Abstract
The signal transducer and activator of transcription STAT5 plays a major role in the cellular response to cytokines, but the mechanism by which it activates transcription remains poorly understood. We show here that deacetylase inhibitors (trichostatin A, suberoylanilide hydroxamic acid, and sodium butyrate) prevent induction of endogenous STAT5 target genes, implying that a deacetylase activity is required for that process. Microarray analyses revealed that this requirement is common to all STAT5 target genes. Using chromatin immunoprecipitation, we show that, following STAT5 DNA binding, deacetylase inhibitors block transcription initiation by preventing recruitment of the basal transcription machinery. This inhibition is not due to effects on histone H3 and H4 acetylation or chromatin remodeling within the promoter region. This novel mechanism of transactivation by STAT5 provides a rationale for the use of deacetylase inhibitors for therapeutic intervention in STAT5-associated cancers.
Transcription in eukaryotes is a multistep process that requires distinct multiprotein complexes. Histone acetyltransferases and histone deacetylases (HDACs) are chromatin-modifying enzymes that tightly cooperate with chromatin-remodeling enzymes to regulate accessibility of the template to DNA binding factors and RNA polymerase II (43). Beside histone acetylation, a variety of histone and nonhistone protein modifications (acetylation, phosphorylation, and methylation), as well as modification of DNA itself by methylation, regulate transcription initiation (reviewed in references 3, 7, 17, 26, 46, 52, 57, and 59). Orchestration of the events required for transcriptional activation is promoter specific. These events (recruitment of the transcription factor, chromatin modification and remodeling, and assembly of the preinitiation complex) do not follow an obligate order and are coordinated so that each step facilitates the next, eventually resulting in efficient transcription initiation (1, 10, 11, 58).
Transcriptional activation is generally correlated with histone acetylation by histone acetyltransferase complexes, and repression is correlated with deacetylation by HDAC complexes (22, 23, 34, 54, 60). However, the analysis of a variety of Saccharomyces cerevisiae promoters has recently revealed that transcriptional activation is not necessarily associated with increased histone acetylation (13). This is consistent with the observation that expression of a small subset of genes (2%) is affected in response to histone hyperacetylation induced by the deacetylase inhibitor trichostatin A (TSA) (61). In addition, genome-wide genetic studies with yeast clearly demonstrated that HDACs are required in both transcriptional activation and repression (4, 44, 63, 64, 69).
The signal transducer and activator of transcription STAT5 functions as an important downstream effector of cytokine signaling. It plays key roles in regulating immune responses, cell proliferation, differentiation, survival, and oncogenesis. STAT5 proteins are present as inactive monomers in the cytoplasm of unstimulated cells. Following cytokine stimulation, STAT5 is phosphorylated by the JAK kinases, allowing its dimerization and translocation into the nucleus where it can bind to its specific DNA binding sites. STAT5 activation is normally transient, and its inactivation by phosphatases, proteasome-dependent degradation, and a negative feedback loop mediated by proteins of the Cis family is essential for proper regulation of STAT signaling (reviewed in reference 31). Improper regulation, especially constitutive activation of STAT5 and STAT3, directly contributes to oncogenesis through stimulation of cell proliferation and prevention of apoptosis (6, 49, 53). STAT family members are known to interact with a variety of cofactors, including SMRT, p300/CBP, Nmi, MCM5, and PIAS (reviewed in reference 56). As for many other transcription factors, interaction of the C-terminal transactivation domain of STAT5 with the acetylase p300/CBP has been proposed to potentiate STAT5-mediated transactivation. MCM5, a protein involved in DNA replication, interacts with the transactivation domain of STAT1 and enhances its transcriptional activity. Nmi interacts with the coiled-coil domain of STAT5 and has been proposed to facilitate the association of STAT5 with p300/CBP, resulting in enhanced STAT5-dependent transcription. SMRT also interacts with the coiled-coil domain of STAT5 but, in contrast to Nmi, down-modulates expression of STAT5 target genes (41). This inhibitory effect of SMRT is likely to involve the recruitment of an HDAC-containing complex (14, 36). Functional cooperation between STAT5 dimers through tetramerization and with other transcription factors bound on adjacent binding sites also appears to play an important role in transactivation by STAT5 (reviewed in references 31 and 56). Despite the identification of those cofactors, the precise mechanism of transactivation by STAT5 following its binding to DNA remains poorly understood.
We show here that a deacetylase activity is required for transcription activation by STAT5. This deacetylase activity controls the proper assembly and/or stability of the basal transcription machinery. This mechanism is shared by all the STAT5 target genes investigated, therefore opening the attractive possibility of using deacetylase inhibitors for therapeutic intervention in STAT5-associated cancers.
MATERIALS AND METHODS
Cells.
The interleukin-3 (IL-3)-dependent murine pro-B-cell line Ba/F3-β was grown in RPMI 1640 containing 10% fetal bovine serum (FBS) and 10 ng of IL-3/ml. Ba/F3-β stably expresses the IL-2Rβ gene and responds to either IL-2 or IL-3 (37). The Ba/F3 wild-type and Ba/F3-1*6 cell lines stably expressing STAT5A-Flag wild type and the 1*6 constitutively active mutant have been kindly provided by Toshio Kitamura. These cells were grown in RPMI 1640 containing 10% FBS and 0.4 mg of Geneticin (Gibco-BRL)/ml, in the presence (wild type) or absence (1*6) of 10 ng of IL-3/ml, as described previously (45). The IL-2-dependent murine T-cell line CTLL-2 was grown in RPMI 1640 containing 10% FBS, 50 μM 2-mercaptoethanol, and 5 ng of recombinant human IL-2 (R&D)/ml. For cytokine stimulation, cells were washed in RPMI 1640 and rested for 6 h in RPMI 1640 containing 10% FBS before addition of cytokine (IL-2 or IL-3). For drug treatment, cells were preincubated for 30 min prior to cytokine stimulation with TSA (Sigma; 200 nM concentration in dimethyl sulfoxide [DMSO]), suberoylanilide hydroxamic acid (SAHA; a kind gift from Aton Pharma, Inc.; 10 μM concentration in DMSO), sodium butyrate (NaB; Sigma; 10 mM concentration in water), or cycloheximide (CHX; Sigma; 20 μg/ml in ethanol).
Protein analysis.
For Western blot analysis, cells were washed in phosphate-buffered saline (PBS), lysed, and immunoblotted as previously described (37). Antibodies used for immunoblotting were as follows: Cis (a kind gift from Aki Yoshimura), c-Myc (N-262) (Santa Cruz Biotechnology; sc-764), Bcl-x (Transduction Laboratories; B611220), p21 (a kind gift from Dave Parry), phospho-STAT5 (Tyr694) (New England Biolabs; catalog no. 9351), and STAT5b (C-17) (Santa Cruz Biotechnology; sc-835). For nuclear and cytosolic lysates, cells were washed in PBS, gently lysed in buffer A (10 mM HEPES [pH 7.6], 15 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 1 mM Na3VO4, 5 mM NaF, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml, and 0.5 mM phenylmethylsulfonyl fluoride) containing 0.2% NP-40, and centrifuged at 960 × g for 20 s. The supernatant was harvested (cytosolic fraction), and the nuclei were washed in buffer A containing 0.25 M sucrose and centrifuged as before. Nuclear proteins were extracted for 30 min with gentle shaking in 50 mM HEPES (pH 7.9)-400 mM KCl-0.1 mM EDTA-1 mM dithiothreitol-10% glycerol-1 mM Na3VO4-5 mM NaF-10 μg of leupeptin/ml-10 μg of aprotinin/ml-0.5 mM phenylmethylsulfonyl fluoride, and nuclear membranes were eliminated by centrifugation (15 min at 20,800 × g).
mRNA analysis.
For real-time PCR expression analysis, cells were washed in PBS, RNAs were isolated, and cDNAs were synthesized as previously described (16). Real-time PCR was performed with a GeneAmp 5700 sequence detector (Perkin-Elmer). Reactions were performed as described previously (16), with the exception that the final volume was 25 μl of SYBR Green reaction mix (Perkin-Elmer). All data were normalized to S9 cDNAs which remained unchanged upon the various drug and cytokine treatments (data not shown) (2CT S9 − CT X × 10,000). Data are expressed as relative RNA levels. The forward and reverse primers used to amplify mouse cDNAs are as follows: S9, GGGATGTTCACCACCTG and GCAAGATGAAGCTGGATTAC; Cis, CTGGACTCTAACTGCTTGTC and TAGGCAGCACCGAGTCAC; c-Myc, AACAGGAACTATGACCTCG and AGCAGCTCGAATTTCTTC; Pim-1, TCTTCTGGCAGGTGCTG and GGTAGCGAATCCACTCTG; Osm, AGATACCTGAGCCCACACAGACAG and ATCGTCCCATTCCCTGAAGACC; Bcl-x, ATGGCAGCAGTGAAGCAAGC and ACGATGCGACCCCAGTTTACTC; p21, CTGGGAGGGGACAAGAG and GCTTGGAGTGATAGAAATCTG; c-Fos, CGAAGGGAACGGAATAAGATGG and AGACCTCCAGTCAAATCCAGGG; Ier2, TAGTGATGCCGGACTGGTACC and CCTCCCCCTCCACCTCTTC; JunB, CAGCTACTTTTCGGGTCAGGG and GGCTAGCTTCAGAGATGCGC; Spp1/Osteopontin, CACTTTCACTCCAATCGTCCC and AAGCCAAGCTATCACCTCGG; 36b4, GCGTCCTGGCATTGTCTGT and GCCGCAAATGCAGATGG; Id, CGACATGAACGGCTGCTACTC and TCTCCACCTTGCTCACTTTGC; Fra-2, TCAGAGTCCTGCTCCAAGGC and GACTGGTCCCCACTGCTACTG; TCRγ-V4, ACCAAGCCTACAACTTGCTGG and TGATCGTGAACTGGAGCTGC; Dok2, TGAAGCTGCGATGGTCAGG and CTTCTTGCCAAAGGTCTGCTG; Thrombin Receptor/CF2R, GCCAACTTCACTTGCGTGG and TGGCAGGTGGTGATGTTGAG; p21, CTGGGAGGGGACAAGAG and GCTTGGAGTGATAGAAATCTG; Fatty Acid Synthase, CTGGACTCGCTCATGGGTG and CATTTCCTGAAGTTTCCGCAG; NIFK, GACAGCCAGGGTCCCACAC and CCTGCGATTTTCGCCTCTC; TDAG51/PQ, TGGTGCAGTACAAAAATCGCC and TGCCTGGTAGACTTGACCGC; MKP-1, GTGCCTATCACGCTTCTCGG and TGGTTGTCCTCCACAGGGAT; IL-4Rα, GGAGAGCTCACGGGAATCC and GCGTTTCTGCTTTTGACACG; Stra13/Clast5, GTTTCCAGACTTGTGCCCGT and TCTCATGCTTCGCCAGGTACT; Pcsk3/FUR, GCCAAGAGGGACGTGTATCAG and CCTTCACATTCAGGTCTCGCT; Spi-2.1, GGCAGTGCCCTGTTTATTGAA and GCTGGAAATCTGCTGTGAAGG; Ryk, TAGTGACGTGTGGGCCTTTG and ATGTCCACGTAGGGCGTCTG; IL-2Rα, CATAGTACCCAGTTGTCGGGC and GGCTTTGAATGTGGCATTGG; Similar to Phosphatidylinositol Binding Clathrin Assembly Protein, TACACCAACGGGCATGATAGG and GTCTCATGACAGGCTGGCTGT; Btg2/Tis21, AAGTGTCTTACCGCATCGGG and TCTTGCAGGTGAGGAGCCC; Phosphatidylinositol Transfer Protein Beta, TCAAGACCAAGAGAGGACCCC and TTGCCAGCTCCTTCTTCCAG; Cytoskeletal gamma-actin, AAGAGTTACGAGCTGCCCGAC and GAACCGCTCATTGCCAATG; Cytoskeletal beta-actin, CACTATTGGCAACGAGCGG and ATACCCAAGAAGGAAGGCTGG; DEAD/H box polypeptide 21, TTTGTGACCATGATCCTGCG and CAAACCCCCAGTTTTCCTTTG.
Chromatin immunoprecipitation (ChIP) assay.
ChIP assays were performed essentially as described previously (16), with the following modifications. Cross-linked cells (2 × 107) were resuspended in 3 ml of sodium dodecyl sulfate (SDS) buffer (100 mM NaCl, 50 mM Tris [pH 8.0], 5 mM EDTA, 0.5% SDS, and protease inhibitors) and sonicated on ice for 20 s as described previously (16). A 1.5-ml (0.5-volume) quantity of Triton buffer (100 mM NaCl, 100 mM Tris [pH 8.0], 5 mM EDTA, 5% Triton X-100, and protease inhibitors) was added, and lysates (now in immunoprecipitation [IP] buffer) were centrifuged at 1,700 × g for 10 min. The lysates were precleared for 1 h against 400 μl of blocked protein A bead slurry (prepared as described in the work of Frank et al. [16]). Seven hundred fifty microliters of precleared lysate was used per IP (3.3 × 106 cells). Fifty microliters of precleared lysate was kept as the input for the real-time PCR. IPs were performed for 3 h in the presence of the antibody, before 60 μl of blocked protein A slurry was added for 2 additional h. IP mixtures were washed successively in 1 ml of IP buffer (100 mM NaCl, 67 mM Tris [pH 8.0], 5 mM EDTA, 0.33% SDS, 1.7% Triton X-100), buffer 150 (150 mM NaCl, 20 mM Tris [pH 8.0], 5 mM EDTA, 1% Triton X-100, 0.2% SDS), buffer 500 (500 mM NaCl, 20 mM Tris [pH 8.0], 5 mM EDTA, 1% Triton X-100, 0.2% SDS), LiCl wash buffer (250 mM LiCl, 10 mM Tris [pH 8.0], 1 mM EDTA, 0.5% [wt/vol] deoxycholic acid, 0.5% NP-40), and Tris-EDTA (pH 7.5). Elution from beads, cross-link reversion, and DNA purification of IP and input samples were performed as described previously (16). DNA from IPs was resuspended in 300 μl of sterile water. DNA from input was subjected to an RNase A treatment for 30 min at 37°C, and the final volume was adjusted to 900 μl with sterile water. Real-time PCR was performed with 5 μl of DNA, as described above. IP data were normalized to input DNA (2CT input − CT IP × 0.0222 × 100), and amounts of DNA recovered in the IPs were expressed as percentages of input DNA, as described in the work of Frank et al. (16). Antibodies used for ChIP were as follows: STAT5a (L-20) and STAT5b (C-17) (Santa Cruz Biotechnology; sc-1081 and sc-835, respectively; 1.2 μg each), acetylated histone H3 (Upstate Biotechnology; catalog no. 06-599; 3 μg), acetylated histone H4 (Upstate Biotechnology; catalog no. 06-866; 3 μl), RNA polymerase II (N-20) (Santa Cruz Biotechnology, sc-899; 2 μg), and TATA-binding protein (TBP; SI-1; Santa Cruz Biotechnology, sc-273; 2 μg). The forward and reverse primers used to amplify mouse Cis genomic DNA are as follows: STAT5 binding sites (third and fourth), amplicon −184/−102, GTCCAAAGCACTAGACGCCTG and TTCCCGGAAGCCTCATCTT; CAP site amplicon −17/+55, GTTCGCACCACAGCCTTTCAGTCC and GTCCAGGGGTGCGAAGGTCAGG. Identical results were obtained for STAT5 binding with primers specific for the amplicon −256/−195 overlapping the first and second STAT5 binding sites (data not shown). The primers used to amplify Osm genomic DNA (CAP site amplicon −21/+40) are GCTGCCAGCCTGCAGGACAC and GTACTCTGGCCCGTGCCTCTCAG, and those used to amplify c-Fos genomic DNA (open reading frame amplicon +1273/+1325) are ATCGGCAGAAGGGGCAAAGTAG and CCACAAAGGTCCAGAATCGCTG.
Chromatin accessibility by real-time PCR (CHART-PCR).
Cells were washed in PBS and lysed in CHART buffer (10 mM Tris [pH 7.4], 15 mM NaCl, 60 mM KCl, 0.15 mM spermine, 0.5 mM spermidine, and protease inhibitors) containing 0.5% NP-40, and the nuclei were washed in CHART buffer. Nuclei to the number of 106 (3.3 μg of DNA) were resuspended in the recommended 1× restriction enzyme buffer (New England Biolabs) supplemented with 100 μg of bovine serum albumin/ml. Restriction enzyme accessibility assays were performed in a 200-μl final volume in the presence of 0.5 to 1 U of restriction enzyme (New England Biolabs)/μl for 1 h at 37°C. A nondigested control was included in the assay, as well as a positive control corresponding to purified genomic (naked) DNA (3.3 μg). Reactions were stopped, and genomic DNA was purified with the QIAamp DNA blood minikit (Qiagen; catalog no. 51104) according to the manufacturer's recommendations. Elution was performed with 200 μl of AE buffer (Qiagen). Real-time PCR was performed with 5 μl of DNA (40 ng), as described above. Primers overlapping the region targeted by restriction enzymes as well as primers of identical efficiencies amplifying an intact region of genomic DNA were used on each digested and nondigested sample. Restriction digest data were normalized to the intact region (2CT intact − CT cut × 100), and data were expressed as percentages of nondigested DNA (percent protection). The forward and reverse primers used to amplify mouse Cis genomic DNA are as follows. SacI (−245), amplicon −438/−195, AGAAGTAGAGGGAAGACAATCTGGTC and AACACCTTTGACAGATTTCCAAGAAC; AvaII (−184), amplicon −256/−99, CAACTCTAGGAGCTCCCGCC and CCCTTCCCGGAAGCCTCATC; SacII (−133), amplicon −184/−102, GTCCAAAGCACTAGACGCCTG and TTCCCGGAAGCCTCATCTT; AluI (−17), amplicon −133/+81, CCGCGGTTCTAGGAAGATGAGG and GGGATGGAAGGAGAAAGGAGCC. SacI data were normalized to amplicon −826/−749, AGGGCTGTCTGGGAGCTGA and TCTCTGAGTGGACCGACAGTTG; AvaII, SacII, and AluI data were normalized to amplicon +878/+944, TACCCCTTCCAACTCTGACTGAGC and TTCCCTCCAGGATGTGACTGTG.
DNA microarray hybridization.
Ba/F3-β cells were washed in PBS, total RNAs were isolated with the RNeasy Maxi kit (Qiagen), and poly(A) mRNAs were isolated from 500 to 800 μg of total RNA with Oligotex resin (Qiagen). cDNA labeling and microarray hybridization were performed at Incyte (Fremont, Calif.) with their proprietary technology, as previously described (70). The arrays contained approximately 16,700 mouse cDNA elements (Incyte Genomics, Palo Alto, Calif., and Schering-Plough Research Institute, Kenilworth, N.J., unpublished data), one half corresponding to known genes and the other half corresponding to unknown expressed sequence tags. Most of the known genes are disease-related genes of the immunology area, as well as other oncogenes or cell signaling, proliferation, differentiation, or transcription-related genes. Most of the cDNAs are present in duplicate or triplicate on the arrays. The 16,700 cDNA elements were hybridized with five probe pairs: unstimulated cells in the absence versus the presence of TSA, cells stimulated with IL-3 for 30 min in the absence versus the presence of TSA, cells stimulated with IL-3 for 2 h in the absence versus the presence of TSA, cells not stimulated versus being stimulated with IL-3 for 30 min, and cells not stimulated versus being stimulated with IL-3 for 2 h. Genes were chosen for further study by being upregulated by IL-3 at 30 min or 2 h by at least threefold compared to unstimulated cells. In total, 89 cDNA elements were identified, representing a total of 40 genes (see text and Table 1).
TABLE 1.
DNA microarray analysis of gene expression in IL-3-stimulated Ba/F3-β cells in response to TSA
| Protein or gene (gene designation)a | Accession no. | Relative microarray expression (fold) after 0.5 and 2 h of IL-3 stimulationc
|
STAT5 target genesd | Group | |||
|---|---|---|---|---|---|---|---|
| −TSA
|
+TSA
|
||||||
| 0.5 h | 2 h | 0.5 h | 2 h | ||||
| Downregulated by TSA | |||||||
| Cytokine-inducible SH2-containing protein (Cis)b | D31943 | 10.1 | 8 | 3 | 2.3 | + | A |
| Myelocytomastosis oncogene (c-Myc)b | X01023 | 8.4 | 5.5 | 1.3 | 1.2 | + | A |
| Oncostatin M (Osm)b | D31942 | 8.1 | 2.5 | 0.9 | 1.2 | + | A |
| Helix-loop-helix DNA binding protein (Id) | M31885 | 5 | 9.3 | 9.2 | 4.9 | + | A |
| p21 (WAF1/CIP1)b | NM_007669 | 4.1 | 1.1 | 2.3 | 1.8 | + | A |
| Fos-like antigen 2 (FosL2/Fra-2) | NM_008037 | 3.8 | 1.2 | 1.1 | 0.7 | + | A |
| Serine protease inhibitor 2-1 (Spi-2.1)b | M64085 | 3 | 0.8 | 1.4 | 1.9 | + | A |
| Fatty acid synthase | AF127033 | 1.7 | 4.9 | 2.6 | 3.8 | + | A |
| IL-2 receptor alphab | NM_008367 | 1.1 | 4 | 0.9 | 1.5 | + | A |
| NIFK (nucleolar RNA binding protein) | AB056870 | 1.5 | 3.5 | 2.1 | 2 | + | A |
| T-cell receptor gamma variable 4 (TCRγ-V4)b | NM_011558 | 1.3 | 3.4 | 0.5 | 0.3 | + | A |
| Thrombin receptor (Cf2r) | U36757 | 2.1 | 3.4 | 1.2 | 1.9 | + | A |
| Downstream of tyrosine kinase 2 (dok2) | NM_010071 | 1.3 | 3 | 1 | 0.5 | + | A |
| TDAG51/PQ | NM_009344 | 5.2 | 2.8 | 2.6 | 2.3 | − | C |
| Mitogen-activated protein kinase phosphatase 1 (MKP-1) | X61940 | 4 | 1 | 0.9 | 1 | − | C |
| IL-4 receptor alpha | M29854 | 3.4 | 6.5 | 2.6 | 3.6 | − | C |
| Stimulated by retinoic acid 13 (stra13/clast5) | AF010305 | 3.2 | 3.7 | 0.9 | 0.7 | − | C |
| Proprotein convertase subtilisin/kexin, type 3 (Furin) | NM_011046 | 2.2 | 4 | 1.5 | 1.4 | − | C |
| Receptor-like tyrosine kinase (ryk)e | M98547 | 1.7 | 6.2 | 1.2 | 1 | ? | D |
| Similar to phosphatidylinositol binding clathrin assembly proteine | BC021491 | 1.3 | 4.1 | 1.7 | 1.6 | ? | D |
| Unaffected by TSA | |||||||
| Immediate-early response 2 (ier2) | NM_010499 | 7.4 | 2.4 | 10.4 | 5.3 | − | B |
| B-cell translocation gene 2 (btg2/tis21) | NM_007570 | 5.2 | 0.6 | 5.7 | 2 | − | B |
| Phosphatidylinositol transfer protein beta | U46934 | 4.6 | 4 | 6.9 | 6 | − | B |
| JunB | XM_125115 | 4.4 | 2.2 | 6.2 | 2 | − | B |
| Cytoskeletal gamma-actin | M21495 | 4.1 | 3.7 | 3.1 | 3.1 | − | B |
| Cytoskeletal beta-actin | X03672 | 3.2 | 3.4 | 2.1 | 2.4 | − | B |
| DEAD/H box polypeptide 21 (RNA helicase II/Gu) | NM_019553 | 1.2 | 4.1 | 1.9 | 4.5 | − | B |
| Secreted phosphoprotein 1 (Spp1/Osteopontin) | NM_009263 | 1.3 | 3.3 | 1.4 | 2.8 | − | B |
Genes induced 3- to 10-fold by IL-3 at 30 min and/or 2 h (n = 28). Twenty genes were downregulated 1.8- to 10.1-fold, by TSA, and eight genes were unaffected by TSA.
Known STAT5 target genes from the literature.
Induction relative to that of untreated unstimulated cells. No significant differences were found between the untreated and the TSA-treated unstimulated cells.
STAT5 targets were identified by real-time PCR from unstimulated and IL-3-stimulated Ba/F3-1*6 cells that express a constitutively active form of STAT5A (see text).
Genes that lost IL-3 inducibility in the Ba/F3-1*6 cell line and thus could not be identified as STAT5 or non-STAT5 targets.
RESULTS
Cytokine induction of STAT5 target genes is inhibited by TSA.
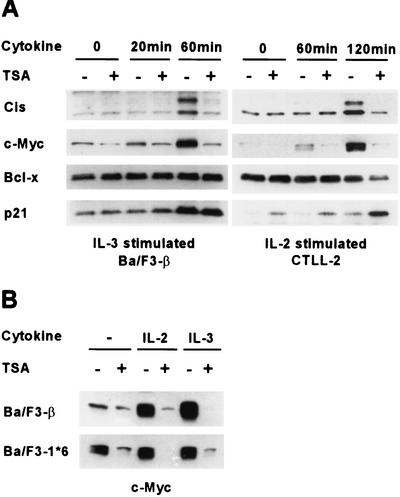
To investigate the role of protein acetylation in the regulation of cytokine-responsive genes, the effect of the deacetylase inhibitor TSA (68) on the cytokine response of murine B and T lymphocytes was examined. Ba/F3-β, an IL-3- and IL-2-dependent murine pro-B-cell line, was pretreated with TSA at a 200 nM concentration for 30 min prior to IL-3 stimulation. Cytokine induction of the murine Cis protein, a STAT5-dependent gene product (35), was totally abolished in TSA-treated cells (Fig. 1A). c-Myc induction showed similar inhibition by TSA. The same effects were observed upon IL-2 stimulation of the murine T-cell line CTLL-2 (Fig. 1A) and of Ba/F3-β (Fig. 1B and data not shown). By contrast, the level of Bcl-x protein, which is barely increased upon cytokine stimulation, was unaffected by TSA in Ba/F3-β and slightly decreased at later time points in CTLL-2 (Fig. 1A). Expression of p21 was previously shown to be activated by TSA and other deacetylase inhibitors in various cell lines (21, 42, 51, 66). We confirmed that p21 is upregulated by TSA in CTLL-2, while p21 protein levels remained unaffected in Ba/F3-β (Fig. 1A).
FIG. 1.
Cytokine induction of Cis and c-Myc proteins is abolished in TSA-treated murine lymphocytes. (A) Effects of TSA on protein expression in cytokine-stimulated murine B and T lymphocytes. Ba/F3-β and CTLL-2 were stimulated with IL-3 and IL-2, respectively, for the indicated times, as described in Materials and Methods. Cells were pretreated with 200 nM TSA (+) or DMSO (−) for 30 min prior to cytokine stimulation. Protein lysates were subjected to a Western blot analysis with the indicated antibodies. (B) c-Myc constitutive expression in a STAT5Aca cell line is repressed by TSA. Ba/F3-β and Ba/F3-1*6 were stimulated with IL-2 or IL-3 for 2 h in the presence of 200 nM TSA or DMSO, as described for panel A. Protein lysates were subjected to a Western blot analysis with a polyclonal antibody specific for c-Myc.
While previous data demonstrated that expression of Cis is dependent on STAT5 (35), expression of c-Myc can be regulated by various transcription factors, including STAT family members (24, 32). To evaluate whether cytokine induction of c-Myc in Ba/F3-β and CTLL-2 and its inhibition by TSA were STAT5 dependent, c-Myc protein levels were analyzed in Ba/F3-1*6 cells, which stably express a constitutively active form of STAT5A. As previously described (45), c-Myc was constitutively expressed in unstimulated Ba/F3-1*6, supporting the idea that its expression is mediated by STAT5 (Fig. 1B). This constitutive expression of c-Myc was strongly inhibited by TSA, suggesting that TSA can block STAT5-mediated activation of c-Myc (Fig. 1B).
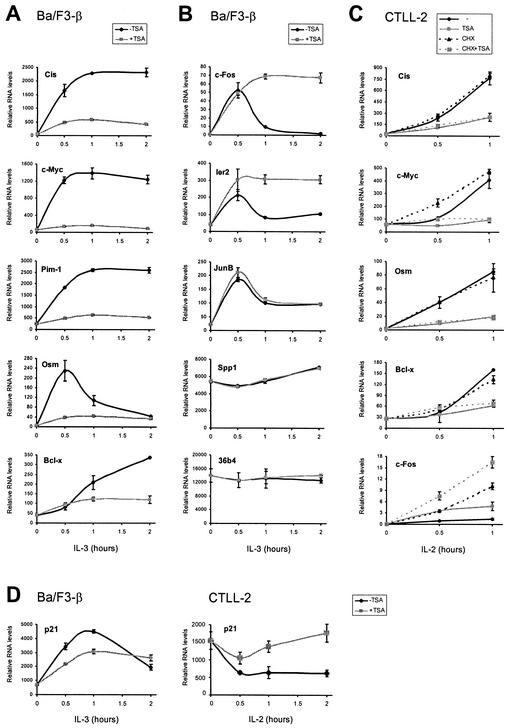
To determine whether the inhibitory effect of TSA occurred at the RNA level and could affect other STAT5 target genes, RNAs were isolated from IL-3-stimulated Ba/F3-β and IL-2-stimulated CTLL-2 cells. Following a reverse transcription reaction, the cDNAs were analyzed by real-time PCR. In both cell lines, cytokine induction of all STAT5 targets tested, including Cis, c-Myc, Pim-1, Osm, and Bcl-x, was abolished by TSA (Fig. 2A and C and data not shown). Genes like Cis, c-Myc, Pim-1, and Osm, which are rapidly induced by cytokines, especially by IL-3, responded immediately to the inhibitory effect of TSA. Bcl-x, which exhibits delayed cytokine inducibility, showed significant inhibition by TSA only at a later time point (2 h), in agreement with protein data (Fig. 1A). In contrast to STAT5 target genes, expression of other cytokine-inducible genes (c-Fos, Ier2, JunB, and Spp1/Osteopontin) or of a housekeeping gene (36b4) was either unaffected or in some cases upregulated by TSA (Fig. 2B and C and data not shown). While certain genes (Osm and c-Fos) showed different expression patterns in Ba/F3-β and CTLL-2 in response to cytokine, the effects of TSA were similar in the two cell lines (Fig. 2A to C). One exception is p21. p21 has been shown elsewhere to be a STAT5 target gene in Ba/F3 cells (45). Accordingly, p21 RNA levels decreased upon TSA treatment, although the apparent inhibition was partial (37 and 32% at 30 min and 1 h poststimulation, respectively) (Fig. 2D). This weak inhibition at the RNA level probably explains the absence of effect observed at the protein level (Fig. 1A). In contrast, the p21 RNA level decreased upon IL-2 stimulation in CTLL-2 (Fig. 2D), indicating that it is not a STAT5 target in those cells. Interestingly, TSA treatment resulted in an increased p21 RNA level, in agreement with protein data (Fig. 1A) and previous observations in other systems (21, 42, 51, 66).
FIG. 2.
TSA inhibits induction of STAT5 target genes at the RNA level, through a primary effect. TSA specifically inhibits transcriptional induction of STAT5 target genes. (A and B) Ba/F3-β cells were stimulated with IL-3 in the absence or presence of TSA, as described in the legend to Fig. 1. mRNA levels were analyzed by real-time PCR, as described in Materials and Methods, with primers specific for STAT5 target genes (A) or control genes (B). (C) Inhibition of transcription by TSA does not require de novo protein synthesis. CTLL-2 cells were stimulated with IL-2 in the absence or presence of TSA and/or CHX, as described in Materials and Methods. mRNA levels were analyzed by real-time PCR as described above. (D) p21 expression is differentially affected by TSA in Ba/F3-β and CTLL-2 cells. Cells were treated, and mRNA levels were analyzed with primers specific for mouse p21, as described above.
To address whether the inhibitory effect of TSA required de novo protein synthesis, IL-2-stimulated CTLL-2 cells were pretreated with CHX. Inhibition of STAT5 target genes by TSA was not affected by CHX treatment (Fig. 2C). In contrast, expression of the control gene c-Fos was upregulated in CHX-treated cells, suggesting that it is controlled by a de novo-synthesized repressor (Fig. 2C). These data show that TSA prevents induction of STAT5 target genes at the RNA level, without requiring protein neosynthesis.
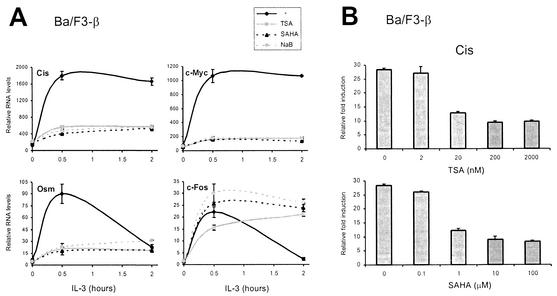
To verify the specificity of the inhibitory effect of TSA and further test the hypothesis that a deacetylase activity is required for transcriptional activation by STAT5, the effects of SAHA, a potent deacetylase inhibitor structurally related to TSA (50), and NaB, a structurally unrelated deacetylase inhibitor (28), were tested. Ba/F3-β cells were pretreated with 200 nM TSA, 10 μM SAHA, or 10 mM NaB prior to IL-3 stimulation, and RNA levels were analyzed by real-time PCR as described above. Treatment with all three deacetylase inhibitors prevented induction of STAT5 target genes to a similar extent, while control genes were unaffected or slightly upregulated as observed before (Fig. 3A and data not shown). Full inhibition of gene expression was reached with concentrations as low as 20 nM TSA and 1 μM SAHA (Fig. 3B and data not shown). These results therefore strongly suggest that a deacetylase activity is required for STAT5-dependent transcription.
FIG. 3.
A deacetylase activity is required for cytokine induction of STAT5 target genes. (A) Three unrelated inhibitors of deacetylases block induction of STAT5 target genes. Ba/F3-β cells were stimulated with IL-3 in the presence of 200 nM TSA, 10 μM SAHA, 10 mM NaB, or DMSO (−), and mRNA levels were analyzed by real-time PCR, as described for Fig. 2. (B) Titration of TSA and SAHA. Ba/F3-β cells were stimulated with IL-3 for 30 min in the presence of increasing concentrations of TSA or SAHA, and mRNA levels were analyzed by real-time PCR, as described above. Data are expressed as the fold induction relative to untreated unstimulated cells.
TSA blocks induction of STAT5 target genes at the transcription initiation level.
We next examined which step of transactivation by STAT5 is inhibited by TSA. STAT5 is found in the cytoplasm of unstimulated cells. Upon cytokine stimulation, it becomes phosphorylated and translocates into the nucleus. Hence, the phosphorylation status and cellular localization of STAT5 were compared in TSA-treated and untreated CTLL-2 cells stimulated with IL-2. A phosphospecific STAT5 antibody detected a signal of equal intensity in the nucleus of stimulated cells whether or not those cells were pretreated with TSA (Fig. 4A, upper panel). Similarly, the total level of STAT5 protein in the cell was not significantly changed upon TSA treatment, as was the amount of STAT5 translocated into the nucleus in response to cytokine (Fig. 4A, lower panel). Identical results were obtained in Ba/F3-β cells (data not shown). We then monitored STAT5 DNA binding activity by electrophoretic mobility shift assay, using either a consensus STAT5 binding site from the β-casein promoter or STAT5 binding sites from the murine Cis promoter. These assays revealed no changes upon TSA treatment (data not shown). To directly investigate STAT5 DNA binding in vivo, ChIP assays were performed in IL-3-stimulated Ba/F3-β cells. Antibodies specific for STAT5a and STAT5b were used to immunoprecipitate cross-linked chromatin. Purified genomic DNA was then subjected to real-time PCR with primers amplifying regions −184/−102 (Fig. 4C) and −256/−195 (data not shown) of the Cis promoter, which overlap four STAT5 binding sites (Fig. 4B) that are essential for induction by cytokines (35). Within 5 min of IL-3 stimulation, STAT5 was associated with the Cis promoter (Fig. 4C). This rapid kinetics is very similar to that recently reported for the TCRγ locus in response to IL-7 (67). In cells pretreated with TSA, STAT5 recruitment was similar to that of untreated cells (Fig. 4C), in agreement with the in vitro data. Thus, neither activation of STAT5 nor its binding to DNA is affected by TSA.
FIG. 4.
TSA blocks transcription initiation by preventing recruitment of the basal transcription machinery to the Cis promoter. (A) TSA does not interfere with STAT5 phosphorylation or its nuclear translocation upon cytokine stimulation. CTLL-2 cells were stimulated with IL-2 for 1 h in the absence or presence of TSA, as described for Fig. 2. Nuclear and cytosolic lysates were subjected to Western blot analysis with antibodies specific for phospho-STAT5 and STAT5b, as described in Materials and Methods. Identical results were obtained with a STAT5a-specific antibody (data not shown). (B) Structure of the murine Cis promoter. Gray boxes represent STAT5 binding sites, together with their positions relative to the transcription start site or CAP site (+1). Positions and sizes of the amplicons analyzed by ChIP in panels C to F are shown. (C to F) TSA does not interfere with STAT5 DNA binding but prevents histone acetylation and recruitment of TBP and RNA polymerase II at the CAP site of the Cis promoter. Ba/F3-β cells were stimulated with IL-3 in the absence or presence of TSA, as described for Fig. 1. At the times indicated, cells were harvested and analyzed by ChIP, with antibodies specific for STAT5a and -b (STAT5) (C), RNA polymerase II (RNA Pol II) and TBP (E), acetylated histone H3 (Ac-H3) and histone H4 (Ac-H4) (F), or no antibody as a control (D), as described in Materials and Methods. DNA samples were analyzed by real-time PCR, with primers amplifying the −184/−102 region (STAT5) or the −17/+55 CAP site region (RNA Pol II, TBP, Ac-H3, Ac-H4, and No Antibody). Identical results were obtained for STAT5 binding when the −256/−195 region was amplified (data not shown). (G and H) Effect of TSA on histone acetylation at the Osm and c-Fos genes. Ba/F3-β cells were treated, and histone acetylation was analyzed by ChIP, as described above, with primers specific for the CAP site (−21/+40, Osm) or the open reading frame (+1273/+1325, c-Fos). Histone acetylation in panels F to H is represented at identical scales for better comparison.
Given that TSA did not affect activation of STAT5 itself, we tested its effect on in vivo recruitment of components of the transcription machinery (TBP and RNA polymerase II) to the CAP site of the Cis promoter. As revealed by ChIP analysis, TBP and RNA polymerase II were progressively recruited to the CAP site (−17/+55) following IL-3 stimulation (Fig. 4E), while the no-antibody control yielded no enrichment (Fig. 4D). RNA polymerase II levels at the CAP site peaked at 20 min poststimulation. In TSA-treated cells, recruitment of TBP and RNA polymerase II to the Cis promoter was abolished (Fig. 4E). Since TSA is an HDAC inhibitor, we monitored changes in histone acetylation. ChIP assays were performed with antibodies specific to acetylated forms of histones H3 and H4. High levels of histone H3 and H4 acetylation were detected along the Cis promoter, both in unstimulated and in stimulated cells (Fig. 4F and data not shown). Concomitant with STAT5 binding, transient hyperacetylation of both histone H3 and histone H4, peaking at 5 min, was detected at the CAP site (−17/+55) (Fig. 4F). This hyperacetylation was localized at the CAP site, and histone acetylation in other regions of the promoter remained unchanged (data not shown). Unexpectedly, the histone H3 and H4 acetylation peak detected at the CAP site was absent from TSA-treated cells (Fig. 4F). In addition, global histone acetylation along the Cis promoter was not increased following TSA treatment and remained essentially at the level observed in unstimulated cells or slightly below (Fig. 4F and data not shown). In contrast, histone acetylation at the Osm gene (Fig. 4G), another STAT5 target, or at the control gene c-Fos (Fig. 4H) was increased upon TSA treatment, indicating that TSA was acting through inhibition of an HDAC activity at those sites. In addition, the acetylation peak observed at the CAP site of the Cis gene was not detected on the Osm gene (Fig. 4G), suggesting that it is specific for Cis and therefore may not be a common feature among STAT5 target genes. Since the histone acetylation pattern and the effect of TSA on histone acetylation are distinct on Cis and Osm STAT5 target genes, this suggests that the inhibitory effect of TSA on STAT5-mediated transcription occurs independently of histone acetylation. In addition, it further suggests that the disappearance of the acetylation peak on the Cis promoter upon TSA treatment is a consequence of an upstream inhibitory event, rather than the cause of transcription inhibition by TSA.
Interestingly, the same effects of TSA on STAT5, TBP, and RNA polymerase II recruitment were detected on the Osm promoter (data not shown), confirming that the inhibitory effect of TSA occurs downstream of STAT5 binding to the promoter and results in preventing assembly of the transcription machinery.
To determine whether TSA is inhibiting a process required for the preinitiation step of transcription or for the following initiation and reinitiation events, Ba/F3-β cells were stimulated with IL-3 for 30 min before TSA was added. Cis RNA levels as monitored by real-time PCR rapidly decreased following addition of TSA (Fig. 5A). At the same time, TBP and RNA polymerase II rapidly dissociated from the Cis promoter, while STAT5 binding and histone H3 and H4 acetylation patterns remained unchanged, as monitored by ChIP (Fig. 5B and data not shown). These observations suggest that TSA prevents reinitiation events by preventing reloading of the transcription machinery. In addition, the observations that TSA can inhibit transcription at a time following the acetylation peak and that histone acetylation is not affected by TSA further support the idea that the inhibitory effect of TSA does not target histone acetylation. Altogether, our data demonstrate that TSA inhibits transcription initiation of STAT5 target genes by preventing recruitment of the basal transcription machinery to the promoter and suggest that this effect is independent of histone acetylation.
FIG. 5.
TSA inhibits transcription reinitiation by preventing reloading of the transcription machinery. (A) TSA rapidly inhibits Cis transcription in cytokine-stimulated cells. Ba/F3-β cells were stimulated with IL-3 in the absence of TSA for 30 min before either DMSO (−TSA) or 200 nM TSA (+TSA) was added to the cells (indicated by an arrowhead). Cells were harvested at the indicated times, and Cis mRNA levels were analyzed by real-time PCR. (B) TBP and RNA polymerase II are rapidly released from the Cis promoter upon TSA treatment of cytokine-stimulated cells. Ba/F3-β cells were treated as described for panel A, and cells were harvested at the indicated times and analyzed by ChIP, as described for Fig. 4C to F.
Cytokine-induced chromatin remodeling at the Cis promoter is not affected by TSA.
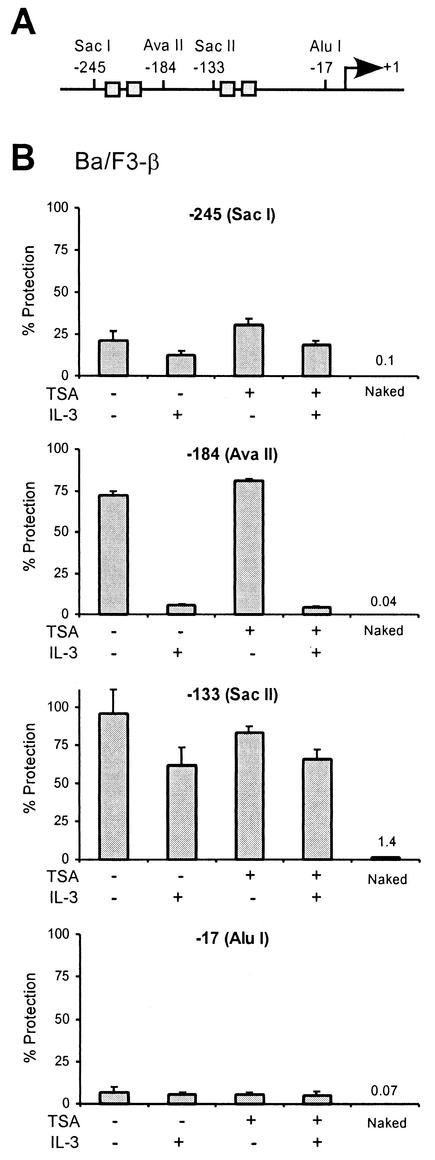
Chromatin remodeling is an essential step during transcription regulation. In concert with covalent histone modification (acetylation, phosphorylation, and methylation), it controls accessibility to DNA binding factors (43). Multiple ATP-dependent chromatin-remodeling complexes have been described. Among them, NuRD (Mi-2) is an HDAC-containing complex, the deacetylase activity of which is required for chromatin remodeling (reviewed in references 2 and 43). This raises the possibility that TSA might interfere with chromatin remodeling at STAT5-targeted promoters and hence inhibit assembly of the transcription machinery. We investigated this possibility by performing chromatin accessibility by real-time PCR (CHART-PCR) assays (48). Nuclei from unstimulated and IL-3-stimulated Ba/F3-β cells were incubated with restriction enzymes cutting at various positions along the Cis promoter, in the vicinity of the STAT5 binding sites and the transcription start site (Fig. 6A). Genomic DNA was then isolated and analyzed by real-time PCR with primers amplifying the region targeted by the enzyme. Accessibility to restriction digest is measured as the percentage of undigested DNA (percent protection) (Fig. 6B). The region of the Cis promoter located upstream of the first two STAT5 binding sites (−245), as well as the region located just upstream of the CAP site (−17), was accessible to restriction digest, whether or not cells were stimulated with IL-3. In contrast, the region located upstream of the next two STAT5 binding sites (−133) was found to be protected from restriction digest both in unstimulated and in IL-3-stimulated cells, although accessibility slightly increased upon IL-3 stimulation. This suggests that this region of the promoter is masked by a nucleosome in both unstimulated and stimulated cells. Interestingly, the region situated upstream (−184) was protected from restriction digest in unstimulated cells but became clearly accessible upon IL-3 stimulation (Fig. 6B), indicating that a chromatin remodeling event occurs downstream of the first two STAT5 binding sites. When CHART-PCR was performed on TSA-treated cells, similar accessibility patterns were observed (Fig. 6B). Therefore, TSA does not appear to interfere with chromatin remodeling at the Cis promoter.
FIG. 6.
TSA does not affect chromatin remodeling on the Cis promoter. (A) Structure of the Cis promoter, indicating the positions of the restriction sites analyzed for panel B. Gray boxes represent STAT5 binding sites. (B) IL-3-mediated chromatin remodeling on the Cis promoter is not affected by TSA. Ba/F3-β cells were stimulated with IL-3 for 30 min in the absence or presence of 200 nM TSA, as described for Fig. 1. Unstimulated and stimulated cells were harvested, nuclei were prepared, and chromatin accessibility was analyzed by CHART-PCR, as described in Materials and Methods. Purified genomic (naked) DNA was included in a parallel reaction as a control for the cutting efficiency of the restriction enzyme. Data are expressed as the percentages of nondigested DNA (% Protection).
The requirement for a deacetylase activity is shared by all STAT5 target genes.
We have shown that a deacetylase activity is required for transcription activation by STAT5. To address whether this requirement was general among all STAT5 target genes and whether other cytokine-regulated genes might also require a deacetylase for their activation, microarray analyses were performed. Ba/F3-β cells were stimulated with IL-3 for 30 min and 2 h in the absence and presence of 200 nM TSA. Arrays containing 16,700 cDNA elements were hybridized with five probe pairs (see Materials and Methods). Twenty-nine known genes and 11 expressed sequence tags were upregulated at least threefold by IL-3 at 30 min or 2 h poststimulation. Of the known genes, 28 were validated by real-time PCR and are shown in Table 1. Of those, 20 were inhibited by TSA and 8 were considered unaffected by TSA (less than twofold variation up or down). These data were confirmed by real-time PCR (data not shown). Of the IL-3-inducible genes inhibited by TSA, seven are known STAT5 targets (Table 1), among which Cis, c-Myc, Osm, and p21 had already been identified in this study (Fig. 1 and 2). In contrast, of the eight IL-3-inducible genes unaffected by TSA, none were known STAT5 targets (Table 1).
To test the specificity and better evaluate the global effect of TSA on gene expression, we undertook the identification of the STAT5 target genes among the IL-3-inducible genes isolated on the array. Ba/F3-1*6 cells which stably express a constitutively active form of STAT5A (45) were used to analyze expression of these genes by real-time PCR. It was previously shown that STAT5 target genes are upregulated in response to IL-3 or even constitutively expressed in unstimulated Ba/F3-1*6 cells, compared to Ba/F3 wild type cells stably expressing the wild-type form of STAT5A, whereas STAT5-independent genes remain unaffected (45). This experimental system allowed us to confirm the known STAT5 target genes and to identify six additional putative STAT5 targets (Id, Fra-2, Dok2, Thrombin receptor/Cf2r, Fatty acid synthase, and NIFK) (Table 1 and data not shown). All 13 known and putative STAT5 targets were inhibited by TSA (Table 1, group A). Genes unaffected by TSA were confirmed as non-STAT5 targets (Table 1, group B, and data not shown). Five genes inhibited by TSA were not putative STAT5 targets based on our criteria (TDAG51, MKP-1, IL-4Rα, Stra-13, and Furin) (Table 1, group C, and data not shown), suggesting that pathways other than STAT5 might also require a deacetylase activity for transcriptional activation. Finally, two genes could not be categorized as their expression was lost in the Ba/F3-1*6 cells (Table 1, group D). These expression data thus demonstrate that all the STAT5 target genes identified are also targeted for inhibition by TSA. Taken together, our present data show that transcriptional activation of all STAT5 target genes analyzed requires a deacetylase activity, which controls recruitment of the basal transcription apparatus.
DISCUSSION
We have shown that a deacetylase activity is required for transcriptional activation by STAT5 and that this requirement is common to all STAT5 target genes tested. We further demonstrate that the function of the putative deacetylase is to control proper assembly of the basal transcription machinery and that this function is independent of histone acetylation.
Analysis of cDNA microarrays revealed that the requirement for a deacetylase activity during transcription activation is shared by all cytokine-induced STAT5 target genes. This requirement was found in all cell lines analyzed, including IL-3- and IL-2-stimulated murine B and T cells, as shown in Results, but also in an IL-3-dependent murine myeloid cell line and IL-2-stimulated human peripheral blood lymphocytes (data not shown). Moreover, serum-mediated induction of c-Myc in Rat1 fibroblasts was also inhibited by TSA (data not shown). As STAT3 is the main STAT molecule induced in those cells in response to serum, this raises the possibility that transactivation by STAT3 might also involve a deacetylase activity. Our microarray analysis also revealed some STAT5-independent genes that were inhibited by TSA (Table 1, group C). Among those genes, TDAG51 and MKP-1 are Ras/Raf-responsive genes (5, 18). Thus, the requirement of a deacetylase activity for transactivation might be a general mechanism utilized by STAT5 and other signaling pathways.
Unexpectedly, very few genes were upregulated by TSA in our microarray analysis. This is probably due to the short time course of cytokine stimulation and TSA treatment performed here (up to 2 h), in comparison to previous work. This is particularly clear for a gene like p21 that was shown to be upregulated by deacetylase inhibitors in numerous studies (21, 42, 51, 66). We found that p21 is moderately upregulated by TSA in T cells (Fig. 2D). In B cells in contrast, where it is induced by STAT5 (Table 1), p21 is initially partially inhibited by TSA (37 and 44% by real-time PCR and microarray, respectively; Fig. 2D and Table 1). At 2 h poststimulation, when the p21 RNA level normally goes down, a slight positive effect of TSA starts to be detected (Fig. 2D and Table 1). We might predict the p21 RNA levels to be further increased at later time points. The intermediate level of inhibition by TSA observed in B cells might be the result of contradictory signals received by the p21 promoter: the inhibitory effect on STAT5 transactivation and the stimulatory effect mediated through histone hyperacetylation.
Deacetylase function has previously been associated with transcriptional repression, through deacetylation of histones. HDACs can bind in a nontargeted manner to DNA as part of the histone-binding SIN3 and NuRD complexes, to promote global chromatin repression. HDACs are also recruited in a targeted manner to promoters by DNA binding factors and corepressors, through SIN3- and NuRD-dependent as well as independent mechanisms. HDACs can also be recruited to promoters by DNA methylases and methyl-CpG binding proteins (reviewed in references 2, 7, 12, and 52). In this context, our finding that deacetylase inhibitors can abolish transcriptional activation by STAT5 was unexpected. However, multiple genome-wide studies have suggested that HDACs are also involved in transcriptional activation (4, 44, 63, 64, 69). More recently examples of genes downregulated by HDAC inhibitors, including some IL-2-inducible genes, have been reported (25, 27, 29, 55, 62, 65), but it remained unclear from these studies whether it was the result of a direct effect.
Our data demonstrate that the requirement for a deacetylase activity lies downstream of STAT5 activation, nuclear translocation, and DNA binding. Indeed, STAT5 phosphorylation was not affected by TSA and STAT5 was recruited to the Cis promoter within minutes of cytokine stimulation. Instead, TSA prevented the recruitment of components of the basal transcription machinery, leading to inhibition of transcription initiation. Histone H3 and H4 acetylation at the Cis and Osm promoters, two STAT5 targets, was differentially affected by TSA, suggesting that inhibition by TSA does not involve histone acetylation. Additional observations support the hypothesis that TSA does not target histones but rather another factor. First, chromatin remodeling at the Cis promoter was not affected by TSA. Second, inhibition by TSA was recapitulated when the Cis promoter was taken out of its natural context, by use of a reporter construct (data not shown), arguing against an essential role of histone acetylation or nucleosomal organization in STAT5-mediated transcription. Third, the rapid kinetics of TSA action on STAT5 response (within minutes of treatment) is inconsistent with the delayed effect of TSA-mediated histone hyperacetylation on gene expression (21, 42, 51, 66), rather suggesting an alternate mechanism involving deacetylation of an unrelated factor. This unidentified factor could be STAT5 itself, another transcription factor, or any other component of the initiation complex. It is tempting to propose that acetylation of this factor might disrupt its interaction with a crucial component of the transcription apparatus. Such a loss of protein interaction upon acetylation has been proposed elsewhere for transcriptional attenuation of estrogen-regulated promoters (8, 9) and transcription downregulation from the beta interferon enhanceosome (38, 39).
Recently, SMRT was shown to interact with STAT5 and to down-modulate expression of STAT5 target genes (41), probably through recruitment of an HDAC-containing complex (15, 19, 20, 30, 40). Interestingly, overexpression of SMRT does not abolish the initial induction of Cis or Osm but rather accelerates their subsequent downregulation. In agreement with a role of SMRT at a later time following cytokine stimulation, an SMRT-STAT5 interaction was detected several hours after stimulation (41). In addition, the constitutively active form of STAT5A expressed in the Ba/F3-1*6 cell line has a mutation in the coiled-coil domain (*6) that is sufficient to abolish its interaction with SMRT in a two-hybrid assay (41) and yet was still sensitive to TSA (Fig. 1B and data not shown). It is therefore unlikely that the rapid inhibition of the STAT5 responses by TSA reported here targets an HDAC complex recruited by STAT5 through SMRT. At least 11 TSA-sensitive deacetylases have been identified to date (HDAC1 to HDAC11). Our attempts to identify the deacetylase involved by knocking down individual deacetylases by a short interfering RNA-mediated approach have so far failed.
With the ever-growing understanding of the molecular mechanisms of transcription regulation, the characterization of the human genome, and its aberrations in cancer, the concept of transcription therapy for cancer has become more attractive over the past few years (47). So far, deacetylase inhibitors represent the most promising cancer drugs due to their strong potency in inducing growth arrest, differentiation, or apoptosis. Butyrates are already in use in the clinic, while the new generation of hydroxamic acid-based deacetylase inhibitors such as SAHA or pyroxamide are in clinical trials (33, 47). It has been proposed elsewhere that these drugs exert their effects through upregulation of gene expression, as shown for p21 (21, 42, 51, 66). However, our data suggest that downregulation of STAT5 target genes may be as important for the clinical effects of these compounds.
The functions of STAT family members of transcription factors, especially STAT3 and STAT5, have been directly associated with oncogenesis. Their constitutive activation directly contributes to the development and progression of many blood and solid tumors in humans, and strategies are under way to target STAT5 and STAT3 activity (6, 49, 53). Thus, our finding that deacetylase inhibitors can specifically block the STAT5 pathway and hence downregulate expression of STAT5 target genes, such as c-Myc, has wide implications in the immune-mediated disease area and validates the use of deacetylase inhibitors as a strategy for therapeutic intervention in STAT5-associated cancers. Identification of the deacetylase(s) involved and of its target substrate(s) will help to better elucidate the mechanism of transactivation by STAT5. This will also provide the possibility of designing a more specific deacetylase inhibitor, to selectively target STAT5 activation in relevant cancers.
ADDENDUM
While the manuscript was under revision, Xu et al. showed that STAT5-mediated transcription of Id-1 involves recruitment of HDAC1 by STAT5 and deacetylation of C/EBPβ (66a).
Acknowledgments
We are very grateful to Emma Lees, Dave Parry, and Joachim Griesenbeck for thoughtful discussions and helpful comments on the manuscript. We gratefully thank Aton Pharma, Inc., for generously providing SAHA. We thank Marianne Schroeder, Scott Frank, and Terri McClanahan and her group for their guidance with real-time PCR, ChIP, and microarray assays, respectively.
DNAX Research Institute is fully supported by Schering-Plough Corporation.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Ahringer, J. 2000. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 16:351-356. [DOI] [PubMed] [Google Scholar]
- 3.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, B. E., J. K. Tong, and S. L. Schreiber. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97:13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brondello, J. M., A. Brunet, J. Pouyssegur, and F. R. McKenzie. 1997. The dual specificity mitogen-activated protein kinase phosphatase-1 and -2 are induced by the p42/p44MAPK cascade. J. Biol. Chem. 272:1368-1376. [DOI] [PubMed] [Google Scholar]
- 6.Buettner, R., L. B. Mora, and R. Jove. 2002. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin. Cancer Res. 8:945-954. [PubMed] [Google Scholar]
- 7.Burgers, W. A., F. Fuks, and T. Kouzarides. 2002. DNA methyltransferases get connected to chromatin. Trends Genet. 18:275-277. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 9.Chen, H., R. J. Lin, W. Xie, D. Wilpitz, and R. M. Evans. 1999. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98:675-686. [DOI] [PubMed] [Google Scholar]
- 10.Cosma, M. 2002. Ordered recruitment. Gene-specific mechanism of transcription activation. Mol. Cell 10:227.. [DOI] [PubMed] [Google Scholar]
- 11.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 12.Cress, W. D., and E. Seto. 2000. Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 184:1-16. [DOI] [PubMed] [Google Scholar]
- 13.Deckert, J., and K. Struhl. 2001. Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol. Cell. Biol. 21:2726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong, S., and D. J. Tweardy. 2002. Interactions of STAT5b-RARα, a novel acute promyelocytic leukemia fusion protein, with retinoic acid receptor and STAT3 signaling pathways. Blood 99:2637-2646. [DOI] [PubMed] [Google Scholar]
- 15.Fischle, W., F. Dequiedt, M. J. Hendzel, M. G. Guenther, M. A. Lazar, W. Voelter, and E. Verdin. 2002. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9:45-57. [DOI] [PubMed] [Google Scholar]
- 16.Frank, S. R., M. Schroeder, P. Fernandez, S. Taubert, and B. Amati. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamble, M. J., and L. P. Freedman. 2002. A coactivator code for transcription. Trends Biochem. Sci. 27:165-167. [DOI] [PubMed] [Google Scholar]
- 18.Gomes, I., W. Xiong, T. Miki, and M. R. Rosner. 1999. A proline- and glutamine-rich protein promotes apoptosis in neuronal cells. J. Neurochem. 73:612-622. [DOI] [PubMed] [Google Scholar]
- 19.Guenther, M. G., O. Barak, and M. A. Lazar. 2001. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, E. Y., J. Zhang, E. A. Miska, M. G. Guenther, T. Kouzarides, and M. A. Lazar. 2000. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 14:45-54. [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, L., Y. Sowa, T. Sakai, and A. B. Pardee. 2000. Activation of the p21WAF1/CIP1 promoter independent of p53 by the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) through the Sp1 sites. Oncogene 19:5712-5719. [DOI] [PubMed] [Google Scholar]
- 22.Kadosh, D., and K. Struhl. 1998. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 12:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadosh, D., and K. Struhl. 1998. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18:5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiuchi, N., K. Nakajima, M. Ichiba, T. Fukada, M. Narimatsu, K. Mizuno, M. Hibi, and T. Hirano. 1999. STAT3 is required for the gp130-mediated full activation of the c-myc gene. J. Exp. Med. 189:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kostyniuk, C. L., S. M. Dehm, D. Batten, and K. Bonham. 2002. The ubiquitous and tissue specific promoters of the human SRC gene are repressed by inhibitors of histone deacetylases. Oncogene 21:6340-6347. [DOI] [PubMed] [Google Scholar]
- 26.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koyama, Y., M. Adachi, M. Sekiya, M. Takekawa, and K. Imai. 2000. Histone deacetylase inhibitors suppress IL-2-mediated gene expression prior to induction of apoptosis. Blood 96:1490-1495. [PubMed] [Google Scholar]
- 28.Kruh, J. 1982. Effects of sodium butyrate, a new pharmacological agent, on cells in culture. Mol. Cell. Biochem. 42:65-82. [DOI] [PubMed] [Google Scholar]
- 29.Laribee, R. N., and M. J. Klemsz. 2001. Loss of PU.1 expression following inhibition of histone deacetylases. J. Immunol. 167:5160-5166. [DOI] [PubMed] [Google Scholar]
- 30.Li, J., J. Wang, Z. Nawaz, J. M. Liu, J. Qin, and J. Wong. 2000. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, J. X., and W. J. Leonard. 2000. The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene 19:2566-2576. [DOI] [PubMed] [Google Scholar]
- 32.Lord, J. D., B. C. McIntosh, P. D. Greenberg, and B. H. Nelson. 2000. The IL-2 receptor promotes lymphocyte proliferation and induction of the c-myc, bcl-2, and bcl-x genes through the trans-activation domain of Stat5. J. Immunol. 164:2533-2541. [DOI] [PubMed] [Google Scholar]
- 33.Marks, P. A., V. M. Richon, R. Breslow, and R. A. Rifkind. 2001. Histone deacetylase inhibitors as new cancer drugs. Curr. Opin. Oncol. 13:477-483. [DOI] [PubMed] [Google Scholar]
- 34.Martinez-Balbas, M. A., A. J. Bannister, K. Martin, P. Haus-Seuffert, M. Meisterernst, and T. Kouzarides. 1998. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 17:2886-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsumoto, A., M. Masuhara, K. Mitsui, M. Yokouchi, M. Ohtsubo, H. Misawa, A. Miyajima, and A. Yoshimura. 1997. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood 89:3148-3154. [PubMed] [Google Scholar]
- 36.Maurer, A. B., C. Wichmann, A. Gross, H. Kunkel, T. Heinzel, M. Ruthardt, B. Groner, and M. Grez. 2002. The Stat5-RARα fusion protein represses transcription and differentiation through interaction with a corepressor complex. Blood 99:2647-2652. [DOI] [PubMed] [Google Scholar]
- 37.Migone, T. S., M. Humbert, A. Rascle, D. Sanden, A. D'Andrea, and J. A. Johnston. 2001. The deubiquitinating enzyme DUB-2 prolongs cytokine-induced signal transducers and activators of transcription activation and suppresses apoptosis following cytokine withdrawal. Blood 98:1935-1941. [DOI] [PubMed] [Google Scholar]
- 38.Munshi, N., T. Agalioti, S. Lomvardas, M. Merika, G. Chen, and D. Thanos. 2001. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science 293:1133-1136. [DOI] [PubMed] [Google Scholar]
- 39.Munshi, N., M. Merika, J. Yie, K. Senger, G. Chen, and D. Thanos. 1998. Acetylation of HMG I(Y) by CBP turns off IFN β expression by disrupting the enhanceosome. Mol. Cell 2:457-467. [DOI] [PubMed] [Google Scholar]
- 40.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 41.Nakajima, H., P. K. Brindle, M. Handa, and J. N. Ihle. 2001. Functional interaction of STAT5 and nuclear receptor co-repressor SMRT: implications in negative regulation of STAT5-dependent transcription. EMBO J. 20:6836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakano, K., T. Mizuno, Y. Sowa, T. Orita, T. Yoshino, Y. Okuyama, T. Fujita, N. Ohtani-Fujita, Y. Matsukawa, T. Tokino, H. Yamagishi, T. Oka, H. Nomura, and T. Sakai. 1997. Butyrate activates the WAF1/Cip1 gene promoter through Sp1 sites in a p53-negative human colon cancer cell line. J. Biol. Chem. 272:22199-22206. [DOI] [PubMed] [Google Scholar]
- 43.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 44.Nawaz, Z., C. Baniahmad, T. P. Burris, D. J. Stillman, B. W. O'Malley, and M. J. Tsai. 1994. The yeast SIN3 gene product negatively regulates the activity of the human progesterone receptor and positively regulates the activities of GAL4 and the HAP1 activator. Mol. Gen. Genet. 245:724-733. [DOI] [PubMed] [Google Scholar]
- 45.Nosaka, T., T. Kawashima, K. Misawa, K. Ikuta, A. L. Mui, and T. Kitamura. 1999. STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells. EMBO J. 18:4754-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 47.Pandolfi, P. P. 2001. Transcription therapy for cancer. Oncogene 20:3116-3127. [DOI] [PubMed] [Google Scholar]
- 48.Rao, S., E. Procko, and M. F. Shannon. 2001. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J. Immunol. 167:4494-4503. [DOI] [PubMed] [Google Scholar]
- 49.Ren, S., H. R. Cai, M. Li, and P. A. Furth. 2002. Loss of Stat5a delays mammary cancer progression in a mouse model. Oncogene 21:4335-4339. [DOI] [PubMed] [Google Scholar]
- 50.Richon, V. M., S. Emiliani, E. Verdin, Y. Webb, R. Breslow, R. A. Rifkind, and P. A. Marks. 1998. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc. Natl. Acad. Sci. USA 95:3003-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richon, V. M., T. W. Sandhoff, R. A. Rifkind, and P. A. Marks. 2000. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc. Natl. Acad. Sci. USA 97:10014-10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robertson, K. D. 2002. DNA methylation and chromatin—unraveling the tangled web. Oncogene 21:5361-5379. [DOI] [PubMed] [Google Scholar]
- 53.Schwaller, J., E. Parganas, D. Wang, D. Cain, J. C. Aster, I. R. Williams, C. K. Lee, R. Gerthner, T. Kitamura, J. Frantsve, E. Anastasiadou, M. L. Loh, D. E. Levy, J. N. Ihle, and D. G. Gilliland. 2000. Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol. Cell 6:693-704. [DOI] [PubMed] [Google Scholar]
- 54.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 55.Sheldon, L. A., M. Becker, and C. L. Smith. 2001. Steroid hormone receptor-mediated histone deacetylation and transcription at the mouse mammary tumor virus promoter. J. Biol. Chem. 276:32423-32426. [DOI] [PubMed] [Google Scholar]
- 56.Shuai, K. 2000. Modulation of STAT signaling by STAT-interacting proteins. Oncogene 19:2638-2644. [DOI] [PubMed] [Google Scholar]
- 57.Soutoglou, E., N. Katrakili, and I. Talianidis. 2000. Acetylation regulates transcription factor activity at multiple levels. Mol. Cell 5:745-751. [DOI] [PubMed] [Google Scholar]
- 58.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901-1904. [DOI] [PubMed] [Google Scholar]
- 59.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 61.Van Lint, C., S. Emiliani, and E. Verdin. 1996. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 5:245-253. [PMC free article] [PubMed] [Google Scholar]
- 62.Van Reeth, T., P. Gabant, C. Szpirer, and J. Szpirer. 2002. Stimulation of the alpha-fetoprotein promoter by unliganded thyroid hormone receptor in association with protein deacetylation. Mol. Cell. Endocrinol. 188:99-109. [DOI] [PubMed] [Google Scholar]
- 63.Vidal, M., and R. F. Gaber. 1991. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:6317-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vidal, M., R. Strich, R. E. Esposito, and R. F. Gaber. 1991. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol. Cell. Biol. 11:6306-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilson, M. A., A. R. Ricci, B. J. Deroo, and T. K. Archer. 2002. The histone deacetylase inhibitor trichostatin A blocks progesterone receptor-mediated transactivation of the mouse mammary tumor virus promoter in vivo. J. Biol. Chem. 277:15171-15181. [DOI] [PubMed] [Google Scholar]
- 66.Xiao, H., T. Hasegawa, and K. Isobe. 2000. p300 collaborates with Sp1 and Sp3 in p21(waf1/cip1) promoter activation induced by histone deacetylase inhibitor. J. Biol. Chem. 275:1371-1376. [DOI] [PubMed] [Google Scholar]
- 66a.Xu, M., L. Nie, S. H. Kim, and X. H. Sun. 2003. STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPβ. EMBO J. 22:893-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ye, S. K., Y. Agata, H. C. Lee, H. Kurooka, T. Kitamura, A. Shimizu, T. Honjo, and K. Ikuta. 2001. The IL-7 receptor controls the accessibility of the TCRγ locus by Stat5 and histone acetylation. Immunity 15:813-823. [DOI] [PubMed] [Google Scholar]
- 68.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174-17179. [PubMed] [Google Scholar]
- 69.Yoshimoto, H., M. Ohmae, and I. Yamashita. 1992. The Saccharomyces cerevisiae GAM2/SIN3 protein plays a role in both activation and repression of transcription. Mol. Gen. Genet. 233:327-330. [DOI] [PubMed] [Google Scholar]
- 70.Zou, J., S. Young, F. Zhu, F. Gheyas, S. Skeans, Y. Wan, L. Wang, W. Ding, M. Billah, T. McClanahan, R. L. Coffman, R. Egan, and S. Umland. 2002. Microarray profile of differentially expressed genes in a monkey model of allergic asthma. Genome Biol. 3:research0020.1-research0020.13. [DOI] [PMC free article] [PubMed]