Abstract
Epstein-Barr virus (EBV) episomal genomes are stably maintained in human cells and are partitioned during cell division by mitotic chromosome attachment. Partitioning is mediated by the viral EBNA1 protein, which binds both the EBV segregation element (FR) and a mitotic chromosomal component. We previously showed that the segregation of EBV-based plasmids can be reconstituted in Saccharomyces cerevisiae and is absolutely dependent on EBNA1, the EBV FR sequence, and the human EBNA1-binding protein 2 (EBP2). We have now used this yeast system to elucidate the functional contribution of human EBP2 to EBNA1-mediated plasmid partitioning. Human EBP2 was found to attach to yeast mitotic chromosomes in a cell cycle-dependent manner and cause EBNA1 to associate with the mitotic chromosomes. The domain of human EBP2 that binds both yeast and human chromosomes was mapped and shown to be functionally distinct from the EBNA1-binding domain. The functionality and localization of human EBP2 mutants and fusion proteins indicated that the attachment of EBNA1 to mitotic chromosomes is crucial for EBV plasmid segregation in S. cerevisiae, as it is in humans, and that this is the contribution of human EBP2. The results also indicate that plasmid segregation in S. cerevisiae can occur through chromosome attachment.
Most of the human population worldwide is infected with Epstein-Barr virus (EBV), a gamma herpesvirus which persists in B lymphocytes for the life of the host. Due to the ability of EBV to efficiently immortalize cells as part of its latent infection, EBV infections predispose the host to the development of a variety of cancers. EBV genomes are maintained at a constant copy number in the nucleus of latently infected cells as double-stranded circular DNA episomes (reviewed in references 9 and 20). The stable persistence of the episomes in dividing cells results from replication of the episomes once per cell cycle and partitioning of the episomes to daughter cells during mitotic cell division (1, 32). The replication and partitioning of the episomes require the latent origin of replication, oriP, and the viral Epstein-Barr nuclear antigen 1 (EBNA1) protein (49).
oriP consists of two functional elements, the dyad symmetry (DS) and the family of repeats (FR), containing four and 20 EBNA1-binding sites, respectively (37, 39). Replication from oriP initiates within the DS element and requires EBNA1 binding to the DS (10). The FR element governs the stable partitioning of EBV episomes and oriP-containing constructs and also facilitates the transcription of several viral latency genes; both of these processes require EBNA1 binding to the FR element and can be carried out in the absence of the DS (26, 38). The replication, segregation, and transcriptional activation functions of EBNA1 require the DNA-binding and dimerization domain located in the EBNA1 C terminus, between amino acids 459 and 607 (3, 6). In addition to this domain, the replication function of EBNA1 requires the N-terminal half of the protein, the segregation function requires a Gly-Arg rich region between amino acids 325 and 376, and the transcriptional activation function requires both residues 325 to 376 and a second region between amino acids 61 and 83 (7, 24, 40, 47).
Several observations indicate that EBV episomes segregate by attaching to host mitotic chromosomes. Specifically, the episomes are tethered to the chromosomes through EBNA1, which binds both the EBV FR element and the condensed mitotic chromosomes, thus enabling the episomes to segregate along with the host genome. This model was first suggested by the observations that EBNA1, EBV, and oriP-containing constructs associate with mitotic chromosomes (12, 13, 36, 41). Like EBV segregation, association of EBV plasmids with metaphase chromosomes requires both FR and EBNA1, suggesting that segregation and mitotic chromosome association activities are coupled (18). EBNA1 mutational studies also indicate that mitotic chromosome attachment is important for plasmid segregation. An EBNA1 mutant that lacks amino acids 325 to 376 is functional for replication but defective in oriP-plasmid partitioning and binding to mitotic chromosomes (40, 46). Another EBNA1 mutant lacking N-terminal residues 8 to 67 exhibits a partial defect in segregation and associates with mitotic chromosomes less tightly than wild-type EBNA1 (47). Finally, fusion proteins containing the DNA-binding and dimerization domain of EBNA1 fused to histone H1 or high-mobility group I have been shown to bind mitotic chromosomes, recruit oriP plasmids to the chromosomes, and maintain these plasmids under long-term selection (16).
Insights into the mechanism by which EBNA1 attaches to chromosomes in mitosis came from the identification of human EBNA1-binding protein 2 (EBP2), a cellular protein that was found to interact with both the FR-bound and unbound forms of EBNA1 (40). Human EBP2 is highly conserved in eukaryotes and is localized to the nucleolus in interphase (8, 40). The conserved cellular function of human EBP2 appears to be in ribosome biogenesis, as the Saccharomyces cerevisiae homolog of EBP2 (yeast EBP2) plays an essential role in rRNA processing (15, 44).
Several pieces of evidence point to a role for human EBP2 in EBNA1-mediated segregation. First, human EBP2 associates with the condensed mitotic chromosomes with a distribution that is much like that of EBNA1 (46). Second, EBNA1 mutants with decreased ability to bind human EBP2 exhibit defects in mitotic chromosome attachment and oriP plasmid segregation (40, 46, 47). Third, a role for human EBP2 in EBNA1-mediated plasmid segregation was directly demonstrated in a reconstituted system in S. cerevisiae (19). In this system, an unstable S. cerevisiae replicating plasmid containing an S. cerevisiae origin of replication (ARS) and the EBV FR element was stably maintained only when both EBNA1 and human EBP2 were present. As in human cells, the partitioning of these plasmids by EBNA1 required the FR element and the EBNA1 region that binds human EBP2 (amino acids 325 to 376) and was moderately decreased by deletion of EBNA1 amino acids 8 to 67. In addition, the EBNA1-binding domain of human EBP2 (amino acids 220 to 306) was required for human EBP2 to support the partitioning function of EBNA1 in the S. cerevisiae system.
To further investigate the mechanism of EBNA1-mediated segregation and the validity of the reconstituted S. cerevisiae system as a model for EBV segregation in human cells, we have used the EBV-based plasmid partitioning system in S. cerevisiae to examine the functional contributions of human EBP2. Here we show that human EBP2 attaches to yeast mitotic chromosomes in a cell cycle-dependent manner, as in human cells, and causes EBNA1 to attach to the chromosomes. We also identified the chromosome attachment domain of human EBP2 and present evidence that the tethering of EBNA1 to mitotic chromosomes is crucial for EBNA1-mediated plasmid partitioning in S. cerevisiae, as it is in human cells.
MATERIALS AND METHODS
Plasmids for S. cerevisiae assays.
The construction of the YRp7FR segregation test plasmid, the human EBP2 expression plasmid pR425/PGK.hEBP2 and the EBNA1 expression plasmid p416MET.EBNA1 have been described previously (19). Plasmids that express yeast EBP2 residues 95 to 306 or human EBP2 residues 220 to 306 were generated by PCR amplification of the appropriate EBP2 sequences and insertion into the BglII site of pR425/PGK. To construct plasmids expressing human EBP2, human EBP2 fragments or yeast EBP2 fragments fused at the C terminus to the EBNA1 nuclear localization signal (NLS) and DNA-binding region (termed 21K), oligonucleotides coding for the EBNA1 NLS (residues 376 to 386) were annealed and inserted into the filled-in BglII site of pR425/PGK. The EBNA1 21K fragment (residues 452 to 641) was then inserted into the StuI site of this plasmid to yield p425PGK.21KNLS. Human EBP2, human EBP2 residues 1 to 100, 1 to 220, 95 to 220, 95 to 306, and 220 to 306, and yeast EBP2 residues 232 to 358 were PCR amplified and inserted into the BglII site of p425PGK.21KNLS upstream of the NLS.
To construct plasmids that express yeast EBP2 or yeast EBP2 amino acids 1 to 347, 179 to 347, or 179 to 427 fused to the N terminus of human EBP2 residues 220 to 306 (F2, F1, F3, and F4, respectively), human EBP2 residues 220 to 306 were PCR amplified and inserted into the filled-in BglII site of pR425/PGK. The resulting plasmid was linearized at the BglII site introduced upstream of human EBP2 residues 220 to 306 during PCR and ligated to PCR-amplified yeast EBP2 or yeast EBP2 fragment 1 to 347, 179 to 347, or 179 to 427.
Two-hybrid assay plasmids pACT63 and pACT.yEBP2 express human EBP2 amino acids 21 to 306 and full-length yeast EBP2, respectively, as fusions to the GAL4 DNA-binding domain. pAS2.EBNA1 expresses EBNA1 as a fusion to the GAL4 activation domain. Construction of these plasmids has been described previously (19, 40). To generate plasmids that express hybrid proteins F1, F2, F3, and F4 fused to the GAL4 activation domain, the appropriate fusion protein was PCR amplified from the pR425/PGK plasmids and inserted into the SmaI site of pACTII (29).
Immunofluorescence on whole S. cerevisiae cells.
S. cerevisiae strain KY320 was transformed with a pR425/PGK plasmid expressing human EBP2, yeast EBP2, or a yeast EBP2 hybrid protein (F1 to F5) or with p416MET.EBNA1. Transformants and KY320 cells not transformed with a plasmid (used to determine localization of endogenous yeast EBP2) were grown until early log phase, fixed in YPD containing 3.7% formaldehyde for 1 h, spheroplasted, and spotted onto poly-lysine-treated coverslips. The coverslips were stained with mouse antibodies against NOP1 (provided by J. Aris) and rabbit antibodies against human EBP2 (45). Yeast EBP2 or EBNA1 (K67 serum, provided by Jaap Middeldorp), followed by Texas Red-conjugated goat anti-mouse and fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin antibodies. The cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) (1 μg/ml). The coverslips were mounted onto slides and observed at 1,000× magnification with a Leica DMR microscope, and immunofluorescence images were photographed and processed with the OpenLab software.
Immunofluorescence on S. cerevisiae DNA spreads.
S. cerevisiae KY320 expressing the protein of interest was obtained by transformation with the appropriate pR425/PGK-based and/or p416MET.EBNA1 construct. Transformed and nontransformed cells were grown until early log phase and then blocked for 3 h in YPD containing 5 μM α-factor (G1 block) or 15 μg of nocodazole per ml (mitotic block). Blocked cells were spheroplasted, and the DNA from the cells was spread on slides and prepared for immunofluorescence microscopy as described previously (35). DNA spreads of cells expressing human EBP2 or yeast EBP2 (endogenous and from a plasmid, respectively) were stained with anti-NOP1 mouse and anti-EBP2 rabbit antibodies; spreads from cells expressing EBNA1 or human EBP2 and EBNA1 were stained with anti-human EBP2 rabbit and anti-EBNA1 mouse (OT1x; supplied by J. Middledorp) antibodies; spreads from cells expressing the 21K fusion proteins were stained with anti-EBNA1 K67 rabbit serum; and spreads from cells expressing the F1 to F4 fusion proteins were stained with anti-human EBP2 rabbit antibodies. The DNA was counterstained with DAPI, and immunofluorescence microscopy was conducted on the spreads as for whole yeast cells.
Plasmid loss assays.
S. cerevisiae strain KY320 was transformed with the YRp7FR segregation plasmid, with a pR425/PGK vector that did or did not express EBP2, or with an EBP2-based fusion protein and, when required, with p416MET.EBNA1. Transformants were used in plasmid loss assays and plated to determine plasmid stability as described previously (19).
S. cerevisiae two-hybrid assays.
S. cerevisiae strain Y190, which contains integrated HIS3 and lacZ reporter genes under the control of the GAL4 binding sites, was transformed with pAS2.EBNA1 and pACTII expressing human EBP2, yeast EBP2, or a yeast EBP2-based fusion protein (F1 to F4). Two-hybrid assays were conducted with the transformants as described previously (19).
GFP fluorescence.
Plasmids expressing enhanced green fluorescent protein (EGFP) fused at the C terminus to human EBP2 or human EBP2 residues 1 to 100, 1 to 220, 95 to 220, 95 to 306, and 220 to 306 were constructed by PCR amplification of the appropriate human EBP2 fragments and insertion into the XmaI site of pEGFP-C1 (Clonetech). HeLa cells were transfected with these plasmids or with pEGFP-C1 alone (negative control) and grown under selection for the plasmid. To obtain metaphase spreads, the cells were blocked in mitosis by incubation with 0.1 μg of colcemid per ml for 16 h, swollen in hypotonic buffer, fixed with methanol-acetic acid (70:30), and dropped onto cooled slides. Slides were washed with 70% acetic acid and phosphate-buffered saline, stained with DAPI (25 ng/μl), and observed for green fluorescent protein (GFP) fluorescence at 630× magnification as described for S. cerevisiae.
RESULTS
Human EBP2 tethers EBNA1 to chromosomes in S. cerevisiae.
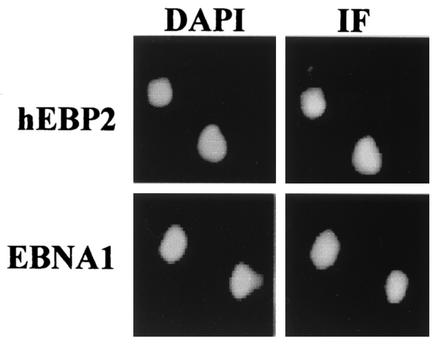
We have previously shown that EBNA1 can efficiently partition plasmids containing the EBV segregation element in S. cerevisiae but can only do so in the presence of human EBP2 (19). To understand the essential function of human EBP2 in EBNA1-mediated segregation, we first examined the localization of EBNA1 and human EBP2 in S. cerevisiae. When log-phase yeast cells expressing either human EBP2 or EBNA1 were stained with antibodies specific for human EBP2 or EBNA1, respectively, both proteins were found to be nuclear in all yeast cells, giving a staining pattern similar to that of DAPI (Fig. 1). The same results were obtained when the localization of human EBP2 and EBNA1 was examined in yeast cells blocked in G1 or mitosis with α-factor or nocodazole, respectively, and in yeast cells coexpressing human EBP2 and EBNA1 (data not shown). The nuclear staining pattern of EBNA1 in S. cerevisiae is much like that observed in human cells (47), whereas the nuclear staining pattern of human EBP2 in S. cerevisiae differs from that in interphase human cells, where human EBP2 is localized only to the nucleolus (8).
FIG. 1.
Nuclear localization of human EBP2 and EBNA1 in S. cerevisiae. Log phase S. cerevisiae expressing either human EBP2 (hEBP2) or EBNA1 were stained with antibodies against human EBP2 or EBNA1 (right column) and counterstained with DAPI (left column). Immunofluorescence (IF) microscopy images were captured under identical conditions.
Studies in human cells have shown that, during mitosis, both EBNA1 and human EBP2 bind all over the condensed chromosomes, giving very similar staining patterns (46). The attachment of human EBP2 to mitotic chromosomes occurs independently of EBNA1, suggesting that EBNA1 might attach to mitotic chromosomes by binding human EBP2. However, the possible requirement of human EBP2 for EBNA1 binding to mitotic chromosomes could not be tested in human cells because human EBP2 is expressed in all proliferating human cells and appears to be essential for cell viability (8, 15). The similar requirements for EBNA1-mediated plasmid partitioning in S. cerevisiae and human cells suggest that EBNA1-mediated segregation in the yeast system occurs through attachment to mitotic chromosomes, as it does in human cells. To test this possibility, we first examined whether human EBP2 and EBNA1 associate with yeast chromosomes in mitosis, using the chromatin spreading technique that has been used to assess other protein interactions with yeast chromosomes (25, 27, 34, 35, 43). In this technique, S. cerevisiae cells are lysed, fixed, and spread on slides so that the DNA and associated proteins remain bound to the slide while other proteins are washed away. As controls, we initially stained the chromatin spreads for the yeast cohesin subunit Mcd1 as well as for the nuclear export protein Cse1, and, as previously reported by others (35, 43), we observed that Mcd1 bound to the yeast chromosomes, while Cse1 did not (data not shown).
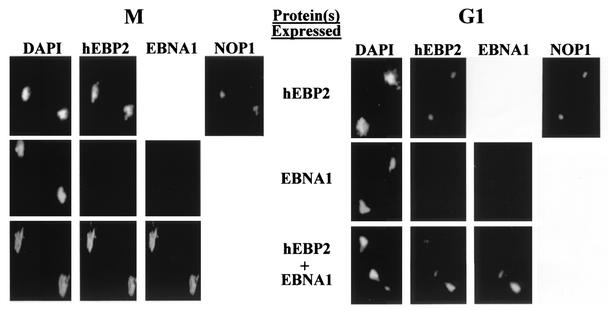
To examine the possible association of human EBP2 and EBNA1 with yeast mitotic chromosomes, S. cerevisiae cells expressing human EBP2 and/or EBNA1 were blocked in mitosis with nocodazole, and then chromatin spreads were prepared and stained with the respective antibodies. Immunofluorescence microscopy clearly showed that, when expressed individually, human EBP2 associated with the yeast chromosomes, whereas EBNA1 did not (Fig. 2, M panels). Human EBP2 localized to chromosomal regions stained by DAPI and to the nucleolar region, which does not stain well with DAPI but is indicated by staining for the Nop1 nucleolar marker. To ensure the specificity of the human EBP2 antibody, we also used this antibody to stain mitotic chromosomes from S. cerevisiae cells that were expressing EBNA1 but not expressing human EBP2; as expected, staining of these chromosomes was not observed (Fig. 2, panel M, middle row). When mitotic chromatin spreads were prepared from S. cerevisiae cells expressing both EBNA1 and human EBP2 and stained for both proteins, both human EBP2 and EBNA1 were observed all over the mitotic chromosomes, with identical staining patterns (Fig. 2, panel M, bottom row). The results indicate that human EBP2 is required for EBNA1 to attach to yeast mitotic chromosomes.
FIG. 2.
Human EBP2 causes EBNA1 to attach to yeast chromosomes. Chromatin from G1 or mitotic yeast cells expressing human EBP2 (hEBP2), EBNA1, or human EBP2 and EBNA1 were spread on slides, washed to remove proteins not bound to DNA, and stained with antibodies against human EBP2 and either EBNA1 or NOP1. DNA was visualized by DAPI staining. Each image contains chromatin from two yeast cells. Immunofluorescence microscopy was performed under identical conditions.
We also asked whether human EBP2 and EBNA1 associate with yeast chromosomes during G1 (Fig. 2, G1 panels). S. cerevisiae cells expressing human EBP2, EBNA1, or both proteins were blocked in G1 with α-factor, and then the chromatin was spread and stained with antibodies against human EBP2 and EBNA1. In both the presence and absence of EBNA1, human EBP2 was found to associate with a small region of the yeast DNA that colocalized with Nop1 and stained poorly with DAPI, indicating localization to nucleolar DNA. Since EBNA1 and Nop1 were both detected with mouse antibodies, we could not stain the chromatin spreads for Nop1 and EBNA1 at the same time. We did, however, stain chromatin spreads from cells expressing both human EBP2 and EBNA1 with human EBP2 and Nop1 antibodies to confirm that EBNA1 expression did not alter the nucleolar localization of human EBP2 (data not shown). The association of human EBP2 with the nucleolar DNA in G1 was quite different from the attachment of human EBP2 over the whole DNA mass observed in M and indicates a cell cycle-dependent localization of human EBP2 on yeast chromosomes. As in mitosis, EBNA1 did not associate with the yeast G1 chromatin in the absence of human EBP2, but when human EBP2 was present, EBNA1 attached to the G1 nucleolar DNA, exhibiting a staining pattern identical to that of human EBP2. Thus, in both interphase and mitosis, EBNA1 requires human EBP2 to attach to yeast chromosomes.
Functional contributions of human EBP2 domains.
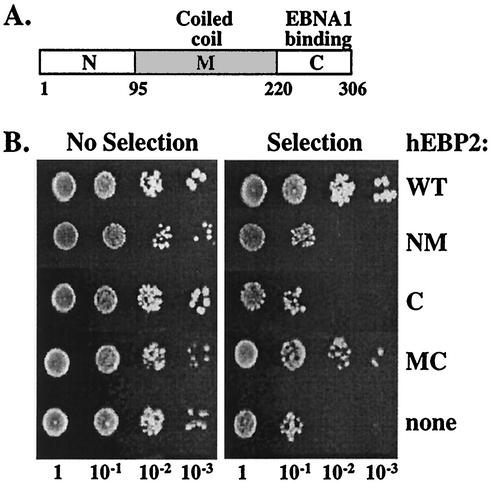
We have previously shown that the EBNA1-binding region of human EBP2 maps to the C-terminal domain (amino acids 220 to 306; Fig. 3A) and that this domain is required for EBNA1 to partition plasmids containing the FR element in S. cerevisiae (19) (Fig. 3B, NM). To determine whether the EBNA1-binding domain of human EBP2 was sufficient for the segregation activity of human EBP2, we tested the ability of this domain to support EBNA1-mediated plasmid partitioning in S. cerevisiae with a plasmid loss assay. S. cerevisiae cells were transformed with the S. cerevisiae replicating plasmid containing the FR segregation element (YRp7FR), with an EBNA1 expression plasmid (p416MET.EBNA1), and with a plasmid expressing either human EBP2 (positive control), the human EBP2 C-terminal domain (amino acids 220 to 306), or no human EBP2 (negative control). Transformants were grown for 11 generations without selection for the YRp7FR segregation plasmid, and serial dilutions of the cultures were plated on selective and nonselective plates with respect to YRp7FR (Fig. 3B).
FIG. 3.
Human EBP2 domains required for EBNA1-mediated plasmid segregation. (A) Schematic representation of human EBP2 (hEBP2), showing the N-terminal (N), middle coiled-coil (M), and C-terminal (C) domains. Amino acid numbers are indicated. (B) Assays of loss of plasmid YRp7FR conducted in the presence of EBNA1 and either full-length human EBP2 (wt), human EBP2 residues 1 to 220 (NM), human EBP2 residues 220 to 306 (C), human EBP2 residues 95 to 306 (MC), or no human EBP2 (none). After 11 generations in the absence of selection for YRp7FR, serially diluted cultures were plated on selective and nonselective plates for YRp7FR.
As expected, YRp7FR was stably maintained in the presence of EBNA1 and human EBP2, as indicated by the similar number of colonies formed on selective and nonselective plates, and was rapidly lost in the absence of human EBP2, as indicated by the decrease in the number of colonies formed on the selective plates. When the C-terminal domain of human EBP2 was expressed with EBNA1, pronounced loss of YRp7FR equivalent to that of the negative control was observed, indicating that the EBNA1-binding domain of human EBP2 is not sufficient to support EBNA1-mediated segregation (Fig. 3B and C).
We then asked which human EBP2 domain was required in addition to the C-terminal domain to rescue EBNA1-mediated plasmid partitioning. Sequence analysis indicates that the middle region of human EBP2 between amino acids 100 and 200 is likely a coiled-coil domain (40). A human EBP2 fragment containing this domain and the C-terminal domain (amino acids 95 to 306) was found to maintain YRp7FR with an efficiency similar to that of wild-type human EBP2 (Fig. 3B, MC). The results indicate that, in addition to the EBNA1-binding domain, the middle coiled-coil domain of human EBP2 plays an essential role in EBNA1-mediated plasmid partitioning and that the N-terminal region (amino acids 1 to 95) is not required for this process.
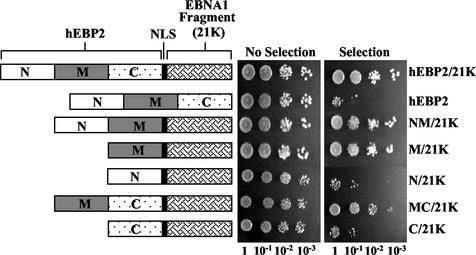
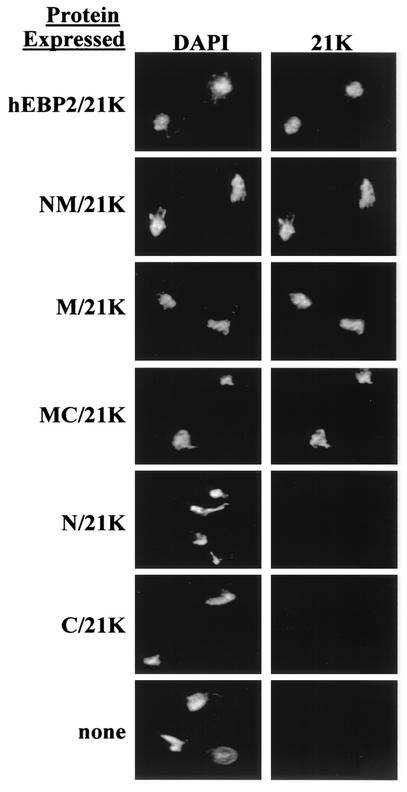
To confirm an essential role for the coiled-coil domain of human EBP2 in EBNA1-mediated segregation and to assess whether this role is distinct from that of the EBNA1-binding domain, we generated human EBP2-EBNA1 fusion proteins and asked which human EBP2 domains were required in this context. To begin with, we fused full-length human EBP2 to an EBNA1 fragment referred to as 21K (amino acids 452 to 641), which contains the DNA-binding and dimerization domain, and tested its ability to partition YRp7FR in the absence of any exogenous EBNA1 or human EBP2 (Fig. 4). A nuclear localization signal (NLS) was also included in the fusion protein to ensure that the protein entered the nucleus. In making the human EBP2-21K fusion protein, we made the assumptions that the human EBP2-binding region of EBNA1 (amino acids 325 to 376) would not be required when the two proteins were fused if its only function was to bind human EBP2 and that the EBNA1 21K fragment would be required in order for the fusion protein to bind and partition the FR-containing plasmid. Both assumptions were confirmed, as the human EBP2-21K fusion protein efficiently partitioned YRp7FR, while human EBP2 alone did not (Fig. 4).
FIG. 4.
Ability of human EBP2 domains fused to 21K to mediate plasmid partitioning. Plasmid loss assays were used to determine the stability of YRp7FR in the presence of a plasmid expressing human EBP2 (hEBP2), human EBP2 fused to 21K, or the indicated human EBP2 domains fused to 21K. After 11 generations in the absence of selection for YRp7FR, diluted cultures were plated on selective and nonselective plates with respect to YRp7FR. Schematic diagrams of the proteins are shown on the left.
We then replaced human EBP2 in the fusion protein with fragments spanning one or two of the human EBP2 domains (Fig. 4). We reasoned that any human EBP2 sequences whose sole contribution to EBNA1-mediated segregation is in binding EBNA1 would not be required in the context of the fusion protein, while sequences that contribute to plasmid partitioning in other ways would be required. In keeping with this hypothesis, a fusion protein containing the N-terminal and middle domains of human EBP2 but lacking the C-terminal EBNA1 binding domain (NM-21K) partitioned YRp7FR as efficiently as human EBP2-21K, indicating that EBNA1 binding is the only contribution of the human EBP2 C-terminal domain to plasmid partitioning (Fig. 4). When fusion proteins containing the human EBP2 N-terminal domain (amino acids 1 to 100; N-21K), middle coiled-coil domain (amino acids 95 to 220; M-21K), or C-terminal domain (amino acids 220 to 306; C-21K) were tested individually for the ability to stably maintain YRp7FR, only M-21K was found to do so. Fusion proteins containing the middle human EBP2 domain in combination with the N- or C-terminal domain also efficiently partitioned YRp7FR. These results support those in Fig. 3 and indicate that the middle coiled-coil domain of human EBP2 makes an important contribution to EBNA1-mediated plasmid partitioning which is not involved in EBNA1 binding.
Since we have shown that human EBP2 attaches to yeast chromosomes in mitosis and causes EBNA1 to bind the chromosomes, we asked whether the contribution of the middle human EBP2 domain to EBNA1-mediated plasmid partitioning was in chromosome attachment. To this end, we prepared mitotic chromosome spreads from yeast cells expressing the 21K fusion proteins used in the partitioning assays and stained the chromosome spreads with an antibody against the EBNA1 portion of the fusion proteins. More than 50 different chromosome spreads from four separate experiments were examined for each fusion protein, and representative images are shown in Fig. 5. All of the fusion proteins that contained the middle domain of human EBP2 consistently bound to yeast mitotic chromosomes, while the fusion proteins that lacked this domain were never observed on the chromosomes. The results indicate that the middle coiled-coil domain of human EBP2 is responsible for chromosome attachment. Furthermore, since all of the fusion proteins that functioned in plasmid partitioning attached to the yeast chromosomes and all of the fusion proteins that were nonfunctional for partitioning did not, the results strongly suggest that mitotic chromosome binding is crucial for plasmid segregation by EBNA1 in S. cerevisiae.
FIG. 5.
Middle coiled-coil domain of human EBP2 binds yeast mitotic chromosomes. Mitotic chromosome spreads from S. cerevisiae expressing human EBP2 (hEBP2) or human EBP2 domains fused to 21K were prepared as in Fig. 2. The spreads were stained with antibody against 21K, counterstained with DAPI, and observed by immunofluorescence microscopy.
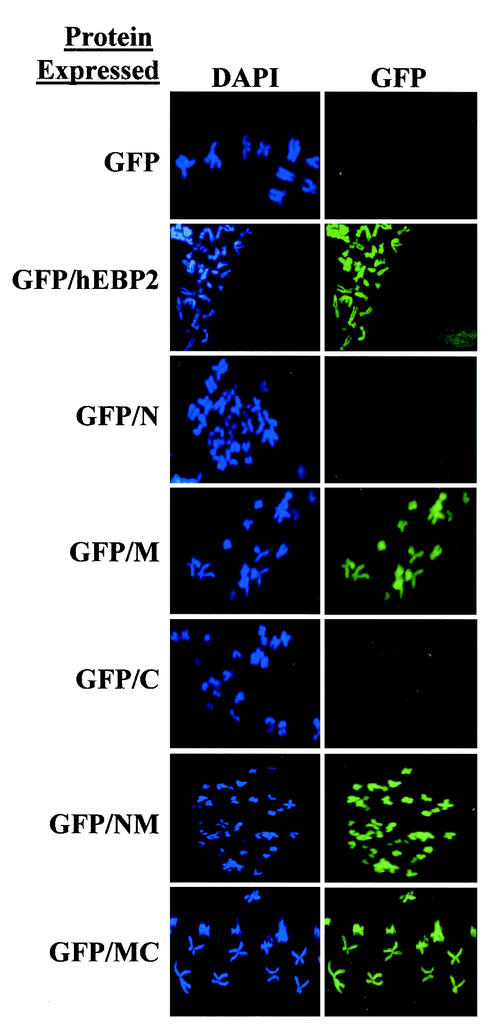
To verify that the results that we obtained in the yeast segregation system were relevant for EBNA1-mediated segregation in human cells, we tested whether the middle coiled-coil domain of human EBP2 was responsible for binding human mitotic chromosomes. To this end, we expressed human EBP2 or human EBP2 fragments fused to GFP in human HeLa cells and verified that the GFP fusion proteins were expressed in the majority of the cells by fluorescence microscopy of the log-phase cells (data not shown). We then blocked the cells in mitosis and prepared chromosome spreads for microscopy (Fig. 6). As expected, GFP alone did not bind to human mitotic chromosomes, while full-length human EBP2 fused to GFP (GFP-human EBP2) did, giving a staining pattern very similar to that of endogenous human EBP2 (46) (Fig. 6). When individual human EBP2 domains were fused to GFP, the middle domain but not the N- or C-terminal domain was found to associate with the mitotic chromosomes, as did human EBP2 fragments containing the middle domain in combination with the N- and C-terminal domains. More than 15 different chromosome spreads were examined for each fusion protein, with consistent results. The results indicate that the middle coiled-coil domain of human EBP2 is responsible for attachment to human chromosomes in mitosis, as it is in S. cerevisiae.
FIG. 6.
Middle coiled-coil domain of human EBP2 binds human mitotic chromosomes. GFP, GFP fused to human EBP2 (GFP/hEBP2), and GFP fused to human EBP2 fragments 1 to 100 (GFP/N), 95 to 220 (GFP/M), 220 to 306 (GFP/C), 1 to 220 (GFP/NM), and 95 to 306 (GFP/MC) were tested for their ability to bind mitotic chromosomes in HeLa cells. The mitotic chromosomes were visualized with DAPI, and the localization of GFP-containing proteins was determined by fluorescence microscopy. Images were captured with similar exposure times.
Functional differences between yeast EBP2 and human EBP2.
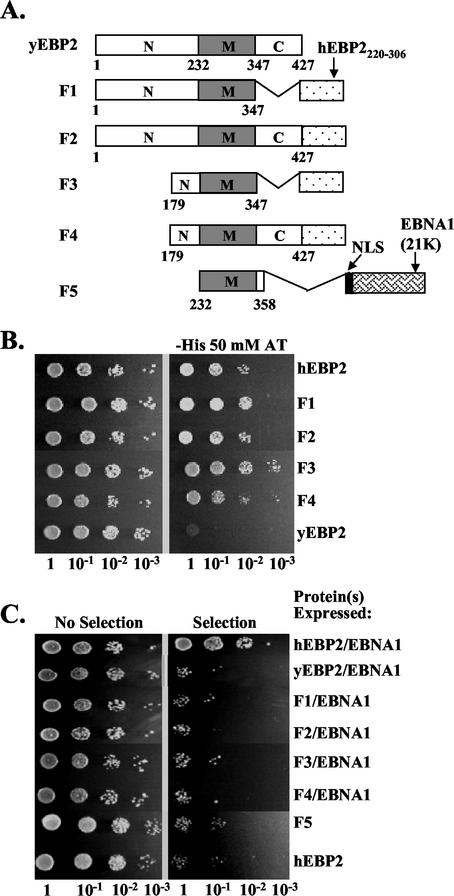
Yeast EBP2, the yeast homologue of human EBP2, plays an essential role in ribosome biogenesis and, like human EBP2, resides in the nucleolus (8, 15, 44). Yeast EBP2 is highly homologous to human EBP2 throughout the central and C-terminal domains but contains a nonessential N-terminal extension that is not present in human EBP2 (for an alignment of yeast and human EBP2 sequences, see reference 40), resulting in a larger N-terminal domain than that of human EBP2 (Fig. 7A). Like that of human EBP2, the central region of yeast EBP2 is predicted to be a coiled-coil domain (40). While yeast EBP2 and human EBP2 likely fulfill the same cellular functions, yeast EBP2 does not detectably interact with EBNA1 in S. cerevisiae (19) (Fig. 7B), and neither endogenous yeast EBP2 nor yeast EBP2 overexpressed from a plasmid could functionally replace human EBP2 in EBNA1-mediated plasmid segregation assays (Fig. 7C).
FIG. 7.
Plasmid segregation assays with yeast EBP2 fusion proteins. (A) Schematic representation of yeast EBP2 (yEBP2) and yeast EBP2-based fusion proteins (F1 to F5). The middle region (M) of yeast EBP2 corresponds to human EBP2 (hEBP2) residues 95 to 220. F2, F1, F3, and F4 are fusion proteins containing full-length yeast EBP2 or yeast EBP2 amino acids 1 to 347, 179 to 347, and 179 to 427, respectively, fused to the EBNA1-binding domain of human EBP2 (hEBP2220-306). F5 contains the middle region of yeast EBP2 (residues 232 to 358) fused to an NLS and the 21K fragment of EBNA1. (B) The ability of human EBP2 (positive control), yeast EBP2 (negative control), and EBP2 fusion proteins F1 through F4 to bind EBNA1 was determined in a yeast two-hybrid assay, where activation of a HIS3 reporter gene indicated an interaction. Dilutions of the two-hybrid assay cultures were grown on plates containing histidine (left panel) or lacking histidine and containing 50 mM aminotriazole (right panel). (C) Plasmid loss assays as described for Fig. 3 were conducted to determine the loss of YRp7FR in the presence of EBNA1 and human EBP2, yeast EBP2, or EBP2 fusion proteins F1 to F4. YRp7FR plasmid loss assays are also shown for F5 and human EBP2 alone.
To gain insight into what properties of human EBP2 were important for its ability to function with EBNA1 to partition plasmids, we took a domain-swapping approach with yeast EBP2. We reasoned that if the failure of yeast EBP2 to support EBNA1-mediated plasmid partitioning was due solely to its inability to bind EBNA1, the addition of the EBNA1-binding domain of human EBP2 to yeast EBP2 should enable yeast EBP2 to function with EBNA1 to partition plasmids. To test this hypothesis, we constructed fusion proteins containing either full-length yeast EBP2 or the yeast EBP2 N-terminal and middle domains fused to the C-terminal domain of human EBP2 (F2 and F1, respectively, in Fig. 7A). We also constructed fusion proteins F3 and F4 by removing the first 178 amino acids from F1 and F2, respectively (Fig. 7A). These were constructed in case the extended N terminus of yeast EBP2, which is not present in human EBP2, interfered with the ability of F1 and F2 to mediate plasmid partitioning.
The ability of the F1 to F4 fusion proteins to bind EBNA1 was tested in a yeast two-hybrid assay, where F1 to F4 were expressed fused to the GAL4 activation domain and EBNA1 was expressed fused to the GAL4 DNA-binding domain. Activation of a HIS3 reporter gene under the control of GAL4 binding sites was then measured by the ability of the S. cerevisiae cells to grow on plates lacking histidine and containing aminotriazole. As shown in Fig. 7B, the F1, F2, F3, and F4 fusion proteins bound EBNA1, resulting in growth on the plates equivalent to that seen with the human EBP2-EBNA1 positive control. However, when tested in the plasmid loss assay, none of these fusion proteins supported the segregation of YRp7FR when coexpressed with EBNA1 (Fig. 7C). This indicated that EBNA1 binding is not the only functional difference between human and S. cerevisiae EBP2 and that the properties of either the middle domain or the remaining portion of the N-terminal region of yeast EBP2 (in F3 and F4) were not suited for plasmid partitioning by EBNA1.
Since we had shown that the middle coiled-coil domain of human EBP2 was crucial for plasmid partitioning and partitioned plasmids when fused to 21K, we tested whether the equivalent middle coiled-coil domain of yeast EBP2 would function to partition plasmids when fused to 21K. According to sequence alignments, amino acids 95 to 220 of human EBP2 (which were functional for partitioning when fused to 21K) correspond to amino acids 232 to 358 of yeast EBP2 (40), and therefore this yeast EBP2 fragment was fused to 21K through an NLS to generate the F5 fusion protein (Fig. 7A). The expression of F5 was confirmed by Western blot (data not shown), and this protein was tested for its ability to partition YRp7FR in the plasmid loss assay (Fig. 7C). Unlike the human EBP2 M-21K construct (Fig. 4), F5 did not support the stable segregation of YRp7FR, indicating that there is an important functional difference in the middle coiled-coil domains of human and yeast EBP2.
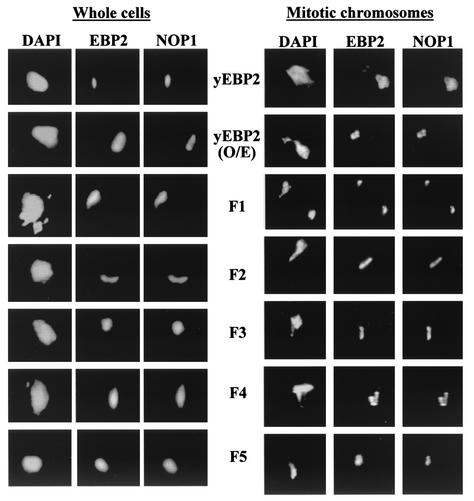
To gain insight into the differences in the yeast EBP2 and human EBP2 middle domains that would affect their ability to support plasmid partitioning, we examined the localization of all of the yeast EBP2-based fusion proteins both in whole yeast cells and on mitotic chromosome spreads. Immunofluorescence microscopy on whole yeast cells from a log-phase culture showed that yeast EBP2 (whether endogenous or overexpressed from a plasmid) and the F1 to F5 fusion proteins were localized to the nucleolus, giving staining patterns similar to that of the NOP1 nucleolar marker (Fig. 8, left panel). The results with yeast EBP2 are consistent with the nucleolar localization of this protein reported previously (15, 44). Immunofluorescence microscopy of mitotic chromosome spreads prepared from S. cerevisiae expressing yeast EBP2 or the yeast EBP2 fusion proteins revealed that all of these proteins were bound to the chromatin at regions that did not stain well with DAPI but did stain with the Nop1 antibody (Fig. 8, right panel), indicating that these proteins bind chromatin in the nucleolar region. Very little or no association of the yeast EBP2 proteins was detected with nonnucleolar DNA. These results indicate that yeast EBP2 and yeast EBP2 fusion proteins only associate with a very localized portion of the mitotic chromosome mass (which corresponds to the nucleolus) and that the yeast EBP2 middle coiled-coil domain is sufficient for the nucleolar localization and chromatin attachment. This pattern of chromosome attachment differs from that of human EBP2 and human EBP2 middle domain-containing proteins, which exhibit much more extensive staining over the mitotic chromatin (Fig. 2). The results suggest that chromosome attachment that is limited to the nucleolar region is not sufficient for EBNA1-mediated plasmid partitioning and that the spreading of human EBP2 over the chromatin mass in mitosis is important for its ability to support plasmid segregation by EBNA1.
FIG. 8.
Nucleolar localization of yeast EBP2 and yeast EBP2-based fusion proteins. Log-phase yeast cells (left panel) and spreads of yeast mitotic chromosomes (right panel) are shown for S. cerevisiae expressing endogenous yeast EBP2 (yEBP2), overexpressed yeast EBP2 (yEBP2 O/E), or a yeast EBP2-based fusion protein (F1 to F5). Cells and chromosome spreads were stained for the EBP2 proteins and for Nop1 (as a nucleolar marker) and counterstained with DAPI. Images were captured by immunofluorescence microscopy under identical conditions.
DISCUSSION
Our previous studies revealed that human EBP2 is required for EBNA1 to partition EBV-based plasmids in S. cerevisiae, suggesting an important role for human EBP2 in EBV segregation in human cells (19). In the present study, we demonstrated that human EBP2 causes EBNA1 to attach to yeast chromosomes. This attachment is limited to chromatin in the nucleolus in G1 but occurs over much of the chromatin mass in mitosis. We also demonstrated that the attachment of EBNA1 over the mitotic chromosomes is a requirement for plasmid partitioning by EBNA1, indicating that this is the essential contribution of human EBP2 to partitioning. The most likely reason why human EBP2 is required for EBNA1 to attach to the chromosomes is that human EBP2 tethers EBNA1 to the chromosomes. A tethering model is supported by the observations that human EBP2 and EBNA1 physically interact (40), the EBNA1-binding region of human EBP2 and the human EBP2-binding region of EBNA1 are required for the two proteins to work together to partition plasmids (19), and human EBP2 and EBNA1 colocalize on the yeast chromosomes.
Previously, we mapped the EBNA1-binding domain of human EBP2 to C-terminal amino acids 220 to 306 (19), and in the present study we have demonstrated that the middle region of human EBP2 (amino acids 95 to 220), which corresponds to a putative coiled-coil domain, is responsible for chromatin attachment in mitosis. It is not yet clear how this domain attaches to the chromosomes, but for the following reasons, we believe that it is likely to attach through a protein component of the chromosomes rather than binding directly to the DNA. First, the interaction of human EBP2 with chromatin is more extensive in mitosis than G1, when the DNA should be less accessible. Second, the chromosome attachment region of human EBP2 corresponds to a predicted coiled-coil domain, and coiled-coil domains in other proteins mediate protein interactions (reviewed in reference 2). Since coiled coils tend to interact with other coiled coils, human EBP2 may attach to chromosomes by binding to a coiled-coil chromosomal protein.
All of the data indicate that EBNA1-mediated plasmid segregation in our reconstituted yeast system reflects that which occurs in human cells. We previously demonstrated that the two segregation systems have the same requirements for the FR element and EBNA1 and are affected to the same degree by mutations in EBNA1 that disrupt or decrease human EBP2 binding (19, 47). A large body of evidence on the segregation of EBV episomes and EBV-based plasmids in human cells indicates that segregation occurs by the EBNA1-mediated tethering of these molecules to the cellular mitotic chromosomes. Our present data indicate that this same segregation mechanism is occurring in our reconstituted yeast system. Since human EBP2 is required for EBNA1 to attach to yeast chromosomes, it is likely that human EBP2 also fulfills this role for EBV segregation in human cells. Unfortunately, the requirement for human EBP2 for plasmid partitioning in human cells cannot easily be tested, since it is expressed in all proliferating cells and is essential for cell viability (8, 15).
The data obtained in both yeast and human cells point to a model for EBV segregation that is likely to be true in both cell systems. In this model, human EBP2 interacts with mitotic chromosomes through its coiled-coil domain (amino acids 95 to 220) and interacts with EBNA1 through its C-terminal domain (amino acids 220 to 306). EBNA1 interacts with human EBP2 through amino acids 325 to 376 and binds to the FR element in the EBV episome or plasmid via its DNA-binding and dimerization domain (amino acids 452 to 607), thus tethering the plasmid to host chromosomes.
In addition to the 325 to 376 region of EBNA, two other EBNA1 sequences (amino acid 8 to 54 and 72 to 84) have been shown to bind mitotic chromosomes when excised from EBNA1 (33). Subsequent deletion analysis of EBNA1 showed that the 8 to 54 region modestly enhanced plasmid partitioning, mitotic chromosome binding, and human EBP2 binding by EBNA1, suggesting that it stabilizes the interaction of the 325 to 376 region to human EBP2 (47). Deletion of the 72 to 84 sequence within EBNA1, however, had no detectable effect on plasmid partitioning, chromosome binding, or human EBP2 binding (47). While it is not clear how the excised 72 to 84 peptide bound mitotic chromosomes, there is presently no evidence that this interaction has any functional consequences.
In human cells, human EBP2 is localized to the nucleolus in interphase but binds throughout the chromosomes during mitosis. Such behavior has been observed for several nucleolar proteins, in particular those involved in rRNA processing, which is likely to be the cellular function of human EBP2 (14, 15, 44). In human cells, the nucleolus breaks down in mitosis, and some nucleolar proteins associate with the surface of the chromosomes, ensuring equal partitioning between dividing cells (14). How the relocalization of these proteins is triggered is not known. In S. cerevisiae, cell cycle-dependent nucleolar alterations are much less extensive, and most yeast nucleolar proteins (including yeast EBP2) remain in the nucleolus during mitosis (11). Given these fundamental differences in the nucleolus in human and yeast cells, we were interested in determining whether human EBP2 retained the ability to relocalize in mitosis when expressed in S. cerevisiae. Interestingly, we observed that human EBP2 exhibited similar cell cycle-dependent relocalization in S. cerevisiae as in human cells, in that human EBP2 associated only with the nucleolar portion of yeast chromatin during G1 but bound all over the chromatin during mitosis. Thus, the signal that triggers human EBP2 to bind all over the mitotic chromosomes occurs in both human and yeast cells.
Our localization studies indicate that there are two pools of human EBP2 in interphase yeast cells; one is bound to the chromatin in the nucleolus (revealed in chromatin spreads shown in Fig. 2, G1 panel), and another is found throughout the nucleus but is not chromatin associated (indicated by nuclear staining of whole cells in Fig. 1). Presently, it is not clear which of these human EBP2 pools becomes bound over the chromosomes in mitosis. It may be that human EBP2 in the nucleolus remains bound to the nucleolar chromatin throughout the cell cycle and that human EBP2 molecules that are not associated with the chromatin in interphase attach to additional regions of the chromosomes in mitosis. Alternatively, human EBP2 bound to the nucleolar chromatin in interphase may redistribute over all of the chromatin in mitosis. While it is the coiled-coil domains of both human EBP2 and yeast EBP2 that are responsible for chromosome attachment, the mechanisms by which these two proteins attach to chromosomes in mitosis is likely different. Unlike human EBP2, yeast EBP2 and fusion proteins containing the chromosome attachment domain of yeast EBP2 do not relocalize in mitosis but instead remain bound to the nucleolar portion of the chromatin. These yeast EBP2 proteins are not able to support EBNA1-mediated plasmid segregation, suggesting that more extensive interaction with the mitotic chromosomes is necessary for this function.
Like EBV, the low-copy-number episomal genomes of bovine papillomavirus and Kaposi's sarcoma-associated herpesvirus (KSHV) also appear to be partitioned in dividing cells by attaching to the cellular mitotic chromosomes (reviewed in reference 9). The E2 protein of bovine papillomavirus and the LANA1 protein of Kaposi's sarcoma-associated herpesvirus, which are the functional counterparts of EBNA1, tether their viral genomes to the mitotic chromosomes by binding to both the chromosomes and the cis-acting segregation element of their respective viruses (5, 17, 28, 42). The components of the mitotic chromosomes to which E2 and LANA attach to partition plasmids are not yet clear but are unlikely to include human EBP2, since we have been unable to detect any interaction between human EBP2 and the chromosome-binding regions of E2 and LANA (P. Kapoor, K. Shire, and L. Frappier, unpublished data). The fact that we have been able to reconstitute EBV plasmid segregation in S. cerevisiae suggests that S. cerevisiae may also be a useful system for identifying the human chromosomal components important for bovine papillomavirus and Kaposi's sarcoma-associated herpesvirus plasmid partitioning.
While there is considerable evidence that stable segregation can occur through chromosome attachment in mammalian cells, it has not been clear whether this segregation mechanism occurs in S. cerevisiae. Plasmids that contain an ARS element and telomeric or HMR E silencer sequences segregate stably in S. cerevisiae in the presence of Rap1 and Sir2/3/4 proteins and are postulated to do so through attachment to structural components of the nucleus that partition equally during mitosis (22, 23, 30, 31). It has not been determined, however, whether these nuclear components are chromosomal. Similarly, plasmids containing an ARS element and the LexA operator sequence are partitioned by a LexA/Sir4 fusion protein, which anchors the plasmids to an unidentified nuclear component (4). Numerous studies have also been conducted on the partitioning mechanism of the S. cerevisiae 2μm plasmid, which requires the cis-acting STB locus and the Rep1/Rep2 proteins (21, 48). 2μm and cellular chromosome segregation have very similar kinetics and cellular protein requirements, indicating that 2μm plasmid partitioning is closely tied to that of cellular chromosomes (34, 45). It is not clear, however, whether the similarities reflect the tethering of the plasmids to the chromosomes or the possibility that the 2μm plasmids, like cellular chromosomes, attach to the mitotic spindle. Our results with the EBV-based plasmid segregation system provide strong evidence that plasmids can be partitioned in S. cerevisiae by mitotic chromosome attachment, as occurs in human cells.
Acknowledgments
We gratefully acknowledge Brigitte Lavoie for providing technical expertise on chromatin spreading and J. Aris and J. Middeldorp for antibodies. We also thank Brigitte Lavoie and Barbara Funnell for critical reading of the manuscript.
This work was supported by a grant to L.F. from the Canadian Institutes of Health Research (CIHR). L.F. is a recipient of the CIHR Scientist award. P.K. is a research student of the National Cancer Institute of Canada, supported with funds provided by the Terry Fox run.
REFERENCES
- 1.Adams, A. 1985. Replication of latent Epstein-Barr virus genomes in Raji cells. J. Virol. 61:1743-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson, J. G., N. E. Zhou, and R. S. Hodges. 1993. Structure, function and application of the coiled-coil protein-folding motif. Curr. Opin. Biotechnol. 4:428-437. [DOI] [PubMed] [Google Scholar]
- 3.Ambinder, R. F., M. Mullen, Y. N. Chang, G. S. Hayward, and S. D. Hayward. 1991. Functional domains of Epstein-Barr virus nuclear antigen EBNA-1. J. Virol. 65:1466-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ansari, A., and M. R. Gartenberg. 1997. The yeast silent information regulator Sir4p anchors and partitions plasmids. Mol. Cell. Biol. 17:7061-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 6.Bochkarev, A., J. A. Barwell, R. A. Pfuetzner, E. Bochkareva, L. Frappier, and A. M. Edwards. 1996. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell. 84:791-800. [DOI] [PubMed] [Google Scholar]
- 7.Ceccarelli, D. F. J., and L. Frappier. 2000. Functional analyses of the EBNA1 origin DNA binding protein of Epstein-Barr virus. J. Virol. 74:4939-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee, A., J. W. Freeman, and H. Busch. 1987. Identification and partial characterization of a Mr 105,000 nucleolar antigen associated with cell proliferation. Cancer Res. 47:6329-6334. [PubMed] [Google Scholar]
- 9.Frappier, L. 2003. Viral plasmids in mammalian cells. In B. E. Funnell and G. Phillips (ed.), The biology of plasmids, in press. ASM Press, Washington, D.C.
- 10.Gahn, T. A., and C. L. Schildkraut. 1989. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 58:527-535. [DOI] [PubMed] [Google Scholar]
- 11.Granot, D., and M. Snyder. 1991. Segregation of the nucleolus during mitosis in budding and fission yeast. Cell Motil. Cytoskel. 20:47-54. [DOI] [PubMed] [Google Scholar]
- 12.Grogan, E. A., W. P. Summers, S. Dowling, D. Shedd, L. Gradoville, and G. Miller. 1983. Two Epstein-Barr viral nuclear neoantigens distinguished by gene transfer, serology, and chromosome binding. Proc. Natl. Acad. Sci. USA 80:7650-7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris, A., B. D. Young, and B. E. Griffin. 1985. Random association of Epstein-Barr virus genomes with host-cell metaphase chromosomes in Burkitt's lymphoma-derived cell-lines. J. Virol. 56:328-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandez-Verdun, D., and T. Gautier. 1994. The chromosome periphery during mitosis. Bioessays 16:179-185. [DOI] [PubMed] [Google Scholar]
- 15.Huber, M. D., J. H. Dworet, K. Shire, L. Frappier, and M. A. McAlear. 2000. The budding yeast homolog of the human EBNA1-binding protein 2 (Ebp2p) is an essential nucleolar protein required for pre-rRNA processing. J. Biol. Chem. 275:28764-28773. [DOI] [PubMed] [Google Scholar]
- 16.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 73:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanda, T., M. Otter, and G. M. Wahl. 2001. Coupling of mitotic chromosome tethering and replication competence in Epstein-Barr virus-based plasmids. Mol. Cell. Biol. 21:3576-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapoor, P., K. Shire, and L. Frappier. 2001. Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast. EMBO J. 20:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In D. M. Knipe and P. M. Howley (ed.), Field's virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 21.Kikuchi, Y. 1983. Yeast plasmid requires a cis-acting locus and 2 plasmid proteins for its stable maintenance. Cell 35:487-493. [DOI] [PubMed] [Google Scholar]
- 22.Kimmerly, W., A. Buchman, R. Kornberg, and J. Rine. 1988. Roles of two DNA-binding factors in replication, segregation and transcriptional repression mediated by a yeast silencer. EMBO J. 7:2241-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimmerly, W. J., and J. Rine. 1987. Replication and segregation of plasmids containing cis-acting regulatory sites of silent mating-type genes in Saccharomyces cerevisiae are controlled by the SIR genes. Mol. Cell Biol. 7:225-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchmaier, A. L., and B. Sugden. 1997. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J. Virol. 71:1766-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein, F., T. Laroche, M. E. Cardenas, J. F. X. Hofmann, D. Schweizer, and S. M. Gasser. 1992. Localization of Rap1 and topoisomerase-II in nuclei and meiotic chromosomes of yeast. J. Cell Biol. 117:935-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krysan, P. J., S. B. Haase, and M. P. Calos. 1989. Isolation of human sequences that replicate autonomously in human cells. Mol. Cell. Biol. 9:1026-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavoie, B. D., E. Hogan, and D. Koshland. 2002. In vivo dissection of the chromosome condensation machinery: reversibility of condensation distinguishes contributions of condensin and cohesin. J. Cell Biol. 156:805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehman, C. W., and M. R. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 95:4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, L., S. J. Elledge, C. A. Peterson, E. S. Bales, and R. J. Legerski. 1994. Specific association between the human DNA repair proteins XPA and ERCC1. Proc. Natl. Acad. Sci. USA 91:5012-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longtine, M. S., S. Enomoto, S. L. Finstad, and J. Berman. 1992. Yeast telomere repeat sequence (TRS) improves circular plasmid segregation, and TRS plasmid segregation involves the RAP1 gene product. Mol. Cell. Biol. 12:1997-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longtine, M. S., S. Enomoto, S. L. Finstad, and J. Berman. 1993. Telomere-mediated plasmid segregation in Saccharomyces cerevisiae involves gene products required for transcriptional repression at silencers and telomeres. Genetics 133:171-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lupton, S., and A. J. Levine. 1985. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol. Cell. Biol. 5:2533-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Pilot, M. Coppey-Moisan, and J.-C. Nicolas. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mehta, S., X. M. Yang, C. S. Chan, M. J. Dobson, M. Jayaram, and S. Velmurugan. 2002. The 2μm plasmid purloins the yeast cohesin complex: a mechanism for coupling plasmid partitioning and chromosome segregation? J. Cell Biol. 158:625-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35-45. [DOI] [PubMed] [Google Scholar]
- 36.Petti, L., C. Sample, and E. Kieff. 1990. Subnuclear localization and phosphorylation of Epstein-Barr-virus latent infection nuclear proteins. Virology 176:563-574. [DOI] [PubMed] [Google Scholar]
- 37.Rawlins, D. R., G. Milman, S. D. Hayward, and G. S. Hayward. 1985. Sequence-specific DNA-binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell 42:859-868. [DOI] [PubMed] [Google Scholar]
- 38.Reisman, D., and B. Sugden. 1986. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen-1. Mol. Cell. Biol. 6:3838-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reisman, D., J. Yates, and B. Sugden. 1985. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol. Cell. Biol. 5:1822-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shire, K., D. F. J. Ceccarelli, T. M. Avolio-Hunter, and L. Frappier. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 73:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson, K., A. McGuigan, and C. Huxley. 1996. Stable episomal maintenance of yeast artificial chromosomes in human cells. Mol. Cell. Biol. 16:5117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 72:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka, T., D. Knapp, and K. Nasmyth. 1997. Loading of a Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90:649-660. [DOI] [PubMed] [Google Scholar]
- 44.Tsujii, R., K. Miyoshi, A. Tsuno, Y. Matsui, A. Toh-e, T. Miyakawa, and K. Mizuta. 2000. Ebp2p, yeast homologue of a human protein that interacts with Epstein-Barr virus nuclear antigen 1, is required for pre-rRNA processing and ribosomal subunit assembly. Genes Cells 5:543-553. [DOI] [PubMed] [Google Scholar]
- 45.Velmurugan, S., X. M. Yang, C. S. M. Chan, M. Dobson, and M. Jayaram. 2000. Partitioning of the 2μm circle plasmid of Saccharomyces cerevisiae: functional coordination with chromosome segregation and plasmid-encoded rep protein distribution. J. Cell Biol. 149:553-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, H., D. F. J. Ceccarelli, and L. Frappier. 2000. The DNA segregation mechanism of Epstein-Barr virus nuclear antigen 1. EMBO Rep. 1:140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, H., P. Kapoor, and L. Frappier. 2002. Separation of the DNA replication, segregation, and transcriptional activation functions of Epstein-Barr nuclear antigen 1. J. Virol. 76:2480-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, L. C. C., P. A. Fisher, and J. R. Broach. 1987. A yeast plasmid partitioning protein is a karyoskeletal component. J. Biol. Chem. 262:883-891. [PubMed] [Google Scholar]
- 49.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 13:812-815. [DOI] [PubMed] [Google Scholar]