Abstract
The capsid proteins of two flaviviruses, yellow fever virus and dengue virus, were expressed in Escherichia coli and purified to near homogeneity suitable for biochemical characterization and structure determination by nuclear magnetic resonance. The oligomeric properties of the capsid protein in solution were investigated. In the absence of nucleic acid, both proteins were predominately dimeric in solution. Further analysis of both proteins with far-UV circular dichroism spectroscopy indicated that they were largely alpha-helical. The secondary structure elements of the dengue virus capsid were determined by chemical shift indexing of the sequence-specific backbone resonance assignments. The dengue virus capsid protein devoid of its C-terminal signal sequence was found to be composed of four alpha helices. The longest alpha helix, 20 residues, is located at the C terminus and has an amphipathic character. In contrast, the N terminus was found to be unstructured and could be removed without disrupting the structural integrity of the protein.
The Flaviviridae family of enveloped RNA viruses causes significant disease in both humans and agriculturally important animals. Flavivirus, the largest of the three genera of Flaviviridae, comprises over 70 viruses, mostly arthropod transmitted, including yellow fever virus (YF), dengue virus (DEN), West Nile virus, and tick-borne encephalitis virus (TBE) (15). The mature flavivirus particle is spherical with a diameter of 50 nm and contains multiple copies of three different structural proteins (C, M, and E), a host-derived membrane bilayer, and a single copy of a positive-sense RNA genome of approximately 11,000 nucleotides. The RNA genome is translated from a single open reading frame generating a polyprotein that is processed by viral and host proteases to yield the three structural proteins located at the N terminus followed by at least seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (23). The nonstructural proteins associate to form the viral replicase complex, for which several enzymatic functions have been identified, including protease, helicase, methyltransferase, and RNA-dependent RNA polymerase (15). The replicase complex is associated with intracellular membranes of the infected host and induces discrete membrane structures (16). Recent evidence suggests that the process of particle assembly may be coupled to genome replication (6, 12). This coupling of replication and virus assembly has made analysis of flavivirus assembly difficult, and thus, little is known about this important aspect of the virus life cycle.
The structure of DEN, recently determined by cryoelectron microscopy (cryo-EM) and three-dimensional image reconstruction, has elucidated the molecular organization of the end product of the flavivirus assembly pathway (11). The organization of the E protein within the DEN particle was determined by modeling the atomic resolution structure of TBE E protein (22) into the outer density of the cryo-EM reconstruction. The model consists of a herringbone arrangement of E proteins, with three E protein monomers in the asymmetric unit of the virus, but lacking the anticipated T=3 icosahedral symmetry. The M protein is thought to reside just below E with both M and E being associated with the host-derived lipid bilayer. Internal to the lipid bilayer is the nucleocapsid core (NC) of the virus, consisting of multiple copies of capsid (C) protein that surround a single copy of the viral RNA genome. The density observed for the NC is roughly half of that observed for the outer density and suggests disorder or movement of the NC within the virus particle. The location of the NC within the virus particle indicates that the C protein directly contacts the genome RNA.
A common phenomenon during flavivirus infection is the production and release into the extracellular medium of virus-like particles (VLPs) (24). NC is not present in VLPs, rendering them noninfectious. VLPs have also been produced in several heterologous expression systems in which only prM and E are present (1, 4, 9, 20, 21). Thus, the ability of E, in the presence of prM, to form a closed spherical protein shell is an intrinsic property of these two proteins and is independent of NC formation. However, the production of infectious particles requires the formation of NC, suggesting that an early stage of virus assembly involves the interaction of C protein with the genome RNA (7). The NC presumably then interacts with prM and/or E, resulting in the formation of a virus particle upon concomitant budding into the lumen of the endoplasmic reticulum (15). Unfortunately, details concerning the interactions of C protein with either the RNA genome or prM and E are not discernible in the cryo-EM reconstruction of DEN. Given the role of C in promoting encapsidation of the viral RNA and subsequent assembly of infectious virus particles, it is important to understand the structural and biophysical properties of the C protein that allow it to function in the flavivirus life cycle.
Cloning, expression, and purification of DEN2C and YFC.
C protein has been suggested previously to be a rather flexible molecule and predicted to be largely alpha-helical in structure (8). As a first step toward understanding the structural and biophysical properties of the C protein, we sought to express and purify the C proteins from two distantly related flaviviruses, DEN and YF. The full-length C protein includes a carboxy-terminal hydrophobic sequence (15 residues in DEN and 20 residues in YF) that serves as a signal sequence during viral polyprotein synthesis and as a membrane anchor following signalase cleavage in the lumen of the endoplasmic reticulum. A proteolytic cleavage event by the viral protease NS2B-3 on the cytoplasmic side of the C protein (DEN2C R100 and YFC R101) results in the mature form of C found in the virus. To minimize any potential solubility problems associated with expressing full-length C protein, the carboxy-terminal hydrophobic signal sequence was excluded from the DEN2C and YFC expression constructs. Therefore, the cDNAs encoding the mature forms of DEN2C (PR-159S1 strain of DEN2 [DEN type 2] from the plasmid pC2 [5]) and YFC (YF strain 17D from the plasmid pYF5′3′ [23]) were cloned into the pET30a (Novagen, Madison, Wis.) expression plasmid for protein expression in Escherichia coli. C protein expression was performed with the E. coli strain BL21(DE3)[RIL] “codon-plus” (Stratagene, La Jolla, Calif.) due to the presence of several rare arginine codons in both the DEN2C and YFC coding sequences.
The optimal conditions for bacterial expression of DEN2C and YFC were determined by varying time, temperature, cell density, and the concentration of isopropyl-β-d-thiogalactopyranoside (IPTG) used for induction. Optical induction of DEN2C and YFC protein expression required the addition of IPTG to a final concentration of 1 mM to cells at an optical density at 600 nm (OD600) of 1.0. Incubation temperature during induction was found to have a significant effect on protein expression. Whereas DEN2C was found to be expressed optimally at 30°C (Fig. 1A, left panel), YFC was expressed optimally at 25°C (Fig. 1B, left panel) with little or no expression at higher temperatures (data not shown).
FIG. 1.
Expression and purification of DEN2C and YFC in E. coli. (Left panels) Optimized expression for DEN2C (A) and YFC (B). Shown are results of Coomassie blue-stained SDS-PAGE of uninduced (−IPTG) and induced (+IPTG) whole-cell extracts of BL21(DE3)[RIL] expressing either DEN2C or YFC protein. (Right panels) Coomassie blue-stained SDS-PAGE of DEN2C and YFC purification. Shown are insoluble (InSol) and soluble (Sol) protein extracts following cell lysis by a French pressure cell and clarification by centrifugation, pooled fractions containing partially purified capsid protein following cation-exchange chromatography with a HiTrap SP column (SP), and purified and buffer-exchanged capsid following size-exclusion chromatography with a Superose 6 column (Sizing). Arrows indicate the presence of the C protein.
The highly basic nature of the C protein facilitated the development of a rather simple purification scheme. Soluble protein extracts containing C protein were first subjected to ammonium sulfate fractionation, followed by cation-exchange chromatography, yielding highly enriched, nearly homogenous preparations of C protein (>90% by Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]). Following cation exchange, C protein was subjected to size-exclusion chromatography, resulting in protein that was greater than 95% pure, as determined by silver stain SDS-PAGE. Preparations of DEN2C and YFC had less than 1% (wt/wt) nucleic acid contamination, based on the ratio of OD260 to OD280. Size-exclusion chromatography could not be used to estimate the molecular mass of C due to significant nonspecific interactions with the column matrix. However, excellent resolution from larger contaminating proteins was achieved as a result of the long retention time of the C protein on the Superose 6 column matrix. The final yield of purified DEN2C and YFC was approximately 3 to 5 mg and 1 to 2 mg from 1 liter of induced E. coli cells in Luria-Bertani medium, respectively (Fig. 1, right panels).
During the initial attempts to purify DEN2C and YFC, it was observed that the addition of protease inhibitor was necessary to prevent C protein degradation. Purification of C protein in the absence of protease inhibitor resulted in the presence of multiple smaller C-protein-specific protein species. Amino-terminal sequencing of these smaller bands revealed that they were the result of N-terminal truncations. The introduction of a general protease inhibitor cocktail during the course of protein purification eliminated the appearance of N-terminally truncated forms of C protein, suggesting that the N-terminal truncations were the result of proteolysis during purification. The susceptibility of the N terminus to proteolysis suggested that the N terminus of the flavivirus C protein might be flexible or structurally disordered. Thus, N- and C-terminal truncations of DEN2C were constructed and assayed for expression in bacteria. Truncated constructs of DEN2C included N-terminal truncations of either the first 13 (DEN2CΔ1-13) or 20 (DEN2CΔ1-20) residues or C-terminal truncations that removed either the last 10 (DEN2CΔ90-100) or 20 (DEN2CΔ80-100) residues. Interestingly, only DEN2CΔ1-13 was found to be expressed in E. coli (data not shown). The conditions for optimal expression of DEN2CΔ1-13 were found to be identical to those for DEN2C. DEN2CΔ1-13 was purified by the same procedure as that for DEN2C, although the yield of purified protein was less (2 to 3 mg of purified protein from 1 liter of induced E. coli cells in Luria-Bertani medium), and this was the result of lower levels of DEN2CΔ1-13 bacterial expression (data not shown).
Analysis of the solution oligomeric state of the C protein.
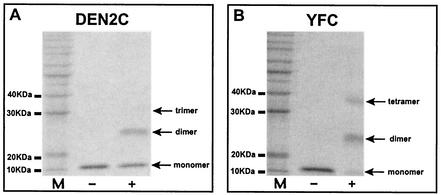
As a first step in understanding the oligomeric properties of the C protein, the solution oligomeric state of the protein was determined in the absence of other viral components by chemical cross-linking, analytical ultracentrifugation, and nuclear magnetic resonance (NMR). Treatment of either DEN2C or YFC with the lysine-specific cross-linking reagent disuccinimidyl suberate (DSS) resulted in the formation of oligomers that could be resolved by SDS-PAGE and visualized by Coomassie blue staining (Fig. 2). DSS cross-linking of DEN2C (Fig. 2A) resulted in the formation of a dimeric species and small amounts of a trimeric species. YFC (Fig. 2B) treated under similar cross-linking conditions resulted in the formation of dimeric and tetrameric species. Moreover, the cross-linking efficiency, judged by the disappearance of the monomeric species in cross-linked samples, was significantly higher for YFC than for DEN2C. This suggested that YFC may have lysine residues that are more favorably positioned than those of DEN2C to allow DSS cross-linking. Formation of a trimeric cross-linked species of YFC could be increased by the addition of nonionic detergents such as Triton X-100 and Tween 20 (data not shown). This suggested that the treatment of YFC with nonionic detergent disrupts a dimer of C, indicating that the native oligomeric state of YFC was likely to be a dimer in solution. However, based on these cross-linking results, the oligomeric state of DEN2C could not be clearly established.
FIG. 2.
Cross-linking of DEN2C and YFC. Shown are results of Coomassie blue-stained SDS-PAGE of purified DEN2C (A) and YFC (B) subjected to cross-linking (+ lanes) with the lysine-specific cross-linking reagent DSS. DSS cross-linking of DEN2C results in the appearance of dimers and trace amounts of trimers. DSS cross-linking of YFC results in dimers and tetramers. The efficiency of DSS cross-linking, estimated by the disappearance of monomer in cross-linking reactions, was routinely higher for YFC. M, 10-kDa protein molecular mass markers.
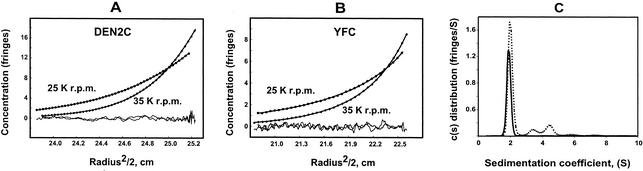
The oligomeric state of DEN2C and YFC was examined further by both equilibrium and velocity sedimentation with analytical ultracentrifugation. Data from equilibrium experiments were analyzed with WINMATCH and WINNL106 software (obtained from The National Analytical Ultracentrifugation Facility [http://www.ucc.uconn.edu/∼wwwbiotc/uaf.html]). Data from velocity sedimentation experiments were analyzed with Sedphat1.1 and Sedfit8.52 obtained from Peter Schuck at the Molecular Interactions Resource, Division of Bioengineering and Physical Science, Office of Research Services, Office of the Director, National Institutes of Health, Bethesda, Md. (www.analyticalultracentrifugation.com). Figure 3 shows representative data from both equilibrium (Fig. 3A and B) and velocity (Fig. 3C) sedimentation analysis of DEN2C and YFC. The plots in Fig. 3A and B show the change in concentration as a function of radial position at two of the different speeds used for DEN2C (Fig. 3A) and YFC (Fig. 3B). The distribution of molecules with a given sedimentation coefficient resulting from velocity sedimentation analysis of DEN2C and YFC is shown in Fig. 3C. Initial sedimentation velocity analysis of YFC was done by using a variation of YFC (YFCN) that included an N-terminal polyhistidine tag followed by approximately 5 kDa of protein sequence resulting from the multiple cloning site within the pET30a expression plasmid. The majority of DEN2C (solid line) and YFCN (dotted line) traveled as a single species, with sedimentation coefficients consistent with a dimeric protein species in either case. In addition, small amounts of additional oligomeric species were also detected. These additional oligomeric species were readily apparent in the velocity sedimentation profile of YFCN (dotted line in Fig. 3C). The results from equilibrium sedimentation analysis of DEN2C and YFC were also consistent with additional oligomeric species. Taken together, these results indicated that, at moderate protein concentrations (0.2 to 5 mg/ml), the major oligomeric state of both DEN2C and YFC is a dimer in solution (Table 1). In addition, a less significant percentage of both proteins exists in higher oligomeric states. The identities of the higher oligomer species for both DEN2C and YFC are currently under investigation.
FIG. 3.
Analytical ultracentrifugation of flavivirus C proteins. (A and B) Equilibrium sedimentation analysis of DEN2C (A) and YFC (B). The change in protein concentration versus radial position is shown for each protein at two different speeds. Circles indicate the observed experimental data; solid lines indicate calculated data. Residuals are also shown. (C) Velocity sedimentation analysis of DEN2C and YFCN. The distribution of molecules with a given sedimentation coefficient is shown for DEN2C (solid line) and YFCN (dotted line). The majority of DEN2C and YFCN sediments as a single species. Additional oligomeric species with larger sedimentation coefficient values can be seen in YFCN.
TABLE 1.
Capsid protein sedimentation values
| Type of value | YFC | DEN2C |
|---|---|---|
| s20,w0 (S)a | 2.19b | 2.06 |
| Be | NSd | 0.0038 |
| f/f0 | 1.69 | 1.55 |
| Calculated mass (kDa) | 11.5 | 11.8 |
| Observed mass (kDa)c | 22.96 | 21.9 |
Determined from velocity sedimentation analysis.
Determined for YFCN.
Determined from equilibrium sedimentation analysis.
NS, not significant.
B, second virial coefficient.
The oligomeric state of DEN2C in solution was also examined by using an NMR relaxation measurement. The molecular rotational correlation time was obtained from a trimmed average ratio of the backbone 15N longitudinal (R1) and transverse (R2) relaxation rates at 27°C. The estimated correlation time of DEN2C is 13 ns, assuming isotropic rotational diffusion. This value is approximately twice the expected value for an 11.8-kDa protein (2) in solution at 27°C, suggesting a predominantly dimeric state of DEN2C.
Attempts were made to establish an in vitro assembly system that could be used to study the effects of nucleic acid addition on the oligomeric state of the C protein. Development of an assembly system would be a useful tool to probe NC formation, as similar systems have been successfully developed to study the in vitro assembly of alphavirus and hepatitis C virus capsid proteins into core-like particles (13, 27). Attempts to establish such a system with either DEN2C or YFC proved unsuccessful, as formation of NC could not be detected by a variety of methods including negative-stain electron microscopy, sucrose gradient sedimentation, and agarose gel assay (data not shown). However, unlike alphaviruses, the flavivirus NC does not form in the cytoplasm of infected cells (see reference 18 for possible exceptions), and therefore, preformed flavivirus NCs may not readily form or may only transiently exist as part of the virus assembly pathway.
Secondary structure analysis of the flavivirus C protein.
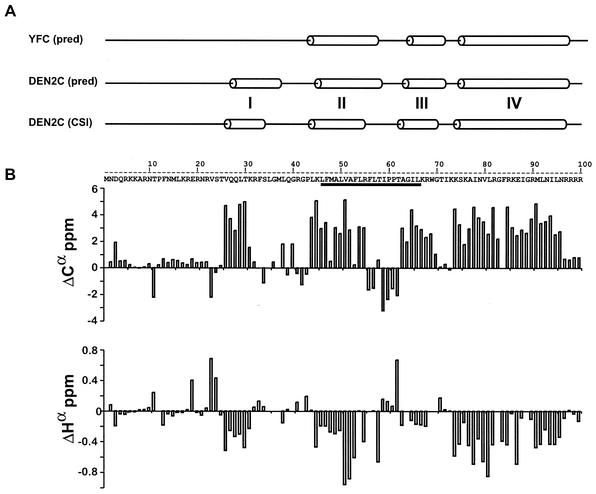
The flavivirus C protein is the least conserved protein of all the flavivirus proteins, with less than 40% sequence identity between most genus members (Table 2). DEN2C and YFC share the least sequence identity between sequenced mosquito-borne flaviviruses (∼22%), while YFC shares closer sequence identity to the C proteins of tick-borne flaviviruses. Comparison of the secondary structure predictions of DEN2C and YFC, with the PredictProtein server (http://www.embl-heidelberg.de/predictprotein/predictprotein.html), indicates that they are both predominately alpha-helical. Moreover, the secondary structure elements that are predicted to exist for DEN2C and YFC are very similar in both their extent and position within their respective primary amino acid sequences (Fig. 4A), with the exception of a short N-terminal alpha helix in DEN2C that is not predicted for YFC. Not surprisingly, the secondary structure elements of YFC are very similar to those for TBE, which is also predicted to have only three helices (data not shown).
TABLE 2.
Percent identities of flavivirus C proteinsa
| Virus | YFV | DEN2 | KUN | WNV | MVE | SLE | TBE |
|---|---|---|---|---|---|---|---|
| DEN2 | 22.5 | ||||||
| KUN | 25.9 | 34.9 | |||||
| WNV | 25.7 | 34.9 | 90.7 | ||||
| MVE | 22.9 | 32.1 | 71.3 | 71.4 | |||
| SLE | 29.4 | 35.2 | 62.0 | 57.9 | 60.7 | ||
| TBE | 33.3 | 14.0 | 20.4 | 14.3 | 25.7 | 24.3 | |
| POW | 32.4 | 18.6 | 19.4 | 15.2 | 23.8 | 23.4 | 59.4 |
KUN, Kunjin virus; WNV, West Nile virus; MVE, Murray Valley encephalitis virus; SLE, St. Louis encephalitis virus; POW, Powassan virus.
FIG. 4.
Flavivirus capsid is an alpha-helical protein. (A) Comparison of the secondary structure predictions of YFC and DEN2C. Secondary structure predictions for both proteins [YFC (pred) and DEN2C (pred)] were generated individually with ProteinPredict. The secondary structure predictions that are depicted are not sequence aligned. The secondary structure of DEN2C determined by a consensus of Cα, Cβ, Hα, and C′ chemical shift indexing [DEN2C (CSI)] is shown below the predicted secondary structures of YFC and DEN2C and is based on the data shown in panel B. Non-alpha-non-beta secondary structures are represented by a straight line. Alpha-helical secondary structures are represented by cylinders. The four alpha helices of DEN2C are indicated by the roman numerals I to IV. The underlined residues within the amino acid sequence of DEN2C indicate the internal hydrophobic sequence. The C-terminal signal sequence is not shown. (B) NMR analysis of the DEN C protein. The differences between observed sequence-specific chemical shift values and the corresponding predicted sequence corrected random-coil values were plotted against the corresponding residue number within DEN2C. Plots of Cα (top) and Hα (bottom) are shown.
The alpha-helical nature of the C protein was confirmed experimentally by far-UV circular dichroism (CD). The far-UV CD spectra of both YFC and DEN2C demonstrated characteristics of alpha-helical proteins with local minima at 222 and 208 and a maximum at 190 nm (data not shown). In addition, the far-UV CD spectra for DEN2CΔ1-13 was essentially identical to that for DEN2C, indicating that removal of the first 13 residues did not significantly perturb the secondary structure of DEN2C (data not shown). This observation is consistent with the secondary structure prediction of DEN2C.
Chemical shift indexing and secondary structure assignment.
The 1H, 13C, and 15N chemical shifts of DEN2C were assigned by using a standard set of two- and three-dimensional heteronuclear NMR experiments (to be described elsewhere) as a first step toward determining the three-dimensional solution structure of DEN2C by NMR. Data were processed with the software NMRPipe (3) and analyzed with the programs ANSIG (10) and SPARKY (5). The differences between the observed 1Hα, 13Cα, 13C′, and 13Cβ chemical shifts and the sequence-corrected random-coil values (25) were used in the secondary structure analysis. Chemical shift differences for 13Cα and 1Hα, the most distinguishing values, are shown in Fig. 4B. The positions of the secondary structure elements of DEN2C identified from the consensus of chemical shift differences with the program CSI (30) are shown in Fig. 4A. Overall, the CSI analysis indicated that DEN2C is 48% alpha-helical. The position and length of the alpha helices within DEN2C determined by chemical shift indexing [DEN2C (CSI) in Fig. 4A] are in good agreement with those predicted with PredictProtein [DEN2C (pred) in Fig. 4A]. The secondary structure of DEN2C is composed of four alpha helices. The most N-terminal helix (helix I, amino acids 26 to 31) is followed by a 14-residue loop and a second helix (helix II, amino acids 45 to 55). Helix II contains the start of an internal hydrophobic region that has been previously implicated in membrane association (underlined residues of DEN2C sequence in Fig. 4B). Following helix II is a loop region (amino acids 56 to 62) that is followed by helix III (amino acids 63 to 69) and then a fourth short loop region (amino acids 70 to 73). The longest helix in DEN2C (helix IV) occurs at the C terminus of the protein (amino acids 74 to 96). The N terminus of DEN2C (amino acids 1 to 21) does not appear to possess secondary structure.
NMR spectroscopy analysis of DEN2C by HSQC.
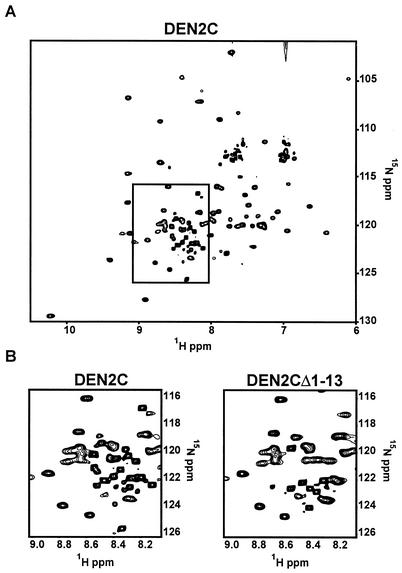
The presence of tertiary structure was evaluated from the 15N heteronuclear spin quantum coherence (HSQC) spectrum of singly labeled DEN2C (Fig. 5A). The pattern of well-dispersed peaks in an HSQC spectrum is a “fingerprint” of the folded protein in solution. The majority of peaks show good chemical shift dispersion, indicating a well-defined fold. However, ∼20% of the chemical shifts approximate random-coil values and possess significantly higher than average signal intensity, indicating an unstructured region of the protein (boxed region in Fig. 5A and left box in Fig. 5B).
FIG. 5.
HSQC spectrum of DEN2C. The folded state of DEN2 was accessed by NMR (see text). (A) The boxed region indicates a region of the HSQC spectrum that contains several peaks corresponding to residues within DEN2C that have random-coil chemical shift values. These residues are thought to be disordered in the solution structure of DEN2C. (B) Comparison of the boxed region in panel A with the same region within the HSQC spectrum of DEN2CΔ1-13, in which the first 13 amino acids have been removed.
To examine whether any of the peaks within this random-coil region of the HSQC spectrum of DEN2C corresponded to residues within its N terminus, 15N DEN2CΔ1-13 was prepared and analyzed by HSQC. Most of the HSQC peaks for DEN2CΔ1-13 were identical to those for DEN2C, indicating that removal of the first 13 residues did not alter the overall fold of the protein. However, the differences that were observed between the two HSQC spectra occurred in the random-coil region of the DEN2CΔ1-13 HSQC spectrum (Fig. 5B). Subsequent complete residue assignment of DEN2C (to be described elsewhere) has confirmed that the high-intensity peaks absent in the DEN2CΔ1-13 HSQC spectrum correspond to the truncated N-terminal residues.
Structure and function of the flavivirus capsid protein.
The N terminus of DEN2C is the least structured region of the protein, and at least the first 13 amino acid residues of the protein are dispensable for the overall fold of the protein. Secondary structure prediction of flavivirus C proteins for which sequence information is available indicates that the N terminus of flavivirus C proteins does not possess regular secondary structure (data not shown). Excluding the possibility of structured loop regions, the absence of any predicted regular secondary structure within the N terminus of flavivirus C proteins may be an indication that the N terminus of the C protein is unstructured (as reported here for DEN) and may be a common feature of flavivirus C proteins. The unstructured N terminus of the C protein may play a role in RNA binding, possibly becoming structured upon interaction with RNA.
The alphaviruses are arthropod-borne viruses and occupy an ecological niche very similar to that of the flaviviruses (26). Interestingly, and quite surprisingly, the flavivirus E protein and the alphavirus E1 protein, one of the two major glycoproteins that make up the outer protein shell of the alphavirus, share remarkable structural similarity, despite less than 20% amino acid sequence identity (14). Moreover, both proteins are responsible for the fusogenic activity of the respective viruses. Recently, Kofler et al. speculated that the N-terminal domain of the C protein from alphaviruses may be structurally and functionally homologous to flavivirus C protein (8). Although there are some functional similarities in the C proteins of these two different virus groups, our results indicate that there are fundamental differences between the flavivirus and alphavirus C proteins.
The N-terminal domain of the alphavirus C protein is similar to the flavivirus C protein in that they are approximately the same size (100 residues) and both contain a large percentage of basic residues, indicative of the role that both C proteins play in binding the viral RNA. In addition, both C proteins possess an internal sequence of hydrophobic residues surrounded by charged residues. However, the role of this sequence appears to be quite different. In alphaviruses, this sequence (amino acids 38 to 55 in Sindbis virus) is predicted to form a coiled-coil interaction with adjacent alphavirus C proteins, providing stability to the alphavirus NC (19). Certain point mutations and small deletions within this region of the alphavirus C protein decrease core stability and virus replication (19). Since the alphavirus C protein is a monomer in the absence of nucleic acid (27), the coiled-coil interaction of the alphavirus C protein appears only to stabilize a nucleic acid-dependent dimerization (28). By contrast, the internal hydrophobic sequence of the flavivirus C protein is not predicted to form coiled-coil interactions, and our studies show that the dimerization of flavivirus C protein is independent of nucleic acid being present. The internal hydrophobic sequence of flavivirus C has, instead, been implicated in tethering C protein to membranes (17). This tethering is thought to play a role in localizing the flavivirus C protein to sites of virus assembly on intracellular membranes. It is possible that the internal hydrophobic sequence, the nature of which is conserved in flavivirus C proteins, may have a role in flavivirus C protein dimerization. However, in the case of TBE, most of the internal hydrophobic sequence is apparently not required for the function of C protein (8).
The recruitment of genome RNA and flavivirus C proteins and their formation into an NC are not understood. However, the interaction between C proteins and RNA appears to be critical for the production of authentic virus particles, for in the absence of C protein, small T=1 VLPs lacking RNA are produced (24). It is anticipated that at some step in assembly the C protein would also interact with the viral transmembrane proteins to promote the formation and incorporation of the NC with the budding particle. Following release of the C protein from its signal sequence membrane anchor, the formation of a protein dimer may be the first step required to facilitate these interactions. The dimeric nature of the C protein may play a role in other aspects of the virus life cycle. Several groups have observed the C protein in the nucleus of infected cells, and at least one group has implicated the C protein in apoptotic pathways (29, 31). Dissecting the function of the oligomeric state of the C protein may provide interesting insights into these various aspects of the flavivirus life cycle.
Acknowledgments
We thank Suchetana Mukhopadhyay for help in reviewing the manuscript and Chinmay Patkar for initial work on protein purification. We also thank Charles Rice for providing the pYF5′3′ plasmid and James Strauss for providing the DEN pC2 plasmid.
This work was supported by a Public Health Service Program Project Grant (AI45976) from the National Institute of Allergy and Infectious Diseases to R.J.K. and C.B.P. C.T.J. was supported by a Purdue Research Foundation Fellowship and an NIH Biophysics Training Grant (GM08296). T.D.G. was also supported by the NIH Biophysics Training Grant.
REFERENCES
- 1.Allison, S. L., K. Stadler, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J. Virol. 69:5816-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantor, C. R., and P. R. Schimmel. 1980. Techniques for the study of biological structure and function. Part II. W. H. Freeman and Co., San Francisco, Calif.
- 3.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277-293. [DOI] [PubMed] [Google Scholar]
- 4.Fonseca, B. A., S. Pincus, R. E. Shope, E. Paoletti, and P. W. Mason. 1994. Recombinant vaccinia viruses co-expressing dengue-1 glycoproteins prM and E induce neutralizing antibodies in mice. Vaccine 12:279-285. [DOI] [PubMed] [Google Scholar]
- 5.Goddard, T. D., and D. G. Kneller. 1993. SPARKY3. University of California, San Francisco.
- 6.Hahn, Y. S., R. Galler, T. Hunkapiller, J. M. Dalrymple, J. H. Strauss, and E. G. Strauss. 1988. Nucleotide sequence of dengue 2 RNA and comparison of the encoded proteins with those of other flaviviruses. Virology 162:167-180. [DOI] [PubMed] [Google Scholar]
- 7.Khromykh, A. A., and E. G. Westaway. 1996. RNA binding properties of core protein of the flavivirus Kunjin. Arch. Virol. 141:685-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kofler, R. M., F. X. Heinz, and C. W. Mandl. 2002. Capsid protein C of tick-borne encephalitis virus tolerates large internal deletions and is a favorable target for attenuation of virulence. J. Virol. 76:3534-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konishi, E., S. Pincus, E. Paoletti, R. E. Shope, T. Burrage, and P. W. Mason. 1992. Mice immunized with a subviral particle containing the Japanese encephalitis virus prM/M and E proteins are protected from lethal JEV infection. Virology 188:714-720. [DOI] [PubMed] [Google Scholar]
- 10.Kraulis, P. J. 1989. ANSIG—a program for the assignment of protein H-1 2D-NMR spectra by interactive computer-graphics. J. Magn. Reson. 84:627-633. [Google Scholar]
- 11.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kummerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunkel, M., M. Lorinczi, R. Rijnbrand, S. M. Lemon, and S. J. Watowich. 2001. Self-assembly of nucleocapsid-like particles from recombinant hepatitis C virus core protein. J. Virol. 75:2119-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lescar, J., A. Roussel, M. W. Wein, J. Navaza, S. D. Fuller, G. Wengler, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 15.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 16.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 75:10787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markoff, L., B. Falgout, and A. Chang. 1997. A conserved internal hydrophobic domain mediates the stable membrane integration of the dengue virus capsid protein. Virology 233:105-117. [DOI] [PubMed] [Google Scholar]
- 18.Ng, M. L., S. H. Tan, and J. J. H. Chu. 2001. Transport and budding at two distinct sites of visible nucleocapsids of West Nile (Sarafend) virus. J. Med. Virol. 65:758-764. [DOI] [PubMed] [Google Scholar]
- 19.Perera, R., K. E. Owen, T. L. Tellinghuisen, A. E. Gorbalenya, and R. J. Kuhn. 2001. Alphavirus nucleocapsid protein contains a putative coiled-coil alpha-helix important for core assembly. J. Virol. 75:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pincus, S., P. W. Mason, E. Konishi, B. A. Fonseca, R. E. Shope, C. M. Rice, and E. Paoletti. 1992. Recombinant vaccinia virus producing the prM and E proteins of yellow fever virus protects mice from lethal yellow fever encephalitis. Virology 187:290-297. [DOI] [PubMed] [Google Scholar]
- 21.Pugachev, K. V., P. W. Mason, and T. K. Frey. 1995. Sindbis vectors suppress secretion of subviral particles of Japanese encephalitis virus from mammalian cells infected with SIN-JEV recombinants. Virology 209:155-166. [DOI] [PubMed] [Google Scholar]
- 22.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature (London) 375:291-298. [DOI] [PubMed] [Google Scholar]
- 23.Rice, C. M., E. M. Lenches, S. R. Eddy, S. J. Shin, R. L. Sheets, and J. H. Strauss. 1985. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229:726-733. [DOI] [PubMed] [Google Scholar]
- 24.Russell, P. K., W. E. Brandt, and J. M. Dalrymple. 1980. Chemical and antigenic structure of flaviviruses, p. 503-529. In R. W. Schlesinger (ed.), The togaviruses. Biology, structure, replication. Academic Press, New York, N.Y.
- 25.Schwarzinger, S., G. J. A. Kroon, T. R. Foss, P. E. Wright, and H. J. Dyson. 2000. Random coil chemical shifts in acidic 8 M urea: implementation of random coil shift data in NMRView. J. Biomol. NMR 18:43-48. [DOI] [PubMed] [Google Scholar]
- 26.Strauss, J. H., and E. G. Strauss. 2001. Virus evolution: how does an enveloped virus make a regular structure? Cell 105:5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tellinghuisen, T. L., A. E. Hamburger, B. R. Fisher, R. Ostendorp, and R. J. Kuhn. 1999. In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in Escherichia coli. J. Virol. 73:5309-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tellinghuisen, T. L., and R. J. Kuhn. 2000. Nucleic acid-dependent cross-linking of the nucleocapsid protein of Sindbis virus. J. Virol. 74:4302-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westaway, E. G., A. A. Khromykh, M. T. Kenney, J. M. Mackenzie, and M. K. Jones. 1997. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology 234:31-41. [DOI] [PubMed] [Google Scholar]
- 30.Wishart, D. S., and B. D. Sykes. 1994. The 13C chemical shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR 4:171-180. [DOI] [PubMed] [Google Scholar]
- 31.Yang, J. S., M. P. Ramanathan, K. Muthumani, A. Y. Choo, S. H. Jin, Q. C. Yu, D. S. Hwang, D. K. Choo, M. D. Lee, K. Dang, W. Tang, and J. J. Kim. 2002. Induction of inflammation by West Nile virus capsid through the caspase-9 apoptotic pathway. Emerg. Infect. Dis. 8:1379-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]