Abstract
The 3′ ends of metazoan histone mRNAs are generated by specialized processing machinery that cleaves downstream of a conserved stem-loop structure. To examine how this reaction might be influenced by transcription, we used a Drosophila melanogaster in vitro system that supports both processes. In this system the complete synthesis of histone mRNA, including transcription initiation and elongation, followed by 3′ end formation, occurred at a physiologically significant rate. Processing of free transcripts was efficient and occurred with a t1/2 of less than 1 min. Divalent cations were not required, but nucleoside triphosphates (NTPs) stimulated the rate of cleavage slightly. Isolated elongation complexes encountered a strong arrest site downstream of the mature histone H4 3′ end. In the presence of NTPs, transcripts in these arrested complexes were processed at a rate similar to that of free RNA. Removal of NTPs dramatically reduced this rate, potentially due to concealment of the U7 snRNP binding element. The arrest site was found to be a conserved feature located 32 to 35 nucleotides downstream of the processing site on the H4, H2b, and H3 genes. The significance of the newly discovered arrest sites to our understanding of the coupling between transcription and RNA processing on the one hand and histone gene expression on the other is discussed.
The replication-dependent histone genes of metazoans encode a unique class of pre-mRNAs that are neither spliced nor polyadenylated. Rather, the mature transcripts arise from an endonucleolytic cleavage on the 3′ side of a conserved stem-loop sequence, just downstream of the translation stop codon (5). An activity that could accomplish this cleavage event in vitro was examined and partially purified from Drosophila cultured cell extract nearly 20 years ago (37). Since then, several elements important for this processing reaction, including both trans-acting factors and sequence elements of the pre-mRNAs themselves, have been identified and characterized (5).
The replication-dependent histone pre-mRNAs all have a 3′ untranslated region that includes a stem-loop forming sequence and a purine-rich region just downstream, called the histone downstream element (HDE). These two sequence elements have been conserved throughout metazoan evolution, in organisms as diverse as sea urchins and mammals (28). In addition to these sequences, 3′-end processing requires the stem-loop binding protein (SLBP) (6, 15, 52, 53, 59), U7 snRNP (30, 44, 49), and a heat-labile factor (13). The U7 snRNP interacts with the HDE through the 5′ end of the U7 RNA and is thought to act as a “molecular ruler,” determining how far upstream of the HDE cleavage will occur (2, 42, 43). The SLBP binds to the stem-loop in the nascent RNA and stabilizes the interaction between the HDE and the U7 snRNP (8, 47, 48). The HDE sequence is somewhat variable, particularly in mammals (23), and from in vitro studies we know that a weaker interaction between the HDE and the U7 snRNP results in a more stringent requirement for SLBP (8, 47, 48). Histone protein levels are tightly regulated in accordance with the cell cycle, exhibiting a marked increase in S phase (28). It has been shown that this regulation is predominantly posttranscriptional and that SLBP plays a major role (16, 24, 32, 52). SLBP remains bound to the stem-loop after cleavage and is important for subsequent events, including nucleocytoplasmic transport, translation, and degradation (5, 53).
The organization of the histone genes on their chromosomes has been conserved in metazoans. Histone genes appear in tightly linked clusters, and it has been suggested that the maintenance of this linkage reflects a particular subnuclear localization of their biosynthesis. In Drosophila melanogaster, there are five different replication-dependent histone genes, all contained in a 4.8-kb element which is repeated in tandem about a hundred times (22). Each of these genes has its own promoter, and they are arranged in both convergent and divergent orientations; in some cases adjacent genes are transcribed from the same strand, and in some cases they are not. It is therefore widely assumed that an efficient termination mechanism must exist between the genes to avoid transcriptional interference. Chodchoy et al. (3) characterized an apparent termination signal between the histone H2a and H3 genes in mice that required an intact 3′-end-processing signal to function (3). Other groups have demonstrated termination sites for polyadenylated genes that are dependent on intact poly(A) addition signals (40). Metazoan histone genes contain cryptic polyadenylation signals located 3′ to their processing sites (50) and, when the SLBP of Drosophila is mutated so that the normal endonucleolytic cleavage cannot occur, the histone mRNAs become polyadenylated (21, 50).
Whereas the various aspects of RNA metabolism (transcription, capping, 3′ end formation, decay, etc.) are separable in vitro, an accumulation of data over the last decade has led to a largely integrated view of these processes (1, 4, 40). Many published studies point to the C-terminal domain (CTD) of RNA polymerase II as a focal point in the interconnection of these events, and a cotranscriptional paradigm has come to dominate contemporary thinking, with the CTD being indispensable for coordination of RNA metabolism (31, 40). Although it is assumed that RNA processing is coupled to transcription, exactly what this means is not universally agreed upon. Clearly, one process depends on the other and the machinery of both processes are physically connected, at least through the RNA and perhaps through the polymerase. However, a more significant requirement for functional coupling requires one of the processes to affect the progression of the other. The CTD has been found to stimulate the rate of splicing in vitro (17, 18, 60), but in those experiments free RNA was used as substrate for the processing machinery. Several studies have demonstrated that splicing (12) or polyadenylation (57, 58) can take place in a transcription reaction. Functional coupling of 5′ capping and transcription has recently been demonstrated by using a human in vitro transcription system (29), but in that study the CTD played only a minor role in the coupling event.
Significant progress has been made in understanding events controlling transcription by RNA polymerase II after initiation (36). The elongating polymerase is first slowed by negative transcription elongation factors (N-TEFs), which restrict the polymerase to the promoter proximal region of genes (26). The negative elongation factor (56), and the DRB (5,6-dichloro-1-β-d-ribofuranosylbenzimidizolesensitivity)-inducing factor(51), are two such N-TEFs that have been demonstrated to slow the rate of elongation of RNA polymerase II in a defined system (41). Positive transcription elongation factor b (P-TEFb) is a cyclin-dependent kinase comprised of Cdk9 and cyclin T in Drosophila (34) and Cdk9 and cyclin T1, T2, or K in humans (35) that phosphorylates the CTD of the large subunit of RNA polymerase II (25) (27) and allows the polymerase to enter productive elongation (36). The function of P-TEFb to reduce the appearance of short promoter proximal transcript and promote the generation of long runoff transcripts is inhibited by the cyclin-dependent kinase inhibitor DRB (25).
In the present study, we describe an in vitro system, with Drosophila nuclear extract and an immobilized DNA template, that has enabled us to further investigate the biochemical requirements and kinetics of the histone 3′-end-processing reaction in the context of transcription. The kinetic data we collected defied our expectations by demonstrating that the rate of 3′-end processing was not enhanced by the presence of a transcription complex. Our experiments identified a strong arrest site about 15 nucleotides (nt) beyond the HDE, which was present in all three of the histone genes that we examined. Transcripts in complexes stalled at this site were processed less efficiently than free RNA, a result potentially due to a conformational change in the polymerase.
MATERIALS AND METHODS
Preparation of extracts.
Drosophila Kc cells were grown and nuclear extracts prepared for transcription reactions as described previously (26, 38).
DNA constructs.
All minigene constructs were derived from the Dm3000 plasmid, which contains the entire Drosophila histone gene cluster (33). For the H4 construct, an upstream fragment of the gene, including the TATA box and transcription start site, was generated by PCR. The primers included restriction sites (BamHI 5′ and XbaI 3′) for subsequent cloning. A downstream fragment from the H4 gene, including the stem-loop and HDE, was also generated from PCR primers that included XbaI (5′) and AvaI (3′) sites. These two fragments were then ligated with BamHI- and AvaI-cut pET21a(+) to form a minigene that had a histone H4 promoter, start site, and processing site but from which most of the intervening sequence had been eliminated. The H2b and H3 minigenes were fashioned similarly, but both used the H4 upstream fragment and so were actually hybrid minigenes, differing only in their 3′ sequences. All three minigene transcription templates were generated by PCR from the pET21a(+) constructs described above, with the same biotinylated 5′ primer and unique 3′ primers, well downstream of the processing signals. The resulting 5′ biotinylated templates were purified with the UltraClean 15 DNA purification kit (MO BIO Labs, Inc.) and incubated with streptavidin-conjugated Dynabeads M280 (Dynal) as previously described (26) to form immobilized templates.
Transcription and processing reactions.
All transcription reactions were carried out in 20 mM HEPES (pH 7.6), 5 mM MgCl2, and 60 mM KCl and included a 15-min preincubation with Kc cell nuclear extract under these conditions to allow the formation of preinitiation complexes. Pulses were done for 45 s in the presence of limiting [α-32P]CTP (10 μCi per reaction), with ATP, GTP, and UTP at 600 μM. Individual preincubation reactions were in a 12-μl total volume, and pulses and chases were in 15 μl. For simple pulse-chase protocols, the pulse was ended by the addition of a chase mixture that contained 1.2 mM cold CTP. For add-back experiments with an immobilized template, the pulse was ended by addition of EDTA to a final concentration 12 to 16 mM. The resulting early elongation complexes were isolated by three washes with high-salt Sarkosyl solution (1 M KCl, 0.3% Sarkosyl, 20 mM HEPES, 5 mM MgCl2), followed by two additional washes and resuspension in transcription buffer (20 mM HEPES, 5 mM MgCl2, 60 mM KCl, 200 μg of bovine serum albumin/ml). A typical “wash” was accomplished in less than 60 s and involved concentrating the beads, removing the wash supernatant, and adding the next wash, with subsequent pipetting up and down to fully resuspend the concentrated beads. For time course experiments, identical reactions were done in bulk, with single reactions stopped by removal into tRNA-Sarkosyl solution (1% Sarkosyl, 0.1 M Tris [pH 8.0], 0.1 M NaCl, 10 mM EDTA, 200 μg of tRNA/ml) at the indicated time points. All individual reactions were ultimately ended by the addition of tRNA-Sarkosyl solution to a final volume of 210 μl, after which they were phenol extracted with ethanol precipitation before being loaded onto the gel. The isolation of free RNA for some experiments was accomplished by the same extraction procedure, except that the RNA was extensively washed with 70% ethanol before being dried and dissolved in water.
RESULTS
Histone genes are transcribed, and the resulting RNA is rapidly processed in nuclear extracts from Drosophila cells.
Early experiments examining 3′-end processing of the Drosophila histone H3 pre-mRNA, performed on dC-tailed templates, indicated that these transcripts could be accurately processed in Drosophila Kc cell nuclear extracts (37). Study of the formation of the 3′ end of the mature histone mRNAs progressed thereafter in sea urchins and mammals and, more recently, in Drosophila (5). There is still much to be learned about the connection of this processing event to transcription because this information is difficult to obtain from in vivo experiments, and most in vitro processing experiments have utilized presynthesized transcripts. To begin to understand how transcription and histone mRNA 3′-end formation are connected, we utilized a Drosophila in vitro transcription system that supports transcription and efficient processing of Drosophila histone pre-mRNAs.
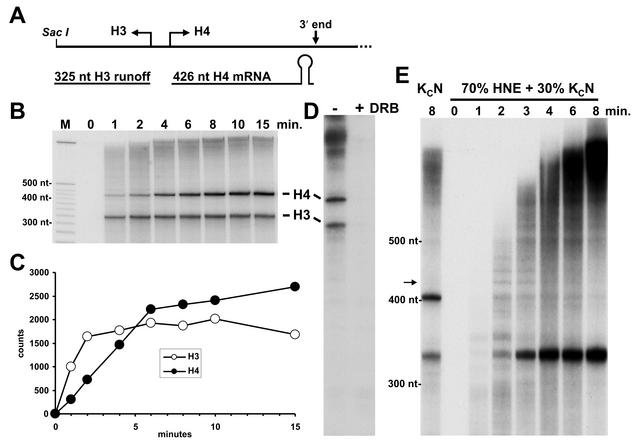
We first performed a pulse-chase experiment with a plasmid containing a full repeat of the Drosophila histone gene cluster that supports strong initiation from the H3 and H4 promoters. The plasmid was cut with SacI, which cleaves between the H3 promoter and the H3 processing signal, thereby generating an unprocessed, runoff transcript (Fig. 1A). The entire H4 gene is left intact by the restriction digest and generates transcripts that can be cleaved 4 nt 3′ of the stem-loop to give the mature mRNA. The labeled transcripts were analyzed by denaturing polyacrylamide gel electrophoresis (Fig. 1B), and the accumulation of the H3 runoff and the processed H4 mRNA was quantified (Fig. 1C). The H3 runoff reached its maximum level after about 2 min and remained constant over the rest of the 15-min time course. The kinetics of accumulation of runoff is dictated by the elongation rate of RNA polymerase II and by the kinetics of the function of P-TEFb (26, 36). Accumulation of the H4 mRNA lagged behind that of the H3 runoff and reached a plateau after about 5 min. The lag can be explained by the added dependence of accurate 3′-end processing on the appearance of the H4 pre-mRNA. To prove that P-TEFb was required for the appearance of long transcripts, transcription in the absence or presence of 50 μM DRB, a nucleoside analog that inhibits P-TEFb, was compared (Fig. 1D). Because in this experiment the transcripts were continuously labeled, there is an accentuation of the long transcripts above the two major species seen with a pulse-chase protocol (Fig. 1D). As was found earlier (26), inhibition of P-TEFb by DRB eliminated all transcripts seen in the portion of the gel shown (Fig. 1D). We conclude from these experiments that generation of the 3′ end of histone H4 requires P-TEFb for the synthesis of appropriately long transcripts to be used as substrates for the processing machinery and that the processing occurs efficiently and rapidly.
FIG. 1.
Drosophila histone RNAs are transcribed and rapidly processed in nuclear extracts from Drosophila cells. (A) Transcribed region of the Dm3000 plasmid showing H3 runoff transcript and H4 processed mRNA. (B) Pulse-chase assay using Dm3000 template and Kc cell nuclear extract. RNA was isolated at the indicated times and analyzed on a 6% denaturing gel followed by autoradiography. M, DNA markers of indicated sizes. H3 runoff and H4 mRNA are indicated. (C) Quantitation of H3 and H4 transcripts. Transcripts in the dried gel were quantitated by using a Packard InstantImager and the relative counts plotted. (D) Continuous-labeling assay. The Dm3000 template was transcribed in Kc cell nuclear extract in the absence or presence of 50 μM DRB, and the resulting transcripts were analyzed on a 6% denaturing gel. (E) Inhibition of RNA processing in a mixture of HeLa and Kc cell nuclear extracts. Reactions similar to those shown in panel B were carried out except that 70% HeLa and 30% Kc cell nuclear extract were used. Samples were collected at the indicated time points. For comparison purposes, one reaction was carried out for 8 min in the presence of only Kc cell nuclear extract. The transcripts were analyzed in a 6% denaturing gel as described above except that the gel was run longer to better separate long transcripts.
Although the Kc cell nuclear extract used here has been shown to accurately carry out 3′-end processing of histone H3 mRNA (37), to confirm that the H4 transcript seen here was generated by a processing event, we transcribed the same template in a mixture of HeLa and Kc cell nuclear extracts that allows efficient transcription but does not allow 3′-end processing (D. H. Price, unpublished result). Human SLBP has been shown to bind to the Drosophila histone mRNA stem-loop and inhibit efficient processing by the fly factors in a dominant-negative fashion (7). A comparison of products generated in 8 min by Drosophila extract alone with those generated in a time course by the mixture of human and Drosophila extracts shows that appearance of the H3 runoff was increased, but the transcript attributed to processing of H4 was significantly decreased (Fig. 1E). This decrease in the H4 transcript was not due to lack of initiation from the H4 promoter because a template containing only that promoter generated a similar amount of runoff transcript compared to the H3 promoter (data not shown). When both extracts were present, other longer transcripts appeared transiently, but these were chased into very long transcripts at the later time points. The significance of paused transcripts longer than processed H4 mRNA will be discussed later. The results from the transcription reactions with a mixture of extracts strongly support the contention that the H4 transcript seen with only Drosophila extract is generated by a processing event.
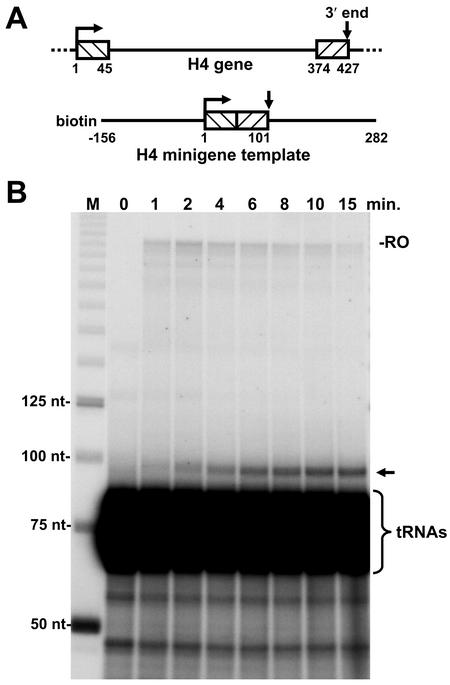
To further investigate the potential connection between transcription and 3′-end processing, we created a histone minigene that should generate a processed transcript of 101 nt. After construction of a minigene plasmid, the template was PCR amplified by using a biotinylated upstream primer for immobilization and a downstream primer that yielded a run off transcript of 282 nt (Fig. 2A). We reasoned that this template would allow the polymerase to reach the processing site faster and provide better resolution of transcripts synthesized. To determine whether removal of the bulk of the coding region affected the kinetics of the processing, a pulse-chase experiment similar to that shown in Fig. 1 was performed. A transcript of the predicted size for the processed transcript appeared with similar kinetics to that seen with the intact H4 gene and is visible above the tRNAs that are extensively labeled during the reactions by CCA addition at their 3′ ends (Fig. 2B).
FIG. 2.
Construction and transcription of an H4 minigene. (A) Diagram of H4 gene and derived minigene template. The minigene template generated by PCR contained a biotin molecule on the upstream end and was missing 328 nt from the coding region of the H4 gene. (B) Pulse-chase assay with H3 minigene template and Kc cell nuclear extract. RNA was isolated at the indicated times and analyzed on a 6.7% denaturing gel, followed by autoradiography. M, DNA markers of indicated sizes. Runoff (RO), end-labeled tRNAs, and processed transcript (arrow) are indicated.
The immobilized H4 minigene template was then utilized to examine transcription through the processing site in the presence or absence of extract. The biotinylated template was first bound to streptavidin-coated, paramagnetic beads. Then preinitiation complexes were formed by incubation with Kc cell nuclear extract. Early elongation complexes formed during a short pulse under limiting CTP conditions were washed with 1 M salt and 0.3% Sarkosyl to remove labeled tRNAs, all unassociated factors, and even factors potentially associated with the polymerase. The isolated elongation complexes were washed into normal transcription conditions, and transcripts were chased by addition of all four nucleoside triphosphates (NTPs), with or without extract. Reactions were stopped at the indicated times, and the RNA was subjected to gel analysis (Fig. 3A). We were surprised to find that, in the absence of extract, ca. 90% of the polymerases arrested at one particular site downstream of the processing site (Fig. 3A, transcript A). A different pattern of transcripts resulted when nuclear extract was added during the chase. In general, fewer long transcripts were seen due to the negative effect of N-TEFs and the incomplete action of P-TEFb (Fig. 3A). The transcript generated by the major arrest site is seen but is now a minor species that disappears in the longer time points. Figure 3 shows that the processed transcript (Fig. 3A, arrow) appeared with kinetics similar to those seen when full-length or minigene constructs were used in experiments without isolation of elongation complexes. We conclude that it is not necessary to load factors onto the transcription complex during initiation to achieve rapid 3′-end processing. Our results are consistent with most processing occurring on transcripts in complexes paused or arrested about 32 nt downstream from the processing site.
FIG. 3.
Kinetic analysis of transcription and processing from the H4 minigene template. (A) Transcription from isolated early elongation complexes. Early elongation complexes were generated and isolated as described in Materials and Methods and then allowed to elongate with or without Kc cell nuclear extract as indicated. RNA was isolated at the indicated times and analyzed on a 6.7% denaturing gel, followed by autoradiography. A, arrested transcript; arrow, processed H4 minigene RNA. (B) Quantitation of processed transcript. The processed transcript was quantified by using a Packard InstantImager, and the relative counts were plotted.
Free RNA is processed more efficiently and more rapidly than RNA in a transcription complex.
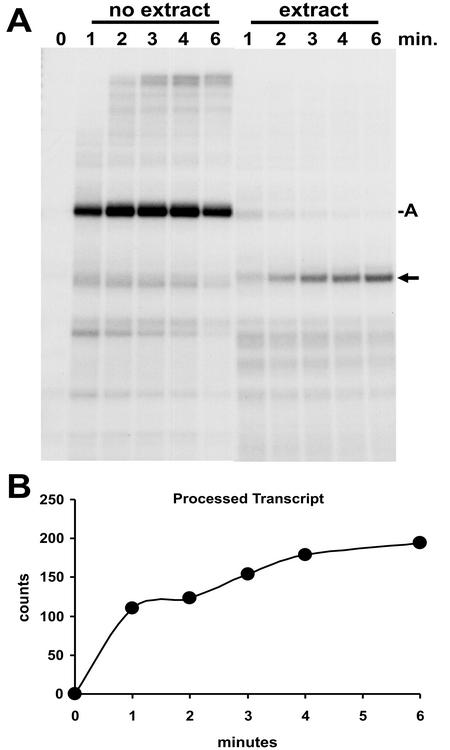
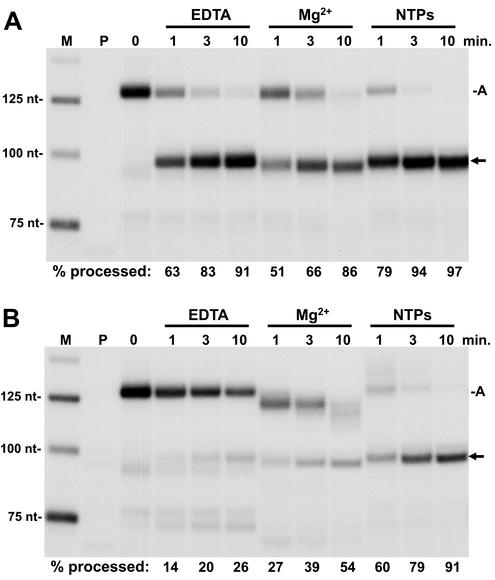
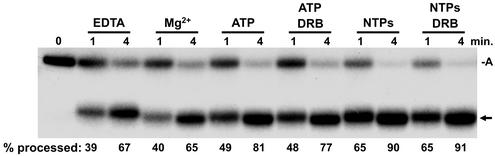
We utilized the immobilized minigene template to generate elongation complexes that were predominantly arrested at one site downstream of the processing signal to study the effect of the elongation complexes on the rate of 3′-end processing. Early elongation complexes were isolated by using the 1 M salt and 0.3% Sarkosyl wash protocol. The transcripts were then extended with an 8-min chase in the absence of extract. As expected, the RNA in the arrested complex was the primary transcript (Fig. 4, 0 min). These complexes were then split, and half were phenol extracted to yield the free RNA. The other half were left as intact elongation complexes. We then compared the kinetics of processing of the two preparations of otherwise identical RNAs. Since there have been conflicting published results on the requirement for divalent cations for processing, processing was carried out with the addition of nuclear extract in the presence of EDTA or Mg. The free RNA efficiently yielded processed transcripts with or without a divalent cation (Fig. 4A). The percent processing was calculated by quantifying the processed and arrested transcripts and dividing the amount of the processed transcript by the sum of both processed and arrested transcripts and multiplying this value by 100. These values are reported below each lane (Fig. 4). When free RNA was used, >50% processing occurred by the 1-min time point regardless of the presence of a divalent cation. The inclusion of NTP at concentrations that are used during normal transcription reactions had a slight positive effect. This could be due to activation of one of the components of the processing reaction by phosphorylation. When RNA in elongation complexes was subjected to identical processing conditions, the results were different (Fig. 4B). In the presence of EDTA or Mg, there was very little processing, even after 10 min. Only in the presence of NTPs was the extent and rate of processing equal to that seen with free RNA.
FIG. 4.
Comparison of processing of free RNA to RNA in a transcription complex. Early elongation complexes generated during initiation with a pulse-labeling on the H4 minigene template (lane marked P) were isolated and washed with a high salt concentration and Sarkosyl as described in Materials and Methods. These complexes were chased for 8 min, reisolated, and phenol extracted to obtain free RNA or used directly as elongation complexes. (A) Processing of free RNA. Free RNA containing predominantly the arrested transcript was subjected to processing by nuclear extract for the indicated times with EDTA, Mg, or NTPs with Mg added. (B) Processing of RNA in elongation complexes. Reactions were identical to those in panel A except that the RNA was in a transcription complex. For both panels A and B, RNA was isolated at the indicated times and analyzed on a 6.7% denaturing gel, followed by autoradiography and quantitation with a Packard InstantImager. The percentage of total RNA processed is given under each lane. A, arrested transcript; arrow, processed H4 minigene RNA.
A clue to what is responsible for this inhibition of processing by the elongation complex comes in looking at the arrested transcripts in the presence of Mg (Fig. 4B). Instead of being processed, the bulk of the arrested transcripts are shortened, presumably through the Mg-dependent function of the elongation factor S-II (14). The mechanism of S-II cleavage is thought to require the polymerase to backslide along the template before the transcript is cleaved by the active site of the polymerase (11). Inhibition of processing in the presence of EDTA can be explained by backsliding of the polymerase without cleavage occurring. In both cases (with or without divalent cation), the polymerase may physically interfere with the binding of U7 snRNP by masking the required HDE. In the presence of NTPs, the backsliding and cleavage are quickly followed by elongation back to the arrest site, resulting in efficient processing. In the reactions with EDTA and elongation complexes, another set of processed transcripts can be seen around 75 nt in length (Fig. 4B). These transcripts appear with kinetics similar to those of the properly processed transcripts and are of the appropriate length to represent cleavage of the arrested transcript upstream of the stem-loop. We do not know why these products are only seen in the elongation complex reactions with EDTA, but perhaps SLBP is directing cleavage with lower specificity due to the lack of U7 function.
Nucleotides stimulate the rate of processing of free RNA.
To follow up on the result in Fig. 4A that indicated that NTPs have a stimulatory effect on processing of free RNA, processing reactions were set up to determine whether ATP alone could account for this effect and if the kinase activity of P-TEFb was involved. The stimulation of processing by NTPs was unexpected, since the effect of S-II seen with elongation complexes should not be a factor in the processing of a transcript that was not associated with a polymerase. Because the source of processing activity is a nuclear extract and RNA polymerase II is present, it was not possible to rule out involvement of the polymerase in processing of even free RNA. Transcripts present in the arrested complex were isolated by phenol extraction (Fig. 5, 0 min), and processing reactions were carried out for 1 or 4 min (Fig. 5). In this experiment the rate of processing was similar in the presence of EDTA or Mg. The percent processing at both the 1-min and the 4-min time points was greater when ATP was included in the reactions. The effect of ATP is apparently not due to the kinase activity of P-TEFb because inclusion of DRB in the ATP-containing reaction did not inhibit the ATP stimulation. When all four NTPs were present, there was a further stimulation of the processing activity, and again DRB had no effect. Overall, these results are consistent with a non-P-TEFb-driven phosphorylation of one of the components of the processing reaction, resulting in a positive effect on the rate of processing; however, we cannot explain the additional small effect of NTPs over ATP alone.
FIG. 5.
Effect of nucleotides on processing of free RNA. Transcripts were isolated from elongation complexes as in Fig. 4A and processed with nuclear extract under the indicated conditions. RNA was isolated at the indicated times and analyzed on a 6.7% denaturing gel, followed by autoradiography. A, arrested transcript; arrow, processed H4 minigene RNA. The percentage of total RNA processed is given under each lane.
3′-end-processing machinery does not associate strongly with the elongation complex.
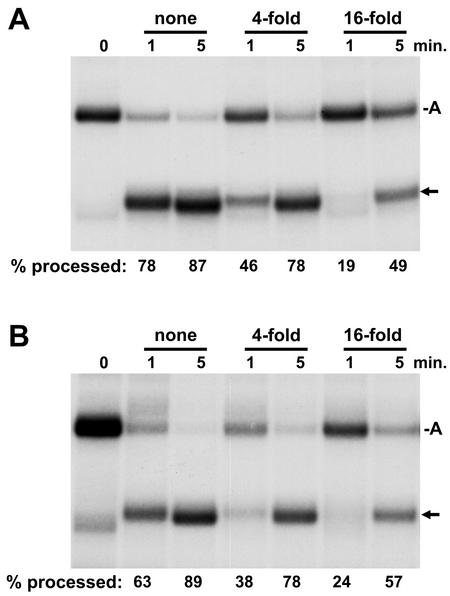
Although the results from preceding experiments indicate that the elongation complex does not have a positive influence on 3′-end processing, we performed a dilution experiment to examine this possibility in another way. Processing of free RNA was again compared to that of RNA in transcription complexes, but this time the processing machinery (extract) was diluted 0-, 4-, or 16-fold before being added to the RNA or transcription complexes. If the processing machinery associated with the elongation complex, the processing of transcripts in complexes might be expected to have less dependence on the concentration of the processing factors. Because processing was done in the presence of NTPs, the rate of processing was similar when the extract was at the normal concentration (Fig. 6). Dilution of the extract causes a slowing of the rate of processing for both free RNA (Fig. 6A) and RNA in elongation complexes (Fig. 6B). The magnitude of the reduction was similar for both. Although it is likely that SLBP binds to the nascent transcript, these results provide evidence that at least one other component of the histone 3′-end processing machinery does not strongly associate with the elongation complex. The dependence of the rate of processing on the concentration of the machinery could explain the differences in the rate of processing seen in different studies and suggests that the nuclear extracts we are using have a relatively high concentration of the required factors.
FIG. 6.
Dependence of processing on extract concentration. Free RNA (A) or isolated elongation complexes (B) were processed by using nuclear extract for the indicated times. A constant amount of extract was used for each reaction, but the concentration was changed by dilution as indicated. RNA was isolated at the indicated times and analyzed on a 6.7% denaturing gel, followed by autoradiography. A, arrested transcript; arrow, processed transcript. The percentage of total RNA processed is given under each lane.
Histone H2b and H3 genes also have arrest sites downstream of the processing site.
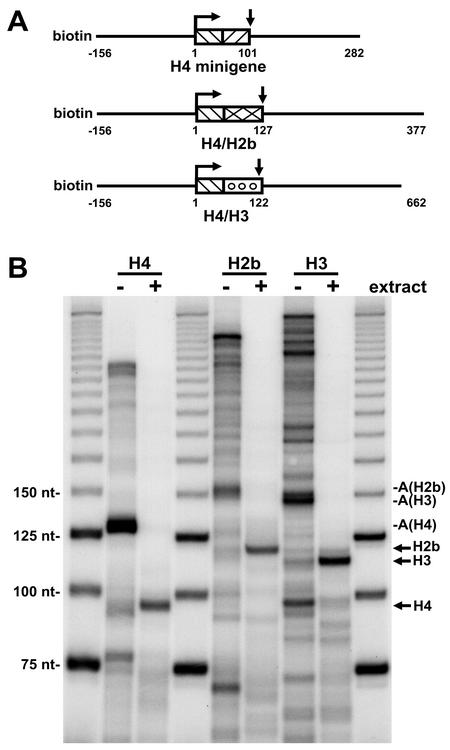
To determine whether the strong arrest site directly downstream from the H4 processing site was unique to H4 or was a more general feature of histone genes, transcription through the 3′ ends of H2b and H3 was examined. Minigenes with the H4 promoter driving H2b (H4/H2b) and H3 (H4/H3) 3′ ends were constructed (Fig. 7A). The templates were immobilized to paramagnetic beads and used to generate early elongation complexes as was done with H4 earlier. The early elongation complexes were washed with high concentrations of salt and Sarkosyl and then allowed to elongate in the absence or presence of nuclear extract. Transcription of the H4 minigene gave the expected major arrest downstream of the processing site in the absence of extract and the properly processed transcript in the presence of extract (Fig. 7B). Both the H4/H2b and the H4/H3 minigenes gave rise to transcripts indicative of a strong pause or arrest site downstream from their respective processing sites (Fig. 7B). The two new minigenes also efficiently gave rise to the expected processed transcripts (Fig. 7B). The sizes of the arrested and processed transcripts were determined by comparing the mobility of the transcripts to that of markers. The difference between the arrested and processed transcripts for all three constructs was between 32 and 35 nt.
FIG. 7.
Comparison of H4, H2b, and H3 genes. (A) Minigene constructs. The 3′ ends of the H2b and H3 genes were cloned downstream of the H4 promoter to give processed transcripts of the indicated sizes. Templates were generated by PCR by using a biotinylated upstream primer and a downstream primer that resulted in a runoff transcript of the indicated size. (B) Transcription and processing of the minigenes. Early elongation complexes were formed during a pulse-labeling step with each of the three minigene templates. The early elongation complexes were washed with a high salt concentration and Sarkosyl and chased in the absence (−) or presence (+) of extract for 10 min. A, arrested transcript; arrow, processed transcript.
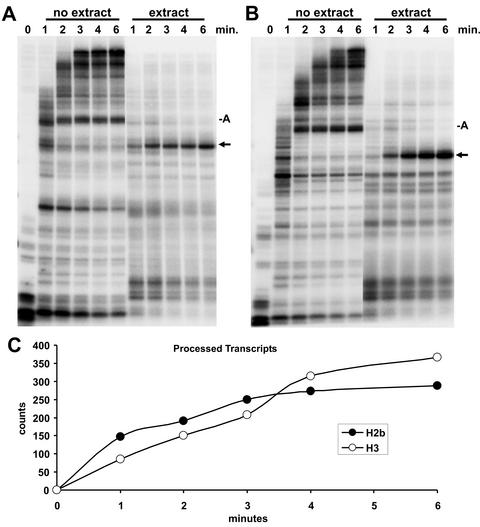
Transcription through the 3′ regions of H2b and H3 was examined more thoroughly by taking time points during a pulse-chase experiment similar to that shown in Fig. 7B. Transcription by RNA polymerase II in the absence of factors (no extract) gave rise to a progressive pattern of transcripts for transcription through the H3 (Fig. 8A) and H2b (Fig. 8B) 3′ regions. The major transcript found to extend downstream of the 3′-processing site on both templates was clearly an arrest site because the levels of those transcripts rose early during the time course and then remained constant during the later time points. There was a relative reduction of transcripts of intermediate length between the processed transcript and the transcript in the arrested complexes for both templates (Fig. 8A and B), as was found for the H4 template (Fig. 3A). This suggests that the polymerase proceeds relatively quickly through this region and then abruptly stops at the arrest site. In the presence of nuclear extract, both templates gave rise to transcripts that were efficiently processed. The transcripts due to arrest were seen in the early time points when transcription was carried out in the presence of extract, but they disappeared during the later time points, suggesting that the transcripts in arrested complexes are substrates for the processing machinery. The fact that all three genes have arrest sites at such similar positions downstream from the processing site suggests that this is a conserved feature of histone genes that might have physiological significance to histone gene expression.
FIG. 8.
Kinetics of transcription and processing on the H4/H2b and H4/H3 minigenes. Transcription and processing as described in Fig. 7 was carried out on the H4/H2b (A) and the H4/H3 (B) minigenes. RNA was isolated at the indicated time points and analyzed on a 6.7% denaturing gel, followed by autoradiography. (C) The processed transcripts were quantitated by using a Packard InstantImager and plotted versus time. A, arrested transcript; arrow, processed transcript.
DISCUSSION
We have examined the kinetics of histone 3′-end processing by using an in vitro system that supports both transcription and processing. When the full-length H4 gene was used, the rate of appearance of mature H4 mRNA was influenced by a number of different processes, including initiation of transcription, the P-TEFb-dependent transition into productive elongation, elongation, and finally the processing reaction itself. Initiation is very fast and was separated from the rest of the reaction during a pulse-labeling. The t1/2 for all postinitiation events was about 3 min. Since the t1/2 for the actual processing event was less than 60 s, the rate-limiting step must be elongation of the transcript or the P-TEFb-dependent transition into productive elongation. Because the H4 transcript is fewer than 500 nt long and because elongation proceeds at about, 1,000 nt/min in the presence of the factors found in Kc cell nuclear extract (46), it should take the polymerase less than 30 s to reach the 3′ end of the gene (20, 39, 45). This leaves the function of P-TEFb as the likely rate-limiting step. This factor has been shown to function with a t1/2 in the range of 1 to 2 min on a variety of genes (26). The rate of appearance of full-length H4 mRNA during transcription is appropriate for the combination of the rates of the sequentially required steps. The production of histone mRNAs in vivo is coupled to the cell cycle, and during the maximal transcription found during S phase the initiation rate is very rapid. Since transcription of the histone genes occurs to variable extent throughout the cell cycle (28), it is possible that the processing event is rate-limiting during other (non-S) phases of the cell cycle due to limiting amounts of SLBP (54). However, it is difficult to prove this is the case because the level of histone mRNAs is dramatically affected by cell cycle-controlled stability of the transcripts (19, 21).
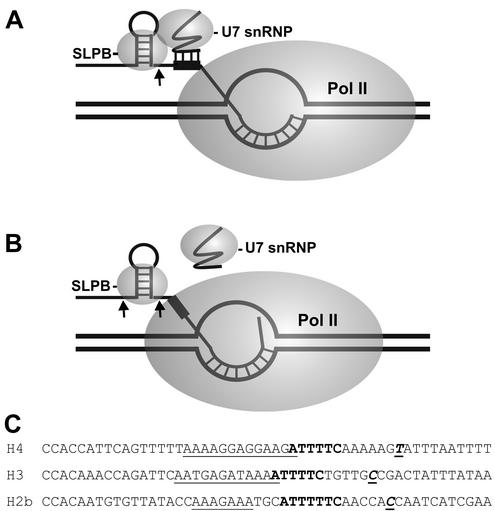
To determine whether histone 3′-end processing is coupled to transcription, we compared the rate of processing of transcripts that are in arrested elongation complexes to that of free RNA. Functional coupling of the two processes would result in an increase or decrease in the rate of processing of transcripts in elongation complexes. When the concentration of NTPs was maintained at transcription levels, processing was only slightly affected by the elongation complex. However, in the absence of NTPs, the processing of transcripts in elongation complexes is severely inhibited, with many transcripts remaining unprocessed. Because of this we suggest that under some conditions processing is negatively coupled to transcription, and we present a model for how this might occur (Fig. 9). When the polymerase encounters the arrest site 32 to 35 nt downstream from the processing site, it is likely that the HDE is just barely extruded from the RNA exit site of the polymerase (Fig. 9A). If the polymerase backslides (Fig. 9B), the HDE is masked, and processing is disrupted because the U7 snRNP cannot bind. In the presence of NTPs and S-II, such a polymerase may cleave and reextend the nascent transcript to the arrest site. U7 snRNA can then associate, and the transcript can be processed. This model is supported by the finding that in the presence of Mg, but without NTPs the transcripts in arrested complexes were shortened by S-II and were not substrates for the processing machinery. Our results are consistent with the polymerase backsliding into a processing-resistant conformation in the absence of NTPs. Further evidence for this processing-resistant conformation came from the experiments done in the presence of EDTA, which inhibits S-II mediated transcript cleavage. Although no shortening of transcripts was observed, processing was still inefficient.
FIG. 9.
Negative coupling of processing to transcription. The model explains how processing might be inhibited by RNA polymerase II arrested downstream of the processing site. (A) RNA polymerase II paused at the arrest site leaves just enough RNA exposed to allow binding of SLBP and the U7 snRNP and processing of the transcript. (B) When the polymerase backslides and enters the arrested conformation, the HDE (black box) is no longer accessible to the U7 snRNP, and processing is inhibited. (C) Comparison of the 3′ regions of H4, H3, and H2b. Starting with the last two nucleotides of the stem-loop RNA, sequences from H4, H3, and H2b were aligned. Underlined text, purine-rich region; boldface text, T-rich region; boldface underlined text, pause-arrest site.
Our results indicate that the transcription complex does not stimulate histone 3′-end processing but do not rule out the possibility that the transcription complex plays an important role in processing. On the contrary, it is not likely to be coincidental that all histone genes examined (H4, H2b, and H3) have strong arrest sites similar distances downstream from the stem-loop. If the arrest site was merely present to stop polymerases from transcribing downstream into nearby histone genes, it would not have to be so precisely positioned but rather could be present anywhere between the variably spaced genes. Inspection of the sequences of the three histone genes used indicates that there is a sequence element, AT4C or AT5C, immediately following the purine-rich HDE (Fig. 9C, boldface sequence). It is possible that this sequence may contribute to the location of the pause-arrest site. Similar stretches of T's are found in analogous locations in the Drosophila H1 and H2a genes. Pausing transcription a specific distance downstream from the HDE may be important for 3′-end formation. One possibility is that the transcription complex plays a negative role, stopping processing under certain conditions allowing control of 3′-end formation. Another possibility is that the arrest site facilitates termination. Since processing of histone mRNA occurs rapidly on free RNA, termination should not impede processing. It is possible that termination might actually stimulate 3′-end formation. The reduction in processing activity seen in the absence of NTPs might be due to inhibition of the ATP-dependent activity of transcription termination factor 2 (55) or some other nucleotide-dependent termination event. 3′-End formation and termination seem to be linked during production of mRNAs with poly(A) tails (9, 10, 40), and an intact 3′-processing site was required for termination to occur close to the processing site of the mouse histone H2a gene (3).
When processing was inhibited by carrying out transcription with a mixture of HeLa and Drosophila nuclear extract (Fig. 1E), a transcript was detected the predicted size of the arrested transcript. This appearance of this transcript and others that were slightly longer was transient, indicating that polymerases do not appear to reside at those sites for a long time in the presence of extract. This is likely due to the extensively studied action of DmS-II to reactivate arrested complexes (14) and because of the elongation stimulatory activity of TFIIF (39). However, since the t1/2 for processing is about 1 min and the t1/2 for the duration of the pause sites downstream of the processing site is 2 or 3 min (see Fig. 1E), the processed transcripts are likely primarily derived from the transcripts in paused elongation complexes.
It is widely argued that RNA processing is coupled to transcription. We have recently shown, by using an in vitro system in which the rate of capping of transcripts is stimulated 2 to 4 orders of magnitude by the elongation complex, that capping is functionally coupled to transcription (29). Most evidence for the coupling of processing and transcription focuses on the CTD and is based on in vitro association studies or chromatin immunoprecipitation-cross-linking studies. In vitro systems in which transcription and splicing or 3′-end cleavage at polyadenylation sites take place in the same reaction have been reported (12, 57, 58). However, the rate at which processing occurs in these systems does not seem compatible with in vivo requirements for RNA processing, and in the splicing study (12) it was not made clear if processing occurred on transcripts that were in elongation complexes. In the studies from both groups, the effect of elongation complexes was not examined by comparing the rates of processing of nascent and free transcripts. Our results indicate that 3′-end cleavage of the histone mRNAs occurs with a reasonable rate and occurs on complexes engaged in transcription. However, we do not see an enhancement of the rate of processing by transcription. This can be rationalized if polyadenylation is assumed to be the default processing pathway. To form a histone mRNA 3′ end, it is necessary to block polyadenylation by factors that might be associated with the elongating polymerase. Since at least some of the histone 3′-end factors are not associated with the elongation complex (our results), a physical disruption of the polyadenylation-polymerase complex does not seem likely. It is more likely that histone 3′-end formation is accomplished by stopping the polymerase downstream of the processing site and then rapidly processing the RNA in an RNA/SLPB/U7snRNP-driven reaction. In support of this idea, if histone 3′-end processing is blocked by removal of a required RNA element or by reduction of SLBP, polyadenylated histone mRNAs result (3, 21). It is possible that histone 3′-end processing may be coupled to transcription by a mechanism that increases transcription termination downstream of the processing site (3). Further work is needed to examine the connection between 3′-end formation and termination and the possible role of the T-rich sequence found following the HDE.
Acknowledgments
This work was supported by the NIH grant GM35500.
We thank Dan Cash and Hannah Jones for technical help with the construction of the histone minigenes.
REFERENCES
- 1.Bentley, D. 2002. The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14:336-342. [DOI] [PubMed] [Google Scholar]
- 2.Cho, D. C., E. C. Scharl, and J. A. Steitz. 1995. Decreasing the distance between the two conserved sequence elements of histone pre-messenger RNA interferes with 3′ processing in vitro. RNA 1:905-914. [PMC free article] [PubMed] [Google Scholar]
- 3.Chodchoy, N., N. B. Pandey, and W. F. Marzluff. 1991. An intact histone 3′-processing site is required for transcription termination in a mouse histone H2a gene. Mol. Cell. Biol. 11:497-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cramer, P., A. Srebrow, S. Kadener, S. Werbajh, M. de la Mata, G. Melen, G. Nogues, and A. R. Kornblihtt. 2001. Coordination between transcription and pre-mRNA processing. FEBS Lett. 498:179-182. [DOI] [PubMed] [Google Scholar]
- 5.Dominski, Z., and W. F. Marzluff. 1999. Formation of the 3′ end of histone mRNA. Gene 239:1-14. [DOI] [PubMed] [Google Scholar]
- 6.Dominski, Z., J. Sumerel, R. J. Hanson, and W. F. Marzluff. 1995. The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. RNA 1:915-923. [PMC free article] [PubMed] [Google Scholar]
- 7.Dominski, Z., X. C. Yang, C. S. Raska, C. Santiago, C. H. Borchers, R. J. Duronio, and W. F. Marzluff. 2002. 3′ end processing of Drosophila melanogaster histone pre-mRNAs: requirement for phosphorylated Drosophila stem-loop binding protein and coevolution of the histone pre-mRNA processing system. Mol. Cell. Biol. 22:6648-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominski, Z., L. X. Zheng, R. Sanchez, and W. F. Marzluff. 1999. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 19:3561-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dye, M. J., and N. J. Proudfoot. 1999. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol. Cell 3:371-378. [DOI] [PubMed] [Google Scholar]
- 10.Dye, M. J., and N. J. Proudfoot. 2001. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell 105:669-681. [DOI] [PubMed] [Google Scholar]
- 11.Feng, G., D. Lee, D. Wang, C. Chan, and R. Landick. 1994. GreA-induced transcript cleavage in transcription complexes containing Escherichia coli RNA polymerase is controlled by multiple factors, including nascent transcript location and structure. J. Biol. Chem. 269:22282-22294. [PubMed] [Google Scholar]
- 12.Ghosh, S., and M. A. Garcia-Blanco. 2000. Coupled in vitro synthesis and splicing of RNA polymerase II transcripts. RNA. 6:1325-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gick, O., A. Kramer, A. Vasserot, and M. L. Birnstiel. 1987. Heat-labile regulatory factor is required for 3′ processing of histone precursor mRNAs. Proc. Natl. Acad. Sci. USA 84:8937-8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo, H., and D. H. Price. 1993. Mechanism of DmS-II-mediated pause suppression by Drosophila RNA polymerase II. J. Biol. Chem. 268:18762-18770. [PubMed] [Google Scholar]
- 15.Hanson, R. J., J. Sun, D. G. Willis, and W. F. Marzluff. 1996. Efficient extraction and partial purification of the polyribosome-associated stem-loop binding protein bound to the 3′ end of histone mRNA. Biochemistry 35:2146-2156. [DOI] [PubMed] [Google Scholar]
- 16.Harris, M. E., R. Böhni, M. H. Schneiderman, L. Ramamurthy, D. Schümperli, and W. F. Marzluff. 1991. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol. 11:2416-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirose, Y., and J. L. Manley. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395:93-96. [DOI] [PubMed] [Google Scholar]
- 18.Hirose, Y., R. Tacke, and J. L. Manley. 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 13:1234-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann, I., and M. L. Birnstiel. 1990. Cell cycle-dependent regulation of histone precursor mRNA processing by modulation of U7 snRNA accessibility. Nature 346:665-668. [DOI] [PubMed] [Google Scholar]
- 20.Kephart, D. D., N. F. Marshall, and D. H. Price. 1992. Stability of Drosophila RNA polymerase II elongation complexes in vitro. Mol. Cell. Biol. 12:2067-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanzotti, D. J., H. Kaygun, X. Yang, R. J. Duronio, and W. F. Marzluff. 2002. Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3′ end processing in vivo. Mol. Cell. Biol. 22:2267-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lifton, R. P., M. L. Goldberg, R. W. Karp, and D. S. Hogness. 1978. The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. Cold Spring Harbor Symp. Quant. Biol. 42(Pt. 2):1047-1051. [DOI] [PubMed] [Google Scholar]
- 23.Liu, T. J., B. J. Levine, A. I. Skoultchi, and W. F. Marzluff. 1989. The efficiency of 3′-end formation contributes to the relative levels of different histone mRNAs. Mol. Cell. Biol. 9:3499-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luscher, B., and D. Schumperli. 1987. RNA 3′ processing regulates histone mRNA levels in a mammalian cell cycle mutant: a processing factor becomes limiting in G1-arrested cells. EMBO J. 6:1721-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall, N. F., J. Peng, Z. Xie, and D. H. Price. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271:27176-27183. [DOI] [PubMed] [Google Scholar]
- 26.Marshall, N. F., and D. H. Price. 1992. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol. Cell. Biol. 12:2078-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall, N. F., and D. H. Price. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270:12335-12338. [DOI] [PubMed] [Google Scholar]
- 28.Marzluff, W. F. 1992. Histone 3′ ends: essential and regulatory functions. Gene Expr. 2:93-97. [PMC free article] [PubMed] [Google Scholar]
- 29.Moteki, S., and D. H. Price. 2002. Functional coupling of capping and transcription of mRNA. Mol. Cell 10:599-609. [DOI] [PubMed] [Google Scholar]
- 30.Muller, B., J. Link, and C. Smythe. 2000. Assembly of U7 small nuclear ribonucleoprotein particle and histone RNA 3′ processing in Xenopus egg extracts. J. Biol. Chem. 275:24284-24293. [DOI] [PubMed] [Google Scholar]
- 31.Oelgeschlager, T. 2002. Regulation of RNA polymerase II activity by CTD phosphorylation and cell cycle control. J. Cell Physiol. 190:160-169. [DOI] [PubMed] [Google Scholar]
- 32.Pandey, N. B., and W. F. Marzluff. 1987. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol. 7:4557-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker, C. S., and J. Topol. 1984. A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell 36:357-369. [DOI] [PubMed] [Google Scholar]
- 34.Peng, J., N. F. Marshall, and D. H. Price. 1998. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem. 273:13855-13860. [DOI] [PubMed] [Google Scholar]
- 35.Peng, J., Y. Zhu, J. T. Milton, and D. H. Price. 1998. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 12:755-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price, D. H., and C. S. Parker. 1984. The 3′ end of Drosophila Histone H3 mRNA is produced by a processing activity in vitro. Cell 38:423-429. [DOI] [PubMed] [Google Scholar]
- 38.Price, D. H., A. E. Sluder, and A. L. Greenleaf. 1987. Fractionation of transcription factors for RNA polymerase II from Drosophila Kc cell nuclear extracts. J. Biol. Chem. 262:3244-3255. [PubMed] [Google Scholar]
- 39.Price, D. H., A. E. Sluder, and A. L. Greenleaf. 1989. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol. Cell. Biol. 9:1465-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 41.Renner, D. B., Y. Yamaguchi, T. Wada, H. Handa, and D. H. Price. 2001. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 276:42601-42609. [DOI] [PubMed] [Google Scholar]
- 42.Scharl, E. C., and J. A. Steitz. 1994. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J. 13:2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scharl, E. C., and J. A. Steitz. 1996. Length suppression in histone messenger RNA 3′-end maturation: processing defects of insertion mutant premessenger RNAs can be compensated by insertions into the U7 small nuclear RNA. Proc. Natl. Acad. Sci. USA 93:14659-14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaufele, F., G. M. Gilmartin, W. Bannwarth, and M. L. Birnstiel. 1986. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature 323:777-781. [DOI] [PubMed] [Google Scholar]
- 45.Sluder, A. E., A. L. Greenleaf, and D. H. Price. 1989. Properties of a Drosophila RNA polymerase II elongation factor. J. Biol. Chem. 264:8963-8969. [PubMed] [Google Scholar]
- 46.Sluder, A. E., D. H. Price, and A. L. Greenleaf. 1988. Elongation by Drosophila RNA polymerase II. J. Biol. Chem. 263:9917-9925. [PubMed] [Google Scholar]
- 47.Spycher, C., A. Streit, B. Stefanovic, D. Albrecht, T. H. Koning, and D. Schumperli. 1994. 3′ end processing of mouse histone pre-mRNA: evidence for additional base-pairing between U7 snRNA and pre-mRNA. Nucleic Acids Res. 22:4023-4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Streit, A., T. W. Koning, D. Soldati, L. Melin, and D. Schumperli. 1993. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone pre-mRNA in vitro. Nucleic Acids Res. 21:1569-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strub, K., and M. L. Birnstiel. 1986. Genetic complementation in the Xenopus oocyte: coexpression of sea urchin histone and U7 RNAs restores 3′ processing of H3 pre-mRNA in the oocyte. EMBO J. 5:1675-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan, E., C. Santiago, E. D. Parker, Z. Dominski, X. Yang, D. J. Lanzotti, T. C. Ingledue, W. F. Marzluff, and R. J. Duronio. 2001. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 15:173-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, Z. F., T. C. Ingledue, Z. Dominski, R. Sanchez, and W. F. Marzluff. 1999. Two Xenopus proteins that bind the 3′ end of histone mRNA: implications for translational control of histone synthesis during oogenesis. Mol. Cell. Biol. 19:835-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, Z. F., M. L. Whitfield, T. C. 3. Ingledue, Z. Dominski, and W. F. Marzluff. 1996. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10:3028-3040. [DOI] [PubMed] [Google Scholar]
- 54.Whitfield, M. L., L. X. Zheng, A. Baldwin, T. Ohta, M. M. Hurt, and W. F. Marzluff. 2000. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20:4188-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie, Z., and D. H. Price. 1997. Drosophila factor 2, an RNA polymerase II transcript release factor, has DNA-dependent ATPase activity. J. Biol. Chem. 272:31902-31907. [DOI] [PubMed] [Google Scholar]
- 56.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 57.Yonaha, M., and N. J. Proudfoot. 1999. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell 3:593-600. [DOI] [PubMed] [Google Scholar]
- 58.Yonaha, M., and N. J. Proudfoot. 2000. Transcriptional termination and coupled polyadenylation in vitro. EMBO J. 19:3770-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zanier, K., I. Luyten, C. Crombie, B. Muller, D. Schumperli, J. P. Linge, M. Nilges, and M. Sattler. 2002. Structure of the histone mRNA hairpin required for cell cycle regulation of histone gene expression. RNA 8:29-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeng, C., and S. M. Berget. 2000. Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol. Cell. Biol. 20:8290-8301. [DOI] [PMC free article] [PubMed] [Google Scholar]