Abstract
We have identified a region near the C terminus of capsid (CA) of murine leukemia virus (MLV) that contains many charged residues. This motif is conserved in various lengths in most MLV-like viruses. One exception is that spleen necrosis virus (SNV) does not contain a well-defined domain of charged residues. When 33 amino acids of the MLV motif were deleted to mimic SNV CA, the resulting mutant produced drastically reduced amounts of virions and the virions were noninfectious. Furthermore, these viruses had abnormal sizes, often contained punctate structures resembling those in the cell cytoplasm, and packaged both ribosomal and viral RNA. When 11 or 15 amino acids were deleted to modify the MLV CA to resemble those from other gammaretroviruses, the deletion mutants produced virions at levels comparable to those of the wild-type virus and were able to complete one round of virus replication without detectable defects. We generated 10 more mutants that displayed either the wild-type or mutant phenotype. The distribution of the wild-type or mutant phenotype did not directly correlate with the number of amino acids deleted, suggesting that the function of the motif is determined not simply by its length but also by its structure. Structural modeling of the wild-type and mutant proteins suggested that this region forms α-helices; thus, we termed this motif the “charged assembly helix.” This is the first description of the charged assembly helix motif in MLV CA and demonstration of its role in virus budding and assembly.
All retroviruses encode at least the gag, pro, pol, and env genes, which are translated into polyproteins and subsequently cleaved into individual virion proteins (38). The gag gene products form the structure of the virions, pro gene products process the viral proteins, pol gene products carry out important enzymatic reactions, and env gene products serve as the viral surface envelope (38). Although all four gene products are essential in the generation of wild-type, infectious virions, the gag gene product, Gag polyprotein, drives the formation of the virus particles. When Gag polyproteins are expressed in host cells, they can form virus-like particles without other retroviral components such as pro, pol, or env gene products or viral RNA containing the retroviral packaging signals (35; J. W. Wills and R. C. Craven, Editorial, AIDS 5:639-654, 1991). Purified Gag polyproteins, or portions of the polyproteins, can also form virus-like particles in vitro (6, 7, 13, 19). Together, these data indicate that Gag polyproteins are necessary and sufficient for the formation of the virus-like particles.
Gag polyproteins from all retroviruses contain at least three domains that are later cleaved by viral protease into mature proteins: matrix (MA), capsid (CA), and nucleocapsid (NC) (38). Most retroviruses also contain other cleavage products that differ in size and in the location of the domain on the polyprotein. For example, murine leukemia virus (MLV) Gag has an additional 12-kDa cleavage product termed p12; the p12 domain is located between MA and CA in the Gag polyprotein. In human immunodeficiency virus type 1 (HIV-1), Gag polyproteins have three additional cleavage products, p2, p1, and p6. The p2 domain is located between CA and NC, whereas p1 and p6 are located C terminal to NC in the Gag polyprotein (32).
Different domains of Gag polyproteins contain distinct signals to direct virus assembly and budding. Signals in MA target Gag to the cell membrane, motifs in the NC domains mediate RNA binding, and motifs in CA and NC are important in the multimerization of Gag polyproteins (35, 38). After formation of the virus-like particles, host cell factors recruited by the late domains, which are located at various positions in Gag of different viruses, interact with the cell membrane to allow the release of the viral particles (16). During or soon after virus budding, viral proteases are activated and cleave the polyproteins into smaller proteins (35). Upon protease cleavage, the virions undergo morphological changes and become mature particles (35). The mature particle, but not the immature particle, contains an electron-dense core, which can be identified by electron microscopy (EM) (38).
CA is involved in multiple stages in the retroviral replication cycle. As a cleaved, mature viral protein, CA forms the condensed core of the mature virus particle and constitutes part of the reverse transcription complex in certain viruses (4, 14, 38). As a part of the Gag polyprotein, CA plays a role in virus assembly. This is supported by genetic studies, in which mutations in CA often lead to assembly defects (15, 20, 35). However, the exact mechanism of CA's involvement in virus assembly remains unclear. Structural analyses of CA from several viruses illustrate that these proteins consist of two α-helix-rich domains, the N-terminal and C-terminal domains, connected by a flexible linker region (3, 18, 23, 24, 39). The primary sequences of the CA from different retroviruses are divergent; the most conserved region in CA is located in the C-terminal domain (QX3EX7R) and is termed the major homology region (MHR) (Wills and Craven, editorial, 1991). Mutations in this region often lead to defects in the viral replication cycle (2, 5, 10, 28).
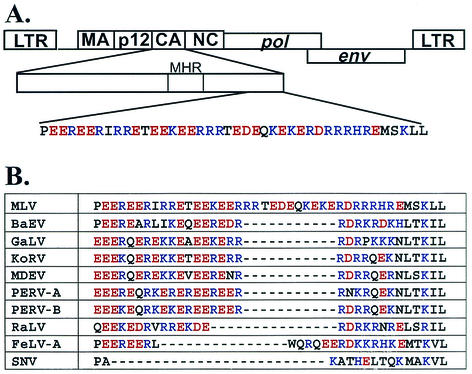
The amino acid sequence at the C-terminal region of MLV CA has a stretch of 41 amino acids rich in charged residues (Fig. 1A). Of these 41 amino acids, 33 are charged residues with similar numbers of acidic and basic residues (16 and 17, respectively). The function of this motif rich in charged residues is unknown. In this report, we examined the effects of mutations on this motif and found that disruption of this motif can result in assembly defects. Furthermore, mutants lacking functional motifs generated virions with highly differing particle sizes and packaged both ribosomal and viral RNAs. These results indicated that this motif rich in charged residues is an important region in the assembly of MLV particles.
FIG. 1.
Location and sequence of the motif rich in charged residues. (A) Location and sequence of the motif in MLV. (B) Comparison of sequences in the motif among several gammaretroviruses. BaEV, baboon endogenous virus; GaLV, gibbon ape leukemia virus; KoRV, Phascolarctos cinereus (koala) retrovirus; MDEV, Mus dunni endogenous virus; PERV-A, porcine endogenous virus class A; PERV-B, porcine endogenous virus class B; RaLV, rat leukemia virus; FeLV-A, feline leukemia virus subgroup A; SNV, spleen necrosis virus. Acidic and basic residues are shown in red and blue, respectively.
MATERIALS AND METHODS
Plasmids and construction of mutant gag-pol expression constructs.
All vectors were constructed using standard cloning procedures (34). Mutations were generated using overlapping PCR and cloning of the resulting DNA fragments into the MLV-based gag-pol expression construct pWZH30 (41), a generous gift from Vinay Pathak. Plasmid pWZH30 is derived from pLGPS (30), which expresses gag-pol from a truncated 5′ long terminal repeat (LTR) and does not contain the packaging signal or the 3′ LTR. The overall structures of the mutant gag-pol constructs were determined by restriction enzyme analyses; the portions of DNA generated by PCR were further characterized by DNA sequencing. Detailed cloning procedures are available upon request. Plasmids pEW22U and pEW22D also contained an additional H114R mutation in the CA region; this mutation was located 108 amino acids N terminal to the motif described in this report.
Plasmid pSV-a-MLV-env expresses amphotropic MLV env (27). MLV-based vector pSR2 (8) contains all the cis-acting elements essential for replication; pSR2 does not express viral proteins, but expresses the hygromycin phosphotransferase B gene (hygro), which confers resistance to hygromycin (22). pSR2 also has two modified LTRs, each of which contains a copy of the green fluorescent protein gene.
Cell culture, DNA transfection, virus propagation, and virus infection.
D17 cells are dog osteosarcoma cells permissive to amphotropic MLV infection (33); 293T cells are human embryonic kidney cells (12). Both cell lines were maintained at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium supplemented with serum and antibiotics (8, 12). Using a CalPhos transfection kit (ClonTech) or MBS transfection kit (Stratagene) with a mixture of DNA containing the gag-pol expression construct, pSR2, and pSV-a-MLV-env at a 2:2:1 weight ratio, DNA transfection was performed by the calcium phosphate precipitation method (34).
Dilutions (10-fold) of the viruses were used to infect D17 cells in the presence of Polybrene (50 μg/ml final concentration). Infected cells were placed on hygromycin selection at a final concentration of 120 μg/ml, and virus titers were determined by the numbers of hygromycin-resistant colonies. In general, viral titers measured in these assays agreed with the reverse transcriptase (RT) activities. The virus titers presented were not standardized to RT activities, because some of the mutants had low RT activities which were not significantly higher than background.
RT assay, Western analyses, and EM analyses.
Using a Surespin 630 rotor (Sorvall) in a Discovery 100S ultracentrifuge (Sorvall), viruses were harvested and concentrated by centrifugation at 25,000 rpm for 90 min at 4°C. RT activities of the viral samples were determined using standard procedures (8, 36). Proteins from cell lysates or virus lysates were examined by Western blotting analyses performed using standard procedures (34). Polyclonal rabbit anti-MLV-CA and anti-MLV-MA antibodies were kind gifts from the AIDS Vaccine Program (Science Applications International Corporation—Frederick).
For EM analyses, transfected 293T cells were washed with phosphate-buffered saline, pelleted by low-speed centrifugation, and fixed in 2% glutaraldehyde in cacodylate buffer (0.1 M, pH 7.4). EM analyses were performed as previously described (37) (Hitachi H7000), and images were captured by a digital camera system (Gatan).
RNA analyses.
To examine the RNA encapsidated in wild-type and mutant viruses, gag-pol expression constructs were used to separately transfect SR2-293T cells. SR2-293T cells are 293T cells containing integrated SR2 proviruses. These cells were generated by infecting 293T cells with SR2-containing virions, subjecting the infected cells to hygromycin selection, and pooling the resulting hygromycin-resistant cell colonies. Cell-free viruses were harvested from transfected SR2-293T cells, and virion RNAs were isolated using previously described protocols (17). The amounts of encapsidated SR2 RNA and rRNA were determined using quantitative real-time RT PCR (ABI 7700 sequence detector). Primers and probe sequences derived from hygro were used to detect SR2 vector RNA. The primers and probe had the following sequences: 5′ ACGAGGTCGCCAACATCTTC 3′ (primer), 5′ AGCGCGTCTGCTGCTCC 3′ (primer), and 5′ 6-carboxyfluorescein (FAM)-TCTGGAGGCCGTGGTTGGCTTGTA-6-carboxy-N,N,N′,N′-tetramethylrhodamine (TAMRA)-3′ (probe). Primers and a probe derived from an 18S rRNA gene were used to detect 18S rRNA. The primers and probe had the following sequences: 5′ GCCGCTAGAGGTGAAATTCTTG 3′ (primer), 5′ CATTCTTGGCAAATGCTTTCG 3′ (primer), and 5′-FAM-ACCGGCGCAAGACGGACCAGA-TAMRA-3′ (probe). RNA transcripts containing hygro sequences and purified rRNAs were used as templates to generate standard curves in the real-time RT PCR for SR2 and rRNA detection, respectively. RNA isolated from mock-transfected samples was used as a negative control.
RESULTS
A motif rich in charged residues is conserved in the CA domain of most gammaretroviruses.
To determine whether the motif rich in charged residues is unique to MLV CA, the primary amino acid sequences of CA from multiple retroviruses were compared. Most of the gammaretroviruses have similar motifs, albeit with different lengths. A notable exception is spleen necrosis virus, which does not have a region with many charged residues in its CA (Fig. 1B). Several viruses, including gibbon ape leukemia virus, have motifs that are 11 amino acids shorter than that of MLV. Rat leukemia virus and feline leukemia virus both have motifs that are 15 amino acids shorter than that of MLV (Fig. 1B). All of the motifs rich in charged residues have nearly balanced numbers of acidic and basic residues.
We also examined the primary sequences of several other groups of more distant retroviruses and found that the CA proteins of these viruses do not have motifs rich in charged residues at the C-terminal region of their CA and sequences between CA and NC (data not shown). These viruses include Rous sarcoma virus (RSV), an alpharetrovirus; Mason-Pfizer monkey virus, a betaretrovirus; human T-cell leukemia virus type 1, a deltaretrovirus; and HIV-1, a lentivirus.
The motif rich in charged residues is essential for efficient MLV replication and assembly.
To test whether this motif is essential for MLV replication and to examine its possible roles, we constructed and characterized a mutant containing a large deletion that encompassed almost the entire motif. Plasmid pSR33 was derived from the MLV gag-pol expression construct pWZH30 (41) and contained a 33-amino-acid deletion in the motif (Fig. 2A). To examine the phenotype of pSR33, we used the following protocols. The MLV gag-pol expression construct pWZH30 or pSR33 was introduced into 293T cells by transfection along with pSV-a-MLV-env (27) and pSR2 (8). pSV-a-MLV-env expresses amphotropic MLV env. pSR2 is an MLV-based vector that does not express gag-pol or env but instead expresses hygro (22), which confers resistance to hygromycin. Supernatants were harvested from the transfected 293T cells, a portion of the sample was used to infect target D17 cells to determine the virus titer, and the other portion was subjected to biochemical analyses. Infected D17 cells were subjected to hygromycin selection, and the SR2 virus titers were determined by the number of hygromycin-resistant cell colonies. The amounts and properties of the viral protein released in the cell-free supernatants were determined by Western blotting analyses and RT assays, and the morphology of the released virions was examined by EM.
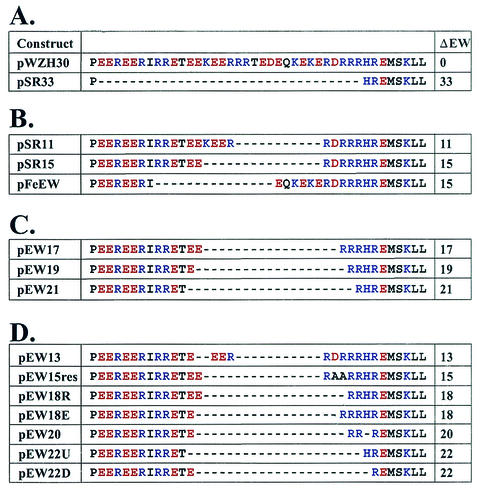
FIG. 2.
Amino acid sequences of the motif rich in charged residues in wild-type and mutant gag-pol expression constructs. ΔEW, the numbers of amino acids deleted.
Viral titers generated from six sets of independent experiments are summarized in Table 1. Supernatants from pWZH30-transfected cells on average generated a titer of 6.0 × 103 hygromycin-resistant CFU/ml. However, supernatants harvested from pSR33-transfected cells did not generate detectable titers in any of the six experiments.
TABLE 1.
Virus titers generated by gag-pol expression constructs with wild-type or mutant motifs
| Construct | Viral titer (103)a | |
|---|---|---|
| pWZH30 | 6.0 ± 2.1 | 7.5 ± 1.9 |
| pSR33 | <0.001 | ND |
| pSR11 | 2.5 ± 1.7 | ND |
| pSR15 | 10.0 ± 8.0 | ND |
| pFeEW | 12.3 ± 11.4 | ND |
| pEW17 | <0.001 | ND |
| pEW19 | 7.3 ± 6.4 | ND |
| pEW21 | <0.001 | ND |
| pEW13 | ND | <0.001 |
| pEW15-res | ND | 10.8 ± 5.1 |
| pEW18E | ND | 7.0 ± 3.9 |
| pEW18R | ND | 3.9 ± 2.8 |
| pEW20 | ND | <0.001 |
| pEW22U | ND | 4.5 ± 2.4 |
| pEW22D | ND | 6.8 ± 1.6 |
Viral titers are shown as CFU/ml (means ± standard errors). The data from the center and right columns were generated from four to six and two to five sets of independent experiments, respectively. ND, not determined.
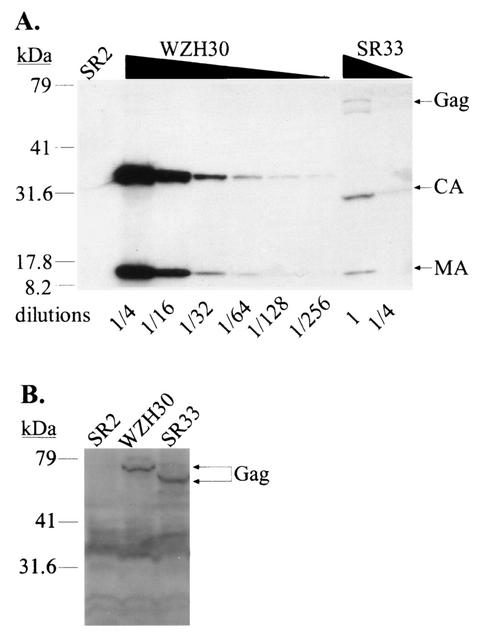
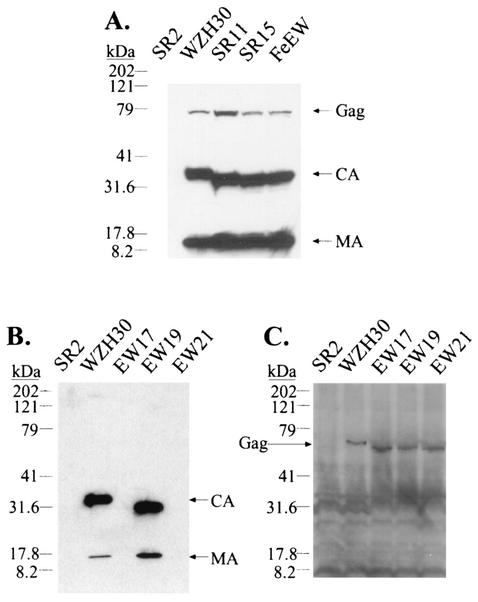
Representative analyses of viral proteins from transfected cells and harvested supernatants are shown in Fig. 3. Virions generated from pWZH30-transfected cells are referred to as wild type. Virions were released from pSR33-transfected cells at an approximately 30- to 100-fold-reduced level compared with that of wild type (Fig. 3A and data not shown). Western analyses also showed that Gag polyproteins expressed by pSR33 were mostly processed; CA from pSR33-derived virions migrated faster than CA from wild-type viruses, consistent with the 33-amino-acid deletion, whereas MA from both viruses had the same mobility. Western analyses from transfected cell lysates showed that Gag polyproteins were expressed in pWZH30- and pSR33-transfected cells at similar levels (Fig. 3B), indicating that the 33-amino-acid deletion did not significantly affect the amount of Gag polyprotein expressed in the transfected cells. Therefore, viruses generated from pSR33-transfected cells had a defect in virus assembly and/or release.
FIG. 3.
Characterization of pWZH30- and pSR33-transfected cells and virions derived from these cells. (A) Western analyses of cell-free virions. (B) Western analyses of cell lysates. Lanes labeled SR2 show results for supernatants or cell lysates from cells transfected with pSR2 and pSV-a-MLV-env; other lane labels specify supernatants or cell lysates from cells transfected with the indicated gag-pol expression construct plus pSR2 vector and pSV-a-MLV-env. Both anti-MLV CA and anti-MLV MA antibodies were used for the detection of virion lysates, whereas only anti-MLV CA antibody was used to detect cell lysates. The larger molecular-size bands (near the 79-kDa marker) detected in the SR33 virion lysates are probably caused by incomplete processing. A wide smear (between 30 to 40 kDa) present in the cell lysate analyses is presumably background created by nonspecific antibody binding. The migration of molecular size standards (in kilodaltons) and Gag, CA, and MA is shown.
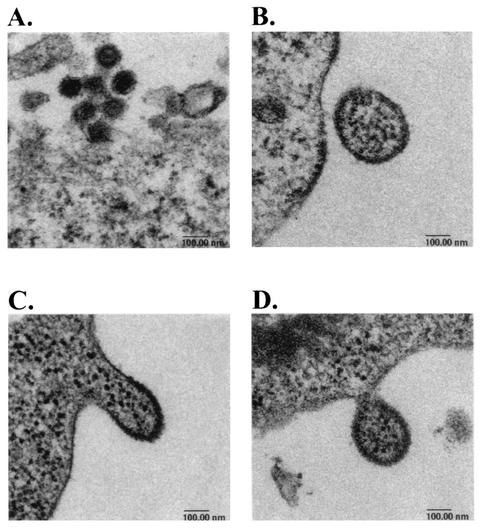
To examine the morphology of the viruses generated from the pWZH30- or pSR33-transfected cells, EM analyses were performed. Representative images of the wild-type and pSR33-derived virions are shown in Fig. 4; all of these images are shown with the same magnification. These analyses indicated that pSR33-derived viruses differed from wild-type viruses in both size and morphology. Wild-type virions were close to 100 nm in diameter, and both immature particles and mature particles with condensed cores were observed (Fig. 4A and data not shown). In contrast, pSR33-derived virions were often much larger than 100 nm and all appeared to be immature (Fig. 4B to D and data not shown). In addition, punctate structures similar to those in the cell cytoplasm were observed in these virions. Virions with a wide range of particle sizes containing punctate structures were observed only in pSR33-transfected cells and not in untransfected cells or pWZH30-transfected cells (data not shown).
FIG. 4.
Electron micrographs of pWZH30-derived virions (A) and pSR33-derived virions (B, C, and D). All four micrographs are shown in the same magnification, with a 100-nm bar indicated at the bottom of each graph.
To further quantify the distributions of virion particle sizes, we measured the sizes of 100 wild-type virions and 100 pSR33-derived virions (Table 2). The particle sizes of the wild-type virions were tightly clustered; of the 100 virions, 98 were 90 to 130 nm in diameter. In contrast, the particle sizes of pSR33-derived virions were distributed through a wide range. Of the 100 pSR33-derived virions, only 31 were 90 to 130 nm in diameter; the other 69 viral particles were smaller or larger than this size range, with some viruses more than 400 nm in diameter.
TABLE 2.
Distribution of particle sizes from pWZH30- and pSR33-derived virions
| Construct | No. of virions with particles of diameter (nm):
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <100
|
100-150
|
150-200 | 200-250 | 250-300 | 300-350 | 350-400 | 400-450 | 450-500 | ||||||
| <90 | 90-100 | 100-110 | 110-120 | 120-130 | 130-140 | 140-150 | ||||||||
| pWZH30 | 0 | 10 | 32 | 41 | 15 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| pSR33 | 5 | 8 | 5 | 10 | 8 | 7 | 6 | 17 | 21 | 6 | 2 | 2 | 2 | 1 |
The electron micrographs shown in Fig. 4C and D captured the images of pSR33-derived viruses in the process of budding out of the cells, and the contents of the mutant virions bear a striking resemblance to the cytoplasm. This observation led us to examine the RNA content of pSR33-derived virions. To avoid the potential complication caused by contamination of transfected MLV vector pSR2 DNA, we used 293T cells containing integrated SR2 proviruses (referred to as SR2-293T cells) in these experiments. SR2-293T cells were transiently transfected with pWZH30 or pSR33, and cell-free viruses were harvested. For each virus sample, an aliquot was used to isolate RNA and the other aliquot was used to generate lysates for Western analyses. Using Western analyses on serially diluted lysates, the amounts of viruses in these samples were determined by measuring the quantity of MA in cell-free viral lysates. The amounts of SR2 RNA and 18S rRNA in each sample were determined using quantitative real-time RT PCR with probes and primers derived from hygro and 18S rRNA sequences, respectively. Data summarized from three sets of independent experiments are shown in Fig. 5A and B.
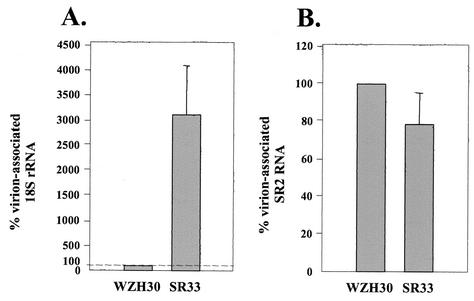
FIG. 5.
Comparison of the RNA contents in pWZH30- and pSR33-derived virions. (A) Virion-associated 18S rRNA. (B) Virion-associated SR2 RNA. Western analyses were performed to estimate the amounts of virions, and RNAs from the same amounts of pWZH30- and pSR33-derived virions were compared. The amount of RNAs from pWZH30-derived virions was set as 100%. Data from three independent experiments are shown as means ± standard errors.
Virions derived from pSR33 encapsidated SR2 RNA at an efficiency of 79% of the wild-type virions. More strikingly, pSR33-derived virions packaged 3,100% of the 18S rRNA relative to wild-type virions. To ensure that the RNA measured in these experiments was derived from virions rather than from cellular debris, we also measured the amounts of SR2 RNA and 18S rRNA in the supernatants from mock-transfected cells and compared the amounts of RNA detected in the same volume of supernatant from pWZH30-transfected cells. In two independent experiments, we were not able to detect SR2 RNA (less than 0.01% of the signals from wild-type virus) and we detected low amounts of rRNA (an average of 1.6% of the amount detected from wild-type virus controls) in the supernatant from mock transfected. These results confirmed that pSR33-derived virions contained rRNA; therefore, the punctate structures inside the virions observed by EM were most likely ribosomes.
Taken together, these data indicate that deletion of the motif rich in charged residues can result in defects in virus assembly, which would affect both the numbers of virions released from the cells and the properties of the released viral particles.
The MLV motif rich in charged residues can tolerate relatively large deletions: phenotypes of mutants with motifs mimicking those of other gammaretroviruses.
As shown in Fig. 1B, many gammaretroviruses have shorter motifs compared with that of MLV. To test whether a shorter motif can function in the context of MLV, we generated three MLV-based mutants containing motifs that were the same lengths as those from gibbon ape leukemia virus, rat leukemia virus, and feline leukemia virus. These mutants (pSR11, pSR15, and pFeEW) contained 11-, 15-, and 15-amino-acid deletions, respectively (Fig. 2B). The phenotypes of these mutants were examined using the protocol described above. Western analyses of the supernatants from cells transfected with these plasmids indicated that the levels of virions released and the processing of Gag polyproteins were similar to those of the wild type (Fig. 6A). Virions from these deletion mutants were infectious, generating titers similar to those from wild-type virus (Table 1). EM analyses indicated that these virions had the wild-type virion morphology (data not shown). These data established that these deletion mutants were phenotypically similar to wild-type viruses. Therefore, the functional requirement of the motif rich in charged residues could be fulfilled by shorter-than-wild-type sequences.
FIG. 6.
Western analyses of the wild type and the deletion mutants. (A and B) Western analysis of cell-free virions. (C) Western analysis of cell lysates. The lane labels, antibodies used, and explanations are the same as those described for Fig. 3. The migration of molecular size standards (in kilodaltons) and Gag, CA, and MA is shown.
The function of the motif rich in charged residues is not solely determined by its length and primary sequences.
Because the virions generated from pSR11-, pSR15-, and pFeEW-transfected cells were similar to wild-type viruses in all aspects examined, we questioned whether the MLV motif can tolerate even larger deletions. To investigate this question, we generated mutants that contained 17-, 19-, and 21-amino-acid deletions in the motif rich in charged residues (pEW17, pEW19, and pEW21, respectively) (Fig. 2C). To maintain the balance in acidic and basic residues, we used pSR15 as a template and deleted equal numbers of acidic and basic residues in these mutants. The phenotypes of these constructs were examined using the protocols described above. Western analyses indicated that pEW17- and pEW21-transfected cells released very little virus in their supernatants (Fig. 6B). In contrast, pEW19-transfected cells released amounts of virus similar to those of the wild type into their supernatants (Fig. 6B); furthermore, the Gag polyproteins from pEW19-generated viruses were processed similarly to the wild-type viruses (Fig. 6B). The reduction of the virion production in pEW17- and pEW21-transfected cells was not due to lack of Gag expression, as indicated by Western analyses of the cell lysates (Fig. 6C). EM studies of these samples revealed that viruses generated from pEW17 resembled those from pSR33, whereas pEW19 generated virions with morphology similar to that of wild-type viruses (data not shown). In four to six sets of independent infection experiments, pEW17 and pEW21 did not generate detectable titers; however, pEW19 and pWZH30 generated similar titers (Table 1). Phenotypically wild-type virions were generated by pEW19 but not by pEW17, which contained a smaller deletion. This result indicates that the length of the primary sequence is not a sufficient criterion for predicting the function of the mutant motif and suggests that a secondary or tertiary structure is important to the function of the motif rich in charged residues.
Testing the hypothesis that the MLV motif rich in charged residues forms a functionally important α-helical structure.
Deletion analyses implied a periodicity to the correlation between the numbers of amino acids deleted and the phenotypes of the resulting mutants. For example, deletion of 11, 15, or 19 amino acids resulted in functional proteins but deletion of 17 or 21 amino acids did not. The differences in the numbers of amino acids among pSR11, pSR15, and pEW19 are four and eight, which approximate one and two complete turns in α helices, respectively. In addition, analyses of the primary sequences of the MLV motif indicate that this region forms an α-helical structure (Chou-Fasman prediction, PepPlot, and PeptideStructure; GCG program, University of Wisconsin). We hypothesized that the MLV motif forms an α-helical structure and that maintenance of the overall structure and preservation of the helical phases are important to the function of this motif. We also hypothesized that deletion of one or more complete turns of the α-helices can allow the preservation of the structure and lead to the production of phenotypically wild-type virions and that deletions of incomplete turns can drastically alter the properties of the α-helices and result in phenotypically altered virions similar to those generated by pSR33. To test this hypothesis, we generated additional mutants and examined their phenotypes by using the same protocols as described above.
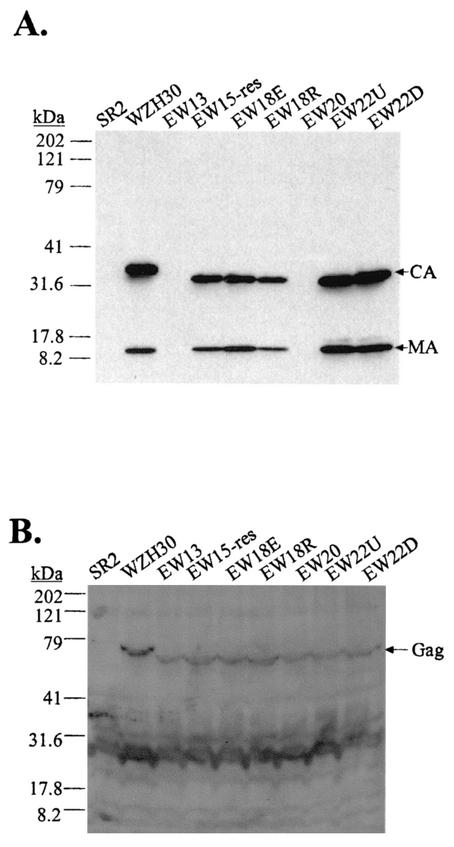
A 13-amino-acid deletion should delete approximately 3.6 turns of the α-helices and should result in the formation of phenotypically altered viruses. Similarly, deletion of 20 amino acids (5.6 turns) should also result in an altered phenotype. Therefore, we generated pEW13 and pEW20 containing 13- and 20-amino-acid deletions in the motif, respectively (Fig. 2D), and we examined their properties. As predicted, Western analyses indicated that these two mutants released drastically reduced levels of virions (Fig. 7A) and these virions generated titers less than 1 CFU/ml in two to five independent experiments (Table 1). Western analyses of cell lysates indicated that Gag polyproteins were expressed in pEW13- and pEW20-transfected cells (Fig. 7B).
FIG. 7.
Western analyses of mutants generated to test the proposed α-helical structure. (A) Western analysis of cell-free virions. (B) Western analysis of cell lysates. The lane labels, antibodies used, and explanations are the same as those described for Fig. 3. The migration of molecular size standards (in kilodaltons) and Gag, CA, and MA is shown.
Deletion of 18 and 22 amino acids of the motif should remove 5 and 6.1 turns of the α-helices, respectively, which should result in functional motifs. Two 18-amino-acid deletion mutants were generated based on pEW17: one (pEW18E) had an acidic residue deleted, and the other (pEW18R) had a basic residue deleted (Fig. 2D). Two 22-amino-acid deletion mutants were also generated based on pEW21: one (pEW22U) had a glutamic acid deleted, and the other (pEW22D) had a histidine deleted (Fig. 2D). Our results indicated that all four of these mutants had wild-type phenotypes in the amounts of virus released (Fig. 7A) and generated infectious viruses with titers similar to those from wild-type virus (Table 1). These data also demonstrated that we were able to restore the infectivity of the virions by deleting one amino acid in the motif from the two parental constructs pEW17 and pEW21, which had generated noninfectious virions with mutant phenotypes. Different numbers of acidic and basic residues were deleted from pEW18E and EW18R. Eight acidic and eight basic residues were deleted from pEW18R, whereas seven acidic and nine basic residues were deleted from pEW18E. Since both constructs generated infectious virions, these data suggested that the motif can tolerate minor fluctuations in the numbers of charged residues. pEW22U and pEW22D also contained an additional mutation elsewhere in the CA region (H114R). This mutation probably did not affect CA function, since viruses derived from these two plasmids had the wild-type phenotype.
We also hypothesized that the individual amino acids do not affect the function of the motif as much as the maintenance of the helical phases. We postulated that the mutant phenotype of pEW17-derived virions was caused by alteration of the helical phase and not by the loss of charged residues. To test this hypothesis, we generated pEW15-res, in which two alanines were inserted into pEW17 to restore the phase of the helices (Fig. 2D). Western analyses and viral titers data indicated that pEW15-res-derived viruses had a wild-type phenotype (Fig. 7A and Table 1).
Taken together, these data strongly support the idea that the MLV motif rich in charged residues forms an α-helical structure; this structure and the phases of the helices play important roles during virus assembly.
DISCUSSION
In this report, we describe a novel motif in MLV CA that is rich in charged residues, affects virus assembly, and most likely forms a helical structure. Thus, we termed this motif the “charged assembly helix.” Many deletions in this motif cause an assembly defect with a new phenotype, i.e., virions with a wide range of particle sizes and contents that morphologically resemble the cell cytoplasm; in addition, these virions package both ribosomal and viral RNAs. Virus with the charged assembly helix motif mutant phenotype exhibits drastically reduced particle production and even more greatly reduced infectivity. We were able to capture EM images with viruses budding out of the cells (Fig. 4C and D). However, the charged assembly helix mutant phenotype is distinct from the late-domain mutants that have virions tethered on the cell surface or viral particles connected to one another via uncleaved membranes (16, 21, 40).
In a previous study, MLV NC mutants that were unable to efficiently package viral RNA were shown to package ribosomes (31). The phenotype of the charged assembly helix mutants differs from that of the NC mutants in two aspects: the packaging of both ribosomal and viral RNA and the widely differing particle sizes. At least two possibilities can explain why charged assembly helix mutants but not NC mutants can package both ribosomal and viral RNA. One possibility is that particle sizes play a critical role in the encapsidation of both types of RNA. Compared with a 100-nm-diameter virion, virions with a 200-, 300-, or 400-nm diameter have 8, 27, and 64 times more volume in the particle, respectively. The larger particles in charged assembly helix mutants might simply have more volume in which to accommodate both ribosomal and viral RNAs, whereas the 100-nm-diameter NC mutants or wild-type viruses do not. The other possibility is that the charged assembly helix mutants have lost a function(s) that prevents the incorporation of rRNA into the virion. It was previously proposed that one of the roles of NC in wild-type viruses is to exclude ribosomes (31). It is possible that Gag-RNA interaction has two distinct functions: to specifically package viral RNA and to exclude ribosomes and possibly other cytoplasmic elements. NC mutants may have lost both functions, whereas the charged assembly helix mutants have lost only the ability to exclude ribosomes and other elements. Further studies are needed to distinguish these possibilities.
Wild-type MLV virions are fairly uniform in size (Table 2), indicating that Gag multimerization is a regulated event. One of the most striking features of the charged assembly helix mutant phenotype is the wide range of virion particle size distribution, indicating defects in this regulation. In the MLV Gag polyprotein, CA and NC are immediately adjacent to each other and the charged assembly helix motif is located near their junction. Substitution mutations in the CA-NC junction in MLV can cause defects in virus assembly (A. Rein, personal communication). In some retroviruses, CA and NC are separated by other cleavage products, e.g., spacer peptides (SP) for RSV and p2 for HIV-1 (32). Extensive mutational analyses revealed that the RSV CA-SP region is critical in the regulation of virion particle sizes; deletions in this region caused the formation of virions with heterogeneous sizes (26). In HIV-1, mutations in p2 have been shown to cause assembly defects that generate virions with heterogeneous sizes and aberrant morphology (25). Interestingly, the CA-p2 junction was proposed to be an α-helical structure that is crucial for virus assembly (1). Together with data from this report, these results suggest that the regions near CA-NC junctions are important in virus assembly and particle size regulation in many retroviruses. An intriguing possibility is that the charged assembly helix motif, SP, and p2 fulfill similar functions during retroviral assembly.
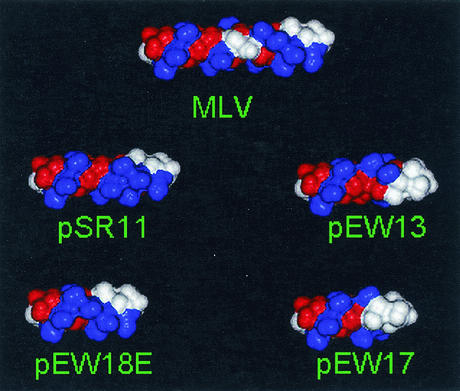
Structural studies of the C-terminal domain of HIV-1 CA revealed the presence of four α-helices; these helices play critical roles in maintaining the overall globular structure of the region as well as promoting CA-CA dimerization (18). However, the structure of the last 11 amino acids of HIV-1 CA and the putative α-helix in p2 is presently unknown and their roles in Gag-Gag interactions during assembly are not determined. Although nuclear magnetic resonance studies were performed on peptides derived from MLV MHR (9), similar to that of HIV-1, the structure of the very C terminus of MLV CA, including the charged assembly helix motif, remains unknown. We performed molecular modeling to simulate the putative structure of the MLV charged assembly helix motif (Fig. 8). The charge distribution in the wild-type motif was such that positively and negatively charged bands (shown in Fig. 8 in blue and red, respectively) were formed; in addition, a basic-residue cluster and a neutral cluster (shown in white) were located near the C terminus of CA. We observed that mutants with deletions of complete α-helical turns had the wild-type phenotypes, whereas mutants with deletions of incomplete turns, which drastically altered the helical phases, had the mutant phenotype. The effects of deleting incomplete turns can be visualized in the modeled structures. For example, in pSR11 and pEW18E, the basic clusters and neutral clusters near the C terminus of CA were oriented in a direction similar to that of the wild type. In contrast, in pEW13 and pEW17, the basic clusters and neutral clusters were oriented drastically differently than that of the wild type. The alteration of the helical phases probably changed the property of the overall structure and thus interfered with the charged assembly helix function.
FIG. 8.
Space-filling models of the predicted helical structure of the wild-type and mutant MLV charged assembly helix. N termini are located on the left. Amino acids are colored by charge: blue, positive; red, negative; and white, neutral. The wild-type charged assembly helix (labeled MLV) is shown in the center, two functional mutants (labeled pSR11 and pEW18E) are located at the left, and the two nonfunctional mutants (labeled pEW13 and pEW17) are located at the right. The basic clusters and neutral clusters near the C terminus of CA were oriented in the same direction in the wild type and the two functional mutants. In contrast, the basic clusters and neutral clusters were oriented drastically differently than that of the wild type in the two nonfunctional mutants.
Currently, we do not know how this putative α-helical structure affects MLV assembly. It is possible that protein-protein interactions mediated by the charged assembly helix motif facilitate and regulate virus assembly. The charged assembly helix may interact with another domain(s) in Gag, a cellular factor(s), or other charged assembly helix motifs in other Gag molecules. It is also possible that the charged assembly helix serves as a structural element to orient NC during Gag-Gag interactions, although we consider this hypothesis less likely. Analyses of HIV-1 NC structure indicate that the regions flanking the Cys-His boxes, including the region immediately adjacent to the α-helical structure, are very flexible (11). Given this flexibility, it is likely that NC can compensate for some structural variations and does not impose such a stringent requirement on the phases of the α-helices in this motif. Of the other three aforementioned possibilities for the protein-protein interactions involving the charged assembly helix motif, we favor the hypothesis that intermolecular interactions among the charged assembly helix motifs facilitate the Gag multimerization process. We find this hypothesis attractive because of the alternating positively and negatively charged bands on the proposed α-helices. In addition, cross-linking experiments indicated that the extreme C termini of the MLV CA domains are in close proximity with one another in immature virions (29). This observation is in agreement with the possibility that charged assembly helix motifs from different Gag polyproteins interact with one another. Studies are under way to probe the structure of the proposed α-helices and their relation to the function of the charged assembly helix motif in MLV CA.
Acknowledgments
S.R.C. and D.T.K.P. contributed equally to this work.
We thank Vinay K. Pathak for many helpful discussions, continuous intellectual input in this project, critical reading of the manuscript, and coining the term “charged assembly helix”; Alan Rein for communicating unpublished results, encouragement, and critical reading of the manuscript; John Coffin for encouragement, discussions, and critical reading of the manuscript; Anne Arthur for her expert editorial help on the manuscript; Sook-Kyung Lee for generating SR2-293T cells; Delphine Muriaux for rRNA used as a control in RNA analyses; Jane Mirro for help with Western analyses of cell lysates; and the NCI's Frederick Biomedical Supercomputing Center for time provided.
This work is supported by the HIV Drug Resistance Program, NCI, NIH, and in part by NIH under SAIC contract N01-CO-12400.
REFERENCES
- 1.Accola, M. A., S. Höglund, and H. G. Göttlinger. 1998. A putative α-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alin, K., and S. P. Goff. 1996. Amino acid substitutions in the CA protein of Moloney murine leukemia virus that block early events in infection. Virology 222:339-351. [DOI] [PubMed] [Google Scholar]
- 3.Birkett, A. J., B. Yelamos, I. Rodriguez-Crespo, F. Gavilanes, and D. L. Peterson. 1997. Cloning, expression, purification, and characterization of the major core protein (p26) from equine infectious anemia virus. Biochim. Biophys. Acta 1339:62-72. [DOI] [PubMed] [Google Scholar]
- 4.Bowerman, B., P. O. Brown, J. M. Bishop, and H. E. Varmus. 1989. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 3:469-478. [DOI] [PubMed] [Google Scholar]
- 5.Cairns, T. M., and R. C. Craven. 2001. Viral DNA synthesis defects in assembly-competent Rous sarcoma virus CA mutants. J. Virol. 75:242-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, S., and V. M. Vogt. 1997. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J. Virol. 71:4425-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheslock, S. R., J. A. Anderson, C. K. Hwang, V. K. Pathak, and W. S. Hu. 2000. Utilization of nonviral sequences for minus-strand DNA transfer and gene reconstitution during retroviral replication. J. Virol. 74:9571-9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clish, C. B., D. H. Peyton, and E. Barklis. 1998. Solution structures of human immunodeficiency virus type 1 (HIV-1) and moloney murine leukemia virus (MoMLV) capsid protein major-homology-region peptide analogs by NMR spectroscopy. Eur J. Biochem. 257:69-77. [DOI] [PubMed] [Google Scholar]
- 10.Craven, R. C., A. E. Leure-duPree, R. A. Weldon, Jr., and J. W. Wills. 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 69:4213-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Guzman, R. N., Z. R. Wu, C. C. Stalling, L. Pappalardo, P. N. Borer, and M. F. Summers. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279:384-388. [DOI] [PubMed] [Google Scholar]
- 12.DuBridge, R. B., P. Tang, H. C. Hsia, P.-M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrlich, L. S., B. E. Agresta, and C. A. Carter. 1992. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 66:4874-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fassati, A., and S. P. Goff. 1999. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol. 73:8919-8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 16.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu, W., R. J. Gorelick, and A. Rein. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamble, T. R., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 19.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 20.Gottlinger, H. G. 2001. The HIV-1 assembly machine. AIDS 15(Suppl. 5):S13-S20. [DOI] [PubMed] [Google Scholar]
- 21.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gritz, L., and J. Davies. 1983. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene 25:179-188. [DOI] [PubMed] [Google Scholar]
- 23.Khorasanizadeh, S., R. Campos-Olivas, and M. F. Summers. 1999. Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J. Mol. Biol. 291:491-505. [DOI] [PubMed] [Google Scholar]
- 24.Kingston, R. L., T. Fitzon-Ostendorp, E. Z. Eisenmesser, G. W. Schatz, V. M. Vogt, C. B. Post, and M. G. Rossmann. 2000. Structure and self-association of the Rous sarcoma virus capsid protein. Structure 8:617-628. [DOI] [PubMed] [Google Scholar]
- 25.Krausslich, H. G., M. Facke, A. M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishna, N. K., S. Campbell, V. M. Vogt, and J. W. Wills. 1998. Genetic determinants of Rous sarcoma virus particle size. J. Virol. 72:564-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landau, N. R., K. A. Page, and D. R. Littman. 1991. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J. Virol. 65:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mammano, F., A. Ohagen, S. Höglund, and H. G. Göttlinger. 1994. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J. Virol. 68:4927-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDermott, J., S. Karanjia, Z. Love, and E. Barklis. 2000. Crosslink analysis of N-terminal, C-terminal, and N/B determining regions of the Moloney murine leukemia virus capsid protein. Virology 269:190-200. [DOI] [PubMed] [Google Scholar]
- 30.Miller, A. D., and C. Buttimore. 1986. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol. Cell. Biol. 6:2895-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muriaux, D., J. Mirro, K. Nagashima, D. Harvin, and A. Rein. 2002. Murine leukemia virus nucleocapsid mutant particles lacking viral RNA encapsidate ribosomes. J. Virol. 76:11405-11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petropoulos, C. 1997. Retroviral taxonomy, protein structures, sequences, and genetic maps. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 33.Riggs, J. L., R. M. McAllister, and E. H. Lennette. 1974. Immunofluorescent studies of RD-114 virus replication in cell culture. J. Gen. Virol. 25:21-29. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 36.Temin, H. M., and S. Mizutani. 1970. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 226:1211-1213. [DOI] [PubMed] [Google Scholar]
- 37.Tobin, G. J., K. Nagashima, and M. A. Gonda. 1996. Immunologic and ultrastructural characterization of HIV pseudovirions containing Gag and Env precursor proteins engineered in insect cells. Methods 10:208-218. [DOI] [PubMed] [Google Scholar]
- 38.Vogt, V. M. 1997. Retroviral virions and genomes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 39.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, W.-H., C. K. Hwang, W.-S. Hu, R. J. Gorelick, and V. K. Pathak. 2002. Zinc finger domain of murine leukemia virus nucleocapsid protein enhances the rate of viral DNA synthesis in vivo. J. Virol. 76:7473-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]