Abstract
The replication-associated protein (Rep) of geminiviruses is involved in several biological processes brought about by the presence of distinct functional domains. Recently, we have exploited the multifunctional character of the Tomato yellow leaf curl Sardinia virus (TYLCSV) Rep to develop a molecular interference strategy to impair TYLCSV infection. We showed that transgenic expression of its N-terminal 210 amino acids (Rep-210) confers resistance to the homologous virus by inhibiting viral transcription and replication. We have now used biochemical and transgenic approaches to carry out a fuller investigation of the molecular resistance mechanisms in transgenic plants expressing Rep-210. We show that Rep-210 confers resistance through two distinct molecular mechanisms, depending on the challenging virus. Resistance to the homologous virus is achieved by the ability of Rep-210 to tightly inhibit C1 gene transcription, while that to heterologous virus is due to the interacting property of the Rep-210 oligomerization domain. Furthermore, we present evidence that in Rep-210-expressing plants, the duration of resistance is related to the ability of the challenging virus to shut off transgene expression by a posttranscriptional homology-dependent gene silencing mechanism. A model of Rep-210-mediated geminivirus resistance that takes transgene- and virus-mediated mechanisms into account is proposed.
Geminiviruses are a large family of plant viruses possessing a genome of one or two circular single-stranded DNA (ssDNA) molecules, each of about 2.7 kb, encapsidated in a paired particle (50). They replicate in the nuclei of the infected cells through double-stranded intermediates (50, 52). The replication-associated protein (Rep) is encoded by the C1 gene and is the only protein absolutely required for replication (18, 19). Rep is a multifunctional protein involved in several biological processes: (i) initiation and termination of rolling circle replication (RCR) by nicking and religating the replication origin of viral DNA (35, 51); (ii) repression of its own gene transcription (16, 53); and (iii) interaction with host cell factors to interfere, inter alia, with control of cell cycle and DNA replication in the infected cells (22, 25). It forms oligomers (36, 45), and mutations in its oligomerization domain affect both replication and Rep-mediated transcription repression (44). The nicking and religating, DNA binding, and repression functions are located in the N-terminal part of the protein (9, 10, 20, 27, 31, 45), whereas the oligomerization domain and the ATPase activity are located in its central (45) and C-terminal portions (15, 26), respectively.
Several geminiviruses causing tomato yellow leaf curl described in the last 10 years are responsible for one of the world's most important tomato diseases (38). Two species with a single genomic component have been extensively studied: Tomato yellow leaf curl Sardinia virus (TYLCSV), originally from Italy (32), and Tomato yellow leaf curl virus (TYLCV), originally from Israel (40). The TYLCSV genome contains six partially overlapping open reading frames (ORFs) organized in two divergent transcriptional units separated by an intergenic region (IR). ORFs V1 and V2 are on the virion sense strand, and ORFs C1 to C4 are on the complementary strand. C4 is completely contained within C1 (32).
Transgenic expression of pathogen-derived sequences has been extensively used to obtain virus-resistant plants. These strategies have variously explored and exploited the general idea that transgenic expression of virus-derived sequences may interfere with the viral life cycle (3). However, the observation that the predicted molecular interference mechanisms have not always coincided with those operating in resistant transgenic plants has revealed the complexity of the interaction between the transgene and the challenging virus. It has become clear that a given transgenic sequence can act via a protein-mediated mechanism or by posttranscriptional gene silencing (PTGS), depending on its molecular fate, and that infection can induce transgene silencing (virus-induced gene silencing [VIGS]), resulting in a recovery phenotype (2, 3). In both VIGS and PTGS, double-stranded RNAs (dsRNAs) produced during infection or synthesized from aberrant transgene mRNAs are processed into 21- to 25-nucleotide (nt)-long small interfering RNAs (siRNAs) that direct ribonucleases to target homologous transgene and viral RNAs (23, 24). Geminiviruses, which possess a DNA genome and do not replicate through dsRNA intermediates, encode, like RNA viruses, suppressors of PTGS (56, 58), suggesting that they may both induce and probably also be targets of PTGS (58). Moreover, geminiviruses can silence via VIGS trans- and endogenes when homologous sequences to these genes are expressed from their genomes (33, 47).
We have previously shown that transgenic expression of a truncated form of the TYLCSV C1 gene encoding Rep's first 210 amino acids (Rep-210), and potentially coexpressing C4, confers resistance to the homologous virus in Nicotiana benthamiana and tomato plants (4, 41). We used an antisense strategy to demonstrate that high Rep-210 levels in the transgenic tissues are required to confer TYLCSV resistance (4, 41). We established that C4, if expressed, did not contribute to this resistance (5). Moreover, we showed that Rep-210 repressed C1 transcription and TYLCSV replication, but not that of the related TYLCSV Murcia strain (TYLCSV-ES[1]), suggesting that transcriptional repression plays an active role in TYLCSV resistance (5). Rep-210 does not contain the NTP binding domain and its ATPase-associated activity required for replication (15), but it does retain both the transcriptional repression activity (5) and the three conserved motifs characteristic of RCR proteins (29). The first N-terminal 116 amino acids of the TYLCSV Rep protein are sufficient to confer specific recognition of its own origin of replication (31). Moreover, by analogy with the Rep proteins of Tomato golden mosaic virus (TGMV) and Tomato leaf curl virus-New Delhi virus (ToLCNDV), TYLCSV Rep-210 is also expected to contain an intact oligomerization domain that may be involved in Rep-210-mediated resistance (8, 45).
We have now carried out a fuller investigation of the molecular resistance mechanisms in transgenic plants expressing Rep-210. The ability of Rep-210 to interfere with a heterologous virus and the contributions of the oligomerization domain and transcriptional repression to resistance have been assessed. A series of C-terminal deletion mutants of TYLCSV Rep-210 were tested to determine their ability to inhibit homologous and heterologous C1 transcription, confer resistance on transgenic plants, inhibit viral replication in protoplasts, and interact in in vivo systems. We show that Rep transcription repression does not require the oligomerization domain and that Rep-210 confers resistance through two distinct molecular mechanisms, depending on the challenging virus. We present data showing that Rep-210 acts as a transdominant-negative mutant that inhibits the homologous virus by tightly repressing the viral C1 promoter, whereas it forms dysfunctional complexes with the Rep of the heterologous TYLCV. Moreover, we show that in Rep-210-expressing plants, the duration of resistance is related to the ability of the challenging virus to silence Rep-210 transgene. A model of Rep-210-mediated geminivirus resistance that takes transgene- and virus-mediated mechanisms into account is proposed.
MATERIALS AND METHODS
Biomaterials.
Nicotiana benthamiana 102.22 and tomato BC1 transgenic lines have been previously described (4, 41). Two transgenic tomato lines transformed with the pSW9 construct carrying the N gene of Tomato spotted wilt virus (TSWV) were kindly provided by A. Vaira (A. Vaira and G. Nervo, unpublished data). Tomato plants naturally infected by TYLCSV were collected in two greenhouses in Sicily, southern Italy.
Plasmids.
The constructs used are listed in Table 1. Viral sequences were PCR amplified with Pfu DNA polymerase (Stratagene) and specific primers containing restriction site sequences at the extremities. The exact length and position of each amplified sequence are reported in Table 1. All final clones were sequenced to confirm the fidelity of PCR amplification and the vector-insert junctions. The entire series of Rep mutants was amplified from pTOM100C4(−), a plasmid that encodes Rep-210, but contains a stop codon for the internal C4 protein (5). Briefly, for TYLCSV expression cassettes, amplified fragments were digested with BamHI and EcoRI and ligated to the corresponding sites of pJIT60 (kindly provided by J. Mullineaux) to generate the pJTR series. pTOM130 was obtained by ligating the KpnI-BglII fragment of pJTR130 to the KpnI-BamHI sites of pBIN19. For two-hybrid system (THS) vectors, cloning into pAS2 and pACT2 (CLONTECH) was performed with NcoI and BamHI sites. Plasmids pREP41HA N and pREP42MH N used for the expression of tagged proteins in Schizosaccharomyces pombe are described in reference 12. The amplified viral sequences carrying an XhoI site at the 5′ end and a BamHI site at the 3′ end were ligated to SalI-BamHI-digested pREP41HA N and pREP42MH N vectors. The β-glucuronidase (GUS) reporter vector pIntPT/GUS was obtained as follows. The NdeI-BamHI fragment from pPORT2 (39) was blunt ended at the NdeI extremity and cloned in the HincII-BamHI sites of pGEM4Z (Promega); the KpnI-PstI fragment of this subclone was ligated to KpnI-PstI-digested pJIT61GUS (5).
TABLE 1.
Constructs used in this study
| Name | Viral sequencesa | Viral DNA coordinatesb |
|---|---|---|
| TYLCSV expression cassettes | ||
| pJTR359 (previously pTOM120)c | 42-bp UTR + C1 ORF | 1538-2656 (1) |
| pJTR210a (previously pTOM100)c | 42-bp UTR + truncated C1 ORF (630 nt) containing C4 coding region | 1985-2656 (1) |
| pJTR210 (previously pTOM100C4-)c | 42-bp UTR + truncated C1 ORF (630 nt) | 1985-2656 (1) |
| pJTR181 | 42-bp UTR + truncated C1 ORF (543 nt) | 2072-2556 (1) |
| pJTR156 | 42-bp UTR + truncated C1 ORF (468 nt) | 2147-2656 (1) |
| pJTR130 | 42-bp UTR + truncated C1 ORF (390 nt) | 2225-2656 (1) |
| pJTR120 | 42-bp UTR + truncated C1 ORF (360 nt) | 2255-2656 (1) |
| pJTR80 | 42-bp UTR + truncated C1 ORF (240 nt) | 2375-2656 (1) |
| pJTR54 | 42-bp UTR + truncated C1 ORF (162 nt) | 2453-2656 (1) |
| pTOM130 | KpnI-BglII cassette of pJTR130 in the binary vector pBIN19 | 2225-2656 (1) |
| Yeast two-hybrid plasmids | ||
| pASPTRep-359 | GAL4 BD fused to TYLCV-[PT] Rep | 1551-2630 (2) |
| pASRep-359 | GAL4 BD fused to TYLCSV Rep | 1538-2614 (1) |
| pASRep-210 | GAL4 BD fused to TYLCSV Rep210 | 1985-2614 (1) |
| pASRep-181 | GAL4 BD fused to TYLCSV Rep181 | 2072-2614 (1) |
| pASRep-156 | GAL4 BD fused to TYLCSV Rep156 | 2147-2614 (1) |
| pASRep-130 | GAL4 BD fused to TYLCSV Rep130 | 2225-2614 (1) |
| pASRep-120 | GAL4 BD fused to TYLCSV Rep120 | 2255-2614 (1) |
| pASRep-80 | GAL4 BD fused to TYLCSV Rep80 | 2375-2614 (1) |
| pASSNF1 | GAL4 BD fused to SNF1 | |
| pACTPTRep-359 | GAL4 AD fused to TYLCV-[PT] Rep | 1551-2630 (2) |
| pACTRep-359 | GAL4 AD fused to TYLCSV Rep | 1538-2614 (1) |
| pACTRep-210 | GAL4 AD fused to TYLCSV Rep210 | 1985-2614 (1) |
| pACTRep-181 | GAL4 AD fused to TYLCSV Rep180 | 2072-2614 (1) |
| pACTRep-156 | GAL4 AD fused to TYLCSV Rep156 | 2147-2614 (1) |
| pACTRep-130 | GAL4 AD fused to TYLCSV Rep130 | 2225-2614 (1) |
| pACTRep-120 | GAL4 AD fused to TYLCSV Rep120 | 2255-2614 (1) |
| pACTRep-80 | GAL4 AD fused to TYLCSV Rep80 | 2375-2614 (1) |
| pACTSNF4 | GAL4 AD fused to SNF4 | |
| S. pombe plasmids | ||
| p42Rep-359 | HIS tag fused to TYLCSV Rep | 1538-2614 (1) |
| p42Rep-210 | HIS tag fused to TYLCSV Rep210 | 1985-2614 (1) |
| p42Rep-181 | HIS tag fused to TYLCSV Rep181 | 2072-2614 (1) |
| p42Rep-156 | HIS tag fused to TYLCSV Rep156 | 2147-2614 (1) |
| p42Rep-130 | HIS tag fused to TYLCSV Rep130 | 2225-2614 (1) |
| p41Rep-359 | HA tag fused to TYLCSV Rep | 1538-2614 (1) |
| p41Rep-210 | HA tag fused to TYLCSV Rep210 | 1985-2614 (1) |
| p41Rep-181 | HA tag fused to TYLCSV Rep181 | 2072-2614 (1) |
| p41Rep-156 | HA tag fused to TYLCSV Rep156 | 2147-2614 (1) |
| p41Rep-130 | HA tag fused to TYLCSV Rep130 | 2225-2614 (1) |
| GUS reporter plasmids | ||
| pTOM202c | IR of TYLCSV; translational fusion of C1 promoter to GUS gene | 2600-152 (1) |
| pIntS/GUSc | IR of TYLCSV; transcriptional fusion of C1 promoter to GUS gene | 2615-152 (1) |
| pIntS-ES[1]/GUSc | IR of TYLCSV-ES[1]; transcriptional fusion of C1 promoter to GUS gene | 2620-154 (3) |
| pIntPT/GUS | IR of TYLCV-[PT]; transcriptional fusion of C1 promoter to GUS gene | 2629-163 (2) |
| Infectious TYLCSV DNA clones | ||
| pTOM6 | SacI tandem dimer of TYLCSV menome | |
| pSP97 | SpeI-BamHI 1.8-mer of TYLCSV-ES[1] genome | |
| pPORT | SphI-BamHI 1.7-mer of TYLCV-[PT] genome |
To in vitro transcribe a TYLCV-[PT] V1-V2 antisense RNA probe, the EcoRI-BamHI fragment (nt 162 to 1739) of pPORT2 (39) was cloned in the corresponding sites of pBSK (Stratagene) to obtain pPort. pTOM202, pIntS/GUS, pInt-ES[1]/GUS, pTOM6, pSP97, pGEM102, and pGEM-P have been described previously (5).
Transgenic plants.
N. benthamiana was transformed with the recombinant Agrobacterium tumefaciens strain C58(pGV2260) harboring the plasmid pTOM130, and kanamycin-resistant plants were regenerated as described previously (41). The primary transformants were checked for the presence of Rep-130 transgene by PCR and for the expression of the Rep-130 protein by Western blotting with the anti-TYLCSV-C1 antibody as described previously (41). The chemiluminescent reaction was detected with either the Renaissance kit (NEN Du Pont) or the Supersignal West Pico chemiluminescent substrate (Pierce).
A specific polyclonal antibody described in reference 55 was used to detect TSWV N protein. The antibody reaction was evaluated with bromo-chloro-indolyl-phosphate and nitroblue tetrazolium salts as substrate.
Protoplast assays.
All protoplast transfection, GUS assay, nucleic acid extraction, and Southern analysis procedures were performed as previously described (5), unless otherwise stated. For the GUS assay using pIntPT/GUS and pIntS/GUS, incubation with substrate was prolonged to 24 h.
For Western blot analysis, protoplast samples (5 × 105) were resuspended in 100 μl of Laemmli buffer, boiled for 5 min, centrifuged for 5 min, and loaded onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS PAGE) (15% polyacrylamide) gels. Rep and its mutants were detected as described above, but with the Lumi-LightPLUS Western blotting kit (Roche).
Probes utilized for Southern blotting were either digoxigenin-labeled RNA transcripts obtained as specified from the supplier (Roche) or radioactive DNA probes (High Prime DNA Labeling kit; Roche) for quantitative replication analysis. The hybridization conditions with radioactively labeled DNA probe were as follows: 50% formamide, 10% dextran sulfate, 1 M NaCl, 1% SDS, and 100 μg of salmon sperm DNA per ml at 42°C. Washes were performed at 60°C in 0.1% SDS-0.1% SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). For quantitative analysis of DNA replication in protoplasts, each construct was assayed in duplicate in three independent experiments. After washes, hybridized filters were analyzed with an Istant Imager (Canberra, Packard).
THS.
Cotransformation of Saccharomyces cerevisiae strain PJ69-4A (30) was performed with lithium acetate-polyethylene glycol. For interaction assays, cotransformants were streaked on selective medium lacking adenine to score the activity of the reporter GAL2-ADE2 gene, lacking histidine to score the activity of the reporter GAL1-HIS3 gene, or lacking both. When testing the prototrophy for histidine, 2 mM 3-aminotriazole was added to the medium to prevent HIS3 gene leakage.
For Western blot analysis, S. cerevisiae transformants were grown and harvested as described in the Clontech Yeast Protocols Handbook (Clontech, Palo Alto, Calif.), and proteins were extracted as described in reference 44. SDS-PAGE, electrotransfer to polyvinylidene difluoride (PVDF), and reaction with GAL4 binding domain monoclonal antibodies from Clontech were all performed according to the manufacturer's instructions. Secondary antibody (antimouse immunoglobulin G [IgG] conjugated with peroxidase) and the chemiluminescent substrate were obtained with the Lumi-LightPLUS Western blotting kit (Roche).
Coimmunoprecipitation.
The standard medium and genetic procedures with S. pombe have been described previously (37). For tagged protein expression, a haploid wild-type S. pombe strain, h− leu1.32 ura4.d18, was used. Exponential liquid cultures of S. pombe cotranformants were harvested by centrifugation 19 h after the induction of the promoter (no thiamine on the medium) driving the expression of histidine (HIS)- and hemagglutinin (HA)-tagged fusion proteins. Cells were lysed in NIB buffer (25 mM Tris-HCl [pH 8], 1 mM EDTA, 0.014 mM β-mercaptoethanol, 0.01% Triton X-100) with protease inhibitors (10 μg of pepstatin per ml, 50 μg of leupeptin per ml, 1 mM phenylmethylsulfonyl fluoride) and 500 μl of glass beads (425 to 600 μm in diameter; Sigma). Samples were vortexed in FastPrep FP120 (BIO 101) at a power setting of 5.5 for two 15-s intervals separated by 1-min intervals on ice. After 30 min of centrifugation (12,000 × g, 4°C) the supernatant was recovered, and the protein concentration was determined with Bradford assays. Immunoprecipitation was performed by incubating 1 mg of protein extract with anti-HA monoclonal antibody overnight at 4°C in the extraction buffer. Protein-antibody complexes were mixed for 2 h at 4°C with protein A-Sepharose CL-4B (Amersham Pharmacia Biotech AB) in TBS-Tween (TBST) and then washed with TBST five times. Bound proteins were eluted with SDS-PAGE sample buffer at 100°C. Coimmunoprecipitation was monitored by SDS-PAGE followed by transfer to nitrocellulose membrane (Hybond ECL; Amersham Pharmacia Biotech AB) and immunoblotting with the ECL enchanced chemiluminescence detection system (Amersham Pharmacia Biotech AB). Primary antibodies were monoclonal anti-His or anti-HA antibody.
Virus resistance assay.
Plants were inoculated by agroinoculation or, in a few cases, with the whitefly vector. For agroinoculation, the A. tumefaciens strain LBA4404 carrying infectious clones of TYLCSV (32), TYLCSV-ES[1] (42), and TYLCV-[PT], was used (39). For whitefly infection, plants were inoculated with the natural vector of TYLCSV transmission (Bemisia tabaci) as previously described (4). Infection was monitored weekly, or every 2 weeks, by tissue print with a digoxigenin-labeled probe specific for the coat protein gene, obtained by PCR. The probe specific for TYLCSV was also used to detect TYLCSV-ES[1], while TYLCV-[PT] was detected with a fully homologous coat protein probe.
The plant apex was excised at the time of agroinoculation (sample defined at 0 weeks postinoculum [p.i.]) and used for molecular analysis of Rep-210 protein and mRNA expression. For assays performed following agroinoculation, the petiole of the second or third leaf immediately below the main new apex were used (sample at x weeks p.i.).
Northern blotting.
Total RNA was extracted from plants with the RNAwiz reagent (Ambion) according to the manufacturer's instructions. Fifteen micrograms of total RNA was denatured with formamide-glyoxal and separated in a 1% agarose gel in 1× TPE buffer as previously described (41). RNA was run at 70 V for 5 h, photographed after ethidium bromide staining, and capillary blotted overnight onto nylon membranes (Roche). Membranes were hybridized with a C1-specific digoxigenin-labeled RNA probe, transcribed from plasmid pGEM103 (41) at 68°C according to the Roche manual. For nptII mRNA detection, a digoxigenin-labeled DNA probe synthesized by PCR from plasmid pBin19 with nptII-specific primers, was used. Hybridization was performed at 50°C in a 50% formamide buffer. Following overnight hybridization, membranes were washed at room temperature in 2× SSC-0.1% SDS and at the hybridization temperature in 0.1× SSC-0.1% SDS. Labeling was detected with CDPstar (Roche) as a chemiluminescent substrate.
Analysis of 21- to 25-nt RNAs.
Total RNA was extracted from plant tissue as described previously (41). Fifty micrograms of total RNA was resuspended in 15 μl of formamide and denatured for 5 min at 65°C; after addition of one-third volume of 4× loading solution (4× TBE, 0.08% bromophenol blue [BPB]) samples were separated by 8% PAGE. The upper portion of the gel was removed, stained with ethidium bromide, and photographed to verify normalization. The lower part of the gel was blotted and fixed to Hybond-N membranes as described in reference 54. Lanes containing oligonucleotide size markers were cut and stained with methylene blue (49). Radiolabeled probes were transcribed from EcoRI-linearized pGEM102 and pGEM103 plasmids (41) and hydrolyzed to lengths averaging 75 nt (11). Hybridization was performed overnight at 39°C, as described in reference 13. After hybridization, filters were washed in 2× SSC-0.2% SDS at 50°C.
RESULTS
Transcription repression by TYLCSV Rep does not require Rep-Rep interaction.
To distinguish the contribution of Rep oligomerization and transcription repression in Rep-210-mediated TYLCSV resistance, we first asked whether repression requires TYLCSV Rep-Rep interaction by assessing the ability of a series of Rep C-terminal deletion mutants to inhibit TYLCSV C1 transcription and self-interact in in vivo and in vivo-in vitro systems.
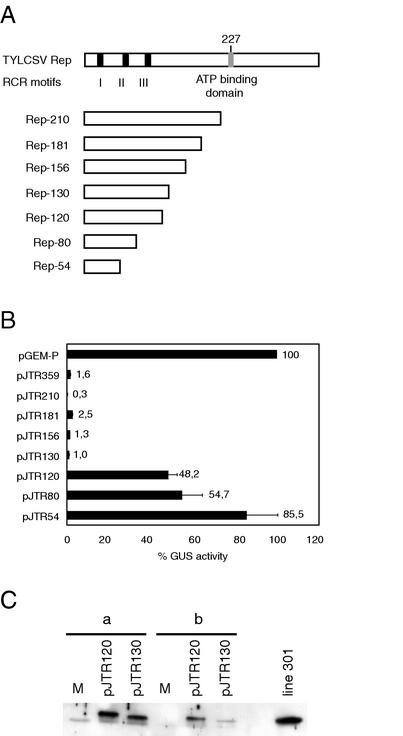
For the repression assay, wild-type N. benthamiana protoplasts were cotransfected with the plasmid pTOM202, containing the GUS reporter gene under the control of the TYLCSV C1 promoter, together with one of the following constructs (Table 1 and Fig. 1A): pJTR359 (Rep), pJTR210 (Rep-210), pJTR181 (Rep-181), pJTR156 (Rep-156), pJTR130 (Rep-130), pJTR120 (Rep-120), pJTR80 (Rep-80), pJTR54 (Rep-54), or pGEM-P (negative control, see Materials and Methods), all containing the Rep-encoding sequences under the control of an enhanced 35S (E35S) Cauliflower mosaic virus promoter. The C-terminal deletion mutants expressing Rep-210, -181, -156, and -130 repressed TYLCSV C1 promoter similarly to the wild-type Rep (Fig. 1B). Deletion of a further 10 amino acids (pJTR120) drastically impaired the ability of Rep-120 to repress the C1 transcription. To determine whether this was merely due to the absence of Rep-120 accumulation, total proteins were extracted from pJTR130- and pJTR120-transfected protoplasts and analyzed by Western blotting with anti-Rep antibodies. As shown in Fig. 1C, Rep-120 was expressed at levels similar to or higher than those of Rep-130. Unexpectedly, Rep-120 somehow migrated more slowly than Rep-130 (Fig. 1C). The same migration pattern was also seen when Rep-130 and Rep-120 were expressed in yeast as fusion products (Fig. 2B). The overall data suggest that transcription repression by TYLCSV Rep is confined to its 130 N-terminal amino acids.
FIG. 1.
Transcriptional repression activity of TYLCSV Rep mutants. (A) Scheme of TYLCSV Rep wild-type and mutant proteins. Solid bars indicate the conserved motifs characteristic of RCR proteins (I, II, and III); a gray bar indicates the ATP binding domain. Rep mutants are C-terminal deletions; the number of retained amino acids is indicated on the left. (B) GUS activity in N. benthamiana protoplasts cotransfected with pTOM202 together with the plasmid indicated on the left. GUS activity in protoplasts cotransfected with pTOM202 and pGEM-P was taken as 100%. Each value is the average of three cotransfections of three independent experiments. Error bars indicate the standard error of the mean. (C) Western blot of protein extracts of N. benthamiana protoplasts transfected with the plasmids indicated above each lane or mock transfected (M). Lane groups a and b represent two independent transfection experiments. Protein extracts from transgenic N. benthamiana protoplasts of line 301 expressing Rep-130 are on the right. The primary antibody was a rabbit polyclonal anti-TYLCSV Rep antibody.
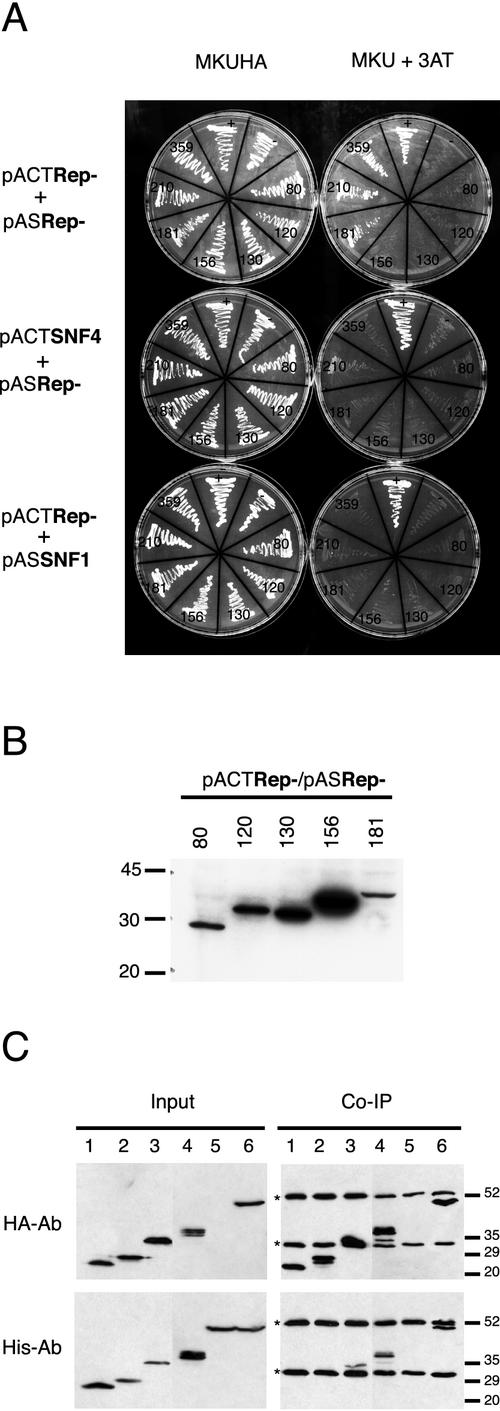
FIG. 2.
Rep-181 is the shortest self-interacting mutant. (A) Testing interaction in S. cerevisiae THS. Growth of strain PJ69-4A cotransformed with the plasmids indicated on the left, in medium selecting for the presence of plasmids (MKUHA [left]) or selecting for the interaction of expressed proteins (MKU + 3AT [right]), is shown. Symbols on plates: −, negative control strain coexpressing AgT and lamin; +, positive control strain coexpressing SNF1 and SNF4. Numbers indicate the number of N-terminal amino acids of each Rep protein. The TYLCSV wild-type Rep protein is 359 amino acids in length. (B) Western blot of protein extracts from S. cerevisiae coexpressing the GAL4 activation domain (AD)- and GAL4 binding domain (BD)-fused Rep proteins as indicated above each lane. The primary antibody was a monoclonal anti-GAL4 binding domain antibody. Migration of molecular mass markers is indicated on the left. (C) Testing interaction in vitro by coimmunoprecipitation. Western blots of total protein extracts from S. pombe coexpressing the HIS-tagged and HA-tagged Rep proteins are shown. Extracts were loaded on SDS-PAGE gel directly (Input) or after immunoprecipitation with anti-HA monoclonal antibodies (Co-Ip). The primary antibody (HA-Ab, monoclonal antibody against HA; His-Ab, monoclonal antibody against His) used in Western blotting is shown to the left. Lanes: 1, HIS-Rep130+HA-Rep130; 2, HIS-Rep156+HA-Rep156; 3, HIS-Rep181+HA-Rep181; 4, HIS-Rep210+HA-Rep210; 5, HIS-Rep; 6, HIS-Rep+HA-Rep. Molecular mass markers are indicated on the right. Asterisks mark the subunits of mouse IgG.
To determine the relationship between Rep-Rep interaction and repression, the ability of wild-type and mutated Rep proteins to self-interact in a yeast THS was investigated. The Rep proteins were expressed in both the GAL4 activation domain (pACT2) and the DNA binding domain (pAS2) plasmids (Table 1), and the interaction was evaluated in terms of the ability of the interacting protein to activate GAL2-ADE2 and LYS2::GAL1-HIS3 gene transcription and make the transformed yeast cells able to grow in a medium without adenine and histidine and containing 3-aminotriazole (Fig. 2A). As controls, both series of pACT2 and pAS2 Reps were tested to assess their interaction with an unrelated pair (SNF4 and SNF1) of interacting proteins (7). The results established that Rep-181 was the last deletion mutant able to self-interact. Rep-156, -130, -120, and -80 proteins did not self-interact, although they accumulated in the yeast cells at amounts similar to or higher than those of Rep-181 (Fig. 2B). In yeast protein extracts, Rep-120 migrated slightly more slowly than Rep-130, a pattern already observed in Western blot analysis of pJTR120- and pJTR130-transfected protoplasts (Fig. 1C).
To validate the results obtained with the THS, we carried out a coimmunoprecipitation assay. Rep, Rep-210, -181, -156, and -130 were coexpressed in S. pombe as HIS- or HA-tagged proteins. Protein extracts from yeast cells coexpressing the same truncated Reps fused to HIS or to HA epitope (Fig. 2C, input), were incubated overnight with monoclonal anti-HA antibody and then with protein A-Sepharose. After extensive washing, we used Western blotting to see whether the HA-tagged proteins (Fig. 2C, upper panel) coimmunoprecipitated the HIS-tagged proteins (Fig. 2C, bottom panel). HIS-Rep and the truncated forms HIS-Rep210 and HIS-Rep181 were coimmunoprecipitated (Fig. 2C, bottom panel, lanes 6, 4, and 3, respectively). In contrast, HIS-Rep156 and HIS-Rep130 were not detected (Fig. 2C, bottom panel, lanes 2 and 1). As a negative control, protein extracts from cells expressing only full-length HIS-Rep (Fig. 2C upper panel, lane 5) were incubated with anti-HA antibody and protein A-Sepharose. No protein was immunoprecipitated (Fig. 2C, bottom panel, lane 5), indicating that HIS-tagged Rep proteins need a HA-tagged Rep protein to immunoprecipitate.
These results confirm those obtained with the THS and indicate that the last interacting deletion mutant is Rep-181. Collectively, they indicate that TYLCSV Rep-Rep interaction is not required for transcription repression of the C1 promoter.
The oligomerization domain of TYLCSV Rep is not required in Rep-210-mediated inhibition of TYLCSV replication.
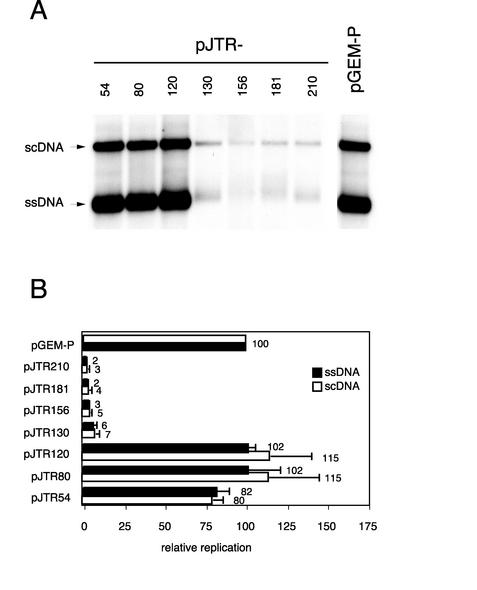
The finding that the oligomerization domain is not required for transcriptional repression of the TYLCSV C1 gene gave us the opportunity to analyze its contribution in TYLCSV Rep-210-mediated resistance. Inhibition of replication was tested by cotransfecting wild-type N. benthamiana protoplasts with an infectious clone of TYLCSV (pTOM6) together with one of the following constructs (Table 1): pJTR210, pJTR181, pJTR156, pJTR130, pJTR120, pJTR80, pJTR54, or pGEM-P employed in the repression assay. Transiently expressed Rep-210, -181, -156, and -130 inhibited TYLCSV replication similarly, whereas no impact was observed with Rep-120, -80, and -54 (Fig. 3). Therefore, the molecular mechanism by which truncated Rep-210 interferes with TYLCSV replication appears to be based on its ability to tightly repress the homologous C1 promoter.
FIG. 3.
Rep-210-mediated inhibition of TYLCSV replication is not dependent on the oligomerization domain. (A) Southern blot of TNA extracts from wild-type N. benthamiana protoplasts cotransfected with pTOM6 together with the plasmid expressing the Rep mutant indicated above each lane; pGEM-P was used as a control. The blot was hybridized with a digoxigenin-labeled Rep-210 sense RNA probe. scDNA, super coiled DNA. (B) Quantitative analysis of TYLCSV replication in protoplasts cotransfected with pTOM6 together with the plasmid expressing the Rep mutant indicated on the left. Blots were hybridized with a 32P-labeled DNA fragment corresponding to the Rep-54 coding sequence. Solid and open bars indicate the amount of ssDNA and scDNA respectively. The replication of TYLCSV in protoplasts cotransfected with pTOM6 and pGEM-P was taken as 100%. Each value is the average of two or three cotransfections of three independent experiments. Error bars indicate the standard error of the mean.
Rep-210-expressing plants are resistant to heterologous TYLCV.
We have previously shown that transgenic Rep-210-expressing N. benthamiana plants from line 102.22 are not resistant to TYLCSV-[ES1] due to the inability of Rep-210 to repress TYLCSV-[ES1] C1 transcription and viral replication (5). To further extend our analysis, we determined whether Rep-210 plants interfere with TYLCV, another geminivirus species similar to, but distinct from, TYLCSV. Line 102.22 plants were agroinoculated with TYLCSV or the Portuguese strain of TYLCV (TYLCV-[PT]), and infection was monitored weekly (Table 2). At 2 weeks p.i., only 19 and 25% of plants were infected by TYLCSV and TYLCV-[PT], respectively. Surprisingly, at 4 weeks p.i., the resistance was mostly overcome in the TYLCSV-inoculated plants only (Table 2). To evaluate if resistance to TYLCV-[PT] acts at the single-cell level, similarly to TYLCSV, transgenic protoplasts were transfected with the infectious clones of TYLCV-[PT] and TYLCSV (pPORT and pTOM6, respectively). Southern blot analysis of total nucleic acids (TNA) extracted from these protoplasts showed that the replication levels of both viruses were inhibited to a similar extent (Fig. 4A).
TABLE 2.
Analysis of resistance of N. benthamiana line 102.22 challenged with TYLCSV and TYLCV-[PT]
| Inoculum | Plant type | No. of wk p.i. | Infected/ inoculated plants
|
|
|---|---|---|---|---|
| No. | % | |||
| TYLCSV | Rep-210 expressing | 2 | 4/21 | 19 |
| 3 | 11/21 | 52 | ||
| 4 | 18/21 | 86 | ||
| Wild type | 2 | 6/6 | 100 | |
| TYLCV-[PT] | Rep-210 expressing | 2 | 4/16 | 25 |
| 3 | 4/16 | 25 | ||
| 4 | 5/16 | 31 | ||
| Wild type | 2 | 4/4 | 100 | |
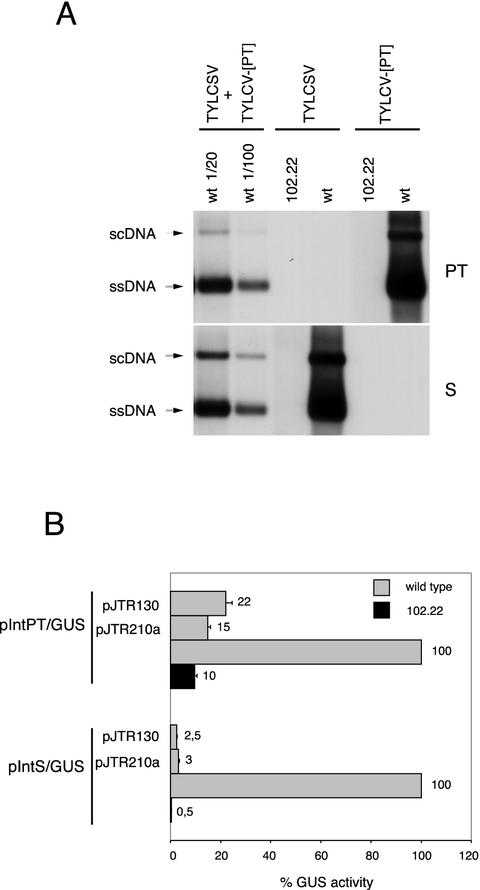
FIG. 4.
Rep-210-mediated resistance to TYLCV-[PT] and TYLCSV involves two different mechanisms.(A) Southern blot of TNA extracts from wild-type (wt) or transgenic Rep-210 expressing (line 102.22) N. benthamiana protoplasts transfected with infectious clones of TYLCSV or TYLCV-[PT], as indicated. The values 1/20 and 1/100 represent the dilution factors of loaded samples. The blot was probed with a digoxigenin-labeled TYLCSV Rep-210 sense RNA probe (S) and reprobed with a digoxigenin-labeled TYLCV-[PT] V1 antisense RNA probe (PT). (B) GUS activity in transgenic Rep-210 expressing N. benthamiana protoplasts (line 102.22 [solid bars]) transfected with the leftmost indicated plasmids or wild-type N. benthamiana protoplasts (gray bars) cotransfected with the leftmost indicated plasmids together with the plasmids indicated on the left. For each GUS-fused plasmid, the GUS activity in wild-type protoplasts was taken as 100%. Each value is the average of three transfections of three independent experiments. Error bars indicate the standard error of the mean.
We then looked to see whether inhibition of TYLCV-[PT] replication correlated with the ability of Rep-210 to tightly repress C1 gene transcription, as observed for TYLCSV. Line 102.22 and wild-type protoplasts were transfected with pIntPT/GUS and pIntS/GUS plasmids (Table 1), which contain the GUS gene fused to the C1 promoter of TYLCV-[PT] and TYLCSV, respectively. In transgenic protoplasts, the GUS activity of pIntS/GUS was repressed 200-fold compared to that of wild-type protoplasts, while pIntPT/GUS showed only a 10-fold reduction (Fig. 4B). These data suggest that in Rep-210-expressing protoplasts, an additional mechanism operates to tightly inhibit TYLCV-[PT] replication, besides transcriptional repression of the heterologous C1 promoter.
The Rep-210 oligomerization domain is required to interfere with TYLCV-[PT] replication.
As we have shown above, Rep-130 retained the ability to inhibit TYLCSV C1 gene transcription and replication in a transient assay, although it lost the ability to self-interact. We speculated that if the oligomerization domain plays a key role in the interference with TYLCV-[PT] replication, then Rep-130-expressing transgenic plants should show only a limited interference with the heterologous virus. To test this hypothesis, we asked whether Rep-130 was able to inhibit TYLCV-[PT] C1 gene transcription and whether Rep-210, but not Rep-130, was able to interact with the wild-type TYLCV-[PT] Rep.
For the C1 transcription assay, wild-type N. benthamiana protoplasts were cotransfected with pIntPT/GUS or pIntS/GUS together with pJTR130 or pJTR210a, which contains the expression cassette used in plant transformation (Table 1). Transiently expressed Rep-210 and Rep-130 repressed TYLCV-[PT] C1 transcription to a similar extent and about twofold less than transgenically expressed Rep-210 (Fig. 4B), indicating that Rep-130 and Rep-210 retained the same ability to repress the heterologous TYLCV-[PT] C1 promoter.
The THS was then used to test the ability of Rep-210 to interact with the homologous or heterologous full-length Rep proteins. Rep-210, but not Rep-130, was able to interact with both TYLCSV and TYLCV-[PT] Reps, indicating that Rep-130 can be used to investigate the role of the oligomerization domain in Rep-210-mediated resistance to TYLCV-[PT] (Fig. 5A).
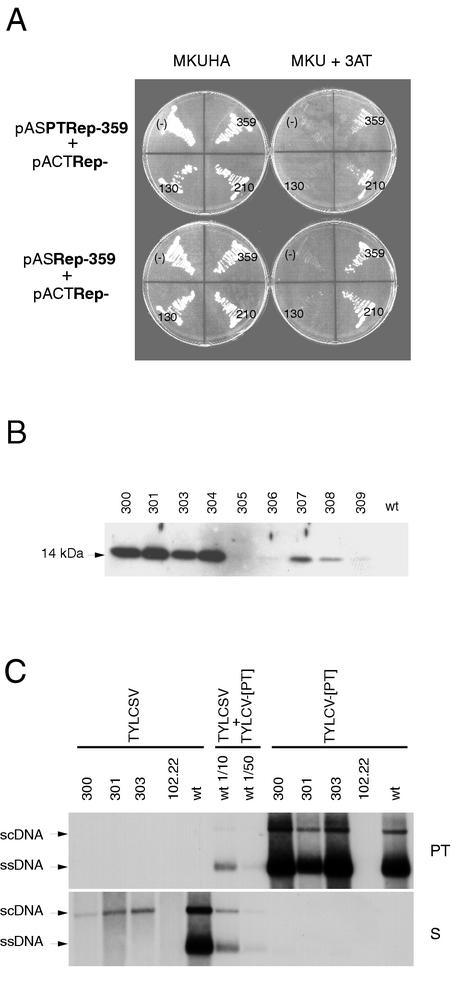
FIG. 5.
The oligomerization domain is required for the inhibition of TYLCV-[PT] replication. (A) Testing interaction in the S. cerevisiae THS. Growth of PJ69-4A strain cotransformed with plasmids indicated on the left in medium selecting for the presence of plasmids (MKUHA, left) or selecting for the interaction of expressed proteins (MKU + 3AT, right). On the plates, (−) indicates the control strain coexpressing SNF4 and either TYLCV-[PT] Rep or TYLCSV Rep, and the numbers indicate the number of N-terminal amino acids of each TYLCSV Rep protein. The TYLCSV wild-type Rep protein is 359 amino acids in length. (B) Western blot analysis of protein extracts from wild-type (wt) or transgenic (no. 300 to 309) N. benthamiana plants transformed with pTOM130. Primary antibody was a rabbit polyclonal anti-TYLCSV Rep antibody. (C) Southern blot analysis of TNA extracts from wild-type (wt) or transgenic N. benthamiana protoplasts expressing Rep-130 (lines 300, 301, and 303) or Rep-210 (line 102.22) transfected with infectious clones of TYLCSV or TYLCV-[PT], as indicated. The values 1/10 and 1/50 indicate the dilution factors of loaded samples. The blot was probed with a digoxigenin-labeled TYLCSV Rep-210 sense RNA probe (S) and reprobed with a digoxigenin-labeled TYLCV-[PT] V1 antisense RNA probe (PT).
Nine N. benthamiana plants, transformed with the binary plasmid pTOM130 harboring the Rep-130 coding sequence under the transcriptional control of the E35S promoter, were obtained. Fragments of the predicted size were amplified by PCR from all plants (data not shown). Rep-130 with an apparent molecular mass of 14 kDa was detected by Western blotting in eight plants (Fig. 5B). To evaluate the impact of the oligomerization domain on virus resistance, protoplasts isolated from plants 300, 301, and 303, expressing high levels of Rep-130, were transfected with TYLCSV or TYLCV-[PT] infectious clones. TYLCSV, but not TYLCV-[PT], replication was inhibited (Fig. 5C). Similar results were obtained with TYLCSV- and TYLCV-[PT] -transfected protoplasts derived from transgenic F1 plants of line 303 (data not shown). Overall, the data establish that the oligomerization domain plays a key role in the Rep-210 resistance to TYLCV-[PT], whereas it probably makes only a secondary contribution to the resistance to TYLCSV, as previously observed in transient expression assays (Fig. 3).
Virus infection in Rep-210-expressing plants correlates with the down-regulation of Rep-210.
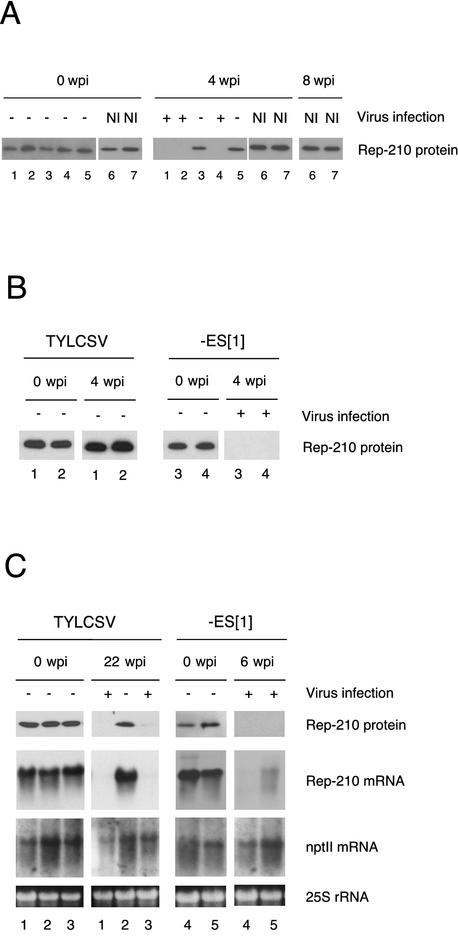
As shown in the time course experiment in Table 2, between 2 and 4 weeks p.i., 14 of the 17 initially TYLCSV-resistant plants became infected compared with only 1 of the 12 TYLCV-[PT]-resistant-plants. The difficulty of reconciling these findings with the tight control of TYLCSV replication observed at the single-cell level prompted us to find out how TYLCSV overcomes Rep-210-mediated resistance. The accumulation of Rep-210 at different times after inoculation was evaluated in transgenic N. benthamiana plants. All plants expressed Rep-210 at the time of inoculation, but not after the infection (Fig. 6A). Plants that were not challenged or not infected continued to express Rep-210, even when assayed up to 8 weeks p.i. These data suggest that the ability of TYLCSV to quickly overcome resistance may be the result of its ability to shut off Rep-210 transgene expression. The complex transgene organization of line 102.22, characterized by incomplete cosegregation of the nptII and the Rep-210 transgenes (41), made further control analysis difficult. To assess the value of this observation and extend its biological significance, therefore, we took advantage of genetically well-characterized TYLCSV-resistant BC1 tomato plants (4), which are heterozygous for the Rep-210 transgene and express large amounts of Rep-210. BC1 plants were challenged with TYLCSV and a related isolate, TYLCSV-[ES1], which overcomes resistance in 102.22 N. benthamiana plants (5). Fourteen of 15 BC1 plants challenged with TYLCSV-[ES1] were infected at 4 weeks p.i., just like the controls, indicating that tomato and N. benthamiana Rep-210-expressing plants show the same reaction to different isolates. Between 8 and 35 weeks p.i., 10 of the 25 BC1 plants challenged with TYLCSV became infected. Five TYLCSV- and 12 TYLCSV-[ES1]-infected plants were analyzed to determine the presence of Rep-210 (data not shown). In all of these plants, Rep-210 either was not detected or was present at very low levels, as revealed by Western blot analysis. Similarly, Rep-210 accumulation was no longer detectable in BC1 plants infected by a viruliferous B. tabaci challenge (Fig. 6B). Moreover, similar results were obtained with other Rep-210 transgenic tomato lines (data not shown). Thus, the shutoff of Rep-210 transgene expression is neither confined to the N. benthamiana host nor dependent on the inoculation procedure and the transgenic line utilized. To test the specificity of the down-regulation of Rep-210, several TYLCSV- and TYLCSV-[ES1]-infected plants were examined to determine the presence of Rep-210 mRNA and protein, as well as mRNA transcribed from nptII, another transgene linked to the Rep-210 transgene. In all virus-infected plants, Rep-210 mRNA and protein were almost undetectable, while the unrelated nptII transgene did not show any remarkable modification of its expression (Fig. 6C). Collectively, these data suggest that repression was specifically directed against the Rep-210 transgene and occurred through down-regulation of the specific transcripts.
FIG. 6.
TYLCV infection correlates with Rep-210 down-accumulation. (A) Expression of transgenic Rep-210 protein in line 102.22 N. benthamiana plants following TYLCSV challenge. Plants 1 to 5 were agroinoculated, and infection was assayed weekly by tissue printing with a TYLCSV-specific probe. Plants infected by the virus are indicated with +, while − denotes noninfected plants. NI indicates noninoculated control plants (lanes 6 and 7). Total protein extracts were prepared with samples collected before inoculation (0 weeks p.i. [wpi]) and at the fourth tissue print assay (4 weeks p.i.). The primary antibody used in Western blots was a rabbit anti-TYLCSV Rep polyclonal antibody. Rep-210 protein is indicated on the side. Plants 1 to 5 are representative cases from Table 2. (B) Accumulation of Rep-210 in BC1 tomato plants following challenge by viruliferous B. tabaci. Plants 1 and 2 were inoculated with TYLCSV, while plants 3 and 4 were inoculated with TYLCSV-[ES1]. Total protein extracts were prepared with samples collected before inoculation (0 weeks p.i.) and at the fourth tissue print assay (4 weeks p.i.). Filter was probed with anti-TYLCSV Rep antibody as in panel A. (C) Expression of transgenic proteins and transcripts in BC1 tomato plants following challenge by TYLCSV and TYLCSV-[ES1]. Agroinoculated plants were analyzed for virus infection weekly with tissue print assays as described above. Infected plants are indicated with +, while − denotes plants that were not infected at the indicated time. Total protein extracts were prepared with samples collected before (0 weeks p.i.) and after virus challenge. Filter was probed with anti-TYLCSV Rep antibody as in panel A. The same plants were analyzed in Northern blots with a C1-specific riboprobe and an nptII-specific DNA probe. Rep-210 and nptII mRNAs are indicated alongside. Ethidium bromide staining for the 25S rRNA is shown to demonstrate similar RNA loadings. Representative examples of uninfected and infected plants are shown. Plants 1 to 3 were inoculated with TYLCSV. Of these, plant 2 remained uninfected throughout the experiment (35 wpi), while plants 1 and 3 became infected at 22 weeks p.i. Plants 1 and 3 are representative of 10 out of 25 BC1 plants infected by TYLCSV between 8 and 35 weeks p.i. Plants 4 and 5 were challenged with TYLCSV-[ES1]. Both were already infected at 3 weeks p.i., as were the nontransgenic controls (not shown) and are representative examples of 12 other TYLCSV-[ES1]-infected plants analyzed similarly.
TYLCSV infection does not influence transgene expression driven by the E35S promoter, whereas it induces the production of Rep-specific 21- to 25-nt siRNAs.
To rule out the possibility that TYLCSV infection prevents the expression of transgenes driven by the E35S promoter, transgenic tomato plants transformed with the N gene of Tomato spotted wilt virus (TSWV) under the transcriptional control of the E35S promoter employed to express Rep-210 were challenged with TYLCSV. Similar amounts of N protein were detected by Western blotting before and after infection (Fig. 7A), showing that Rep-210-mediated repression did not act at the transcriptional level.
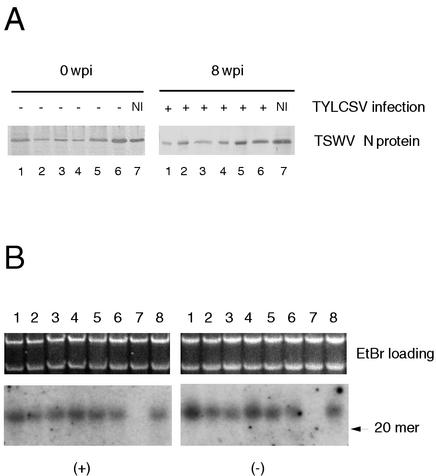
FIG. 7.
TYLCSV infection does not influence the expression of a heterologous transgene, although it induces Rep gene-specific siRNAs. (A) Western blot analysis of TYLCSV-challenged tomato plants expressing the N gene of TSWV. Virus infection and transgenic N protein expression were monitored for 8 weeks. Total protein extracts were prepared before (0 weeks p.i. [wpi]) and following (8 weeks p.i.) inoculation, and TSWV-N transgenic protein was detected with specific polyclonal antibodies. Plants 1 to 4 are from one transgenic line, and plants 5 to 7 are from another. NI indicates a noninoculated plant. All inoculated plants were infected by TYLCSV at 4 weeks p.i. (B) Northern blot of RNA extracts from tomato plants infected by TYLCSV either naturally under field conditions (lanes 1 to 6) or via agroinoculation under laboratory conditions (lane 8). Lane 7 contained RNA extracts from a healthy plant. Two identical filters were produced and probed with 32P-labeled transcripts corresponding to Rep-210 antisense RNA (+) or Rep-210 sense RNA (−). The upper panels show RNA loading. The migration of a 20-nt oligonucleotide is indicated on the right.
To study the mechanism of repression, we investigated the possibility that natural TYLCSV infection induces the production of 21- to 25-nt siRNAs homologous to the viral Rep gene. We analyzed RNA extracted from TYLCSV-infected wild-type tomato plants, derived either by agroinoculation (Fig. 7B, lane 8) or by natural vector transmission in the field (Fig. 7B, lanes 1 to 6), to look for the presence of Rep-specific siRNAs. Interestingly, a band migrating just above a 20-nt-long oligonucleotide DNA and hybridizing with both plus- and minus-strand Rep-210 probes was visible in Northern blots of total RNA extracted from both agroinoculated and naturally infected tomato plants (Fig. 7B). These data indicate that during natural virus infection, an RNA duplex homologous to the Rep gene is probably produced and specifically degraded by the host plant machinery to generate the siRNAs. In addition, they suggest that RNA sequences homologous to Rep mRNA can be the target of PTGS during viral infection.
DISCUSSION
The replication-associated viral proteins of small DNA viruses are involved in several biological processes due to the presence of distinct functional domains and different states of protein aggregation. The multifunctional character of these proteins is well suited to development of molecular interference strategies to impair viral infection. In this respect, we have previously shown that transgenic expression of the N-terminal 210 amino acids of the TYLCSV Rep, Rep-210, confers resistance to TYLCSV in both N. benthamiana and tomato plants (4, 41) and that Rep-210 acts as a transdominant-negative mutant repressing C1 gene transcription (5).
In the present work, TYLCSV-resistant Rep-210-expressing transgenic plants were challenged with the heterologous TYLCV-[PT] virus, and the mechanisms of resistance to both viruses were dissected by biochemical and transgenic approaches. Our data underline that a complex interaction between the challenging virus and the transgene exists. We show that Rep-210 confers resistance to the homologous and heterologous virus through two different strategies and that this resistance can be overcome by the ability of the challenging virus to silence the homologous transgene.
Rep-210 mediates homologous virus resistance by tightly repressing C1 gene transcription.
To further dissect the mechanism of resistance operating in TYLCSV-resistant transgenic plants, we asked whether the formation of dysfunctional Rep-210/Rep oligomers contributes to resistance and designed experiments to illustrate the relationship between oligomerization and transcription repression by determining the ability of a series of Rep-210 C-terminal deletion mutants to self-interact and repress the homologous C1 promoter. The transcriptional repression assay revealed that Rep-130 was the shortest deleted Rep protein that represses C1 transcription to a similar extent (about 100-fold) as the wild-type Rep protein and identified the C-terminal limit of the repression domain between residues 120 and 130. Interestingly, Rep-156 and Rep-130, which retained repression activities, were unable to self-interact in both THS and coimmunoprecipitation assays (Fig. 2A and C). This inability is in agreement with results obtained with TGMV and ToLCNDV, where the C-terminal limits of the oligomerization domain were mapped between residues 160 and 180 (8, 45). Our data suggest that TYLCSV Rep-Rep interaction is not needed to down-regulate C1 transcription, although the threefold-higher repression activity of Rep-210 compared to that of Rep-130 (Fig. 1B) could suggest a partial contribution of amino acids 130 to 210 to the full transcriptional repression activity. In this respect, our data differ from those obtained with TGMV Rep, whose in vitro ability to bind the DNA sequences required for transcriptional repression of the AL1 gene is dependent on amino acids 1 to 130 for protein-DNA contact and on the oligomerization domain, indicating that dimerization of Rep is a prerequisite for DNA binding in vitro (43). Conversely, the ability of Wheat dwarf virus (WDV) Rep to bind DNA requires the presence of monomer Rep molecules in solution (36), suggesting that at least in this case, Rep complexes are formed by the assembly of individual monomers, similarly to the large DNA protein complexes formed by the large T antigen of simian virus 40 (14). We do not know if repression of the C1 promoter requires binding of two molecules of Rep on DNA. However, the lack of Rep-Rep interaction does not have a large influence on repression. Notably, when the ability of TGMV Rep oligomerization mutants to repress the AL1 promoter was evaluated, most of them unexpectedly displayed an enhanced repression activity (44). A possible explanation to reconcile the TYLCSV and TGMV results is that in vivo the Rep-DNA complex is stabilized by other protein or DNA contacts. However, the difference observed could reflect a dissimilar ability of TYLCSV and TGMV Rep to contact their cognate DNAs and repress transcription, as substantiated by the finding that deletion of only 39 amino acids from the C-terminal portion of TGMV Rep abolishes repression (25). Moreover, a truncated Rep of African cassava mosaic virus (ACMV) containing only the first 57 amino acids retained full repression activity (28). Thus, TYLCSV, TGMV, and ACMV Reps appear to deeply differ in the amino acid sequences required to repress their own genes.
The ability of Rep-156 and Rep-130 to tightly inhibit C1 gene transcription in the absence of the oligomerization domain illustrated the contribution of Rep oligomerization to resistance. Interestingly, we found a perfect correlation between the repression results and replication interference assays, since all of the clones that tightly down-regulate C1 transcription also repressed TYLCSV replication (Fig. 1B, 3, and 5C). These findings, coupled with the inability of Rep-130 to interact in THS experiments with the wild-type Rep of the homologous virus (Fig. 5), strongly suggest that the formation of Rep-210/Rep dysfunctional complexes makes only a modest contribution, if any, to homologous resistance. It has recently been shown that residues 4 to 121 of TYLCSV Rep retain the ability to cut the viral DNA at the origin (6). Our results show that Rep-120 was unable to inhibit TYLCSV replication, suggesting that this Rep activity is not involved in resistance. Previous data showed that TYLCSV Rep cuts both homologous and heterologous (WDV) DNA origins, although the sequences constituting the stem of the loop are rather different in the two viruses (27). In our case, TYLCSV and TYLCV-[PT] share the same stem-loop sequences, but, as shown in the replication interference assay, Rep-130 represses TYLCSV replication only (Fig. 5C). Taken together, these data provide strong evidence that repression of TYLCSV C1 gene transcription is the master mechanism of resistance in Rep-210 plants.
Rep-210 oligomerization domain is absolutely required to confer resistance to TYLCV-[PT].
As shown above, the Rep-210 oligomerization domain does not make any appreciable contribution to TYLCSV resistance. Yet, even though Rep-210 transgenic plants are susceptible to TYLCSV-[ES1] (5), Rep-210 may interfere with the accumulation of heterologous viruses through its oligomerization domain, as recently suggested for a deletion mutant form of ToLCNDV Rep (Rep-160) (8). Interestingly, when Rep-210-expressing plants were challenged with either TYLCV-[PT] or TYLCSV and tested soon after infection (2 weeks p.i. [Table 2]), similar levels of resistance were observed. Moreover TYLCV-[PT] replication was highly inhibited in Rep-210-expressing transgenic protoplasts, showing that TYLCV-[PT] resistance works at the single-cell level, as for TYLCSV (Fig. 4A). However, although Rep-210 conferred similar levels of resistance to both viruses, it was 20-fold less active in repressing the heterologous TYLCV-[PT] C1 promoter (Fig. 4B), suggesting the existence of a second resistance mechanism. The10-fold reduction of heterologous TYLCV-[PT] C1 gene transcription by Rep-210 was unexpected, considering the differences in the iteron sequences between the two viruses (1). The ability of Rep-210 to interact with the TYLCV-[PT] Rep (Fig. 5A) suggests that resistance may be achieved through the formation of Rep-210/PTRep dysfunctional complexes. To formally prove the involvement of the oligomerization domain in TYLCV-[PT] resistance, we tested the ability of deletion mutant Rep proteins, lacking the oligomerization activity but active in repressing heterologous C1 transcription, to interfere with replication. Interestingly, while Rep-130 and Rep-210 inhibited TYLCV-[PT] C1 promoter to a similar extent (Fig. 4B), Rep-130 was unable to interfere with TYLCV-[PT] replication (Fig. 5C), suggesting that transcriptional repression does not make an appreciable contribution to the inhibition of TYLCV-[PT] replication. Like Rep-130, Rep-156, which lacks oligomerization activity (Fig. 2A and C), was unable to confer resistance to TYLCV-[PT] (G. P. Accotto and M. Tavazza, unpublished results). Collectively our data strongly suggest that Rep-210 oligomerization domain plays a key role in TYLCV-[PT] resistance. Recently it was shown that the deleted ToLCNDV Rep-160, containing both the DNA binding and the oligomerization domains, interferes with the homologous and heterologous viruses in a transient assay (8). Based on an in vitro DNA binding competition assay, the authors suggested that interference with heterologous virus could be the result of the oligomerization activity of the ToLCNDV Rep-160. Unfortunately, the ToLCNDV Rep DNA binding and oligomerization domains seem to overlap, preventing direct assessment of their respective contributions to the homologous and heterologous virus interference.
In our case, due to the biological characteristics of TYLCSV Rep, we were able to dissect the contribution of the transcriptional repression and oligomerization domains in homologous and heterologous virus resistance. Our results show that the Rep-210-mediated resistance to the heterologous virus TYLCV-[PT] relies on the interaction property of the oligomerization domain. To our knowledge, this is the first demonstration that the same protein, acting as a transdominant-negative mutant, confers resistance to distinct viruses through the employment of different molecular mechanisms.
Transgene expression shutoff correlates with the ability of TYLCSV to overcome Rep-210-mediated resistance.
We have shown that Rep-210 inhibits TYLCSV and TYLCV-[PT] replication to a similar extent (Fig. 4A). However TYLCSV, but not TYLCV-[PT], rapidly overcomes Rep-210-mediated resistance (Table 2). A drastic reduction of Rep-210 accumulation was constantly observed in TYLCSV-infected 102.22 plants (Fig. 6A). Conversely, plants that were challenged with TYLCSV or TYLCV-[PT], but remained virus free, expressed high levels of Rep-210 as well as unchallenged 102.22 plants during all their life cycle (Fig. 6A) (data not shown). Interestingly, the single TYLCV-[PT]-challenged plant that became infected between 2 and 4 weeks p.i. (Table 2) no longer accumulated Rep-210 (data not shown). The down-regulation of Rep-210 appears to be strictly linked to the ability of the viruses to overcome resistance, but independent of the actual resistance mechanism. This is consistent with our previous observations that high levels of Rep-210 are required to confer resistance (4) and that Rep-210 is no longer detectable in old (15 weeks p.i.) TYLCSV-infected transgenic plants (41).
If down-regulation of transgene expression is specifically directed to the Rep-210 gene, it should be demonstrated that expression of an unrelated gene present in the same chromosomal location, as well as that of a gene possessing the same regulatory sequences, is not affected by TYLCSV infection. To prove this, we took advantage of the tomato line BC1, which expresses Rep-210 (4). A number of observations showed that the two biological systems, i.e., plants of lines 102.22 (N. benthamiana) and BC1 (tomato), are in fact interchangeable: (i) TYLCSV resistance is protein mediated and active at the single-cell level (4, 5), (ii) some plants initially resistant to TYLCSV became infected later (4, 41), (iii) both are susceptible to TYLCSV-[ES1] (5), and (iv) late infections were always associated with down-regulation of Rep-210 protein and mRNA (Fig. 6) (data not shown). In addition, we showed that expression of the Rep-210-linked nptII gene was unaffected by TYLCSV infection (Fig. 6C). Similar results were observed when BC1 plants were challenged with TYLCSV-[ES1] (Fig. 6C), but they were susceptible to viral infection, and Rep-210 was silenced soon after infection. Furthermore, we used an unrelated transgenic line to show that TYLCSV infection itself is not able to down-regulate transgene expression driven by the E35S promoter (Fig. 7A). Collectively these data demonstrate that infection specifically down-regulates Rep-210 transgene expression and suggest that virus-mediated interference acts at a posttranscriptional level. The finding that TYLCSV was more efficient than TYLCV-[PT] in overcoming resistance and switching off Rep-210 transgene expression is in accordance with a virus-induced homology-dependent mechanism of gene silencing. The evidence that monopartite geminiviruses causing yellow leaf curl in tomato are mostly restricted to the vascular tissue (48) suggests that extensive Rep-210 silencing relies on a diffusible silencing signal.
It is well established that RNA and DNA viruses silence transgenes and endogenes (21, 33, 34, 47) and that diffusible signals are produced during the silencing process (46, 57). However VIGS of transgenes triggered by RNA viruses normally leads to systemic silencing, which in turn limits infection in the new silenced tissue (2). A possible explanation to reconcile our observation with previous findings is that during natural TYLCSV infection, the RNA encoding viral Rep is converted in or present as an RNA duplex that triggers homologous RNA silencing. As a consequence, the virus should have evolved a mechanism or mechanisms to escape Rep silencing, because Rep is absolutely required for its replication. The presence of 21- to 25-nt siRNAs homologous to a target gene is the hallmark of an activated RNA silencing process in all living organisms studied so far (17, 23). Interestingly, and in agreement with our assumption, siRNAs homologous to the Rep gene were detected in total RNA extracts of TYLCSV-infected wild-type tomato plants (Fig. 7B). The presence of Rep-specific siRNAs was also detected in TYLCSV-infected wild-type N. benthamiana and in tomato plants infected by TYLCSV-[ES1] and -Sic (A. Lucioli and M. Tavazza, unpublished results). These data strongly suggest that the production of siRNAs homologous to the Rep gene is a well-conserved step in TYLCSV infection.
We do not know whether and to what extent viral Rep mRNA is a target of RNA silencing, nor do we know the molecular mechanism that allows TYLCSV to evade silencing. Geminiviruses, unlike all other plant viruses studied in this regard, do not have a genomic RNA phase. Thus, although they trigger silencing, they may be inherently less susceptible to its effect than RNA viruses. Thus, the ability of TYLCSV to spread in silenced transgenic plants could reflect a balance between silencing, silencing suppression (56, 58), and virus replication. Natural infections, after all, are the result of a balance between these processes, and the balance points might differ for DNA and RNA viruses.
The ability of TYLCSV to overcome resistance thus seems deeply interconnected with its ability to down-regulate Rep-210 transgene expression via an RNA-mediated PTGS mechanism.
Conclusions.
Our studies of both homologous and heterologous viruses reveal complex interactions between the challenging virus and transgene. Based on previous (5) and present data, we propose a working model of Rep-210-mediated virus resistance.
After the virus enters the cell and is uncoated, its genome is converted into a duplex circular DNA. At this time, if the challenging virus is the homologous TYLCSV, Rep-210 can efficiently recognize and bind to its DNA, tightly inhibiting but not completely abolishing C1 transcription (Fig. 1B). The reduced amount of Rep will be in competition with Rep-210 for the viral sequence required for plus-strand viral replication. The combined action of Rep-210 reduces the replication rate (Fig. 3). The THS data (Fig. 5A) show that Rep-210 forms dysfunctional Rep-210/Rep complexes, though the impact of this mechanism on TYLCSV resistance is limited (Fig. 3 and 5C). The reduced but not abolished viral expression triggers, via an RNA-mediated mechanism (Fig. 7B), down-regulation of Rep-210 (Fig. 6), and this in turn releases the transcriptional control of the viral C1 gene and leads to productive viral infection.
When the challenging virus is TYLCSV-[ES1], Rep-210 is unable to repress the heterologous C1 promoter (5), resulting in the rapid production of TYLCSV-[ES1] Rep-specific siRNAs, which due to their extensive homology with the Rep-210 transgene (Fig. 8), leads to the down-regulation of Rep-210 and prevents the formation of dysfunctional Rep-210/Rep-[ES1] complexes.
FIG. 8.
Alignment of TYLCSV-[ES1] and TYLCV-[PT] sequences with TYLCSV Rep-210 transgene sequence. The box indicates the start codon of the Rep genes. Dotted and solid lines indicate fragments of TYLCV-[PT] and TYLCSV-[ES1], respectively, longer than 22 nt, which show 100% homology with the TYLCSV Rep-210 transgene sequence.
In TYLCV-[PT]-challenged plants, Rep-210 moderately represses the heterologous C1 promoter (Fig. 4B). However, transcriptional repression alone is not sufficient to substantially reduce replication (Fig. 5C). TYLCV-[PT] gene expression induces the accumulation of TYLCV-[PT] Rep-specific siRNAs. However, in this case, the low homology (Fig. 8) between the transgene and PT Rep gene (only one fragment with 100% homology is longer than 22 nt) does not activate or weakly activates transgene silencing. Therefore, Rep-210 that is expressed at a high level in the cell productively interacts with PT Rep (Fig. 5A) and efficiently interferes with replication (Fig. 4A).
In conclusion, the resistant or susceptible phenotype can be regarded as the result of a battle in which the plant uses a transgenic protein to repress viral transcription and to assemble dysfunctional oligomers, while the virus induces silencing of the transgene.
Acknowledgments
A. Lucioli, E. Noris, and A. Brunetti contributed equally to this work.
We thank A. Fammartino and E. Vecchiati for excellent technical assistance and D. Marian and P. Caciagli for whitefly transmission. We are also grateful to M. Davino for kindly providing samples of tomato plants naturally infected with TYLCSV.
A. Brunetti was the recipient of an Accademia Nazionale dei Lincei fellowship. This work was partially funded by the Programma Nazionale di Ricerca, Biotecnologie Avanzate II, and the Italian Ministry for Agriculture, special project Horticulture.
REFERENCES
- 1.Arguello-Astorga, G. R., and R. Ruiz-Medrano. 2001. An iteron-related domain is associated with motif 1 in the replication proteins of geminiviruses: identification of potential interacting amino acid-base pairs by a comparative approach. Arch. Virol. 146:1465-1485. [DOI] [PubMed] [Google Scholar]
- 2.Baulcombe, D. 1999. Viruses and gene silencing in plants. Arch. Virol. Suppl. 15:189-201. [DOI] [PubMed] [Google Scholar]
- 3.Beachy, R. N. 1997. Mechanisms and applications of pathogen-derived resistance in transgenic plants. Curr. Opin. Biotechnol. 8:215-220. [DOI] [PubMed] [Google Scholar]
- 4.Brunetti, A., M. Tavazza, E. Noris, R. Tavazza, P. Caciagli, G. Ancora, S. Crespi, and G. P. Accotto. 1997. High expression of truncated viral Rep protein confers resistance to Tomato Yellow Leaf Curl Virus in transgenic tomato plants. Mol. Plant-Microbe Interact. 10:571-579. [Google Scholar]
- 5.Brunetti, A., R. Tavazza, E. Noris, A. Lucioli, G. P. Accotto, and M. Tavazza. 2001. Transgenically expressed T-Rep of tomato yellow leaf curl Sardinia virus acts as a trans-dominant-negative mutant, inhibiting viral transcription and replication. J. Virol. 75:10573-10581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campos-Olivas, R., J. M. Louis, D. Clerot, B. Gronenborn, and A. M. Gronenborn. 2002. The structure of a replication initiator unites diverse aspects of nucleic acid metabolism. Proc. Natl. Acad. Sci. USA 99:10310-10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Celenza, J. L., F. J. Eng, and M. Carlson. 1989. Molecular analysis of the SNF4 gene of Saccharomyces cerevisiae: evidence for physical association of the SNF4 protein with the SNF1 protein kinase. Mol. Cell. Biol. 9:5045-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterji, A., R. N. Beachy, and C. M. Fauquet. 2001. Expression of the oligomerization domain of the replication-associated protein (Rep) of Tomato leaf curl New Delhi virus interferes with DNA accumulation of heterologous geminiviruses. J. Biol. Chem. 276:25631-25638. [DOI] [PubMed] [Google Scholar]
- 9.Chatterji, A., U. Chatterji, R. N. Beachy, and C. M. Fauquet. 2000. Sequence parameters that determine specificity of binding of the replication-associated protein to its cognate site in two strains of tomato leaf curl virus-New Delhi. Virology 273:341-350. [DOI] [PubMed] [Google Scholar]
- 10.Choi, I. R., and D. C. Stenger. 1995. Strain-specific determinants of beet curly top geminivirus DNA replication. Virology 206:904-912. [DOI] [PubMed] [Google Scholar]
- 11.Cox, K. H., D. V. DeLeon, L. M. Angerer, and R. C. Angerer. 1984. Detection of mRNAs in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev. Biol. 101:485-502. [DOI] [PubMed] [Google Scholar]
- 12.Craven, R. A., D. J. Griffiths, K. S. Sheldrick, R. E. Randall, I. M. Hagan, and A. M. Carr. 1998. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene 221:59-68. [DOI] [PubMed] [Google Scholar]
- 13.Dalmay, T., A. Hamilton, E. Mueller, and D. C. Baulcombe. 2000. Potato virus X amplicons in arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell 12:369-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean, F. B., J. A. Borowiec, T. Eki, and J. Hurwitz. 1992. The simian virus 40 T antigen double hexamer assembles around the DNA at the replication origin. J. Biol. Chem. 267:14129-14137. [PubMed] [Google Scholar]
- 15.Desbiez, C., C. David, A. Mettouchi, J. Laufs, and B. Gronenborn. 1995. Rep protein of tomato yellow leaf curl geminivirus has an ATPase activity required for viral DNA replication. Proc. Natl. Acad. Sci. USA 92:5640-5644. (Erratum, 92:11322.) [DOI] [PMC free article] [PubMed]
- 16.Eagle, P. A., B. M. Orozco, and L. Hanley-Bowdoin. 1994. A DNA sequence required for geminivirus replication also mediates transcriptional regulation. Plant Cell 6:1157-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elmer, J. S., L. Brand, G. Sunter, W. E. Gardiner, D. M. Bisaro, and S. G. Rogers. 1988. Genetic analysis of the tomato golden mosaic virus. II. The product of the AL1 coding sequence is required for replication. Nucleic Acids Res. 16:7043-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Etessami, P., K. Saunders, J. Watts, and J. Stanley. 1991. Mutational analysis of complementary-sense genes of African cassava mosaic virus DNA A. J. Gen. Virol. 72:1005-1012. [DOI] [PubMed] [Google Scholar]
- 20.Gladfelter, H. J., P. A. Eagle, E. P. Fontes, L. Batts, and L. Hanley-Bowdoin. 1997. Two domains of the AL1 protein mediate geminivirus origin recognition. Virology 239:186-197. [DOI] [PubMed] [Google Scholar]
- 21.Guo, H. S., and J. A. Garcia. 1997. Delayed resistance to plum pox potyvirus mediated by a mutated RNA replicase gene: involvement of a gene-silencing mechanism. Mol. Plant-Microbe Interact. 10:160-170. [Google Scholar]
- 22.Gutierrez, C. 2000. Geminiviruses and the plant cell cycle. Plant Mol. Biol. 43:763-772. [DOI] [PubMed] [Google Scholar]
- 23.Hamilton, A. J., and D. C. Baulcombe. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950-952. [DOI] [PubMed] [Google Scholar]
- 24.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 25.Hanley-Bowdoin, L., S. B. Settlage, B. M. Orozco, S. Nagar, and D. Robertson. 1999. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 18:71-106. [PubMed] [Google Scholar]
- 26.Hanson, S. F., R. A. Hoogstraten, P. Ahlquist, R. L. Gilbertson, D. R. Russell, and D. P. Maxwell. 1995. Mutational analysis of a putative NTP-binding domain in the replication-associated protein (AC1) of bean golden mosaic geminivirus. Virology 211:1-9. [DOI] [PubMed] [Google Scholar]
- 27.Heyraud-Nitschke, F., S. Schumacher, J. Laufs, S. Schaefer, J. Schell, and B. Gronenborn. 1995. Determination of the origin cleavage and joining domain of geminivirus Rep proteins. Nucleic Acids Res. 23:910-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong, Y., and J. Stanley. 1995. Regulation of African cassava mosaic virus complementary-sense gene expression by N-terminal sequences of the replication-associated protein AC1. J. Gen. Virol. 76:2415-2422. [DOI] [PubMed] [Google Scholar]
- 29.Ilyina, T. V., and E. V. Koonin. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 20:3279-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast1425-36. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jupin, I., F. Hericourt, B. Benz, and B. Gronenborn. 1995. DNA replication specificity of TYLCV geminivirus is mediated by the amino-terminal 116 amino acids of the Rep protein. FEBS Lett. 362:116-120. [DOI] [PubMed] [Google Scholar]
- 32.Kheyr-Pour, A., M. Bendahmane, V. Matzeit, G. P. Accotto, S. Crespi, and B. Gronenborn. 1991. Tomato yellow leaf curl virus from Sardinia is a whitefly-transmitted monopartite geminivirus. Nucleic Acids Res. 19:6763-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kjemtrup, S., K. S. Sampson, C. G. Peele, L. V. Nguyen, M. A. Conkling, W. F. Thompson, and D. Robertson. 1998. Gene silencing from plant DNA carried by a geminivirus. Plant J. 14:91-100. [DOI] [PubMed] [Google Scholar]
- 34.Kumagai, M. H., J. Donson, G. della-Cioppa, D. Harvey, K. Hanley, and L. K. Grill. 1995. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. USA 92:1679-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laufs, J., W. Traut, F. Heyraud, V. Matzeit, S. G. Rogers, J. Schell, and B. Gronenborn. 1995. In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc. Natl. Acad. Sci. USA 92:3879-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Missich, R., E. Ramirez-Parra, and C. Gutierrez. 2000. Relationship of oligomerization to DNA binding of wheat dwarf virus RepA and Rep proteins. Virology 273:178-188. [DOI] [PubMed] [Google Scholar]
- 37.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 38.Moriones, E., and J. Navas-Castillo. 2000. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 71:123-134. [DOI] [PubMed] [Google Scholar]
- 39.Navas-Castillo, J., S. Sanchez-Campos, E. Noris, D. Louro, G. P. Accotto, and E. Moriones. 2000. Natural recombination between Tomato yellow leaf curl virus-is and Tomato leaf curl virus. J. Gen. Virol. 81:2797-2801. [DOI] [PubMed] [Google Scholar]
- 40.Navot, N., E. Pichersky, M. Zeidan, D. Zamir, and H. Czosnek. 1991. Tomato yellow leaf curl virus: a whitefly-transmitted geminivirus with a single genomic component. Virology 185:151-161. [DOI] [PubMed] [Google Scholar]
- 41.Noris, E., G. P. Accotto, R. Tavazza, A. Brunetti, S. Crespi, and M. Tavazza. 1996. Resistance to tomato yellow leaf curl geminivirus in Nicotiana benthamiana plants transformed with a truncated viral C1 gene. Virology 224:130-138. (Erratum, 227:519, 1997.) [DOI] [PubMed]
- 42.Noris, E., E. Hidalgo, G. P. Accotto, and E. Moriones. 1994. High similarity among the tomato yellow leaf curl virus isolates from the west Mediterranean basin: the nucleotide sequence of an infectious clone from Spain. Arch. Virol. 135:165-170. [DOI] [PubMed] [Google Scholar]
- 43.Orozco, B. M., and L. Hanley-Bowdoin. 1998. Conserved sequence and structural motifs contribute to the DNA binding and cleavage activities of a geminivirus replication protein. J. Biol. Chem. 273:24448-24456. [DOI] [PubMed] [Google Scholar]
- 44.Orozco, B. M., L. J. Kong, L. A. Batts, S. Elledge, and L. Hanley-Bowdoin. 2000. The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J. Biol. Chem. 275:6114-6122. [DOI] [PubMed] [Google Scholar]
- 45.Orozco, B. M., A. B. Miller, S. B. Settlage, and L. Hanley-Bowdoin. 1997. Functional domains of a geminivirus replication protein. J. Biol. Chem. 272:9840-9846. [DOI] [PubMed] [Google Scholar]
- 46.Palauqui, J. C., T. Elmayan, J. M. Pollien, and H. Vaucheret. 1997. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16:4738-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peele, C., C. V. Jordan, N. Muangsan, M. Turnage, E. Egelkrout, P. Eagle, L. Hanley-Bowdoin, and D. Robertson. 2001. Silencing of a meristematic gene using geminivirus-derived vectors. Plant J. 27:357-366. [DOI] [PubMed] [Google Scholar]
- 48.Rojas, M. R., H. Jiang, R. Salati, B. Xoconostle-Cazares, M. R. Sudarshana, W. J. Lucas, and R. L. Gilbertson. 2001. Functional analysis of proteins involved in movement of the monopartite begomovirus, Tomato yellow leaf curl virus. Virology 291:110-125. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Saunders, K., A. Lucy, and J. Stanley. 1991. DNA forms of the geminivirus African cassava mosaic virus consistent with a rolling circle mechanism of replication. Nucleic Acids Res. 19:2325-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanley, J. 1995. Analysis of African cassava mosaic virus recombinants suggests strand nicking occurs within the conserved nonanucleotide motif during the initiation of rolling circle DNA replication. Virology 206:707-712. [DOI] [PubMed] [Google Scholar]
- 52.Stenger, D. C., G. N. Revington, M. C. Stevenson, and D. M. Bisaro. 1991. Replicational release of geminivirus genomes from tandemly repeated copies: evidence for rolling-circle replication of a plant viral DNA. Proc. Natl. Acad. Sci. USA 88:8029-8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sunter, G., M. D. Hartitz, and D. M. Bisaro. 1993. Tomato golden mosaic virus leftward gene expression: autoregulation of geminivirus replication protein. Virology 195:275-280. [DOI] [PubMed] [Google Scholar]
- 54.Szittya, G., A. Molnar, D. Silhavy, C. Hornyik, and J. Burgyan. 2002. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 14:359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaira, A. M., M. Vecchiati, V. Masenga, and G. P. Accotto. 1996. A polyclonal antiserum against a recombinant viral protein combines specificity with versatility. J. Virol. Methods 56:209-219. [DOI] [PubMed] [Google Scholar]
- 56.van Wezel, R., X. Dong, H. Liu, P. Tien, J. Stanley, and Y. Hong. 2002. Mutation of three cysteine residues in tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and posttranscriptional gene-silencing suppression. Mol. Plant-Microbe Interact. 15:203-208. [DOI] [PubMed] [Google Scholar]
- 57.Voinnet, O., and D. C. Baulcombe. 1997. Systemic signalling in gene silencing. Nature 389:553.. [DOI] [PubMed] [Google Scholar]
- 58.Voinnet, O., Y. M. Pinto, and D. C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]