Abstract
We used feline immunodeficiency virus (FIV) protease (PR) as a mutational framework to define determinants for the observed substrate and inhibitor specificity distinctions between FIV and human immunodeficiency virus (HIV) PRs. Multiple-substitution mutants were constructed by replacing the residues in and around the active site of FIV PR with the structurally equivalent residues of HIV-1 PR. Mutants included combinations of three critical regions (FIV numbering, with equivalent HIV numbering in superscript): I3732V in the active core region; N5546M, M5647I, and V5950I in the flap region; and L9780T, I9881P, Q9982V, P10083N, and L10184I in the 90s loop region. Significant alterations in specificity were observed, consistent with the involvement of these residues in determining the substrate-inhibitor specificity distinctions between FIV and HIV PRs. Two previously identified residues, I35 and I57 of FIV PR, were intolerant to substitution and yielded inactive PRs. Therefore, we attempted to recover the activity by introducing secondary mutations. The addition of G6253F and K6354I, located at the top of the flap and outside the active site, compensated for the activity lost in the I5748G substitution mutants. An additional two substitutions, D10588N and N8874T, facilitated recovery of activity in mutants that included the I3530D substitution. Determination of Ki values of potent HIV-1 PR inhibitors against these mutants showed that inhibitor specificity paralleled that of HIV-1 PR. The findings indicate that maintenance of both substrate and inhibitor specificity is a function of interactions between residues both inside and outside the active site. Thus, mutations apparently peripheral to the active site can have a dramatic influence on inhibitor efficacy.
Retroviral aspartic protease plays a critical role in processing the viral Gag and Gag-Pol polyproteins into individual functional proteins during the budding process as the virus leaves the host cell. This highly ordered step is essential for producing mature and infectious virions to initiate the next cycle of replication (24, 25, 39). Therefore, human immunodeficiency virus type 1 (HIV-1) protease (PR) has been an important target for successful design of antiretroviral drugs. However, the continuous development of drug-resistant strains of the virus has posed a serious and challenging problem for the design of a new generation of PR inhibitors (22, 29, 33). Thus, development of drugs less prone to resistance development is an important area of research.
Feline immunodeficiency virus (FIV) PR is responsible for cleaving the FIV Gag and Gag-Pol polyproteins into nine separate, functional proteins, including matrix, capsid, P1, nucleocapsid, P2, protease (PR), reverse transcriptase (RT), RNase H, dUTPase, and integrase (9). The cleavage sites for PR are similar in character to those identified in HIV-1 but are distinct in actual sequences (9, 40). Our focus has been to use FIV and HIV-1 PRs as a mutational analysis system to study the molecular basis of specificity of retroviral PRs (7, 8). The structure-based approach has led to the development of TL-3, a competitive inhibitor that is capable of inhibiting FIV, simian immunodeficiency virus (SIV), and HIV-1 and several HIV-1 drug-resistant strains ex vivo (18, 19).
FIV PR is structurally very similar to HIV-1 PR but is only 23% identical at the amino acid level (Fig. 1) and exhibits distinct substrate and inhibitor specificities. Furthermore, most residues in the active site of FIV and HIV-1 PRs are different despite striking similarity in the three-dimensional structures of the two proteases (17, 40, 41). Interestingly, 27 mutations in HIV-1 PR have been identified in response to drug treatment (31) that are either identical or highly similar to the equivalent residues of FIV PR. Among these, 10 mutations (K20I, V32I, M36R, I47 M, I50V, L63H, A71I, N88D, L90M, and I93F) are thought to contribute to drug resistance. Therefore, the FIV and HIV-1 PR comparative model is an attractive system to use in the analysis of the molecular interactions with substrate and inhibitor as well as for defining the specificity determinants of retroviral PRs. Studies of the two lentivirus systems can help establish the structural basis of the observed specificity distinctions and, in turn, further aid the development of broad-based inhibitors against retroviral PRs and drug-resistant PRs.
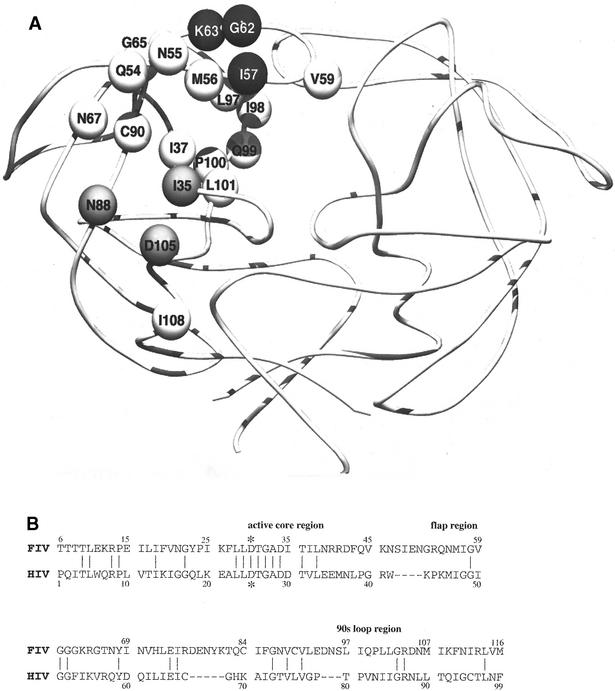
FIG. 1.
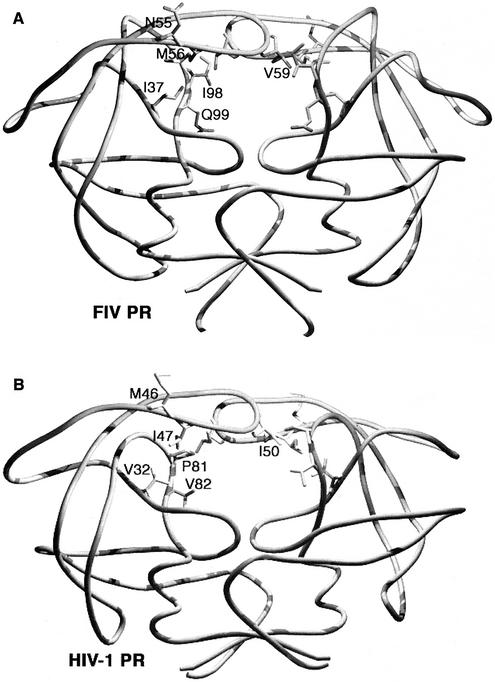
(A) Residues in and around the substrate binding pocket of FIV PR, shown on one chain of the homodimer. These residues were the focus of the substitutions in this study. Earlier studies had identified the I3530D and I5748G substitutions as intolerant to change in the background of FIV PR, whereas other substitutions allow maintenance of activity and contribute to the substrate and inhibitor specificity significantly. Residues outside the substrate-binding pocket but in close proximity to I3530D or I5748G are shown in gray and black, respectively. (B) Amino acid sequence alignment of FIV and HIV-1 PRs based on crystal structures (41). *, catalytic aspartic acids (D30 of FIV PR and D25 of HIV-1PR). Numbers indicate the positions of structurally equivalent residues.
We previously performed extensive mutagenesis of residues associated with the S4 to S4′ subsites of FIV PR in order to attempt to identify residues associated with substrate and inhibitor specificities for FIV and HIV-1 PRs (4, 21). Substrate specificity was analyzed by assaying the cleavage efficiency of the mutant PRs on peptides representing FIV and HIV-1 viral cleavage junctions and phage display library peptides which were selected with HIV-1 PR (3). In the present study, we expanded on these studies by constructing mutant FIV PRs in which as many as 24 amino acid residues (in a single chain), in and around the active site of FIV PR, have been substituted for the structurally equivalent residues of HIV-1 PR. These mutants were expressed and purified and their substrate and inhibitor specificities were determined. In addition, inhibitor specificities were analyzed by determining the Ki values of potent HIV-1 PR inhibitors, including the Food and Drug Administration (FDA)-approved drugs (12) saquinavir, ritonavir, and nefinavir, against the chimeric FIV PRs. Our findings show that residues outside the active site of PR act to stabilize pivotal residues in direct contact with substrate and inhibitor, which offers a structural explanation as to how changes in certain distal residues can influence binding in the active site.
MATERIALS AND METHODS
Mutagenesis of chimeric FIV PRs.
Chimeric FIV PRs were constructed by substituting the residues of FIV PR for the structurally equivalent residues of HIV-1 PR with PCR-mediated megaprimer site-directed mutagenesis as described previously (1, 28). The chimeric PR genes were digested with NdeI and HindIII and cloned into pET-21a and pET-28a (Novagen, Inc.). The pET expression vectors were originally constructed by Studier and Moffatt (32). The substitutions were verified by dideoxy DNA sequencing.
Expression and purification of PRs.
The chimeric PR constructs were transformed into the BL21(DE3)/pLysS strain of Escherichia coli for protein expression (32). PR expression was induced with 1 mM isopropylthiogalactopyranoside (IPTG) for 3 h at 37°C. Inclusion bodies containing PR were isolated by centrifugation, solubilized in 8 M urea containing 20 mM Tris and 5 mM EDTA, pH 8, and subsequently purified by ion exchange chromatography as described previously (18). The denatured PR was dialyzed and refolded in 25 mM phosphate buffer containing 150 mM NaCl, 5 mM EDTA, and 2 mM dithiothreitol. The purified PRs were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and verified by immunoblot with a specific antibody against FIV PR. HIV-1 PR of the SF2 strain was purified and verified as described previously (19).
Peptide synthesis and purification.
Viral junction peptides were synthesized with Fmoc (9-fluorenylmethoxy carbonyl) solid-phase chemistry as described previously (2). N-Methylpyrrolidinone was used instead of dimethylformamide as the coupling solvent. The FIV and HIV-1 junction peptides represent the amino acid sequences from the viral cleavage sites (9, 21). Phage library peptides are based on the primary sequences of the random hexamer region of the phage, which were specifically cleaved by either HIV-1 or FIV PR (3). Phage peptides were synthesized by the protein core facility of the Scripps Research Institute with standard Boc (1-butoxycarbonyl) coupling protocols. All peptides were acetylated at the N-terminal nitrogen and contained C-terminal amides. They were purified on a Vydac C-4 column with reverse-phase high-pressure liquid chromatography (HPLC), and the mass was verified with an electrospray mass spectrometer.
Cleavage assay with fluorogenic substrates or fluorescamine.
The cleavage efficiency of PR was assayed in 0.05 M sodium citrate-0.1 M sodium phosphate buffer, pH 5.25, containing 1 mM dithiothreitol and 0.2 M NaCl (18). Enzyme kinetics of PRs on FIV substrates were analyzed with the FIV fluorogenic substrate A-L-T-(Abz)K-V-Q/(p-NO2)F-V-Q-S-K-G, which mimics the FIV capsid/NC2 cleavage junction (11). The fluorogenic substrate contains a self-quenching pair which become separated upon PR hydrolysis and generate increased fluorescence. At least six different concentrations (2 to 150 μM) of substrate were used. The data were collected by continuously monitoring changes in fluorescence for 6 min at an excitation at 325 nm and an emission at 410 nm with an F-2000 fluorescence spectrophotometer (Hitachi Inc.). Continuous absence of increased fluorescence for 6 min was considered no detectable activity in this study.
The proteolytic activity of FIV PRs on HIV-1 substrates was first assayed on three HIV-1 fluorogenic substrates, Abz-T-I-Nle/(p-NO2)F-Q-R (excitation at 325 nm and emission at 420 nm), analogous to the HIV-1 P2/nucleocapsid cleavage junction (36); Abz-R-V-Nle/(p-NO2)F-E-A-Nle (excitation at 330 nm and emission at 430 nm) analogous to the HIV-1 capsid/P2 cleavage junction; and I-R-(Abz)K-I-L/(p-NO2)F-L-D-G (excitation at 325 nm and emission at 410 nm), which represents the HIV-1 reverse transcriptase-integrase cleavage junction. The data were plotted, and the Km and Vmax values were calculated with Grafit 3 (Erithacus Software Ltd.).
The cleavage efficiency of PR was also analyzed with a fluorescamine assay (14). Fluorescamine readily reacts with amines in aqueous solution, and the products are highly fluorescent (38). The cleavage was normalized to a cleavage junction peptide (acelyl [Ac]-I-R-K-I-L/F-L-D-G-NH2) as described previously (21).
Cleavage assay with synthetic peptides and reverse-phase HPLC.
Two FIV peptides, Ac-K-R-S-T-G-V-F/S-S-W-V-D-R-K-NH2 and Ac-K-R-L-T-K-V-Q/V-V-Q-S-K-R-K-NH2, representing the FIV dUTPase/integrase and capsid/NC2 cleavage junctions, and one HIV-1 peptide, Ac-R-K-I-L/F-L-D-G-NH2, representing the HIV-1 RT/integrase cleavage junctions, were tested in order to determine the relative cleavage efficiencies of wild-type and mutant FIV PRs. In addition, four nonviral peptides selected from a phage display library were also employed (3). The final concentration of peptide substrate was 100 μM. The PR concentration (100 nM to 1,000 nM) and incubation time (10 min to 1 h) varied depending on the PR and peptide used. The reaction was terminated by mixing in an equal volume of 6 M guanidine HCl. The products were analyzed by reverse-phase HPLC with a Vydac C18 analytical column with a linear gradient of 0% to 67% acetonitrile in 0.1% trifluoroacetic acid aqueous solution at a flow rate of 1 ml/min. Cleavage products were collected, based on their absorbance at 214 nm, and analyzed by electrospray mass spectrometry to verify the correct cleavage site and cleaved fragments. The cleavage efficiencies (percent cleavage) by PRs were calculated from integrated areas of the remaining uncleaved substrate and the total uncleaved control.
Determination of active-site concentration of PR and Ki values for inhibitors.
The concentration of PR was titrated by using TL-3, a tight-binding inhibitor of FIV, SIV, and HIV-1 PRs (18). Different concentrations of TL-3 were mixed with PR and incubated for 30 s. The reaction was initiated by mixing in the FIV capsid/NC2 fluorogenic substrate and monitoring initial velocity for 3 min with a fluorimeter. The active-site concentrations of PRs were obtained from the extrapolated intercepts from the plots of It/(1 − vi/v0) against v0/vi as described previously (15), where It is the total concentration of inhibitor; vi is the velocity in the presence of inhibitor, and v0 is the velocity in the absence of inhibitor. The IC50 was defined as the concentration of inhibitor that inhibited the activity of a given PR by 50%. The inhibition constant (Ki) value was derived from the IC50 value with the following equation for the competitive inhibitor: Ki = IC50/(1 + [S]/Km) (4), where [S] is the concentration of substrate.
Computer modeling.
The quadruple-mutant FIV PR I3732V/N5546M/M5647I/V5950I (the equivalent HIV numbering is in superscript) was built with the Accelrys package InsightII 2000 (Accelrys, San Diego, Calif.), with the crystal structure of the wild-type FIV PR solved by Wlodawer et al. with the HIV/FIV PRI TL-3 (20) (Protein Data Bank code 1B11). In the biopolymer module, the residues at positions 37, 55, 56, and 59 were replaced with the corresponding wild-type residues in HIV PR (I3732V, N5546M, M5647I, and V5950I, respectively). Each replaced side chain underwent a rotameric search to find its lowest energy conformation. The crystal structure of ritonavir with wild-type HIV PR is also available (16) (Protein Data Bank code 1HXW) and was used to superimpose the common catalytic aspartates from HIV PR onto the corresponding FIV PR aspartates. With this structural alignment, the coordinates of ritonavir were merged with those of the protein chains of both wild-type FIV PR and the quadruple-mutant of FIV PR. Both complexes retained the five active-site water molecules, one between the tips of the flaps and two pairs of water molecules adjacent to the R13 residue in each chain of the FIV PR.
One of the catalytic aspartic acids was protonated, while the other was left negatively charged. The two new complexes, ritonavir with wild-type FIV PR and ritonavir with the quadruple-mutant FIV PR, were assigned potentials with the CVFF force field parameters of Discover 3, and the complexes were subjected to short energy minimization with 100 iterations. The energy-minimized coordinates of ritonavir were discarded from each of the minimized complexes. The ritonavir coordinates from the cocrystallized ritonavir-HIV PR crystal structure, Protein Data Bank code 1HXW, were used to set up a flexible ligand for docking with AutoDock version 3.0.5 (23) to each of the FIV PR structures, either the wild type or the quadruple mutant. The backbone conformation of ritonavir was kept as in the HIV PR complex throughout all the dockings, but all of its side chains and the terminal thiozole and isopropylthiozolidine groups were allowed to change conformation (this gave 11 rotatable bonds).
A grid box of 71 by 61 by 61 grid points with a spacing of 0.375 angstroms centered on the PR active site was set up around each of the two FIV PRs with AutoDockTools, while the atomic affinity grid maps and the electrostatic potential grid maps were computed with AutoGrid version 3.0 (23). One hundred dockings of ritonavir to wild-type FIV PR and 100 dockings of ritonavir to the quadruple mutant of FIV PR were carried out with 50,000,000 energy evaluations of the Lamarckian genetic algorithm of AutoDock 3.0. Every docking experiment began with a random population of 150 different candidate binding modes. These 200 dockings were carried out in parallel with 40 nodes of an SGI Origin 2400 and 3800 server with a total 256 500-Mhz R14000 CPUs and 128 gigabytes of memory. The resulting AutoDock docked conformations were clustered into families of similar conformations, and their energies and cluster sizes were compared.
RESULTS
Construction and kinetic analysis of FIV chimeric mutants.
A series of mutant FIV PRs were constructed containing four (3732, 5546, 5647, and 5950 or 5546, 5647, 5950, and 9982) (the numbering of structurally equivalent position of HIV-1 PR is in superscript), five (3732, 5546, 5647, 5950, and 9982), six (3732, 5546, 5647, 5950, 9881, and 9982), or nine (F9s) substitutions (Fig. 1A and B). Previously reported mutant FIV PR constructs containing single or double substitutions were shown to change some substrate and inhibitor specificity toward that of HIV-1 PR (18, 21). However, they were insufficient to change the specificity dramatically, and no single or double mutation would cleave the HIV-1 fluorogenic substrate. Therefore, mutants that combined four, five, six, and nine substitutions were constructed.
The new multiple substitutions were composed mainly of combinations of mutations that had previously been shown to generate reasonable enzyme activity as well as contribute to changes in substrate and inhibitor specificity. These substitutions included I3732V of the active core region; N5546M, M5647I, and V5950I of the flap region; and I9881P and Q9982V of the 90s loop (Fig. 2A and B), all associated with the substrate binding pocket. Other relevant positions for substitutions in FIV PR are also shown in Fig. 1A. The structurally equivalent residues of HIV-1 PR are shown in the sequence alignment of Fig. 1B. We generated mutants with these multiple substitutions and purified the enzymes to homogeneity. The activity of these mutants was then evaluated against both FIV and HIV fluorogenic substrates, and enzyme kinetic parameters were determined (Table 1). The mutants had good enzyme activities, ranging from about 25% to 70% of that of wild-type FIV PR, based on the Kcat/Km values with the FIV fluorogenic substrate. However, the activities of these mutant PRs against the HIV fluorogenic substrate were not detectable in this assay. The HIV fluorogenic substrate contains Gln at P2′, which is known to be preferred by HIV-1 PR but not FIV PR (3). Apparently, the above substitutions failed to alter this specificity distinction between the two PRs.
FIG. 2.
Comparison of six structurally equivalent residues with side chains between FIV PR and HIV-1 PR. The roles of these residues in the determination of substrate and inhibitor specificity were analyzed extensively in this study. (A) I37, N55, M56, V59, I98, and Q99 of FIV PR. (B) The structurally equivalent V32, M46, I47, I50, P81, and V82 of HIV-1 PR, respectively.
TABLE 1.
Relative kinetic parameters of mutant FIV PRs with fluorogenic substrates
| Proteasea | FIV FSb
|
HIV FSc
|
||||
|---|---|---|---|---|---|---|
| Km (μM) | Kcat (min−1) | Kcat/Km | Km (μM) | Kcat (min−1) | Kcat/Km | |
| Wild-type FIV | 20.4 ± 1.8 | 3.01 ± 0.11 | 0.147 | —d | — | — |
| Mutants | ||||||
| 37/55/56/59 | 8.7 ± 0.4 | 0.89 ± 0.02 | 0.102 | — | — | — |
| 55/56/59/99 | 30.2 ± 2.2 | 1.32 ± 0.03 | 0.044 | — | — | — |
| 37/55/56/59/99 | 35.9 ± 1.9 | 1.33 ± 0.05 | 0.037 | — | — | — |
| 37/55/56/59/98/99 | 38.3 ± 3.2 | 2.25 ± 0.08 | 0.059 | — | — | — |
| F9s | 12.4 ± 0.6 | 0.53 ± 0.02 | 0.043 | 96.1 ± 17.7 | 0.05 ± 0.005 | 0.0005 |
| Wild-type HIV-1 | 8 ± 2 | 0.026 ± 0.006 | 0.003 | 2.2 ± 0.7 | 0.9 ± 0.2 | 0.4 |
37,13732V; N5546M; 56,M5647I; 59,V5950I; 98,19881P; 99,Q9982V. The equivalent HIV numbering is in superscript. F9s: 13732V/N5546M/M5647I/V5950I/L9780T/19881P/Q9982V/P10083N/L10184I.
FIV FS, fluorogenic substrate derived from the FIV CA/NC2 cleavage junction (ALTKVQ*VVQSKG).
HIV FS, fluorogenic substrate derived from the HIV-1 P2/NC cleavage junction (TIM*MQR).
—, no detectable activity in this assay.
Since the L9780T/I9881P/Q9982V/P10083N/L10184I mutant PR was shown to be reasonably active in earlier studies (21), this mutant was used as a basis for the preparation of an additional mutant (F9s) containing nine substitutions (3732, 5546, 5647, 5950, 9780, 9881, 9982, 10083, and 10184). The F9s mutant demonstrated low but detectable enzyme activity on the HIV fluorogenic substrate (Table 1) and was thus the first in the series of FIV PR mutants to demonstrate any activity against the HIV fluorogenic substrate.
Cleavage efficiency on the HIV-1 RT/integrase junction (RKIL/FLDG) and phage library peptides (SGIM/FESN) by FIV chimeric mutants.
To further test the degree to which these mutants had changed their substrate specificity to that of HIV-1 PR, cleavage of two specific peptides were assayed. One peptide corresponded to the HIV-1 RT/integrase junction, which is the most efficiently cleaved peptide among all the HIV-1 junction peptides (37). A second peptide selected from a phage library screen was also examined (3). This peptide was more efficiently cleaved than any natural HIV-1 junction peptide, including the HIV-1 RT/integrase junction peptide (3). Neither of these peptides was cleaved efficiently by wild-type FIV PR. The 3732/5546/5647/5950 and 3732/5546/5647/5950/9881/9982 mutant FIV PRs cleaved the HIV-1 RT/integrase peptide almost as efficiently as HIV-1 PR (Table 2). The 3732/5546/5647/5950/9982 mutant also showed improved cleavage efficiency. However, the 5546/5647/5950/9982 mutant did not show increased cleavage activity significantly on this peptide.
TABLE 2.
Altered substrate specificity of chimeric FIV PRs on peptidesa
| Proteaseb | Mean Cleavage efficiency (%) ± SD
|
|
|---|---|---|
| HIV RT/IN peptide (RKIL/FLDG) | Phage library peptide (SGIM/FESN) | |
| Wild-type FN | 16 ± 2 | 8 ± 1 |
| Mutants | ||
| 37/55/56/59 | 76 ± 9 | 33 ± 4 |
| 55/56/59/99 | 28 ± 5 | 30 ± 6 |
| 37/55/56/59/99 | 60 ± 8 | 51 ± 7 |
| 37/55/56/59/98/99 | 82 ± 8 | 42 ± 5 |
| F9s | 72 ± 11 | 36 ± 6 |
| Wild-type HIV-1 | 81 ± 10 | 93 ± 7 |
The substrate specificity was analyzed by assaying the cleavage efficiency of mutants on two peptides. These two peptides were very efficiently cleaved by HIV-1 PR but not by FIV PR. The assay was done with 100 nM PR and 10 min of incubation for the HIV RT/IN junction peptide and 75 nM PR and 5 min of incubation for the phage library peptide. The data are the means ± standard deviation of three independent experiments.
See Table 1, footnote a.
The RT/integrase junction (RKIL/FLDG) contains relatively large Ile and Leu residues at P2 and P2′, which are preferred by HIV-1 PR but not by FIV PR. Therefore, it is probable that the I37V substitution, which creates more space within the S2 and S2′ subsites, contributes significantly to the cleavage efficiency of the RT/integrase junction peptide. In cleavage attempts against the phage peptide (SGIM/FESN), all the mutants showed significantly increased cleavage activity relative to wild-type FIV PR (Table 2). However, in no case was cleavage as efficient as with HIV-1 PR. The phage peptide contains Glu at P2′ and thus is not cleaved by wild-type FIV PR (3, 4). These results are consistent with the observation that the HIV fluorogenic substrate, containing Gln at P2′, is not cleaved by FIV PR. The data again indicate that the preference at P2′ of the substrate appears to be mainly determined by the D30 in the S2 and S2′ sites of HIV-1 PR (equivalent to I35 of FIV PR).
Inhibition constant (Ki) of saquinavir, ritonavir, nefinavir, and JE-2147 against FIV chimeric PRs.
Three FDA-approved HIV-1 PR inhibitors (saquinavir, ritonavir, and nefinavir) and JE-2147 (42) were used to probe the inhibitor specificity of FIV PR. The inhibition constants (Ki) of these inhibitors were assayed against the mutant FIV PRs that contained single, double, or multiple substitutions (Table 3). None of the potent HIV-1 PR inhibitors were good inhibitors for wild-type FIV PR. The Ki values of all four drugs could not be determined because of their poor potency against wild-type FIV PR and poor solubility at high concentration. All inhibitors showed some drop in Ki values against the Q9982V single mutant, indicating that Q99 (equivalent to V82 of HIV-1 PR) is important in inhibitor selectivity for FIV PR. A more significant decrease in Ki value was observed with ritonavir against the I3732V/M5647I double mutant. These two substitutions are mainly involved in interactions within the S2 and S2′ subsites and generate more space that might result in the better binding of the P2 and P2′ moieties of ritonavir.
TABLE 3.
Ki values of potent HIV-1 PR inhibitors against mutant FIV PRsa
| Proteaseb |
Ki (nM)
|
|||
|---|---|---|---|---|
| Saquinavir | Ritonavir | Nelfinavir | JE-2147 | |
| Wild-type FIV | >45,000 | >45,000 | >30,000 | >45,000 |
| Mutants | ||||
| Q99V | 16,000 | 8,500 | 14,000 | 10,000 |
| I37V/M56I | >45,000 | 668 | 23,000 | 2,200 |
| I37V/V59I | >45,000 | 4,500 | 8,700 | 2,000 |
| 37/55/56/59 | >45,000 | 25 | 3,200 | 104 |
| 55/56/59/99 | 31,000 | 54 | 7,900 | 185 |
| 37/55/56/59/99 | 44,000 | 33 | 2,800 | 53 |
| 37/55/56/59/98/99 | 5,600 | 27 | 354 | 37 |
| F9s | 1,300 | 29 | 238 | 70 |
Data were obtained in the assay described in Materials and Methods. Ki values were derived from IC50 values with the equation Ki = IC50/(1 + [S]/[Km]).
37,13732V; 55,M5546M; 56,M5647I; V5950I; 98,19881P; 99,Q9982V. For F9s, see Table 1, footnote a.
The Ki values for most other inhibitors remained in the millimolar range against the single and double mutants. The Ki values against multiple-substitution mutants were consistent with a progressive decrease in value as a function of the number of substitutions. Saquinavir, with Asn at the P2 position, was a poor inhibitor for most of the mutants, consistent with the previous observation that Asn at P2 is preferred by HIV-1 PR but not FIV PR (26, 35). However, ritonavir was a good inhibitor, particularly against the 3732/5546/5647/5950 mutant, with a Ki value of 25 nM (an 1,800-fold decrease of value relative to wild-type FIV PR). This indicates that a mutant FIV PR with only four substitutions showed inhibitor specificity very similar to that of HIV-1 PR. JE-2147 was also a good inhibitor, with a low (nanomolar) Ki against several mutant FIV PRs. The results showed that the inhibitor specificity of FIV PR could be altered drastically by substantially fewer changes than required for alteration of substrate specificity.
Basis for preference for either Gln or Asn at P1 position in a substrate.
Previous studies on substrate specificity with a phage peptide display library showed that FIV PR tolerates a polar Gln or Asn residue at P1 and a Ser residue at P1′ in substrates, whereas HIV-1 PR prefers hydrophobic amino acids at these sites (4). A similar amino acid preference was observed in the FIV capsid/NC2 and dUTPase/integrase cleavage junctions (TKVQ/VVQS and TGVF/SSWV, respectively), which are cleaved efficiently by FIV PR but poorly by HIV-1 PR. Computer modeling suggested that the Gln at position 99 of FIV PR (Val 82 of HIV-1 PR) might play an essential role in determining the selectivity at the P1 and P1′ positions in a substrate. The molecular determinants of specificity were assessed by assaying the cleavage efficiency of a panel of mutant FIV PRs on three synthetic peptides which contained either Gln or Asn at P1(Table 4). These peptides were cleaved very efficiently by FIV PR but very poorly by HIV-1 PR. One peptide represents the FIV capsid/NC2 cleavage junction (TKVQ/VVQS). Two others were selected from a phage display library (4). The results showed that Q9982V alone or mutant PRs containing Q9982V reduced cleavage efficiency against all three substrates, whereas cleavage by the I3732V/N5546M/M5647I/V5950I mutant was more efficient. The data indicate that Q99 (V82 of HIV-1 PR) is the major determinant in FIV PR for the residue preference at the P1 position.
TABLE 4.
Determinant of preference for Gln or Asn at the P1 position in a substratea
| Proteaseb | Mean cleavage efficiencya (%) ± SD
|
||
|---|---|---|---|
| FIV CA/NC2 peptide (TKVQ/VVQS) | P1-Gln peptide (SRVQ/VVNG) | P1-Asn peptide (SRVN/VVNG) | |
| Wild-type FIV | 70.8 ± 6.2 | 76.2 | 87.8 |
| Mutants | |||
| Q99V | 37.1 ± 3.0 | 19.2 | 25.7 |
| 37/55/56/59 | 63.1 ± 4.9 | 78.6 | 87.0 |
| 55/56/59/99 | 16.9 ± 2.1 | 5.8 | 17.3 |
| 37/55/56/59/99 | 23.8 ± 4.6 | 14.3 | 26.8 |
| 37/55/56/59/98/99 | 27.6 ± 3.1 | 10.5 | 28.0 |
| Wild-type HIV | 15.8 ± 5.1 | 10.1 | 9.7 |
The determinant was probed by assaying the cleavage efficiency of Q9982V and a panel of chimeric mutants on three peptides. These peptides contained either Gln or Asn at P1 and were cleaved very efficiently by FIV PR but poorly by HIV-1 PR. The data from three different peptides showed similar results; the Q9982V mutant and any mutant containing the Q9982V substitution drastically decreased the cleavage efficiency on these three peptides. All assays were done with 150 nM PR. The incubation time was 45 min for both the FIV CA/NC2 junction peptide and the P1-Gln peptide and 30 min for the P1-Asn peptide.
See Table 1, footnote a.
Basis for preference for Ser at the P1′ position in a substrate.
The determinant of P1′ specificity was assayed with the same panel of mutant FIV PRs in Table 4 as well as two synthetic peptides containing Ser at P1′ as substrates (Table 5). One peptide represents the FIV dUTPase/integrase cleavage junction (TGVF/SSWV), and the other peptide was selected from a phage display library for specificity for FIV PR. The results showed that Q9982V did not reduce the cleavage efficiency against these peptides. However, mutant PRs containing N5546M, M5647I, and V5950I had drastically decreased cleavage efficiencies. The determinants of the P1′ specificity of substrates were further probed with another panel of mutants, including I3732V, I3732V/N5546M, I3732V/M5647I, and I3732V/V5950I. The data showed that the preference was determined mainly by M56 (I47 of HIV-1 PR) and, to a lesser degree, V59 (I50 of HIV-1 PR) (Table 6).
TABLE 5.
Determinant of preference for Ser at P1′ position in a substratea
| Proteaseb | Mean cleavage efficiency (%) ± SD
|
|
|---|---|---|
| FIV DU/IN peptide (TGVF/SSWV) | P1′-S peptide (SGVF/SVNG) | |
| Wild-type FIV | 76.3 ± 11.9 | 80.4 |
| Mutants | ||
| Q99V | 67.9 ± 6.6 | 82.2 |
| 37/55/56/59 | 2.8 ± 0.9 | 29.9 |
| 55/56/59/99 | 1.8 ± 1.0 | 24.8 |
| 37/55/56/59/99 | 1.6 ± 0.8 | 31.9 |
| 37/55/56/59/98/99 | 2.3 ± 1.1 | 25.7 |
| Wild-type HIV | 1.5 ± 1.0 | 5.0 |
The determinant was probed by assaying the cleavage activity of the panel of mutants as described in Table 4 on two peptides that contain Ser at the P1′ position. These peptides were cleaved cleaved efficiently by FIV PR but poorly by HIV-1 PR. All assays were done with 150 nM PR. The incubation time for the FIV dUTPase/integrase (DU/IN) junction peptide was 25 min and for phage peptide (SGVF/SVNG) was 30 min.
See Table 1, footnote a.
TABLE 6.
Determinant of preference for Ser at P1′ position in a substratea
| Protease | Mean cleavage efficiency (%) ± SD with the FIV DU/IN peptide (TGVF/SSWV) |
|---|---|
| Wild-type FIV | 63.3 ± 8.3 |
| Mutants | |
| I37V | 68.5 ± 7.1 |
| I37V/N55M | 57.4 ± 7.7 |
| I37V/M56I | 31.5 ± 5.1 |
| I37V/V59I | 41.5 ± 5.2 |
| Wild-type HIV | 3.1 ± 1.0 |
The determinant was further probed with another panel of mutants that included I3732V, I3732V/N5546M, I3732V/M5647I, and I3732V/V5950I. The cleavage efficiency of these mutants on the FIV dUTPase/integrase (DU/IN) junction peptide was assayed. The data showed that the mutants containing the M5647I or V5950I substitution decreased the cleavage efficiency significantly.
Basis for loss of activity of I57G-containing mutants.
Previous results have shown that the activity of mutant FIV PRs containing I5748G or I5748G and G6253F were barely measurable (21). The I57 (G48 of HIV-1 PR) residue is located in the S1 and S1′ subsites and interacts directly with the substrate. In order to further explore the possible role of this determinant in specificity, a K6354I substitution was added (Fig. 3). The addition of G6253F and K6354I in combination with I5748G to F9s (now F12s) restored activity against the FIV fluorogenic substrate (Table 7). The kinetic analysis of the F12s mutant on the FIV fluorogenic substrate showed a Km of 12.9 ± 1.5 mM and a Kcat of 1.07 ± 0.05 min−1, which is less active than wild-type FIV PR. However, it had a relatively low activity on the HIV fluorogenic substrate (data not shown). Residue I63 is conserved among HIV-1 PRs as well as HIV-2, SIV, and Rous sarcoma virus PRs but not FIV PR. Furthermore, a major mutation of HIV-1 associated with resistance to saquinavir, G48V (position 57 of FIV PR), is frequently associated with an I54V (position 63 of FIV PR) mutation, indicating the close interaction between residues 48 and 54 (positions 57 and 63 of FIV PR).
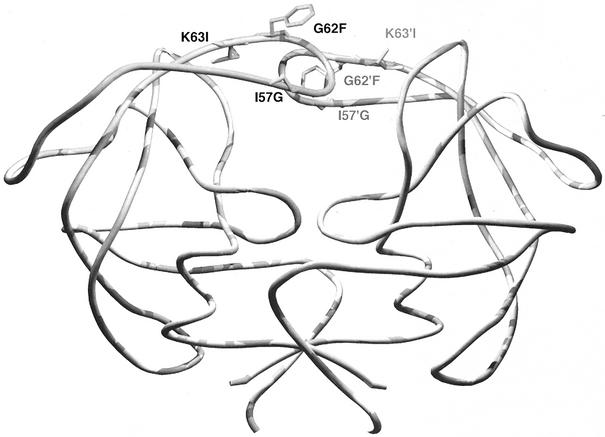
FIG. 3.
Addition of G6253F and K6354I substitutions restored activity to I5748G-containing mutants. G6253F and K6354I are located at the top of the flap and outside the binding pocket (see Fig. 1). The I5748G substitution resulted in loss of a β-branched side chain and gain of a flexible Gly around the tip of the flap. Computer modeling indicated that the substitution resulted in a disordered flap, likely responsible for the observed loss of activity. However, the two substitutions provide a large phenyl ring and β-branched side chain that could stabilize the flap and, in turn, restore the activity.
TABLE 7.
Substitutions and activities of 15748G and 13530D mutant FIV PRs
| Protease | Position and amino acid
|
Activitya
|
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FIV FS | HIV FS | VI | ||||||||||||||||||||||||||
| FIV | 35 | 37 | 53 | 54 | 55 | 56 | 57 | 59 | 62 | 63 | 64 | 65 | 66 | 67 | 88 | 90 | 97 | 98 | 99 | 100 | 101 | 105 | 106 | 107 | 108 | |||
| I | I | R | Q | N | M | I | V | G | K | R | G | T | N | N | C | L | I | Q | P | L | D | N | M | I | ||||
| HIV | D | V | P | K | M | I | G | I | F | I | K | V | R | Q | T | L | T | P | V | N | I | N | L | L | T | |||
| 30 | 32 | 44 | 45 | 46 | 47 | 48 | 50 | 53 | 54 | 55 | 56 | 57 | 58 | 74 | 76 | 80 | 81 | 82 | 83 | 84 | 88 | 89 | 90 | 91 | ||||
| Mutants | ||||||||||||||||||||||||||||
| F9s | V | M | I | I | T | P | V | N | I | ND | ND | |||||||||||||||||
| F12s | V | M | I | G | I | F | I | T | P | V | N | I | +++ | + | ||||||||||||||
| F12s′ | D | V | K | M | I | G | I | T | P | V | N | I | ND | ND | ||||||||||||||
| F13s | D | V | M | I | G | I | F | I | T | P | V | N | I | ND | ND | |||||||||||||
| F14s | D | V | K | M | I | G | I | F | I | T | P | V | N | I | ND | ND | ||||||||||||
| F15s | D | V | P | K | M | I | G | I | F | I | T | P | V | N | I | ND | ND | |||||||||||
| F15s′ | D | V | K | M | I | G | I | F | I | L | T | P | V | N | I | ND | ND | |||||||||||
| F16s | D | V | K | M | I | G | I | F | I | L | T | P | V | N | I | N | ND | ± | ||||||||||
| F17s | D | V | K | M | I | G | I | F | I | T | L | T | P | V | N | I | N | ± | + | + | ||||||||
| F20s | D | V | K | M | I | G | I | F | I | K | V | R | Q | L | T | P | V | N | I | N | ND | ± | ||||||
| F21s | D | V | K | M | I | G | I | F | I | K | V | R | Q | T | L | T | P | V | N | ND | + | + | ||||||
| F23s | D | V | K | M | I | G | I | F | I | K | V | R | Q | L | T | P | V | N | I | N | L | L | T | ND | ND | |||
| F24s | D | V | K | M | I | G | I | F | I | K | V | R | Q | T | L | T | P | V | N | I | N | L | L | T | ND | ± | ||
Activity of mutants was assayed on both FIV and HIV-1 fluorogenic substrates (FS), which represent FIV CA/NC2 and HIV-1 P2/NC cleavage junctions. VI, fluorogenic substrate representing the HIV-1 CA/P2 cleavage junction. ND, activity not detectable after monitoring cleavage for 6 min; ±, marginal activity; +, <5% of wild-type activity; +++, >50% of wild-type activity.
Basis for loss of activity in I3530D-containing mutants.
Previous attempts to prepare an active FIV PR containing an I3530D substitution, alone and in several combinations, failed. Upon examination of the three-dimensional structure, it was noted that several residues in close proximity to I35 of FIV PR (D30 of HIV-1 PR) are also unique in HIV-1 PR. These residues are not in the active site or directly interactive with inhibitors or substrates and are located mainly at three regions of PR (Fig. 1). One region is at the base of the flap, involving primarily an interaction with Q54 (K45 of HIV-1 PR), and to a lesser degree, with the G65 to N67 region. The second region is located around N88 to C90. The final region, D105 to I108, is near the dimer interface. R5344P, Q5445K, C9076L, and D10588N were first introduced into the inactive F13s mutant to generate F14s, F15s, F15s′, and F16s, respectively (Table 7). The β-carbon distances between Q54, C90, D105, and I35 are 7.6 Å, 5.7 Å, and 5.2 Å, respectively. The R5344P mutant was attempted because of the close interaction with Q54K.
The addition of R5344P, Q5445K, and C9076L generated mutant PRs with no detectable activity (Table 7). However, a marginal activity against the HIV fluorogenic substrate was observed in F16s, which resulted from the addition of D10588N. Nevertheless, the data suggested that additional surrounding secondary substitutions might be needed for further recovery of activity. N8874T was then introduced into F16s to generate F17s. The β-carbon distance between N88, I35, and D105 were 7.2 Å and 5.2 Å, respectively.
The substitutions R6455K, G6556V, T6657R, and N6758Q, which are located in the C-terminal part of the flap region, were further introduced to generate F20s. Despite close interaction, particularly between N67 and I35, only marginal activity was observed against the HIV fluorogenic substrate. N8874T was also introduced into F20s to generate the F21s mutant. Again, this substitution restored some activity on the HIV fluorogenic substrate, similar to that observed with the F17s mutant. Further attempts to recover additional activity by introduction of N10689L, M10790L, or I10891T were not successful. The results indicated that addition of D10588N or N8874T restored detectable activity in mutants containing the I3530D substitution. However, the difficulty of further recovery of activity highlights the importance of interaction between residues outside the substrate-binding pocket.
Inhibitor specificity of F12s and F21s mutants.
Three FDA-approved drugs (saquinavir, ritonavir, and nefinavir) and JE-2147 were used to analyze the inhibitor specificity of the F12s and F21s chimeric mutants. The Ki values of all inhibitors improved markedly against these two mutants (Table 8). The findings indicated that these two mutant FIV PRs have inhibitor specificities that are very similar but not identical to that of HIV-1 PR.
TABLE 8.
Ki values of potent HIV-1 PR inhibitors against the F12s and F21s mutantsa
| Proteaseb |
Ki (nM)
|
|||
|---|---|---|---|---|
| Saquinavir | Ritonavir | Nelfinavir | JE-2147 | |
| FIV | ||||
| Wild type | >45,000 | >45,000 | >30,000 | >45,000 |
| F12s | 37 | 15 | 29 | NAc |
| F21s | 25 | 44 | 16 | 18 |
| HIV-1 | 3 | 2 | 3 | 3 |
Data were obtained at pH 5.6 in 0.1 M NaH2PO4-0.1 M sodium citrate-1.5 M NaCl-0.1 mM dithiothreitol-5% glycerol. The Ki values were derived from IC50 values with the equation Ki = IC50/(1 + [S]/Km).
F12s, 3732/5546/5647/5748/5950/6253/6354/9780/9881/9982/10083/10184; F21s, F12s plus 3530/5445/6455/6556/6657/6758/8874/9076/10588.
NA, not assayed.
DISCUSSION
We used the FIV and HIV-1 PR system as a tool for mutational analysis in order to study the substrate and inhibitor specificities of retroviral PRs. With a similar strategy, a series of studies on the substrate specificity of Rous sarcoma virus PR have been reported (5, 13, 27). These studies showed that Rous sarcoma virus PR can be engineered to drastically alter the substrate and inhibitor specificity. In an early study, we identified certain residues within the substrate binding pocket that are important in contributing to the distinct specificity of FIV PR (21). In this study, we further explored the role of these residues combined with peripheral substitutions in determining substrate and inhibitor specificity. The findings show that the degree to which the substrate specificity of FIV PR can be changed completely depends on the sequence context of the substrate assayed.
In this study, two of the chimeric FIV PRs with I3732V, N5546M, M5647I, and V5950I or I3732V, N5546M, M5647I, V5950I, I9881P, and Q9982V substitutions showed the same cleavage efficiency as HIV-1 PR on the HIV-1 RT/integrase (RKIL/FLDG) junction peptide. The peptide consists mainly of hydrophobic residues from the P2 to the P2′ position. The hydrophobicity and van der Waals volume of the residues appear to play a major role in the determination of specificity. However, these two FIV chimeric PRs were unable to efficiently cleave an HIV PR-selected phage peptide (SGIM/FESN) that contained a charged Glu at P2′. The results suggest that other substitutions are required to facilitate selectivity at P2′. Neither wild-type FIV PR nor any of the chimeric FIV PRs tested cleaved any substrate efficiently if it contained charged Glu or polar Gln at the P2′ position. Two fluorogenic substrates representing the HIV-1 capsid/P2 (ARVL/AEAM) and P2/nucleocapsid (ATIM/MQRG) junctions were not cleaved efficiently by any of the chimeric FIV PRs. The results are consistent with previous observations that FIV PR has distinct P2′ specificity (4, 21).
Previous observations indicated that FIV PR prefers a hydrophobic P2 or P2′ residue, such as Val, as opposed to the Asn at P2 and Gln at P2′ that are favored by HIV-1 PR (4, 35). The observations indicate the S2 and S2′ subsites play a critical role in distinguishing the substrate specificities of FIV from HIV-1 PR. The preferences for distinct P2 and P2′ residues were confirmed by assaying the cleavage efficiency of FIV and HIV-1 PRs on a panel of peptides that contained various amino acid substitutions, including Val or Asn at P2 and Val, Gln, or Glu at P2′ (data not shown). Modeling indicated that the I3530D substitution, which is located within the S2 and S2′ subsites, might be the major determinant for the P2 and P2′ preference. The FIV I35/HIV-1 D30 residue has been implicated as being involved in defining the binding specificity between FIV and HIV-1 PRs in a protein-ligand study (6). I3530D is the only substitution inside the substrate-binding pocket that failed to be engineered successfully into FIV PR and generate active enzyme. Findings in the present study support the role of position 35 (30 of HIV-1 PR) in determining P2 and P2′ preference.
We further investigated the role of combined substitutions in the determination of inhibitor specificity. The results show that the inhibitor specificity of FIV PR could be drastically altered with only four substitutions in the active site. The Ki values of ritonavir and JE-2147 against the I3732V/N5546M/M5647I/V5950I mutant decreased from unmeasurable to 25 nM and 104 nM, respectively. The findings indicate that these residues play an essential role in the alteration of specificity for a particular type of inhibitor. Interestingly, the same four substitutions were responsible for the complete alteration of substrate specificity on the HIV-1 RT/integrase junction peptide. The observation indicates that some overlap between residues contributes to both inhibitor and substrate specificity.
The resulting AutoDock (23) docked conformations of ritonavir with wild-type FIV PR and ritonavir with the quadruple-mutant FIV PR were clustered into families of similar binding modes, with a root mean squares deviation (RMSD) clustering tolerance of 1.50 angstroms. In other words, all conformations within a given family or cluster had a positional RMSD of all the atoms of 1.50 angstroms or less. For ritonavir with wild-type FIV PR, 36% of the dockings clustered into the lowest energy family; this cluster had a mean energy of −18.78 kcal/mol, with the lowest energy in this cluster being −18.94 kcal/mol (lower energies mean tighter binders). For ritonavir with the quadruple mutant (37/55/56/59) of FIV PR, 52% of the dockings clustered into the lowest energy family, and this cluster had a mean energy of −20.16 kcal/mol, while the lowest energy was −20.55 kcal/mol. Thus, it was clear from the repeated AutoDock experiments that AutoDock consistently ranked ritonavir as preferential to bind to the “HIV-inized” quadruple mutant of FIV PR, rather than the wild-type FIV PR (Fig. 4). Furthermore, it is interesting that the bound conformation of the ritonavir in the wild-type PR was more variable than when bound to the mutant FIV PR. As can be seen from the experimentally determined Ki values (Table 3), that for ritonavir-wild type was >45,000 nM, while that for ritonavir-37/55/56/59 was 25 nM. Thus, AutoDock's ranking agreed with the experimental values.
FIG. 4.
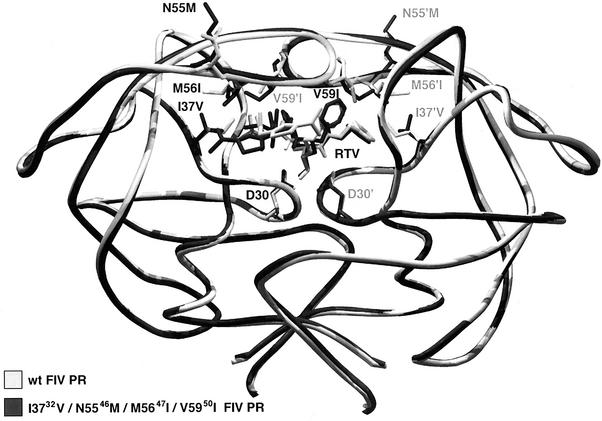
Results of docking ritonavir (RTV), an HIV PR inhibitor, to wild-type and “HIV-inized” FIV PR. The conformation with the best docking energy in each case is shown: the wild-type (wt) FIV PR is in light grey, while the HIV-inized quadruple mutant of FIV PR (I3732V/N5546M/M5647I/V5950I) is shown in dark grey, as are the corresponding conformations of ritonavir. Note that the catalytic aspartic acids are also visible at the base of the active site. The conformation of ritonavir is similar from sites P2′ to P2 but differs significantly at the P3′ site.
The side chain of N55 is located at the outer face of the S4 and S4′ subsites and is not in direct contact with ritonavir, which lacks a P4 or P4′ group. The V59 side chain has potential interaction with the P1 and P1′ and P2 and P2′ sites of the inhibitor. The I37 and M56 residues are adjacent to one another and are mainly involved in the interactions of the S2 and S2′ subsites. The mutation of M56 to isoleucine introduces a β-branched side chain, and the added methyl at the β-branch ends up at the optional distance to stabilize the valine side chain at P2 and the thiozole ring at P2′ of the asymmetric ritonavir. Previous studies suggested that I3732V and M5647I could potentially influence the interaction between the thiozole moiety at P2′ of VL-346 and the S2′ subsite of FIV PR (21). Ritonavir has the same chemical group at P2′ as VL-346. Computer modeling indicated that there is a subtle but noticeable change of overall conformation of the chimeric I3732V/N5546M/M5647I/V5950I mutant compared to the wild-type FIV PR. The change made the interaction between the mutant PR and ritonavir, particularly that between the thiozolidine group and I3732V and M5647I, tighter. This might explain the much lower Ki value of ritonavir. Ritonavir extends from the P3 to P2′ positions and is longer than any other inhibitor used in this study, which occupies four positions. This result is consistent with the previous observation that FIV PR prefers longer substrates (11, 30).
The introduction of adjacent substitutions G6253F and K6354I resulted in a mutant FIV PR (F12s) with good activity (Table 7). G62 and K63 are located on the top of the flap but remain outside the binding pocket and are very different from the corresponding F53 and I54 of HIV-1 PR in size and polarity (Fig. 3). In addition, the G62 and K63 of FIV PR are very close and interact with N55, M56, and I57 to form the central part of the flap and may affect the vital interaction of I57 in the binding pocket. Importantly, the two corresponding positions of HIV-1 PR are associated with drug resistance. F12s is the first I5748G-containing mutant PR that is active against both the FIV and HIV-1 fluorogenic substrates. The I5748G change resulted in loss of a β-branched side chain and added one more Gly to the flap tip region in which there are already four glycines in FIV PR.
Retroviral PRs usually have three to four but not five glycines around the flap tip (40). Loss of Ile could destabilize the already dynamic flap and result in the loss of interaction between the flap and substrate. This could explain why no activity was detected in the I5748G-containing mutants. Molecular simulation confirmed the likelihood of disordered flaps, and addition of G6253F and K6354I could stabilize the flap. In particular, K6354I adds a β-branched residue that might compensate for the loss of the other β-branched I57. Interestingly, in the HIV-1 drug resistance database, G48V (I57 of FIV PR) often occurs in conjunction with mutations at I54 (K63 of FIV PR). I54 in the HIV-1 PR is one of the hot spots for mutations associated with drug resistance, and I54V and I54T are the most frequent mutations associated with saquinavir treatment (31). They are all β-branched amino acids, consistent with the need for a β-branched amino acid to maintain the stability of the local structure.
A future use for certain chimeric FIV PRs will be for construction of additional mutants containing amino acid residues associated with drug resistance, in order to study the molecular basis of resistance. Broad-based mutants that are capable of cleaving both FIV and HIV-1 viral junction peptides and support the production of infectious and mature viral particles can be employed in a cell-based assay to act as targets for secondary and tertiary drug screens. Better understanding of the specificity of both PRs will facilitate the design and generation of a panel of FIV mutant PRs that can be used for screening inhibitors both in vitro (cell free) and ex vivo (cell-based assay systems). The data from the mutagenesis studies indicate that I3530D and Q9982V might be the most important residues that dictate the specificity differences between FIV and HIV-1 PRs. These positions largely determine the residue preference at the S1 and S1′ and at the S2 and S2′ subsites of these two PRs. FIV PR appears to favor a hydrophobic nature at the S2 and S2′ subsites, as opposed to the hydrophilic nature of HIV-1 PR. In contrast, HIV-1 PR maintains a hydrophobic nature at the S1 and S1′ subsites, as opposed to the polar residues tolerated by FIV PR.
The introduction of I5748G, G6253F, and K6354I into F9s not only restored the activity of the I5748G mutant (F12s) but also altered its inhibitor specificity toward saquinavir and nefinavir. In particular, the Ki value of saquinavir decreased from 1,300 nM to 37 nM (about a 35-fold change). Interestingly, a drug resistance mutation (G48V) in HIV-1 PR (equivalent to I57 of FIV PR) occurs primarily in patients receiving saquinavir (10). Mutations often occur at positions 54 and 82, equivalent to positions 63 and 99 of FIV PR. The data suggest that G48 of HIV-1 PR, which has no side chain, might tolerate the bulky decahydro-isoquinoline group at the P1′ position of saquinavir, whereas Val at position 48 will not. Therefore, the I5748G substitution at the equivalent position in FIV PR drastically changed the inhibitor specificity of FIV PR toward that of wild-type HIV-1 PR.
The low Ki values against all the inhibitors tested indicated a dramatic change in the inhibitor specificity of F21s. The specificity was very similar to that of HIV-1 PR but not identical. The data also indicated that the specificity of PR can be dramatically altered by exchanging the residues in the substrate-binding pocket. However, in order to completely alter the specificity as well as retain the catalytic activity, other secondary substitutions outside the binding pocket are important. Because of the complexity of the substitutions of F21s, it is difficult to explain the recovery of lost activity resulting from the I3530D mutation. However, the N8874T and D10588N substitutions appear to help in recovering some activity. The I3530D substitution might result in a disordered local structure due to the drastic change to charged Asp (D35), which results in a close interaction with D105. The addition of the D10588N substitution might thus counteract the charge. The hydroxyl group of T88 might provide both a hydrogen bond donor and an acceptor, resulting in stronger interaction with nearby residues. Interestingly, the majority of mutations in HIV PR that are associated with inhibitors at this position involve serine, which also has a hydroxyl group (31). In addition, the nefinavir-resistant D30N (I35 of FIV PR) mutation of HIV-1 PR occurs frequently in association with N88D (D105 of FIV PR) (31, 34), indicating a strong association between these residues.
The study of FIV and HIV-1 PR chimeras has provided novel insights into the structural basis of substrate and inhibitor specificity. The changes that have a drastic influence on inhibitor binding may have relatively little effect on substrate specificity. Considerable overlap exists between residues associated with substrate specificity and residues involved in drug resistance development. Changes distal to the active site may also influence substrate and inhibitor specificity. These results offer valuable information for refining inhibitor specificity and may therefore greatly assist in the development of broad-based PR inhibitors against drug-resistant mutants.
Acknowledgments
We thank Alex Wlodawer and Alla Gustchina of the National Cancer Institute, Frederick, Md., for crystal structures as well as valuable discussion and advice; Michael Fitzgerald, Duke University, for synthesis of fluorogenic substrates; Chi-Huey Wong, Philip Dawson, and Bruce Torbett of the Scripps Research Institute for use of certain laboratory facilities as well as valuable discussions; and Jackie Wold for administrative assistance.
This work was supported by grants P01 GM48870 (A.J.O.) and R01 AI40882 (J.H.E.) from the National Institutes of Health.
REFERENCES
- 1.Aiyar, A., and J. Leis. 1993. Modification of the megaprimer method of PCR mutagenesis: improved amplification of the final product. BioTechniques 14:366-369. [PubMed] [Google Scholar]
- 2.Alewood, P., D. Alewood, L. Miranda, S. Love, W. Meutermans, and D. Wilson. 1997. Rapid in situ neutralization protocols for Boc and Fmoc solid phase chemistries. Methods Enzymol. 289:14-29. [DOI] [PubMed] [Google Scholar]
- 3.Beck, Z. Q., L. Hervio, P. E. Dawson, J. H. Elder, and E. L. Madison. 2000. Identification of efficiently cleaved substrates for HIV-1 protease with a phage display library and use in inhibitor development. Virology 274:391-401. [DOI] [PubMed] [Google Scholar]
- 4.Beck, Z. Q., Y. C. Lin, and J. H. Elder. 2001. Molecular basis for the relative substrate specificity of human immunodeficiency virus type 1 and feline immunodeficiency virus proteases. J. Virol. 75:9458-9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cameron, C. E., T. W. Ridky, S. Shulenin, J. Leis, I. T. Weber, T. Copeland, A. Wlodawer, H. Burstein, D. Bizub-Bender, and A. M. Skalka. 1994. Mutational analysis of the substrate binding pockets of the Rous sarcoma virus and human immunodeficiency virus-1 proteases. J. Biol. Chem. 269:11170-11177. [PubMed] [Google Scholar]
- 6.Dominy, B. N., and C. L. Brooks 3rd. 1999. Methodology for protein-ligand binding studies: application to a model for drug resistance, the HIV/FIV protease system. Proteins 36:318-331. [DOI] [PubMed] [Google Scholar]
- 7.Elder, J. H., G. A. Dean, E. A. Hoover, J. A. Hoxie, M. H. Malim, L. Mathes, J. C. Neil, T. W. North, E. Sparger, M. B. Tompkins, W. A. Tompkins, J. Yamamoto, N. Yuhki, N. C. Pedersen, and R. H. Miller. 1998. Lessons from the cat: feline immunodeficiency virus as a tool to develop intervention strategies against human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 14:797-801. [DOI] [PubMed] [Google Scholar]
- 8.Elder, J. H., and T. R. Phillips. 1995. Feline immunodeficiency virus as a model for development of molecular approaches to intervention strategies against lentivirus infections. Adv. Virus Res. 45:225-247. [DOI] [PubMed] [Google Scholar]
- 9.Elder, J. H., M. Schnolzer, C. S. Hasselkus-Light, M. Henson, D. A. Lerner, T. R. Phillips, P. C. Wagaman, and S. B. Kent. 1993. Identification of proteolytic processing sites within the Gag and Pol polyproteins of feline immunodeficiency virus. J. Virol. 67:1869-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ermolieff, J., X. Lin, and J. Tang. 1997. Kinetic properties of saquinavir-resistant mutants of human immunodeficiency virus type 1 protease and their implications in drug resistance in vivo. Biochemistry 36:12364-12370. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald, M. C., G. S. Laco, J. H. Elder, and S. B. Kent. 1997. A continuous fluorometric assay for the feline immunodeficiency virus protease. Anal. Biochem. 254:226-230. [DOI] [PubMed] [Google Scholar]
- 12.Flexner, C. 1998. HIV-protease inhibitors. N. Engl. J. Med. 338:1281-1292. [DOI] [PubMed] [Google Scholar]
- 13.Grinde, B., C. E. Cameron, J. Leis, I. T. Weber, A. Wlodawer, H. Burstein, D. Bizub, and A. M. Skalka. 1992. Mutations that alter the activity of the Rous sarcoma virus protease. J. Biol. Chem. 267:9481-9490. [PubMed] [Google Scholar]
- 14.Grinde, B., C. E. Cameron, J. Leis, I. T. Weber, A. Wlodawer, H. Burstein, and A. M. Skalka. 1992. Analysis of substrate interactions of the Rous sarcoma virus wild-type and mutant proteases and human immunodeficiency virus-1 protease with a set of systematically altered peptide substrates. J. Biol. Chem. 267:9491-9498. [PubMed] [Google Scholar]
- 15.Henderson, P. J. F. 1972. A linear equation that describes the steady state kinetics of enzymes and subcellular particles interacting with tightly bound inhibitors. Biochem. J. 127:321-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kempf, D. J., K. C. Marsh, J. F. Denissen, E. McDonald, S. Vasavanonda, C. A. Flentge, B. E. Green, L. Fino, C. H. Park, and X. P. Kong. 1995. ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc. Natl. Acad. Sci. USA 92:2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laco, G. S., C. Schalk-Hihi, J. Lubkowski, G. Morris, A. Zdanov, A. Olson, J. H. Elder, A. Wlodawer, and A. Gustchina. 1997. Crystal structures of the inactive D30N mutant of feline immunodeficiency virus protease complexed with a substrate and an inhibitor. Biochemistry 36:10696-10708. [DOI] [PubMed] [Google Scholar]
- 18.Lee, T., G. S. Laco, B. E. Torbett, H. S. Fox, D. L. Lerner, J. H. Elder, and C. H. Wong. 1998. Analysis of the S3 and S3′ subsite specificities of feline immunodeficiency virus (FIV) protease: development of a broad-based protease inhibitor efficacious against FIV, SIV, and HIV in vitro and ex vivo. Proc. Natl. Acad. Sci. USA 95:939-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, T., V. D. Le, D. Lim, Y. C. Lin, G. M. Morris, A. L. Wong, A. J. Olson, J. E. Elder, and C. H. Wong. 1999. Development of a new type of protease inhibitors, efficacious against FIV and HIV variants. J. Am. Chem. Soc. 121:1145-1155. [Google Scholar]
- 20.Li, M., G. M. Morris, T. Lee, G. S. Laco, C. H. Wong, A. J. Olson, J. H. Elder, A. Wlodawer, and A. Gustchina. 2000. Structural studies of FIV and HIV-1 proteases complexed with an efficient inhibitor of FIV protease. Proteins 38:29-40. [DOI] [PubMed] [Google Scholar]
- 21.Lin, Y. C., Z. Beck, T. Lee, V. D. Le, G. M. Morris, A. J. Olson, C. H. Wong, and J. H. Elder. 2000. Alteration of substrate and inhibitor specificity of feline immunodeficiency virus protease. J. Virol. 74:4710-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, V. 2001. International perspectives on antiretroviral resistance. Resistance to protease inhibitors. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):S34-S50. [DOI] [PubMed] [Google Scholar]
- 23.Morris, G. M., D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew, and A. J. Olson. 1998. Automated docking with a lamarckian genetic alogorithm and an empirical binding free energy. J. Comput. Chem. 19:1639-1662. [Google Scholar]
- 24.Pettit, S. C., M. D. Moody, R. S. Wehbie, A. H. Kaplan, P. V. Nantermet, C. A. Klein, and R. Swanstrom. 1994. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 68:8017-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettit, S. C., N. Sheng, R. Tritch, S. Erickson-Viitanen, and R. Swanstrom. 1998. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv. Exp. Med. Biol. 436:15-25. [DOI] [PubMed] [Google Scholar]
- 26.Poorman, R. A., A. G. Tomasselli, R. L. Heinrikson, and F. J. Kezdy. 1991. A cumulative specificity model for proteases from human immunodeficiency virus types 1 and 2, inferred from statistical analysis of an extended substrate data base. J. Biol. Chem. 266:14554-14561. [PubMed] [Google Scholar]
- 27.Ridky, T. W., D. Bizub-Bender, C. E. Cameron, I. T. Weber, A. Wlodawer, T. Copeland, A. M. Skalka, and J. Leis. 1996. Programming the Rous sarcoma virus protease to cleave new substrate sequences. J. Biol. Chem. 271:10538-10544. [DOI] [PubMed] [Google Scholar]
- 28.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 29.Schinazi, R. F., B. A. Larder, and J. W. Mellors. 1997. Mutations in retroviral genes associated with drug resistance. Int. Antiviral News 5:129-142. [Google Scholar]
- 30.Schnolzer, M., H. R. Rackwitz, A. Gustchina, G. S. Laco, A. Wlodawer, J. H. Elder, and S. B. Kent. 1996. Comparative properties of feline immunodeficiency virus (FIV) and human immunodeficiency virus type 1 (HIV-1) proteinases prepared by total chemical synthesis. Virology 224:268-275. [DOI] [PubMed] [Google Scholar]
- 31.Shafer, R. W., D. R. Jung, B. J. Betts, Y. Xi, and M. J. Gonzales. 2000. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 28:346-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 33.Swanstrom, R., and J. Erona. 2000. Human immunodeficiency virus type-1 protease inhibitors: therapeutic successes and failures, suppression and resistance. Pharmacol. Ther. 86:145-170. [DOI] [PubMed] [Google Scholar]
- 34.Tebas, P., A. K. Patick, E. M. Kane, M. K. Klebert, J. H. Simpson, A. Erice, W. G. Powderly, and K. Henry. 1999. Virologic responses to a ritonavir-saquinavir-containing regimen in patients who had previously failed nelfinavir. AIDS 13:F23-F28. [DOI] [PubMed]
- 35.Tomasselli, A. G., and R. L. Heinrikson. 1994. Specificity of retroviral proteases: an analysis of viral and nonviral protein substrates. Methods Enzymol. 241:279-301. [DOI] [PubMed] [Google Scholar]
- 36.Toth, M. V., and G. R. Marshall. 1990. A simple, continuous fluorometric assay for HIV protease. Int. J. Peptide Protein Res. 36:544-550. [DOI] [PubMed] [Google Scholar]
- 37.Tozser, J., I. Blaha, T. D. Copeland, E. M. Wondrak, and S. Oroszlan. 1991. Comparison of the HIV-1 and HIV-2 proteinases with oligopeptide substrates representing cleavage sites in Gag and Gag-Pol polyproteins. FEBS Lett. 281:77-80. [DOI] [PubMed] [Google Scholar]
- 38.Udenfriend, S., S. Stein, P. Bohlen, W. Dairman, W. Leimgruber, and M. Weigele. 1972. Fluorescamine: a reagent for assay of amino acids, peptides, proteins, and primary amines in the picomole range. Science 178:871-872. [DOI] [PubMed] [Google Scholar]
- 39.von der Helm, K. 1996. Retroviral proteases: structure, function and inhibition from a non anticipated viral enzyme to the target of a most promising HIV therapy. Biol. Chem. 377:765-774. [PubMed] [Google Scholar]
- 40.Wlodawer, A., and A. Gustchina. 2000. Structural and biochemical studies of retroviral proteases. Biochim. Biophys. Acta 1477:16-34. [DOI] [PubMed] [Google Scholar]
- 41.Wlodawer, A., A. Gustchina, L. Reshetnikova, J. Lubkowski, A. Zdanov, K. Y. Hui, E. L. Angleton, W. G. Farmerie, M. M. Goodenow, D. Bhatt, et al. 1995. Structure of an inhibitor complex of the proteinase from feline immunodeficiency virus. Nat. Struct. Biol. 2:480-488. [DOI] [PubMed] [Google Scholar]
- 42.Yoshimura, K., R. Kato, K. Yusa, M. F. Kavlick, V. Maroun, A. Nguyen, T. Mimoto, T. Ueno, M. Shintani, J. Falloon, H. Masur, H. Hayashi, J. Erickson, and H. Mitsuya. 1999. JE-2147: a dipeptide protease inhibitor (PI) that potently inhibits multi-PI-resistant HIV-1. Proc. Natl. Acad. Sci. USA 96:8675-8680. [DOI] [PMC free article] [PubMed] [Google Scholar]