accumulation of cross-links to Prp22p at intron positions −3 and −4 but not at exon position +2 (30). In contrast, only very weak cross-linking of Prp22p to substrates with functional, wild-type 3′ splice site sequences could be detected, suggesting that Prp22p may normally associate only transiently with the 3′ splice site prior to the second step (30). This interpretation is also consistent with the observed weak cross-linking of Prp22p at position −8 (Fig. 2B and C). To more accurately assess the extent of the association of Prp22p with intron sequences in the lariat intermediate, cross-linking of Prp22p at positions +2, −4, −8, and −11 to both wild-type and mutant (CAG↓→ACG↓) ACM13 substrates was examined (Fig. 7). Although each of the mutant substrates fails to undergo the second step, as evidenced by the accumulation of the lariat-3′ exon intermediate (Fig. 7A, lanes 2, 4, 7, and 10), strong cross-linking of Prp22p is observed with the substrates containing s4U at positions −8 and −4 but not at positions −11 and +2 (Fig. 7B, lanes 2, 4, 7, and 10). These results suggest that Prp22p is closely associated with at least the last eight intron nucleotides of the lariat-3′ exon intermediate and not with either position −11 or position +2.
It was previously reported that the strong cross-linking of Prp22p to a mutant 3′ splice site requires Prp16p and therefore occurs following the first step but prior to the second step (30). However, because the accumulation of cross-links to Prp22p was observed only with mutant 3′ splice sites, it is possible that the strong cross-linking of Prp22p is associated with a failure to properly bind to the active site for the second step and therefore does not reflect an authentic interaction of Prp22p with the 3′ end of the intron (30). To address this possibility, we used four-way in vitro RNA ligations to create wild-type versions of the ACM13 substrate with s4U at position −4, −8, or −11 and containing an Rp phosphorothioate substitution at the 3′ splice site to block the second step without altering the primary sequence (28, 32). Although the phosphorothioate substitution at the 3′ splice site effectively blocks the second step of splicing with each of the s4U-containing pre-mRNA substrates (Fig. 7A, lanes 5, 8, and 11), strong cross-linking of Prp22p is observed at positions −4 and −8 but not at position −11 (Fig. 7B, lanes 5, 8, and 11). These results demonstrate that the cross-linking of Prp22p to the lariat intron-3′ exon intermediate represents an authentic transient intermediate prior to the second catalytic step. In agreement with the known involvement of the U2 and U5 snRNPs in the second step (53), the phosphorothioate substitution results in dramatically increased cross-linking of Hsh155p at positions −4 and −11 and increased cross-linking of Prp8p at all three positions. The failure of the phosphorothioate substitution to result in the accumulation of cross-links to Prp16p at position −4 supports the previous finding that the normal interaction of Prp16p with the spliceosome occurs transiently (44).
DISCUSSION
We have used a series of closely related s4U-containing pre-mRNA cross-linking substrates to construct a detailed map of the associations between splicing factors and the region encompassing the branch site, 3′ splice site region, and adjacent 3′ exon sequences established during spliceosome assembly and the subsequent catalytic steps. Although photochemical cross-linking analysis unambiguously establishes the close spatial relationships between the pre-mRNA, splicing intermediates, and products with these factors, caution must be exercised in interpreting the results. Importantly, because photochemical cross-linking requires that both specific geometrical and chemical constraints be satisfied (17), the absence of a particular cross-linked species must be interpreted in a subjective manner, and cannot be evaluated conclusively. In addition, although the cross-linking of two components reflects their close proximity to each other, it does not attest to either the strength or specificity of this association.
Whereas our analysis provides an approximate apparent molecular mass for each cross-linked species, the difference in this value from the actual molecular mass of the uncross-linked form of a protein is unpredictable, varying from ∼2 kDa (Mud2p) to ∼17 kDa (Prp16p). This variation, coupled with a lack of specific information about other properties of a cross-linked species, hinders the identification of novel cross-linked species. For example, for the ∼80-kDa species that cross-links at position −8 following the second step of splicing (Fig. 3), a large number of known yeast proteins have molecular masses of between 63 and 78 kDa, and of these, eight represent previously identified splicing factors. Of these eight, we have tested three so far by epitope tagging without success (unpublished results).
BBP and Mud2p are the earliest factors known to associate specifically with the branch site-3′ splice site region.
Using extracts prepared from yeast strains expressing 13-Myc-tagged versions of either BBP or Mud2p, we have observed the close association of both of these factors with the branch site-3′ splice site region. Although BBP was previously shown to specifically bind and recognize the pre-mRNA branch site sequence (5), we have been unable to detect s4U cross-linking to native (untagged) BBP in our experiments; in all cases where we have observed either weak or strong cross-linking to BBP-13Myc, no visible difference in the intensity of lower-molecular-mass cross-linked species has been detected (Fig. 2, 4, and 5). These results suggest that despite the extensive cross-linking of BBP-13Myc observed in our experiments, cross-linking at all positions likely involves the 13-Myc tag and not BBP itself. Similarly, although cross-linking to Mud2p-13Myc is observed from positions −16 to +13, cross-linking to native Mud2p is detected only at positions −11, −8, −4, −3, and +2 (Fig. 2, 4, and 5; also, data not shown), suggesting that the cross-linking observed at other positions (−16, +10, and +13) most likely involves cross-linking to the 13-Myc tag and not to Mud2p.
Interestingly, although the A259C branch site mutation prevents prespliceosome formation as detected by native gel analysis (11) and eliminates the cross-linking of the U2 snRNP component Hsh155p, the weak cross-linking of both BBP-13Myc and Mud2p-13Myc appears largely unaffected (Fig. 4B). These results suggest that the defect in prespliceosome assembly caused by the A259C mutation must affect the association of some other factor(s) with the branch site sequence, or some specific aspect of interaction of the branch site sequence with BBP/Mud2p. Consistent with the A259C mutation having other effects on very early splicing complex formation, this mutation also results in significant changes in the cross-linking of a large number of unidentified species in U1-depleted extracts (Fig. 4B).
Hsh155p is extensively associated with the entire branch site-3′ splice site-3′ exon region.
Our studies have shown that, during assembly of the prespliceosome, extensive interactions are established between the U2 snRNP associated protein, Hsh155p, and the branch site-3′ splice site-3′ exon region of the ACM13 substrates (Fig. 2, 4, and 5). Our results are in general agreement with those obtained for the human ortholog of Hsh155p, SAP155, a component of the multimeric U2-snRNP complex SF3b (19, 38, 56). SAP155 was previously reported to cross-link very strongly just downstream of the branch site, at a position equivalent to −11 in our experiments (Fig. 2D). Cross-linking of SAP155 upstream of the branch site sequence has also been reported, where it is thought to function in part by anchoring the U2 snRNP tightly to the pre-mRNA (19, 39). However, no cross-linking of SAP155 to either the 3′ splice site or to 3′ exon sequences was previously reported (Fig. 2 and 4) and may be due to either differences in the sequence or structure of SAP155/Hsh155p or the cross-linking techniques used. The C termini of both Hsh155p and SAP155 contain a large number of nonidentical tandem repeats, which are similar to those found in the A subunit of protein phosphatase 2A (56). In protein phosphatase 2A, these repeats are organized into an elongated, rod-like structure that acts as a scaffolding molecule to coordinate the assembly of the catalytic C subunit and the regulatory B subunit (20). Such an extended structure in Hsh155p may also explain the extensive cross-linking in the branch site-3′ splice site-3′ exon region observed with the ACM13 substrates (Fig. 2), as well as its cross-linking to the much more distant 3′ splice site of the ACT47 substrate (30). It is tempting to speculate that the nonidentical tandem repeats in Hsh155p serve as a scaffold to organize factors involved in 3′ splice site recognition. Systematic purification of protein complexes from yeast has revealed that Hsh155p is associated with several different multicomponent splicing-related complexes, including one containing Prp8p and another containing Prp22p, although there is no evidence supporting direct interactions between Hsh155p and these factors (18). We also note that cross-links to Hsh155p accumulate in our experiments with substrates containing either a 3′ splice site mutation or a phosphorothioate substitution at the 3′ splice site (Fig. 7), indicating that Hsh155p remains in very close proximity to the branch site-3′ splice site-3′ exon region through the second catalytic step.
The 3′ splice site and adjacent 3′ exon sequences define a ligand for Prp16p.
Our cross-linking studies have shown that, following the first step of splicing, the DEAH box protein Prp16p is closely associated with the last four nucleotides of the intron and at least 13 nucleotides of the adjacent 3′ exon. Our failure to observe an increase in cross-linking to Prp16p at position −4 when the second step is blocked by either a 3′ splice site mutation or phosphorothioate substitution is also consistent with release of Prp16p from the spliceosome prior to the second step (45) (Fig. 7). Although initial genetic analyses demonstrated that mutations in Prp16p could partially restore splicing of a yeast actin intron containing a branch point mutation (16), no cross-linking of Prp16p to either the branch site sequence, or the sequences immediately downstream of the branch site sequence is observed (Fig. 2 and 4). Whereas it is possible that the observed association of Prp16p with the 3′ splice site-3′ exon region is a consequence of the short interval between the branch site and the 3′ splice site in our ACM13 substrates, this possibility seems unlikely, since cross-linking of Prp16p to the 3′ splice sites of pre-mRNAs with much longer intervals between the branch site and the 3′ splice site has also been observed (30, 52). An alternative possibility is that Prp16 directly or indirectly interacts with the 3′ splice site-3′ exon region in a specific manner. Genetic analyses so far have failed to uncover any direct role for Prp16 in 3′ splice site recognition (8). However, other genetic analyses have suggested a complex (but poorly understood) relationship between Prp16p, branch site recognition, 3′ splice site recognition, and mutations at nucleotide U57 in the yeast U6 snRNA (9, 29).
Two hybrid and coimmunoprecipitation experiments support the existence of either direct or indirect interactions of the U5 snRNP associated DEXH box protein Brr2p and Prp16p, and it has been proposed that Brr2p may mediate the recruitment of Prp16p to the spliceosome (54). In addition, synthetic lethal interactions have been found between the prp16-301 and prp8-101 mutants and imply a possible physical interaction between these two proteins (52). In agreement with our results (Fig. 2, 4, 5, and 6), previous cross-linking studies have shown that Prp8p is extensively associated with the branch site-3′ splice site-3′ exon region (48). However, because cross-linking of Prp8p is not apparently affected by either mutation or duplication of the conserved 3′ splice site sequence, its binding does not appear overly sequence specific (30, 48). The extensive binding of Prp8p has been proposed to stabilize the weak interactions between the conserved loop of the U5 snRNA and exon sequences adjacent to the splice sites (48). Mutants that affect 3′ splice site selection have been isolated in both Prp8p (13) and the conserved loop of the U5 snRNA (34). In addition, following the first step of splicing, the first nucleotide of the 3′ exon can be cross-linked to the conserved loop of the U5 snRNA (35). If the association of Prp16p with the 3′ splice site region is directed by an initial recognition step involving the U5 snRNA, the cross-linking between the conserved loop of U5 and the first nucleotide of the 3′ exon may also be detectable in Prp16-depleted extracts, since the cross-linking of Prp8p to the 3′ splice site appears completely unaffected in Prp16p-depleted extracts (Fig. 6).
The interaction of Prp22p with the intron represents an authentic transient intermediate.
Following the first catalytic step of splicing, binding and subsequent ATP hydrolysis by Prp16p has been found to decrease the accessibility of a mutant 3′ splice site to oligonucleotide-mediated RNase H cleavage (43). We previously demonstrated that either mutation of the 3′ splice site or sequence context changes near the 3′ splice site that block the second step lead to a very dramatic increase in the cross-linking of Prp22p to the 3′ splice site. Because of the strong correlation between the change in accessibility of a mutant 3′ splice site to oligonucleotide-mediated RNase H cleavage (43) and Prp22p cross-linking, we have suggested that the observed protection likely arises as a direct consequence of the interaction of Prp22p with the 3′ splice site region (30). We have also determined that the strong cross-linking of Prp22p to a mutant 3′ splice is dependent on Prp16p and does not extend into the adjacent 3′ exon sequence (30). Using a series of substrates containing either 3′ splice site mutations or site specific phosphorothioate substitutions to block the second step, we have now demonstrated that Prp22p is very closely associated with at least the last eight intron nucleotides in the intron lariat-3′ exon intermediate. Because site-specific phosphorothioate substitution, unlike a 3′ splice site mutation, should be less likely to affect any interactions involved in the specific recognition of the conserved 3′ splice site sequence, our results strongly suggest that the association of Prp22p represents a bona fide transient intermediate that exists prior to the second step. Our data also suggest that Prp22p resides in very close proximity to the active site just prior to the second step and that Prp22p may function, at least in part, to bind to and organize the structure of the last eight intron nucleotides for catalysis (see also reference 42).
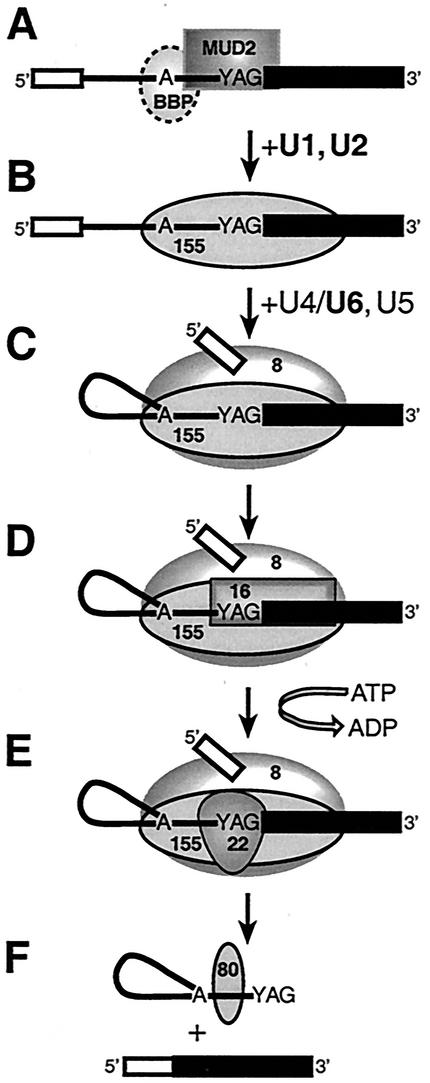
A model for the temporal and spatial organization of RNA-protein interactions in the branch site-3′ splice site region of yeast pre-mRNAs.
The earliest known steps involved in the recognition of the branch site-3′ splice site region are the U1- and ATP-independent association of BBP and Mud2p (1, 5). Although our experiments have failed to detect cross-links to native (untagged) BBP, the specific interaction of BBP with the conserved branch site sequence has been rigorously demonstrated, as have the cooperative interactions in the binding of BBP and the mammalian ortholog of Mud2p, U2AF65 (4, 5). In our system, cross-linking to native Mud2p in commitment complexes extends from positions −11 to +2, and is depicted in Fig. 8A together with the specific interaction of BBP with the branch site sequence (5). Upon prespliceosome formation, extensive associations of the U2-snRNP associated splicing factor Hsh155p with the entire branch site-3′ exon region are established (Fig. 8B). Previous studies showed that the conversion of commitment complexes to prespliceosomes involves the release of both Mud2p and BBP (41). Following the first catalytic step of splicing, we detect the formation of extensive interactions involving the U5 snRNP associated protein, Prp8p (Fig. 8C). Using depleted extracts, we have shown that the interaction of Prp8p with the 3′ splice site region can occur independently of the DEAH box splicing factor, Prp16p. The subsequent interaction of Prp16p (44) occurs with the very 3′ end of the intron together with the adjacent 3′ exon sequences (Fig. 8D) and is required for the subsequent cross-linking of Prp22p (30). Our studies have established that the cross-linking of Prp22p reflects an authentic transient intermediate that occurs just prior to the second catalytic step and that this interaction appears to be restricted to the 3′ end of the lariat intermediate (Fig. 8E). Following completion of the second step, Prp22p plays a second, distinct role in splicing required for the ATP-dependent release of mRNA from the spliceosome (42); we have previously observed that the ATP-dependent role of Prp22p does not appear to involve its prior association with the 3′ terminus of the intron (30). Following completion of the second step, an as-yet-unidentified ∼80-kDa protein interacts with sequences near branch site and may be involved in the later steps of intron metabolism (Fig. 8F). Whether the interaction of the ∼80-kDa protein requires ATP as well as the exact disposition of Prp8p and Hsh155p following completion of the second step remains unknown.
FIG. 8.
Model for the temporal and spatial organization of RNA-protein interactions in the branch site-3′ splice site region during pre-mRNA splicing in S. cerevisiae. The 5′ and 3′ exons are depicted as open and closed boxes, respectively, and a line depicts the intron. Shown are proteins associated with the pre-mRNA branch site-3′ splice site region in commitment complexes (A), prespliceosomes (B), active spliceosomes (C to E), and intron-containing postsplicing complexes (F). The approximate positions of cross-linked proteins are indicated as follows: 8, Prp8p; 155, Hsh155p; 16, Prp16p; 22, Prp22p; and 80, unidentified ∼80-kDa species. Note that ATP hydrolysis is shown only for the Prp16p-dependent step and that the presence or absence of other proteins in postsplicing complexes has not been verified. See the text for details. Boldfacing indicates the snRNAs that were depleted from the extracts in many of the experiments presented here.
Acknowledgments
We thank Jim Bruzik, Jonatha Gott, Wes Kroeze, Tim Nilsen, and Jo Ann Wise for critical reading of the manuscript. We especially thank Mike Harris for help with the high-pressure liquid chromatography separation of the phosphorothioate stereoisomers and George Perry for patience. We also thank Manuel Ares, Jr., and Pat Maroney for helpful suggestions and Beate Schwer for generous gifts of Prp16 and Prp22 antibodies.
This work was supported by grant GM64682 to D.S.M. from the National Institutes of Health.
REFERENCES
- 1.Abovich, N., X. Liao, and M. Rosbash. 1994. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 8:843-854. [DOI] [PubMed] [Google Scholar]
- 2.Abovich, N., and M. Rosbash. 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89:403-412. [DOI] [PubMed] [Google Scholar]
- 3.Ansari, A., and B. Schwer. 1995. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J. 14:4001-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berglund, J. A., N. Abovich, and M. Rosbash. 1998. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 12:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berglund, J. A., K. Chua, N. Abovich, R. Reed, and M. Rosbash. 1997. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell 89:781-787. [DOI] [PubMed] [Google Scholar]
- 5a.Brys, A., and B. Schwer. 1996. Requirement for SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3′ splice site. RNA 2:707-717. [PMC free article] [PubMed]
- 6.Burge, C. B., T. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosomes, p. 525-560. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world, 2nd ed., vol. 37. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 7.Burgess, S., J. R. Couto, and C. Guthrie. 1990. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell 60:705-717. [DOI] [PubMed] [Google Scholar]
- 8.Burgess, S. M., and C. Guthrie. 1993. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 73:1377-1391. [DOI] [PubMed] [Google Scholar]
- 9.Chang, J. S., and D. S. McPheeters. 2000. Identification of a U2/U6 helix la mutant that influences 3′ splice site selection during nuclear pre-mRNA splicing. RNA 6:1120-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, S., K. Anderson, and M. J. Moore. 2000. Evidence for a linear search in bimolecular 3′ splice site AG selection. Proc. Natl. Acad. Sci. USA 97:593-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, S. C., and J. Abelson. 1987. Spliceosome assembly in yeast. Genes Dev. 1:1014-1027. [DOI] [PubMed] [Google Scholar]
- 12.Clark, T. A., C. W. Sugnet, and M. Ares, Jr. 2002. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science 296:907-910. [DOI] [PubMed] [Google Scholar]
- 13.Collins, C. A., and C. Guthrie. 1999. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev. 13:1970-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins, C. A., and C. Guthrie. 2001. Genetic interactions between the 5′ and 3′ splice site consensus sequences and U6 snRNA during the second catalytic step of pre-mRNA splicing. RNA 7:1845-1854. [PMC free article] [PubMed] [Google Scholar]
- 15.Collins, C. A., and C. Guthrie. 2000. The question remains: is the spliceosome a ribozyme? Nat. Struct. Biol. 7:850-854. [DOI] [PubMed] [Google Scholar]
- 16.Couto, J. R., J. Tamm, R. Parker, and C. Guthrie. 1987. A trans-acting suppressor restores splicing of a yeast intron with a branch point mutation. Genes Dev. 1:445-455. [DOI] [PubMed] [Google Scholar]
- 17.Favre, A., C. Saintome, J. L. Fourrey, P. Clivio, and P. Laugaa. 1998. Thionucleobases as intrinsic photoaffinity probes of nucleic acid structure and nucleic acid-protein interactions. J. Photochem. Photobiol. 42:109-124. [DOI] [PubMed] [Google Scholar]
- 18.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 19.Gozani, O., J. Potashkin, and R. Reed. 1998. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 18:4752-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groves, M. R., N. Hanlon, P. Turowski, B. A. Hemmings, and D. Barford. 1999. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell 96:99-110. [DOI] [PubMed] [Google Scholar]
- 21.Horowitz, D. S., and J. Abelson. 1993. Stages in the second reaction of pre-mRNA splicing: the final step is ATP independent. Genes Dev. 7:320-329. [DOI] [PubMed] [Google Scholar]
- 22.Kandels-Lewis, S., and B. Seraphin. 1993. Role of U6 snRNA in 5′ splice site selection. Science 262:2035-2039. [DOI] [PubMed] [Google Scholar]
- 23.Lesser, C. F., and C. Guthrie. 1993. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science 262:1982-1988. [DOI] [PubMed] [Google Scholar]
- 24.Lin, R. J., A. J. Newman, S. C. Cheng, and J. Abelson. 1985. Yeast mRNA splicing in vitro. J. Biol. Chem. 260:14780-14792. [PubMed] [Google Scholar]
- 25.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle.1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 26.Luukkonen, B. G., and B. Seraphin. 1997. The role of branchpoint-3′ splice site spacing and interaction between intron terminal nucleotides in 3′ splice site selection in Saccharomyces cerevisiae. EMBO J. 16:779-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maroney, P. A., C. M. Romfo, and T. W. Nilsen. 2000. Nuclease protection of RNAs containing site-specific labels: a rapid method for mapping RNA-protein interactions. RNA 6:1905-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maschhoff, K. L., and R. A. Padgett. 1992. Phosphorothioate substitution identifies phosphate groups important for pre-mRNA splicing. Nucleic Acids Res. 20:1949-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McPheeters, D. S. 1996. Interactions of the yeast U6 RNA with the pre-mRNA branch site. RNA 2:1110-1123. [PMC free article] [PubMed] [Google Scholar]
- 30.McPheeters, D. S., B. Schwer, and P. Muhlenkamp. 2000. Interaction of the yeast DExH-box RNA helicase prp22p with the 3′ splice site during the second step of nuclear pre-mRNA splicing. Nucleic Acids Res. 28:1313-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore, M. J., C. C. Query, and P. A. Sharp. 1993. Splicing of precursor to messenger RNAs by the spliceosome, p. 303-358. In R. Gesteland and J. Atkins (ed.), The RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Moore, M. J., and P. A. Sharp. 1993. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature 365:364-368. [DOI] [PubMed] [Google Scholar]
- 33.Moore, M. J., and P. A. Sharp. 1992. Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice sites. Science 256:992-997. [DOI] [PubMed] [Google Scholar]
- 34.Newman, A. J., and C. Norman. 1992. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell 68:743-754. [DOI] [PubMed] [Google Scholar]
- 35.Newman, A. J., S. Teigelkamp, and J. D. Beggs. 1995. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA 1:968-980. [PMC free article] [PubMed] [Google Scholar]
- 36.Parker, R., and P. G. Siliciano. 1993. Evidence for an essential non-Watson-Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature 361:660-662. [DOI] [PubMed] [Google Scholar]
- 37.Patterson, B., and C. Guthrie. 1991. A U-rich tract enhances usage of an alternative 3′ splice site in yeast. Cell 64:181-187. [DOI] [PubMed] [Google Scholar]
- 38.Pauling, M. H., D. S. McPheeters, and M. Ares, Jr. 2000. Functional Cus1p is found with Hsh155p in a multiprotein splicing factor associated with U2 snRNA. Mol. Cell. Biol. 20:2176-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reed, R. 1996. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr. Opin. Genet. Dev. 6:215-220. [DOI] [PubMed] [Google Scholar]
- 40.Reed, R., and E. Hurt. 2002. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108:523-531. [DOI] [PubMed] [Google Scholar]
- 41.Rutz, B., and B. Seraphin. 1999. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA 5:819-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwer, B., and C. H. Gross. 1998. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 17:2086-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwer, B., and C. Guthrie. 1992. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 11:5033-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwer, B., and C. Guthrie. 1991. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature 349:494-499. [DOI] [PubMed] [Google Scholar]
- 45.Siliciano, P. G., and C. Guthrie. 1988. 5′ splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 2:1258-1267. [DOI] [PubMed] [Google Scholar]
- 46.Slim, G., and M. J. Gait. 1991. Configurationally defined phosphorothioate-containing oligoribonucleotides in the study of the mechanism of cleavage of hammerhead ribozymes. Nucleic Acids Res. 19:1183-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spingola, M., L. Grate, D. Haussler, and M. Ares, Jr. 1999. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA 5:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teigelkamp, S., A. J. Newman, and J. D. Beggs. 1995. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 14:2602-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teigelkamp, S., E. Whittaker, and J. D. Beggs. 1995. Interaction of the yeast splicing factor PRP8 with substrate RNA during both steps of splicing. Nucleic Acids Res. 23:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Umen, J. G., and C. Guthrie. 1996. Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics 143:723-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Umen, J. G., and C. Guthrie. 1995. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes Dev. 9:855-868. [DOI] [PubMed] [Google Scholar]
- 52.Umen, J. G., and C. Guthrie. 1995. Prp16p, Slu7p, and Prp8p interact with the 3′ splice site in two distinct stages during the second catalytic step of pre-mRNA splicing. RNA 1:584-597. [PMC free article] [PubMed] [Google Scholar]
- 53.Umen, J. G., and C. Guthrie. 1995. The second catalytic step of pre-mRNA splicing. RNA 1:869-885. [PMC free article] [PubMed] [Google Scholar]
- 54.van Nues, R. W., and J. D. Beggs. 2001. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 157:1451-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vijayraghavan, U., R. Parker, J. Tamm, Y. Iimura, J. Rossi, J. Abelson, and C. Guthrie. 1986. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 5:1683-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, C., K. Chua, W. Seghezzi, E. Lees, O. Gozani, and R. Reed. 1998. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 12:1409-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Will, C. L., and R. Lührmann. 1997. snRNP structure and function, p. 132-173. In A. R. Krainer (ed.), Eukaryotic mRNA processing. IRL Press, New York, N.Y.
- 58.Zhang, X., and B. Schwer. 1997. Functional and physical interaction between the yeast splicing factors Slu7 and Prp18. Nucleic Acids Res. 25:2146-2152. [DOI] [PMC free article] [PubMed]