Abstract
Human immunodeficiency virus (HIV)-specific CD8+ T-lymphocyte pressure can lead to the development of viral escape mutants, with consequent loss of immune control. Antiretroviral drugs also exert selection pressures on HIV, leading to the emergence of drug resistance mutations and increased levels of viral replication. We have determined a minimal epitope of HIV protease, amino acids 76 to 84, towards which a CD8+ T-lymphocyte response is directed. This epitope, which is HLA-A2 restricted, includes two amino acids that commonly mutate (V82A and I84V) in the face of protease inhibitor therapy. Among 29 HIV-infected patients who were treated with protease inhibitors and who had developed resistance to these drugs, we show that the wild-type PR82V76-84 epitope is commonly recognized by cytotoxic T lymphocytes (CTL) in HLA-A2-positive patients and that the CTL directed to this epitope are of high avidity. In contrast, the mutant PR82A76-84 epitope is generally not recognized by wild-type-specific CTL, or when recognized it is of low to moderate avidity, suggesting that the protease inhibitor-selected V82A mutation acts both as a CTL and protease inhibitor escape mutant. Paradoxically, the absence of a mutation at position 82 was associated with the presence of a high-avidity CD8+ T-cell response to the wild-type virus sequence. Our results indicate that both HIV type 1-specific CD8+ T cells and antiretroviral drugs provide complex pressures on the same amino acid sequence of the HIV protease gene and, thus, can influence viral sequence evolution.
Cell-mediated immune responses can exert significant selection pressures on pathogens (7, 33). One of the best-studied models of cytotoxic T lymphocyte (CTL) pressure is in human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) infection, where escape viruses have been identified in primary (1, 5, 31, 34) and chronic (6, 11, 13, 23, 30, 32, 37) infection. Further support for CTL-mediated pressure comes from the study of monkeys vaccinated and infected with pathogenic SIV, where the frequency of viral sequence mutations within CTL epitopes correlated with the level of viral replication (4). Two recent papers also demonstrated evidence of HIV adaptation to HLA-restricted CTL responses at a population level (27, 38). However, the characteristics of the CTL response that lead to viral escape are not well understood. It is apparent that a strong response directed towards an epitope does not always lead to escape but sometimes appears to constrain evolution. In HIV-infected individuals with the HLA-B*2705 allele, an immunodominant CTL response is made to an epitope in Gag (28), and this strong response is maintained until late in disease, when mutations within the epitopic sequence can occur and are associated with an increase in viremia (13, 19). Thus, a strong dominant CTL response against an epitopic region can suppress viral CTL epitopic escape until late in disease.
In addition to immune-mediated pressure, antiretroviral drugs also select for drug escape mutations (15). Although some drugs select for single one-step mutations (i.e., lamivudine and the M184V mutation), the evolutionary pathway for most antiretroviral drugs, including the protease inhibitors (PIs), is complex and requires multistep mutations (8, 26). The pathways of viral evolution for any given drug can be diverse and difficult to predict, suggesting that host factors may affect viral evolution under drug pressure.
During prolonged treatment failure of PI-based combination antiretroviral therapy, plasma HIV RNA levels often remain well below the off-treatment viral load set point. This occurs despite the emergence of highly PI-resistant HIV variants (10). The selective maintenance of a drug-resistant variant of a lower replication capacity partially accounts for this altered set point (3), but it does not fully account for durable partial viral suppression, suggesting that other factors such as the host response are exerting virologic control (35, 36). Given the complex nature of viral evolution under drug pressure and the partial control of some drug-resistant variants, we reasoned that HIV-specific cellular immune responses directed at epitopes within protease could constrain viral evolution and replication during antiretroviral therapy. We tested this hypothesis in a group of 29 chronically HIV-infected patients with PI-resistant HIV, all of whom had detectable plasma viremia and at least one known primary mutation within protease (15).
MATERIALS AND METHODS
Study subjects and samples.
We sampled 29 HIV-infected subjects participating in a cohort study of the long-term effects of antiretroviral therapy (the “Study of the Consequences of the Protease Inhibitor Era”) who met the following inclusion criteria: (i) current or prior use of indinavir, ritonavir, and/or lopinavir of the PI class; (ii) current plasma HIV RNA level of greater than 200 copies/ml; and (iii) presence of one or more primary mutations associated with multi-PI resistance (M46I/L, V82A/F/T/S, I84V, and L90M) (15) (Table 1). Peripheral blood mononuclear cell (PBMC) samples from two HLA-A2-positive seronegative volunteers were used as negative controls. This study was approved by the local Institutional Review Board, and all subjects provided written informed consent.
TABLE 1.
HIV-1 genotype and clinical characteristics of HLA-A2-positive subjects
| Patient | Current ARVa | Previous PI historyb | Mutationc | VLd | CD4e | Nadir CD4 |
|---|---|---|---|---|---|---|
| 2081 | LPV/RTV, 3TC, ZDV, TFV | K201, L33L/F, M36M/1, A71V, 184V, L90M | 356 | 310 | 65 | |
| 3011 | NFV, SQV, d4T | RTV, NFV, SQV | M36I/M/V, 1541/V, A71A/V, G73S, I84I/V, L90M | 3,584 | 237 | 50 |
| 3012 | IDV, RTV, 3TC/ZDV, ABC | SQV | M46I, 154V, A71V, V77I, V82A, L90M | 8,267 | 425 | 110 |
| 3013 | IDV, 3TC, d4T, EFV | L10I/L, M461 | 40,611 | 285 | 177 | |
| 3023 | RTV, APV, d4T, EFV | RTV, SQV | L10I, M36I, M46L, I54V, V82A, 184V | 158,576 | 395 | 240 |
| 3037 | RTV, SQV, ABC | L10I, G48V, A71T/I, V82A, L90M | 2,222 | 420 | 172 | |
| 3040 | LPV/RTV, d4T, 3TC | IDV, RTV, LPV | L10I, M46I, G48V, V82A, L90M | 500 | 296 | 96 |
| 3043 | IDV, 3TC, d4T. | SQV | M46I, A71T/A, N88D, L90M/L | 1,982 | 239 | 84 |
| 3050 | LPV/RTV, 3TC, d4T, NVP | SQV, NFN | K20R, M36I, I54V, V82A | 7,511 | 203 | 9 |
| 3057 | IDV, d4T, 3TC | L10I, M461, A71T, N88S, L90M/L | 500 | 416 | 143 | |
| 3084 | d4T, ABC, NVP | IDV, RTV | V32I, M46I, A71V, V82A/V | 5,351 | 411 | 193 |
| 3088 | LPV/RTV, d4T, ABC | IDV, SQV, NFV | L10I, K20R, L24I, M36I, M46I, 147I/V, F53L, 154V, V82A | 21,637 | 44 | 37 |
| 3151 | RTV, APV, d4T, ddI, 3TC, EFV | SQV, IDV, RTV, APV | I54M, A71V, I84V, L90M | 79,800 | 271 | 50 |
| 3153 | RTV, APV, d4T, ddI, 3TC | SQV, IDV, RTV, APV | L10I, M46I, I54I/L, I84V, L90M | 2,500 | 466 | 198 |
| 3156 | NFV, ABC, 3TC | RTV, IDV, NFV | L10I, M36I/M/L, I54I/V, A71T/A, 184V, N88N/S | 160,750 | 766 | 599 |
ARV, antiretroviral therapy.
LPV/RTV, lopinavir-ritonavir; 3TC, lamivudine; ZDV, zidovudine; TFV, tenofovir; NFV, nelfinavir; SQV, saquinavir; d4T, stavudine; IDV, indinavir; RTV, ritonavir; APV, amprenavir; ddI, didanosine; EFV, efavirenz; ABC, abacavir; NVP, nevirapine.
Mutations in the protease gene associated with reduced susceptibility to PIs (15). The presence of detectable heterogeneity within the viral sequence population is indicated. e.g., L33L/F points out the presence of both a leucine (L) and phenylalanine (F) at position 33.
VL, viral load (copies per milliliter) at time of study.
CD4+ cell count (cells per cubic millimeter) at time of study.
Reagents.
Recombinant vaccinia virus (rVV) constructs expressing HIV Pol, Gag, Env, and Nef or no antigen (deletion in the tk gene) were obtained from Therion, Inc. (Cambridge, Mass.). Peptides were synthesized using Fmoc-protected amino acids and were obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, National Institute of Allergy and Infectious Diseases [NIAID], National Institutes of Health [NIH]), from ResGen (Invitrogen Corporation, Carlsbad, Calif.), and from G. S. Ogg (John Radcliffe Hospital, Oxford, United Kingdom). Peptide pools were made up using 20-mer peptides corresponding to the HIV Pol gene (HIV HXB2 strain amino acid position 41 to 390) obtained through the NIH AIDS Research and Reference Reagent Program. The amino acid sequences of the HLA-A2-restricted HIV protease peptides were as follows: PR82V65-84, EICGHKAIGTVLVGPTPVNI; PR82V76-84, LVGPTPVNI; PR84V76-84, LVGPTPVNV; and PR82A76-84, LVGPTPANI. The amino acid position given corresponds to the position within the protease gene of HIV strain HXB2. The sequences of additional peptides were as follows: RT158-166, AIFQSSMTK (restricted by HLA-A3 supertype A3, A11, A68, and B*0301); Gag162-172 (p55), KAFSPEVIPMF (restricted by HLA-B57 and -B58); and cytomegalovirus (CMV) pp65, NLVPMVATV (restricted by HLA-A2). Monoclonal antibodies for anti-gamma interferon (IFN-γ; fluorescein isothiocyanate [FITC] conjugated), anti-CD4 (R-phycoerythrin conjugated), anti-CD3 (peridinin chlorophyll protein conjugated), and anti-CD8 (allophycocyanin [APC] conjugated) were purchased from BD PharMingen (San Jose, Calif.).
HLA typing and genotyping.
DNA was extracted from PBMC using a QIAamp DNA Mini kit (QIAGEN Inc., Valencia, Calif.). HLA typing was carried out by using an amplification refractory mutation system with sequence-specific primers as described by the manufacturer (Pel-Freeze, Madison, Wis.).
The genotypic resistance profile of the protease gene was determined by population-based sequencing using the TRUEGENE HIV-1 genotyping kit on viral RNA derived from plasma (Visible Genetics, Toronto, Canada).
Intracellular cytokine flow cytometry (CFC).
PBMC (3 × 105 to 5 × 105 cells) were prestimulated with peptides (5 μg/ml) for 1 h or infected with an rVV construct expressing HIV Pol (multiplicity of infection, 2:1) for 4 h. Purified anti-CD28 (3 μg/ml) (BD PharMingen) was included for additional costimulation. Brefeldin A (Sigma-Aldrich, St. Louis, Mo.) was added (10 μg/ml) to the cells. Cells with no antigen added or with a vaccinia virus construct with a deletion in the tk gene served as negative controls, while staphylococcal enterotoxin B was used as a positive control. After 5.5 h of antigen stimulation, the cells were washed, fixed with 4% paraformaldehyde, and permeabilized with FACS perm solution (Becton Dickinson). After washing, the cells were stained with monoclonal antibody against intracellular IFN-γ and the surface markers CD4, CD3, and CD8 (BD PharMingen), washed, and analyzed by flow cytometry on a FACSCalibur instrument using CellQuest software (BD Biosciences, Mountain View, Calif.). Data were gated on viable CD3+ T cells, and 105 lymphocytes in each sample were acquired. Analysis of the percentage of IFN-γ-producing CD8+ T cells was subsequently carried out using FlowJo (San Carlos, Calif.) gating on viable CD3+ CD4− CD8+ T cells. The background (response to no antigen) was subtracted in each experiment (median, 0.024%; interquartile range, 0.012 to 0.032%) (data not shown).
Expansion of antigen-specific CD8+ T cells.
To detect low frequencies of memory PR82V76-84-specific CD8+ T cells, 1 to 4 million PBMC were cultured in the presence of 1 μM peptide and 1 ng of interleukin-7 and interleukin-15 per ml for 7 days. The cells were extensively washed and then subjected to a standard CFC assay in the presence or absence of peptide. To exclude the possibility of expansion of naive cells, PMBC from a seronegative HLA-A2-positive individual were analyzed in parallel. No expansion was detected. As a positive control, cells from patients 3040 and 3156, who had a detectable response to both the wild-type and mutant peptide, were included. A 1-log expansion of the magnitude of both the wild-type and mutant-specific responses was obtained. The background (response to no antigen) was subtracted in each experiment.
Functional avidity assay.
A standard CFC assay was performed using 10-fold dilutions of peptides ranging from 10 μM to 1 pM on fresh or frozen PBMC. Functional avidity was expressed as the peptide concentration at which 50% of the maximal IFN-γ response in each individual experiment was reached, as described by O'Connor et al. (31).
Antagonism assay.
A standard CFC assay was performed using agonist peptide (autologous sequence) at a suboptimal concentration with the addition of antagonist peptide (mutant peptide) in 10-fold dilutions, ranging from a ratio (antagonist/agonist) of 100:1 to 0.1:1. The suboptimal concentration of peptide was defined in the functional avidity assay as the concentration at which 50% of the maximal IFN-γ response was reached. The addition of agonist and antagonist alone to the PBMC served as positive and negative controls.
Lytic assay.
HLA-matched or mismatched Epstein-Barr virus-transformed B-cell lines (BCLs) were used as target cells in a standard 51Cr-release cytotoxic T-lymphocyte assay with antigen-specific CTL lines established by peptide stimulation (1 μM) for 14 days. The CTL lines were restimulated with peptide at day 7. The target cells were incubated with synthetic peptide or no antigen for 1 h, washed, and plated together with CTL lines for 4 h. The percentage of specific lysis was determined as follows: (experimental lysis − spontaneous lysis)/(total lysis − spontaneous lysis) × 100. The lysis by the CTL lines (effectors) of unpulsed target cells was used as a negative control (range, 0 to 5%).
RESULTS
Study subjects.
Twenty-nine chronically infected patients with PI-resistant HIV were studied. All of the subjects had detectable plasma viremia (median, 7,511 copies/ml; interquartile range, 1,854 to 82,073) and at least one known primary mutation within protease (15). The median CD4+ T-cell count was 310 cells/mm3 (interquartile range, 235 to 398), and the median nadir CD4+ T-cell count was 59 cells/mm3 (interquartile range, 21 to 137). HLA typing showed that 15 of the patients were HLA-A2 positive (Table 1) while 14 were negative.
Mapping of a new HLA-A2-restricted CD8+ T-cell epitope in HIV protease.
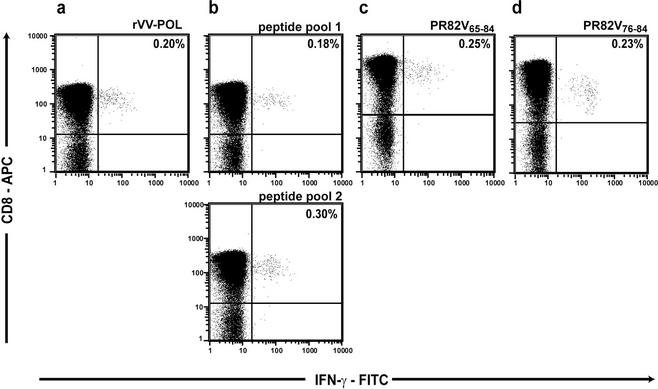
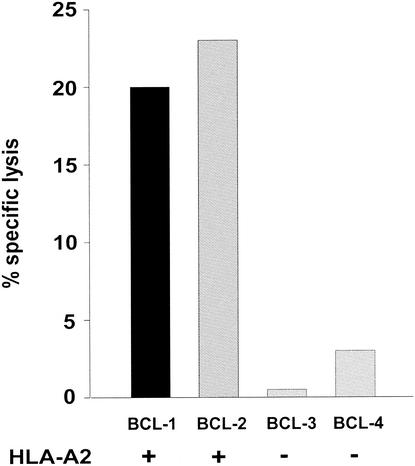
To determine potential competitive or synergistic pressures from drug and CTL responses, we initially mapped a new CTL epitope in protease recognized in HIV-infected subjects. To map CTL epitopes in protease, PBMC were exposed to an rVV expressing HIV Pol in a CFC assay, which detects the production of IFN-γ after specific stimulation. In several patients, HIV Pol-specific IFN-γ production was detected. A representative example from subject 3011 is shown in Fig. 1a. In order to map the CTL epitope in protease, a matrix peptide pool (20) was used with 20-mer peptides (Fig. 1b), and a specific 20-mer peptide was identified (Fig. 1c). The minimal peptide was then defined with short peptides and was shown to have the sequence LVGPTPVNI (PR82V76-84) (Fig. 1d). HLA restriction analysis was performed by testing CTL on HLA-matched or -mismatched BCLs in a 51Cr-release assay, and this peptide was identified as HLA-A2 restricted (Fig. 2). Two previous studies had implicated this region as containing an HLA-A2-restricted epitope (2, 24). Interestingly, this peptide covered the region of the protease gene in which two commonly occurring mutations, V82A and I84V, are present. No responses to this peptide were measured in two HIV-seronegative controls (HLA-A2-positive) or 14 HIV-seropositive (HLA-A2-negative) individuals (Table 2).
FIG. 1.
Mapping of a new epitope (PR82V76-84) in the HIV-1 protease gene. HIV-1-specific CD8+ T-cell responses were detected by using CFC ex vivo on fresh PBMC. (a) An antigen-specific response was detected to the Pol gene using an rVV construct expressing HIV-1 Pol. (b) Using a peptide matrix made of pools of 20-mer peptides, where each peptide was present in two different pools, a response was mapped to two of the pools. (c) It was confirmed that the specific response was directed to the PR82V65-84 peptide contained within the two pools. (d) The minimal peptide was defined with short peptides to the PR82V76-84 peptide. The percentages of IFN-γ-producing CD8+ T cells correspond to raw data obtained in the CFC assay; background (0.01 to 0.07%) has not been subtracted.
FIG. 2.
The epitope PR82V76-84 is restricted by HLA-A2. The HLA-A2 restriction of PR82V76-84 was confirmed in a killing assay using HLA-A2-matched and -mismatched BCL presenting PR82V76-84. An antigen-specific CTL line had been established from patient 3156 by peptide stimulation of fresh PBMC for 14 days. The class I HLA haplotypes of the matched BCL were BCL-1 (HLA-A2/30 [19]), B7/65 (Cw7/8 [15]), and BCL-2 (HLA-A2/11, B35/53, Cw4/6), and for the mismatched BCL-3 they were HLA-A1, B7, Cw7, and BCL-4 A30 (19)/74 (19), Cw2/7. The effector-to-target ratio was 30:1. The background killing ranged from 0 to 5% (median 0%) as determined by lysis of unpulsed BCL.
TABLE 2.
Frequency of antigen-specific CD8+ T cells recognizing the PR82V76-84 and/or PR82A76-84 peptide in the HIV-1 protease genea
| Patient | Amino acid at position 82 | % IFN-γ-producing CD8+ T cells
|
HLA-A2 | |
|---|---|---|---|---|
| PR82V76-84 | PR82A76-84 | |||
| SNC | <0.050 | <0.050 | + | |
| SNC | <0.050 | <0.050 | + | |
| 3005 | Val | <0.050 | <0.050 | − |
| 3085 | Val | <0.050 | <0.050 | − |
| 3148 | Val | <0.050 | <0.050 | − |
| 3152 | Val | <0.050 | <0.050 | − |
| 2004 | Ala | <0.050 | <0.050 | − |
| 3004 | Ala | <0.050 | <0.050 | − |
| 3025 | Ala | <0.050 | <0.050 | − |
| 3028 | Ala/Val | <0.050 | <0.050 | − |
| 3047 | Ala | <0.050 | <0.050 | − |
| 3062 | Ala | <0.050 | <0.050 | − |
| 3071 | Ala | <0.050 | <0.050 | − |
| 3072 | Ala | <0.050 | <0.050 | − |
| 3075 | Ala | <0.050 | <0.050 | − |
| 3102 | Ala | <0.050 | <0.050 | − |
| 2081 | Val | 0.166 | <0.050 | + |
| 3011 | Val | 0.204 | <0.050 | + |
| 3013 | Val | 0.070 | <0.050 | + |
| 3043 | Val | <0.050 | <0.050 | + |
| 3057 | Val | 0.056 | <0.050 | + |
| 3151 | Val | 0.104 | 0.075 | + |
| 3153 | Val | 0.280 | 0.100 | + |
| 3156 | Val | 0.330 | 0.180 | + |
| 3012 | Ala | <0.050 | <0.050 | + |
| 3023 | Ala | 0.130 | 0.360 | + |
| 3037 | Ala | <0.050 | 0.061 | + |
| 3040 | Ala | 0.054 | 0.123 | + |
| 3050 | Ala | <0.050 | <0.050 | + |
| 3084 | Ala/Val | <0.050 | <0.050 | + |
| 3088 | Ala | <0.050 | <0.050 | + |
Cells gated on IFN-γ-producing CD3+ CD8+ CD4− T cells were detected by intracellular CFC. The proportion of patients with the V82A mutation is greater in the group of HLA-A2-negative patients than in the HLA-A2-positive group, but the difference does not reach statistical significance (P = 0.17; chi square test). Levels above 0.05 with the background subtracted (mean background + 2SD = 0.05) are considered positive. SNC, seronegative control; Ala/Val, presence of viral sequences with both Ala and Val at position 82.
Relationship between HLA status and viral genotype.
We next assessed the impact of drug exposure, HLA-A2 haplotype, and Pol-specific CD8+ T-cell response on genotypic evolution in the 29 PI-treated patients. Notably, all 29 patients had received and failed a PI regimen containing indinavir, ritonavir, and/or lopinavir that primarily selects for the V82A mutation (18). The median duration on any of these PIs was 30 months (interquartile range, 15 to 49). There were no significant differences in duration between the HLA-A2-negative and HLA-A2-positive groups (P = 0.708; t test) or within the HLA-A2-positive group between those who maintained wild-type V82V or had developed the V82A mutation (P = 0.854; t test). However, only 7 of the 15 HLA-A2-positive individuals had developed the V82A mutation (47%), while 10 of the 14 HLA-A2-negative subjects (71%) were found with the V82A mutation, giving an odds ratio of 0.65 (95% confidence interval, 0.35 to 1.23) (P = 0.26).
Association between CD8+ T-cell response and viral genotype.
Assessments were performed in order to determine the relationship between PI-related mutations and peptide-specific CTL responses. We initially tested peptide-specific CD8+ T-cell responses using the CFC assay to the wild-type (PR82V76-84) or mutant (PR82A76-84) peptides to test whether mutation of valine (Val) to alanine (Ala) at position 82 would affect recognition in our HLA-A2-positive study subjects. Ten of 15 HLA-A2-positive patients recognized either the wild-type or mutant peptide, while none of 14 HLA-A2-negative subjects recognized these two peptides. Of the 15 HLA-A2-positive subjects, 9 recognized the wild-type peptide and 6 recognized the mutant peptide (Table 2).
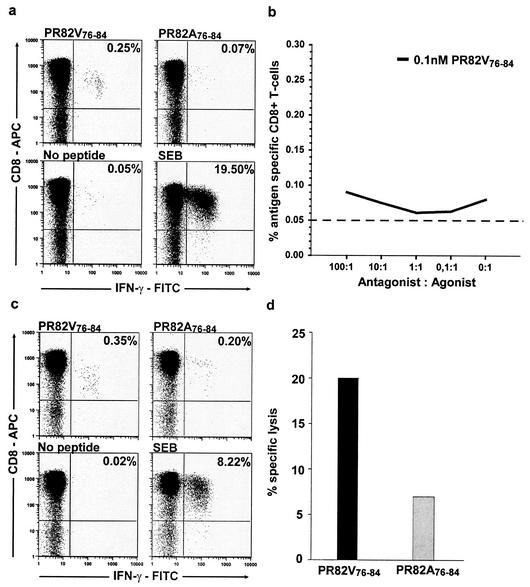
The wild-type peptide stimulated weak to moderate IFN-γ production by CD8+ T cells (median, 0.166%; range, 0.056 to 0.330%) in seven of the eight HLA-A2-positive individuals with a Val (wild type) at position 82 (Table 2). Four of these individuals did not recognize (above background) the mutant peptide containing Ala at position 82 (representative data from subject 3011 are shown in Fig. 3a).
FIG. 3.
Antigen-specific CTL for the epitope PR82V76-84 do not recognize and kill targets presenting the mutant PR82A76-84 peptide. (a) In four out of the seven HLA-A2-positive subjects who remained wild type at position 82 (V82V), antigen-specific responses detected by CFC were only seen to the wild-type peptide PR82V76-84 but not to the PR82A76-84 mutant peptide, as illustrated with data from patient 3011. (b) To investigate if the PR82A76-84 mutant had antagonist properties, PBMC were stimulated with a suboptimal concentration (0.1 nM) of the agonist peptide (wild-type PR82V76-84) in the absence or presence of antagonist peptide (mutant PR82A76-84) in the CFC assay. The dashed line corresponds to the cutoff of the assay (mean background plus 2 standard deviations = 0.05). (c) In the three remaining patients, who were wild type at position 82 (V82V), reactivity was detected to the mutant PR82A76-84 but was always of a lower magnitude than to the wild-type peptide PR82V76-84, as illustrated in patient 3156. (d) In one of the patients, number 3156, with a reactivity to both peptides, CTL specific for the wild-type (PR82V76-84) epitope were not able to recognize and kill target cells presenting the mutant (PR82A76-84) epitope and thus confirmed that the V82A mutation induces viral escape from wild-type-specific CTL.
To determine if the emergence of the mutant V82A could potentially inhibit the wild-type-specific CD8+ T-cell response in vivo, an antagonism experiment was performed (17, 21). Different ratios of the mutant peptide were added together with suboptimal concentrations of the autologous PR82V76-84 peptide directly ex vivo and assayed in a CFC assay. No significant inhibition of the response was detected by the potential antagonist PR82A76-84 peptide, as shown for patient 3011 in Fig. 3b. As subject 3011 also had the I84V mutation, peptides containing Val at position 84 were made and tested in the CFC assay. A similar response was seen with both peptides, suggesting that the I84V mutation did not affect recognition (data not shown).
We carefully studied the sequences of patients who participated in this study. Only 2 of the 15 HLA-A2-positive subjects showed mutations other than V82A or I84V within the PR82V76-84 epitope. Subject 3012 had a Val-to-Ile mutation at position 77, and subject 3013 had a Leu-to-Val mutation at position 76. These mutations are rarely observed in either treated or untreated patients; we believe that these mutations are either uncommon secondary “compensatory” mutations or reflect polymorphisms. Given the small numbers, no further analysis was carried out.
While seven of the eight HLA-A2-positive patients with wild-type Val at position 82 had a detectable response to the autologous PR82V76-84 peptide, only three showed reactivity against the mutant PR82A76-84 peptide, and these responses were all of a lower magnitude compared to responses seen to the autologous wild-type peptide (Fig. 3c). In a killing assay, it was confirmed that CTL specific for the wild-type peptide (PR82V76-84) did not kill target cells presenting the peptide containing the V82A mutation above the threshold for significant killing (10% specific lysis) (Fig. 3d).
Three of the seven HLA-A2-positive patients with the V82A mutation had a low to moderate CD8+ T-cell response to the mutant peptide (Table 2). Two of them had a detectable response to the wild-type peptide, compared to seven out of eight subjects with Val at position 82 (Table 2) (P = 0.04; Fisher's exact test). Only in the patient with the lowest response to the mutant peptide (subject 3037; 0.061%) were we unable to detect any response to the wild-type peptide. The other four patients with the V82A mutation failed to make any detectable responses to any of these peptides, wild type or mutant (Table 2). In order to test whether subjects with V82A who failed to make a response to the wild-type PR82V76-84 peptide had memory CTL to V82V, we cultured PBMC from six patients (3156, 3040, 3012, 3037, 3050, and 3084) and two seronegative controls with the PR82V76-84 peptide for 7 days (data not shown). Subjects 3156 and 3040, who had a very low frequency of response to the wild-type peptide, had specific CD8+ T cells that were readily expandable in vitro. In contrast, the other four subjects and the seronegative controls had no response even after in vitro peptide stimulation. Thus, although there are exceptions, these data suggest that the emergence of V82A under selective pressure of a PI occurred primarily in those subjects who have no detectable response to V82V.
We next assessed the potential influence of the response to the single autologous PR82V76-84 or PR82A76-84 peptide in relation to the total HIV-specific CD8+ T-cell responses. In seven of the HLA-A2-positive individuals (six V82V subjects and one V82A subject), the total HIV-specific CD8+ T-cell response to the major HIV proteins (Gag, Pol, Env, and Nef) was assayed in the CFC assay by using rVV constructs expressing the individual proteins. The responses to the autologous PR82V76-84 peptide corresponded to 6 to 44% (median, 33%) of the major HIV responses and 27 to 100% (median, 55%) of the total HIV Pol response in these individuals (data not shown). Thus, in these chronically infected individuals with PI-resistant HIV, the magnitude of the response to the PR8276-84 epitope was a substantial fraction of the total CD8+ T-cell response to the major HIV Gag, Pol, Env, and Nef proteins.
Functional avidity of the CD8+ T-cell responses.
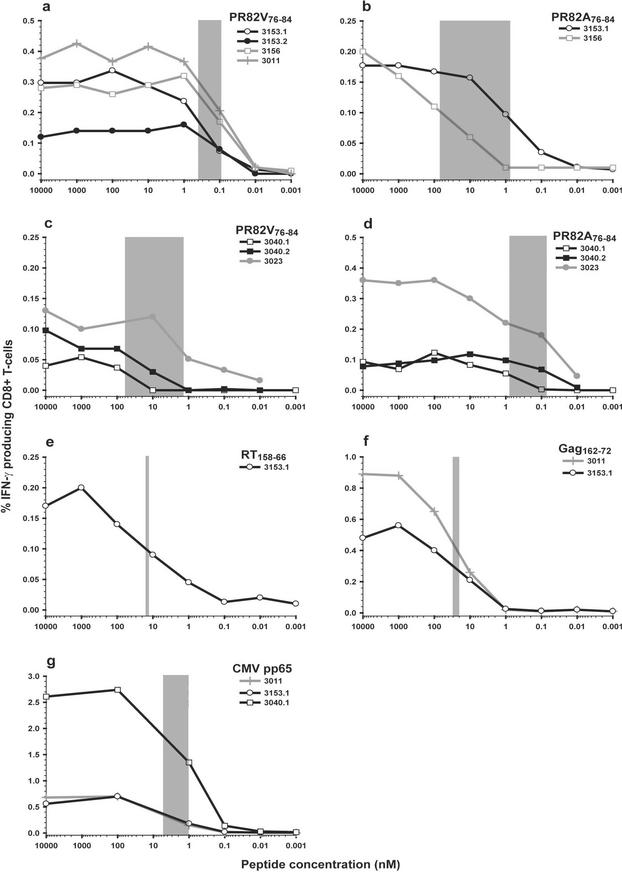
It has recently been shown that T cells with high functional avidity are associated with rapid CTL escape in acute SIV infection (31). Such rapid selection for escape mutations is often observed with antiretroviral therapy, particularly when lamivudine and non-nucleoside reverse transcriptase inhibitors are used. In order to test the functional avidity of wild-type or mutant responses, we performed avidity measurements on PBMC from several subjects. Log-fold dilutions of peptides were used to determine reactivity to different epitopes, and the avidity was determined by the 50% maximal IFN-γ response. As described by O'Connor et al. (31), we divided the functional avidity into three categories, high (0 to 5 nM), intermediate (5 to 50 nM), and low (>50 nM) (31). The functional avidity was analyzed in three of the seven subjects with Val at position 82 who had a detectable response to the autologous wild-type (PR82V76-84) peptide (Fig. 4a). To determine the reproducibility of the assay, two individual samples were analyzed from patient 3153 drawn 16 weeks apart (sample 3153.1 drawn at baseline and 3153.2 drawn 16 weeks later). The functional avidity of T-cell responses was consistent and of high functional avidity (range, 0.10 to 0.40 nM) among the intra- and interpatient samples (Fig. 4a). Three of the patients, with wild-type Val at position 82, also had low-magnitude responses to the mutant peptide. To investigate the functional avidity of the cross-reactive response, samples from two patients (3153 and 3156) were analyzed for avidity. In both cases a lower functional avidity was detected to the mutant than to the wild-type peptides, although the difference was more pronounced in 3156 (70 versus 0.09 nM) than in 3153 (0.80 versus 0.1 nM) (Fig. 4b). Thus, V82V appeared to be maintained in these subjects despite high avid CTL responses and the presence of a strong drug-associated selective pressure.
FIG.4.
High functional avidity of antigen-specific responses to the autologous epitope, PR82V76-84, or PR82A76-84 detected in PBMC of HLA-A2-positive individuals. The range of peptide concentrations at which 50% maximal IFN-γ responses were reached is indicated with gray bars illustrating the interpatient range of functional avidity detected to an epitope. (a) A high functional avidity was detected to the autologous epitope PR82V76-84 epitope in three patients with a wild-type Val at position 82. (b) An extremely low functional avidity was detected to the mutant PR82A76-84 peptide in one of these patients. In the other patient (3153.1) the difference between the avidities was less pronounced, with functional avidities of 0.40 nM to the wild-type peptide and 0.80 nM to the mutant peptide. (c) In patients with the Val-to-Ala mutation at position 82, the response to the PR82V76-84 epitope ranged from high to extremely low. (d) These patients had developed a high-avidity response to the mutant peptide. To further measure the functional avidity to other epitopes of different HLA restrictions, we studied avidity to other epitopes. (e to g) The functional avidity was shown to be of intermediate strength to the RT158-66 epitope (50% maximal response, 15 nM) (e), intermediate to the Gag162-172 epitope (50% maximal response range, 20 to 30 nM) (f), and high to the CMV pp65 epitope (50% maximal response range, 1 to 5 nM) (g).
We next assessed the responses in patients who had developed the V82A mutation. In the two patients (3040 and 3023) with a response to the wild-type peptide, the functional avidity ranged between 1.30 and 60 nM (high to low) but was lower than in patients maintaining the wild type at position 82, where the avidity was below 0.40 nM (Fig. 4c). From one of the patients (3040), two individual samples drawn 24 weeks apart from each other (3040.1 at baseline and 3040.2 drawn 24 weeks later) were assayed, and the avidity to each sample was low (sample 3040.1, 60 nM; sample 3040.2, 30 nM). Looking at the responses to the mutant peptide corresponding to the autologous sequence in patients with the V82A mutation, a high functional avidity was detected, ranging between 0.80 and 0.07 nM (Fig. 4d), comparable to the functional avidity detected to the wild-type sequence in patients who maintained V82V.
To be able to evaluate the functional avidity data to the epitope in protease-spanning amino acids 76 to 84, we included other peptides corresponding to epitopes within HIV and CMV that are restricted by different HLA molecules. The strength of the response to epitopes in reverse transcriptase (RT) and Gag of HIV was found to be of intermediate functional avidity (15, 20, and 30 nM [Fig. 4e and f]). Finally, a response of high functional avidity, ranging from 1 to 5 nM, was detected to an epitope within the CMV pp65 matrix protein (Fig. 4g). These data confirm that the response detected to the autologous PR82V76-84 epitope (ranging from 0.10 to 0.40 nM) can be considered to be of a high functional avidity.
DISCUSSION
Both antiretroviral therapy and the cellular immune system act to control viral replication. In this cross-sectional study, we identified a new HLA-A2-restricted HIV-specific CD8+ T-cell epitope, LVGPTPVNI (PR82V76-84), in protease-spanning amino acids 76 to 84. In this peptide, at position 82 valine (Val) is the consensus wild-type amino acid in drug-naive patients. In subjects with PI failure, a Val-to-Ala mutation at position 82 is associated with resistance to several PIs. We showed that the wild-type epitope is commonly recognized by CTL in HLA-A2-positive patients and that the CTL responses directed to this epitope are of high avidity. The V82V-specific CTL response is only weakly cross-reactive with the mutant peptide containing V82A. Paradoxically, and in contrast to our original hypothesis, there was no increased risk of developing a V82A mutation during PI failure in HLA-A2-positive patients compared to HLA-A2-negative patients. Indeed, there was a nonsignificant trend suggesting that HLA-A2-positive patients are less likely to develop V82A than HLA-A2-negative subjects. Among HLA-A2-positive patients, a strong high-avidity CTL response to V82V was associated with the lack of V82A during virologic failure of a PI-based regimen. The V82A mutation was more often observed in HLA-A2-negative patients and in HLA-A2-positive patients who had only a weak or moderate CTL response to V82V. Interestingly, a memory V82V-specific CTL response could not be rescued in most patients with V82A, and when the V82V CTL response was present in patients with V82A it was of relatively low avidity.
Among the eight HLA-A2-positive subjects harboring wild-type V82V, seven had a detectable CTL response to PR82V76-84, while only two of the seven subjects with the V82A mutation had a detectable response to this epitope. It has been shown that viral escape from CTL requires high viral replication and compensatory mutations (19). Only after prolonged periods of time and extensive viral replication would V82A eventually emerge. It is noteworthy that the PR82V76-84 responses were of high avidity, which has been shown to be associated with rapid selection of CTL escape mutants during acute infection (31). However, in these patients the viral replication was limited through partially effective antiretroviral suppression and likely by lowered replicative capacity of a drug-resistant virus (3). Prior studies have suggested that an immunodominant response can act in a positive manner to maintain an epitope during chronic infection (13). In addition, we were unable to detect a response to the wild-type peptide after restimulation in four of the five HLA-A2-positive patients who had developed V82A. Given the complex nature of viral evolution under drug pressure and the partial control of some drug-resistant variants, one interesting hypothesis is that high-avidity HIV-1-specific cellular immune responses directed at epitopes within protease may delay or prevent the emergence of drug resistance mutations during antiretroviral treatment. Testing this hypothesis will require longitudinal observation of patients receiving and ultimately failing antiretroviral therapy.
Three of the eight HLA-A2-positive subjects who remained wild type at position 82 (V82V) had a weak reactivity to the mutant peptide. We do not know if these three subjects had V82A viral variants present at low frequency or whether PR82V76-84-specific T cells can be cross-reactive. Since it has been shown that altered peptide ligands can function as competitive antagonists (12, 17, 21), this hypothesis was tested directly in vivo. In an antagonist experiment it was shown that the addition of PR82A76-84 peptide could not inhibit the wild-type-specific responses and, thus, did not function as an antagonist.
In three of the seven HLA-A2-positive patients who had developed the V82A mutation during treatment, a mutant-specific response could be measured and is likely to represent a response to the new antigenic stimuli from the mutant viral sequence. In two of these individuals, we observed the persistence of a weak but detectable CTL response to the wild-type PR82V76-84 epitope. This could reflect the presence of cross-reactive cells, which have been shown to be selectively expanded in mice infected with pairs of influenza A virus variants, indicating that memory T-cell populations can provide protection against antigenic variants (14). It is also possible that the response only reflects persistence of the wild-type viral variant as a minority sequence. Alternatively, the persistence of this response could reflect “original antigenic sin.” Original antigenic sin is a phenomenon in which the antibody or T-cell response elicited in an individual after secondary viral infection reacts more strongly to the viral variant that originally infected the individual (22). If this was applicable here, one would expect that the development of mutations would preferably lead to an expansion of wild-type-specific CTL over mutant-specific CTL. However, although antigen-specific IFN-γ-producing CD8+ T cells were detected both in response to the wild-type and mutant peptides, the functional avidity and magnitude of the responses were consistently higher to the mutant epitope in patients who had developed the V82A mutation. Taken together, these results do not support original antigenic sin.
There are several explanations for V82V maintenance in patients with a potent CTL response to this peptide. First, it is possible that CTL-mediated immunologic pressure on other viral proteins may indirectly affect protease sequence evolution during treatment (9, 25, 40), thus preventing the rapid emergence of V82A. Second, the protease-specific CD8+ T-cell responses observed in some HLA-A2 individuals may be of insufficient magnitude to select for the V82A mutation. Immune responses can be maintained during high viral load without evidence for escape mutations (16). However, it has been thought that such maintenance is due to a low “efficiency” of the CD8+ T-cell response, which seemed unlikely given the high avidity demonstrated in this study. Third, the lack of CTL-mediated selective pressure in vivo may reflect impaired CD8+ T-cell function (cytokine production and cytotoxicity) and/or the lack of effective CD4+ T-cell help (29, 39). Their persistent detection may simply reflect continued antigenic exposure. Finally, a dysfunctional cellular immune response directed at V82V may result in generalized cytokine-mediated local T-cell activation and the recruitment of susceptible, activated target CD4+ T cells, thus resulting in the selective maintenance of the wild-type V82V epitope over the mutant V82A epitope.
In conclusion, we have shown that HLA-A2-positive individuals recognize a specific epitope within HIV-1 protease that contains two common PI-related mutations, V82A and I84V. We demonstrated that the V82A mutation can function as a CTL escape mutation. However, the impact of the CTL response against this specific epitope is complex. Although PI therapy clearly favors the selective advantage of V82A over V82V (the mutation is rarely observed prior to PI therapy and rapidly wanes after therapy is interrupted), it is not clear from our data that a potent CTL response to V82V favors the emergence of the escape mutation. Indeed, our data point to the opposite effect, in that V82V was more common among those able to recognize this epitope. Collectively, our results indicate that both HIV-1-specific CD8+ T cells and antiretroviral drugs provide complex pressures on the same amino acid sequence of the HIV protease gene and, thus, can influence viral sequence evolution.
Acknowledgments
This work was supported by grants from the NIH, AI44595 and AI46254 (to D.F.N.) and ID01-SF-049 (to S.G.D.), the California Universitywide AIDS Research Program (grants CC99-SF-001 and PTI), the UCSF-Gladstone Institute of Virology and Immunology Center for AIDS Research (P30 MH59037), the General Clinical Research Center at San Francisco General Hospital (grant 5-M01-RR00083-37), and the Center for AIDS Prevention Studies (P30 MH62246). Douglas F. Nixon is an Elizabeth Glaser scientist of the Elizabeth Glaser Pediatric AIDS Foundation.
HIV peptides were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. We thank J. M. McCune for helpful discussions.
REFERENCES
- 1.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 2.Altfeld, M. A., B. Livingston, N. Reshamwala, P. T. Nguyen, M. M. Addo, A. Shea, M. Newman, J. Fikes, J. Sidney, P. Wentworth, R. Chesnut, R. L. Eldridge, E. S. Rosenberg, G. K. Robbins, C. Brander, P. E. Sax, S. Boswell, T. Flynn, S. Buchbinder, P. J. Goulder, B. D. Walker, A. Sette, and S. A. Kalams. 2001. Identification of novel HLA-A2-restricted human immunodeficiency virus type 1-specific cytotoxic T-lymphocyte epitopes predicted by the HLA-A2 supertype peptide-binding motif. J. Virol. 75:1301-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbour, J. D., T. Wrin, R. M. Grant, J. N. Martin, M. R. Segal, C. J. Petropoulos, and S. G. Deeks. 2002. Evolution of phenotypic drug susceptibility and viral replication capacity during long-term virologic failure of protease inhibitor therapy in human immunodeficiency virus-infected adults. J. Virol. 76:11104-11112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 5.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Z. W., A. Craiu, L. Shen, M. J. Kuroda, U. C. Iroku, D. I. Watkins, G. Voss, and N. L. Letvin. 2000. Simian immunodeficiency virus evades a dominant epitope-specific cytotoxic T lymphocyte response through a mutation resulting in the accelerated dissociation of viral peptide and MHC class I. J. Immunol. 164:6474-6479. [DOI] [PubMed] [Google Scholar]
- 7.Ciurea, A., L. Hunziker, M. M. Martinic, A. Oxenius, H. Hengartner, and R. M. Zinkernagel. 2001. CD4+ T-cell-epitope escape mutant virus selected in vivo. Nat. Med. 7:795-800. [DOI] [PubMed] [Google Scholar]
- 8.Condra, J. H., D. J. Holder, W. A. Schleif, O. M. Blahy, R. M. Danovich, L. J. Gabryelski, D. J. Graham, D. Laird, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, T. Yang, J. A. Chodakewitz, P. J. Deutsch, R. Y. Leavitt, F. E. Massari, J. W. Mellors, K. E. Squires, R. T. Steigbigel, H. Teppler, and E. A. Emini. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cote, H. C., Z. L. Brumme, and P. R. Harrigan. 2001. Human immunodeficiency virus type 1 protease cleavage site mutations associated with protease inhibitor cross-resistance selected by indinavir, ritonavir, and/or saquinavir. J. Virol. 75:589-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 11.Evans, D. T., D. H. O'Connor, P. Jing, J. L. Dzuris, J. Sidney, J. da Silva, T. M. Allen, H. Horton, J. E. Venham, R. A. Rudersdorf, T. Vogel, C. D. Pauza, R. E. Bontrop, R. DeMars, A. Sette, A. L. Hughes, and D. I. Watkins. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270-1276. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Peydro, M., A. Paradela, J. P. Albar, and J. A. Castro. 2000. Antagonism of direct alloreactivity of an HLA-B27-specific CTL clone by altered peptide ligands of its natural epitope. J. Immunol. 165:5680-5685. [DOI] [PubMed] [Google Scholar]
- 13.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 14.Haanen, J. B., M. C. Wolkers, A. M. Kruisbeek, and T. N. Schumacher. 1999. Selective expansion of cross-reactive CD8+ memory T cells by viral variants. J. Exp. Med. 190:1319-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA Panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 16.Islam, S. A., C. M. Hay, K. E. Hartman, S. He, A. K. Shea, A. K. Trocha, M. J. Dynan, N. Reshamwala, S. P. Buchbinder, N. O. Basgoz, and S. A. Kalams. 2001. Persistence of human immunodeficiency virus type 1-specific cytotoxic T-lymphocyte clones in a subject with rapid disease progression. J. Virol. 75:4907-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jameson, S. C., F. R. Carbone, and M. J. Bevan. 1993. Clone-specific T cell receptor antagonists of major histocompatibility complex class I-restricted cytotoxic T cells. J. Exp. Med. 177:1541-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kantor, R., W. J. Fessel, A. R. Zolopa, D. Israelski, N. Shulman, J. G. Montoya, M. Harbour, J. M. Schapiro, and R. W. Shafer. 2002. Evolution of primary protease inhibitor resistance mutations during protease inhibitor salvage therapy. Antimicrob. Agents Chemother. 46:1086-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kern, F., I. P. Surel, N. Faulhaber, C. Frommel, J. Schneider-Mergener, C. Schonemann, P. Reinke, and H. D. Volk. 1999. Target structures of the CD8+-T-cell response to human cytomegalovirus: the 72-kilodalton major immediate-early protein revisited. J. Virol. 73:8179-8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klenerman, P., S. Rowland-Jones, S. McAdam, J. Edwards, S. Daenke, D. Lalloo, B. Koppe, W. Rosenberg, D. Boyd, A. Edwards, et al. 1994. Cytotoxic T-cell activity antagonized by naturally occurring HIV-1 Gag variants. Nature 369:403-407. [DOI] [PubMed] [Google Scholar]
- 22.Klenerman, P., and R. M. Zinkernagel. 1998. Original antigenic sin impairs cytotoxic T lymphocyte responses to viruses bearing variant epitopes. Nature 394:482-485. [DOI] [PubMed] [Google Scholar]
- 23.Koenig, S., A. J. Conley, Y. A. Brewah, G. M. Jones, S. Leath, L. J. Boots, V. Davey, G. Pantaleo, J. F. Demarest, C. Carter, et al. 1995. Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat. Med. 1:330-336. [DOI] [PubMed] [Google Scholar]
- 24.Konya, J., G. Stuber, A. Bjorndal, E. M. Fenyo, and J. Dillner. 1997. Primary induction of human cytotoxic lymphocytes against a synthetic peptide of the human immunodeficiency virus type 1 protease. J. Gen. Virol. 78:2217-2224. [DOI] [PubMed] [Google Scholar]
- 25.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 27.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 28.Nixon, D. F., A. R. Townsend, J. G. Elvin, C. R. Rizza, J. Gallwey, and A. J. McMichael. 1988. HIV-1 gag-specific cytotoxic T lymphocytes defined with recombinant vaccinia virus and synthetic peptides. Nature 336:484-487. [DOI] [PubMed] [Google Scholar]
- 29.Nowak, M. A., and C. R. Bangham. 1996. Population dynamics of immune responses to persistent viruses. Science 272:74-79. [DOI] [PubMed] [Google Scholar]
- 30.Nowak, M. A., R. M. May, R. E. Phillips, S. Rowland-Jones, D. G. Lalloo, S. McAdam, P. Klenerman, B. Koppe, K. Sigmund, C. R. Bangham, et al. 1995. Antigenic oscillations and shifting immunodominance in HIV-1 infections. Nature 375:606-611. [DOI] [PubMed] [Google Scholar]
- 31.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493-499. [DOI] [PubMed] [Google Scholar]
- 32.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, et al. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453-459. [DOI] [PubMed] [Google Scholar]
- 33.Pircher, H., D. Moskophidis, U. Rohrer, K. Burki, H. Hengartner, and R. M. Zinkernagel. 1990. Viral escape by selection of cytotoxic T cell-resistant virus variants in vivo. Nature 346:629-633. [DOI] [PubMed] [Google Scholar]
- 34.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samri, A., G. Haas, J. Duntze, J. M. Bouley, V. Calvez, C. Katlama, and B. Autran. 2000. Immunogenicity of mutations induced by nucleoside reverse transcriptase inhibitors for human immunodeficiency virus type 1-specific cytotoxic T cells. J. Virol. 74:9306-9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt, M., E. Harrer, A. Goldwich, M. Bauerle, I. Graedner, J. R. Kalden, and T. Harrer. 2000. Specific recognition of lamivudine-resistant HIV-1 by cytotoxic T lymphocytes. AIDS 14:653-658. [DOI] [PubMed] [Google Scholar]
- 37.Wilson, C. C., S. A. Kalams, B. M. Wilkes, D. J. Ruhl, F. Gao, B. H. Hahn, I. C. Hanson, K. Luzuriaga, S. Wolinsky, R. Koup, S. P. Buchbinder, R. P. Johnson, and B. D. Walker. 1997. Overlapping epitopes in human immunodeficiency virus type 1 gp120 presented by HLA A, B, and C molecules: effects of viral variation on cytotoxic T-lymphocyte recognition. J. Virol. 71:1256-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yusim, K., C. Kesmir, B. Gaschen, M. M. Addo, M. Altfeld, S. Brunak, A. Chigaev, V. Detours, and B. T. Korber. 2002. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J. Virol. 76:8757-8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zajac, A. J., J. N. Blattman, K. Murali-Krishna, D. J. Sourdive, M. Suresh, J. D. Altman, and R. Ahmed. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 188:2205-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, Y. M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. P. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 71:6662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]