Abstract
Ref-1 participates in DNA repair as well as in redox regulation of transcription factor function. The redox function of Ref-1 involves reduction of oxidized cysteine residues within the DNA binding domains of several transcription factors, including Fos and Jun. Reduction of these residues is required for DNA binding, providing a redox-dependent mechanism for regulation of target gene expression. Previous in vitro studies implicated cysteine 65 of human Ref-1 (cysteine 64 of mouse Ref-1) as the redox catalytic site. We analyzed the in vivo role of cysteine 64 in redox regulation of AP-1 activity by introducing a cysteine-to-alanine point mutation into the endogenous mouse Ref-1 gene (ref-1C64A). Unlike Ref-1 null mice, which die very early in embryonic development, homozygous ref-1C64A mice are viable, they survive to normal life expectancy, and they display no overt abnormal phenotype. Although Ref-1 provides the major AP-1-reducing activity in murine cells, ref-1C64A cells retain normal levels of endogenous AP-1 DNA binding activity in vivo as well as normal Fos- and Jun-reducing activity in vitro. These results demonstrate that Ref-1 cysteine 64/65 is not required for redox regulation of AP-1 DNA binding in vivo, and they challenge previous hypotheses regarding the mechanism by which Ref-1 regulates the redox-dependent activity of specific transcription factors.
The DNA binding activity of the AP-1 transcription factor complex is subject to reduction-oxidation (redox) regulation. This regulation involves posttranslational modification of a conserved cysteine residue in the DNA binding domains of Fos and Jun (2). The critical cysteine residue is highly conserved, and it is flanked by one or two basic amino acids in the four fos family members, the three jun family members, and at least four members of the ATF-CREB family of transcription factors. Chemical oxidation of the cysteines inhibits the DNA binding activity of Fos and Jun, and this inhibition is alleviated by reduction of the cysteines by thiol compounds or by a cellular protein, Ref-1 (43). Loss of redox regulation of AP-1 family members has biological effects. For example, in the oncogenic v-Jun protein, the cysteine has been replaced with a serine (26). Furthermore, serine substitution of the cysteine in Fos results in loss of redox regulation and enhancement of DNA binding and transforming potential (27). Ref-1 is also implicated in redox regulation of DNA binding of several other transcription factors, including NF-κB (43), Egr-1 (16), HIF-1α (7, 15, 25), HLF (7, 22), Pax-5 and Pax-8 (34-36), p53 (10, 18), and others.
Ref-1 (also known as HAP1, Ape1, and APEX1) is a bifunctional enzyme. It was independently identified by its AP-1 redox activity (41, 42) and by its sequence identity to class II hydrolytic apurinic/apyrimidinic (AP) DNA repair endonucleases (4, 30, 31). These enzymes function in base excision repair of abasic DNA lesions generated by spontaneous hydrolysis or by exposure to reactive oxygen radicals (6). The DNA repair and redox regulatory activities of Ref-1 are carried out by physically separable domains of the protein (44). The dual activities of Ref-1 suggest that the repair of DNA lesions induced by reactive oxygen species and the regulation of the transcriptional response to an oxidizing cellular environment are tightly coupled.
Immunodepletion studies using affinity-purified antibodies specific for Ref-1 demonstrated that Ref-1 is the major redox regulator of AP-1 DNA binding in HeLa cells (43). Furthermore, the essential role of Ref-1 in mammalian development was demonstrated by genetic inactivation of Ref-1 in mice (45). Embryos lacking a functional Ref-1 gene fail to develop beyond embryonic day 6. Death occurs following blastocyst formation, shortly after implantation. These embryos lack both the DNA repair and redox regulatory activities of Ref-1. Therefore, it is not possible to determine whether Ref-1 DNA repair activity, redox regulatory activity, or both are essential for viability.
Ref-1 contains two cysteine residues within the redox-active domain (cysteines 64/65 and 93). Previous in vitro studies using recombinant human Ref-1 proteins suggested that cysteine 65 (cysteine 64 of mouse Ref-1) is the redox-active site of Ref-1 (39). These results suggested a sulfhydryl switch mechanism in which cysteine 64/65 of Ref-1 interacts with the conserved cysteines within the DNA binding domains of Fos and Jun and serves as a reductant. Here we report the first genetic analysis of the role of cysteine 64 in Ref-1 redox activity in vivo. Our findings demonstrate that Ref-1 cysteine 64 is not required for Ref-1 redox regulation of AP-1.
MATERIALS AND METHODS
Gene targeting.
A 9.6-kb SacI-MunI genomic fragment containing the entire Ref-1 gene was subcloned into pBluescript II to generate pG1. A 1.3-kb NotI-SpeI fragment containing exon 2 was subcloned into pBluescript II to generate pC64. The C64A (TG→GC) and S65S (TC→AG) mutations were introduced into this clone with the Chameleon in vitro mutagenesis kit (Stratagene). The NotI-SpeI fragment harboring the mutations was subcloned into pG1 to generate pRefC64A. A 2.8-kb neo-thymidine kinase (NeoTK) cassette flanked by loxP sites was ligated into the unique SpeI site located 236 bp downstream of the C64A codon in the second intron of Ref-1 to create pRefC64ANeo. The gene-targeting vector was linearized with SalI prior to electroporation into WE9.5 embryonic stem (ES) cells (45). Neor clones were selected by using 100 μg of G418/ml. After 9 days, 432 neomycin-resistant colonies were isolated and screened by Southern analysis. ES cell DNA was digested with SacI and BglII and probed with an external 3′ 1.1-kb MunI-BglII fragment. Two independent ES cell clones with normal karyotypes were electroporated with the pMC-Cre plasmid (12) and selected in medium containing 0.2 μM FIAU [1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil]. FIAU-resistant ES clones were screened for loss of the NeoTK cassette via PCR with primers that flank the insertion site of the NeoTK cassette. All correct clones also tested negative by PCR with primers specific to the NeoTK cassette. Two independently targeted and excised ES cell clones were microinjected into blastocysts to produce germ line-transmitting chimeric mice. Gene-targeted mice were genotyped by PCR amplification of DNA extracted from tail biopsy specimens (37) by using primers flanking the loxP site within intron 3. Primer sequences are 5′GGTTGGTGGAGGGCTCCTAAAACGG3′ (upstream) and 5′GTAGTTACTGGGATTGAGGATATGC3′ (downstream).
Generation of primary embryonic fibroblasts and preparation of nuclear extracts.
Embryos derived from a cross of two ref-1C64A/+ mice were harvested at embryonic day 13.5. Heads and viscera were removed, and DNA was extracted for genotyping as described above. The remaining embryonic tissue was triturated by repeated passage through an 18-gauge needle in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (FCS), l-glutamine, penicillin, and streptomycin. Dissociated cells were seeded onto 100-mm-diameter dishes and incubated at 37°C and 5% CO2. After two passages, cells were harvested by trypsinization and pelleted by centrifugation at 3,000 rpm (Sorvall T6000D) for 10 min at 4°C. Nuclear extracts were prepared essentially as previously described (5). Briefly, cells were washed with 2.5 ml (10 volumes) of ice-cold phosphate-buffered saline and pelleted at 420 × g for 10 min at 4°C. Cells were resuspended in 1.25 ml (5 volumes) of ice-cold buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT]) and incubated on ice for 10 min. Cells were pelleted at 420 × g for 10 min at 4°C, resuspended in 0.5 ml (2 volumes) of buffer A, and homogenized with 10 strokes in a Dounce homogenizer. Nuclei were pelleted at 2,000 rpm for 10 min at 4°C and resuspended in 0.2 ml of buffer C (20 mM HEPES [pH 7.9], 20% glycerol, 420 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 mM DTT). Samples were homogenized as described above and incubated on ice for 30 min with frequent vortexing. Samples were centrifuged at 20,000 × g for 30 min at 4°C. Supernatants were dialyzed against buffer D (20 mM HEPES [pH 7.9], 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 5 mM MgCl2, 0.5 mM PMSF, 0.5 mM DTT) for 4 h at 4°C. Samples were again centrifuged at 20,000 × g for 20 min at 4°C. Protein concentrations were determined by using the Bio-Rad protein assay kit. Samples were adjusted to 1 mg/ml, aliquoted, and snap-frozen on dry ice. Nuclear extracts were stored at −80°C. For serum induction experiments, cells were cultured in Dulbecco modified Eagle medium supplemented with 0.5% FCS, l-glutamine, penicillin, and streptomycin for 4 days at 37°C and 5% CO2. Medium supplemented with 20% FCS was added, and cells were harvested at the indicated time points. Nuclear extracts were prepared as described above.
Purification of recombinant proteins.
Human Ref-1 cDNA was amplified by reverse transcriptase PCR and ligated into the ClaI site of pSRα (pSRα-Ref-1). A C65A point mutation was introduced by in vitro mutagenesis with the Chameleon mutagenesis kit (Stratagene) to create pSRα-C65A. Wild-type and C65A mutant cDNAs were excised from pSRα-Ref-1 and pSRα-C65A, respectively, and subcloned into the pQE30 bacterial expression vector (Qiagen). Other bacterial expression constructs used in these experiments were generated as previously described (1, 22). Recombinant protein expression was induced in M15 cells with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 37°C (500-ml culture volume). Cells were harvested by centrifugation and resuspended in 5 ml of ice-cold lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 10 mM DTT, 0.5 mM PMSF, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml). Five milligrams of lysozyme was added, and cells were incubated for 30 min on ice followed by sonication. Lysates were centrifuged at 10,000 × g for 30 min at 4°C. Supernatants were transferred to fresh tubes, 2 ml of 50% nickel-nitrilotriacetic acid (Ni-NTA) agarose slurry (Qiagen) was added, and mixtures were rocked for 1 h at 4°C. Ni-NTA agarose was pelleted by centrifugation at 10,000 × g for 10 min at 4°C, washed three times with ice-cold wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 10 mM DTT, 0.5 mM PMSF, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml), resuspended in 2 ml of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, 1 mM DTT, 0.5 mM PMSF, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml), and rocked for 10 min at 4°C. Ni-NTA agarose was recovered by centrifugation at 10,000 × g for 10 min at 4°C. Supernatants were dialyzed against storage buffer (20 mM Tris [pH 7.5], 50 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5 mM DTT, 0.5 mM PMSF) for 4 h at 4°C. Recombinant Ref-1 proteins were concentrated by using Millipore Ultrafree-4 centrifugal filter devices. Protein concentrations were determined by the Bio-Rad protein assay kit and adjusted to 10 mg/ml. Aliquots were snap-frozen on dry ice. All recombinant proteins were stored at −80°C.
AP endonuclease assay.
AP endonuclease activity was assayed as previously described (17). Briefly, to induce AP lesions, 50 μg of pBluescript II KS(+) plasmid DNA was treated with 3 volumes of 50 mM sodium citrate (pH 3.5) at 60°C for 15 min. Total volume was adjusted to 0.5 ml with 50 mM Tris (pH 7.4), and the reaction mixture was chilled on ice. Treated DNA was dialyzed against 50 mM Tris (pH 7.4) overnight at 4°C. Recombinant proteins and nuclear extracts were diluted to 10 ng/ml in 1× assay buffer (10 mM Tris [pH 8.0], 5 mM MgCl2, 1 mM EDTA, 0.01% NP-40). Five hundred nanograms of acid-treated or mock-treated plasmid was incubated with the indicated quantity of recombinant protein or nuclear extract at 37°C for 15 min in a 20-μl total volume. Reaction mixtures were immediately analyzed by electrophoresis in a 0.8% agarose-Tris-acetate-EDTA gel.
EMSAs.
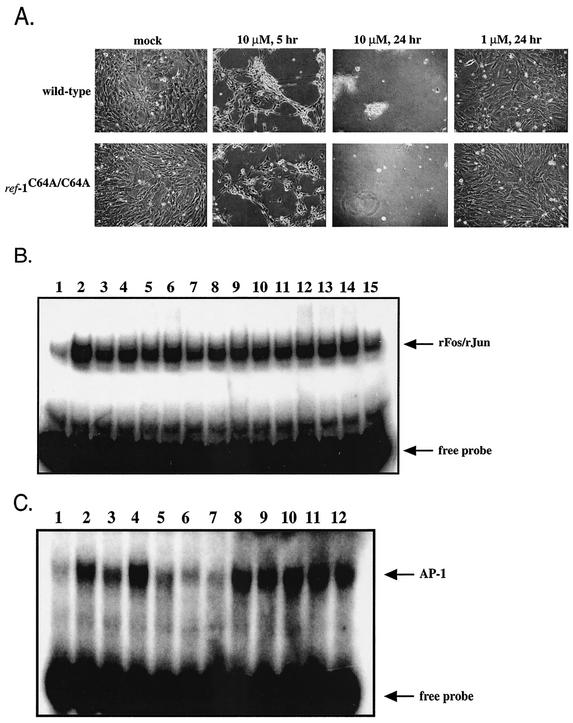
Oxidized Fos (wbFos) (1) and Jun (TK550) (22) peptides were prepared as described above. An electrophoretic mobility shift assay (EMSA) was performed as previously described (41). Recombinant proteins and nuclear extracts were diluted in binding buffer (10 mM Tris [pH 7.5], 50 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 5% glycerol, 5% sucrose, 0.01% NP-40, 0.2 mg of bovine serum albumin/ml) prior to addition to the EMSA reaction mixture. For assays including recombinant Fos and Jun peptides, 58 nM (each) peptides and the indicated recombinant protein or extract were incubated at 37°C for 15 min in binding buffer (18-μl total volume). One microgram of poly(dI-dC) · poly(dI-dC) (Pharmacia) was added, and the reaction mixtures were incubated for 5 min at room temperature. Twenty-five femtomoles (∼1 × 105 to 4 × 105 cpm) of [α-32P]-labeled 25-bp double-stranded oligonucleotide including the human metallothionein IIA promoter AP-1 site (41) was added, and the reaction mixtures were incubated for 15 min at room temperature. Samples were immediately analyzed by electrophoresis in a nondenaturing 5% acrylamide-Tris-glycine (pH 8.5) gel (240 V for 1 h at 4°C), dried onto Whatman paper, and visualized by autoradiography. For assays of endogenous AP-1 DNA binding activity, EMSA reactions were performed as described above, except that no recombinant Fos or Jun was added. For serum induction experiments, primary embryonic fibroblasts were cultured in medium containing 0.5% FCS for 4 days. Cultures were induced by the addition of medium containing 20% FCS, and extracts were prepared for EMSA at the indicated time points. For H2O2 treatment experiments, primary embryonic fibroblasts were cultured in medium containing 0.5% FCS for 48 h. H2O2 was added to a final concentration of 1 μM, and extracts were prepared for EMSA at the indicated time points.
RESULTS
Production of mice with gene-targeted ref-1C64A mutation.
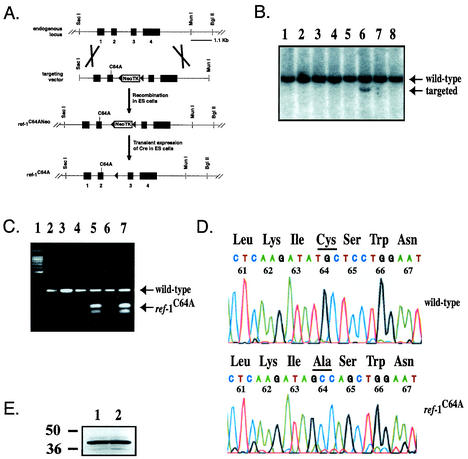
A 9.6-kb fragment including the entire Ref-1 gene was used to construct a gene-targeting vector. A C64A point mutation was introduced into exon 2 by in vitro mutagenesis (see Materials and Methods). A NeoTK cassette including a phosphoglycerate kinase promoter and flanked by loxP sites was ligated into the SpeI restriction enzyme site in the second intron of Ref-1 (Fig. 1A). The linearized vector was electroporated into WE9.5 ES cells (45), and neomycin-resistant clones were isolated and screened by Southern blot analysis (Fig. 1B). A total of 18 independent gene-targeted ES cell clones were identified among 289 clones screened. The gene-targeting strategy was designed to insert an additional silent mutation within the serine codon immediately 3′ to the cysteine 64 codon, thus creating a PvuII restriction enzyme site linked to the C64A mutation. To verify the C64A mutation in gene-targeted ES cell clones, Ref-1 exon 2 was amplified by PCR and amplicons were digested with PvuII. Amplicons from ref-1C64A alleles, but not from wild-type alleles, were digested into two smaller fragments (Fig. 1C). The transient Cre expression vector pMC-Cre (12) was electroporated into two independent ES cell clones with normal karyotypes. FIAU-resistant clones were isolated, and Cre-mediated excision of the NeoTK cassette was confirmed by PCR (data not shown). Two independent ES cell clones were injected into blastocysts to produce germ line-transmitting chimeric mice. There were no observable differences between ref-1C64A mice derived from the two ES cell clones. Sequencing of PCR products including exon 2 amplified from wild-type or ref-1C64A/C64A mouse genomic DNA unequivocally verified that ref-1C64A mice carry the C64A mutation (Fig. 1D). Analysis of Ref-1 protein levels in wild-type and ref-1C64A/C64A cell extracts demonstrated that the point mutation did not affect the expression or stability of Ref-1 protein (Fig. 1E).
FIG. 1.
(A) Diagram of gene-targeted mutagenesis strategy. See the text for details. ref-1C64ANeo, ref-1C64A with neomycin resistance cassette. (B) Southern blot screen for gene-targeted ES cell clones. DNA from neomycin-resistant ES cell clones was digested with SacI and BglII and hybridized to an external probe as shown in panel A. The NeoTK cassette includes a SacI site, allowing detection of homologous recombination by the presence of a 9.1-kb fragment in addition to the wild-type 10.8-kb fragment. Lane 6 represents a gene-targeted clone. (C) Confirmation of gene targeting by PCR followed by restriction enzyme digestion. Exon 2 was PCR amplified from ES cell DNA, and amplicons were digested with PvuII. Lanes: 1, DNA ladder; 2, wild-type undigested amplicons; 3, wild-type PvuII-digested amplicons; 4 and 6, ref-1C64A/+ undigested amplicons; 5 and 7, ref-1C64A/+ PvuII-digested amplicons. Arrows mark bands amplified from wild-type alleles and digested bands amplified from ref-1C64A alleles as indicated. (D) Confirmation of the endogenous ref-1C64A mutation by genomic sequencing. Ref-1 exon 2 was PCR amplified from genomic DNA extracted from wild-type and ref-1C64A/C64A primary embryonic fibroblasts, and amplicons were sequenced. Codons are labeled by mouse Ref-1 amino acid number and identity. Cysteine/alanine 64 is underlined. Sequencing verified the TG-GC mutation at codon 64 and the silent TC-AG mutation at codon 65. (E) Western blot analysis of Ref-1 protein in wild-type and ref-1C64A/C64A cells. Lanes: 1, 20 μg of wild-type embryonic fibroblast nuclear extract; 2, 20 μg of ref-1C64A/C64A embryonic fibroblast nuclear extract. Western blot analysis was performed with a polyclonal antibody specific for Ref-1 (43).
Mice homozygous for ref-1C64A are viable and display no overt abnormalities.
In contrast to the early embryonic lethality associated with the complete loss of Ref-1 (45), homozygous ref-1C64A mice were generated in expected Mendelian ratios. Of 168 offspring of heterozygous crosses, 41 (24.4%) were wild type, 85 (50.6%) were ref-1C64A/+, and 42 (25%) were ref-1C64A/C64A. Casual observations revealed no differences among homozygous mutants and their heterozygous or wild-type littermates at birth, and homozygous mutants developed no consistent physical or behavioral abnormalities over a typical 1.5- to 2-year observation period. Extensive histological analyses of two ref-1C64A/C64A and two wild-type mice at 6 months of age revealed no apparent cellular abnormalities. Three ref-1C64A/C64A mice over 2 years of age developed large masses on the flank, suggestive of tumorigenesis. Histological analyses identified papillary carcinomas in lungs and multifocal tumors in livers, spleens, and skin with cytologic morphology and immunocytochemistry profiles consistent with angiosarcomas. Tumor cells stained intensely with antibodies to VEGF-R2 and CD34 and weakly to moderately with antibodies to CD31 and cytokeratin AE1, and they were negative for Factor 8 and cytokeratin (data not shown). However, most ref-1C64A/C64A mice over 2 years of age did not develop tumors. Histological analysis of two additional ref-1C64A/C64A mice and three wild-type mice over 2 years of age revealed no remarkable abnormalities. Therefore, the tumors in the three aged mutant mice are unlikely to be a direct consequence of the ref-1C64A mutation.
These results demonstrate that although Ref-1 function is essential for mouse embryonic development, mutation of the putative Ref-1 redox-active cysteine residue did not impair mouse development or survival. Furthermore, the establishment of this mutant mouse line provided the first opportunity to test the in vivo role of Ref-1 cysteine 64 in redox regulation of transcription factor activity.
Mutation of Ref-1 cysteine 64 does not affect AP-1 DNA binding in vivo or in vitro.
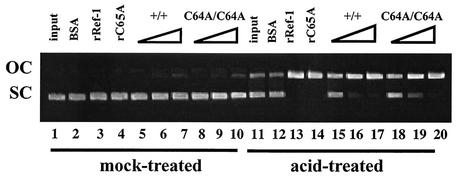
To determine whether the ref-1C64A mutation compromises the DNA repair activity of endogenously expressed Ref-1, we subjected nuclear extracts from wild-type and ref-1C64A/C64A primary embryonic fibroblasts to an in vitro AP endonuclease assay (17). The DNA repair activity of Ref-1 introduces an endonucleolytic cleavage of the phosphodiester backbone 5′ to AP sites within plasmid DNA, thus converting plasmid DNA from supercoils to relaxed circles. Purified recombinant Ref-1 with the C64A mutation (Ref-1C64A) retains this endonuclease activity (Fig. 2). Furthermore, nuclear extracts from wild-type and ref-1C64A/C64A cells display identical dose-dependent levels of AP endonuclease activity. Therefore, mutation of Ref-1 cysteine 64 does not affect its DNA repair activity in vivo.
FIG. 2.
The C64A mutation does not affect Ref-1 AP endonuclease activity. Supercoiled pBluescript II plasmid was mock-treated or acid-treated to induce AP lesions as indicated. Plasmids were reacted with 50 ng of bovine serum albumin (BSA; lanes 2 and 12), 50 ng of recombinant human Ref-1 (rRef-1; lanes 3 and 13), 50 ng of recombinant human C65A Ref-1 (rC65A; lanes 4 and 14), nuclear extract from wild-type primary embryonic fibroblasts (10, 50, and 100 ng [lanes 5 to 7 and 15 to 17, respectively]), or nuclear extract from ref-1C64A/C64A primary embryonic fibroblasts (10, 50, and 100 ng [lanes 8 to 10 and 18 to 20, respectively]). Ref-1 AP endonuclease activity was assayed by the conversion of supercoiled plasmid (SC) to an open circular form (OC).
The normal phenotype of ref-1C64A/C64A mice raises three potentially exclusive hypotheses: (i) Ref-1 is not essential for appropriate redox regulation of transcription factor activity in vivo, (ii) redox regulation of these transcription factors is not necessary for normal development or survival, and (iii) cysteine 64 is not essential for the redox regulatory function of Ref-1 in vivo. To distinguish among these hypotheses, we analyzed the effect of Ref-1C64A on the DNA binding activity of Fos and Jun, the heterodimeric AP-1 transcription factor complex first shown to be regulated by the redox activity of Ref-1 (2, 42).
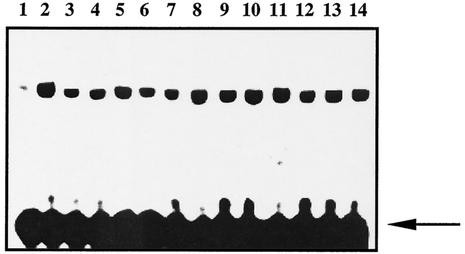
In EMSAs, nuclear extracts from wild-type and ref-1C64A/C64A hearts and lungs demonstrated equal levels of Fos- and Jun-reducing activity (Fig. 3). The oxidized Fos and Jun peptides used in these experiments can heterodimerize but are unable to bind to DNA without prior reduction. As expected, mock-treated recombinant Fos and Jun are unable to bind an oligonucleotide containing an AP-1 target sequence. Addition of 10 mM DTT dramatically stimulates DNA binding (Fig. 3). Similarly, addition of extracts from cells expressing wild-type Ref-1 or Ref-1C64A results in a dose-dependent reduction of Fos and Jun.
FIG. 3.
Ref-1C64A/C64A tissues retain normal Fos/Jun-reducing activity. Oxidized recombinant Fos and Jun peptides were incubated to allow heterodimerization. Extracts were added and Fos/Jun DNA binding was determined by EMSA by using a radiolabeled oligonucleotide probe containing an AP-1 DNA binding site. Lanes: 1, dilution buffer alone; 2, 10 mM DTT; 3 to 5, 10, 25, and 50 ng of wild-type lung extract, respectively; 6 to 8, 10, 25, and 50 ng of ref-1C64A/C64A lung extract, respectively; 9 to 11, 10, 25, and 50 ng of wild-type heart extract, respectively; 12 to 14, 10, 25, and 50 ng of ref-1C64A/C64A heart extract, respectively. Free probe is indicated by an arrow.
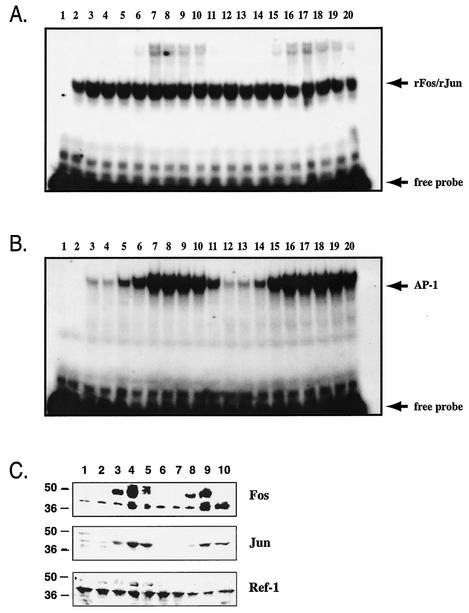
To determine whether Ref-1 regulates the DNA binding activity of induced, but not basally expressed, AP-1, we cultured wild-type and ref-1C64A/C64A primary embryonic fibroblasts under serum-deprived conditions (0.5% FCS) for 4 days. Immediate-early gene expression was induced by addition of 20% FCS, and cells were harvested at several time points up to 24 h. AP-1 DNA binding activity in nuclear extracts was analyzed by EMSA. Serum induction had no effect on the level of Fos- and Jun-reducing activity in either wild-type or ref-1C64A/C64A extracts (Fig. 4A). Furthermore, under these conditions, the ref-1C64A mutation did not compromise induction of endogenous AP-1 DNA binding activity (Fig. 4B). Conversely, the maximal level of endogenous AP-1 DNA binding activity in ref-1C64A/C64A extracts appeared to persist into later time points following serum induction than that in wild-type extracts. This effect is not a result of increased steady-state levels of Fos or Jun protein in mutant cells (Fig. 4C).
FIG. 4.
(A) Serum induction does not affect Fos/Jun-reducing activity in wild-type or ref-1C64A/C64A primary embryonic fibroblast extracts. Fos/Jun-reducing activity was determined by EMSA as described in the legend to Fig. 3. Primary embryonic fibroblasts were cultured in medium containing 0.5% FCS for 4 days and then stimulated with medium containing 20% FCS. Cells were harvested at various time points throughout the 24-h stimulation. Lanes: 1, dilution buffer alone; 2, 10 mM DTT; 3 to 11, 1-μg wild-type nuclear extracts harvested at 0, 0.2, 0.5, 1, 2, 3, 5, 10, and 24 h poststimulation, respectively; 12 to 20, 1-μg ref-1C64A/C64A nuclear extracts harvested at 0, 0.2, 0.5, 1, 2, 3, 5, 10, and 24 h poststimulation, respectively. Free probe and recombinant Fos/Jun-bound probe (rFos/rJun) are indicated by arrows. (B) Serum-induced endogenous AP-1 DNA binding is not compromised in ref-1C64A/C64A cells. Nuclear extracts were analyzed by EMSA as described for panel A except without added recombinant Fos and Jun peptides. Lanes are identical to those described for panel A. Free probe and recombinant Fos/Jun- or endogenous AP-1-bound probe are indicated by arrows. (C) Fos, Jun, and Ref-1 protein levels at selected time points during serum induction. Extracts described for panels A and B were subjected to Western analysis with antibodies for the indicated proteins. Lanes: 1 to 5, wild-type extracts at 0, 0.5, 1, 3, and 5 h poststimulation, respectively; 6 to 10, ref-1C64A/C64A extracts at 0, 0.5, 1, 3, and 5 h poststimulation, respectively. Antibodies are indicated on the right of the panel. Molecular masses in kilodaltons are indicated on the left on the panel.
AP-1 activity is induced under oxidative stress conditions in vitro (3, 24, 38). These findings underscore the importance of redox mechanisms for maintaining AP-1 in a reduced state capable of binding DNA. We analyzed the redox regulation of AP-1 under oxidative stress conditions by treating ref-1C64A/C64A and wild-type primary embryonic fibroblasts with H2O2. Cells were first treated with various concentrations of H2O2 (0 to 1 mM) to determine the highest dose that did not result in substantial cell death within the duration of the experiment. Addition of 10 μM H2O2 resulted in significant death of both ref-1C64A/C64A and wild-type embryonic fibroblasts within 5 h and complete loss of viable cells within 24 h (Fig. 5A). However, both mutant and wild-type cells survived for 24 h following challenge with 1 μM H2O2. Mutant and wild-type cells displayed identical susceptibilities to H2O2-induced cell death in dose-response experiments (Fig. 5A and data not shown). Similar to the results obtained during serum induction (Fig. 4A), ref-1C64A/C64A and wild-type extracts exhibited equivalent levels of recombinant Fos/Jun-reducing activity throughout the H2O2 exposure (Fig. 5B). However, ref-1C64A/C64A cells exhibited an increased level of endogenous AP-1 DNA binding activity relative to wild-type cells under oxidative stress conditions, consistent with the increased duration of maximal AP-1 DNA binding following serum stimulation (Fig. 4B). As previously reported (3, 24, 38), H2O2-induced oxidative stress resulted in an increase in AP-1 DNA binding activity. In wild-type embryonic fibroblasts, induction peaked at 3 h following addition of H2O2 and returned to baseline level within 24 h (Fig. 5C, lanes 1 to 6). However, in ref-1C64A/C64A fibroblasts, peak induction was reached within 30 min following treatment and the elevated level of endogenous AP-1 DNA binding was maintained up to 24 h posttreatment (Fig. 5C, lanes 7 to 12).
FIG. 5.
(A) Wild-type and ref-1C64A/C64A embryonic fibroblasts are equally susceptible to H2O2 treatment. Primary embryonic fibroblast cultures were treated with H2O2 at concentrations ranging from 100 nM to 1 mM for 24 h. Mock-treated cultures and cultures treated with 1 μM and 10 μM H2O2 are shown at 5 and 24 h posttreatment, as indicated. (B) H2O2 treatment does not affect Fos/Jun-reducing activity in wild-type or ref-1C64A/C64A primary embryonic fibroblast extracts. Recombinant Fos/Jun-reducing activity was determined by EMSA as described in the legend to Fig. 3. Lanes: 1, mock-treated sample; 2, 10 mM DTT; 3 to 8, wild-type embryonic fibroblast extracts at 0, 0.5, 1, 3, 6, and 24 h following 1 μM H2O2 challenge, respectively; 9 to 14, ref-1C64A/C64A embryonic fibroblast extracts at 0, 0.5, 1, 3, 6, and 24 h following 1 μM H2O2 challenge, respectively; 15, extract dilution buffer alone. The recombinant Fos/Jun complex (rFos/rJun) and free probe are indicated by arrows. (C) H2O2-induced AP-1 DNA binding is enhanced in ref-1C64A/C64A cells. Endogenous AP-1 DNA binding activity was determined as described in the legend to Fig. 3. Lanes: 1 to 6, wild-type embryonic fibroblasts assayed at 0, 0.5, 1, 3, 6, and 24 h following challenge with 1 μM H2O2; 7 to 12, ref-1C64A/C64A embryonic fibroblasts assayed at 0, 0.5, 1, 3, 6, and 24 h following challenge with 1 μM H2O2. Free probe and endogenous AP-1-bound probe are indicated by arrows.
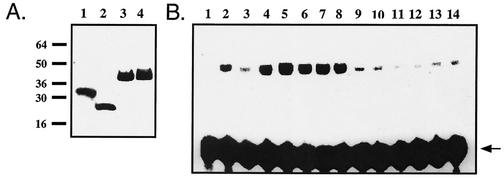
These results suggest either that cells express an AP-1 redox regulatory factor that compensates for loss of Ref-1 redox function or that cysteine 64 is dispensable for Ref-1 redox activity. Previous experiments have shown that immunodepletion of Ref-1 from mammalian cell extracts results in the elimination of Fos/Jun-reducing activity, indicating that Ref-1 is the major redox regulatory factor for AP-1 (43). Therefore, we compared the in vitro redox regulatory activities of recombinant wild-type and C65A human Ref-1 proteins. Histidine-tagged recombinant proteins were expressed in M15 Escherichia coli and purified under native conditions by affinity chromatography by using Ni-NTA matrices. This strategy provided highly concentrated pure preparations of recombinant wild-type Ref-1 and Ref-1C65A (Fig. 6A). Recombinant Disabled-1 (mDab-1) (provided by Hee-Won Park, St. Jude Children's Research Hospital) was purified by the same procedure for use as a negative control. Surprisingly, in contrast to the findings from a previous report (39), wild-type Ref-1 and Ref-1C65A exhibited comparable levels of Fos/Jun-reducing activity in vitro. Stimulation of Fos/Jun DNA binding is not a nonspecific consequence of adding recombinant protein to the reaction because equivalent amounts of recombinant mDab-1 did not induce the dramatic increase in Fos/Jun DNA binding activity seen with addition of Ref-1 and Ref-1C65A. The storage buffer for recombinant proteins must include 0.5 mM DTT for Ref-1 to maintain reducing activity (42). However, induction of Fos/Jun DNA binding was not a result of DTT carried over into the reaction mixture, because equal volumes of storage buffer alone did not induce Fos/Jun DNA binding activity. Together with our in vivo findings, these results demonstrate that cysteine 64/65 is not essential for Ref-1 redox regulation of AP-1. Interestingly, recombinant C65A Ref-1 appears to have a somewhat elevated reducing activity relative to that of the wild-type recombinant protein (Fig. 6B). In addition, the decrease in endogenous AP-1 DNA binding activity between 10 and 24 h following serum induction is more dramatic in wild-type fibroblasts than in ref-1C64A/C64A fibroblasts (Fig. 4B) and the induction of AP-1 DNA binding activity under oxidative stress conditions occurs within a shorter time frame in ref-1C64A/C64A fibroblasts than in wild-type fibroblasts (Fig. 5C). Based on in vitro studies of recombinant human Ref-1, it was previously suggested that Ref-1 cysteine 64/65 is the redox-active site but that cysteine 95 and cysteine 64/65 cooperate in a negative regulatory mechanism mediated by a disulfide bridge between the two residues (39). Our in vivo studies demonstrate that mutation of cysteine 64/65 does not affect Ref-1 redox activity and suggest that mutation of either cysteine 64/65 or cysteine 95 results in increased Ref-1 redox activity. These results indicate that hypotheses regarding the mechanism of Ref-1 redox regulation of transcription factor activity must be reevaluated.
FIG. 6.
(A) All recombinant proteins were expressed as six-His-tagged fusion proteins and purified as described in Materials and Methods. Ten micrograms of each recombinant protein was run on a sodium dodecyl sulfate-polyacrylamide gel and stained with Coomassie blue. Lanes: 1, wbFos (1); 2, TK550 Jun (22); 3, wild-type Ref-1; 4, C65A Ref-1. (B) Recombinant C65A Ref-1 retains Fos/Jun-reducing activity. Recombinant Ref-1 proteins were assayed for redox activity by EMSA as described in the legend to Fig. 3. Recombinant protein stocks were adjusted to 10 mg/ml to ensure equal carryovers of storage buffer. Lanes: 1, dilution buffer alone; 2, 10 mM DTT; 3 to 5, 10, 25, and 50 ng of recombinant Ref-1, respectively; 6 to 8, 10, 25, and 50 ng of recombinant C65A Ref-1, respectively; 9 to 11, 10, 25, and 50 ng of recombinant mDab-1, respectively; 12 to 14, storage buffer diluted 1:1,000, 1:400, and 1:200, respectively. Free probe is indicated by an arrow.
DISCUSSION
The identification of Ref-1 as the protein responsible for redox regulation of Fos and Jun revealed a molecular crossroad between the processes of oxidative DNA damage repair and regulation of transcriptional responses to oxidative conditions. Subsequently, Ref-1 was found to regulate redox-dependent DNA binding of several transcription factors, including NF-κB, Egr-1, HIF-1α, HLF, Pax-5, Pax-8, and others (reviewed in reference 8). Ref-1 also regulates p53 activity through both redox-dependent and redox-independent mechanisms (10, 18). However, the precise mechanism of these regulatory interactions has remained elusive.
Redox reactions are notoriously difficult to control and characterize in vitro or in the intact cell. Previous in vitro studies of recombinant Ref-1 proteins implicated cysteine 65 (cysteine 64 of mouse Ref-1) as the Ref-1 redox-active site (39). However, Ref-1 appears to participate in a redox signaling pathway in conjunction with thioredoxin, thioredoxin reductase, and possibly other factors (43). Therefore, the mechanism by which Ref-1 mediates redox regulation of various transcription factors is difficult to address using isolated in vitro assay systems. Here, we report the first genetic analysis of the effect of mutation of the proposed Ref-1 redox-active residue in vivo.
In contrast to embryos homozygous for a null ref-1 allele, mice homozygous for the ref-1C64A mutation are born in normal Mendelian ratios and survive to a normal life expectancy. Histological analyses of tissues from ref-1C64A/C64A mice did not detect any consistent abnormalities. Furthermore, under 10% FCS culture conditions, nuclear extracts from wild-type and ref-1C64A/C64A tissues exhibited equal levels of endogenous AP-1 DNA binding activity, as well as equal levels of exogenous Fos- and Jun-reducing activity. Given that immunodepletion of Ref-1 from cell extracts results in nearly complete loss of Fos- and Jun-reducing activity in vitro (43), these results suggest that Ref-1C64A retains sufficient redox activity to maintain normal levels of basal AP-1 DNA binding activity in vivo. We examined the effect of the ref-1C64A mutation on induced AP-1 DNA binding by analyzing nuclear extracts from wild-type or ref-1C64A/C64A primary embryonic fibroblasts following serum stimulation. Stimulated mutant fibroblasts exhibited a normal induction of endogenous AP-1 DNA binding activity and maintained exogenous Fos- and Jun-reducing activity throughout the stimulation time course. Interestingly, peak levels of endogenous AP-1 DNA binding appear to be extended into later time points in ref-1C64A/C64A cells than in wild-type cells, suggesting that mutation of Ref-1 cysteine 64 results in a subtle increase in Ref-1 redox activity rather than elimination of activity. We also investigated the effects of the ref-1C64A mutation on AP-1 DNA binding under oxidative stress conditions induced by H2O2 treatment. Again, mutation of Ref-1 cysteine 64 appears to enhance rather than inhibit the ability of the enzyme to redox regulate AP-1 activity.
The lack of an overt phenotype in ref-1C64A/C64A mice and the inability to detect a decrease in AP-1 DNA binding potential prompted us to reexamine the redox activity of purified recombinant mutant Ref-1 protein in vitro. A potential explanation for the lack of an effect of the cysteine 64 mutation in vivo is that an independent redox regulatory factor may be induced specifically in the context of loss of Ref-1 redox function, thereby compensating for the loss of Ref-1 regulation of AP-1 in mutant cells but not in normal cells. However, our analyses of recombinant Ref-1C65A support the conclusion that cysteine 64/65 is not essential for Ref-1 redox activity in vitro or in vivo. Again, Ref-1C65A appeared to have a slightly elevated rather than eliminated ability to reduce oxidized Fos and Jun. These findings are in direct contradiction with those in a previous report (39), and they underscore the difficulty of analyzing oxidation-reduction mechanisms in vitro. In our study, we expressed human wild-type and mutant Ref-1 as His6-tagged proteins in E. coli and purified the recombinant proteins to homogeneity under native conditions by affinity chromatography. In the previous study, wild-type and mutant recombinant human Ref-1 proteins were overexpressed in E. coli and total proteins were precipitated with ammonium sulfate, followed by chromatographic purification with phosphocellulose P11 and phenyl Superose columns. Purification was monitored through enzymatic activity, and final preparations were >90% pure (39). It is possible that the discrepancies in the activities of the C65A mutant Ref-1 proteins in the two studies are a result of the different purification strategies utilized.
Structural studies indicate that Ref-1 cysteines 64/65 and 93 (both within the redox-active domain of Ref-1) are embedded within the interior of the native protein and inaccessible to proteolytic digestion (33). This suggests that the two residues could act as direct chemical reductants only if Ref-1 undergoes a major conformational change upon interaction with its target transcription factor molecules. Alternatively, it has been suggested that these residues contribute to redox activity by maintaining the tertiary structure of the Ref-1 redox domain, thus permitting another residue(s) to act as a reductant (39). However, the in vivo and in vitro findings reported here demonstrate that Ref-1 cysteine 64/65 is not essential for the direct reduction of cysteines within the DNA binding domains of Fos and Jun or for the maintenance of a protein conformation critical for either the redox regulatory or the DNA repair activities of Ref-1.
The conclusion that cysteine 64/65 is not required for Ref-1 redox activity is supported by the severe phenotypes of mice lacking transcription factors regulated by Ref-1. For example, genetic inactivation of c-jun results in lethality by embryonic day 12.5 (14, 20). Mice lacking c-Fos are viable but develop bone disease due to a defect in the osteoclast lineage (11, 19, 40). Inactivation of Ref-1 redox activity would not necessarily result in complete inactivation of the transcription factors that it regulates. However, considering the variety of transcription factors regulated by Ref-1, it is likely that loss of Ref-1 redox regulation of these molecules would result in detectable adverse consequences.
Interestingly, some ref-1C64A/C64A mice older than 2 years of age developed tumors. Since this was seen in only a small number of mutant mice over 2 years of age, we concluded that the tumors were not a direct effect of the Ref-1 mutation. However, it remains possible that the ref-1C64A mutation contributed to an increased susceptibility to tumorigenesis in aged mice. Altered expression or subcellular localization of Ref-1 in some human tumor types has been reported. For example, overexpression of Ref-1 has been detected in prostate tumors (21), nuclear expression of Ref-1 is directly associated with resistance to chemotherapy and poor survival with head-and-neck cancer (23), elevated expression of Ref-1 in testicular cancer cell lines results in resistance to therapeutic agents (29), and cytoplasmic expression of Ref-1 was predictive of poor prognosis in non-small cell lung carcinomas with lymph node involvement (28). In addition, a recent study identified Ref-1 as a member of the SET complex targeted by granzyme A (GzmA) during cytotoxic-T-lymphocyte-mediated cell death and demonstrated that Ref-1 is a direct proteolytic substrate of GzmA (9). GzmA cleaves Ref-1 at lysine 31 and blocks both its DNA repair and redox regulatory functions, suggesting that GzmA promotes cytotoxic-T-lymphocyte-mediated cell death, at least in part by inactivating an antiapoptotic function of Ref-1. Given its ubiquitous expression, Ref-1 may participate in similar antiapoptic mechanisms in many cell types, and the subtle increase in Ref-1C64A redox activity shown in this study could eventually tip the intricate balance between normal cell survival and abnormal cell death in favor of resistance to apoptosis, resulting in low-penetrance tumorigenesis in aged animals. Interestingly, overexpression of a noncleavable Ref-1 mutated at both lysine 31 and cysteine 64 in HeLa cells resulted in decreased protection against GzmA-induced cytotoxicity relative to that in cells with overexpression of wild-type Ref-1 (9). The authors suggested that the redox regulatory function of Ref-1 is involved in protection against GzmA-mediated cell death. However, the results reported here suggest that the role of Ref-1 cysteine 64 in GzmA-mediated cytotoxicity is independent of redox regulation of transcription factor binding. Additional experiments will be required to understand the role of Ref-1 in cytotoxic-T-lymphocyte cytotoxicity and possibly tumorigenesis.
In conclusion, the findings reported here constitute the first genetic study of the proposed mechanism of Ref-1 redox regulation of AP-1 DNA binding activity. Surprisingly, we have found that Ref-1 cysteine 64/65 is not essential for the redox regulatory function of Ref-1. This result dictates that the proposed mechanism for reduction of critical cysteines within the target molecules of Ref-1 be reevaluated. At this point, we can only speculate on the true mechanism. For example, it is possible that either cysteine 64/65 or cysteine 93 (both included within the redox-active domain of Ref-1) can serve as a redox-active site in the absence of the other. Given the location of these residues within the interior of the predicted Ref-1 protein structure, we must also consider the possibility that neither residue is essential for redox activity. Finally, additional posttranslational modifications of Ref-1 may be involved in the redox mechanism. For example, NO was recently demonstrated to S-nitrosylate thioredoxin at cysteine 69, a residue distinct from the direct redox-active residues (13). This modification is essential for maintaining the redox regulatory and antiapoptotic functions of thioredoxin in endothelial cells, possibly via a transnitrosylation mechanism. Futhermore, selenomethionine was recently demonstrated to activate DNA binding and transactivation activities of p53 via a Ref-1-dependent mechanism (32). Further work is necessary to define the mechanisms of Ref-1 redox regulatory function.
Acknowledgments
J.M.O. and D.E. contributed equally to this work.
We thank Peter McKinnon and Suzanne Baker for supplying critical reagents, Wanyun Zhong and Karen Forbes for technical assistance, Jerold Rehg and the St. Jude Children's Research Hospital Animal Diagnostic Laboratory for expert histological analyses, and Lakhu Keshvara and David Benhayon for their critiques of the manuscript. ES cell microinjections were performed by the staff of the St. Jude Children's Research Hospital Genetics Core Facility.
This work was supported by National Institutes of Health grant RO1 CA 84139 (T.C.), Training in the Biology of Cancer grant CA 09346-20 (J.M.O.), American Cancer Society postdoctoral fellowship PF-98-006-01-CCG (D.E.), and the American Lebanese Syrian Associated Charities.
REFERENCES
- 1.Abate, C., D. Luk, R. Gentz, F. J. Rauscher III, and T. Curran. 1990. Expression and purification of the leucine zipper and DNA-binding domains of Fos and Jun: both Fos and Jun contact DNA directly. Proc. Natl. Acad. Sci. USA 87:1032-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abate, C., L. Patel, F. J. Rauscher III, and T. Curran. 1990. Redox regulation of fos and jun DNA-binding activity in vitro. Science 249:1157-1161. [DOI] [PubMed] [Google Scholar]
- 3.Beiqing, L., M. Chen, and R. L. Whisler. 1996. Sublethal levels of oxidative stress stimulate transcriptional activation of c-jun and suppress IL-2 promoter activation in Jurkat T cells. J. Immunol. 157:160-169. [PubMed] [Google Scholar]
- 4.Demple, B., T. Herman, and D. S. Chen. 1991. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc. Natl. Acad. Sci. USA 88:11450-11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doetsch, P. W., and R. P. Cunningham. 1990. The enzymology of apurinic/apyrimidinic endonucleases. Mutat. Res. 236:173-201. [DOI] [PubMed] [Google Scholar]
- 7.Ema, M., K. Hirota, J. Mimura, H. Abe, J. Yodoi, K. Sogawa, L. Poellinger, and Y. Fujii-Kuriyama. 1999. Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 18:1905-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans, A. R., M. Limp-Foster, and M. R. Kelley. 2000. Going APE over ref-1. Mutat. Res. 461:83-108. [DOI] [PubMed] [Google Scholar]
- 9.Fan, Z., P. J. Beresford, D. Zhang, Z. Xu, C. D. Novina, A. Yoshida, Y. Pommier, and J. Lieberman. 2003. Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nat. Immunol. 4:145-153. [DOI] [PubMed] [Google Scholar]
- 10.Gaiddon, C., N. C. Moorthy, and C. Prives. 1999. Ref-1 regulates the transactivation and pro-apoptotic functions of p53 in vivo. EMBO J. 18:5609-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grigoriadis, A. E., Z. Q. Wang, M. G. Cecchini, W. Hofstetter, R. Felix, H. A. Fleisch, and E. F. Wagner. 1994. c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science 266:443-448. [DOI] [PubMed] [Google Scholar]
- 12.Gu, H., Y. R. Zou, and K. Rajewsky. 1993. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell 73:1155-1164. [DOI] [PubMed] [Google Scholar]
- 13.Haendeler, J., J. Hoffmann, V. Tischler, B. C. Berk, A. M. Zeiher, and S. Dimmeler. 2002. Redox regulatory and anti-apoptotic functions of thioredoxin depend on S-nitrosylation at cysteine 69. Nat. Cell Biol. 4:743-749. [DOI] [PubMed] [Google Scholar]
- 14.Hilberg, F., A. Aguzzi, N. Howells, and E. F. Wagner. 1993. c-jun is essential for normal mouse development and hepatogenesis. Nature 365:179-181. [DOI] [PubMed] [Google Scholar]
- 15.Huang, L. E., Z. Arany, D. M. Livingston, and H. F. Bunn. 1996. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J. Biol. Chem. 271:32253-32259. [DOI] [PubMed] [Google Scholar]
- 16.Huang, R. P., and E. D. Adamson. 1993. Characterization of the DNA-binding properties of the early growth response-1 (Egr-1) transcription factor: evidence for modulation by a redox mechanism. DNA Cell Biol. 12:265-273. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda, S., S. Seki, S. Watanabe, M. Hatsushika, and K. Tsutsui. 1991. Detection of possible DNA repair enzymes on sodium dodecyl sulfate-polyacrylamide gels by protein blotting to damaged DNA-fixed membranes. Anal. Biochem. 192:96-103. [DOI] [PubMed] [Google Scholar]
- 18.Jayaraman, L., K. G. Murthy, C. Zhu, T. Curran, S. Xanthoudakis, and C. Prives. 1997. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 11:558-570. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, R. S., B. M. Spiegelman, and V. Papaioannou. 1992. Pleiotropic effects of a null mutation in the c-fos proto-oncogene. Cell 71:577-586. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, R. S., B. van Lingen, V. E. Papaioannou, and B. M. Spiegelman. 1993. A null mutation at the c-jun locus causes embryonic lethality and retarded cell growth in culture. Genes Dev. 7:1309-1317. [DOI] [PubMed] [Google Scholar]
- 21.Kelley, M. R., L. Cheng, R. Foster, R. Tritt, J. Jiang, J. Broshears, and M. Koch. 2001. Elevated and altered expression of the multifunctional DNA base excision repair and redox enzyme Ape1/ref-1 in prostate cancer. Clin. Cancer Res. 7:824-830. [PubMed] [Google Scholar]
- 22.Kerppola, T. K., and T. Curran. 1997. The transcription activation domains of Fos and Jun induce DNA bending through electrostatic interactions. EMBO J. 16:2907-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koukourakis, M. I., A. Giatromanolaki, S. Kakolyris, E. Sivridis, V. Georgoulias, G. Funtzilas, I. D. Hickson, K. C. Gatter, and A. L. Harris. 2001. Nuclear expression of human apurinic/apyrimidinic endonuclease (HAP1/Ref-1) in head-and-neck cancer is associated with resistance to chemoradiotherapy and poor outcome. Int. J. Radiat. Oncol. Biol. Phys. 50:27-36. [DOI] [PubMed] [Google Scholar]
- 24.Lakshminarayanan, V., E. A. Drab-Weiss, and K. A. Roebuck. 1998. H2O2 and tumor necrosis factor-alpha induce differential binding of the redox-responsive transcription factors AP-1 and NF-kappaB to the interleukin-8 promoter in endothelial and epithelial cells. J. Biol. Chem. 273:32670-32678. [DOI] [PubMed] [Google Scholar]
- 25.Lando, D., I. Pongratz, L. Poellinger, and M. L. Whitelaw. 2000. A redox mechanism controls differential DNA binding activities of hypoxia-inducible factor (HIF) 1alpha and the HIF-like factor. J. Biol. Chem. 275:4618-4627. [DOI] [PubMed] [Google Scholar]
- 26.Nishimura, T., and P. K. Vogt. 1988. The avian cellular homolog of the oncogene jun. Oncogene 3:659-663. [PubMed] [Google Scholar]
- 27.Okuno, H., A. Akahori, H. Sato, S. Xanthoudakis, T. Curran, and H. Iba. 1993. Escape from redox regulation enhances the transforming activity of Fos. Oncogene 8:695-701. [PubMed] [Google Scholar]
- 28.Puglisi, F., G. Aprile, A. M. Minisini, F. Barbone, P. Cataldi, G. Tell, M. R. Kelley, G. Damante, C. A. Beltrami, and C. Di Loreto. 2001. Prognostic significance of Ape1/ref-1 subcellular localization in non-small cell lung carcinomas. Anticancer Res. 21:4041-4049. [PubMed] [Google Scholar]
- 29.Robertson, K. A., H. A. Bullock, Y. Xu, R. Tritt, E. Zimmerman, T. M. Ulbright, R. S. Foster, L. H. Einhorn, and M. R. Kelley. 2001. Altered expression of Ape1/ref-1 in germ cell tumors and overexpression in NT2 cells confers resistance to bleomycin and radiation. Cancer Res. 61:2220-2225. [PubMed] [Google Scholar]
- 30.Robson, C. N., and I. D. Hickson. 1991. Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in E. coli xth (exonuclease III) mutants. Nucleic Acids Res. 19:5519-5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robson, C. N., A. M. Milne, D. J. Pappin, and I. D. Hickson. 1991. Isolation of cDNA clones encoding an enzyme from bovine cells that repairs oxidative DNA damage in vitro: homology with bacterial repair enzymes. Nucleic Acids Res. 19:1087-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seo, Y. R., M. R. Kelley, and M. L. Smith. 2002. Selenomethionine regulation of p53 by a ref1-dependent redox mechanism. Proc. Natl. Acad. Sci. USA 99:14548-14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strauss, P. R., and C. M. Holt. 1998. Domain mapping of human apurinic/apyrimidinic endonuclease. Structural and functional evidence for a disordered amino terminus and a tight globular carboxyl domain. J. Biol. Chem. 273:14435-14441. [DOI] [PubMed] [Google Scholar]
- 34.Tell, G., L. Pellizzari, D. Cimarosti, C. Pucillo, and G. Damante. 1998. Ref-1 controls pax-8 DNA-binding activity. Biochem. Biophys. Res. Commun. 252:178-183. [DOI] [PubMed] [Google Scholar]
- 35.Tell, G., A. Scaloni, L. Pellizzari, S. Formisano, C. Pucillo, and G. Damante. 1998. Redox potential controls the structure and DNA binding activity of the paired domain. J. Biol. Chem. 273:25062-25072. [DOI] [PubMed] [Google Scholar]
- 36.Tell, G., A. Zecca, L. Pellizzari, P. Spessotto, A. Colombatti, M. R. Kelley, G. Damante, and C. Pucillo. 2000. An ‘environment to nucleus' signaling system operates in B lymphocytes: redox status modulates BSAP/Pax-5 activation through Ref-1 nuclear translocation. Nucleic Acids Res. 28:1099-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Truett, G. E., P. Heeger, R. L. Mynatt, A. A. Truett, J. A. Walker, and M. L. Warman. 2000. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). BioTechniques 29:52, 54.. [DOI] [PubMed] [Google Scholar]
- 38.Vollgraf, U., M. Wegner, and C. Richter-Landsberg. 1999. Activation of AP-1 and nuclear factor-kappaB transcription factors is involved in hydrogen peroxide-induced apoptotic cell death of oligodendrocytes. J. Neurochem. 73:2501-2509. [DOI] [PubMed] [Google Scholar]
- 39.Walker, L. J., C. N. Robson, E. Black, D. Gillespie, and I. D. Hickson. 1993. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol. Cell. Biol. 13:5370-5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, Z. Q., C. Ovitt, A. E. Grigoriadis, U. Mohle-Steinlein, U. Ruther, and E. F. Wagner. 1992. Bone and haematopoietic defects in mice lacking c-fos. Nature 360:741-745. [DOI] [PubMed] [Google Scholar]
- 41.Xanthoudakis, S., and T. Curran. 1994. Analysis of c-Fos and c-Jun redox-dependent DNA binding activity. Methods Enzymol. 234:163-174. [DOI] [PubMed] [Google Scholar]
- 42.Xanthoudakis, S., and T. Curran. 1992. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 11:653-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xanthoudakis, S., G. Miao, F. Wang, Y. C. Pan, and T. Curran. 1992. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 11:3323-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xanthoudakis, S., G. G. Miao, and T. Curran. 1994. The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains. Proc. Natl. Acad. Sci. USA 91:23-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xanthoudakis, S., R. J. Smeyne, J. D. Wallace, and T. Curran. 1996. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl. Acad. Sci. USA 93:8919-8923. [DOI] [PMC free article] [PubMed] [Google Scholar]