Abstract
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV), also known as human herpesvirus 8, is an etiologic agent of KS, primary effusion lymphoma (PEL), and multicentric Castleman's disease. We recently demonstrated that hypoxia can induce lytic replication of KSHV in PEL cell lines. Hypoxia induces the accumulation of hypoxia-inducible factors (HIF), and we hypothesized that the KSHV genome may respond to hypoxia through functional hypoxia response elements (HREs). Here, we demonstrate the presence of at least two promoters within the KSHV genome that are activated by hypoxia or hypoxia mimics. One is in the promoter region of the gene for Rta, the main lytic switch gene, and the other is within the promoter region of ORF34, a lytic gene of unknown function. The ORF34 promoter contains three putative consensus HREs oriented in the direction of the gene. Dissection and site-directed mutagenesis studies confirmed that one of the HREs of the ORF34 promoter is functional. Under conditions of hypoxia, the ORF34 promoter was strongly upregulated by HIF-1α and HIF-2α. By contrast, the promoter of the gene for Rta appeared to be preferentially upregulated by HIF-2α. Reverse transcription-PCR analysis revealed that specific messages for ORF34 and ORF50 are upregulated in BCBL-1 cells exposed to hypoxia. An HIF-1 binding and competition assay demonstrated that the HRE sequence from the ORF34 promoter can compete for HIF-1α binding to an erythropoietin HRE oligonucleotide while a mutant sequence cannot. Thus, we demonstrated that a viral gene can be activated by hypoxia through activation of a functional viral HRE. To our knowledge, this is the first example of a functional HRE in a viral promoter.
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV), also known as human herpesvirus 8, is a gammaherpesvirus originally identified in KS tissue (5). It is the etiologic agent of KS and plays important roles in the pathogenesis of primary effusion lymphoma (PEL) and multicentric Castleman's disease (4, 28, 32). KSHV has significant sequence homology with herpesvirus saimiri and Epstein-Barr virus (22, 24). Like other herpesviruses, KSHV can establish a latent or lytic infection. Lytic gene expression can be induced by treatment of latently infected cells with chemical agents such as 12-O-tetradecanoylphorbol-13-acetate (TPA) or sodium butyrate (2, 21, 40). However, little is known about the factors responsible for activation of KSHV under physiologic conditions.
It was recently demonstrated that hypoxia can induce lytic replication of KSHV and that it may be a relevant biological activator of this virus (6). However, the mechanism by which KSHV responds to hypoxia is unknown. Cells exposed to hypoxic conditions accumulate hypoxia-inducible factor 1 (HIF-1), which controls transcriptional activation of a number of genes responsive to low cellular oxygen, including those for erythropoietin (EPO) (30), vascular endothelial growth factor (VEGF) (9), and transferrin receptor (17, 34) and others. HIF-1 was originally described as a heterodimer composed of HIF-1α and HIF-1β (37). HIF-1α is a basic helix-loop-helix (bHLH) transcription factor that is constitutively expressed but rapidly degraded in normoxic cells (13, 26). Under hypoxic conditions, HIF-1α degradation is blocked, allowing it to accumulate and bind to HIF-1β. The HIF-1α-HIF-1β dimer then binds to hypoxia-responsive elements (HREs) within the promoter of hypoxia-responsive target genes and serves to upregulate these genes (15, 37). The core consensus sequence that HIF-1 heterodimers bind to has been identified as the 5′-RCGTG-3′ (29). Recently, a second highly related HIF, termed endothelial PAS domain protein 1 (EPAS1) or HIF-2α, was identified that shares 48% sequence identity with HIF-1α (8, 36). HIF-2α also forms a functional heterodimer with HIF-1β and activates gene transcription from sites with an appropriate HRE (8, 36).
In the present study, we investigated the mechanism of activation of KSHV by hypoxia. In particular, we explored the hypothesis that KSHV may contain one or more functional HREs that are responsive to HIF-1α and/or HIF-2α.
MATERIALS AND METHODS
Cell culture.
Hep3B cells were obtained from the American Type Culture Collection (Manassas, Va.) and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. The source and culture of the KSHV-positive BCBL-1 cell line and the conditions for normoxia and hypoxia have been described elsewhere (6).
Reporter and expression plasmids.
The Rta gene promoter luciferase reporter construct contains the region spanning nucleotides (nt) −1424 to +36 upstream of the gene for Rta, where +1 indicates the transcription start site of the Rta gene promoter (18). The promoter region was amplified from BCBL-1 cellular DNA by PCR with primers RtaP-F1 (5′-CTAGCTAGCTTCCGCGGAAAAATACCCA-3′) and RtaP-R1 (5′-GGAGATCTTTTTGTGGCTGCCTGGACAG-3′), which contain NheI and BglI1 sites, respectively (underlined). The PCR fragment was inserted into the corresponding sites of the reporter vector pGL3-Basic (Promega, Madison, Wis.) to generate the reporter RtaP. The ORF34 wild-type luciferase reporter plasmid contains the region spanning nt −890 to +109 upstream of the methionine initiation codon of ORF34, where +1 indicates the transcription start site of the ORF34 gene, as determined by rapid amplification of 5′ cDNA ends (data not shown). The promoter region was PCR amplified with primers 34P-F1 (5′-CTAGCTAGCCTGGGTCCTCTTACGAAT-3′) and 34P-R1 (5′-GAAGATCTGCTGCTCAGCCGTCACAGG-3′), which contain NheI and BglI1 sites, respectively. The amplified product was cloned into the same sites of pGL3-Basic vectors and designated 34PWT. A series of 5′ deletions of the reporter plasmid, 34PWT, were constructed by the use of different primers at the 5′ end and a common primer, 34P-R1, at the 3′ end. Oligonucleotides 34PD1-F2 (5′-CTAGCTAGCGTAACAGCCGTTCAGAAA-3′), 34PD2-F3 (5′-CTAGCTAGCTTGATAGGGACTCCAATA-3′), and 34PD3-F4 (5′-CTAGCTAGCGTGCCGTGTGCAGTATGT-3′) were used for the amplification of deletions 34PD1, 34PD2, and 34PD3, respectively. The amplified products were cloned into the same sites as described above. The reporter plasmid containing the full-length human VEGF gene promoter (pVEGF-KpnI) (9) and the expression plasmids encoding human HIF-1α (hHIF1α-pcDNA3), HIF-2α (hEPAS1-pcDNA3) (36), and also human HIF-1α (pHAHIF1α-pcDNA3) (13) have been described previously.
Site-directed mutagenesis.
A plasmid expressing a mutagenized HRE was constructed by PCR-based mutagenesis as described previously (12, 35). To construct plasmid 34PD1(HREm), two DNA fragments were amplified by PCR with two sets of primer pairs, 34PD1-F2/34P(HRE)mR and 34P(HRE)mF/34P-R1, and plasmid 34PD1 as the template. The sequence of primer 34P(HRE)mF was 5′-TTGATCGGCCGTGGAGATATACGCGTCCTC-3′, and that of 34P(HRE)mR was 5′-GAGGACGCGTATATCTCCACGGCCGATCAA-3′. These sequences are complementary to each other. The sequences of the other primers have been shown above. The two amplified PCR products were purified with a QIAquick PCR purification kit (Qiagen, Valencia, Calif.), mixed, and annealed. The resulting double-stranded DNA was used as a template for the next PCR with primers 34PD1-F2 and 34P-R1 and cloned into the NheI and BglI1 sites of the pGL3-Basic vector to generate the mutant reporter 34PD1-(HREm). The fidelity of the mutant plasmid was confirmed by sequencing. The mutant plasmid 34PD1(HREm) contains a 3-nt substitution in the HRE.
Reporter assays.
All reporter experiments were performed with Hep3B cells in 12-well plates and transfection with SuperFect transfection reagent (Qiagen) similar to that described previously (11). Transfection of expression plasmids encoding HIF-1α or HIF-2α was performed in a 10-cm-diameter dish. Cells were cotransfected with a constant amount of internal control plasmid pSV-β-gal (Promega) for normalization of transfection efficiency. After transfection, cells were allowed to grow under normoxic conditions for 30 h and then exposed to either 21 or 1% oxygen or treated with 150 μM CoCl2 for 18 h. Cells were then harvested, and luciferase and β-galactosidase (β-gal) activities were assayed with Promega's substrate. The luciferase value was normalized to that of β-gal to correct for transfection efficiency. The normalized value for the reporter incubated at 21% oxygen was set to unity and compared to the results obtained under conditions of hypoxia or other conditions, which are expressed as fold induction.
RT-PCR.
Total cellular RNA was isolated from BCBL-1 cells that were cultured in 21% oxygen (normoxia), exposed to 1% oxygen (hypoxia), or treated with TPA for 92 h with Trizol reagent (Invitrogen, Carlsbad, Calif.) in accordance with the manufacturer's protocol. Total RNA (2 μg) was first treated with RQ1 RNase-free DNase (Promega) to eliminate genomic DNA contamination. The RNA was then directly used for reverse transcription (RT)-PCR analysis with Promega's Access RT-PCR kit with gene-specific primers for ORF34 and ORF50 in accordance with the manufacturer's protocol. The sequences and locations of the primers were as follows: ORF34F, 5′-TAGAATTCATGTTTGCTTTGAGCTCGCT-3′, nt 54675 to 54694; ORF34R, 5′-GTAGAATTCCTCCAGAGCCGACTTAAT-3′, nt 55686 to 55669 (the underlined sequences represents EcoRI restriction linkers); ORF50F, 5′-ATGAAAGAATGTTCCAAGCTTGGTGCG-3′, nt 72734 to 72760; ORF50R, 5′-TTCTCTGCGACAAAACATGCAGCGAC-3′, nt 73758 to 73733. The numbers and sequences are according to Russo et al. (24). The first-strand cDNA synthesis was carried out at 48°C for 45 min. After an initial denaturation step of 94°C for 2 min to inactivate the reverse transcriptase, the reactions were run for 40 cycles of 15 s at 94°C, 15 s at 58°C, and 1 min at 72°C, followed by a 7-min extension at 72°C. PCR products were separated on a 0.8% agarose gel and visualized by ethidium bromide staining.
Immunoblot analysis.
Nuclear extracts were prepared with a nuclear extraction kit (Pierce, Rockford, Ill.) from Hep3B cells transfected with plasmids HIF-1α and HIF-2α. Equal amounts (20 μg) of nuclear proteins were electrophoresed on 4 to 12% bis-tris polyacrylamide gels (Invitrogen), transferred to nitrocellulose membrane, and then incubated with antibodies to HIF-1α or HIF-2α (Novus Biological, Littleton, Colo.). After incubation with a secondary antibody conjugated to alkaline phosphatase, bands were visualized with the appropriate substrate (Promega).
HIF-1α binding and competition assay.
HIF-1 binding and competition assays were performed with a TransAM kit (Active Motif, Carlsbad, Calif.), which measures binding to a 26-bp HRE oligonucleotide from the EPO gene attached to a 96-well plate. Nuclear extracts were prepared from Hep3B and BCBL-1 cells as described above following exposure of cells to hypoxia or 100 μM CoCl2 for 16 h. Five micrograms of nuclear extract was used per well. Synthetic oligonucleotide probes for the ORF34 promoter containing the wild-type and mutated HREs were synthesized and annealed. The following 30-bp double-stranded probes were used for the HIF-1 binding and competition experiment (the sequence of the sense strand is shown): WT, 5′-TTGATCGGCCGTGGAGACGTGCGCGTCCTC-3′; mut, 5′-TTGATCGGCCGTGGAGATATACGCGTCCTC-3′ (the HRE is underlined, and the mutated bases are in bold). For binding and competition experiments, 20 or 100 pmol of either the wild-type or the mutant oligonucleotide of the EPO gene promoter or the ORF34 promoter was first added to the appropriate well before addition of the nuclear extract and incubation for 1 h. After washing three times, HIF-1α binding was assessed by incubation for 1 h each with an anti-HIF-1α antibody and then with a horseradish peroxidase-conjugated secondary antibody. The A450 was then determined.
Statistical analysis.
For certain observations in which data from several experiments were compiled, median values are shown and the signed-rank test was used to compare conditions. For those observations in which results from multiple replicates from one experiment are shown, mean values are shown and the Student t test was used to compare different conditions. All P values are two sided.
RESULTS
KSHV promoters are responsive to hypoxia.
We examined the KSHV genome (24) for sequences similar to identified cellular HREs (16, 17, 29). The genome has multiple potential HREs containing the core HRE sequence (5′-RCGTG-3′) (16, 17, 29). We focused on two promoter regions containing such sequences. The Rta gene promoter region was selected because the gene for Rta has been reported to be the main lytic switch gene for KSHV (10, 18, 19). Also, the ORF34 promoter region was selected because of the presence of four potential HREs spanning this region.
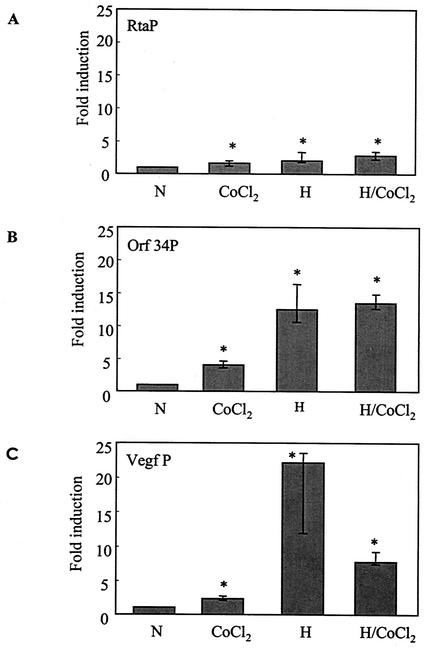
Reporter plasmids were constructed with the pGL3-Basic vector containing the Rta gene or ORF34 promoter region and transfected into Hep3B cells. The pGL3-Basic vector had minimal activity under normoxic conditions, and in agreement with a previous study with the related pGL2 vector (34), there was little or no stimulation of this pGL3-Basic vector in hypoxia (Fig. 1 legend). The Rta reporter activity increased approximately twofold when the cells were exposed to hypoxia (Fig. 1A). However, treatment of cells with CoCl2, a chemical that blocks degradation of HIF-1α and can act as a mimic of hypoxia, induced only minimal activation of Rta. Also, CoCl2 did not increase reporter activity under hypoxic conditions (Fig. 1A). More pronounced (approximately 12-fold) promoter activity was seen when cells were transfected with the ORF34 promoter (34PWT) and exposed to hypoxia (Fig. 1B). Again, treatment with CoCl2 was less effective than hypoxia alone. As a control, we tested a full-length VEGF-kpn1 reporter. Similar to previous reports (9), we found higher induction by hypoxia than by normoxia (Fig. 1C). Here, too, CoCl2 was less effective than hypoxia. These results suggest that the 5′-flanking sequences of Rta and ORF34 contain sequences that are responsible for hypoxic stimulation.
FIG. 1.
Activation of KSHV promoters in response to hypoxia and CoCl2. Cells were transfected with 1.3 μg of a reporter plasmid encoding the Rta gene promoter (RtaP) (A), the ORF34 promoter (Orf 34P) (B), or the VEGF gene promoter (Vegf P) as a positive control (C). Two hundred nanograms of an internal control plasmid, pSV-β-gal, was cotransfected for normalization of transfection efficiency. After exposure of cells to normoxia (N) or hypoxia (H), in the presence or absence of CoCl2 for 18 h, luciferase activity was determined and normalized to β-gal activity. Little activity of the pGL3-Basic reporter vector in normoxia or hypoxia was seen in the absence of inserted promoters. In an early representative experiment, for example, the pGL3-Basic reporter vector in normoxia, in hypoxia, and with the ORF34 promoter in normoxia produced 8,230, 7,800, and 90,430 relative light units, respectively. The β-gal activity from the pSV-β-gal internal control plasmid in this experiment yielded 0.148, 0.129, and 0.320 optical density unit, respectively, yielding normalized luciferase values of 5.56 × 104, 6.04 × 104, and 28.2 × 104 relative light units/optical density unit, respectively. For each promoter, the fold induction was calculated compared to the normalized value in normoxia alone (N), which was set at unity. All values represent the median of six separate experiments, each done in triplicate. Error bars denote the quartiles. *, P < 0.05 compared with normoxia.
ORF34 and ORF50 mRNA induction by hypoxia in KSHV-infected cells.
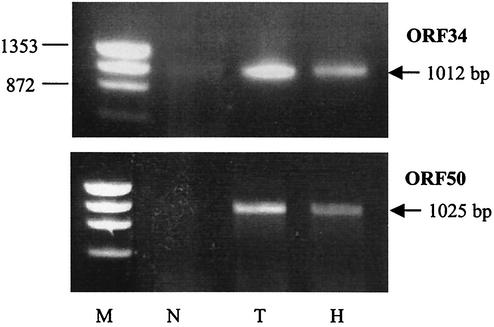
Since hypoxia induced ORF34 and ORF50 promoters in transiently transfected Hep3B cells, we investigated whether these genes are also activated by hypoxia in KSHV-infected cells. To accomplish this, we exposed BCBL-1 cells to normoxia, to hypoxia, or to normoxia with TPA for 92 h, extracted the total RNA, and performed RT-PCR analysis. As expected, TPA strongly induced both genes while little or no expression was observed in untreated cells (Fig. 2). Hypoxia also activated the expression of ORF34 and ORF50 on the basis of detection of the 1,012-bp gene and the 1,025-bp fragment, respectively, which were used to detect these genes (Fig. 2). This result indicates that hypoxia can activate these genes in KSHV-infected cells.
FIG. 2.
Induction of ORF34 and ORF50 mRNAs in hypoxic BCBL-1 cells as evidenced by RT-PCR. Total cellular RNA was isolated from BCBL-1 cells 92 h after treatment. With RT-PCR, cDNA was amplified with gene-specific primers as described in Materials and Methods. PCR products were resolved on 0.8% agarose gel and visualized by ethidium bromide staining. The positions of a 1,012-bp fragment specific for ORF34 and a 1,025-bp fragment specific for ORF50 are indicated on the right. Cells were exposed to normoxia (N; 21% oxygen), TPA (T) treatment, or hypoxia (H; 1% oxygen). DNA molecular size markers (M; HaeIII-digested phage φX174 DNA) are indicated on the left.
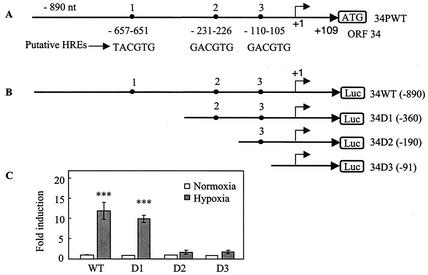
The ORF34 promoter contains a functional HRE.
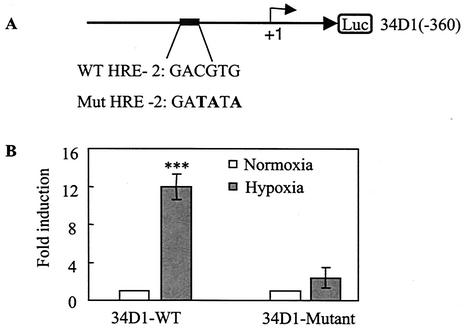
The ORF34 promoter contains three potentials HREs oriented in the direction of the gene (Fig. 3A), as well as one oriented in the opposite direction on the negative strand located between D2 and D3 (Fig. 3B). We constructed a series of deletion mutant forms of 34PWT (Fig. 3B) and tested their response to hypoxia following transfection into Hep3B cells. The reporter plasmid containing the DNA fragment up to −360 (34PD1) retained a strong response to hypoxia similar to that of the wild-type reporter (Fig. 3C, D1). By contrast, the reporter plasmids containing DNA fragments up to positions −190 (34PD2) and −91 (34PD3) both had a substantially reduced response to hypoxia compared to those of the wild-type and 34D1 reporters (Fig. 3C). These results suggest that the second HRE (HRE-2) is the principal functional HRE in the ORF34 promoter. To confirm that the second HRE was, in fact, functional, we performed site-directed mutational analysis on HRE-2. A three-base substitution was made in the HRE-2 sequence of 34PD1 (Fig. 4A) and its response to hypoxia was compared to that obtained with the 34PD1 wild-type reporter plasmid. The wild-type HRE-2-containing reporter, 34PD1, was activated more than 12-fold by hypoxia, while the mutant HRE-2-containing plasmid, 34PD1-HREm, was minimally activated by hypoxia (Fig. 4B). Overall, these data indicate that HRE-2 is the principal HRE in the ORF34 promoter region.
FIG. 3.
The ORF34 promoter contains regulatory elements that mediate the transcriptional response to hypoxia. (A) Diagram showing the upstream putative HRE sequences of the ORF34 promoter and DNA fragments used to generate reporter constructs. (B) 5′ deletion constructs of the ORF34 promoter. The constructs are named according to the 5′-end transcription start site of the ORF34 gene, as determined by rapid amplification of 5′ cDNA ends (data not shown), and are not drawn to scale. (C) Activity of the ORF34 wild-type (WT) and deletion reporters under hypoxic and normoxic conditions. Cells were cotransfected with wild-type and deletion reporters (WT, D1, D2, and D3) as described in the legend to Fig. 1. After normalization, fold induction was calculated as described in the legend to Fig. 1. All values represent means of triplicate determinations in one representative experiment out of two, with error bars denoting the standard deviations. ***, P < 0.001 compared with normoxia.
FIG. 4.
Effect of site-directed mutagenesis on the ORF34 promoter in response to hypoxia. (A) Sequences of the wild-type (WT) and HRE-2 mutant ORF34D1 reporters. The mutant reporter has a 3-bp substitution within the HRE-2 sequence, which is shown in bold. (B) Cells were cotransfected with a wild-type or mutant reporter and normalized as described in the legend to Fig. 1. All values represent means of triplicate determinations in one representative experiment out of three, with error bars denoting the standard deviations. ***, P < 0.0001 compared with normoxia.
Stimulation of Rta gene and ORF34 promoter activity by HIF-1α and HIF-2α.
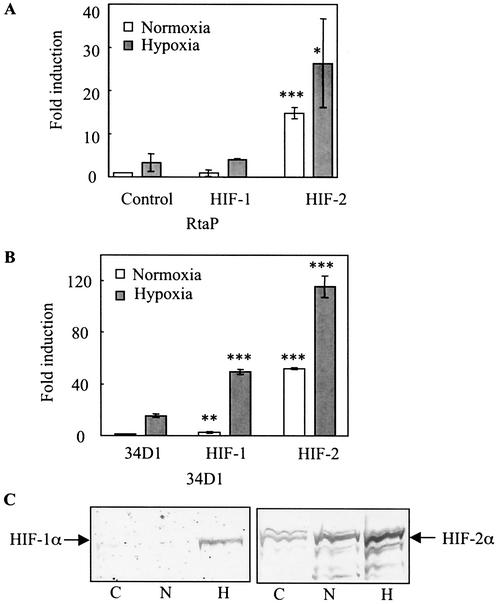
We wanted to assess the differential activation of the HREs by HIF-1α and HIF-2α from the Rta gene and ORF34 promoters. Cells were transiently transfected with the Rta (Fig. 5A) or 34PD1 (Fig. 5B) reporter in the absence or presence of an expression plasmid encoding either HIF-1α or HIF-2α and then maintained at 21 or 1% oxygen. Cotransfection of 250 ng of a plasmid encoding HIF-1α with the Rta reporter did not increase promoter activity in normoxia and had little or no effect in hypoxia (Fig. 5A). By contrast, cotransfection of 250 ng of a plasmid encoding HIF-2α increased Rta gene promoter activity almost 15- and 8-fold compared to that observed with the Rta reporter alone in normoxia and hypoxia, respectively (Fig. 5A). Cotransfection of HIF-1α with 34PD1 in normoxia similarly had little effect on the induction of 34PD1 promoter activity (Fig. 5B). However, 34PD1 promoter activity was increased about threefold by HIF-1α under hypoxic conditions compared with that of (Fig. 5B). With a different HIF-1 plasmid (13), similar induction of 34PD1 was observed (data not shown). An HIF-2α expression plasmid increased the 34PD1 promoter activity about 50- and 7-fold compared to that observed with the reporter alone in normoxia and hypoxia, respectively (Fig. 5B). We further analyzed the effects of HIF-1α and HIF-2α overexpression with different amounts of transfected plasmids up to a maximum of 500 ng. This dose-response experiment revealed that the maximal activation of Rta attained with HIF-1α under normoxic conditions was only 1.4-fold, while the maximum activation attained with HIF-2α was 24-fold compared to that obtained with the Rta gene promoter alone (data not shown). Similarly, under hypoxic conditions, the maximal activation attained with HIF-1α was 3-fold and that attained with HIF-2α was 55-fold. A similar pattern was observed with the ORF34 promoter, although the degree of activation was greater.
FIG. 5.
Activation of reporter genes by HIF-1α and HIF-2α. Cells were cotransfected with 750 ng of reporter plasmids encoding Rta (A) or with the ORF34D1 promoter (B) in the presence or absence of 250 ng of an expression plasmid encoding HIF-1α or HIF-2α. Cells also received 250 ng of an internal control plasmid, and the total amount of DNA was adjusted by addition of an appropriate amount of filler plasmid. Cells were exposed to normoxia or hypoxia and normalized as described in the legend to Fig. 1. The normalized value of each of the reporters with no expression vector incubated with 21% O2 was set at unity and compared to the results obtained with the expression vector, which are expressed as fold induction. All of the values shown represent means of triplicate determinations in one representative experiment out of two, with error bars denoting the standard deviations. For cells cotransfected with HIF-1α or HIF-2α in panels A and B, a statistical comparison was made with control cells cultured under the same oxygen conditions but without HIF transfection. *, P < 0.05; **, P < 0.005; ***, P < 0.0001. (C) Exposure of transfected Hep3B cells to hypoxia induces expression of the HIF-1α and HIF-2α proteins. Cells were transfected with 10 μg of each of these expression plasmids and exposed to either normoxia or hypoxia, and then protein extracts were analyzed by immunoblotting. C, untransfected control cells incubated at 21% oxygen; N and H, cells transfected with an HIF-1α or HIF-2α expression plasmid and incubated with 21 and 1% oxygen, respectively.
To further investigate the potential roles of HIF-1α and HIF-2α in the response of these genes to hypoxia, we analyzed Hep3B cells for HIF-1α and HIF-2α protein expression. HIF-1α was not detected in either untransfected or transfected cells in normoxia, (Fig. 5C). However, it was detected in hypoxic transfected cells. By contrast, HIF-2α was detected in normoxic control cells and the levels increased in transfected cells in normoxia and to an even greater degree in hypoxia. HIF-2α levels paralleled the induction of the ORF34 and Rta gene promoters under these conditions.
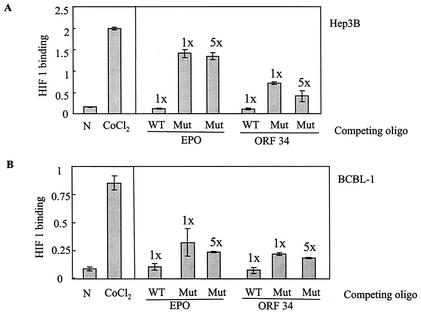
HRE-2 in the ORF34 promoter competes with the EPO HRE for HIF1 binding.
The HIF-1α-HIF-1β heterodimer has been shown to bind to HREs of cellular promoters, leading to the activation of certain genes (8, 36). To determine if the ORF34 promoter could bind to HIF-1α, we carried out an HIF-1 binding and competition ELISA in which the binding of HIF-1α to the HRE of the EPO gene was assessed in the presence or absence of a competing HRE. Nuclear extracts from CoCl2-treated cells showed strong binding to the EPO HRE, indicating the presence of the HIF-1α heterodimer (Fig. 6A and B, left side). We then used these extracts for the competition assay. Pretreatment of the nuclear extracts with 20 pmol of a 30-bp wild-type oligonucleotide spanning the HRE-2 region of the ORF34 promoter (HRE-2W) was able to substantially reduce HIF-1α binding to the EPO HRE (Fig. 6A). However, the mutant HRE-2 was much less effective at competing with the EPO HRE, even at a fivefold higher concentration. As expected, pretreatment of the extract with a 26-bp wild-type EPO oligonucleotide competed for binding, while a mutant EPO oligonucleotide was ineffective (Fig. 6A). We also tested the wild-type and mutant HRE-2s in the ORF34 promoter for competition with extracts from KSHV-infected PEL (BCBL-1) cells. Less HIF-1 binding activity to the EPO HREs was found in the BCBL-1 cells than in the Hep3B cells (Fig. 6B). However, wild-type HRE-2 from the ORF34 promoter was also able to compete for the HIF-1α binding from these cells (Fig. 6B). Again, mutant HRE-2 in the ORF34 promoter was less effective than wild-type HRE-2, even at a fivefold higher concentration (Fig. 6B).
FIG. 6.
HIF-1 binding and competition assay. Nuclear extracts were prepared from Hep3B and BCBL-1 cells exposed to normoxia (N) or CoCl2 for 16 h. Five micrograms each of nuclear extract of Hep3B (A) and BCBL-1 cells (B) was incubated with 20 pmol (1×) or 100 pmol (5×) of wild-type (WT) or mutant (Mut) probe from the 26-bp EPO gene promoter HRE oligonucleotide (oligo) or the 30-bp ORF34 promoter HRE oligonucleotide, as shown. Binding to the 26-bp HRE oligonucleotide from the EPO gene was assessed as described in the text. For competition experiments, the promoter probe was first added to the appropriate well before addition of the nuclear extracts. The left ends of panels A and B show the results of a control experiment with nuclear extracts with an EPO probe but with no competing oligonucleotide. All values represent means of triplicate determinations in one representative experiment, with error bars denoting the standard deviations.
DISCUSSION
These studies show that KSHV contains at least two promoters, of Rta and ORF34, that can be activated by increased levels of HIF-1α and/or HIF-2α in cells exposed to hypoxia. This represents the first identification and characterization of a functional HRE in a viral genome. The ability of the promoter of the Rta gene to respond to hypoxia is of particular interest, as the gene for Rta is the main lytic switch gene of KSHV and activation of this gene is sufficient to induce lytic replication (18, 19, 33). Thus, activation of the Rta gene promoter by hypoxia can account for the induction of lytic KSHV replication observed in PEL cell lines exposed to hypoxia (6).
The Rta gene promoter has seven potential HREs, six of which are oriented opposite to the direction of the gene and only one of which is oriented in the same direction as the gene. We have not shown which HRE may be active in this promoter region. It is noteworthy that while Rta gene promoter activity only increased about twofold in transfection experiments with Hep3B cells, there appeared to be a more pronounced increase in Rta RNA in PEL cells exposed to hypoxia, as assessed by a semiquantitative RT-PCR assay. While it is hard to make quantitative comparisons, it is possible that the previously described autoactivation of the Rta gene (7, 25) may serve to amplify a relatively small effect directly initiated by hypoxia in KSHV-infected cells.
In addition to Rta, the promoter region of ORF34 contains a functional HRE that is strongly activated in cells exposed to hypoxia. ORF34 codes for a late lytic protein of unknown function that shares about 33% identity with the BGLF3 gene of Epstein-Barr virus (14, 23, 24, 27). The genomic organization of KSHV (24) suggests that the ORF34 promoter may also regulate several overlapping downstream genes, including ORF35 to ORF38 (3). Although the ORF34 promoter contains three potential HREs oriented in the direction of the genes, only one (HRE-2) was responsible for most of the hypoxic activation. The single oppositely oriented HRE located between deletions D2 and D3 is not responsive (Fig. 3C). The HRE sequence in the ORF34 promoter is similar to that of other genes such as the human EPO and transferrin receptor genes. Most of the known functional HREs contain at least one sequence termed the hypoxia binding sequence, as well as an additional element, termed the ancillary sequence, that is downstream of and adjacent to the hypoxia binding sequence. Studies have indicated that this ancillary sequence is required for full hypoxic activation (16). The HRE of the ORF34 promoter also contains a potential hypoxia ancillary sequence (CACAC) downstream of HRE-2 that is identical to that found in certain cellular genes (17, 20). Our RT-PCR result indicates that hypoxia not only activated the Rta gene and ORF34 promoters in Hep3B cells but also activated them in KSHV-infected cells.
The first factor identified as mediating the cellular response to hypoxia was HIF-1α (30). A second factor, called HIF-2α or EPAS, was subsequently identified as an alternate mediator of the cellular response to hypoxia (8, 36). Both of these factors can bind to HREs, but the differences in their activities and their different roles in the cellular response to hypoxia are not well understood. The cotransfection experiments performed here with the 34PD1 promoter indicated that it is strongly activated by HIF-2α. Maximal effects were seen in hypoxic cells transfected with HIF-2α, while essentially no increased activity was seen with HIF-1α in normoxic cells. However transfection with HIF-1α increased the activation in hypoxic cells. Subsequent experiments examining the levels of HIF-1α and HIF-2α expression provided evidence that the lack of induction of the promoter by HIF-1α in normoxia was likely due to the low levels of HIF-1α protein under these conditions. These results are consistent with a previous study showing that transfection of Hep3B cells with HIF-2α in normoxia is more effective at activating cellular hypoxia-responsive genes than is transfection with HIF-1α, possibly because of the more rapid degradation of HIF-1α under normoxic conditions (39). It is alternatively possible that these promoters respond better to HIF-2α than to HIF-1α. It is interesting that hypoxia was a better inducer of the ORF34 and Rta gene promoters than was CoCl2. Analysis of nuclear extracts from Hep3B cells by Western blotting indicates that hypoxia induces a higher ratio of HIF-2α to HIF-1α than does CoCl2, and this may explain the higher induction by hypoxia (data not shown).
The Rta gene promoter also responded to HIF-2α, but a clear response to HIF-1α was not seen, even under hypoxic condition. However this promoter was weaker overall and the possibility of a small response to HIF-1α could not be excluded. With regard to the VEGF gene promoter, there are conflicting reports on its activation by HIF-1α and HIF-2α (1, 9). It is possible that these differences are, in part, due to the various conditions used (for example, one study used cotransfection with HIF-1β), and it will be important to sort out how these differences may influence the activities of HIF-1α and HIF-2α.
The present study was initially undertaken in an attempt to understand the mechanism by which hypoxia may induce KSHV replication (6). The finding that the Rta gene promoter can respond to hypoxia is sufficient to explain this process. Interestingly, at least one additional gene of KSHV, ORF34, contains a functional HRE. These observations raise the question of what roles hypoxia plays in the life cycle of KSHV. Endothelial cells are thought to represent a principal target cell population for KSHV, and the growth of these cells is stimulated by a number of factors, several of which (such as VEGF) are stimulated by hypoxia in the body. KSHV itself codes for a number of proangiogenic factors, such as viral interleukin-6 or viral macrophage inflammatory proteins 1 and 2, that stimulate endothelial cell growth. Moreover, ORF74 of KSHV codes for a G-protein-coupled receptor that stimulates VEGF production and also upregulates the activity of HIF-1α (31). KSHV is thus poised to both replicate in and stimulate the growth of an important target cell population under hypoxic conditions. This may provide a mechanism that helps the virus establish infection in the hypoxic tissues found at the edge of a wound. In addition, it may help explain the tendency of Kaposi's sarcoma to occur on the lower extremities (which are often relatively hypoxic) (38). In this light, it is interesting to speculate as to what evolutionary advantages may be afforded KSHV by having certain specific genes, such as ORF34, directly stimulated by hypoxia. A better understanding of the KSHV life cycle will allow us to delineate the role of these genes in virus replication.
Acknowledgments
We thank Steven L. McKnight (University of Texas) for the expression plasmids encoding HIF-1α and HIF-2α, Eric Huang (National Cancer Institute) for the HIF-1α plasmid, and Gregg L. Semenza (Johns Hopkins University) for the VEGF gene reporter. We also thank Zhi-Ming Zheng (National Cancer Institute) for comments on the manuscript.
REFERENCES
- 1.Akeno, N., M. F. Czyzyk-Krzeska, T. S. Gross, and T. L. Clemens. 2001. Hypoxia induces vascular endothelial growth factor gene transcription in human osteoblast-like cells through the hypoxia-inducible factor-2α. Endocrinology 142:959-962. [DOI] [PubMed] [Google Scholar]
- 2.Arvanitakis, L., E. A. Mesri, R. G. Nador, J. W. Said, A. S. Asch, D. M. Knowles, and E. Cesarman. 1996. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 88:2648-2654. [PubMed] [Google Scholar]
- 3.Cannon, J. S., F. Hamzeh, S. Moore, J. Nicholas, and R. F. Ambinder. 1999. Human herpesvirus 8-encoded thymidine kinase and phosphotransferase homologues confer sensitivity to ganciclovir. J. Virol. 73:4786-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 5.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 6.Davis, D. A., A. S. Rinderknecht, J. P. Zoeteweij, Y. Aoki, E. L. Read-Connole, G. Tosato, A. Blauvelt, and R. Yarchoan. 2001. Hypoxia induces lytic replication of Kaposi sarcoma-associated herpesvirus. Blood 97:3244-3250. [DOI] [PubMed] [Google Scholar]
- 7.Deng, H., A. Young, and R. Sun. 2000. Auto-activation of the rta gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 81:3043-3048. [DOI] [PubMed] [Google Scholar]
- 8.Ema, M., S. Taya, N. Yokotani, K. Sogawa, Y. Matsuda, and Y. Fujii-Kuriyama. 1997. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1α regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Natl. Acad. Sci. USA 94:4273-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forsythe, J. A., B. H. Jiang, N. V. Iyer, F. Agani, S. W. Leung, R. D. Koos, and G. L. Semenza. 1996. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell. Biol. 16:4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haque, M., J. Chen, K. Ueda, Y. Mori, K. Nakano, Y. Hirata, S. Kanamori, Y. Uchiyama, R. Inagi, T. Okuno, and K. Yamanishi. 2000. Identification and analysis of the K5 gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 74:2867-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, L. E., J. Gu, M. Schau, and H. F. Bunn. 1998. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 95:7987-7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, B. H., E. Rue, G. L. Wang, R. Roe, and G. L. Semenza. 1996. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J. Biol. Chem. 271:17771-17778. [DOI] [PubMed] [Google Scholar]
- 16.Kimura, H., A. Weisz, T. Ogura, Y. Hitomi, Y. Kurashima, K. Hashimoto, F. D'Acquisto, M. Makuuchi, and H. Esumi. 2001. Identification of hypoxia-inducible factor 1 ancillary sequence and its function in vascular endothelial growth factor gene induction by hypoxia and nitric oxide. J. Biol. Chem. 276:2292-2298. [DOI] [PubMed] [Google Scholar]
- 17.Lok, C. N., and P. Ponka. 1999. Identification of a hypoxia response element in the transferrin receptor gene. J. Biol. Chem. 274:24147-24152. [DOI] [PubMed] [Google Scholar]
- 18.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 20.Madan, A., and P. T. Curtin. 1993. A 24-base-pair sequence 3′ to the human erythropoietin gene contains a hypoxia-responsive transcriptional enhancer. Proc. Natl. Acad. Sci. USA 90:3928-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, G., L. Heston, E. Grogan, L. Gradoville, M. Rigsby, R. Sun, D. Shedd, V. M. Kushnaryov, S. Grossberg, and Y. Chang. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 71:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore, P. S., S.-J. Gao, G. Dominguez, E. Cesarman, O. Lungu, D. M. Knowles, R. Garber, P. E. Pellett, D. J. McGeoch, and Y. Chang. 1996. Primary characterization of a herpesvirus agent associated with Kaposi's sarcoma. J. Virol. 70:549-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulose-Murphy, M., N. K. Ha, C. Xiang, Y. Chen, L. Gillim, R. Yarchoan, P. Meltzer, M. Bittner, J. Trent, and S. Zeichner. 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salceda, S., and J. Caro. 1997. Hypoxia-inducible factor 1α (HIF-1α) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J. Biol. Chem. 272:22642-22647. [DOI] [PubMed] [Google Scholar]
- 27.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schalling, M., M. Ekman, E. E. Kaaya, A. Linde, and P. Biberfeld. 1995. A role for a new herpes virus (KSHV) in different forms of Kaposi's sarcoma. Nat. Med. 1:707-708. [DOI] [PubMed] [Google Scholar]
- 29.Semenza, G. L., B. H. Jiang, S. W. Leung, R. Passantino, J. P. Concordet, P. Maire, and A. Giallongo. 1996. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271:32529-32537. [DOI] [PubMed] [Google Scholar]
- 30.Semenza, G. L., and G. L. Wang. 1992. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 12:5447-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sodhi, A., S. Montaner, V. Patel, M. Zohar, C. Bais, E. A. Mesri, and J. S. Gutkind. 2000. The Kaposi's sarcoma-associated herpes virus G protein-coupled receptor up-regulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1α. Cancer Res. 60:4873-4880. [PubMed] [Google Scholar]
- 32.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 33.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tacchini, L., L. Bianchi, A. Bernelli-Zazzera, and G. Cairo. 1999. Transferrin receptor induction by hypoxia. HIF-1-mediated transcriptional activation and cell-specific post-transcriptional regulation. J. Biol. Chem. 274:24142-24146. [DOI] [PubMed] [Google Scholar]
- 35.Takeda, K., M. Haque, T. Sunagawa, T. Okuno, Y. Isegawa, and K. Yamanishi. 1997. Identification of a variant B-specific neutralizing epitope on glycoprotein H of human herpesvirus-6. J. Gen. Virol. 78:2171-2178. [DOI] [PubMed] [Google Scholar]
- 36.Tian, H., S. L. McKnight, and D. W. Russell. 1997. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 11:72-82. [DOI] [PubMed] [Google Scholar]
- 37.Wang, G. L., B. H. Jiang, E. A. Rue, and G. L. Semenza. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 92:5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webster-Cyriaque, J. 2002. Development of Kaposi's sarcoma in a surgical wound. N. Engl. J. Med. 346:1207-1210. [DOI] [PubMed] [Google Scholar]
- 39.Wiesener, M. S., H. Turley, W. E. Allen, C. Willam, K. U. Eckardt, K. L. Talks, S. M. Wood, K. C. Gatter, A. L. Harris, C. W. Pugh, P. J. Ratcliffe, and P. H. Maxwell. 1998. Induction of endothelial PAS domain protein-1 by hypoxia: characterization and comparison with hypoxia-inducible factor-1α. Blood 92:2260-2268. [PubMed] [Google Scholar]
- 40.Yu, Y., J. B. Black, C. S. Goldsmith, P. J. Browning, K. Bhalla, and M. K. Offermann. 1999. Induction of human herpesvirus-8 DNA replication and transcription by butyrate and TPA in BCBL-1 cells. J. Gen. Virol. 80:83-90. [DOI] [PubMed] [Google Scholar]