Abstract
MAGI-1 is a membrane-associated guanylate kinase protein at tight junctions in epithelial cells. It interacts with various molecules and functions as a scaffold protein at cell junctions. We report here a novel MAGI-1-binding protein that we named junctional adhesion molecule 4 (JAM4). JAM4 belongs to an immunoglobulin protein family. JAM4 was colocalized with ZO-1 in kidney glomeruli and in intestinal epithelial cells. Biochemical in vitro studies revealed that JAM4 bound to MAGI-1 but not to ZO-1, whereas JAM1 did not bind to MAGI-1. JAM4 and MAGI-1 interacted with each other and formed clusters in COS-7 cells when coexpressed. JAM4 mediated calcium-independent homophilic adhesion and was accumulated at cell-cell contacts when expressed in L cells. MAGI-1, ZO-1, and occludin were recruited to JAM4-based cell contacts. JAM4 also reduced the permeability of CHO cell monolayers. MAGI-1 strengthened JAM4-mediated cell adhesion in L cells and sealing effects in CHO cells. These findings suggest that JAM4 together with MAGI-1 provides an adhesion machinery at tight junctions, which may regulate the permeability of kidney glomerulus and small intestinal epithelial cells.
Tight junctions (TJs) play essential roles in the maintenance of a physical barrier between external and internal environments in the body (for reviews, see references 2 and 39). Recent studies have revealed the molecular architecture of TJs. Like other cell junctions, TJs have integral membrane components and membrane-associated proteins. Occludin and claudin are integral membrane proteins involved in the formation of TJ strands (14, 15, 16). In addition, junctional adhesion molecule 1 (JAM1) is an integral membrane protein of TJs (29). JAM1 belongs to the immunoglobulin (Ig) superfamily and does not directly constitute TJ strands. The interaction between membrane-associated proteins and integral membrane proteins might be crucial for the organization of TJs. The most representative membrane-associated proteins at TJs are ZO-1 and its isoforms, ZO-2 and ZO-3 (1, 18, 24). ZO-1, -2, and -3 belong to membrane-associated guanylate kinases (MAGUKs) and bind to the C termini of claudins (5, 22; for reviews, see reference 13). ZO-1 also binds to JAM1 (11). The interactions are mediated by PDZ domains and PDZ-binding motifs. Similarly, MUPP1, another TJ component that has 13 PDZ domains, binds to claudin-1 and JAM1 (17). Mammalian homologue of PAR-3 is also concentrated at TJs, and its PDZ domain binds to JAM1 (12, 23).
TJs have another member of the MAGUK family, MAGUK with inverted domain structure (MAGI-1) (8). MAGI-1 was originally identified as a protein interacting with Ki-ras in rats. It has a unique molecular organization composed of one guanylate kinase domain, two WW domains, and six PDZ domains. The human MAGI-1 is called brain angiogenesis inhibitor 1-associated protein 1 (37). The synaptic scaffolding molecule (S-SCAM) is the neural isoform of MAGI-1 (also called MAGI-2, atrophin-interacting protein, and activin receptor-interacting protein) and interacts with various molecules at synapses (20, 38, 40). MAGI-1 is localized at TJs in epithelial cells (21). Like S-SCAM, MAGI-1 binds various molecules, including β-catenin, mNET1, RapGEP, synaptopodin, α-actinin-4, and megalin (9, 10, 30, 35, 36). β-Catenin is presumably involved in the polarized targeting of MAGI-1 to lateral membranes of epithelial cells (10, 32). Because β-catenin is not localized at TJs in polarized epithelial cells, the interaction with β-catenin is not sufficient for the recruitment of MAGI-1 to TJs. mNET1 and RapGEP are regulators for small GTP-binding proteins, and MAGI-1 may accumulate these molecules to TJs. In kidneys, MAGI-1 is highly expressed in podocytes (36). Synaptopodin and α-actinin-4 are actin-binding proteins and are expressed in podocytes (31, 36). MAGI-1 may be linked to the actin cytoskeleton through these proteins in podocytes. Among MAGI-1-interacting proteins, only megalin is an integral membrane protein. Megalin is a transmembrane endocytic receptor glycoprotein which belongs to the low-density lipoprotein receptor family (for reviews, see reference 6). In kidneys, megalin and MAGI-1 are colocalized in podocytes, where MAGI-1 may modulate the endocytic activity of megalin. However, megalin is localized on apical membranes in polarized epithelial cells and is unlikely to interact with MAGI-1 at TJs.
In this study, we tried to identify an integral membrane component of TJs that interacts with MAGI-1 and found a novel transmembrane protein. The protein is similar to JAM1 in molecular structure, and we call this molecule JAM4. JAM4 has cell adhesion activity. We have also discovered that the cell adhesion activity of JAM4 is regulated by MAGI-1.
MATERIALS AND METHODS
Construction of expression vectors.
Using 5′-acgcgttacagagatttacctgcc-3′ and 5′-gtcgactacactaaagtcacatttct-3′ and 5′-aagcttgtacaaggcaagggttcgg-3′ and 5′-gtcgactacaccaggaacgacgaggt-3′ as primers, cDNAs of mouse JAM1 and mouse JAM4, respectively, were obtained by PCR on mouse lung cDNA (BioChain, Inc.) and kidney cDNA (Clontech). pGex4T-1 and pFLAG-CMV-1 were purchased from Amersham Pharmacia Biotech and Eastman Kodak Company, respectively. A linker (5′-agcttcgacgcgtaggatcctcgcgaatgcatgtcgacgcggccgc-3′ and 5′-gatcgcggccgcgtcgacatgcattcgcgaggatcctacgcgtcga-3′) was ligated into HindIII/BamHI sites of pLGFPN (Clontech) to generate pLGFPN-2, which was subsequently digested with NotI and religated to generate pLN. pMXpuro Myc was generated by ligating a linker (5′-gatccgccccaccatggagcagaaacttatcagcgaggaggacctgacgcgtggattcg-3′ and 5′-aattcgaatccacgcgtcaggtcctcctcgctgataagtttctgctccatggtggggc-3′) into BamHI/EcoRI sites of pMXpuro. PCR was performed on pLGFPC, with 5′-ggatccctaccggtcgccaccatggtg-3′ and 5′-gtcgacggtaccacgcgtcgagatctgagtccggccggact-3′ as primers. The PCR product was digested with BamHI/SalI and ligated into the BglII/SalI site of pShuttle cytomegalovirus (CMV) (a gift of Tong-Chuan He and Bert Vogelstein, Johns Hopkins Oncology Center) to generate pShuttle CMV green fluorescent protein (GFP). The following constructs expressed the corresponding amino acids: pLN JAM4, amino acids 1 to 370 of JAM4; pLN JAM4ΔC, 1 to 361 of JAM4; pFLAG JAM4, 21 to 370 of JAM4; pFLAG JAM4ΔC, 21 to 361 of JAM4; pGex4T-1 JAM4-C, 262 to 370 of JAM4; pFLAG JAM1, 25 to 300 of JAM1; pClneo Myc MAGI-1, pMXpuro Myc MAGI-1, and pShuttle CMV GFP MAGI-1, 1 to 1256 of human MAGI-1; pBTM116 MAGI-1-8, 429 to 1256 of human MAGI-1; pGexKG S-SCAM-1, 303 to 405 of rat S-SCAM; pGex4T-1 S-SCAM-7, 1 to 421 of rat S-SCAM; pClneo Myc CASK, 1 to 909 of rat CASK; pClneo Myc ZO-1-1, 1 to 920 of mouse ZO-1; and pClneo Myc Lin-7, 1 to 207 of rat Lin-7A. Using 5′-acgcgttaattcaccatgtctgcac-3′ and 5′-ctcgagaagctggccgcgtaccc-3′ as primers, PCRs were performed on pFLAG JAM4, pFLAG JAM4ΔC, and pFLAG JAM1. PCR products were digested with MluI/SalI and ligated into MluI/SalI sites of pLN to generate pLN FLAG JAM4, pLN FLAG JAM4ΔC, and pLN FLAG JAM1. MluI/SalI fragments from pLN FLAG JAM4 were ligated into the same sites of pClneo to generate pClneo FLAG JAM4 for in vitro transcription and translation. Throughout this study, we use the term MAGI-1 to describe all constructs that are derived from human brain angiogenesis inhibitor protein 1-associated protein 1. The constructs of JAM4 used in this study are summarized in Fig. 1E.
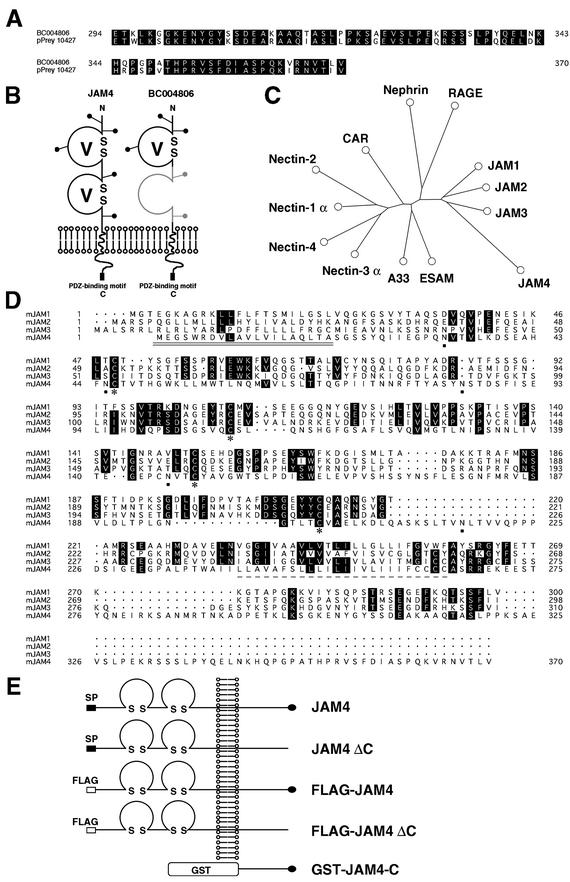
FIG. 1.
Mouse JAM4 amino acid sequence. (A) Alignment of the amino acid sequences of pPrey 10427 and mouse BC004806. Amino acid residues are shown by single-letter codes. pPrey 10427 has 76 amino acids. Among them, 61 amino acids are identical to the C-terminal region of BC004806. (B) Structural model of JAM4 and mouse BC004806. The exrtacellular region of JAM4 contains two domains with intramolecular disulfide bonds typical of Ig-like loops of the V type. BC004806 has only the first loop. JAM4 and BC004806 have the C-terminal PDZ-binding motif. (C) Phylogenetic tree indicating the relationship between JAM4 and other Ig protein family members. Sequences were obtained from GenBank. The alignment was analyzed with Phylotree (http://cbrg.inf.ethz.ch/Server/AllAll.html). Accession numbers are as follows: for Nectin-1α, AAF76195; for Nectin-2, P32507; for Nectin-3α, AAF63685; for Nectin-4, AAL79833; for coxsackievirus and adenovirus receptor (CAR), AAH16457; for intestinal A33 antigen, AAH08528; for endothelial cell-selective adhesion molecule (ESAM), AAK51504; for Nephrin, AAK38483; for receptor for advanced glycation end product (RAGE), AAB82007; for JAM1, AAC32982; for JAM2, NP076333; and for JAM3, NP075766. (D) Amino acid sequence of mouse JAM4 and alignment with mouse JAM1, JAM2, and JAM3. The putative hydrophobic signal peptide (indicated with double solid underlines) and transmembrane (dashed single underline) sequences of mouse JAM4 are marked. Cysteines likely to form disulfide bonds are indicated by asterisks. Putative N-linked glycosylation sites are indicated by boldface dots. Numbers indicate the numbers of amino acid residues of each protein. Amino acid residues conserved among more than three proteins are shown on a black background. (E) Constructs of JAM4 used in this study. SP, signal peptide; FLAG, FLAG tag. Solid ovals correspond to the C-terminal 9 amino acids containing the PDZ-binding motif.
Antibodies.
The rabbit anti-JAM4 antibody was raised against the product of pGex4T-1 JAM4-C. The rabbit antibody against the product of pGexKG S-SCAM-1 was used for the immunoprecipitation of MAGI-1. For immunohistochemistry, the affinity-purified rabbit antibody against the product of pGex S-SCAM-7 was used to detect MAGI-1. The rabbit anti-ERBIN and the sheep anti-Myc antibodies were described previously (33). The rat monoclonal anti-occludin antibody was a gift from Shoichiro Tsukita (Kyoto University). Mouse anti-Myc-tag monoclonal antibody 9E10 was obtained from the American Type Culture Collection. Mouse and rabbit anti-ZO-1 antibodies (Zymed), mouse anti-β-catenin antibody (BD Transduction), mouse anti-GFP antibody (Santa Cruz), mouse anti-FLAG antibody (Sigma-Aldrich Fine Chemicals), rat anti-ZO-1 antibody (Chemicon), and rhodamine-conjugated, fluorescein isothiocyanate (FITC)-conjugated, and Cy5-conjugated second antibodies for dual labeling (Chemicon) were purchased from commercial sources.
Cells.
Phoenix ampho, mouse mammary gland epithelial MTD-1A, Madin- Darby canine kidney II (MDCK), mouse fibroblastic L, COS-7, and porcine kidney LLC-PK1 cells were grown in cultures in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml under 5% CO2 at 37°C. CHO cells were grown in cultures in the medium containing 2 mM l-glutamine. COS-7 cells were transfected with a DEAE-dextran method. L cells were transfected with Effectene transfection reagent (Qiagen). A retrovirus system was used to generate stable transformants of MDCK, L, CHO, and LLC-PK1 cells. Briefly, Phoenix ampho cells were transfected using a calcium phosphate coprecipitation method with pLN FLAG JAM4, pLN FLAG JAM4ΔC, pLN JAM4, pLN JAM4ΔC, or pMXpuro Myc MAGI-1. The medium was collected 48 h later as a virus stock. MDCK, L, CHO, and LLC-PK1 cells were grown in cultures in the medium containing retrovirus for 48 h and then in selective medium with 1 mg of G418 (Calbiochem)/ml or 2 μg of puromycin (Invitrogen)/ml. To generate L-FLAG-JAM4/Myc-MAGI-1 and L-FLAG-JAM4ΔC/Myc-MAGI-1 cells, L-FLAG-JAM4 and L-FLAG-JAM4ΔC cells were infected with retrovirus to express Myc-MAGI-1 and selected by G418 and puromycin. The resulting resistant colonies were tested for the expression of each protein and cloned.
Collagen culture.
L-FLAG-JAM4, L-FLAG-JAM4ΔC, L-FLAG-JAM4/Myc-MAGI-1, and L-FLAG-JAM4ΔC/Myc-MAGI-1 cells were isolated with trypsin and triturated into a single-cell suspension. Cells were diluted to 104 cells/ml in a type I collagen-neutral solution (Cellegen, Koken, Tokyo). The cultures were incubated at 37°C to allow collagen to gel and then added with the medium. Cells were grown in cultures and observed by confocal microscopy (Olympus EV 300-BX).
Yeast two-hybrid screening.
Yeast two-hybrid screening was performed using a human lung cDNA library (Clontech) and yeast strain L40. Histidine selection plates contained 8 mM 3-amino-1,2,4-triazole, 180 mg of isopropyl-β-d-1-thiogalactopyranoside/liter, and 360 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside/liter. After 6 days of incubation, blue colonies were picked up for further analysis.
Immunofluorescence microscopy.
COS-7, L, and MDCK cells were fixed with 4% (wt/vol) formaldehyde in phosphate-buffered saline (PBS) at room temperature for 15 min and blocked with 50 mM glycine in PBS for 30 min. MTD-1A cells were fixed with cold methanol for 15 min at −20°C. The samples were incubated with 0.2% (wt/vol) Triton X-100 in PBS for 15 min and subsequently with 1% (wt/vol) bovine serum albumin (BSA) in PBS at room temperature for 30 min. The samples were incubated with various first antibodies and visualized with rhodamine-, FITC-, or Cy5-conjugated second antibodies. The images were obtained by confocal microscopy (Zeiss LSM 510).
Immunohistochemistry.
All procedures related to the care and treatment of animals were in accordance with institutional and National Institutes of Health guidelines. Wistar rats (4 weeks old) were deeply anesthetized with pentobarbital sodium (60 mg/kg of body weight, intraperitoneally administered) and perfused with 4% (wt/vol) paraformaldehyde in 0.1 M phosphate buffer (PB; pH 7.4). Kidneys and small intestines were removed and postfixed in the same fixative, immersed sequentially with 10% (wt/vol), 20% (wt/vol), and 30% (wt/vol) sucrose in 0.1 M PB (pH 7.4), and frozen at −80°C. Sections (5 μm) were prepared, washed in 0.1 M PB, blocked with 0.1 M PB containing 5% goat serum and 0.2% (wt/vol) Triton X-100 for 2 h, and incubated with the first antibodies at 4°C overnight. After the mixture was washed with 0.1 M PB four times, bound antibodies were detected with the second antibodies at room temperature for 3 h. Images were obtained with a confocal microscope (Olympus FV300-BX or Zeiss LSM 510).
Immunoelectron microscopy.
Kidneys were isolated from anesthetized adult rats, which were fixed by perfusion of 0.5% (wt/vol) glutaraldehyde in PBS. Tissues were cut in small pieces and fixed in the same fixative for 1 h on ice, washed with PBS, and dehydrated with a graded series of ethanol. Cubes of renal cortex (2 mm) were embedded in LR white resin (London Resin Company Ltd., London, United Kingdom) and polymerized with LR white accelerator and stored at −20°C. Ultrathin sections were mounted on 300-mesh nickel grids and treated with 3% H2O2 for 10 min. After blocking with 1% (wt/vol) BSA in PBS for 30 min, grids were incubated overnight at 4°C with the affinity-purified rabbit anti-JAM4 or anti-MAGI-1 antibody, washed with PBS, and incubated with 10-nm gold-conjugated goat anti-rabbit IgG (Amersham) at a dilution of 1:20 for 1 h at room temperature. Grids were washed with distilled water and stained with uranyl acetate for 20 min. After air drying at room temperature, the sections were photographed on a Hitachi 7100 electron microscope at ×10,000 magnification.
Northern blotting.
A multiple-tissue Northern blot of mouse tissues (Seegene) containing 20 μg of total RNA was hybridized with uniformly labeled probe derived from the full coding sequence of JAM4.
Cell aggregation assay.
Wild-type L cells or L-JAM4 cells were treated with 0.2% (wt/vol) trypsin and 1 mM EDTA at 37°C for 5 min and dispersed by pipetting. Cells were resuspended in Hanks balanced salt solution with either 1 mM CaCl2 or 5 mM EDTA at 106 cells/ml, placed in 12-well plates precoated with BSA, and rotated on a gyratory shaker at 37°C for the indicated periods of time. Aggregation was stopped by adding 2% (vol/vol) glutaraldehyde. The extent of aggregation was defined as the ratio of the total particle number after incubation to the initial particle number before incubation. We took the image with an Olympus IX70 apparatus at ×10 magnification with a 1.5× zoom and printed it on paper 29 by 21 cm. We regarded any group of cells that contacted each other as one particle.
Cell dissociation assay.
Cells were plated at 106 cells/60-mm dish and grown in cultures for 3 days to confluency, and the medium was removed. When indicated, cells were treated with dimethyl sulfoxide (DMSO), 1 μg of nocodazole/ml, or 5 μM latrunculin A in fresh medium for 60 min. After being washed with PBS, cells were scraped off by rotating the plate one round under a 3-cm-wide silicone blade cell scraper (Sumitomo Bakelite Co.). Cells were subsequently dissociated in 500 μl of PBS by pipetting with disposable 1-ml plastic tips.
Subcellular fractionation.
Various L cells were collected on one 10-cm-diameter plate and homogenized in 200 μl of 20 mM HEPES-NaOH (pH 7.4) containing 100 mM NaCl and 1% (wt/vol) Triton X-100. The homogenates were centrifuged at 100,000 × g for 15 min. The supernatant and pellet were designated as Triton X-100-soluble and -insoluble fractions, respectively. Comparable amounts of the fractions were immunoblotted with appropriate antibodies.
Permeability assay.
Cells were plated at 5 × 104 cells/well in Transwells (Costar Corp.; polycarbonate filter [0.4-μm pore size, 6.5-mm diameter]) and grown in cultures for 5 days to confluency. The culture medium was then changed to serum-free medium, and 1 mg of FITC-dextran/ml with an average molecular mass of 40,000 Da (Sigma-Aldrich Fine Chemicals) was added to the upper chamber. At 2 h later, 100-μl aliquots were collected from the lower chamber and assayed by fluorimetry (Luminescence Spectrometer LS50B; Perkin-Elmer) (excitation at 492 nm and emission at 520 nm). The value for wild-type CHO cells was set at 100%.
Adenovirus production and infection.
Adenovirus to express GFP-tagged MAGI-1 was prepared using an AdEasy system (19). Briefly, pShuttle CMV GFP-MAGI-1 was transformed into BJ5183 derivatives containing AdEasy-1 plasmid. The recombinant adenoviral plasmid was transfected into 911 cells (Introgene, Leiden, The Netherlands) with Lipofectamine (Invitrogen). Viral supernatants were obtained by freezing and thawing infected cells. The infection of cells was repeated for further amplification. High-titer viral stock was prepared by CsCl gradient. CHO cells were infected to express either control GFP or GFP-MAGI-1. At 24 h later, CHO cells were harvested and plated on Transwells for the permeability assay. Infected cells were grown in cultures on regular plates to examine the cell viability and harvested to confirm the expression of proteins.
Immunoprecipitation.
Using the DEAE-dextran method, COS-7 cells were transfected with pClneo Myc MAGI-1 and pFLAG JAM4, pFLAG JAM4ΔC, or pFLAG JAM1. Cells from two 10-cm-diameter plates were homogenized in 400 μl of lysis buffer A (20 mM HEPES-NaOH [pH 7.4], 100 mM NaCl, 1% [wt/vol] Triton X-100) and centrifuged at 100,000 × g for 15 min at 4°C. The supernatant was incubated with 1.0 μl of the anti-MAGI-1 serum or the preimmune serum fixed on 7.5 μl of protein G-Sepharose 4 fast-flow beads. After the beads were washed, the precipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with either the anti-Myc or the anti-FLAG antibody.
GST pulldown assay.
COS-7 cells were transfected with the indicated constructs using the DEAE-dextran method. Cells from one 10-cm plate were homogenized in 200 μl of the lysis buffer A and centrifuged at 100,000 × g for 15 min at 4°C. The supernatant (175 μl) was incubated with 250 pmol of various glutathione S-transferase (GST) proteins fixed on 7.5 μl of glutathione-Sepharose 4B beads. After the beads were washed, the precipitates were analyzed by SDS-PAGE and immunoblotted with the appropriate antibodies.
In vitro transcription and translation.
Using 5 μg of pClneo FLAG-JAM4 as a template in 50 μl of the reaction buffer according to the manufacturer's protocol, [35S]methionine-labeled FLAG-JAM4 was prepared with in vitro transcription and translation (TnT T7 quick-coupled transcription/translation system; Promega). The product was analyzed by SDS-PAGE, and the image was obtained with an imaging analyzer (FLA-3000; Fujifilm).
Deglycosylation of JAM4.
Stable transformants of L, MDCK, LLC-PK1, and CHO cells expressing FLAG-JAM4 and COS-7 cells transfected with pFLAG JAM4 were harvested. Cell lysates were digested for 60 min at 37°C with N-glycosidase F (New England BioLabs) in the presence of 1% (wt/vol) NP-40 according to the manufacturer's recommendations.
Cell surface biotinylation.
COS-7 cells that were transfected with pFLAG-JAM4 or pFLAG-JAM4ΔC, L-FLAG-JAM4, and L-FLAG-JAM4ΔC were biotinylated with 0.5 mg of sulfo-N-hydroxysuccinimide-biotin (Pierce)/ml in PBS containing 1 mM MgCl2 and 0.1 mM CaCl2 at 4°C for 60 min. After the reaction was stopped by adding 50 mM NH4Cl, membrane fractions were collected from harvested cells and extracted in buffer containing 20 mM HEPES-NaOH (pH 7.4) containing 100 mM NaCl and 1% Triton X-100. The extracts were incubated with avidin-agarose beads (Sigma-Aldrich Fine Chemicals), and the precipitated proteins were immunoblotted with the appropriate antibody.
Nucleotide sequence accession number.
The nucleotide sequence encoding JAM4 has been deposited in GenBank (accession number AF537215).
RESULTS
Identification of JAM4.
Using the PDZ domains of human MAGI-1 as bait to identify a protein interacting with MAGI-1, we performed yeast two-hybrid screening with a human lung cDNA library. We screened 2 × 106 clones and obtained positive results for 65 clones. Sequence analysis of these clones identified previously uncharacterized genes. Among them, we focused on a particular clone, pPrey 10427. The encoded peptide sequence was composed of 76 amino acids and terminated with a PDZ-binding motif (Fig. 1A). A search of the GenBank database revealed that the amino acid sequence had 80% homology to the C terminus sequence of the product of mouse BC004806. Mouse BC004806 encodes a putative type I transmembrane glycoprotein with one V-type Ig loop (Fig. 1B). Using sense and antisense primers that were designed based on the sequence of BC004806, we subsequently performed PCR on mouse lung cDNA. The PCR product encoded a protein composed of 370 amino acids (Fig. 1D). The protein included all amino acids of mouse BC004806 protein and one additional V-type Ig-like loop (Fig. 1B). Because the gene product is similar to JAM1, -2, and -3 among the members of the Ig protein family, we named it JAM4 (Fig. 1C). It shows 14.0, 10.4, and 13.5% amino acid homology to mouse JAM1, mouse JAM2, and mouse JAM3, respectively (Fig. 1D).
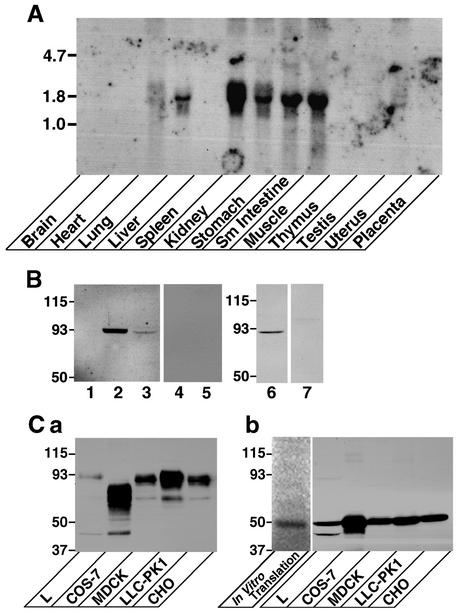
To examine which tissues express JAM4, we hybridized a multitissue mouse RNA blot with a JAM4 cDNA probe. A single 1.6-kb transcript was present in liver, kidney, stomach, small intestine, and skeletal muscle cells (Fig. 2A). The weak signal was also detected in lung cells. We raised a rabbit polyclonal antibody against the intracellular domain of JAM4. The affinity-purified antibody detected a 93-kDa protein in kidney and small intestine cells but not in spleen cells (Fig. 2B). A weak signal of the same size was detected in MTD-1A cells too. All signals disappeared after the antibody was adsorbed by the immunogen (Fig. 2B, lanes 4, 5, and 7). The calculated molecular mass of JAM4 is 40 kDa, but FLAG-JAM4 was detected as a protein of about 93 kDa in various cells (Fig. 2Ca). FLAG-JAM4 was smaller in COS-7 cells. After the treatment with N-glycosidase F to remove N-glycan chains, FLAG-JAM4 from all cells migrated at the same site as the in vitro transcription and translation product (Fig. 2Cb). Therefore, we conclude that the 93-kDa protein recognized by the antibody is JAM4 with N-glycosylation.
FIG. 2.
(A) Northern blot analysis of JAM4. A uniformly labeled probe corresponding to the full length of JAM4 was prepared. Blots with 20 μg of total RNA from each mouse tissue were hybridized with the probe (3,600,000 cpm) and exposed for 8 days. The mobilities of molecular mass standards (in kilobases) are indicated at the left. (B) Western blot with the anti-JAM4 antibody of rat kidney, intestine, spleen, and MTD-1A cells. Homogenates (total protein, 20 μg) from rat kidney, small intestine, spleen, and MTD-1A cells were resolved by SDS-PAGE and immunoblotted with the affinity-purified antibody against JAM4. Lane 1, spleen cells; lanes 2 and 4, kidney cells; lanes 3 and 5, small intestine cells; lanes 6 and 7, MTD-1A cells. For lanes 4, 5, and 7, the antibody was preincubated with 5 μM immunogen. The mobilities of molecular mass standards (in kilodaltons) are indicated at left. (C) FLAG-JAM4 in various cells. (a) Homogenates (total protein, 20 μg) from various stable transformants expressing FLAG-JAM4 were resolved by SDS-PAGE and immunoblotted with the anti-FLAG antibody. (b) Homogenates of stable transformants were treated with N-glycosidase F and charged onto SDS-PAGE gel with the in vitro transcription and translation product. The lane for the in vitro transcription and translation product was separately analyzed with the image analyzer. The remaining lanes were immunoblotted with the anti-FLAG antibody. The mobilities of molecular mass standards (in kilodaltons) are indicated at the left.
JAM4 is localized at TJs in epithelial cells.
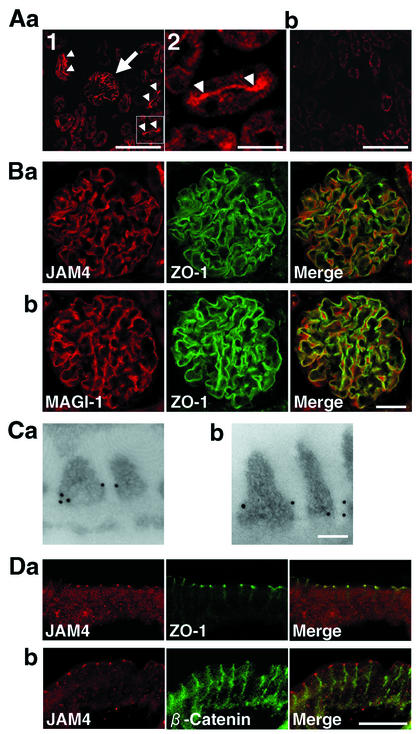
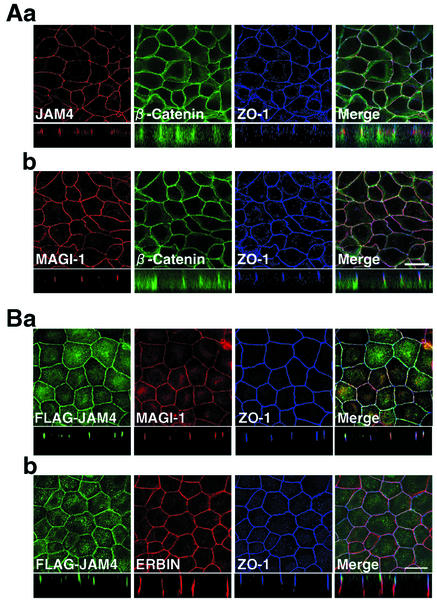
We examined the distribution of JAM4 in rat kidney cells. At low magnification, the image showed the intense signal in glomeruli and at apical membranes of proximal tubules (Fig. 3Aa). After incubation of the antibody with 5 μM antigen, the signals of glomeruli and apical membranes of proximal tubules disappeared, suggesting that both signals were specific (Fig. 3Ab). The double staining with the rabbit anti-JAM4 and mouse anti-ZO-1 antibodies indicated that JAM4 and ZO-1 were colocalized in glomeruli (Fig. 3Ba). MAGI-1 was also concentrated in glomeruli as described previously and colocalized with ZO-1 (Fig. 3Bb) (35). Immunogold labeling of ultrathin sections of glomeruli revealed that JAM4 and MAGI-1 were localized at slit diaphragms of podocytes (Fig. 3C). An immunofluorescence study using small intestine cells showed that JAM4 was colocalized with ZO-1 in intestinal epithelial cells (Fig. 3Da). In contrast, β-catenin, an adherens junction marker protein, showed a more diffuse distribution along the lateral membrane of cells (Fig. 3Db). The signal colocalized with ZO-1 was adsorbed by the excess amount of the immunogen (data not shown). Cytosolic staining was also detected but did not completely disappear even in the presence of the immunogen, suggesting that it was nonspecific (data not shown). JAM4 was also detected at TJs of luminal epithelial cells in ducts of mammary glands (data not shown). The immunofluorescence study revealed that JAM4 and MAGI-1 were colocalized with ZO-1, a TJ marker protein, in mouse mammary gland epithelial MTD-1A cells (Fig. 4A). We also confirmed directly the colocalization of FLAG-JAM4 and MAGI-1 by immunostaining stable transformant MDCK cells expressing FLAG-JAM4 (Fig. 4B). These data indicate that JAM4 is a TJ protein in various epithelial cells and is colocalized with MAGI-1.
FIG. 3.
Immunohistological analysis of JAM4. (A) JAM4 in kidney cells. The sections of rat kidney were stained with the affinity-purified antibody in the absence (a) or the presence (b) of 5 μM immunogen. Signals were detected in glomeruli (arrow) and on apical membranes of proximal tubules (arrowheads). (a2) The demarcated area shown in panel a1 at higher magnification. Bars, 100 μm (a1 and b) and 20 μm (a2). (B) Colocalization of JAM4 and MAGI-1 with ZO-1 in kidney glomeruli. Sections of rat kidney were double stained with various antibodies. (a) JAM4 and ZO-1; (b) MAGI-1 and anti-ZO-1. Bar, 20 μm. (C) Electron microscopy localization of JAM4 in podocytes. (a) JAM4; (b) MAGI-1. Bar, 0.1 μm. (D) Colocalization of JAM4 with ZO-1 in small intestine epithelial cells. (a) JAM4 and ZO-1; (b) JAM4 and β-catenin. Bar, 50 μm.
FIG. 4.
Immunofluorescence analysis of JAM4 in MTD-1A and MDCK cells. (A) MTD-1A cells were fixed and immunostained with anti-JAM4, anti-MAGI-1, anti-β-catenin, and anti-ZO-1 antibodies. JAM4 and MAGI-1 were detected by anti-rabbit antibodies. β-Catenin and ZO-1 were detected by anti-mouse monoclonal and anti-rat monoclonal antibodies, respectively. (a) JAM4, β-catenin, and ZO-1 in MTD-1A cells; (b) MAGI-1, β-catenin, and ZO-1 in MTD-1A cells. Bar, 20 μm. (B) MDCK-FLAG-JAM4 cells were fixed and immunostained with anti-FLAG, anti-MAGI-1, anti-ERBIN, and anti-ZO-1 antibodies. MAGI-1 and ERBIN were detected by anti-rabbit antibodies. FLAG and ZO-1 were detected by anti-mouse monoclonal and anti-rat monoclonal antibodies, respectively. (a) FLAG-JAM4, MAGI-1, and ZO-1 in MDCK cells; (b) FLAG-JAM4, ERBIN, and ZO-1 in MDCK cells. Bar, 20 μm.
JAM4 binds to MAGI-1 in vitro.
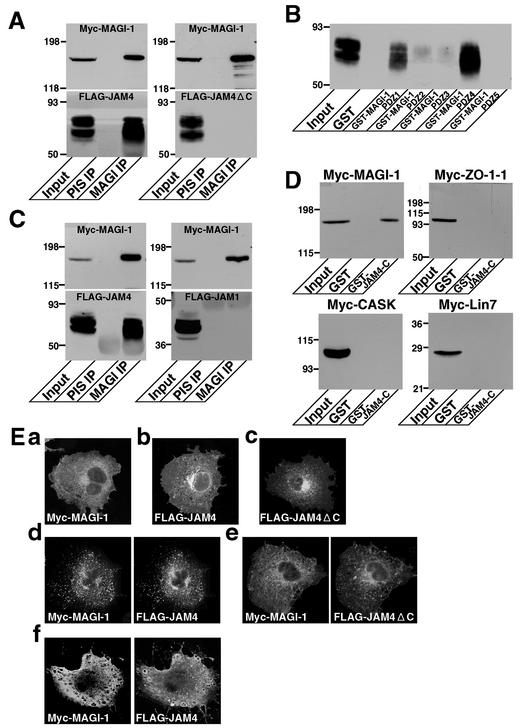
To confirm the interaction between JAM4 and MAGI-1, we first attempted to coimmunoprecipitate JAM4 and MAGI-1 from rat kidney cells. We excised kidneys and extracted glomeruli from minced renal cortex by sieving techniques. MAGI-1 was highly concentrated in the glomerular fraction (data not shown) as described in a previous report (36). JAM4 was also slightly enriched in the same fraction. Therefore, we made use of the glomerular fraction as starting material for the immunoprecipitation. MAGI-1 was, however, highly resistant to detergent extraction, and only a small amount of MAGI-1 was extracted in the detergents including Triton X-100 and deoxycholic acid (data not shown). Even after treatment with 6 M urea, a significant amount of MAGI-1 remained in the insoluble fraction. JAM4 was also Triton X-100 insoluble in kidney, small intestine, and lung cells, although it was soluble in deoxycholic acid (data not shown). Therefore, we overexpressed Myc-MAGI-1 and FLAG-JAM4 in COS-7 cells and immunoprecipitated Myc-MAGI-1 with the anti-MAGI-1 antibody from the lysates. The overexpressed Myc-MAGI-1 and FLAG-JAM4 were partially recovered in Triton X-100-soluble fractions in COS-7 cells. FLAG-JAM4 was coprecipitated with Myc-MAGI-1 (Fig. 5A). In contrast, FLAG-JAM4ΔC lacking the PDZ-binding motif was not coprecipitated, suggesting that JAM4 binds to MAGI-1 by its C-terminal PDZ-binding motif.
FIG. 5.
Interaction of JAM4 with MAGI-1. (A) Coimmunoprecipitation of JAM4 and MAGI-1. Myc-MAGI-1 with either FLAG-JAM4 or FLAG-JAM4ΔC was coexpressed in COS-7 cells. Anti-MAGI-1 antibody coprecipitated FLAG-JAM4 but not FLAG-JAM4ΔC with Myc-MAGI-1. The mobilities of molecular mass standards (in kilodaltons) are indicated at the left. Input, cell lysates; PIS IP, immunoprecipitate with the preimmune serum; MAGI IP, immunoprecipitate with the anti-MAGI-1 serum. (B) GST pulldown assay (using GST fusion proteins containing various PDZ domains of MAGI-1) of FLAG-JAM4. Lysates of COS cells expressing FLAG-JAM4 were incubated with GST alone or GST-MAGI-1-PDZ1, -PDZ2, -PDZ3, -PDZ4, or -PDZ5 immobilized on glutathione-Sepharose beads. The resulting complexes were analyzed by immunoblotting with the anti-FLAG antibody. The mobilities of molecular mass standards (in kilodaltons) are indicated at the left. (C) Specificity of the interaction of MAGI-1 with JAM4. Myc-MAGI-1 was coexpressed with either FLAG-JAM4 or FLAG-JAM1 in COS-7 cells. Lysates were incubated with either preimmune serum or anti-MAGI-1 serum immobilized on protein G-Sepharose beads. The resulting complexes were analyzed with either the anti-Myc or anti-FLAG antibody. MAGI-1 did not bind JAM1. The mobilities of molecular mass standards (in kilodaltons) are indicated at the left. (D) Specificity of the interaction of JAM4 with MAGI-1. Lysates of COS-7 cells expressing Myc-MAGI-1, Myc-ZO-1-1, Myc-CASK, or Myc-Lin-7 were incubated with GST alone or GST-JAM4-C immobilized on glutathione-Sepharose beads. The resulting complexes were analyzed by immunoblotting with the anti-Myc antibody. JAM4 did not bind ZO-1, CASK, or Lin-7. The mobilities of molecular mass standards (in kilodaltons) are indicated at the left. (E) Interaction of JAM4 and MAGI-1 in COS-7 cells. Subcellular localization of Myc-MAGI-1 (a), FLAG-JAM4 (b), and FLAG-JAM4ΔC (c) in COS-7 cells is shown. (d) Myc-MAGI-1 and FLAG-JAM4 were colocalized and formed clusters in COS-7 cells when coexpressed. (e) Myc-MAGI-1 and FLAG-JAM4ΔC did not interact with each other in COS-7 cells. (f) The effect of latrunculin A on the clustering of FLAG-JAM4 and Myc-MAGI-1. COS-7 cells were transfected with pFLAG JAM4 and pClneo Myc MAGI-1 and were treated (before fixation) with 5 μM latrunculin A for 5 min.
To identify the JAM4-binding region of MAGI-1, we prepared GST fusion proteins of MAGI-1-PDZ1, -PDZ2, -PDZ3, -PDZ4, and -PDZ5 and performed pulldown assays. FLAG-JAM4 was isolated with GST-PDZ1 and GST-PDZ4 but not with GST-PDZ5 (Fig. 5B). GST-PDZ2 and GST-PDZ3 trapped a trace amount of FLAG-JAM4. FLAG-JAM4ΔC did not bind to any of GST proteins (data not shown). These findings suggest that JAM4 binds to PDZ1 and PDZ4 of MAGI-1.
JAM4 does not bind to ZO-1 and JAM1 does not bind to MAGI-1.
JAM1 is known to bind to the PDZ domains of several proteins, including ZO-1 and CASK, through its C-terminal PDZ-binding motif (5, 11, 28). We tested whether MAGI-1 specifically bound to JAM4. We coexpressed FLAG-tagged JAM1, as well as FLAG-JAM4, with Myc-MAGI-1 in COS-7 cells and immunoprecipitated Myc-MAGI-1 with the anti-MAGI-1 antibody. FLAG-JAM4 was coimmunoprecipitated with Myc-MAGI-1, while FLAG-JAM1 was not (Fig. 5C). Using GST pulldown assays, we also examined whether the PDZ-binding motif of JAM4 bound to other PDZ proteins. GST-JAM4-C containing the C-terminal PDZ-binding motif did not bind Myc-ZO-1-1 under the conditions in which it bound MAGI-1 (Fig. 5D). GST-JAM4-C did not bind Myc-CASK or Myc-Lin-7 either, suggesting that JAM4 specifically interacts with MAGI-1.
MAGI-1 induces clustering of JAM4.
To obtain independent evidence for the interaction between JAM4 and MAGI-1, an immunofluorescence study was performed with COS-7 cells expressing Myc-MAGI-1 and FLAG-JAM4. MAGI-1 was diffusely distributed in cells when expressed alone (Fig. 5Ea). FLAG-JAM4 and FLAG-JAM4ΔC were also diffusely distributed (Fig. 5Eb and c). In cells expressing both Myc-MAGI-1 and FLAG-JAM4, Myc-MAGI-1 and FLAG-JAM4 formed clusters and were colocalized with each other (Fig. 5Ed). Myc-MAGI-1 did not colocalize with FLAG-JAM4ΔC and did not induce its clustering (Fig. 5Ee). After latrunculin A treatment to disrupt the actin cytoskeleton, the clusters of FLAG-JAM4 and Myc-MAGI-1 were no longer detected (Fig. 5Ef). The cell surface biotinylation revealed that FLAG-JAM4 and FLAG-JAM4ΔC were expressed on cell surfaces (data not shown). This suggests that the clustering of JAM4 takes place on the cell surface.
Transfected JAM4 promotes cell adhesion and recruits MAGI-1 to intercellular contacts.
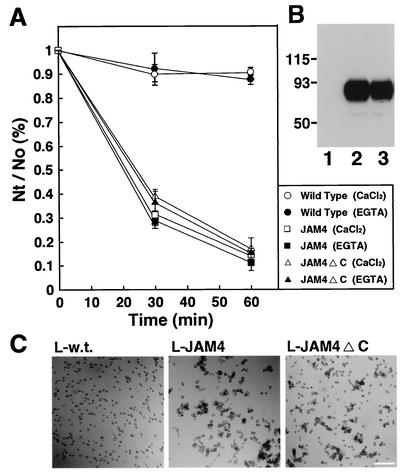
In the next set of experiments, we evaluated the ability of JAM4 to mediate cell adhesion. We performed an aggregation assay with stable transformants of L cells expressing JAM4 (L-JAM4). JAM4 showed cell aggregation activity in a time-dependent manner (Fig. 6A and C). L-JAM4 cells formed aggregates in the presence of 5 mM EGTA, suggesting that this activity did not require Ca2+. We also prepared L-JAM4ΔC cells, which expressed JAM4 without the PDZ-binding motif. The cell surface biotinylation revealed that JAM4 and JAM4ΔC were expressed on the cell surface at similar levels (Fig. 6B). L-JAM4ΔC cells also formed aggregates (Fig. 6A and CFi).
FIG. 6.
Cell aggregation activity of JAM4. (A) Cell aggregation assays were performed using L cells or stable transformants of L cells expressing JAM4 (L-JAM4) or JAM4ΔC (L-JAM4ΔC). The extent of cell aggregation is represented by the ratio of the total particle number at the indicated time point (Nt) to the initial particle number (N0). The Nt/N0 values are representative of the means ± standard errors of three independent experiments. (B) Expression of JAM4 and JAM4ΔC. Cell surface proteins were biotinylated, precipitated by avidin-agarose beads, and then immunoblotted with the anti-JAM4 antibody. Lane 1, wild-type L cells; lane 2, L-JAM4 cells; lane 3, L-JAM4ΔC cells. (C) Cell aggregation of L-JAM4 cells. Results for wild-type L cells (L-w.t.), L-JAM4 cells (L-JAM4), and L-JAM4ΔC cells (L-JAM4ΔC) at 60 min in a cell aggregation assay are shown. Bar, 200 μm.
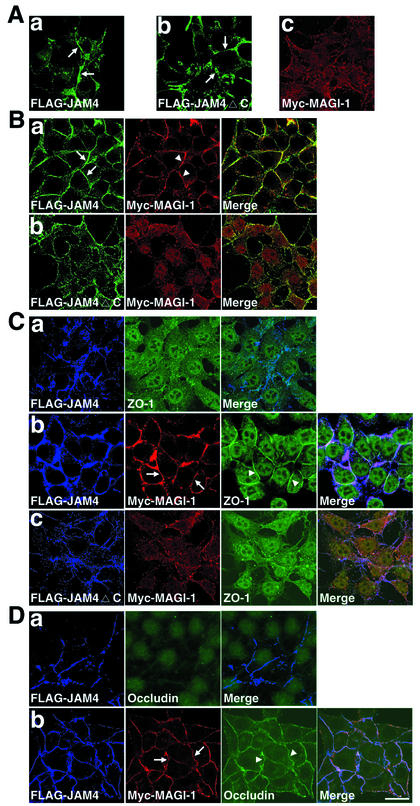
We prepared various single and double stable transformants of L cells, including L-FLAG-JAM4, L-FLAG-JAM4ΔC, L-Myc-MAGI-1, L-FLAG-JAM4/Myc-MAGI-1, and L-FLAG-JAM4ΔC/Myc-MAGI-1, and grew them in cultures to generate cell contacts. FLAG-JAM4 and FLAG-JAM4ΔC were accumulated at cell contacts in L-FLAG-JAM4 and L-FLAG-JAM4ΔC cells (Fig. 7Aa and b). Myc-MAGI-1 was not concentrated anywhere in cells (Fig. 7Ac). In L-FLAG-JAM4/Myc-MAGI-1 cells, Myc-MAGI-1 was concentrated at cell contacts and colocalized with FLAG-JAM4 (Fig. 7Ba). In L-FLAG-JAM4ΔC/Myc-MAGI-1 cells, FLAG-JAM4ΔC did not recruit Myc-MAGI-1 (Fig. 7Bb). We immunostained endogenous ZO-1 in L-FLAG-JAM4, L-FLAG-JAM4/Myc-MAGI-1, and L-FLAG-JAM4ΔC/Myc-MAGI-1 cells. ZO-1 was detected mainly in the nucleus in L-FLAG-JAM4 cells (Fig. 7Ca). ZO-1 was accumulated at JAM4-based intercellular contacts in L-FLAG-JAM4/Myc-MAGI-1 cells but not in L-FLAG-JAM4ΔC/Myc-MAGI-1 cells (Fig. 7Cb and c). These findings imply that MAGI-1 recruits ZO-1 to JAM4-based cell contacts. Moreover, another TJ protein, occludin, was also accumulated by Myc-MAGI-1 at JAM4-based cell contacts in L-FLAG-JAM4/Myc-MAGI-1 cells (Fig. 7D).
FIG. 7.
Recruitment of Myc-MAGI-1, ZO-1, and occludin to JAM4-based cell contacts. (A) Stable transformants of L cells expressing FLAG-JAM4 (L-FLAG-JAM4) (a) and FLAG-JAM4ΔC (L-FLAG-JAM4ΔC) (b) were grown in cultures to generate cell contacts. FLAG-JAM4 and FLAG-JAM4ΔC were accumulated at cell contacts (arrows). (c) L-Myc-MAGI-1 cells. Myc-MAGI-1 was diffusely distributed. (B) Myc-MAGI-1 in L-FLAG-JAM4/Myc-MAGI-1 (a) and L-FLAG-JAM4ΔC/Myc-MAGI-1 (b) cells. FLAG-JAM4 (arrows) and Myc-MAGI-1 (arrowheads) were accumulated at cell contacts in L-FLAG-JAM4/Myc-MAGI-1 cells. (C) (a) L-FLAG-JAM4 cells were double stained with anti-FLAG and anti-ZO-1 antibodies. (b) L-FLAG-JAM4/Myc-MAGI-1 cells; (c) L-FLAG-JAM4ΔC/Myc-MAGI-1 cells. L-FLAG-JAM4/Myc-MAGI-1 cells and L-FLAG-JAM4ΔC/Myc-MAGI-1 cells were triple stained with anti-FLAG, anti-Myc, and anti-ZO-1 antibodies. ZO-1 (arrowheads) was recruited with Myc-MAGI-1 (arrows) to cell contacts in L-FLAG-JAM4/Myc-MAGI-1 cells. (D) (a) L-FLAG-JAM4 cells were double stained with anti-FLAG and anti-occludin antibodies. (b) L-FLAG-JAM4/Myc-MAGI-1 cells were triple stained with anti-FLAG, anti-Myc, and anti-occludin antibodies. Occludin (arrowheads) was recruited with Myc-MAGI-1 (arrows) to cell contacts in L-FLAG-JAM4/Myc-MAGI-1 cells. Bar, 20 μm.
MAGI-1 strengthens JAM4-mediated cell adhesion.
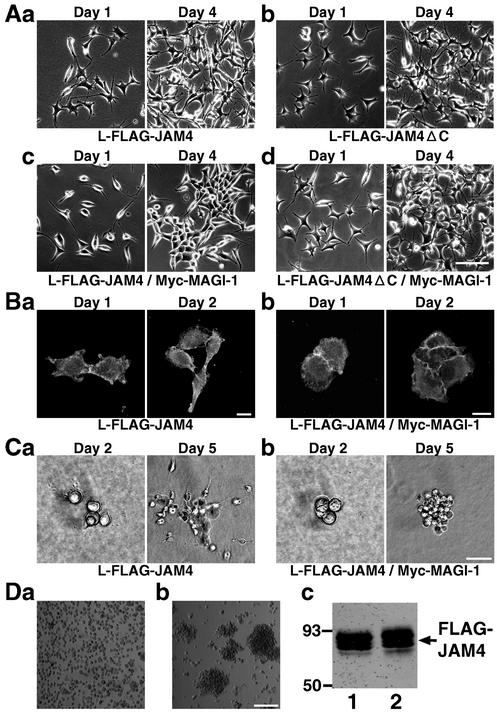
We addressed the question of whether MAGI-1 regulated JAM4-mediated cell adhesion. L-FLAG-JAM4 and L-FLAG-JAM4/Myc-MAGI-1 cells showed different appearances in the cultures. When plated at 105 cells/35-mm-diameter dish, both cell cultures were scattered on day 1 (Fig. 8Aa and c, left panels). On day 4, L-FLAG-JAM4 cells were still scattered while L-FLAG-JAM4/Myc-MAGI-1 cells formed aggregates (Fig. 8Aa and c, right panels). To avoid the bias caused by the difference between clones, we transiently transfected L-FLAG-JAM4 cells with pClneo Myc MAGI-1 and observed that cells expressing both FLAG-JAM4 and Myc-MAGI-1 formed aggregates (data not shown). L-FLAG-JAM4ΔC/Myc-MAGI-1 cells appeared to be similar to L-FLAG-JAM4ΔC cells even on day 4 (Fig. 8Ab and d). L-JAM4 cells at low density contacted each other by cell protrusions, and FLAG-JAM4 was accumulated at cell contacts (Fig. 8Ba). In contrast, L-FLAG-JAM4/Myc-MAGI-1 cells formed wide contacts and FLAG-JAM4 was detected as linear shapes (Fig. 8Bb). L-FLAG-JAM4/Myc-MAGI-1 cells appeared in round shapes. We also confirmed the difference between L-FLAG-JAM4 and L-FLAG-JAM4/Myc-MAGI-1 in collagen cultures. L-FLAG-JAM4 cells dispersed in collagen, but L-FLAG-JAM4/Myc-MAGI-1 cells did not (Fig. 8C). Based on these findings, we reasoned that L-FLAG-JAM4/Myc-MAGI-1 cells displayed greater adhesive activity than L-FLAG-JAM4 cells. To compare the strengths of cell adhesion, we performed a cell dissociation assay. L-FLAG-JAM4 cells became a single cell suspension after being pipetted five times (Fig. 8Da). L-FLAG-JAM4/Myc-MAGI-1 cells remained as cell aggregates after being pipetted five times (Fig. 8Db). We confirmed that both cells expressed FLAG-JAM4 at similar levels (Fig. 8Dc).
FIG. 8.
Effect of MAGI-1 on the cell adhesive activity of JAM4. (A) Phase-contrast images of L-FLAG-JAM4 (a), L-FLAG-JAM4ΔC (b), L-FLAG-JAM4/Myc-MAGI-1 (c), and L-FLAG-JAM4ΔC/Myc-MAGI-1 (d) cells grown in cultures on plastic dishes. Bar, 50 μm. (B) Immunofluorescence of FLAG-JAM4 in L-FLAG-JAM4 (a) and L-FLAG-JAM4/Myc-MAGI-1 (b) cells. Bar, 10 μm. (C) Phase-contrast images of L-FLAG-JAM4 (a) and L-FLAG-JAM4/Myc-MAGI-1 (b) cells in collagen cultures. Bar, 100 μm. (D) Dissociation assay of L-FLAG-JAM4 and L-FLAG-JAM4/Myc-MAGI-1 cells. (a) L-FLAG-JAM4 cells after being pipetted five times; (b) L-FLAG-JAM4/Myc-MAGI-1 cells after being pipetted five times. Bar, 200 μm. (c) Western blot with the anti-FLAG antibody of L-FLAG-JAM4 and L-FLAG-JAM4/Myc-MAGI-1 cells. Lane 1, L-FLAG-JAM4 cells; lane 2, L-FLAG-JAM4/Myc-MAGI-1 cells. The mobilities of molecular mass standards (in kilodaltons) are indicated at the left.
MAGI-1 links JAM4 to Triton X-100-insoluble structures.
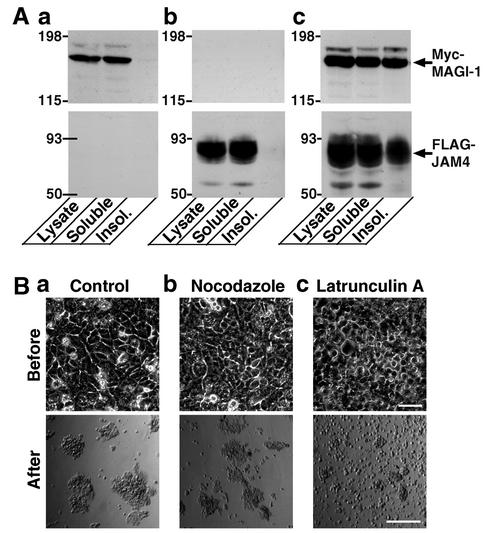
With regard to the mechanism underlying the reinforcement of JAM4-mediated cell adhesion, we hypothesized that MAGI-1 might link JAM4-based cell contacts to the cytoskeleton. First, we performed the subcellular fractionation of L-Myc-MAGI-1, L-FLAG-JAM4, and L-FLAG-JAM4/Myc-MAGI-1 cells (Fig. 9A). Myc-MAGI-1 and FLAG-JAM4 were Triton X-100 soluble when expressed alone. However, in L-FLAG-JAM4/Myc-MAGI-1 cells, both Myc-MAGI-1 and FLAG-JAM4 were recovered in the Triton X-100-soluble and -insoluble fractions. L-FLAG-JAM4/Myc-MAGI-1 cells treated with latrunculin A became easily dispersed in the cell dissociation assay (Fig. 9B). Nocodazole treatment did not show any effect.
FIG. 9.
Effect of disruption of the cytoskeleton on JAM4/MAGI-1-mediated cell adhesion. (A) Subcellular fractionation of L-Myc-MAGI-1, L-FLAG-JAM4, and L-FLAG-JAM4/Myc-MAGI-1 cells. Comparable amounts of the fractions from various L cells were immunoblotted with the anti-Myc or the anti-FLAG antibody. Insol., insoluble. (a) L-Myc-MAGI-1 cells; (b) L-FLAG-JAM4 cells; (c) L-FLAG-JAM4/Myc-MAGI-1 cells. (B) Dissociation assay of L-FLAG-JAM4/Myc-MAGI-1 cells after mock, nocodazole, and latrunculin A treatment. L-FLAG-JAM4/Myc-MAGI-1 cells were treated with DMSO (a), 1 μg of nocodazole/ml (b), or 5 μM latrunculin A (c) for 60 min before the dissociation assay. Upper photos, cells after the treatment and before scraping; lower photos, cells after being pipetted five times. Bar, 200 μm.
JAM4 reduced permeability of CHO cell monolayers and MAGI-1 augmented the reduction.
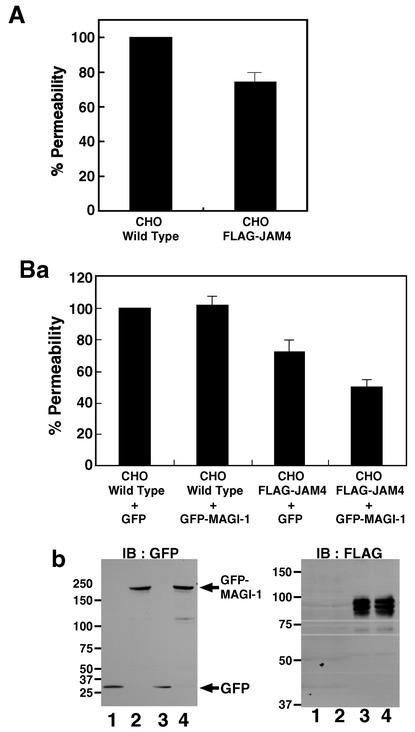
In the last set of experiments, we examined whether JAM4 controls the permeability of CHO cell monolayers as JAM1 does. The permeability of FITC-dextran was reduced by 25% in stable transformant CHO cells expressing FLAG-JAM4 (CHO-FLAG-JAM4 cells) (Fig. 10A). We expressed control GFP or GFP-MAGI-1 using adenovirus in wild-type CHO and CHO-FLAG-JAM4 cells. GFP-MAGI-1 further decreased the permeability in CHO-FLAG-JAM4 cells but not in wild-type CHO cells (Fig. 10Ba). We confirmed the expression of FLAG-JAM4, GFP, and GFP-MAGI-1 in these cells (Fig. 10Bb).
FIG. 10.
Effect of JAM4 and MAGI-1 on the permeability of CHO cell monolayers. (A) Wild-type CHO and CHO-FLAG-JAM4 cells were grown in cultures in Transwells. FITC-dextran was added to the upper chamber, and the permeability was measured. The permeability of wild-type CHO cells was taken as 100%. Data are indicated as the means ± standard errors of three independent experiments. (B) (a) Wild-type CHO and CHO-FLAG-JAM4 cells were infected by adenovirus to express either GFP or GFP-MAGI-1. At 2 h after the infection, these cells were plated on Transwells for the permeability assay. (b) Immunoblots (IB) of CHO and CHO-FLAG-JAM4 cells to confirm the protein expression. Left panel, immunoblot with the anti-GFP antibody; right panel, immunoblot with the anti-FLAG antibody. Lanes 1, wild-type CHO cells with GFP; lanes 2, wild-type CHO cells with GFP-MAGI-1; lanes 3, CHO-FLAG-JAM4 cells with GFP; lanes 4, CHO-FLAG-JAM4 cells with GFP-MAGI-1. The mobilities of molecular mass standards (in kilodaltons) are indicated at the left.
DISCUSSION
For this study, we have reported a novel protein that interacts with MAGI-1. We have named this protein JAM4. Because we had only rabbit polyclonal antibodies against both MAGI-1 and JAM4, we cannot directly show the colocalization of MAGI-1 and JAM4. However, JAM4 is colocalized with ZO-1 in kidney glomeruli and small intestinal epithelial cells, whereas MAGI-1 is colocalized with ZO-1 (21). Moreover, FLAG-JAM4 is colocalized with endogenous MAGI-1 in MDCK cells. Although we could not perform a coimmunoprecipitation study using endogenous proteins, JAM4 is coprecipitated with MAGI-1 from COS-7 cells expressing JAM4 and MAGI-1. An immunofluorescence study showed that MAGI-1 and JAM4 cooperatively form clusters in COS-7 cells. These data suggest that JAM4 interacts with MAGI-1 at TJs.
The amino acid sequence of JAM4 suggests that JAM4 has two domains with intrachain disulfide bonds typical of Ig loops of the V type. JAM4 has five potential sites of N-glycosylation. JAM4 appears as a protein of about 93 kDa in various cells, although the calculated size is 40 kDa. The result from the experiment using N-glycosidase F indicates that JAM4 is indeed modified by N-glycosylation. Cell aggregation assays using L cells showed that JAM4 mediates calcium-independent cell adhesion. L cells are suitable for this assay, because wild-type L cells do not express cadherins and do not form aggregates. JAM4 is similar to JAM1 in the arrangement of V-V-type loops. JAM1 is concentrated at TJs of epithelial and endothelial cells (29, 34). JAM1 has a C-terminal PDZ-binding motif and binds to PDZ domains of ZO-1, CASK, afadin/AF-6, ASIP/PAR-3, and MUPP1 (11, 12, 17, 23, 28). Besides PDZ proteins, cingulin also interacts with JAM1 directly or indirectly (5). All these interactions suggest that JAM1 plays central roles in the assembly of TJ components. By analogy, we speculated that JAM4 is involved in the assembly of TJ components. First, we have shown that MAGI-1 is recruited to JAM4-based cell contacts in L cells. Moreover, we have observed that ZO-1 and occludin are also accumulated to JAM4-based cell contacts in a MAGI-1-dependent manner. Although the molecular mechanism underlying the recruitment of ZO-1 and occludin remains to be clarified, the complex of JAM4 and MAGI-1 may be involved in the assembly of components of TJs. It is necessary to examine how various mutants of JAM4 and MAGI-1 interfere with the formation of intact TJs. In immunofluorescence assays, JAM4 is detected on apical membranes of proximal renal tubules. The molecular constituents that tether JAM4 to TJs in MTD-1A, MDCK, and intestinal epithelial cells may be absent in proximal tubular epithelial cells, with the result that JAM4 is not localized at TJs. It is also important to determine the molecular mechanism that localizes JAM4 at TJs.
JAM4 is slightly enriched in the glomerular fraction, and it appears to be colocalized with ZO-1 and MAGI-1 in kidney glomeruli at the immunofluorescence level. Immunoelectron microscopy revealed the localization of JAM4 in podocytes. JAM1 is known to regulate TJ resealing in epithelial cells (27). Nephrin, which is an Ig-like transmembrane protein localized at the podocyte slit diaphragm, is essential for maintenance of normal glomerular permeability and is involved in congenital nephrotic syndrome (25). We showed that JAM4 reduced the permeability of CHO cell monolayers. JAM4 may be relevant in the control of the physical barrier at epithelial cells and podocytes.
Our data also indicate that MAGI-1 strengthens the cell adhesion and sealing activities of JAM4. A simple explanation is that MAGI-1 may induce the clustering of JAM4 and concentrate JAM4 at cell-cell contacts. We also speculate that cytoskeletons may contribute to the effect of MAGI-1. JAM4 becomes Triton X-100 insoluble when expressed with MAGI-1 in L cells. MAGI-1 also becomes Triton X-100 insoluble, depending on the presence or absence of JAM4. Treatments disrupting the actin cytoskeleton abolish the clustering of MAGI-1 and JAM4 in COS-7 cells and the effect of MAGI-1 on the cell adhesion activity of JAM4 in L cells. The complex of JAM4 and MAGI-1 may form a scaffold which is linked to the cytoskeleton. Consistent with this, endogenous MAGI-1 and JAM4 are Triton X-100 insoluble in kidney, small intestine, and lung cells. The attachment of the actin cytoskeleton may be necessary for MAGI-1 and JAM4 to form tight adhesions.
Besides JAM1, two JAM-related proteins have been identified. JAM2 and JAM3 have one V-type and one C2-type Ig loops (3, 4, 7, 26). JAM2 is expressed in endothelial and lymphatic cells and interacts with JAM3. JAM3 is expressed on T, NK, and dendritic cells. The interaction between JAM2 and JAM3 may be important for the trafficking of these cells across the endothelium (26). JAM1 is also expressed in endothelial cells and is involved in regulating monocyte migration (29, 34). We have not confirmed the localization of JAM4 in endothelial cells. It remains to be determined whether JAM4 plays roles in endothelial cells as well as in epithelial cells.
Acknowledgments
This study was supported by grants-in-aids for Scientific Research and Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science, and Technology and a grant from Yamanouchi Foundation for Research on Metabolic Disorders (2000).
We thank S. Tsukita (Kyoto University) for cDNA of mouse ZO-1, rat anti-occludin antibody, and MDCKII cells, M. Takeichi for MTD-1A cells, Y. Nakamura (University of Tokyo) for cDNA of human MAGI-1, S. Uchida and S. Sasaki (Tokyo Medical and Dental University) for LLC-PK1 cells, T. Kitamura (University of Tokyo) for pMXpuro vector, G. Noran (Stanford University) for Phoenix ampho cells, S. Hollenberg (Oregon Health Sciences University) for reagents for yeast two-hybrid screening, and T.-C. He and B. Vogelstein (Johns Hopkins Oncology Center) for the pAdEasy system. We thank N. Terada, S. Ichinose, and K. Terashima (Tokyo Medical and Dental University) for valuable advice. We also thank C. Rokukawa and M. Miyahara-Tenkatsu for skillful technical assistance.
REFERENCES
- 1.Anderson, J. M., B. R. Stevenson, L. A. Jesaitis, D. A. Goodenough, and M. S. Mooseker. 1988. Characterization of ZO-1, a protein component of the tight junction from mouse liver and Madin-Darby canine kidney cells. J. Cell Biol. 106:1141-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J. M., and C. M. van Itallie. 1999. Tight junctions: closing in on the seal. Curr. Biol. 9:R922-R924. [DOI] [PubMed] [Google Scholar]
- 3.Arrate, M. P., J. M. Rodriguez, T. M. Tran, T. A. Brock, and S. A. Cunningham. 2001. Cloning of human junctional adhesion molecule 3 (JAM3) and its identification as the JAM2 counter-receptor. J. Biol. Chem. 276:45826-45832. [DOI] [PubMed] [Google Scholar]
- 4.Aurrand-Lions, M., L. Dunca, C. Ballestrem, and B. A. Imhof. 2001. JAM-2, a novel immunoglobulin superfamily molecule, expressed by endothelial and lymphatic cells. J. Biol. Chem. 276:2733-2741. [DOI] [PubMed] [Google Scholar]
- 5.Bazzoni, G., O. M. Martinez-Estrada, F. Orsenigo, M. Cordenonsi, S. Citi, and E. Dejana. 2000. Interaction of junctional adhesion molecule with the tight junction components ZO-1, cingulin, and occludin. J. Biol. Chem. 275:20520-20526. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, E. I., and H. Birn. 2002. Megalin and cubilin: multifunctional endocytic receptors. Nat. Rev. Mol. Cell Biol. 3:258-268. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham, S. A., M. P. Arrate, J. M. Rodriguez, R. J. Bjercke, P. Vanderslice, A. P. Morris, and T. A. Brock. 2000. A novel protein with homology to the junctional adhesion molecule. J. Biol. Chem. 275:34750-34756. [DOI] [PubMed] [Google Scholar]
- 8.Dobrosotskaya, I., R. K. Guy, and G. L. James. 1997. MAGI-1, a membrane-associated guanylate kinase with a unique arrangement of protein-protein interaction domains. J. Biol. Chem. 272:31589-31597. [DOI] [PubMed] [Google Scholar]
- 9.Dobrosotskaya, I. Y. 2001. Identification of mNET1 as a candidate ligand for the first PDZ domain of MAGI-1. Biochem. Biophys. Res. Commun. 283:969-975. [DOI] [PubMed] [Google Scholar]
- 10.Dobrosotskaya, I. Y., and G. L. James. 2000. MAGI-1 interacts with beta-catenin and is associated with cell-cell adhesion structures. Biochem. Biophys. Res. Commun. 270:903-909. [DOI] [PubMed] [Google Scholar]
- 11.Ebnet, K., C. U. Schulz, M.-K. M. zu Bickwedde, G. G. Pendl, and D. Vestweber. 2000. Junctional adhesion molecule interacts with the PDZ domain-containing proteins AF-6 and ZO-1. J. Biol. Chem. 275:27979-27988. [DOI] [PubMed] [Google Scholar]
- 12.Ebnet, K., A. Suzuki, Y. Horikoshi, T. Hirose, M.-K. M. zu Brickwedde, S. Ohno, and D. Vestweber. 2001. The cell polarity protein ASIP/PAR-3 directly associates with junctional adhesion molecule (JAM). EMBO J. 20:3738-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fanning, A. S., and J. M. Anderson. 1999. Protein modules as organizers of membrane structure. Curr. Opin. Cell Biol. 11:432-439. [DOI] [PubMed] [Google Scholar]
- 14.Furuse, M., T. Hirase, M. Itoh, A. Nagafuchi, S. Yonemura, S. Tsukita, and S. Tsukita. 1993. Occludin: a novel integral membrane protein localizing at tight junctions. J. Cell Biol. 123:1777-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furuse, M., K. Fujita, T. Hiiragi, K. Fujimoto, and S. Tsukita. 1998. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 141:1539-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furuse, M., H. Sasaki, K. Fujimoto, and S. Tsukita. 1998. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell Biol. 143:391-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamazaki, Y., M. Itoh, H. Sasaki, M. Furuse, and S. Tsukita. 2002. Multi-PDZ domain protein 1 (MUPP1) is concentrated at tight junctions through its possible interaction with claudin-1 and junctional adhesion molecule. J. Biol. Chem. 277:455-461. [DOI] [PubMed] [Google Scholar]
- 18.Haskins, J., L. Gu, E. S. Wittchen, J. Hibbard, and B. R. Stevenson. 1998. ZO-3, a novel member of the MAGUK protein family found at the tight junction, interacts with ZO-1 and occludin. J. Cell Biol. 141:199-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, T.-C., S. Zhou, L. T. Da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirao, K., Y. Hata, N. Ide, M. Takeuchi, M. Irie, I. Yao, M. Deguchi, A. Toyoda, T. C. Sudhof, and Y. Takai. 1998. A novel multiple PDZ domain-containing molecule interacting with N-methyl-D-aspartate receptors and neuronal cell adhesion proteins. J. Biol. Chem. 273:21105-21110. [DOI] [PubMed] [Google Scholar]
- 21.Ide, N., Y. Hata, H. Nishioka, K. Hirao, I. Yao, M. Deguchi, A. Mizoguchi, H. Nishimori, T. Tokino, Y. Nakamura, and Y. Takai. 1999. Localization of membrane-associated guanylate kinase (MAGI)-1/BAI-associated protein (BAP) 1 at tight junctions of epithelial cells. Oncogene 18:7810-7815. [DOI] [PubMed] [Google Scholar]
- 22.Itoh, M., M. Furuse, K. Morita, K. Kubota, M. Saitou, and S. Tsukita. 1999. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J. Cell Biol. 147:1351-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itoh, M., H. Sasaki, M. Furuse, H. Ozaki, T. Kita, and S. Tsukita. 2001. Junctional adhesion molecule (JAM) binds to PAR-3: a possible mechanism for the recruitment of PAR-3 to tight junctions. J. Cell Biol. 154:491-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jesaitis, L. A., and D. A. Goodenough. 1994. Molecular characterization and tissue distribution of ZO-2, a tight junction protein homologous to ZO-1 and the Drosophila discs-large tumor suppressor protein. J. Cell Biol. 124:949-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kestilä, M., U. Lenkkeri, M. Männikkö, J. Lamerdin, P. McCready, H. Putaala, V. Ruotsalainen, T. Morita, M. Nissinen, R. Herva, C. E. Kashtan, L. Peltonen, C. Holmberg, A. Olsen, and K. Tryggvason. 1998. Positionally cloned gene for a novel glomerular protein—nephrin—is mutated in congenital nephrotic syndrome. Mol. Cell 1:575-582. [DOI] [PubMed] [Google Scholar]
- 26.Liang, T. W., H. H. Chiu, A. Gurney, A. Sidle, D. B. Tumas, P. Schow, J. Foster, T. Klassen, K. Dennis, R. A. DeMarco, T. Pham, G. Frantz, and S. Fong. 2002. Vascular endothelial-junctional adhesion molecule (VE-JAM)/JAM2 interacts with T, NK, and dendritic cells through JAM3. J. Immunol. 168:1618-1626. [DOI] [PubMed] [Google Scholar]
- 27.Liu, Y., A. Nustrat, F. J. Schnell, T. A. Reaves, S. Walsh, M. Pochet, and C. A. Parkos. 2000. Human junction adhesion molecule regulates tight junction resealing in epithelia. J. Cell Sci. 113:2363-2374. [DOI] [PubMed] [Google Scholar]
- 28.Marinez-Estrada, O. M., A. Villa, F. Breviario, F. Orsenigo, E. Dejana, and G. Bazzoni. 2001. Association of junctional adhesion molecule with calcium/calmodulin-dependent serine protein kinase (CASK/Lin-2) in human epithelial Caco-2 cells. J. Biol. Chem. 276:9291-9296. [DOI] [PubMed] [Google Scholar]
- 29.Marin-Padura, I., S. Lostaligo, M. Schneemann, L. Williams, M. Romano, P. Fruscella, C. Panzeri, A. Stoppacciaro, L. Ruco, A. Villa, D. Simmons, and E. Dejana. 1998. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 142:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mino, A., T. Ohtsuka, E. Inoue, and Y. Takai. 2000. Membrane-associated guanylate kinase with inverted orientation (MAGI)-1/brain angiogenesis inhibitor 1-associated protein (BAP1) as a scaffolding molecule for Rap small G protein GDP/GTP exchange protein at tight junctions. Genes Cells 5:1009-1016. [DOI] [PubMed] [Google Scholar]
- 31.Mundel, P., H. W. Heid, T. M. Mundel, M. Kruger, J. Reiser, and W. Kriz. 1997. Synaptopodin: an actin-associated protein in telencephalic dendrites and renal podocytes. J. Cell Biol. 139:193-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishimura, W., T. Iizuka, S. Hirabayashi, N. Tanaka, and Y. Hata. 2000. Localization of BAI-associated protein1/membrane-associated guanylate kinase-1 at adherens junctions in normal rat kidney cells: polarized targeting mediated by the carboxyl-terminal PDZ domains. J. Cell. Physiol. 185:358-365. [DOI] [PubMed] [Google Scholar]
- 33.Ohno, H., S. Hirabayashi, T. Iizuka, H. Ohnishi, T. Fujita, and Y. Hata. 2002. Localization of p0071-interacting proteins, plakophilin-related armadillo-repeat protein-interacting protein (PAPIN) and ERBIN, in epithelial cells. Oncogene 21:7042-7049. [DOI] [PubMed] [Google Scholar]
- 34.Ozaki, H., K. Ishii, H. Horiuchi, H. Arai, T. Kawamoto, K. Okawa, A. Iwamatsu, and T. Kita. 1999. Cutting edge: combined treatment of TNF-α and IFN-γ causes redistribution of junctional adhesion molecule in human endothelial cells. J. Immunol. 163:553-557. [PubMed] [Google Scholar]
- 35.Patrie, K. M., A. J. Drescher, M. Goyal, R. C. Wiggins, and B. Margolis. 2001. The membrane-associated guanylate kinase protein MAGI-1 binds megalin and is present in glomerular podocytes. J. Am. Soc. Nephrol. 12:667-677. [DOI] [PubMed] [Google Scholar]
- 36.Patrie, K. M., A. J. Drescher, A. Welihinda, P. Mundel, and B. Margolis. 2002. Interaction of two actin-binding proteins, synaptopodin and α-actinin-4, with the tight junction protein MAGI-1. J. Biol. Chem. 277:30183-30190. [DOI] [PubMed] [Google Scholar]
- 37.Shiratsuchi, T., M. Futamura, K. Oda, H. Nishimori, Y. Nakamura, and T. Tokino. 1998. Cloning and characterization of BAI-associated protein 1: a PDZ domain-containing protein that interacts with BAI1. Biochem. Biophys. Res. Commun. 247:597-604. [DOI] [PubMed] [Google Scholar]
- 38.Shoji, H., K. Tsuchida, H. Kishi, N. Yamakawa, T. Matsuzaki, Z. Liu, T. Nakamura, and H. Sugino. 2000. Identification and characterization of a PDZ protein that interacts with activin type II receptors. J. Biol. Chem. 275:5485-5492. [DOI] [PubMed] [Google Scholar]
- 39.Tsukita, S., M. Furuse, and M. Itoh. 2001. Multifunctional strands in tight junctions. Nat. Rev. Mol. Cell Biol. 2:285-293. [DOI] [PubMed] [Google Scholar]
- 40.Wood, J. D., J. Yuan, R. L. Margolis, V. Colomer, K. Duan, J. Kushi, Z. Kaminsky, J. J. Kleiderlein, A. H. Sharp, and C. A. Ross. 1998. Atrophin-1, the DRPLA gene product, interacts with two families of WW domain-containing proteins. Mol. Cell. Neurosci. 11:149-160. [DOI] [PubMed] [Google Scholar]