Abstract
This study molecularly elucidates the basis for the dominant negative mechanism of the glucocorticoid receptor (GR) isoform hGRβ, whose overexpression is associated with human glucocorticoid resistance. Using a series of truncated hGRα mutants and sequential mutagenesis to generate a series of hGRα/β hybrids, we find that the absence of helix 12 is neither necessary nor sufficient for the GR dominant negative phenotype. Moreover, we have localized the dominant negative activity of hGRβ to two residues and found that nuclear localization, in addition to heterodimerization, is a critical feature of the dominant negative activity. Molecular modeling of wild-type and mutant hGRα and hGRβ provides structural insight and a potential physical explanation for the lack of hormone binding and the dominant negative actions of hGRβ.
Glucocorticoids are among the most widely used classes of drugs in the world. The anti-inflammatory effects of glucocorticoids are routinely exploited in treatment of many pathological conditions ranging from asthma, rheumatoid arthritis, ulcerative colitis, and common eczema. Likewise, the immunosuppressive benefits of glucocorticoids afford them a role in most chemotherapeutic regimes as well as for management of autoimmunity. However, prolonged glucocorticoid treatment, as well as rare genetic dispositions, can result in glucocorticoid resistance (7). An increasing number of studies in the last several years have implicated alternative splicing of the glucocorticoid receptor (GR) gene and subsequent expression of the hGRβ protein isoform as a contributing factor to glucocorticoid resistance in a variety of pathological conditions (17, 21, 28, 38, 39, 41). Since hGRβ does not undergo ligand-induced down regulation and has an increased half-life, the expression of hGRβ becomes more significant (31). Expression of hGRβ is also enhanced by proinflammatory cytokines, such as tumor necrosis factor alpha and interleukin 1; however, a precise physiological role for hGRβ remains elusive (43).
The GR, a member of the nuclear hormone receptor superfamily of ligand-activated transcription factors, participates in numerous signaling pathways leading to altered gene expression in target cells and tissues and is essential for life (8). The GR can modulate gene expression either positively or negatively by directly binding as a homodimer to glucocorticoid response elements (GRE) located in the promoter regions of target genes. Alternatively, the ligand-activated GR is known to repress or antagonize other nuclear factors involved in regulating gene expression, such as NF-κB and AP-1, through direct protein interactions (29). Both of these functions of GR signaling and transactivation and transrepression, appear to involve distinct and separable regions of the receptor and are major components of the physiological response to both natural and synthetic glucocorticoids.
In humans, alternative splicing of the ninth and final GR exon gives rise to hGRα and hGRβ proteins divergent at only the extreme carboxy termini (11, 20). Although the two proteins are 94% identical, the hGRβ isoform fails to bind hormone or activate gene expression and functions as a dominant negative inhibitor of hGRα (3, 31). In addition, hGRβ does not undergo ligand-dependent down regulation and consequently has a half-life longer than that of hGRα (43). As observed among all nuclear receptor superfamily members, the GR carboxy-terminal region encodes the ligand-binding domain (LBD). Extensive structural analysis of numerous nuclear receptor LBDs has revealed a common fold and structural mechanism for hormone action (40, 42). Essentially a 12-helix bundle, nuclear receptor LBDs contain a transactivation function (AF-2) created in large part by ligand-induced conformational changes involving the 12th and final helix (helix 12 [H12]) of the domain. The H12/AF-2 core region is absent in hGRβ and is replaced by a unique carboxy-terminal tail distinct from that of hGRα. It is unclear if the absence of H12 or the presence of unique carboxy-terminal residues gives rise to the dominant negative phenotype of hGRβ.
Our interest lies in understanding the molecular mechanism of the hGRβ dominant negative activity and its potential role in human pathology. Previous studies have proposed that a likely mechanism for the dominant negative activity of hGRβ is the formation of inactive heterodimers with hGRα; however, the precise mechanism and structural basis is unknown. The purpose of the present study was to functionally discriminate between the absence of H12 and the presence of the unique carboxy-terminal end of hGRβ and to determine which residues, if any, were critical for the dominant negative effect. Using carboxy-terminal hGRα truncation mutants and site-directed mutagenesis, we have mapped the dominant negative activity to two amino acids within the unique hGRβ region. Molecular modeling and secondary structure alignments provide further insight and suggest a mechanistic model for the dominant negative action of hGRβ.
MATERIALS AND METHODS
Reagents and plasmids.
Dexamethasone (Dex) was purchased from Steraloids (Wilton, N.H.). trans-35S label (1,108 Ci/mmol) was purchased from ICN Radiochemicals (Irvine Calif.). [14C]Chloramphenicol (40 to 60 mCi/mmol) was obtained from NEN Life Science Products. Acetyl-coenzyme A (CoA) and protease inhibitors were purchased from Roche Molecular Biochemicals (Indianapolis, Ind.). Oligonucleotide primers for mutagenesis and PCR were synthesized by Oligo's Etc. (Bethel, Maine). For all mutagenesis, the QuickChange site-directed mutagenesis kit from Stratagene was utilized. The peroxidase-labeled anti-rabbit immunoglobulin G secondary antibody and the enhanced chemiluminescence detection kit (ECL) were purchased from Amersham Pharmacia Biotech. The TNT coupled reticulocyte lysate translation system was purchased from Promega (Madison, Wis.). Reagents for immunocytochemistry were purchased from BioGenex (San Ramon, Calif.).
The use and construction of the GR expression vectors pCMVhGRα, pCMVhGRβ, pCMVhGRα(1-727), pT7/T3-hGRα, and pT7/T3-hGRβ, were previously described (reference 30 and references therein). Note that pCMVhGRα(1-727) was originally named pCMVhGR728T. For synthesis of pCMVhGRα(1-742) and pCMVhGRα(1-706), stop codons (TAG) were introduced at positions 743 and 707, respectively, of pCMVhGRα. Analogous mutagenesis of pT7/T3 was carried out to create the pT7/T3 hGRα truncation mutants (1-706, 1-727, and 1-742) used for in vitro synthesis with [35S]methionine. Generation of the hGRα/β hybrids was carried out by sequential mutagenesis of pCMVhGRα, substituting the corresponding residues of hGRβ(728-742) for hGRα. All mutants were verified by DNA sequencing at the National Institutes of Environmental Health Sciences core sequencing facility. Other plasmids used in these studies, pGMCS (mouse mammary tumor virus-chloramphenicol acetyltransferase [MMTV-CAT]), the empty pCMV5 vector, and the glutathione S-transferase (GST) fusion protein vectors, pGEX4T and pGEX4T-hGRα(523-777), have been previously described (30).
Cell culture, transfections, and CAT assays.
COS-1 cells were maintained in Dulbecco's minimum essential medium with high glucose containing 2 mM glutamine and a 10% (vol/vol) mixture of heat-inactivated fetal calf-calf serum (1:1). For transactivation assays, cells were incubated for 1 to 2 days in medium containing dextran-coated charcoal-stripped serum to remove endogenous steroids. All cell culture media contained 100 IU of penicillin/ml and 100 mg of streptomycin/ml. Cell cultures were maintained in a 5%-CO2 humidified incubator at 37°C and passaged every 3 to 4 days. The COS-1 cells were transfected with plasmid DNA using the DMRIE-C reagent (Gibco-BRL) or Effectene transfection reagent (Qiagen).
For CAT assays, COS-1 cells were transfected with pGMCS (MMTV-CAT) and hGRα/hGRβ mutants and treated as described above. Cells were harvested in phosphate-buffered saline (PBS), and cell pellets were resuspended in 0.25 M Tris-HCl, pH 8, and 5 mM EDTA. The cells were sonicated for 30 s and then pelleted at 20,800 × g at 4°C for 5 min. The supernatant was removed and heat inactivated for 10 min at 68°C. Protein concentrations were determined using a Bradford protein assay (Bio-Rad). Ten micrograms of cell extract was assayed for CAT activity and quantitated as previously described (1, 30).
For luciferase assays, cells were transfected with pHHLuc (MMTV-Luc) and hGR plasmids as described above. Cells were harvested in 1× cell lysis buffer for reporter assays following the manufacturer's instructions (Roche Applied Science). Duplicate samples of 50 μl were assayed using a MLX-3 luminometer (Dynex Technologies), and numbers of relative light units were adjusted for protein concentration.
GST pull-down assays.
GST pull-down assays were carried out essentially as described previously (22). Briefly, GST or GST-hGRα(523-777) fusion proteins were induced in Escherichia coli DH5α by addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside to log-phase cells. Following a 4 h incubation, the cells were harvested and lysed by sonication in a solution containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 100 mM NaCl, 10% glycerol, 0.2-mg/ml lysozyme, 75 μM PeFa block (Boehringer Mannheim), and 0.2-μg/ml leupeptin. Following centrifugation to remove insoluble protein, the soluble protein extract was incubated overnight with PBS-washed glutathione agarose (Amersham Pharmacia) in the presence of 1% Triton X-100 and 1 mM dithiothreitol. Following extensive washing, the agarose with the immobilized fusion proteins was then incubated with full-length [35S]methionine-labeled hGRα, hGRβ, or hGRα truncation mutants overnight at 4°C with constant rotation in a binding buffer containing Tris-EDTA (pH 8.0) with 100 mM NaCl, 10% glycerol, 1 mM dithiothreitol, 0.01% NP-40, and the protease inhibitors PeFa block and leupeptin. After extensive washing of the agarose beads with the above-described buffer, bound proteins were resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized by autoradiography. Typically, more than 25% of total bacterial expressed hGR(523-777) was recovered in the soluble fraction; however, only 5 to 10% of this fraction bound the glutathione-agarose under the conditions described.
Western blotting and immunocytochemistry.
For Western blotting, cell extracts prepared for CAT assays were separated on precast 8% Tris-glycine gels (Novex, San Diego, Calif.) and transferred to nitrocellulose membranes (0.2 μm). The membranes were washed in Tris-buffered saline with 0.1% Tween 20 (TBST) and blocked in TBST containing 10% nonfat milk for a minimum of 2 h at room temperature. Blots were next incubated in TBST supplemented with affinity-purified primary anti-GR antibody, Ab57 (1:1,000) overnight at 4°C. Following extensive rinsing and washing in TBST (3×, 10 to 15 min), the blots were probed with peroxidase-conjugated goat anti-rabbit secondary antibody (1:10,000) for 2 h at room temperature. Bands were visualized using ECL reagents (Amersham).
Immunocytochemistry was carried out essentially as previously described (31). The day following transfection, cells were split on two-chamber glass slides. Approximately 48 h posttransfection, the cells were treated with 100 nM Dex or vehicle for 2 h. The cells were fixed in 2% paraformaldehyde, washed in PBS, and permeabilized with 0.2% Triton X-100. Cells were again washed in PBS, treated with 2% normal goat serum, washed in PBS, and incubated overnight with a 1:8,000 dilution of Ab57. The following day, the slides were washed and incubated with a secondary antibody at 1:7,500 (Texas Red conjugated antibody from Molecular Probes) for 20 h at 4°C. After washing, fluorescence was measured using a Zeiss LSM 510 laser scanning confocal microscope with a Plan-Apochromat 100× oil immersion objective and the recommended excitation and emission wavelengths for Texas Red (Molecular Probes). Digital images were optimized in Photoshop to reveal both intensity and distribution of immunoreactive GR. The cells stained with peroxidase were processed as described above using the Super sensitive detection kit for mouse and rat (Biogenex) followed by a 6-min reaction with diaminobenzidine-peroxidase solution. Cells were counterstained with hematoxylin.
Structure predictions and molecular modeling.
The following programs were used for secondary structure prediction: DPM (10); DSC (26); GOR1 (13); GOR2 (15); HNN (hierarchal neural network method) and MLRC (16); PHD (35); Predator (12); SOPM (self-optimized prediction method) (14); PSIpred (position-specific iterated prediction) (25); and Sspro (2). The majority of these algorithms can be found and run on the Internet or are available at no charge from the authors.
LBD models were developed for hGRα, hGRβ, the hGRα(1-742) truncation mutant, and the hGRα(β6-7) double mutant using the Molecular Simulations Homology module and associated programs. Models were based on the coordinates for the solved, ligand-bound progesterone receptor LBD structure (Protein Data Bank entry 1A28). The alignment and models developed were checked to ensure that they were in agreement with a variety of secondary structure predictions (19; Wisconsin Package version 10.3 [Accelrys, Inc., San Diego, Calif.]). The models developed were also checked using the programs PROCHECK and WhatIf to insure that psi and phi angles occurred in the expected regions of the Ramachandran map and that there were no steric bumps or overlaps in the structure. Energy minimization and dynamics were performed as part of the homology modeling using the Discover CVFF force field in the Molecular Simulations software. For comparison of our model with the solved structure (5), root mean square deviation was calculated using the program MVP (27).
RESULTS
The domain organization of the full-length wild type (wt) hGRα is shown in Fig. 1A. Nuclear receptor LBDs exhibit a highly conserved three-dimensional architecture (6, 42, 46). The recent solution of the GR LBD further demonstrates the structural homology among nuclear receptors (5). However, the GR LBD coordinates are not yet publicly available. In order to understand the potential structural differences between hGRα and hGRβ, we have modeled the hGRα LBD using the coordinates from the progesterone receptor (hPR) (Protein Data Bank entry 1A28), the most closely related superfamily member whose LBD crystal structure has been solved (Fig. 1B) (45). The tertiary structure of hGRα, (LBD, residues 525 to 777) was predicted using the Molecular Simulations Homology module and contains the structure of Dex bound to the receptor in place of progesterone. Our model was compared with the solved structure of hGRα, and the overall root mean square deviation on all backbone atoms between the reported experimental structure (residues 531 to 776) and the model developed is 1.78 Å (M. Lambert, personal communication).
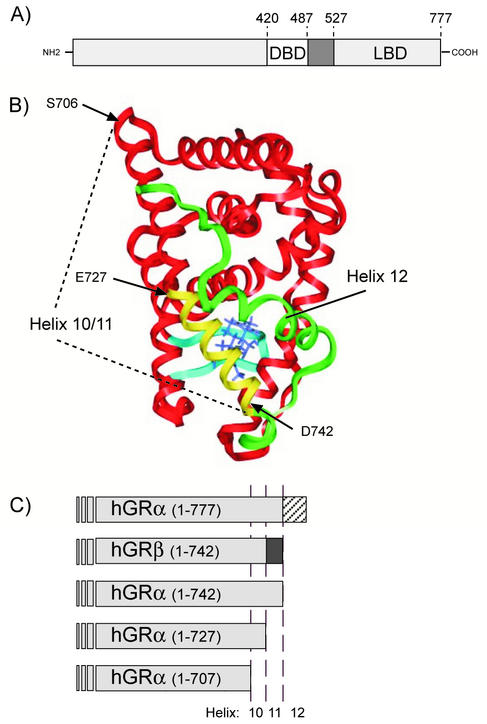
FIG. 1.
(A) Linear representation of the hGRα, indicating the boundaries of the DBD and LBD. (B) Homology model of the hGRα LBD (residues 525 to 777) based on the crystal structure coordinates of the progesterone receptor (PR) LBD (45). The three amino acids labeled, S706, E727, and D742, represent the boundaries of the truncation mutants described below. The residues, which are unique for hGRβ (amino acids 728 through 742), are shown in yellow, while the entire H11-H12 loop and H12 are shown in green (residues 743 to 777). (C) Illustration of hGRα C-terminal deletion constructs used in the experiments described. hGRα(1-742) represents an hGRα truncation that is the same length as hGRβ. hGRα(1-727), previously described by Oakley et al., is truncated at the hGRα and hGRβ divergence point. Finally, in the hGRα(1-706) truncation, the entirety of H10/11 and H12 is removed.
To functionally distinguish between the absence of H12 and the role of the 15 unique residues at the carboxy terminus of hGRβ, we created several hGRα truncation mutants (Fig. 1C). It is noteworthy that the boundaries of the hGRβ unique sequence lie at structurally sensible points: the end of H10 (that is the boundary of two distinct helices in several nuclear receptors) and the end of H11. The hGRα(1-742) construct represents the hGRα homologue to hGRβ in that both proteins are the same length but are composed of 15 distinct amino acids at the carboxy terminus. Our second hGR truncation, hGRα(1-727), is useful for understanding the role of residues 728 to 742 in hGR function (30). Finally, we created a third strategic truncation, hGRα(1-706), which lacks the entire putative H10/11 region and should be severely compromised in both structure and function.
Functional analysis of hGRα, hGRβ, and truncation mutants.
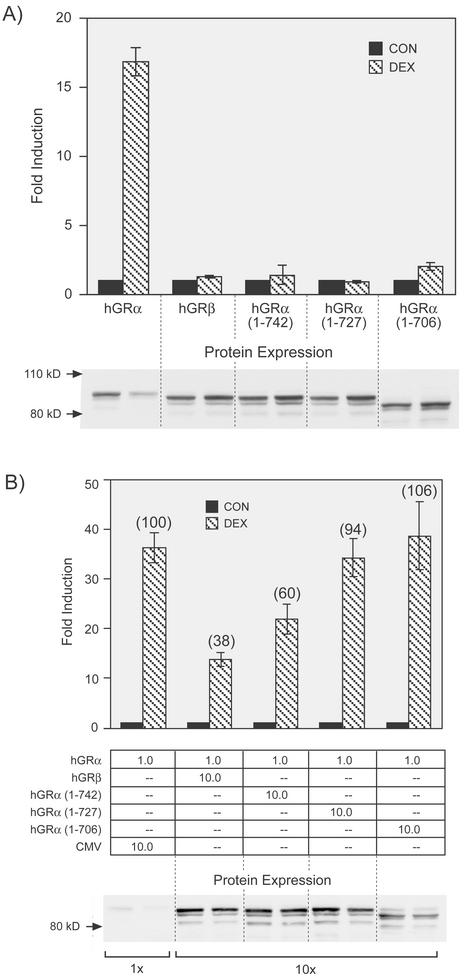
The truncation mutants described in Fig. 1 were evaluated for transactivation and dominant negative activity. Of the five hGR constructs shown in Fig. 1C, only full-length hGRα is transcriptionally active from the glucocorticoid-responsive promoter MMTV-CAT (Fig. 2A). This result is consistent with reports in which less severe carboxy-terminal truncations of the mouse GR result in complete loss of binding and transcriptional activity (48). Western blots shown below the graph indicate that the lack of transactivation is not a result of different protein expression levels. The three hGRα truncations were next compared to hGRβ for dominant negative activity (Fig. 2B). Only hGRα(1-742) exhibited an inhibitory effect on full-length hGRα activity, and that was only partially as strong as that observed for hGRβ [40% inhibition for hGRα(1-742) compared to 62% for hGRβ]. Thus, the lack of H12 alone cannot entirely account for the dominant negative activity of hGRβ. Western blots shown beneath the graph indicate that protein expression levels are equivalent and do not account for the differences in dominant negative activity observed. The Western blots also show that only hGRα undergoes ligand-dependent down regulation; the truncation mutants and hGRβ do not.
FIG. 2.
A) Transactivation of hGRα, hGRβ, and hGRα truncation mutants. The hGRα proteins described in Fig. 1C were analyzed for transactivation potential following transient transfection in GR-deficient COS-1 cells. Cells were cotransfected with the indicated GR expression vector and a glucocorticoid-responsive MMTV-CAT reporter gene (pGMCS). Hormone response was measured as the fold induction of CAT activity in response to 100 nM Dex treatment (DEX, hatched bars) over that of untreated cells (CON, solid bars). Data are an average from two experiments with the indicated standard error of the mean. Protein expression levels, determined by Western blotting for each condition, are shown beneath the graph. (B) Dominant negative activities of hGRβ and the three-hGRα truncation mutants. Dominant negative activity is measured as the decrease in hormone response (fold induction) upon coexpression of hGRα with 10-fold more of the empty vector (pCMV5 [CMV]), hGRβ, or the other hGRα truncation mutants. The percentage of transactivation is shown in parentheses above each bar, where hGRα plus the empty pCMV5 vector is 100%. The relative amounts of transfected expression vectors are indicated below the graph. Transfection and CAT assays were carried out as for panel A. Data shown are an average for four to seven experiments with the indicated standard error of the mean.
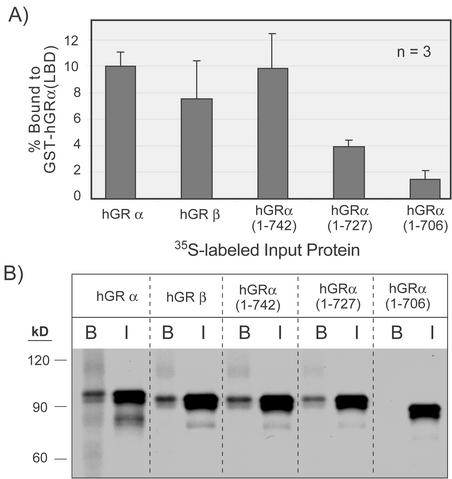
The formation of inactive heterodimers is the likely mechanism for dominant negative activity (30). A possible explanation for the observed difference between hGRβ and hGRα(1-742) and the other truncation mutants is an altered dimerization surface. To ascertain how the hGRα LBD interacts with the other GR constructs, GST fusion protein pull-down assays were developed. The entire molecular surface involved in dimerization of the GR is not completely understood, although regions within both the DNA binding domain (DBD) and the LBD are involved (5, 36). Since all of the constructs used in Fig. 2 contain the DBD, bacterially expressed hGRα LBD (residues 525 to 777) was prepared as a GST fusion protein in order to assess the contribution of only the LBD in dimerization. Glutathione agarose-immobilized hGRα LBD was incubated with 35S-labeled, in vitro-transcribed and -translated hGRα, hGRβ, and hGRα truncations. The radiolabeled hGRα, hGRβ, and hGRα(1-742) interact equivalently with the hGRα LBD in a ligand-independent manner, whereas the hGRα(1-727) truncation exhibits weak interaction while the hGRα(1-706) interaction approaches background levels (Fig. 3A). None of the labeled proteins interacts with GST alone (data not shown). A representative experiment is shown indicating the bound and input protein for each construct (Fig. 3B). Since hGRα(1-742) and hGRβ heterodimerize with equal affinity to hGRα LBD, this suggests that the weaker dominant negative activity of hGRα(1-742) (compared to that of hGRβ) is not a result of a weaker dimerization interface in this region of the LBD.
FIG. 3.
In vitro interaction of hGRα LBD with hGRα, hGRβ, and hGRα C-terminal truncation mutants. A) Summary of GST pull-down assays. The E. coli-expressed hGRα LBD (amino acids 525 to 777) was immobilized on glutathione agarose and incubated with the indicated 35S-labeled, in vitro-translated hGR proteins described in Fig. 1C. The percent of 35S-protein bound by GST-hGRα LBD averaged from three independent experiments was determined by densitometry of autoradiographs. (B) A representative autoradiograph of the data summarized in panel A. I indicates input protein, and B is the bound fraction. Positions of molecular size markers are shown to the left.
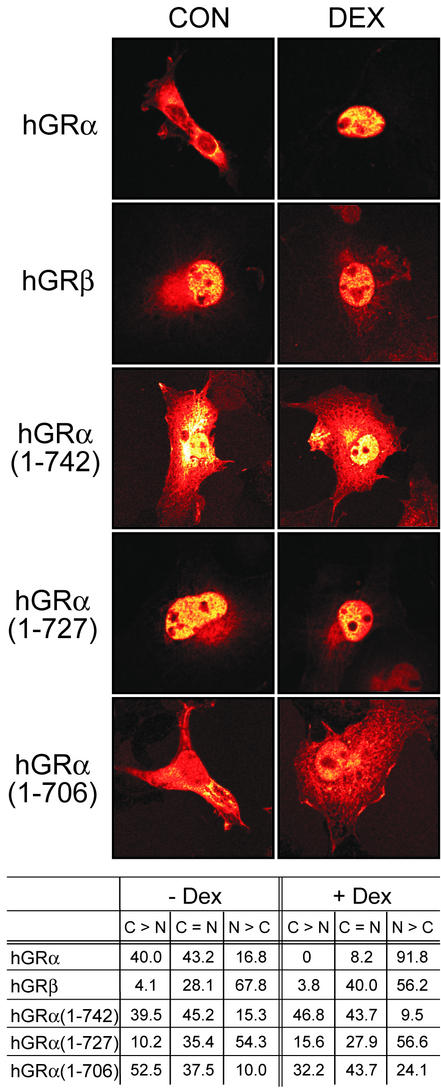
To further investigate the biochemical and functional differences between hGRα, hGRβ, and the truncation mutants, we compared their subcellular distribution by immunocytochemistry. Surprisingly, unlike the constitutively nuclear hGRβ, the hGRα(1-742) truncation remains predominantly cytosolic in the absence or presence of Dex (Fig. 4). Interestingly, the hGRα(1-727) truncation is nuclearly localized, similar to the case for hGRβ. The hGRα(1-706) truncation, which is missing the entire H10-H12 region and putative dimerization interface, exhibits diffuse cellular staining relative to the other proteins studied. Only hGRα changes cellular distribution in response to the agonist Dex, which is consistent with ligand binding and transactivation data. It thus appears that both subcellular localization and physical interaction with hGRα are important prerequisites for hGRβ imparting a dominant negative effect. Although the truncation hGRα(1-727) is mostly nuclear, as is hGRβ, its inability to interact with hGRα likely explains its lack of dominant negative activity.
FIG. 4.
Immunocytochemical analysis of hGRs. The subcellular distribution of hGRα, hGRβ, and the indicated hGRα truncation mutants was measured in the absence (CON) and presence (DEX) of Dex. A representative image for each condition is shown. The data were quantified by ranking cell staining according to their predominantly cytosolic (C > N), predominantly nuclear (N > C), or equivalent (C = N) distribution. The numbers represent the percentages of cells counted which fell into the indicated rank. Approximately 150 cells were counted from two or three individual experiments.
Generation of hGRα/β hybrids.
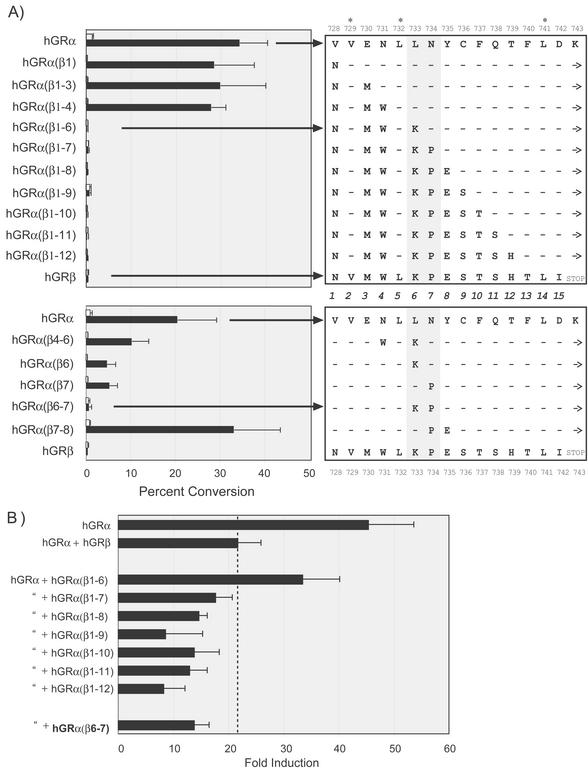
We next asked whether the dominant negative activity of hGRβ could be attributed to any particular amino acid or was dependent on the entire 15-residue sequence of hGRβ. To address this, sequential cumulative mutagenesis of hGRα residues 728 to 742 to the corresponding amino acids in hGRβ was performed. These α/β hybrids were first tested for hormone-induced transactivation. No significant changes in transactivation were observed when each of the first three unique residues (728, 730, and 731) of hGRβ were swapped into hGRα. However, the next amino acid replacement, leucine (L) 733 to lysine (K), totally abolished ligand-dependent transactivation. This loss of activity was maintained for each successive mutation in the α/β hybrids (Fig. 5, top panel). This abrupt all-or-nothing effect on transactivation prompted us to generate single and double point mutations surrounding residues 733 and 734, replacing hGRα residues with the corresponding beta sequence. Although diminished transactivation was observed with several mutants, only one double-mutant hybrid, hGRα(β6-7), in which leucine 733 and asparagine (N) 734 were replaced with lysine and proline (P), as in hGRβ, was completely inactive (Fig. 5, bottom panel).
FIG. 5.
Transactivation and dominant negative activity of hGRα/β hybrids. (A) The amino acids 728 to 742 of hGRα were replaced with the corresponding amino acids of hGRβ in a sequential fashion. The residues from 728 through 742 were numbered 1 to 15, shown in the center. Note that three residues, Val729 (position 2), Leu732 (position 5), and Leu741 (position 14), are conserved in both hGRα and hGRβ (∗). Beginning with the mutation of Val728 to Asp, hGRα(β1), each additional mutant included the one before it, until a complete hGRα/β hybrid was generated. − indicates that no change from wt hGRα was made. Only the first 12 sequential mutants are shown. Dex-induced transactivation was measured as for Fig. 2A with the data expressed as percent conversion of [14C]chloramphenicol to the acetylated form. The top panel shows the sequential mutants, where each residue in hGRα was changed with a corresponding hGRβ residue in succession. The bottom panel shows data from single and double point mutants described in the text. For each data set the wt hGRα is shown as the top sequence and the hGRβ sequence is on the bottom. Note that each hybrid contains the H12 carboxy-terminal extension (→), except hGRβ, which terminates following Ile742. The shaded region through positions 6 and 7 (amino acids 733 and 734) indicates the two amino acid mutations sufficient to completely block transactivation. As in Fig. 2, Western blots indicated similar expression levels for each receptor tested (data not shown). (B) The hybrid receptors from panel A that were completely deficient in Dex-induced transactivation were tested for dominant negative activity following the experimental protocol described for Fig. 2A. The top two bars, hGRα (+ pCMV5) and hGRα + hGRβ, are the controls. The dashed line indicates the level of dominant negative activity observed with hGRβ. All receptors tested for dominant negative activity were transfected at 10-fold-higher levels relative to levels for hGRα. The middle bars are the sequential mutants that lack transactivation, and the double mutant, hGRα(β6-7), is shown at the bottom.
We next evaluated the dominant negative activity of the transactivation-deficient receptors described in Fig. 5. The α/β hybrid, hGRα(β1-6), although completely defective in transactivation, is a poor dominant negative inhibitor of hGRα. However, the next sequential hybrid, hGRα(β1-7), in which the first seven unique residues in hGRβ are swapped into the corresponding hGRα sequence, is a potent dominant negative inhibitor. The remaining sequential hybrids, hGRα(β1-8) to hGRα(β1-12), are also dominant negative proteins. Interestingly, the double mutant, hGRα(β6-7), is also a strong dominant negative inhibitor of hGRα function, illustrating that only two residues from hGRβ, Lys733 and Pro734, are necessary for dominant negative activity (Fig. 5B).
The hGRα(β6-7) mutant.
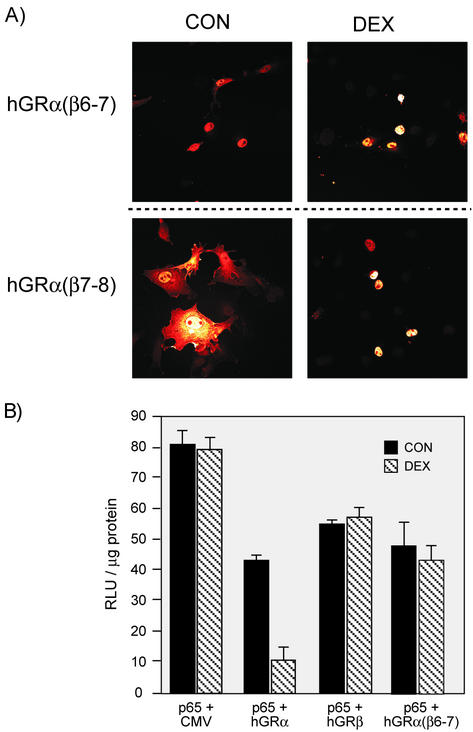
The hGRα(β6-7) mutant was further characterized and compared with hGRβ. Immunocytochemistry of the double mutants hGRα(β6-7) and hGRα(β7-8) supports the hypothesis that constitutive nuclear localization is an important feature in dominant negative activity. The dominant negative hGRα(β6-7) is essentially nuclear in the absence and presence of hormone. However, the hybrid hGRα(β7-8), which transactivates the reporter gene and is nuclearly localized in response to Dex, similar to wt hGRα, is found mainly in the cytosol in the absence of Dex (Fig. 6A). Interestingly, many of the other hGR α/β hybrids exhibit a diffuse cellular GR staining pattern, suggesting that H12 may have an effect on cellular localization and that the complex functions associated with this region of the receptor are not freely dissociated (data not shown).
FIG. 6.
Analysis of hGRα(β6-7). (A) Nuclear localization of double mutants hGRα(β6-7) and hGRα(β7-8). The dominant negative double mutant hGRα(β6-7) was compared with the double mutant hGRα(β7-8), which appears normal in transactivation (see Fig. 5). Immunocytochemistry was carried out as for Fig. 4. (B) Repression of p65 by hGRα but not hGRβ or hGRα(β6-7). COS1 cells were transfected with the 3X MHC-luc reporter plus pCMV5, hGRα, hGRβ, or hGRα(β6-7) and treated with 100 nM Dex or left untreated. Overexpression of hGRβ or hGRα(β6-7) does not repress transactivation of an NF-κB reporter gene in the presence of hormone.
A potentially important distinction between hGRα and hGRβ is the repressive effect on NF-κB-mediated transactivation. wt hGRα will transrepress p65-mediated reporter in response to hormone, whereas hGRβ shows no hormone-dependent repression (30). When tested in this system, the hGRα(β6-7) mutant also failed to further transrepress p65 reporter gene activity in the presence of hormone (Fig. 6B). These data further indicate that these two residues are sufficient for creating GRβ-like activity.
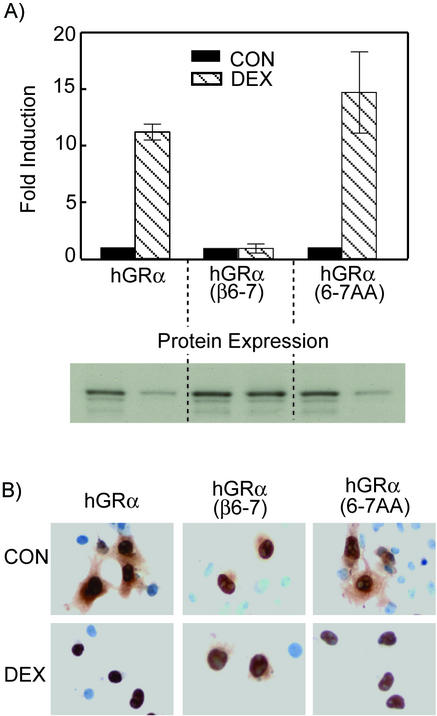
To test whether the biochemical properties of the hGRα(β6-7) mutant were dependent on the particular amino acid side chains or simply a result of changing the existing residues in hGRα, additional mutagenesis of hGRα residues 733 and 734 was carried out. Both amino acids were mutated to alanine, thereby generating the hGRα L733A/N734A double mutant. Functional analysis of this mutant showed transactivation ability similar to that of wt hGRα as well as hormone-induced down regulation and nuclear translocation (Fig. 7). Therefore, it appears that residues K733 and P734 are necessary and sufficient for generation of a dominant negative inhibitor of hGRα that is functionally homologous to hGRβ despite the presence of H12 and the associated AF-2 function.
FIG. 7.
Transactivation and nuclear localization of an hGRα alanine mutant. The amino acids at positions 6 and 7 of hGRα previously described (L733 and N734) were mutated to alanine [hGRα(6-7AA]. (A) Transactivation of MMTV-Luc and corresponding protein expression. (B) Nuclear localization of hGRα, hGRβ, and hGRα(6-7AA) in the absence and presence of Dex. Localization of hGRα(6-7AA) is the same as that of wt hGRα.
Molecular modeling.
Several strategies were employed to elucidate the structural basis for the dominant negative activity of hGRβ and to evaluate the contribution of the side chains of residues 733 and 734. Twelve unique secondary structure prediction algorithms were employed in concert with homology modeling to develop three-dimensional structures and alignments for hGRα, hGRβ, hGRα(β6-7), and the truncation mutants. The secondary structure alignments are shown in Fig. 8A. Not surprisingly, these algorithms clearly predict that the hGRα(728-742) sequence, VVENLLNYCFQTFLD, forms an alpha helix in agreement with its homology to solved LBD structures. However, the unique hGRβ sequence, NVMWLKPESTSHTLI, is predicted to be largely disordered. The same secondary structure predictions carried out with the truncation mutant hGRα(1-742) indicate that the random coil predicted for the unique hGRβ region is not simply a result of truncation. Furthermore, predictions for the hGRα(β6-7) double mutant indicate a kink, break, or distortion at that point in H11 (Fig. 8A).
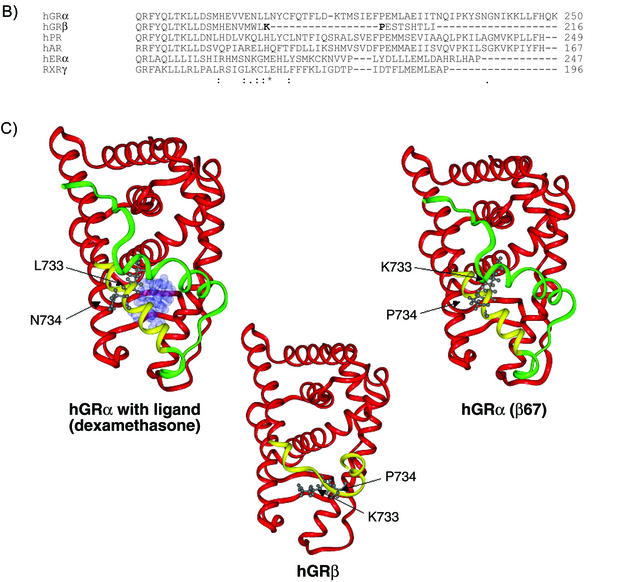
FIG. 8.
Structure predictions and molecular modeling. (A) A total of 12 secondary structure algorithms were run on the entire sequences of hGRα, hGRβ, hGRα(β6-7), and hGRα(1-742). The single-letter amino acid abbreviation for each receptor from residue 701 until the end is shown at the top of each sequence prediction block. Below the wt hGRα sequence is a comparison with the hPR. Below the hPR is the actual secondary structure found for the crystallographic solution of the hPR structure. The solid lines between hGR and hPR indicate a conserved amino acid. (B) Multiple sequence alignment of hGRα, hGRβ, and several other related receptors (hPR, hAR, hERα, and RXRγ). Alignment of hGRβ was done using the program Clustal W. Symbols underneath alignment indicate conserved residues. C) Three-dimensional models of hGRα, hGRβ, and hGRα(β6-7). Modeling was done using Molecular Simulations software. The common helices of each model (residues 525 to 727) are shown in red, the nonhomologous region from amino acid 728 through 742 is colored yellow, and the H12 region (residues 743 to 777), present in hGRα and hGRα(β6,7), is colored green. The hGRα model is shown with ligand occupying the binding pocket. The images also show the side chain orientations of residues 733 and 734 in each molecule.
The solved crystal structure for the ligand-bound progesterone receptor was used as the target structure for the development of a structural homology model of hGRα and hGRβ (45). The basis for the hGRβ structural model is a multiple sequence alignment of several members of the nuclear receptor superfamily ligand binding domains using the Clustal W alignment algorithm (Fig. 8B) (19). An alignment was also performed using the GCG Pileup program (Wisconsin Package version 10.3). Published alignments of the nuclear receptor ligand binding domains (46) and the Pfam protein domain databases were consulted as well. As in Fig. 1B, Dex replaces progesterone in the ligand-binding site of the hGRα model (45). The three-dimensional structure models of hGRα, hGRβ, and hGRα(β6-7) are shown (Fig. 8C). Whereas H10/11 in the truncated hGRα is relatively undisturbed, the unique hGRβ sequence adopts a significantly different structure with the unwinding of H11 beginning near residue 733. This gross unwinding likely reflects the constraints applied from the alignment in Fig. 8B. Nevertheless, these models, along with that of hGRα(β6-7), predict that the disruption of H10/11 in hGRβ is caused by replacing leucine and asparagine in hGRα with lysine and proline, respectively, thereby generating side chain interactions that are both electrostatically and sterically unfavorable.
DISCUSSION
The dominant negative activity of hGRβ results from several cooperating factors associated with the extreme carboxy-terminal ends of hGRα and hGRβ. Hormone binding, transactivation (AF-2), dimerization, and surprisingly, nuclear localization all depend upon sequences within the last 50 or so residues of hGRα. In hGRβ, alternative splicing replaces the final 50 amino acids of hGRα with 15 unique residues. It remains unclear which of the properties listed above are affected by this change and how they contribute to the dominant negative phenotype of hGRβ. We have determined that hGRβ's unique sequence and the missing H12 both contribute to dominant negative activity. First, the absence of H12 renders the receptor unable to bind ligand or transactivate reporter genes. Second, the 15 unique residues function to keep the receptor constitutively nuclear localized. This feature, in addition to physical interaction, is critical for maximum dominant negative activity. Surprisingly, the dominant negative function of hGRβ can be localized to just two amino acids within the unique hGRβ sequence. These data, along with secondary structure predictions and molecular modeling, support an atomic-scale hypothesis describing how the different secondary and tertiary structures of hGRβ and hGRα result in the dominant negative phenotype.
Other dominant negative nuclear receptor mechanisms.
Many nuclear receptors have naturally occurring dominant negative isoforms in which the mutant form blocks the activity of the wt protein when the two are coexpressed in the same cell. Dominant negative forms of the thyroid hormone receptor and the peroxisome proliferator activated receptor gamma are important factors in disease pathology associated with these receptors (4, 9). Similar reports with ER dominant negative mutants (23, 37), as well as other nuclear receptors (24), suggest that H12 and the associated AF-2 contribute to the dominant negative effect and that most dominant negative receptors are generated via alterations in the length, sequence, conformation, or stability of the carboxy-terminal end of the LBD.
The hGRβ dominant negative mechanism contains distinguishing features in comparison to most other nuclear receptors. First, it appears that the GR ligand binding is more sensitive to perturbations of H10/11 and H12 than other receptors, such as ER or RXR, where disruption of H12 does not prevent agonist binding (37, 48). Second, hGRα is a cytosolic protein in the absence of hormone, while most nuclear receptors are constitutively nuclear. Although none of the dominant negative GRs generated bind Dex, whether disruption of binding is necessary for dominant negative activity is unclear. For example, the hGRα(β1-6) receptor is transcriptionally inactive but lacks full dominant negative activity, suggesting that these two features are separable.
Subcellular localization, DNA binding, and dimerization.
The classical view of GR action involves ligand binding-induced conformational changes with concurrent release of cytoplasmic accessory proteins and unmasking of a nuclear localization signal located immediately carboxy-terminal to the DBD (32). However, the immunocytochemistry experiments presented here suggest a role of the carboxy-terminal end of the GR LBD in nuclear or cytoplasmic retention. That hGRβ and hGRα(1-727) are primarily nuclear while hGRα(1-742) is cytoplasmic suggests that the region from amino acid 728 to 742 is somehow involved in cytoplasmic retention and/or nuclear export. Whether this is directly coded within this region of the receptor or relies on other parts of the receptor is not clear. However, Zelko et al. recently reported that the homologous H10/11 region in the nuclear receptor CAR is involved in nuclear localization of that receptor (47). Two conserved regions within H10/11 of hGR somewhat resemble the leucine-rich regions of well-defined nuclear export signals found in human immunodeficiency virus Rev and IκB (18). Whether this region of the GR harbors a functional signaling or localization motif remains to be established.
One possible mechanism for dominant negative activity is the blocking of DNA binding sites by the transcriptionally silent hGRβ. However, several lines of evidence do not support this mechanism. First, overexpression of hGRβ does not significantly affect progesterone or androgen receptor transactivation (30). Since the recognition sites are identical for all three receptors, the lack of an effect of hGRβ suggests a mechanism other than squelching of response elements. In addition, preliminary analysis of mutations in the DNA binding domain shows no effect on the ability of hGRβ to function as a dominant negative inhibitor of hGRα (M. R. Yudt and J. A. Cidlowski, unpublished observations).
Our model for dominant negative action centers on the formation of inactive, or weakly active, heterodimers between hGRα and hGRβ. The recently published crystal structure of the hGRα LBD suggests that the dimerization interface is distinct from other solved nuclear receptor LBD structures. Before the structure of the hGRα was solved, the interface of all nuclear receptor LBD dimers involved H10 and/or H12 (6). However, the hGRα LBD dimer interface involves H4 and beta strands 3 and 4, located on the opposite face of the folded domain from H10 to H12. This suggests that the gross structural changes in H11 and H12 associated with hGRβ or hGRα(β6-7) would not be expected to perturb the dimer interface found in hGRα. The crystal structure also provides a potential explanation for our observations that the hGRα LBD can dimerize with the full-length in vitro-translated receptor in a ligand-independent fashion, since the conformational changes believed to occur upon hormone binding are far away from the published dimer interface (5).
Molecular modeling and the structure of hGRβ.
Despite being the first receptor cloned (44) and its obvious pharmacological interest, the GR LBD structure has only recently been determined, and the structural information is not yet publicly accessible (5). To date, well over 20 different nuclear receptor LBD structures have been published, including apo-, agonist-bound, antagonist-bound, and ternary complexes with coactivator or corepressor peptides (for reviews, see reference 42). Nuclear receptor LBDs conform to a canonical folding architecture (46). The model of the hGRα presented here is similar to previously published homology models of the GR LBD also generated from the structure of the progesterone receptor LBD (33, 34). In addition, comparison of our model with the solved structure (M. Lambert, personal communication) supports the claim that the hGRα model presented here is a valid model and is sufficiently similar to the experimental structure to support our conclusions.
The purpose of our modeling experiments is to identify the structural consequences of alternative splicing in relation to the function of hGRα and hGRβ and to better understand the molecular environment of residues 733 and 734 in particular. The results from 12 structure prediction algorithms strongly indicate a nonhelical or random conformation of the 15 unique residues in hGRβ. Τhe same predictions on hGRα(1-742) show that the disorder of these residues is not merely a result of truncation and the absence of H12. In fact, secondary structure predictions for both full-length hGRα and the truncated hGRα(1-742) concur with homologous solved structures, such as that of progesterone receptor (45), which predict helicity up to residue 741 to 743. Inspection of side chain conformations in hGRα and hGRβ as well as in the hGRα(β6-7) double mutant supports the experimental evidence that disruption of H11 at or near residue 733 is a critical feature for the dominant negative activity of hGRβ. In conclusion, the 15 unique residues of hGRβ, by keeping the receptor nuclearly localized, play an important role in dominant negative activity of this hGR isoform. We have shown that the absence of H12 itself is neither necessary nor sufficient for a full dominant negative phenotype.
.
Acknowledgments
We thank Mill Lambert for comparing our model with the solved hGR X-ray structure.
REFERENCES
- 1.Allgood, V. E., R. H. Oakley, and J. A. Cidlowski. 1993. Modulation by vitamin B6 of glucocorticoid receptor-mediated gene expression requires transcription factors in addition to the glucocorticoid receptor. J. Biol. Chem. 268:: 20870-20876. [PubMed] [Google Scholar]
- 2.Baldi, P., S. Brunak, P. Frasconi, G. Soda, and G. Pollastri. 1999. Exploiting the past and the future in protein secondary structure prediction. Bioinformatics 15:937-946. [DOI] [PubMed] [Google Scholar]
- 3.Bamberger, C. M., A. M. Bamberger, M. de Castro, and G. P. Chrousos. 1995. Glucocorticoid receptor beta, a potential endogenous inhibitor of glucocorticoid action in humans. J. Clin. Investig. 95:2435-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barroso, I., M. Gurnell, V. E. F. Crowley, M. Agostini, J. W. Schwabe, M. A. Soos, G. L. Maslen, T. D. M. Williams, H. Lewis, A. J. Schafer, V. K. K. Chatterjee, and S. O'Rahilly. 1999. Dominant negative mutations in human PPAR gamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature 402:880-883. [DOI] [PubMed] [Google Scholar]
- 5.Bledsoe, R. K., V. G. Montana, T. B. Stanley, C. J. Delves, C. J. Apolito, D. D. McKee, T. G. Consler, D. J. Parks, E. L. Stewart, T. M. Willson, M. H. Lambert, J. T. Moore, K. H. Pearce, and H. E. Xu. 2002. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 110:93-105. [DOI] [PubMed] [Google Scholar]
- 6.Bourguet, W., P. Germain, and H. Gronemeyer. 2000. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol. Sci. 21:381-388. [DOI] [PubMed] [Google Scholar]
- 7.Bronnegard, M., P. Stierna, and C. Marcus. 1996. Glucocorticoid resistant syndromes—molecular basis and clinical presentations. J. Neuroendocrinol. 8:405-415. [DOI] [PubMed] [Google Scholar]
- 8.Cole, T. J., J. A. Blendy, A. P. Monaghan, K. Krieglstein, W. Schmid, A. Aguzzi, G. Fantuzzi, E. Hummler, K. Unsicker, and G. Schutz. 1995. Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell-development and severely retards lung maturation. Genes Dev. 9:1608-1621. [DOI] [PubMed] [Google Scholar]
- 9.Damm, K., C. C. Thompson, and R. M. Evans. 1989. Protein encoded by V-Erba functions as a thyroid-hormone receptor antagonist. Nature 339:593-597. [DOI] [PubMed] [Google Scholar]
- 10.Deleage, G., C. Combet, C. Blanchet, and C. Geourjon. 2001. ANTHEPROT: an integrated protein sequence analysis software with client/server capabilities. Comput. Biol. Med. 31:259-267. [DOI] [PubMed] [Google Scholar]
- 11.Encio, I. J., and S. D. Detera-Wadleigh. 1991. The genomic structure of the human glucocorticoid receptor. J. Biol. Chem. 266:7182-7188. [PubMed] [Google Scholar]
- 12.Frishman, D., and P. Argos. 1997. Seventy-five percent accuracy in protein secondary structure prediction. Proteins Struct. Funct. Genet. 27:329-335. [DOI] [PubMed] [Google Scholar]
- 13.Garnier, J., J. F. Gibrat, and B. Robson. 1996. GOR method for predicting protein secondary structure from amino acid sequence. Comput. Methods Macromol. Sequence Anal. 266:540-553. [DOI] [PubMed] [Google Scholar]
- 14.Geourjon, C., and G. Deleage. 1994. Sopm—a self-optimized method for protein secondary structure prediction. Protein Eng. 7:157-164. [DOI] [PubMed] [Google Scholar]
- 15.Gibrat, J. F., J. Garnier, and B. Robson. 1987. Further developments of protein secondary structure prediction using information-theory—new parameters and consideration of residue pairs. J. Mol. Biol. 198:425-443. [DOI] [PubMed] [Google Scholar]
- 16.Guermeur, Y., C. Geourjon, P. Gallinari, and G. Deleage. 1999. Improved performance in protein secondary structure prediction by inhomogeneous score combination. Bioinformatics 15:413-421. [DOI] [PubMed] [Google Scholar]
- 17.Hamid, Q. A., S. E. Wenzel, P. J. Hauk, A. Tsicopoulos, B. Wallaert, J. J. Lafitte, G. P. Chrousos, S. J. Szefler, and D. Y. M. Leung. 1999. Increased glucocorticoid receptor beta in airway cells of glucocorticoid-insensitive asthma. Am. J. Respir. Crit. Care Med. 159:1600-1604. [DOI] [PubMed] [Google Scholar]
- 18.Henderson, B. R., and A. Eleftheriou. 2000. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 256:213-224. [DOI] [PubMed] [Google Scholar]
- 19.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Comput. Methods Macromol. Sequence Anal. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 20.Hollenberg, S. M., C. Weinberger, E. S. Ong, G. Cerelli, A. Oro, R. Lebo, E. B. Thompson, M. G. Rosenfeld, and R. M. Evans. 1985. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature 318:635-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honda, M., F. Orii, T. Ayabe, S. Imai, T. Ashida, T. Obara, and Y. Kohgo. 2000. Expression of glucocorticoid receptor beta in lymphocytes of patients with glucocorticoid-resistant ulcerative colitis. Gastroenterology 118:859-866. [DOI] [PubMed] [Google Scholar]
- 22.Hong, H., K. Kohli, A. Trivedi, D. L. Johnson, and M. R. Stallcup. 1996. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc. Natl. Acad. Sci. USA 93:4948-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ince, B. A., Y. Zhuang, C. K. Wrenn, D. J. Shapiro, and B. S. Katzenellenbogen. 1993. Powerful dominant negative mutants of the human estrogen receptor. J. Biol. Chem. 268:14026-14032. [PubMed] [Google Scholar]
- 24.Johnson, B. S., R. A. Chandraratna, R. A. Heyman, E. A. Allegretto, L. Mueller, and S. J. Collins. 1999. Retinoid X receptor (RXR) agonist-induced activation of dominant-negative RXR-retinoic acid receptor α403 heterodimers is developmentally regulated during myeloid differentiation. Mol. Cell. Biol. 19:3372-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 26.King, R. D., and M. J. E. Sternberg. 1996. Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci. 5:2298-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambert, M. H. 1997. Docking conformationally flexible molecules into protein binding sites, p. 243-303. In P. S. Charifson (ed.), Practical application of computer-aided drug design. Marcel Dekker, New York, N.Y.
- 28.Leung, D. Y. M., Q. Hamid, A. Vottero, S. J. Szefler, W. Surs, E. Minshall, G. P. Chrousos, and D. J. Klemm. 1997. Association of glucocorticoid insensitivity with increased expression of glucocorticoid receptor beta. J. Exp. Med. 186:1567-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKay, L. I., and J. A. Cidlowski. 1998. Cross-talk between nuclear factor-kappa B and the steroid hormone receptors: mechanisms of mutual antagonism. Mol. Endocrinol. 12:45-56. [DOI] [PubMed] [Google Scholar]
- 30.Oakley, R. H., C. M. Jewell, M. R. Yudt, D. M. Bofetiado, and J. A. Cidlowski. 1999. The dominant negative activity of the human glucocorticoid receptor beta isoform. Specificity and mechanisms of action. J. Biol. Chem. 274:27857-27866. [DOI] [PubMed] [Google Scholar]
- 31.Oakley, R. H., M. Sar, and J. A. Cidlowski. 1996. The human glucocorticoid receptor beta isoform. Expression, biochemical properties, and putative function. J. Biol. Chem. 271:9550-9559. [DOI] [PubMed] [Google Scholar]
- 32.Picard, D., and K. R. Yamamoto. 1987. Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 6:3333-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ray, D. W., C. S. Suen, A. Brass, J. Soden, and A. White. 1999. Structure/function of the human glucocorticoid receptor: tyrosine 735 is important for transactivation. Mol. Endocrinol. 13:1855-1863. [DOI] [PubMed] [Google Scholar]
- 34.Robin-Jagerschmidt, C., J. M. Wurtz, B. Guillot, D. Gofflo, B. Benhamou, A. Vergezac, C. Ossart, D. Moras, and D. Philibert. 2000. Residues in the ligand binding domain that confer progestin or glucocorticoid specificity and modulate the receptor transactivation capacity. Mol. Endocrinol. 14:1028-1037. [DOI] [PubMed] [Google Scholar]
- 35.Rost, B., and C. Sander. 1993. Prediction of protein secondary structure at better than 70-percent accuracy. J. Mol. Biol. 232:584-599. [DOI] [PubMed] [Google Scholar]
- 36.Savory, J. G. A., G. G. Prefontaine, C. Lamprecht, M. M. Liao, R. F. Walther, Y. A. Lefebvre, and R. J. G. Hache. 2001. Glucocorticoid receptor homodimers and glucocorticoid-mineralcorticoid receptor heterodimers form in the cytoplasm through alternative dimerization interfaces. Mol. Cell. Biol. 21:781-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schodin, D. J., Y. Zhuang, D. J. Shapiro, and B. S. Katzenellenbogen. 1995. Analysis of mechanisms that determine dominant negative estrogen receptor effectiveness. J. Biol. Chem. 270:31163-31171. [DOI] [PubMed] [Google Scholar]
- 38.Shahidi, H., A. Vottero, C. A. Stratakis, S. E. Taymans, M. Karl, C. A. Longui, G. P. Chrousos, W. H. Daughaday, S. A. Gregory, and J. M. D. Plate. 1999. Imbalanced expression of the glucocorticoid receptor isoforms in cultured lymphocytes from a patient with systemic glucocorticoid resistance and chronic lymphocytic leukemia. Biochem. Biophys. Res. Commun. 254:559-565. [DOI] [PubMed] [Google Scholar]
- 39.Sousa, A. R., S. J. Lane, J. A. Cidlowski, D. Z. Staynov, and T. H. Lee. 2000. Glucocorticoid resistance in asthma is associated with elevated in vivo expression of the glucocorticoid receptor beta-isoform. J. Allergy Clin. Immunol. 105:943-950. [DOI] [PubMed] [Google Scholar]
- 40.Steinmetz, A. C. U., J. P. Renaud, and D. Moras. 2001. Binding of ligands and activation of transcription by nuclear receptors. Annu. Rev. Biophys. Biomol. Struct. 30:329-359. [DOI] [PubMed] [Google Scholar]
- 41.Strickland, I., K. Kisich, P. J. Hauk, A. Vottero, G. P. Chrousos, D. J. Klemm, and D. Y. M. Leung. 2001. High constitutive glucocorticoid receptor beta in human neutrophils enables them to reduce their spontaneous rate of cell death in response to corticosteroids. J. Exp. Med. 193:585-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weatherman, R. V., R. J. Fletterick, and T. S. Scanlan. 1999. Nuclear-receptor ligands and ligand-binding domains. Annu. Rev. Biochem. 68:559-581. [DOI] [PubMed] [Google Scholar]
- 43.Webster, J. C., R. H. Oakley, C. M. Jewell, and J. A. Cidlowski. 2001. Proinflammatory cytokines regulate human glucocorticoid receptor gene expression and lead to the accumulation of the dominant negative beta isoform: a mechanism for the generation of glucocorticoid resistance. Proc. Natl. Acad. Sci. USA 98:6865-6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinberger, C., S. M. Hollenberg, E. S. Ong, J. M. Harmon, S. T. Brower, J. Cidlowski, E. B. Thompson, M. G. Rosenfeld, and R. M. Evans. 1985. Identification of human glucocorticoid receptor complementary-DNA clones by epitope selection. Science 228:740-742. [DOI] [PubMed] [Google Scholar]
- 45.Williams, S. P., and P. B. Sigler. 1998. Atomic structure of progesterone complexed with its receptor. Nature 393:392-396. [DOI] [PubMed] [Google Scholar]
- 46.Wurtz, J. M., W. Bourguet, J. P. Renaud, V. Vivat, P. Chambon, D. Moras, and H. Gronemeyer. 1996. A canonical structure for the ligand-binding domain of nuclear receptors. Nat. Struct. Biol. 3:87-94. (Erratum, 3:206.) [DOI] [PubMed]
- 47.Zelko, I., T. Sueyoshi, T. Kawamoto, R. Moore, and M. Negishi. 2001. The peptide near the C terminus regulates receptor CAR nuclear translocation induced by xenochemicals in mouse liver. Mol. Cell. Biol. 21:2838-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, S., X. Liang, and M. Danielsen. 1996. Role of the C terminus of the glucocorticoid receptor in hormone binding and agonist/antagonist discrimination. Mol. Endocrinol. 10:24-34. [DOI] [PubMed] [Google Scholar]