Abstract
The nucleus-localized C2 protein of Tomato yellow leaf curl virus-China (TYLCV-C) is an active suppressor of posttranscriptional gene silencing (PTGS). Consistently, infection with TYLCV-C resulted in PTGS arrest in plants. The C2 protein possesses a functional, arginine-rich nuclear localization signal within the basic amino acid-rich region 17KVQHRIAKKTTRRRR31. When expressed from potato virus X, C2-RRRR31DVGG (in which the four consecutive arginine residues 28RRRR31 were replaced with DVGG) that had been tagged with a green fluorescent protein (GFP) failed to transport GFP into nuclei and was dysfunctional in inducing necrosis and suppressing PTGS in plants. Amino acid substitution mutants C2-K17D-GFP, C2-HR21DV-GFP, and C2-KK25DI-GFP localized to nuclei and produced necrosis, but only C2-K17D-GFP suppressed PTGS. The N-terminal portions C21-31 and C217-31 fused in frame to GFP were capable of targeting GFP to nuclei, but neither caused necrosis nor affected PTGS. Our data establish that nuclear localization is likely required for C2 protein to function in C2-mediated induction of necrosis and suppression of PTGS, which may follow diverse pathways in plants. Possible mechanisms of how the C2 protein involves these biological functions are discussed.
Posttranscriptional gene silencing (PTGS), RNA interference, and gene quelling represent a conserved cellular defense system for controlling foreign gene expression across the plant, animal, and fungal kingdoms (2, 6, 8, 17, 32, 42, 45). These silencing systems involve double-stranded RNA (dsRNA) from which a 21- to 26-nucleotide (nt) short interfering RNA (siRNA) is derived by the action of an RNase III-like dicer, and they share a common molecular mechanism in which a target RNA is degraded in an RNA homology-dependent manner by an RNA-induced silencing multisubunit RNase complex under the guidance of siRNA (2, 3, 15-17, 32). In plants, PTGS defends the host against virus infection, down-regulates transgene expression, and may also participate in the control of development (42, 45). To counterattack, plant viruses have evolved the ability to encode proteins (i.e., PTGS suppressors) capable of suppressing PTGS by targeting various stages of the PTGS process, including initiation, propagation, and maintenance (6, 29, 30, 42, 45). For example, the potyvirus protein HC-Pro affects PTGS maintenance by interfering with a step coincident with, or upstream of, the production of siRNAs (25, 27). HC-Pro interacts with a calmodulin-related protein that can suppress PTGS in plants (1). However, the p25 cell-to-cell movement protein of Potato virus X (PVX) and the 2b protein of Cucumber mosaic virus (CMV) preclude the spread of silencing signals (13, 43). On the other hand, the viral PTGS suppressors are often found to be pathogenicity determinants, and their PTGS suppression activity is associated with pathogenicity determination (44). It is worth noting that certain mutants of the CMV 2b protein have been reported to be functional in pathogenesis but dysfunctional in PTGS suppression and vice versa (26).
Geminiviruses are a family of unique small circular single-stranded DNA (ssDNA) viruses that replicate via double-stranded (ds) DNA intermediates by a rolling circle mechanism in plant cell nuclei (18). The transcriptional activator protein (TrAP, also known as AL2, AC2, or C2), one of the proteins encoded by bipartite and monopartite begomoviruses, is multifunctional. The C2 protein (134 amino acid residues) of Tomato yellow leaf curl virus-China (TYLCV-C), one of the monopartite ssDNA begomoviruses, acts as an effective PTGS suppressor when expressed from a PVX vector (39, 40). This protein possesses an N-terminal basic domain, a C-terminal acidic domain, and an intervening core region. A novel zinc-finger, C36-X1-C38-X7-C46-X6-H53, within the central core region was identified for the TYLCV-C C2 protein. The zinc finger domain is indispensable for the C2 protein to bind zinc and DNA and is essential for C2-mediated induction of necrosis and suppression of PTGS in planta (39, 41). Interestingly, the zinc finger mutant C2 proteins, like the wild-type C2 protein, are localized but surprisingly aggregated in the nucleus, suggesting that the zinc finger provided essential structural stability to C2 protein (41). In this study, we report that infection of TYLCV-C, and transient expression of the TYLCV-C C2 protein via agroinfiltration arrested PTGS in plants. We further characterized an arginine-rich nuclear localization signal (NLS) that was essential for targeting C2 protein to nuclei and demonstrated that nuclear localization was likely required for the C2 protein to induce necrosis and suppress PTGS. We have also identified C2 mutants that induced necrosis but were unable to suppress PTGS, indicating a separation of the effects of C2 on silencing and necrosis-mediated host response.
MATERIALS AND METHODS
Plasmid constructs.
In order to identify the NLS, coding sequences for two truncated C2 proteins and four point mutants were fused in frame to the green fluorescent protein (GFP) coding sequence in the vector PVX/GFP (40). The primers used for mutagenesis and the cloning strategy are shown in Tables 1 and 2. Briefly, the sequence encoding the N-terminal 31 amino acids was PCR amplified with primers PP47 and PP115 and digested with ClaI and EagI prior to being cloned into the ClaI-EagI sites of PVX/GFP to produce PVX/C21-31-GFP. The double-stranded oligonucleotide formed by annealing PP125 and PP126 was cloned into the ClaI-EagI sites of PVX/GFP to produce PVX/C217-31-GFP. Sequences corresponding to the 5′ and 3′ regions of the C2 gene were PCR amplified with either PP47 and the appropriate noncoding strand primer or PP49 and the appropriate coding strand primer. PCR products corresponding to the two halves of the gene were digested with either ClaI or EagI, together with AatII, and were cloned into ClaI-EagI-digested PVX/GFP to produce the mutant C2-GFP fusion constructs described in Table 2. The integrity of each construct was confirmed by nucleotide sequencing. PVX/C2-GFP was constructed as described previously (40). Fragments encompassing C2 and mC2 coding sequences were excised from PVX/C2 and PVX/mC2 (40), respectively, and cloned into a plant gene expression vector (12) under a tandem repeat of the 35S promoter and polyadenylation sequences to produce p35S-C2 and p35S-mC2. The C2 gene expression cassettes were then subcloned into pBINPLUS (38) to produce pBin35S-C2 and pBin35S-mC2, which were electrotransformed into Agrobacterium tumefaciens LBA4404 (20). pBin1.3A and pBinTYLCV-C were constructed in A. tumefaciens LBA4404 as described previously (24, 46).
TABLE 1.
Primers used for mutagenesis
| Primer | Sequencea | Modification introduced |
|---|---|---|
| PP47 | 5′ GGCTGTAatcgATGCGATCTTCG 3′b | ClaI (C2 start codon ATG italic) |
| PP49 | 5′ ATTGGcggccgAATACTCTTAAGAAATG 3′ | EagI |
| PP115 | 5′ GTAGATCcggCCGTCGTCGTCTGGTTG 3′ | EagI |
| PP116 | 5′ GTGTTGgACgTCGATAGGAACCTGAG 3′ | AatII |
| PP117 | 5′ CCTATCgAcGTcCAACACCGAATCGCC 3′b | AatII |
| PP118 | 5′ GGCGATgacGTcTTGCACCTTGATAGG 3′ | AatII |
| PP119 | 5′ GTGCAAgACgtcATCGCCAAGAAGACA 3′b | AatII |
| PP120 | 5′ TGGTTGTgacgTcGGCGATTCGGTGTTGCAC 3′ | AatII |
| PP121 | 5′ TCGCCgAcgtcACAACCAGACGACGAC 3′b | AatII |
| PP122 | 5′ TCAACCCcTCcgacgtcGGTTGTCTTCTTGGC 3′ | AatII |
| PP123 | 5′ ACAACCgacgtcgGAgGGGTTGATCTACCTTG 3′b | AatII |
| PP124 | 3′ tATAcTTCCACGTTGTGGCTTAGCGGTTCTTCTGTTGGTCTGCTGCTGCCgg 5′ | ClaI-EagI (introduced ATG boldface) |
| PP125 | 5′ cgaTATgAAGGTGCAACACCGAATCGCCAAGAAGACAACCAGACGACGAC 3′b | ClaI-EagI (introduced ATG boldface) |
Introduced restriction endonuclease sites are underlined, and modified nucleotides are lowercase.
Coding strand primers. The other primers are complementary to the coding strand.
TABLE 2.
Construction of C2 expression cassettes
| Constructa | Primer pair (5′ region/3′ region) | Cloning sites | Mutation |
|---|---|---|---|
| PVX/C21-31-GFP | PP47-PP115 | ClaI-EagI | C21-31 |
| PVX/C217-31-GFP | PP125-PP126 | ClaI-EagI | C217-31 |
| PVX/C2-K17D-GFP | PP47-PP116/PP117-PP49 | ClaI-AatII + AatII-EagI | Lys17 → Asp |
| PVX/C2-HR21DV-GFP | PP47-PP118/PP119-PP49 | ClaI-AatII + AatII-EagI | His20 → Asp, Arg21 → Ala |
| PVX/C2-KK25DI-GFP | PP47-PP120/PP121-PP49 | ClaI-AatII + AatII-EagI | Lys24 → Asp, Lys25 → Ileu |
| PVX/C2-RRRR31DVGG-GFP | PP47-PP122/PP123-PP49 | ClaI-AatII + AatII-EagI | Arg28 → Asp, Arg29 → Val, Arg30 → Gly, Arg31 → Gly |
The PVX/GFP vector was used for all constructs.
Virus infection and gene expression in plants.
PVX RNA transcripts were produced by in vitro transcription and mechanically inoculated onto Nicotiana benthamiana plants as described previously (7). Plants were maintained in an insect-free growth room at 25°C with continuous lighting to give a 12-h photoperiod. Symptom development was photographically recorded with a Nikon Coolpix995 digital camera. To detect viral RNA accumulation, total RNAs were extracted from leaf tissues with an RNeasy plant mini kit (Qiagen) and assayed by Northern blot hybridization. RNA aliquots (5 μg) were fractionated on a 1% formaldehyde agarose gel, transferred to nylon membrane, hybridized with digoxigenin (DIG)-labeled specific probes, and immunodetected with a DIG DNA labeling and detection kit (Roche). To investigate recombinant protein expression in plants, total protein was extracted from leaf tissues as described by Hong et al. (21). Western blot analyses of protein aliquots (10 μg) were performed with a polyclonal antiserum raised against PVX CP or GFP and detected with goat anti-rabbit immunoglobulin G conjugated with alkaline phosphatase (Sigma) and 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium (BCIP/NBT) substrates (Roche).
Plant tissue fixation, embedding, sectioning, and fluorescence microscopy.
N. benthamiana leaves were collected, cut into 3-mm-wide strips, vacuum infiltrated, and fixed overnight in 4% paraformaldehyde-100 mM phosphate buffer (pH 7.0). Tissues were then infiltrated with 15% sucrose-100 mM phosphate buffer (pH 7.0), embedded in 5% low-melting-point agarose, and sectioned in a cryostat at −20°C (Bright Instruments OTS). Ten-micrometer sections were mounted in 50% glycerol containing 1 μg of 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI) per ml and examined with a Zeiss Axiophot microscope equipped with a Nikon Coolpix995 digital camera. Fluorescence was observed with filters for GFP (excitation, 450 to 490 nm; long-pass emission, 520 nm) and DAPI (excitation, 365 nm; long-pass emission, 420 nm).
PTGS suppression assay.
Seedlings of transgenic N. benthamiana line 16c carrying the GFP gene were infiltrated with A. tumefaciens strain C58C1 carrying a functional 35S-GFP expression cassette (4, 16) alone, or together with A. tumefaciens strain LBA4404 carrying pBin1.3A, pBinTYLCV-C, pBin35S-C2 or pBin35S-mC2 respectively. Alternatively, seedlings of transgenic N. benthamiana line 16c were mechanically inoculated with RNA transcripts produced by in vitro transcription from SpeI-linearized PVX/GFP, PVX/C2-GFP, PVX/C21-31-GFP, PVX/C217-31-GFP, PVX/C2-K17D-GFP, PVX/C2-HR21DV-GFP, PVX/C2-KK25DI-GFP, and PVX/C2-RRRR31DVGG-GFP. PTGS of GFP expression and PTGS suppression were routinely examined under long-wavelength UV light and photographically recorded with a Nikon Coolpix990 digital camera through a yellow filter. Leaf samples were taken for RNA extraction and Northern blot analysis as described above.
RESULTS
PVX-based expression vectors for production of C2-GFP fusion proteins in plants.
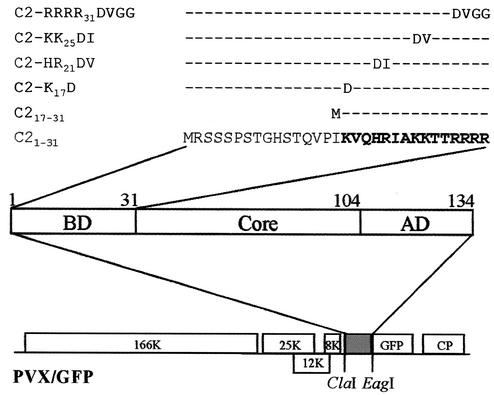
The TYLCV-C C2 protein localizes in the nucleus (40). However, an NLS has not yet been identified. The N-terminal regions (amino acids 1 to 31, especially 17 to 31) of the C2 protein are rich in basic amino acids and may contribute to its NLS. To identify the NLS and determine its biological significance, the aforementioned regions and four other C2 mutants with substitution of highly conserved basic amino acids among begomoviruses were made in frame fused to GFP (Tables 1 and 2 and Fig. 1). Fusion proteins C21-31-GFP and C217-31-GFP, including a portion of the N-terminal 31 residues or 15 residues between amino acids 17 and 31, can be expressed from PVX/C21-31-GFP or PVX/C217-31-GFP. GFP-tagged C2 mutants carrying individual amino acid substitution of K17 with D for C2-K17D, 20HR21 with DV for C2-HR21DV, 24KK25 with DI for C2-KK25DI, and 28RRRR31 with DVGG for C2-RRRR31DVGG can be expressed in plants from PVX/C2-K17D-GFP, PVX/C2-HR21DV-GFP, PVX/C2-KK25DI-GFP, and PVX/C2-RRRR31DVGG-GFP, respectively.
FIG. 1.
PVX-based C2 protein expression cassettes. C2 coding sequences containing truncations and point mutations were fused in-frame with the GFP coding sequence of PVX/GFP by using the ClaI-EagI sites. The basic domain (BD; amino acids 1 to 31), core region (Core; amino acids 32 to 104) and acidic domain (AD; amino acids 105 to 134) of the C2 protein and the mutated amino acids within the basic domain are indicated.
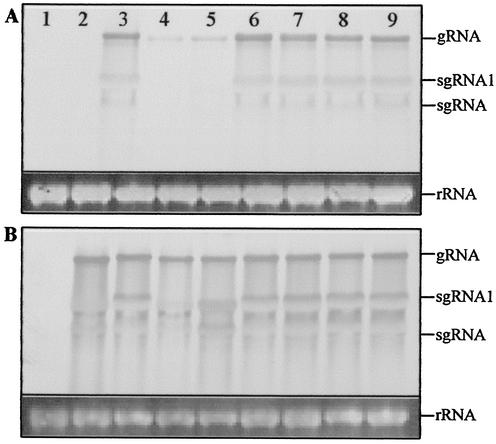
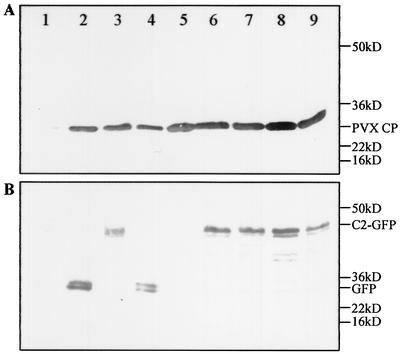
In PVX-infected N. benthamiana plants, viral genomic and subgenomic RNAs accumulated to similar levels irrespective of the inserted C2 gene sequence (Fig. 2). It should be noted that the viral RNAs associated with PVX/C21-31-GFP and PVX/C217-31-GFP infections, containing approximately 1/5 and 1/10 of the C2 sequence, respectively, hybridized only weakly to the full-length C2-specific probe (Fig. 2A, lanes 4 and 5). However, the amount of PVX coat protein expression associated with each infection (Fig. 3A) is consistent with the accumulation of similar levels of viral RNA. In plants infected with PVX/C2-GFP, PVX/C2-K17D-GFP, PVX/C2-HR21DV-GFP, PVX/C2-KK25DI-GFP, and PVX/C2-RRRR31DVGG-GFP, a single predominant protein with the predicted size (42.1 kDa) of the C2-GFP fusion protein was detected by the GFP-specific antiserum (Fig. 3B, lanes 3 and 6 to 9). Free GFP (26.9 kDa) and C21-31-GFP (30.5 kDa) were detected in plants infected with PVX/GFP and PVX/C21-31-GFP, respectively (lanes 2 and 4), but only a trace of C217-31-GFP (28.9 kDa; visible on the original membrane) was detected in plants infected with PVX/C217-31-GFP (lane 5). C217-31-GFP may be less stable; alternatively, the context around the introduced start codon AUG may not be efficient for translation initiation of C217-31-GFP in plants.
FIG. 2.
Northern blot analysis of PVX genomic and subgenomic RNAs in systemically infected N. benthamiana plants. RNA samples (5 μg) were extracted from leaves of mock-inoculated plants (lane 1) and plants infected with PVX/GFP (lane 2), PVX/C2-GFP (lane 3), PVX/C21-31-GFP (lane 4), PVX/C217-31-GFP (lane 5), PVX/C2-K17D-GFP (lane 6), PVX/C2-HR21DV-GFP (lane 7), PVX/C2-KK25DI-GFP (lane 8), and PVX/C2-RRRR31DVGG-GFP (lane 9) and harvested 13 days p.i. Blots were hybridized with DIG-labeled probes specific for C2 (A) and gfp (B) sequences. Equal loading of samples was verified by ethidium bromide staining of host ribosomal rRNA. The positions of PVX genomic (g) and subgenomic (sg) RNAs are indicated.
FIG. 3.
Expression of GFP-tagged C2 protein and PVX coat protein in N. benthamiana plants. Proteins were extracted from leaves of mock-inoculated plants (lane 1) and plants infected with PVX/GFP (lane 2), PVX/C2-GFP (lane 3), PVX/C21-31-GFP (lane 4), PVX/C217-31-GFP (lane 5), PVX/C2-K17D-GFP (lane 6), PVX/C2-HR21DV-GFP (lane 7), PVX/C2-KK25DI-GFP (lane 8), and PVX/C2-RRRR31DVGG-GFP (lane 9) and harvested 13 days p.i. Proteins were detected by Western blot analysis with antibodies specific to (A) PVX coat protein (A) and GFP (B). The positions and sizes of protein markers, the PVX coat protein, GFP, and GFP-tagged C2 protein are indicated.
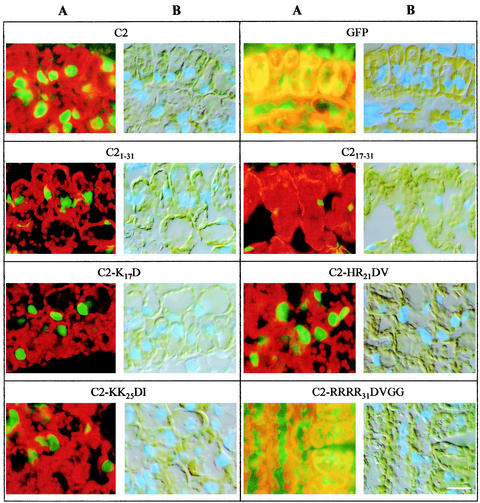
Characterization of the NLS.
Virus-infected young N. benthamiana leaf tissues were collected and examined by fluorescence microscopy; representative sections of leaf mesophyll cells are shown in Fig. 4. Fluorescence was confined in the nuclei of cells infected with PVX/C21-31-GFP, indicating that the N-terminal 31-amino-acid portion, like the entire C2 protein, was able to relocate GFP to the nucleus. Similarly, amino acids 17 to 31 expressed from PVX/C217-31-GFP were also capable of relocating GFP to the nucleus. However, the intensity of fluorescence was much weaker in this case, consistent with the decreased amount of C217-31-GFP detected by Western blotting (Fig. 3B). GFP fluorescence was also restricted to nuclei of cells infected with PVX/C2-K17D-GFP, PVX/C2-HR21DV-GFP, and PVX/C2-KK25DI-GFP, indicating that none of the five basic amino acids K17, 20HR21, 24KK25 located between positions 17 and 25 is essential for nuclear localization. However, fluorescence in cells infected with PVX/C2-RRRR31DVGG-GFP occurred throughout the cytoplasm and was indistinguishable from that associated with free GFP, suggesting that the four arginine residues located between positions 28 and 31 contribute to the C2 protein NLS.
FIG. 4.
Analysis of the C2 protein NLS. Leaf tissues infected with PVX/C2-GFP (C2), PVX-GFP (GFP), PVX/C21-31-GFP (C21-31), PVX/C217-31-GFP (C217-31), PVX/C2-K17D-GFP (C2-K17D), PVX/C2-HR21DV-GFP (C2-HR21DV), PVX/C2-KK25DI-GFP (C2-KK25DI), and PVX/C2-RRRR31DVGG-GFP (C2-RRRR31DVGG) were collected 13 days p.i., fixed, embedded, and sectioned. Sections were examined by microscopy with long-pass filters for GFP fluorescence (A) and under light-field illumination for DAPI blue fluorescence (B). Bar, 10 μm.
Effects of NLS mutation on C2 biological functions.
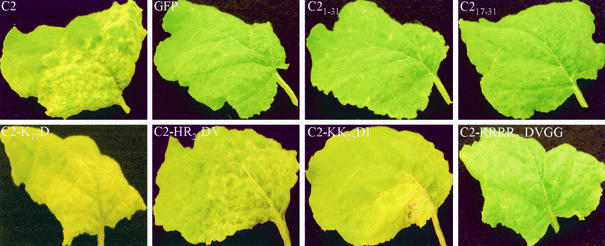
Consistent with the accumulation of viral RNA and coat protein production, N. benthamiana plants challenged with the recombinant viruses developed local and systemic symptoms. It has been established that expression of C2 or C2-GFP protein from PVX/C2 or PVX/C2-GFP induces necrosis, a novel phenotype that is not associated with PVX infection of N. benthamiana (40). A similar necrosis phenotype was produced in plants infected with PVX/C2-K17D-GFP, PVX/C2-HR21DV-GFP, and PVX/C2-KK25DI-GFP (Fig. 5). Not surprisingly, plants infected with PVX/C21-31-GFP or PVX/C217-31-GFP only produced local and systemic chlorosis, indicating that other regions of the C2 protein are required for C2-mediated induction of necrosis in plants. Only C2-RRRR31DVGG-GFP when expressed from PVX/C2-RRRR31DVGG-GFP, which was unable to target GFP green fluorescence into the nucleus, failed to induce necrosis, but instead produced chlorotic phenotypes (Fig. 5).
FIG. 5.
Phenotypes of PVX vectors expressing C2 mutants on systemically infected N. benthamiana leaves. The panels show necrosis on leaves infected with PVX/C2-GFP (C2), PVX/C2-K17D-GFP (C2-K17D), PVX/C2-HR21DV-GFP (C2-HR21DV), and PVX/C2-KK25DI-GFP (C2-KK25DI) and chlorosis on leaves infected with PVX/GFP (GFP), PVX/C21-31-GFP (C21-31), PVX/C217-31-GFP (C217-31), and PVX/C2-RRRR31DVGG-GFP (C2-RRRR31DVGG). Leaves were photographed at 13 days p.i.
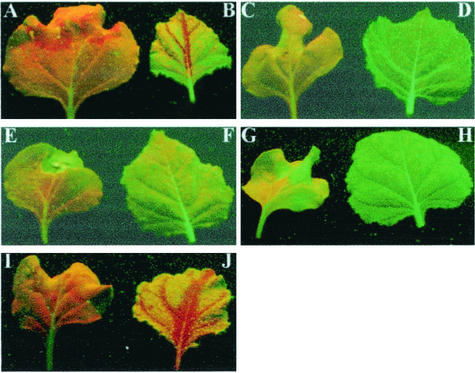
The effect of NLS mutation on C2 biological function was further investigated with respect to C2-mediated PTGS suppression. Agroinfiltration of an additional 35S-GFP transgene into transgenic N. benthamiana line 16c plants already containing a functional GFP transgene resulted in silencing of transient and resident GFP expression at the infiltrated sites, followed by systemic GFP silencing in young and noninfiltrated leaves. These tissues showed red fluorescence from chlorophyll rather than green fluorescence (Fig. 6A and B). However, line 16c GFP plants co-agroinfiltrated with the 35S-GFP transgene and pBinTYLCV-C maintained green fluorescence, indicating that local and systemic silencing of transient and transgenic GFP expression was arrested (Fig. 6C and D). The 16c plants developed curling symptoms associated with the TYLCV-C infection, and accumulation of TYLCV-C DNA was readily detectable in systemically infected young leaves (data not shown). In a parallel control experiment, co-agroinfiltration with the 35S-GFP transgene and pBin1.3A, a partial dimer (1.3A) of the DNA A component of African cassava mosaic virus (ACMV), failed to silence transient and transgenic GFP expression. Consequently, 16c plants constantly showed green fluorescence (Fig. 6E and F), consistent with the finding that ACMV infection blocked PTGS in plants (44). Furthermore, no silencing of GFP expression occurred in plants co-agroinfiltrated with the 35S-GFP and 35S-C2 transgenes, while PTGS was still induced in plants co-agroinfiltrated with the 35S-GFP and 35S-mC2 transgenes (Fig. 6G to J). These data reinforce the finding that the TYLCV-C C2 protein is a PTGS suppressor, concluded previously from PVX-based assay (39). Our data also suggest that TYLCV-C infection, and indeed the C2 protein, may target an early stage of the process of PTGS induction.
FIG. 6.
Local and systemic suppression of PTGS in plants by TYLCV-C infection and transient expression of the C2 protein. GFP transgenic line 16c plants were observed under long-wavelength UV illumination through a yellow filter. Plants were agroinfiltrated with 35S-GFP transgene alone (A and B) or together with pBinTYLCV-C (C and D), pBin1.3A (E and F), pBin35S-C2 (G and H), and pBin35S-mC2 (I and J). Photographs were taken at 30 days post-agroinfiltration for infiltrated leaves (A, C, E, G, and I) and noninfiltrated young leaves (B, D, F, H, and J).
The Agrobacterium-mediated approach has been widely used in assaying the induction and suppression of gene silencing (16). However, we noticed that induction of local and systemic silencing of GFP in plants was a considerably slow process, and establishment of systemic silencing was frequently incomplete in newly emerging, noninfiltrated leaves 30 days post-agroinfiltration. Alternatively, modified plant viruses provide an efficient and effective system to induce gene silencing in plants and to assay PTGS suppression. Moreover, virus-mediated PTGS induction is often associated with a “recovery” phenotype in plants following initial viral infection. Thus, such virus-based systems are particularly valuable to assess how mutations on a given PTGS suppressor affect its function in suppressing PTGS. Therefore, we chose the PVX-based gene silencing induction system to investigate PTGS suppression by C2 NLS mutants. It should be noted that PVX itself encodes a PTGS suppressor, the p25 movement protein (43). It may be arguable that even if the PVX suppressor is unable to interfere with silencing suppression in the transgenic N. benthamiana system alone, the PVX suppressor could have synergistic effects with a partially disabled C2 protein. However, considering the functional mode of the p25 protein in preventing spread of the gene silencing signal in N. benthamiana and the possible mechanism involved in C2-mediated PTGS suppression as discussed below, PVX's own suppressor is unlikely to affect the outcome of our experiments.
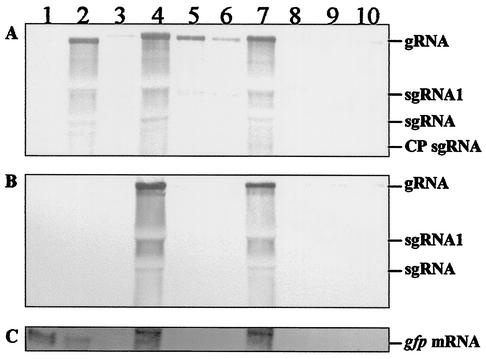
Line 16c plants challenged with PVX/GFP, PVX/C21-31-GFP, PVX/C217-31-GFP, and PVX/C2-RRRR31DVGG-GFP developed systemic symptoms by 7 days postinoculation (p.i), but later recovered and showed only sporadic mosaic symptoms on young leaves. Silencing of GFP expression was apparent approximately 7 days p.i and was almost complete by 15 days p.i, at which time, plants showed no obvious green fluorescence. Surprisingly, transgenic plants infected with PVX/C2-HR21DV-GFP and PVX/C2-KK25DI-GFP also recovered from infection, and GFP expression was silenced. Only those plants challenged with PVX/C2-GFP and PVX/C2-K17D remained symptomatic and showed no obvious GFP silencing. Consistent with these observations, the levels of recombinant viral RNA and GFP transgene mRNA were significantly reduced in transgenic plants infected with PVX/GFP, PVX/C21-31-GFP, PVX/C217-31-GFP, PVX/C2-HR21DV-GFP, PVX/C2-KK25DI-GFP, and PVX/C2-RRRR31DVGG-GFP (Fig. 7, lanes 3, 5, 6, and 8 to 10), but were maintained in plants infected with PVX/C2-GFP and PVX/C2-K17D-GFP (lanes 4 and 7) at levels similar to those associated with PVX infection (lane 2).
FIG. 7.
Northern blot analysis of PTGS induction and suppression in transgenic 16c plants. RNA samples (5 μg) were extracted from leaves of mock-inoculated plants (lane 1) and plants infected with PVX (lane 2), PVX/GFP (lane 3), PVX/C2-GFP (lane 4), PVX/C21-31-GFP (lane 5), PVX/C217-31-GFP (lane 6), PVX/C2-K17D-GFP (lane 7), PVX/C2-HR21DV-GFP (lane 8), PVX/C2-KK25DI-GFP (lane 9), and PVX/C2-RRRR31DVGG-GFP (lane 10) and harvested 15 days p.i. Blots were analyzed with DIG-labeled probes specific to PVX (A), C2 (B), and gfp (C) sequences. The positions of PVX genomic (g) and subgenomic (sg) RNAs (CP, coat protein) and the GFP transgene mRNA are indicated.
DISCUSSION
A C2-GFP fusion protein localizes in plant and insect cell nuclei (40), consistent with its proposed function in transcriptional transactivation as described for other geminivirus TrAPs (18). However, an NLS was not characterized, and its contribution to C2 protein-mediated induction of necrosis and suppression of PTGS remained unknown. We now show that C2 protein nuclear localization is likely required for the induction of a necrotic response and PTGS suppression in plants. The C2 protein (15.2 kDa) consists of 134 amino acids and possesses the N-terminal basic amino acid-rich region 17KVQHRIAKKTTRRRR31 (amino acids in boldface are highly conserved in homologues encoded by other monopartite and bipartite begomoviruses). Our findings indicate that the NLS is located within this region and suggest that it comprises the four consecutive arginine residues 28RRRR31. It resembles the NLS of the simian virus 40 large T antigen (23), implying that all four arginine residues are needed for the nuclear localization function. However, whether this is indeed the case remains to be elucidated. Mutation of the other basic amino acids individually or in pairs within the region (K17, H20, R21, K24, and K25) had no obvious effect on nuclear localization of the C2 protein. A C2 mutant with a modified 28RRRR31 motif was not only unable to localize in the nucleus but also had altered pathogenic characteristics, producing chlorosis rather than necrosis when expressed from a PVX vector. Correlated with this, the mutant failed to suppress PTGS of GFP expression in transgenic line 16c plants when expressed from a PVX vector, and plants recovered from virus infection. However, it is likely that the NLS per se is not directly responsible for these biological functions. In support of this idea, C21-31-GFP and C217-31-GFP both contain an intact NLS and target GFP to the nucleus, although neither protein induces necrosis or suppresses PTGS. In addition, mutants C2-C36R, C2-C38N, and C2-C46I are dysfunctional in necrosis induction and PTGS suppression, although each possesses an unmodified NLS and localizes to the nucleus (39, 41). On the other hand, we cannot role out the possibility that the NLS is directly responsible for PTGS suppression. It could be argued that mutations at C2-C36R, C2-C38N, and C2-C46I could modify C2 in such a manner as to make the role of the NLS in PTGS suppression ineffective. Similarly, mutations in PVX/C2-HR21DV-GFP and PVX/C2-KK25DI-GFP, both of which are near the NLS and do not suppress PTGS, could modify the protein in such a way to make the NLS dysfunctional in PTGS suppression. Nonetheless, it is possible that the triggering of necrosis induction and PTGS suppression by C2 protein may occur in the nucleus.
Pathogenicity determinants encoded by plant viruses are often PTGS suppressors, and mutants that are defective in pathogenesis (necrosis induction) are much less active in PTGS suppression than the wild-type protein (26, 44). Thus, C2-K17D behaved appropriately in suppressing PTGS, because this mutant, like C2 protein, induced a necrotic response. Surprisingly, however, mutants C2-HR21DV and C2-KK25DI also induced necrotic responses, yet both were unable to suppress PTGS. This implies that necrosis induction and PTGS suppression may follow different pathways, a view supported by a previous report that mutants with CMV 2b protein mutation are defective in pathogenesis but retain normal PTGS suppression activity (26). As predicted with a computer program (9), the overall secondary structures of the wild-type C2 protein and C2-K17D, C2-RRRR31DVGG, C2-HR21DV, and C2-KK25DI are very similar, with the exception of a slight conformation alteration around the mutated amino acids in the cases of C2-HR21DV and C2-KK25DI. However, only mutant C2-RRRR31DVGG failed to target a tagged GFP to nucleus. This mutant was also incapable of inducing necrosis and suppressing PTGS in planta. Mutants C2-K17D, C2-HR21DV, and C2-KK25DI, like the wild-type C2 protein, induced necrosis, and their subcellular localization remained unchanged, although C2-HR21DV and C2-KK25DI had a loss of function in PTGS suppression. Thus, it is unlikely that the specificity of effects on nuclear localization and biological functions observed in this report is due to the substituted amino acids that would alter C2 protein structures and bring about dysfunction of its activities.
Clearly, our data suggested that the TYLCV-C C2 protein played an important role in virus infection by suppression of the plant PTGS defense mechanism. It should be interesting to test the direct impact of the NLS mutants on their ability to transactivate virion-sense gene transcription, TYLCV-C replication, and infection. However, C2 protein with truncations or 28RRRR31 substitution would be highly likely to be lethal to the virus, because nuclear localization is likely required for the transcriptional transactivation function of the C2 protein. The other three point mutations may impose a less drastic effect on the viability of TYLCV-C in plants. Nevertheless, several begomoviruses containing mutated C2 or its homologue, TrAP, were unable to establish systemic infection in plants, although these mutant viruses retained the ability to replicate in protoplasts (18). TYLCV-C infection, and indeed transient expression of the C2 protein via agroinfiltration, effectively overcame the local and systemic induction of PTGS in plants, suggesting the C2 protein has targeted an early stage of PTGS. Interestingly, the C2 homologue ACMV TrAP has recently been shown to downregulate the production of 21- and 25-nt siRNAs (16). Similar to TrAPs encoded by other geminiviruses, the TYLCV-C C2 protein is confined to the nucleus, binds zinc, and contributes to sequence-nonspecific DNA binding activity (19, 28, 31, 33, 40, 41). The work presented in this report and our previous findings (39, 41) suggest that the C2 protein would require the nuclear localization, zinc, and DNA binding activities to establish its proper biological function in PTGS suppression. However, how the process of PTGS suppression triggered by the C2 protein correlates with its diverse biochemical and biological function remains to be elucidated. It is likely that C2-mediated PTGS suppression directly results from interplay between the C2 protein and its plant partner or partners. On the other hand, the C2 protein may involve transcriptional regulation of viral and probably host gene expression as shown for other geminiviral TrAPs (5, 10, 11, 14, 19, 21, 22, 34-37). Thus, differential expression of host genes controlled by the C2 protein is another plausible mechanism. However, the C2 protein, similar to other TrAPs, is unlikely a canonical transcriptional activator due to its sequence-nonspecific binding to DNA. In this scenario, a C2-plant protein-interacting complex may need to be formed prior to C2-mediated specific modulation of relevant host gene expression, which would then lead to PTGS suppression in plants.
Acknowledgments
Xiangli Dong and Rene van Wezel contributed equally to this work.
We thank D. Baulcombe for providing the original PVX-based vector and transgenic N. benthamiana line 16c and S. Santa Cruz for providing GFP and PVX CP antibodies. We are grateful to T. M. A. Wilson for his encouragement throughout this work.
This project was partially supported by the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Anandalakshmi, R., R. Marathe, X. Ge, J. M. Herr, C. Mau, A. Mallory, G. Pruss, L. Bowman, and V. B. Vance. 2000. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290:142-144. [DOI] [PubMed] [Google Scholar]
- 2.Bass, B. L. 2000. Double-stranded RNA as a template for gene silencing. Cell 101:235-238. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 4.Brigneti, G., O. Voinnet, W.-X. Li, L.-H. Ji, S.-W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Brough, C. L., G. Sunter, W. E. Gardiner, and D. M. Bisaro. 1992. Kinetics of tomato golden mosaic virus DNA replication and coat protein promoter activity in Nicotiana tabacum protoplasts. Virology 187:1-9. [DOI] [PubMed] [Google Scholar]
- 6.Carrington, J. C., K. D. Kasschau, and L. K. Johansen. 2001. Activation and suppression of RNA silencing by plant viruses. Virology 281:1-5. [DOI] [PubMed] [Google Scholar]
- 7.Chapman, S., T. Kavanagh, and D. Baulcombe. 1992. Potato virus X as a vector for gene expression in plants. Plant J. 2:549-557. [DOI] [PubMed] [Google Scholar]
- 8.Cogoni, C., and G. Macino. 2000. Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev. 10:638-643. [DOI] [PubMed] [Google Scholar]
- 9.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dry, I., L. Krake, P. Mullineaux, and A. Rezaian. 2000. Regulation of tomato leaf curl virus gene expression in host tissues. Mol. Plant-Microbe Interact. 13:529-537. [DOI] [PubMed] [Google Scholar]
- 11.Groning, B. R., R. J. Hayes, and K. W. Buck. 1994. Simultaneous regulation of tomato golden mosaic virus coat protein and AL1 gene expression: expression of the AL4 gene may contribute to suppression of the AL1 gene. J. Gen. Virol. 75:721-726. [DOI] [PubMed] [Google Scholar]
- 12.Guerineau, F., L. Lucy, and P. Mullineaux. 1992. Effect of two consensus sequences preceding the translation initiator codon on gene expression in plant protoplasts. Plant Mol. Biol. 18:815-818. [DOI] [PubMed] [Google Scholar]
- 13.Guo, H. S., and S. W. Ding. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21:398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haley, A., X. C. Zhan, K. Richardson, K. Head, and B. Morris. 1992. Regulation of the activities of African cassava mosaic virus promoters by the AC1, AC2 and AC3 gene products. Virology 188:905-909. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton, A. J., and D. C. Baulcombe. 1999. A novel species of small antisense RNA in post-transcriptional gene silencing. Science 286:950-952. [DOI] [PubMed] [Google Scholar]
- 16.Hamilton, A. J., O. Voinnet, L. Chappell, and D. C. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond, S. M., A. A. Caudy, and G. J. Hannon. 2001. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2:110-119. [DOI] [PubMed] [Google Scholar]
- 18.Hanley-Bowdoin, L., S. B. Settlage, B. M. Orozco, S. Nagar, and D. Robertson. 1999. Geminivirus: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 18:71-106. [PubMed] [Google Scholar]
- 19.Hartitz, M. D., G. Sunter, and D. M. Bisaro. 1999. The tomato golden mosaic virus transactivator (TrAP) is a single-stranded DNA and zinc-binding phosphoprotein with an acidic activation domain. Virology 263:1-14. [DOI] [PubMed] [Google Scholar]
- 20.Hoekema, A., P. R. Hirsch, P. J. J. Hooykaas, and R. A. Schilperoort. 1983. A binary plant vector based on separation of vir and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303:179-180. [Google Scholar]
- 21.Hong, Y., K. Saunders, M. R. Hartley, and J. Stanley. 1996. Resistance of geminivirus infection by virus-induced expression of dianthin in transgenic plants. Virology 220:119-127. [DOI] [PubMed] [Google Scholar]
- 22.Hong, Y., K. Saunders, and J. Stanley. 1997. Transactivation of dianthin transgene expression by African cassava mosaic virus AC2. Virology 228:383-387. [DOI] [PubMed] [Google Scholar]
- 23.Kalderon, D., W. D. Richardson, A. F. Markham, and A. E. Smith. 1984. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311:33-38. [DOI] [PubMed] [Google Scholar]
- 24.Klinkenberg, F. A., S. Ellwood, and J. Stanley. 1989. Fate of African cassava mosaic virus coat protein deletion mutants after agroinoculation. J. Gen. Virol. 70:1837-1844. [DOI] [PubMed] [Google Scholar]
- 25.Llave, C., K. Kasschau, and J. Carrington. 2000. Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. USA 97:13401-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucy, A. P., H. S. Guo, W. X. Li, and S. W. Ding. 2000. Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 19:1672-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mallory, A. C., L. Ely, T. H. Smith, R. Marathe, R. Anandalakshmi, M. Fagard, H. Vaucheret, G. Pruss, L. Bowman, and V. B. Vance. 2001. HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13:571-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noris, E., I. Jupin, G. P. Accotto, and B. Gronenborn. 1996. DNA-binding activity of the C2 protein of tomato yellow leaf curl geminivirus. Virology 217:607-612. [DOI] [PubMed] [Google Scholar]
- 29.Palauqui, J.-C., and H. Vaucheret. 1998. Transgenes are dispensable for the RNA degradation step of cosuppression. Proc. Natl. Acad. Sci. USA 95:9675-9680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruiz, M. T., O. Voinnet, and D. C. Baulcombe. 1998. Initiation and maintenance of virus-induced gene silencing. Plant Cell 10:937-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanderfoot, A. A., and S. G. Lazarowitz. 1995. Cooperation in viral movement: the geminivirus BL1 movement protein interacts with BR1 and redirects it from the nucleus to the cell periphery. Plant Cell 7:1185-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp, P. A. 2001. RNA interference. Genes Dev. 15:485-490. [DOI] [PubMed] [Google Scholar]
- 33.Sung, Y. K., and R. H. A. Coutts. 1996. Potato yellow mosaic geminivirus AC2 protein is a sequence non-specific DNA binding protein. FEBS Lett. 383:51-54. [DOI] [PubMed] [Google Scholar]
- 34.Sunter, G., and D. M. Bisaro. 1991. Transactivation in a geminivirus: AL2 gene product is needed for coat protein expression. Virology 180:416-419. [DOI] [PubMed] [Google Scholar]
- 35.Sunter, G., and D. M. Bisaro. 1992. Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4:1321-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunter, G., and D. M. Bisaro. 1997. Regulation of a geminivirus coat protein promoter by AL2 protein (TrAP): evidence for activation and derepression mechanisms. Virology 232:269-280. [DOI] [PubMed] [Google Scholar]
- 37.Sunter, G., J. L. Sunter, and D. M. Bisaro. 2001. Plants expressing tomato golden mosaic virus AL2 or beet curly top virus L2 transgenes show enhanced susceptibility to infection by DNA and RNA viruses. Virology 285:59-70. [DOI] [PubMed] [Google Scholar]
- 38.Van Engelen, F. A., J. W. Molthoff, A. J. Conner, J.-P. Nap, A. Pereira, and W. J. Stiekema. 1995. PBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res. 4:288-290. [DOI] [PubMed] [Google Scholar]
- 39.Van Wezel, R., X. Dong, H. Liu, P. Tien, J. Stanley, and Y. Hong. 2002. Mutation of three cysteine residues in Tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and posttranscriptional gene silencing suppression. Mol. Plant-Microbe Interact. 15:203-208. [DOI] [PubMed] [Google Scholar]
- 40.Van Wezel, R., H. Liu, P. Tien, J. Stanley, and Y. Hong. 2001. Gene C2 of the monopartite geminivirus Tomato yellow leaf curl virus-China encodes a pathogenicity determinant that is localised in the nucleus. Mol. Plant-Microbe Interact. 14:1125-1128. [DOI] [PubMed] [Google Scholar]
- 41.van Wezel, R., H. Liu, Z. Wu, J. Stanley, and Y. Hong. 2003. Contribution of the zinc finger to zinc and DNA binding by a suppressor of posttranscriptional gene silencing. J. Virol. 77:696-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 43.Voinnet, O., C. Lederer, and D. C. Baulcombe. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157-167. [DOI] [PubMed] [Google Scholar]
- 44.Voinnet, O., Y. M. Pinto, and D. C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waterhouse, P. M., M. B. Wang, and T. Lough. 2001. Gene silencing as an adaptive defence against viruses. Nature 411:834-842. [DOI] [PubMed] [Google Scholar]
- 46.Yin, Q., H. Yang, Q. Gong, H. Wang, Y. Liu, Y. Hong, and P. Tien. 2001. Tomato yellow leaf curl China virus: monopartite genome organization and agroinfection of plants. Virus Res. 81:69-76. [DOI] [PubMed] [Google Scholar]