Abstract
The PML tumor suppressor gene is consistently disrupted by t(15;17) in patients with acute promyelocytic leukemia. Promyelocytic leukemia protein (PML) is a multifunctional protein that plays essential roles in cell growth regulation, apoptosis, transcriptional regulation, and genome stability. Our study here shows that PML colocalizes and associates in vivo with the DNA damage response protein TopBP1 in response to ionizing radiation (IR). Both PML and TopBP1 colocalized with the IR-induced bromodeoxyuridine single-stranded DNA foci. PML and TopBP1 also colocalized with Rad50, Brca1, ATM, Rad9, and BLM. IR and interferon (IFN) coinduce the expression levels of both TopBP1 and PML. In PML-deficient NB4 cells, TopBP1 was unable to form IR-induced foci. All-trans-retinoic acid induced reorganization of the PML nuclear body (NB) and reappearance of the IR-induced TopBP1 foci. Inhibition of PML expression by siRNA is associated with a significant decreased in TopBP1 expression. Furthermore, PML-deficient cells express a low level of TopBP1, and its expression cannot be induced by IR or IFN. Adenovirus-mediated overexpression of PML in PML−/− mouse embryo fibroblasts substantially increased TopBP1 expression, which colocalized with the PML NBs. These studies demonstrated a mechanism of PML-dependent expression of TopBP1. PML overexpression induced TopBP1 protein but not the mRNA expression. Pulse-chase labeling analysis demonstrated that PML overexpression stabilized the TopBP1 protein, suggesting that PML plays a role in regulating the stability of TopBP1 in response to IR. Together, our findings demonstrate that PML regulates TopBP1 functions by association and stabilization of the protein in response to IR-induced DNA damage.
The function of the PML tumor suppressor gene, which encodes a multifunctional protein, is consistently disrupted by t(15;17) in acute promyelocytic leukemia (APL) (26). Promyelocytic leukemia protein (PML) suppresses cell growth (31) and plays an essential role in multiple pathways of apoptosis (37, 49). PML-deficient thymocytes are resistant to programmed cell death induced by many apoptosis-inducing agents (49). PML overexpression induces G1/G0 cell cycle arrest (21) and lengthens the G1 phase of the cell cycle (30). PML-induced G1 cell cycle arrest is associated with increased expression of p53 and p21 and an inhibitor of cell cycle checkpoint kinases and results in hypophosphorylation of Rb (21, 30). PML induces apoptosis through both p53-dependent and p53-independent pathways (11, 35). Recent study showed that PML inhibits A20 protein expression induced by tumor necrosis factor alpha through the NF-κB site (52). That study suggests that PML induces apoptosis by sensitizing the tumor necrosis factor death receptor pathway.
PML is normally modified in vivo by the small ubiquitin-like modifier protein SUMO-1 at three different sites (K65, K160, and K490) (15, 60). This modification is essential for the organization of PML nuclear bodies (NBs) and for PML's role in transcription regulation and apoptosis (14, 42). Transfection of the PML SUMO-1 mutant into a normal human cell line forms normal PML NBs (23) but not in PML-deficient cells (10, 11), indicating that SUMO-1 modification of PML is essential for the formation of PML NBs (14, 60). SUMO-1 modification is essential for many of PML functions, including interaction with p53, Sp100, and Daxx (14, 22, 23, 60).
Substantial evidence demonstrates that PML plays a role in the regulation of gene expression (24, 59). PML interacts with transcription coactivator CREB-binding protein in vivo and activates the trancription of target genes (3, 6, 19). Documented evidence also shows that PML is associated with p53 in vivo and activates the transcription of p53 target genes (8, 9, 34). Activation of transcription of other genes by PML has also been reported (22, 46). Interestingly, evidence that PML plays a role in transcriptional repression has also been documented. When PML is fused downstream to the GAL4 DNA-binding domain, it represses GAL4-mediated transcription (45), possibly through a mechanism involving recruitment of the transcriptional corepressor histone deacetylases (51). PML interacts and associates in vivo with histone deacetylases and represses transactivation by deacetylation of the target promoter. The in vivo association of PML with other transcriptional corepressors has also been reported (16). Another mechanism by which PML represses transactivation is the interaction of PML with transcription factors, which are then prevented from binding to the target sites. PML sequesters and inhibits transcription mediated by Daxx (22, 23). PML interacts with Sp1 and inhibits Sp1-mediated transactivation of the epidermal growth factor receptor promoter (47). In addition, PML interacts with Nur77 (53) and NF-κB (52) and represses transcription mediated by these transcription factors in a promoter-specific manner.
Recent studies suggest that PML may play a role in the maintenance of genome stability (58) and DNA repair (4, 58). PML colocalizes in vivo with BLM, a RecQ DNA helicase deficient in patients with Bloom syndrome (2, 58). Deficiency of BLM results in genome instability (7, 50). PML was also shown to colocalize with the alternative lengthening of telemeres (ALT), suggesting that PML plays a role in maintaining the stability of the telomere ends (10, 56). PML is associated in vivo with several DNA repair proteins, including Mre11, Rad51, and H2AX, and is localized to the single-stranded DNA (ssDNA) repair foci in response to ionizing radiation (IR) (4, 27, 36). These studies suggest that PML plays a role in the organization of the double-strand break (DSB) DNA repair complex.
TopBP1, a topoisomerase IIβ-binding protein, was initially identified by yeast two-hybrid screening as the human homologue of the fission yeast Rad4/Cut5 protein, which involves in cellular responses to DNA damage and replication checkpoint controls (40, 41). TopBP1 is a DNA damage response gene, containing multiple copies of the Brca1 carboxyl-terminal motif, which has been shown to bind DNA (54). TopBP1 is a substrate of ATM kinase and is phosphorylated rapidly after exposure of the cells to IR (55). Specific antisense morpholino oligomers inhibit TopBP1 expression and induce cell death by apoptosis, indicating that TopBP1 function may be required for cell survival against IR (55). There is also evidence that TopBP1 functions in a manner similar to its fission yeast counterpart and likely involve a role in repair of DSB DNA damage and the replication checkpoint in mammalian cells (13, 25).
In response to IR exposure TopBP1 forms IR-induced foci, a nuclear speckled staining pattern similar to that of the PML NBs in vivo (13, 55). Since TopBP1 plays a role in DNA damage response and replication checkpoint controls, we investigated whether PML NBs and TopBP1 foci are functionally associated in vivo. Our study demonstrated that PML colocalized with TopBP1 at the endogenous levels in response to IR. TopBP1 and PML colocalized with the IR-induced bromodeoxyuridine (BrdU) ssDNA foci and several proteins critical for DSB DNA repairs. PML and TopBP1 were coinduced by IR and interferon (IFN), and overexpression of PML significantly induced the expression of TopBP1 via stabilization of the protein. Our study demonstrated a PML-dependent induction of TopBP1 expression. The results presented here suggest that PML plays a role in regulating TopBP1 functions by association and stabilization of the protein in response to IR-induced DNA damage.
MATERIALS AND METHODS
Plasmids.
The expression plasmid pcNDA3/myc-TopBP1 was obtained from Junjie Chen (Division of Oncology Research, Mayo Clinic, Rochester, Minn.) (55). The plasmid pcDNA3/his-PML and its mutants were constructed by using the PMLIV isoform as described elsewhere (53).
Cell cultures and gene transfection.
The human U2OS (osteosarcoma), SiHa (cervical), and NB4 (APL) cell lines were cultured in a humidified incubator containing 5% CO2 at 37°C. U2OS/PML and U2OS/pMEP4 stable cell lines were established, as described previously, by using PMLIV cDNA (8). PML−/− mouse embryo fibroblasts (MEFs) were obtained from P. P. Pandolfi (Department of Human Genetics and Molecular Biology Program, Memorial Sloan-Kettering Cancer Center, New York, NY.). All of the adherent cells were grown as monolayers in Dulbecco modified Eagle medium (Gibco-BRL) containing 10% fetal bovine serum (FBS). The NB4 cells were cultured in RPMI 1640 medium supplemented with 10% FBS. NB4 cells were induced to differentiate with 10−6 M all-trans retinoic acid (ATRA) for 72 h (20). Gene transfection was performed with Fugene 6 transfection reagent (Roche Diagnostics Corp., Indianapolis, Ind.) in accordance with the manufacturer's instructions. Adenovirus-mediated gene transfer with the recombinant PML-adenovirus (Ad-PML) or the antisense PML-adenovirus was carried out as described in a previous study (21)
siRNA inhibition of PML expression.
Two pairs of small interfering RNAs (siRNAs) were selected for the following target sequences of PML cDNA: 5′-AAGCACGAAGACAGACCTCTGG-3′ and 5′-AACGACAGCCCAGAAGAGGAA-3′ (Xeragon-Qiagen, Germantown, Md.). Proliferating U2OS cells were transfected with the two pairs of siRNA by using the Transmessager RNA transfection reagents (Qiagen). A total of 3.2 μg of siRNA was used per well in a six-well plate. Cells were then cultured for 48 h, and total proteins were isolated and analyzed for PML expression by immunofluorescence staining and Western blotting.
Antibodies.
Mouse anti-TopBP1 monoclonal antibody was purchased from BD Biosciences-Transduction Laboratories (Lexington, Ky.). Anti-PML (H-238) and anti-BLM polyclonal antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.). Anti-ATM and anti-Brca1 polyclonal antibodies were obtained from Novus Biologicals, Inc. (Littleton, Colo.). BrdU monoclonal antibody was obtained from BD Biosciences-Pharmingen (San Diego, Calif.). BrdU polyclonal antibody was purchased from RDI Research Diagnostics, Inc. (Flanders, N.J.). Fluorescence- or horseradish peroxidase-conjugated secondary antibodies were obtained from Amersham Biosciences, Inc. (Piscataway, N.J.). The polyclonal PML antibody was raised in rabbits against the glutathione S-transferase-PML fusion protein, as described earlier (47).
Detection of ssDNA after IR-induced DNA damage.
Cells were grown on coverslips in 10% fetal calf serum containing 10 μg of BrdU/ml for 30 h and then exposed to IR in BrdU-free medium, as described previously, with modifications (2, 38). Briefly, cells were fixed with 4% paraformaldehyde for 20 min and then washed three times with ice-cold phosphate-buffered saline (PBS). Cells were then permeabilized in 1% Triton X-100 and 0.5% Nonidet P-40 (NP-40) for 20 min, washed three times in PBS for 5 min each, and blocked in 1% bovine serum albumin containing 20 μg of DNase-free RNase A/ml. Double-color immunofluorescence staining was performed with anti-BrdU and anti-TopBP1 or anti-PML antibodies. Fluorescein isothiocyanate- or Cy3-conjugated secondary antibodies were used. Cells were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) and viewed under a Leica (DM LB) fluorescence microscope. Images were captured by the Kodak digital imaging system.
Northern blot analysis.
Cells were washed once with PBS buffer, and total RNA was isolated by using an RNeasy minikit from Qiagen (Valencia, Calif.). For Northern blot analysis, 30 μg of total RNA was loaded onto a formaldehyde-agarose gel. RNA was transferred onto a positively charged membrane by using the Northern Max-Plus kit from Ambion (Austin, Tex.). Hybridization was performed with a 32P-labeled TopBP1 probe.
IR.
Cells were exposed to IR (3.5 Gy/min) in the presence of 10% fetal calf serum by using a Nasatron generator and then immediately transferred to a humidified incubator at 37°C in 5% CO2. After incubation for the indicated times, cells were harvested for Western blot analysis or immunofluorescence staining as described previously (53).
Immunofluorescence staining.
Double-color immunofluorescence staining was performed in accordance with the procedure described by Mirzoeva and Petrini (27), with some modifications. Cells were cultured on coverslips in six-well plates and fixed with 4% paraformaldehyde at 4°C for 20 min. After fixation, the cells were permeabilized in 1% Triton X-100 and 0.5% NP-40 for 30 min. Samples were blocked with 1% bovine serum albumin and then incubated with primary antibody for 2 h and secondary (fluorescein isothiocyanate-, rhodamine-, or Cy3-labeled) antibody for 1 h. Cells were then stained with DAPI to visualize nuclear DNA. To reduce the background signals in the cytoplasm and nucleoplasm, cells were pretreated with cytoskeletal stripping buffer before fixation. Cells on coverslips were washed twice in PBS, incubated in cytoskeleton buffer {PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)]; pH 6.8], 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100} for 5 min on ice, and then incubated in stripping buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 1% Tween 20, 0.25% sodium deoxycholate) for 3 min on ice. The cells were washed three times in ice-cold PBS and then were fixed and processed as described above. The coverslips were mounted onto glass slides with VectaShield antifade (Vector Laboratories, Burlingame, Calif.) and visualized by using a Leica DM LB fluorescence microscope or a Zeiss LSM 5 confocal microscope. Images were captured with a Kodak digital imaging system.
Pulse-chase labeling of cells with [35S]methionine and [35S]cysteine.
U2OS/PML and U2OS/pMEP4 cells were grown to 60% confluency in 100-mm culture dishes and treated with 100 μM ZnSO4 overnight to induce PML expression. Cells were washed twice with 10 ml of prewarmed pulse-labeling medium (methionine- and cysteine-free Dulbecco modified Eagle medium, dialyzed overnight against saline solution and 25 mM HEPES [pH 7.4; Gibco-BRL]) containing 10% FBS. Next, 5 ml of prewarmed pulse-labeling medium was added to the cells, which were then incubated for 15 min in a humidified incubator at 37°C containing 5% CO2. The medium was removed and replaced with 2 ml of labeling solution containing 0.15 mCi of [35S]methionine and [35S]cysteine (Perkin-Elmer Life Sciences, Boston, Mass.)/ml in prewarmed pulse-labeling medium, and the cells were incubated for 45 min in a CO2 incubator. The labeling solution was removed by two washes with 10 ml of 37°C chase medium (pulse-labeling medium containing 15 mg of unlabeled methionine and 31.5 mg of unlabeled cysteine/liter), and then 10 ml of 37°C chase medium was added. The cells were incubated for 0, 2, 4, 8, and 12 h in a CO2 incubator. Cells were harvested, and total proteins were isolated. Immunoprecipitation was performed with specific antibodies, and the precipitated proteins were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE). Gels were fixed, dried, and autoradiographed. Signals were quantified by densitometry by using the ImageQuant software (Molecular Dynamics).
Immunoprecipitation and Western blot analysis.
U2OS/PML cells were lyzed with cell lysis buffer (50 mM PIPES, pH 7.5; 400 mM NaCl; 1 mM EGTA; 1 mM EDTA; 1% Triton X-100; 0.5% NP-40; 10% glycerol; protease inhibitors). The lysates were precleared by treatment with protein A/G-Sepharose beads (Santa Cruz Biotechnology), antibody-conjugated beads were added, and the lysates were incubated for 4 h. After five washes with buffer (50 mM PIPES, pH 7.5; 100 mM NaCl; 0.25 mM EGTA; 0.25 mM EDTA; 0.25% Triton X-100; 0.125% NP-40; 2.5% glycerol; protease inhibitors) and one wash with ice-cold PBS, precipitated proteins were dissolved in an equal volume of 2× SDS loading buffer and then resolved by SDS-10% PAGE. Western blot analysis was carried out according to standard procedures.
RESULTS
Colocalization of PML with TopBP1 in SiHa cells after exposure to IR.
PML NBs colocalize in vivo with several proteins involved in the repair of IR-induced DNA damage. A recent study showed that the DNA damage response protein TopBP1 localized within the nucleus in a nuclear speckled staining pattern similar to that of PML NBs (13, 55). To determine whether the IR-induced TopBP1 nuclear foci colocalized with the PML NBs, we performed double-color immunofluorescence staining of PML and TopBP1 before and after exposure to IR. The results shown in Fig. 1A demonstrated that only a few of the TopBP1 foci colocalized with the PML NBs before the cells were exposed to IR but that the level of PML and TopBP1 colocalization increased substantially after 1 h of IR exposure (Fig. 1A). The numbers of TopBP1 nuclear foci and PML NBs increased in response to IR and peaked between 4 and 8 h. Both the number of TopBP1 foci and the number of PML NBs declined after 24 h. The in vivo colocalization between the two proteins decreased after 36 h. This observation suggests that PML and TopBP1 colocalize in vivo in response to IR-induced DNA damage.
FIG. 1.
(A) TopBP1 colocalizes in vivo with PML at various time points after IR exposure. SiHa cells were exposed to 15 Gy of IR. Double-color immunofluorescence staining was performed with a monoclonal antibody to TopBP1 (green) and our polyclonal PML antibody (red) at 0, 1, 4, 8, 12, and 36 h after IR treatment. Nuclear DNA in the cells was then counterstained with DAPI (blue). Images were captured by using the Kodak digital imaging system mount on top of a Leica fluorescence microscope. Fluorescent images were recorded and superimposed (Merge), by using the Adobe Photoshop 6.0 software. (B) Colocalization of TopBP1 and PML with Rad 50, Rad9, ATM, Brca1, and BLM after exposure of SiHa cells to 15 Gy of IR. Double-color immunofluorescence staining was performed at 8 h after the SiHa cells were exposed to IR.
Colocalization of PML and TopBP1 with DSB DNA repair enzymes after IR exposure.
The results presented above demonstrated colocalization of PML and TopBP1 in SiHa cells after IR exposure and suggest that PML and TopBP1 are associated with the DNA repair complex in response to IR-induced DNA damage. PML has been shown to colocalize with Rad51, H2AX, BLM, and Mre11. To determine whether PML and TopBP1 colocalize with other enzymes known to be involved in IR-induced DSB DNA repair, we performed double-color immunofluorescence staining with TopBP1 and PML antibodies and antibodies against several other proteins involved in the DSB DNA repair. The results in Fig. 1B show that PML and TopBP1 colocalized with Rad 50, ATM, BLM, Brca1, and Rad9 proteins 8 h after SiHa cells were exposed to IR. These results show that PML and TopBP1 were associated with the IR-induced DNA-repair enzyme foci in SiHa cells.
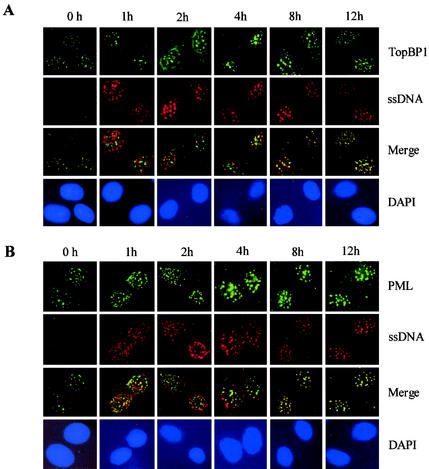
PML and TopBP1 are colocalized with the IR-induced BrdU ssDNA foci.
PML NBs colocalized with ssDNA repair foci in response to IR (30, 31). These ssDNA foci formed primarily in response to DNA DSBs, presumably the sites of late-stage DNA repair (2, 38). Only a low number (1%) of proliferating BrdU-labeled cells were found to contain the ssDNA foci, suggesting that the foci do not represent the sites of active DNA replication. The formation of ssDNA foci may require several kilobases of ssDNA as described by Raderschall et al. (38); such long stretches of ssDNA are rare in normal cultured cells. The ssDNA foci detected after IR exposure were not a result of apoptosis because no PARP degradation or FISEL staining was detected (2, 38). In addition, UV did not induce any significant degree of ssDNA foci (2). Our results show that no significant degree of BrdU foci was found at 0 h after cells were exposed to IR (Fig. 2). This result is in agreement with the previous reports (2, 38).
FIG. 2.
Colocalization of PML and TopBP1 with the ssDNA foci after IR. SiHa cells were labeled with BrdU, as described in Materials and Methods, and exposed to 15 Gy of gamma irradiation. Cells were fixed 0, 1, 2, 4, 8, and 12 h after IR, and double-color immunofluorescence staining was performed with BrdU monoclonal antibody and polyclonal PML antibody (A) or BrdU polyclonal antibody and monoclonal TopBP1 antibody (B), as described in Materials and Methods. Images were recorded as described in the Fig. 1 legend.
TopBP1 is a human homologue of the yeast Rad4/Cut5 protein and is involved in cellular responses to DNA damage and replication block (40, 41). Although TopBP1 has been shown to colocalize with some of the DNA repair proteins, its exact physiological function is not clear. We next examined whether TopBP1 and PML colocalize with the BrdU ssDNA foci after exposure of SiHa cells to IR. The results presented in Fig. 2A show that no significant colocalization between TopBP1 and IR-induced ssDNA repair foci was observed within 2 h after IR. A high degree of colocalization between the two signals was found after 4 h of IR exposure. The results in Fig. 2B show that some degree of colocalization between PML and ssDNA repair foci was detectable as early as 1 h after IR exposure. The degree of colocalization increased progressively with time and peaked at 4 to 8 h. The results of the present study demonstrated that PML and TopBP1 are associated with the BrdU ssDNA foci in response to DNA damage induced by IR.
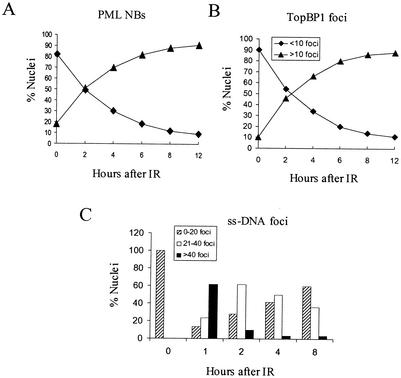
The results shown in Fig. 1A and 3A and B show that cells with higher numbers of PML and TopBP1 foci increased progressively over the 12-h period after IR exposure. However, cells with high numbers of ssDNA foci (i.e., >40) increased dramatically in 1 h, and cells with lower numbers of ssDNA foci predominated after 2 h of IR exposure (Fig. 3C). This result shows that the increase in the number of PML and TopBP1 foci coincided with the decrease in the number of ssDNA foci, suggesting that an increased number of PML and TopBP1 foci promotes repair of DNA DSBs induced by IR.
FIG. 3.
The numbers of PML NBs, IR-induced TopBP1 foci, and ssDNA foci at various time points after IR exposure. Proliferating SiHa cells were exposed to 15 Gy of gamma irradiation, and immunofluorescence staining was performed at 0, 2, 4, 6, 8, and 12 h after irradiation. For detection of IR-induced ssDNA foci, proliferating SiHa cells were incubated with 10 μg of BrdU/ml for 30 h before irradiation. The numbers of PML NBs, TopBP1 foci, and ssDNA foci represent an average of 200 nuclei per time point. (A and B) The percentage of nuclei containing >15 PML NBs and >10 IR-induced TopBP1 foci increased progressively after IR exposure. (C) The percentage of IR-induced ssDNA foci after SiHa cell exposed to IR. Cells were divided into three different groups (i.e., 0 to 20 foci, 21 to 40 foci, and >40 foci) by the number of ssDNA foci formed after IR exposure. The results obtained at each time point represent an average percentages of ssDNA foci in 200 nuclei.
Coinduction of PML and TopBP1 in response to IR.
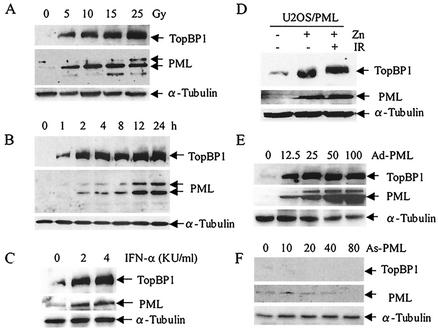
TopBP1 is a substrate of ATM kinase and is rapidly phosphorylated in response to IR (55). The results presented in Fig. 1 and 2 suggest that PML and TopBP1 are colocalized at the site of DSB DNA repair in SiHa cells after treatment with IR. We next examined whether the expression of PML and TopBP1 are coinduced in SiHa cells after treatment with IR. The results shown in Fig. 4A show that increasing IR the dosage from 0 to 25 Gy significantly increased the expression of TopBP1, which coincided with increased expression of PML. The expression levels of both PML and TopBP1 peaked 12 h after treatment with 15 Gy of IR (Fig. 4B). Our results suggest that PML and TopBP1 are coinduced by IR.
FIG. 4.
Coinduction of PML and TopBP1 in vivo. (A) Expression of TopBP1 and PML in response to various dosages of IR in SiHa cells. Cells were treated with the indicated dosages of gamma irradiation, and total proteins were isolated for Western blot analysis 12 h after IR exposure. The same filter was reprobed with anti-α-tubulin antibody to serve as a control. (B) Time course induction of TopBP1 and PML in SiHa cells exposed to 15 Gy of IR. Cells were harvested, total proteins were isolated at the indicated time points, and Western blotting was performed with the respective antibodies. (C to E) Induction of PML significantly increased the expression of TopBP1. (C) PML expression was induced by various concentrations of IFN-α, total protein was isolated after 24 h, and Western blotting was performed with TopBP1 and PML antibodies. (D) PML expression was induced or not induced with 100 μM ZnSO4 and then treated or not treated with 15 Gy of IR. Total proteins were isolated after 24 h, and Western blot analysis was performed as described in Materials and Methods. (E and F) Ad-PML infection increased the expression of TopBP1. SiHa cells were infected with increasing concentrations of Ad-PML and antisense recombinant PML adenovirus (As-PML). Total proteins were isolated after 24 h, and the expression levels of PML and TopBP1 were analyzed by Western blotting.
The PML gene is a direct target of IFN (43); we therefore next investigated whether the induction of PML expression by IFN also coinduced the expression of TopBP1. As demonstrated in Fig. 4C, IFN-α significantly increased PML expression, a finding similar to the result reported previously (43). Interestingly, TopBP1 was also coinduced by IFN-α. We next sought to determine whether the increased TopBP1 expression was directly related to the PML induction by examining the effects of inducible overexpression of PML on TopBP1 expression. Zn- or IR-induced PML expression significantly increased the TopBP1 expression levels (Fig. 4D). This conclusion was further supported in an experiment with Ad-PML. U2OS cells infected with various concentrations of Ad-PML expressed increasing amounts of the PML protein, and the TopBP1 expression levels in these cells increased dramatically in response to PML (Fig. 4E). TopBP1 expression levels did not increase in cells infected with the negative controls.
PML-dependent expression of TopBP1 protein.
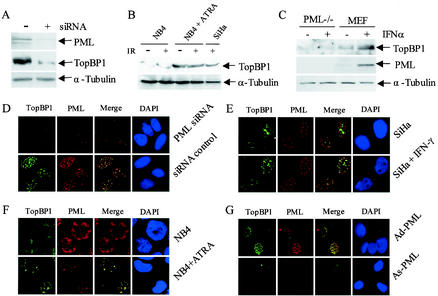
To further investigate whether TopBP1 expression is PML-dependent, siRNA specific for PML mRNA was designed as described in Materials and Methods. The results presented in Fig. 5A and D show that inhibition of PML expression by siRNA was associated with a significant reduction in IR-induced TopBP1 expression. We next investigated the cellular localization of TopBP1 in the PML-deficient APL-derived NB4 cells. In these cells, the normal PML staining pattern has been altered to an abnormal microspeckled pattern as a result of forming a heterodimer with the PML-RARα fusion protein (26). ATRA induced a rapid degradation of the fusion protein and reorganization of the normal PML NBs. The staining pattern and the in vivo localization of TopBP1 was altered in NB4 cells (Fig. 5F). The signal intensity of TopBP1 appeared to be weak, and the staining pattern was similar to the typical microspeckled pattern of PML. In NB4 cells, TopBP1 failed to form IR-induced foci. Interestingly, after treatment of NB4 cells with ATRA, both PML NBs and TopBP1 foci were reorganized, and the two proteins colocalized in vivo 8 h after IR exposure. These results strongly support the concept that a functional in vivo interaction exists between PML and TopBP1. The results of Western blot analysis showed that TopBP1 protein was expressed at low levels even in cells exposed to 10 Gy of IR (Fig. 5B). After the NB4 cells were treated with ATRA, TopBP1 expression levels increased significantly, with or without IR treatment. Finally, we investigated the expression of the TopBP1 protein in PML−/− MEFs and normal MEFs. We found that PML−/− MEFs express low levels of TopBP1 and that its expression was not inducible by treatment with IFN-α (Fig. 5C). When PML expression was reintroduced into the PML−/− MEFs by adenovirus-mediated gene transfer, a significant increase in TopBP1 expression was found, which colocalized with the PML NBs (Fig. 5G). Together, our findings support the conclusion that IR-induced upregulation of TopBP1 is PML dependent. PML either activates TopBP1 expression or, alternatively, recruits TopBP1 to the PML NBs and stabilizes the protein.
FIG. 5.
PML-dependent upregulation of TopBP1 by IR. (A and D) Effects of inhibition of PML expression by siRNA on IR-induced expression of TopBP1. U2OS cells were transfected with siRNA as described in Materials and Methods and cultured for 48 h. Cells were then exposed to 10 Gy of IR and harvested after an additional 8 h. Total proteins were isolated for Western blot analysis (A) or fixed on slides for immunofluorescence staining (D). (B and F) TopBP1 fails to form IR-induced foci in the PML-deficient NB4 cells and express low levels of TopBP1. NB4 cells with or without treatment with 1 μM ATRA for 72 h were irradiated with 10 Gy of IR. (F) Cells were then collected and immobilized on slides by cytospin, and immunofluorescence staining was performed with TopBP1 monoclonal and PML polyclonal antibodies. Cell nuclei were visualized by staining with DAPI. (B) For Western blot analysis, NB4 cells were treated with ATRA and IR as described above, and total proteins were isolated 12 h after IR. Each lane was loaded with 200 μg of protein, except for the SiHa cells, in which only 100 μg of protein per lane was loaded. (C) IFN fails to induce TopBP1 expression in PML−/− MEFs. PML−/− MEFs and normal MEFs were treated with 2,000 U of IFN-α/ml for 24 h. Total proteins were isolated, and Western blot analysis was performed with PML and TopBP1 antibodies. (E) IFN induced colocalization of PML and TopBP1. SiHa cells treated with 2,000 U of IFN-α/ml or untreated were fixed on slides. Double color immunofluorescence staining was performed with polyclonal PML antibody and monoclonal TopBP1 antibody. (G) Adenovirus-mediated reexpression of PML in PML−/− MEFs induced TopBp1 expression. PML−/− MEFs were infected at a multiplicity of infection of 25 with Ad-PML as described previously (12, 21). Double-color immunofluorescence staining was performed with the PML polyclonal antibody and TopBP1 monoclonal antibody.
Stabilization of TopBP1 by inducible overexpression of PML.
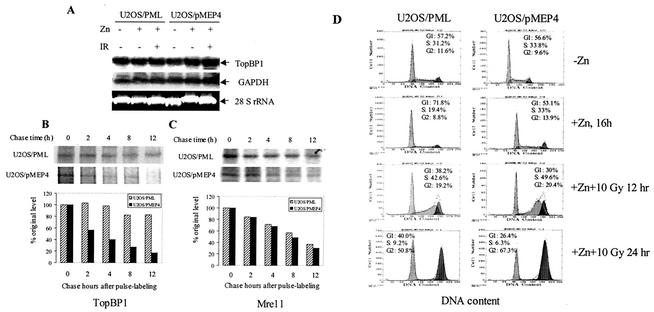
The results presented above suggest that increased TopBP1 expression induced by PML is regulated at the transcriptional or posttranscriptional level. To determine the mechanism by which PML induced the expression of TopBP1, we first performed a Northern blot analysis to examine the effects of PML on TopBP1 mRNA expression. Our results showed that PML overexpression did not alter the level of TopBP1 mRNA (Fig. 6A), indicating that PML affects TopBP1 expression at the posttranscriptional levels. We next examined whether PML overexpression increased the stability of the TopBP1 protein. We performed a pulse-chase labeling experiment to determine the stability of the TopBP1 protein in the presence or absence of PML overexpression. The experiment was performed with the inducible PML stable cell line U2OS/PML and the control cell line U2OS/pMEP4, which was established as described in an earlier study (52). The half-life of the TopBP1 protein in the U2OS/pMEP4 control cells was about 3 h (Fig. 6B). After PML expression in U2OS/PML cells was induced with 100 μM ZnSO4, however, the stability of the TopBP1 protein significantly increased. Overexpression of PML extended the half-life of TopBP1 to more than 12 h. In a Western blot analysis, we found that overexpression of PML did not affect the expression of Mre11 and Brca1 (data not shown). We therefore examined the stability of Mre11 protein under similar experimental conditions. PML overexpression did not affect the half-life of Mre11 (Fig. 5C). Because Mre11 also colocalizes with PML in vivo (4, 27), our observation indicates that PML stabilization of TopBP1 is a specific event. These results demonstrated that PML associated with TopBP1 in vivo and stabilized the TopBP1 IR-induced foci.
FIG. 6.
Inducible expression of PML enhanced the stability of TopBP1. (A) Northern blot analysis of TopBP1 expression in the PML-inducible stable cell line. U2OS/PML and the control U2OS/pMEP4 cells were induced with 100 μM ZnSO4 for 16 h. Cells were then exposed to 15 Gy of IR. Total RNA was isolated and Northern blotting was carried out as described in Materials and Methods. (B and C) Inducible PML expression stabilizes TopBP1 protein in U2OS cells. U2OS/PML and U2OS/pMEP4 cells were induced with 100 μM ZnSO4 for 16 h and pulse-labeled with [35S]methionine and [35S]cysteine for 45 min. Immediately after labeling, the cells were irradiated with 15 Gy of IR and chased for different periods of time as indicated. Total cell lysates were isolated and immunoprecipitated with mouse anti-TopBP1 monoclonal antibody (B) or mouse anti-Mre11 monoclonal antibody (C). The precipitated proteins were resolved by SDS-10% PAGE and autoradiographed. The intensities of the protein bands in the autoradiograph were quantified by scanning with the Alpha Innotech gel documentation system. (D) Effects of PML overexpression and IR exposure on cell cycle distribution in U2OS cells. U2OS/PML and U2OS/pMEP4 cells were induced with 100 μM Zn2+ for 16 h and then exposed to 10 Gy of IR. Cells were then continued to culture for the indicated time points. Cell cycle distribution was determined by flow cytometry.
Since TopBP1 expression is regulated during cell cycle progression and peaks at S phase (25). To rule out the possibility that the overexpression of PML induces TopBP1 as a result of cell cycle effects, U2OS/PML and U2OS/pMEP4 (negative control) cells were treated with Zn2+ to induce PML overexpression, and then the cells were treated or not treated with 10 Gy of IR. The results (Fig. 6D) showed that induced PML expression resulted in a significant increased in the number of cells at the G1 phase of the cell cycle, a finding in agreement with previous studies (21, 30). Exposure of U2OS/PML to 10 Gy of IR also did not significantly increase the cell number at the S phase (Fig. 6D). The results of the present study demonstrated that increased expression and stabilization of TopBP1 by PML overexpression is not a result of a cell cycle effect.
Association of PML and TopBP1 in vivo.
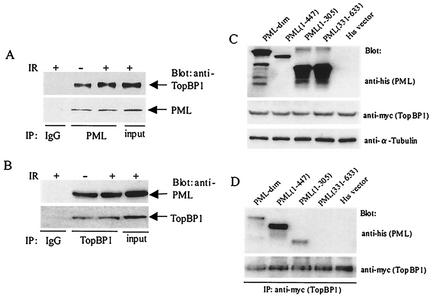
To further support the concept that PML associated with TopBP1 in vivo, we performed a coimmunoprecipitation assay using PML and TopBP1 antibodies. The results of the present study showed that TopBP1 antibody coimmunoprecipitated the PML protein, and that PML antibody coimmunoprecipitated the TopBP1 protein (Fig. 7A and B), supporting the idea that the two proteins physically associate in vivo. To elucidate the domain of PML, which is associated with TopBP1 in vivo, His-tagged PML mutant plasmids were created and cotransfected with the Myc-tagged TopBP1 expression plasmid (55). Total proteins were isolated, and coimmunoprecipitation was performed with Myc-tagged specific antibody (against the Myc-tagged TopBP1 fusion protein). The precipitated proteins were analyzed by Western blot analysis with His-tagged specific antibody (for detecting His-tagged PML protein). The present study showed that the N-terminal domain of PML is required for its in vivo association with TopBP1 (Fig. 7C and D).
FIG. 7.
In vivo association between PML and TopBP1. (A and B) Coimmunoprecipitation of PML and TopBP1 in a U2OS/PML stable cell line. Total protein (800 μg) isolated from the U2OS/PML cells induced with 100 μM of ZnSO4 was used in the immunoprecipitation assay. Coimmunoprecipitation was carried out with the TopBP1 or PML antibody, and then Western blotting was performed with PML or TopBP1 antibody. A nonspecific immunoglobulin G control was also included. (C and D) The N-terminal portion of PML is required for in vivo association with TopBP1. U2OS cells were transfected with pCDNA3/myc-TopBP1 and various pcDNA3/his-PML mutants. At 24 h after transfection, total cell lysates were prepared and immunoprecipitated with nonspecific (i.e., immunoglobulin G) or anti-myc antibody. (D) Precipitated proteins were resolved by SDS-10% PAGE, and Western blot analysis was performed with the His-tagged specific antibody. (C) Expression of the PML mutant plasmids in the cotransfection experiment was confirmed by Western blot analysis. The same filter was reprobed with α-tubulin to serve as an internal control.
DISCUSSION
PML plays a role in cellular responses to DNA damage.
PML is a multifunctional protein and a major component of the macronuclear complex responsible for organization of the PML NBs (26). A great number of proteins involved in numerous cellular functions have been found to be associated with the PML NBs. These proteins include the tumor suppressors proteins p53 (8, 9) and Rb (1); the transcription coactivator CREB-binding protein (3, 6, 19); and the transcription factors Sp1 (47, and our unpublished result), Nur77 (53), and Daxx (22, 23). Recent findings showed that PML colocalizes in vivo with several DNA repair enzymes, including BLM (4, 58), Rad51 (2), Mre11 (4, 27), and H2AX (4). PML was found in the complex containing Rad51, BLM, and the replication protein PR-A during the S/G2 phase of the cell cycle (2). PML and BLM proteins localized to the ssDNA sites in response to IR. PML colocalized with Mre11 8 to 12 h after IR exposure, and the PML/Mre11/p53 complex colocalized with the H2AX protein, suggesting that PML NBs are involved in the DNA damage response (4). Our study here demonstrated that PML is physically and functionally associated with the DNA damage response protein TopBP1 and colocalized at the endogenous levels with many of the proteins involve in DSB DNA repair. PML and TopBP1 also colocalized with the IR-induced BrdU ssDNA foci. Together, our study supports a role of PML in cellular responses to DNA damage induced by IR.
Functional association of PML with TopBP1 in response to IR.
TopBP1 consists of eight Brca1 carboxyl-terminal motifs, a human homologue of the Rad4/Cut5 protein of fission yeast, which is required for DNA damage repair, the regulation of DNA replication checkpoint, or both (13, 40, 41). A recent study demonstrated that a human ubiquitin ligase, hHYD, interacts with TopBP1 and that ubiquitination leads to degradation of the protein through the proteosome pathway (13). IR stabilizes TopBP1 by reducing ubiquitination, probably through phosphorylation by the ATM kinase (55) and increased colocalization with H2AX in the DNA breaks. This study suggests that hHYD regulates TopBP1 in response to IR-induced DNA damage. Our results in the present study showed that PML and TopBP1 (together with other proteins involved in DSB DNA repair) are colocalized at the endogenous level in response to IR. Furthermore, both PML NBs and the IR-induced TopBP1 foci are colocalized with the ssDNA foci, presumably the sites of DSB DNA repair (2, 38). An increase in the number of PML NBs and TopBP1 foci coincided with the rapid decrease in the number of the ssDNA foci, suggesting that these two proteins are probably involved in DSB DNA repair (Fig. 3).
Our study showed that PML and TopBP1 proteins are coinduced by IR. IFN, which induces PML expression at the transcriptional level (43), also induced TopBP1 expression. Overexpression of PML in U2OS cells induced by Zn2+ or by infection with Ad-PML significantly increased TopBP1 expression. Furthermore, inhibition of PML expression by specific siRNA reduced TopBP1 expression in cells exposed to IR and that reintroduction of PML expression in PML−/− MEFs is associated with a dramatic increase in the TopBP1 protein that colocalized with the PML NBs. The results of the present study demonstrated that expression of the TopBP1 protein in response to DNA damage induced by IR is PML dependent. These results strongly support a functional link between the two proteins. Results obtained from the pulse-chase labeling experiment showed that overexpression of PML substantially increased the stability of TopBP1 but not Mre11, which was also found to colocalize with PML (27). PML plays a role in regulating TopBP1 function by recruiting and stabilizing the protein in response to IR.
Our results showed that PML colocalizes with the IR-induced ssDNA foci within 1 h after the cells were exposed to IR; however, it took more than 2 h to detect any significant levels of TopBP1 colocalized with the ssDNA. Our results also showed that inducible overexpression of PML stabilized the Rad51 protein (unpublished result). The results of the present study suggest that PML recruits TopBP1 and other proteins involved in DSB DNA repair to the PML NBs in response to IR-induced DNA damage and stabilized the protein complex.
PML NBs as organizer of the DSB-DNA repair enzyme complex.
The proteins found to colocalize with PML and TopBP1 play important roles in DSB-DNA repair by homologous recombination (5, 44). For example, ATM is the master controller of IR-induced responses (33, 39); Rad51 is a DNA recombinase, the human homologue of the RecA protein in bacteria that plays a critical role in DSB DNA repair (17); the Mre11/Rad50/NBS complex plays a critical role in DSB DNA repair (18); H2AX is the earliest protein found to associate with the site of DSB DNA repair (28); BLM is a RecQ-like DNA helicase, possibly functioning in DNA replication, recombination, and DNA repair (2, 58); and Brca1 function is required in both homologous recombination and nonhomologous end joining DNA repair (29, 48, 57). Although the functions of both TopBP1 and Rad9 are not well understood, there is evidence that these two proteins are involved in the cellular responses following DNA damage (13, 55). The important question to be addressed is how is PML involved in cellular responses to DNA DSBs after IR exposure? It is intriguing that proteins that colocalize with PML, including the upstream regulator ATM, are involved in different steps of the DSB DNA repair mechanism. This argues against PML having a role in a specific step of the DNA repair mechanism. Our pulse-chase analysis showed that overexpression of PML substantially stabilized TopBP1 and that PML-deficient cells were unable to form IR-induced foci of TopBP1, Brca1, BLM, and Rad51 (Fig. 5 and our unpublished results), strongly suggesting that PML plays an essential role in the organization and stabilization of the IR-induced DNA-repair enzyme complex. In addition, recent findings demonstrated a metabolic-energy-dependent movement of the PML NBs in living cells, suggesting that PML NBs act as a nuclear sensor for detecting foreign materials (32). Together, we speculate that PML acts as a nuclear sensor and an organizer, detecting the sites of DNA DSBs during the late phase of DNA repair in response to DNA damage and organizing the DNA repair enzymes complex. PML NBs are macronuclear structures that are up to 0.5 μm in size. Their ability to move rapidly within the nucleus may provide an efficient mechanism for sensing and an anchorage point for assembling and organizing the enzyme complex.
Acknowledgments
We thank Zhaohui Xu for technical assistance and Mariann Crapanzano for editing and critical reading of the manuscript. We thank Pier Paolo Pandolfi and Paolo Salomoni for providing the PML−/− MEFs and Junjie Chen for providing the Myc-tagged TopBP1 expression plasmid. We also thank Michael D. Story and Randy J. Legerski for helpful discussions during the progress of this study.
This study was supported by grant CA55577 from the National Institutes of Health to K.-S.C.
REFERENCES
- 1.Alcalay, M., L. Thomassoni, E. Colombo, S. Stoldt, F. Grignani, M. Gagioli, L. Szekely, K. Helin, and P. G. Pellicci. 1998. The promyelocytic leukemia gene product (PML) forms stable complexes with the retinoblastoma protein. Mol. Cell. Biol. 18:1084-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bischof, O., S. H. Kim, J. Irving, S. Beresten, N. A. Ellis, and J. Campisi. 2001. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J. Cell Biol. 153:367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boisvert, F.-M., M. J. Kruhlak, A. K. Box, M. J. Hendzel, and D. P. Bazett-Jones. 2001. The transcription coactivator CBP is a dynamic component of the promyelocytic leukemia nuclear body. J. Cell Biol. 152:1099-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carbone, R., M. Pearson, S. Minucci, and P. P. Pelicci. 2002. PML NBs associate with hMre11 complex and p53at sites of irradiation-induced DNA damage. Oncogene 21:1633-1640. [DOI] [PubMed] [Google Scholar]
- 5.Coultas, L., and A. Strasser. 2000. The molecular control of DNA damage-induced cell death. Apoptosis 5:491-507. [DOI] [PubMed] [Google Scholar]
- 6.Doucas, V., M. Tini, D. A. Egan, and R. M. Evans. 1999. Modulation of CREB binding protein function by the promyelocytic (PML) oncoprotein suggests a role for nuclear bodies in hormone signaling. Proc. Natl. Acad. Sci. USA 96:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis, N. A., and J. German. 1996. Molecular genetics of Bloom's syndrome. Hum. Mol. Genet. 5:1457-1463. [DOI] [PubMed] [Google Scholar]
- 8.Ferbeyre, G., E. Stanchina, E. Querido, N. Baptiste, C. Prives, and S. W. Lowe. 2000. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 14:2015-2027. [PMC free article] [PubMed] [Google Scholar]
- 9.Fogal, V., M. Gostissa, P. Sandy, P. Zacchi, T. Sternsdorf, K. Jensen, P. P. Pandolfi, H. Will, C. Schneider, and G. Del Sal. 2000. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 19:6185-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grobelny, J. V., A. K. Godwin, and D. Broccoli. 2000. ALT-associated PML bodies are present in viable cells and are enriched in cells in the G2/M phase of the cell cycle. J. Cell Sci. 113:4577-4585. [DOI] [PubMed] [Google Scholar]
- 11.Guo, A., P. Salomoni, J. Luo, A. Shih, S. Zhong, W. Gu, and P. P. Pandolfi. 2000. The function of PML in p53-dependent apoptosis. Nat. Cell Biol. 2:730-736. [DOI] [PubMed] [Google Scholar]
- 12.He, D., Z. M. Mu, X. F. Le, J.-T. Hsieh, R.-C. Pong, L. W. K. Chung, and K. S. Chang. 1997. Adenovirus-mediated expression of PML suppresses growth and tumorigenicity of prostate cancer cells. Cancer Res. 57:1868-1872. [PubMed]
- 13.Honda, Y., M. Tojo, K. Matsuzaki, T. Anan, M. Matsumoto, M. Ando, H. Saya, and M. Nakao. 2002. Cooperation of HECT-domain ubiquitin ligase hHYD and DNA topoisomerase II-binding protein for DNA damage response. J. Biol. Chem. 277:3599-3605. [DOI] [PubMed] [Google Scholar]
- 14.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss II, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamitani, T., K. Kito, H. P. Nyuyen, H. Wada, T. Fukuda-Kamitani, and E. T. Yeh. 1998. Identification of three major sentrinization sites in PML. J. Biol. Chem. 273:26675-26682. [DOI] [PubMed] [Google Scholar]
- 16.Khan, M. M., T. Nomura, H. Kim, S. C. Kaul, R. Wadhwa, T. Shinagawa, E. Ichikawa-Iwata, S. Zhong, P. P. Pandolfi, and S. Ishii. 2001. Role of PML and PML-RAR in Mad-mediated transcriptional repression. Mol. Cell 7:1233-1243. [DOI] [PubMed] [Google Scholar]
- 17.Khanna, K. K., and S. P. Jackson. 2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27:247-254. [DOI] [PubMed] [Google Scholar]
- 18.Labachev, K. S., D. A. Gordenin, and M. A. Resnick. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108:183-193. [DOI] [PubMed] [Google Scholar]
- 19.LaMorte, V. J., J. A. Dyck, R. L. Ochs, and R. M. Evans. 1998. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc. Natl. Acad. Sci. USA 95:4991-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanotte, M., V. Martin-Thouvenin, S. Najman, P. Ballerini, F. Valensi, and R. Berger. 1991. NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3). Blood 77:1080-1086. [PubMed] [Google Scholar]
- 21.Le, X. F., S. Vallian, Z. M. Mu, M. C. Hung, and K. S. Chang. 1998. Recombinant PML adenovirus suppresses growth and tumorigenicity of human breast cancer cells by inducing G1 cell cycle arrest and apoptosis. Oncogene 16:1839-1849. [DOI] [PubMed] [Google Scholar]
- 22.Lehembre, F., S. Muller, P. P. Pandolfi, and A. Dejean. 2001. Regulation of Pax3 transcriptional activity by SUMO-1-modified PML. Oncogene 20:1-9. [DOI] [PubMed] [Google Scholar]
- 23.Li, H., C. Leo, J. Zhu, X. Y. Wu, J. O'Neil, E. J. Park, and J. D. Chen. 2000. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell. Biol. 20:1784-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, R. J., T. Sternsdorf, M. Tini, and R. M. Evans. 2001. Transcriptional regulation in acute promyelocytic leukemia. Oncogene 20:7204-7215. [DOI] [PubMed] [Google Scholar]
- 25.Makiniemi, M. T., J. Hillukkala, K. Reini, M. Vaara, D. Huang, H. Pospiech, I. Majuri, T. Westerling, T. P. Makela, and J. E. Syvaoja. 2001. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J. Biol. Chem. 276:30399-30406. [DOI] [PubMed] [Google Scholar]
- 26.Melnick, J., and D. Licht. 1999. Deconstructing a disease: RARα, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood 93:3167-3215. [PubMed] [Google Scholar]
- 27.Mirzoeva, O. K., and J. H. J. Petrini. 2001. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21:281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Modesti, M., and R. Kanaar. 2001. DNA repair: spot(light)s on chromatin. Curr. Biol. 11:R229-232. [DOI] [PubMed] [Google Scholar]
- 29.Moynahan, M. E., J. W. Chiu, B. H. Koller, and M. Jasin. 1999. Brca1 controls homology-directed DNA repair. Mol. Cell 4:511-518. [DOI] [PubMed] [Google Scholar]
- 30.Mu, Z. M., X. F. Le, S. Vallian, A. B. Glassman, and K. S. Chang. 1997. Stable overexpression of PML alters regulation of cell cycle progression in HeLa cells. Carcinogenesis 18:2063-2069. [DOI] [PubMed] [Google Scholar]
- 31.Mu, Z. M., K. V. Chin, J. H. Liu, G. Lozano, and K. S. Chang. 1994. PML, a growth suppressor disrupted in acute promyelocytic leukemia. Mol. Cell. Biol. 14:6858-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muratani, M. D., Gerlich, S. M. Janicki, M. Gebhard, R. Eils, and D. L. Spector. 2002. Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat. Cell Biol. 4:106-110. [DOI] [PubMed] [Google Scholar]
- 33.Pandita, T. K. 2002. ATM function and telomere stability. Oncogene 21:611-618. [DOI] [PubMed] [Google Scholar]
- 34.Pearson, M., R. Carbone, C. Sebastiani, M. Cioce, M. Fagioli, S. Saito, Y. Higashimoto, E. Appella, S. Minucci, P. P. Pandolfi, and P. G. Pelicci. 2000. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406:207-210. [DOI] [PubMed] [Google Scholar]
- 35.Pearson, M., and P. G. Pelicci. 2001. PML interaction with p53 and its role in apoptosis and replicative senescence. Oncogene 20:7250-7256. [DOI] [PubMed] [Google Scholar]
- 36.Petrini, J. H. J., R. S. Maser, and D. A. Bressan. 2001. The MRE11-RAD50 complex. DNA Damage Repair 3:147-172. [Google Scholar]
- 37.Quignon, F., F. De Bels, M. Koken, J. Feunteun, J. C. Ameisen, and H. de The. 1998. PML induces a novel caspase-independent death process. Nat. Genet. 20:259-265. [DOI] [PubMed] [Google Scholar]
- 38.Raderschall, E., E. I. Golub, and T. Haaf. 1999. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. USA 96:1921-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouse, J., and S. P. Jackson. 2002. Interfaces between the detection, signaling, and repair of DNA damage. Science 297:547-551. [DOI] [PubMed] [Google Scholar]
- 40.Saka, Y., F. Esashi, T. Matsusaka, S. Mochida, and M. Yanagida. 1997. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with cut5 and Chk1. Genes Dev. 11:3387-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saka, Y., P. Fantes, T. Sutani, C. McInerny, J. Creanor, and M. Yanagida. 1994. Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J. 13:5319-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seeler, J.-S., and A. Dejean. 2001. SUMO: of branched proteins and nuclear bodies. Oncogene 20:7243-7249. [DOI] [PubMed] [Google Scholar]
- 43.Stadler, M., M. K. Chelbi-Alix, M. H. M. Koken, L. Venturini, C. Lee, A. Saib, F. Quignon, L. Pelicano, M. C. Guillemin, C. Schindler, and H. de The. 1995. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene 11:2565-2573. [PubMed] [Google Scholar]
- 44.Thompson, L. H., and D. Schild. 2001. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. 477:131-153. [DOI] [PubMed] [Google Scholar]
- 45.Vallian, S., J. A. Gaken, I. D. Trayner, E. B. Gingold, T. Kourizarides, K. S. Chang, and F. Farzaneh. 1997. Transcriptional repression by the promyelocytic leukemia protein. Exp. Cell Res. 237:371-382. [DOI] [PubMed] [Google Scholar]
- 46.Vallian, S., A. J. Gaken, E. B. Gingold, T. Kouzarides, K. S. Chang, and F. Farzaneh. 1998. Regulation of Fos-mediated AP1 transcription by the promyelocytic leukemia protein. Oncogene 16:2843-2853. [DOI] [PubMed] [Google Scholar]
- 47.Vallian, S., K. V. Chin, and K. S. Chang. 1998. The promyelocytic leukemia protein interacts with Sp1 and inhibits its transactivation of the epidermal growth factor receptor promoter. Mol. Cell. Biol. 18:5147-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venkitaraman, A. R. 2002. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108:171-182. [DOI] [PubMed] [Google Scholar]
- 49.Wang, Z. G., D. Ruggero, S. Ronchetti, S. Zhong, M. Gaboli, R. Rivi, and P. P. Pandolfi. 1998. PML is essential for multiple apoptotic pathways. Nat. Genet. 20:266-272. [DOI] [PubMed] [Google Scholar]
- 50.Watts, P. M., and I. D. Hickson. 1996. Failure to unwind causes cancer, genome stability Curr. Biol. 6:265-267. [DOI] [PubMed] [Google Scholar]
- 51.Wu, W. S., S. Vallian, E. Seto, W. M. Yang, D. Edmondson, S. Roth, and K. S. Chang. 2001. The growth suppressor PML represses transcription by functionally and physically interacting with histone deacetylases. Mol. Cell. Biol. 21:2259-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, W. S., Z. X. Xu, and K. S. Chang. 2002. The promyelocytic leukemia protein represses A20-mediated transcription. J. Biol. Chem. 277:31734-31739. [DOI] [PubMed] [Google Scholar]
- 53.Wu, W. S., Z. X. Xu, R. Ran, F. Meng, and K. S. Chang. 2002. Promyelocytic leukemia protein PML inhibits Nur77-mediated transcription through specific functional interactions. Oncogene 21:3925-3933. [DOI] [PubMed] [Google Scholar]
- 54.Yamane, K., and T. Tsuruo. 1999. Conserved BRCT regions of TopBP1 and of the tumor suppressor BRCA1 bind strand breaks and termini of DNA. Oncogene 18:5194-5203. [DOI] [PubMed] [Google Scholar]
- 55.Yamane, K., X. Wu, and J. Chen. 2002. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol. Cell. Biol. 22:555-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeager, T. R., A. A. Neumann, A. Englezou, L. I. Huschtscha, J. R. Noble, and R. R. Reddel. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59:4175-4179. [PubMed] [Google Scholar]
- 57.Zhong, Q., T. G. Boyer, L. P. Chen, and W.-H. Lee. 2002. Deficient nonhomologous end-joining activity in cell-free extracts from Brca1-null fibroblasts. Cancer Res. 62:3966-3970. [PubMed] [Google Scholar]
- 58.Zhong, S., T. Z. Ye, R. Stan, N. A. Ellis, and P. P. Pandolfi. 1999. A role for PML and the nuclear body in genomic stability. Oncogene 18:7841-7847. [DOI] [PubMed] [Google Scholar]
- 59.Zhong, S., P. Solomoni, and P. P. Pandolfi. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell. Biol. 2:E85-E90. [DOI] [PubMed] [Google Scholar]
- 60.Zhong, S., S. Muller, S. Ronchetti, P. S. Freemont, A. Dejean, and P. P. Pandolfi. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748-2752. [PubMed] [Google Scholar]