Abstract
The sequences of the capsid protein VP1 of all minor receptor group human rhinoviruses were determined. A phylogenetic analysis revealed that minor group HRVs were not more related to each other than to the nine major group HRVs whose sequences are known. Examination of the surface exposed amino acid residues of HRV1A and HRV2, whose X-ray structures are available, and that of three-dimensional models computed for the remaining eight minor group HRVs indicated a pattern of positively charged residues within the region, which, in HRV2, was shown to be the binding site of the very-low-density lipoprotein (VLDL) receptor. A lysine in the HI loop of VP1 (K224 in HRV2) is strictly conserved within the minor group. It lies in the middle of the footprint of a single repeat of the VLDL receptor on HRV2. Major group virus serotypes exhibit mostly negative charges at the corresponding positions and do not bind the negatively charged VLDL receptor, presumably because of charge repulsion.
Human rhinoviruses (HRVs), members of the picornavirus family, are small (∼30 nm) icosahedral particles. They are composed of 60 copies each of the capsid proteins VP1, -2, -3, and -4 and a positive-strand (messenger sense) RNA genome of roughly 7,200 bases (for review see reference 35). There are presently 102 distinct serotypes that have been divided into two principal groups according to their specific attachment to intercellular adhesion molecule 1 (ICAM-1) (the major group with 91 serotypes) or to members of the low-density lipoprotein receptor (LDLR) family (the minor group with 10 serotypes) (19, 39). HRV87 is an exception and does not use either of the above receptors (39). On the basis of phylogenetic relationships, now a major principle in virus taxonomy, the HRVs are known to form two species, HRV-A and HRV-B (23). The HRV-A species includes all minor receptor group serotypes and 65 serotypes of the major receptor group (37). Again, HRV87 is an exception, and in spite of being acid sensitive as is typical for HRVs, it is genetically definitely a member of the human enterovirus D species (5). The genetic clustering roughly resembles that based on stronger affinity for one or the other group of antivirally active compounds and consequently on the form of the hydrophobic pocket within the viral capsid to which they bind (3).
Although the high-resolution X-ray structures of HRV1A and HRV2 (minor group) and of HRV3, HRV14, and HRV16 (major group) are available and sequences of capsid proteins of a number of HRVs are known, it was not possible to deduce the receptor specificity from sequence or structure, since it turned out that the intragroup similarity does not exceed the intergroup similarity.
Major group HRVs bind ICAM-1 within the canyon, a cleft encircling the fivefold axes of the viral icosahedral symmetry. The receptor molecule reaches down to the canyon floor, where amino acid residues are somewhat more conserved than at the more accessible viral surface (7). Minor group HRVs bind the LDLR, the very- LDLR (VLDLR), and the LDLR-related protein (LRP); a number of other receptors and signal transducers also belong to the LDLR family (20), but these were hitherto not investigated with respect to viral binding. LDLR is functionally and structurally unrelated to ICAM-1. Its ligand binding domain is composed of seven cysteine-rich repeats, each about 40 amino acids long and containing three disulfide bridges (with the connectivity C1-3, C2-5, and C4-6) required for correct folding (4). VLDLR contains 8 and LRP 31 such repeats. Based on the structures of several single repeats, it is thought that all chelate a Ca2+ ion trapped within an octahedral cage (13). Removal of Ca2+ and/or reduction of the disulfide bonds results in loss of receptor activity. Next to the ligand binding domain is a region with similarity to epidermal growth factor precursor and YWDT β-propeller domains that are believed to take part in the low-pH-dependent release of the ligands in endosomes (10, 27, 34). Some receptors also have a highly O-glycosylated region proximal to the transmembrane sequence. The cytoplasmic domain contains NPXY and YXXL clathrin-coated pit internalization signals (16).
Based on sequence comparison and the known ionic character of the interaction between LDLR family members and their ligands such as apo-E, apo-B, lactoferrin, and a specific chaperon (the receptor-associated protein) among many others, it had been suggested that minor group HRVs also bind to their receptors via positive charges at the viral surface. A lysine within the TEK (Thr-Glu-Lys) sequence present in the HI loop of VP1 of all minor group viruses sequenced so far was proposed to be part of the receptor interaction site (11, 22). Expression of a number of recombinant receptor fragments and analysis of their complexes with HRV2 by cryoelectron microscopy revealed that the footprint of the receptor on the virus indeed covers residues in the BC and HI loops (18). By analysis of complexes formed between HRV2 and small VLDL receptor fragments or artificial concatemers of multiple ligand binding repeats, it was shown that the second and third repeats attach to the viral surface (E. Neumann, R. Moser, L. Snyers, D. Blaas, and E. A. Hewat, submitted for publication). The third repeat binds most strongly with a footprint that includes the residues TEKHI of the HI loop and ANYN of the BC loop of one VP1 molecule, i.e., Thr-222 to Ile-226 and Ala-87 to Asn-90. The footprint of the second domain includes residues HKVH of the HI loop and EVTL of the BC loop of the adjacent VP1, i.e., His-227 to His-230 and Glu-83 to Leu-86. Leu-132 of the DE loop is also probably included. The second repeat is only bound when the neighboring VP1 is not occupied by another receptor molecule.
The present investigation was initiated in order to provide insight into the basis of receptor recognition and discrimination. To this end, the VP1 sequences of all minor group HRVs were determined and compared to presently available sequences of major group HRVs. Our data extend the earlier observation that the similarity within the respective receptor groups is not superior to that between the groups; the only residue strictly conserved in minor group HRVs is a lysine in the HI loop of VP1. There are patches of basic amino acid residues in the close vicinity of the fivefold axes within the site equivalent to the receptor footprint on HRV2; these residues are predominantly acidic in major group HRVs. We thus believe that the basic charge patterns are indeed responsible for receptor recognition, as the other residues in the vicinity are largely divergent in the different minor group serotypes. Similar to the recognition of diverse ligands via distinct repeats in LRP (28, 40), it is also possible that the different viral serotypes attach to different ligand binding repeats or to nonidentical combinations thereof.
MATERIALS AND METHODS
Sequencing and sequence analysis.
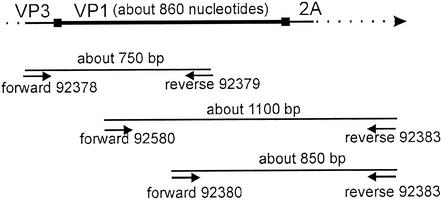
The nucleotide sequences of the VP1 protein-coding region of the minor receptor group HRVs, i.e., HRV29, -30, -31, -44, -47, -49 and -62, were determined. The origin of the prototype strains of these HRVs has been previously described (37). After two passages of the viruses in HeLa Ohio cells, the VP1 sequences were determined by reverse transcriptase PCR amplicon sequencing as described earlier (37) by using the primers given in Table 1 and in references 21 and 30, following the strategy outlined in the Fig. 1 legend. All sequences were determined at least twice. Multiple-sequence alignment of the newly produced VP1 sequences and the sequences retrieved from the picornavirus database (http://www.iah.bbsrc.ac.uk/virus/Picornaviridae/SequenceDatabase/Index.html) was performed with ClustalW version 1.82 by using the molecular biology server (http://www.ebi.ac.uk/clustalw/) with default parameters.
TABLE 1.
Primers used in HRV VP1 sequencing
| No. | Polarity | Positiona | Sequence, 5′→3′b | Reference or source |
|---|---|---|---|---|
| 92378 | Sense | 2060-2084 | ATG ITI GGI ACI CAY GTN GTN TGG G | This study |
| 92379 | Antisense | 2768-2790 | GGI GCI CCI GGI GGN ACA TAC AT | This study |
| 92380 | Sense | 2645-2669 | GAI ATG GTI CAI ATY AGR AGR AAA T | This study |
| 92383 | Antisense | 3497-3518 | CCI CCI CAI TCW CCW GGT TC | This study |
| 92580 | Sense | 2435-2454 | ACI GCI GYI GAR ACI GGN CA | 30 |
According to HRV1B complete genome sequence (21).
I = inosine; N = A, C, G, or T; R = A or G; W = A or T; Y = C or T.
FIG. 1.
Strategy for the reverse transcriptase PCR sequencing of the genomic HRV region encoding VP1. The primers used are listed in Table 1.
Model building.
Based on the available X-ray structures of HRV1A, HRV2, HRV3, HRV14, and HRV16 present in the PDB database, three-dimensional models of VP1 were automatically generated for the remaining HRVs by using SwissModel (15). The coordinates of minor group HRVs thus obtained were superimposed onto the structure of HRV2 with Swiss-PdbViewer (version 3.7) by using “magic fit.” The same was done with model coordinates of major group HRVs but by using HRV16 as a template for superimposition. To estimate the quality of the fit, models of HRV1A (1R1A), HRV2 (1FPN), HRV3 (1RHI), HRV14 (4RH4), and HRV16 (1AYM) were automatically built with SwissModel by using the structures of the four other HRVs as templates. The coordinates of the models were then compared with the X-ray structures, and the root mean square (RMS) deviations were calculated.
For delineating the region containing the receptor binding site, a Ca2+ ion present at the fivefold axis of HRV1A was used as center of a sphere with a 35-Å radius and residues within this range were selected. These were then introduced into the program ROADMAP (6), and amino acids were colored blue, red, green, and yellow for basic, acidic, hydrophilic, and hydrophobic, respectively.
Surface potentials were calculated for pentamers with Swiss-PdbViewer by using only charged residues and a dielectric constant for the solvent of 80 and for the protein of 4 (default). For rendering the region of the receptor binding site for all HRVs, the same color scale, i.e., red, −2.5; white, 0.0; and blue, 2.5, was used.
RESULTS
Alignments.
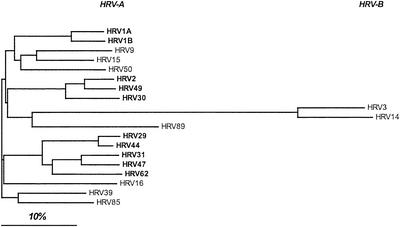
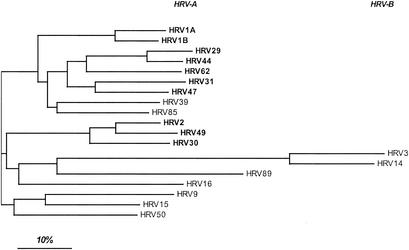
Alignment of all available VP1 amino acid sequences (including the 10 minor group HRVs and 9 major group HRVs) yielded a pairwise sequence similarity of between 35% (HRV14 and HRV50) and 95% (HRV29 and HRV44) (not shown). ClustalW analysis and neighbor-joining tree representation (Fig. 2) showed that the similarities within one receptor group were generally not higher than the similarity of serotypes belonging to different groups. However, the minor group HRVs, i.e., HRV1A and -1B, -29, -31, -44, -47, and -62 as well as -2, -30, and -49, cluster together and are thus less divergent. As anticipated from earlier work in several laboratories, receptor specificity appears to be unpredictable from the sequence. As expected from HRV3 and HRV14 belonging to species B, these are less related to the serotypes that belong to species A. The rhinovirus family might thus have separated into group A and B, still using ICAM-1 as receptor prior to evolution of the minor receptor group from group A, by switching to novel receptor specificity.
FIG. 2.
Neighbor-joining tree of 10 minor and 9 major group HRVs based on VP1 amino acid sequences. The analysis was carried out with ClustalW. Note the grouping of HRV1A and -B; -29, -31, -44, -47, and -62; and -2, -30, and -49. Minor group HRVs are shown in boldface; classification as species A and B is also indicated.
Structure of the receptor binding site.
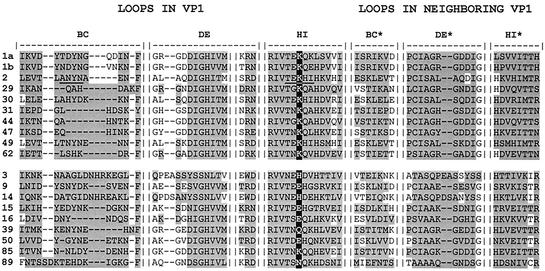
The recent determination of the structure of a complex between a recombinant VLDR fragment encompassing the ligand binding repeats 1 to 3 (V123) and HRV2 by electron cryomicroscopy revealed that the footprint of the receptor essentially covers the BC and HI loops of VP1 (18). Furthermore, use of a number of receptor fragments and artificial concatemers of the repeats expressed in bacteria allowed more exact definition of the binding site of each receptor repeat. This also revealed that a given repeat (e.g., V3) could bind to two distinct sites contributed by the BC and the HI loops, though apparently to a different extent (Neumann et al., submitted). In order to identify residues in the other minor group serotypes that correspond to those covered by the principal footprint of V3 in HRV2 in their three-dimensional context, the known X-ray structures of HRV1A and HRV2 and of models of other serotypes automatically built with SwissModel (14, 15) were structurally aligned. As expected, the sequences were strongly conserved throughout the β-sheet structures but substantially diverged within the loops (entire sequences not shown but deposited in GenBank with the following accession codes: HRV29, AY273202; HRV30, AY273205; HRV31, AY273200; HRV44, AY273203; HRV47, AY273201; HRV49, AY273206; and HRV62, AY273204). An alignment of the amino acid residues in the BC, DE, and HI loops within 12 Å from a lysine found to be present in the HI loop of all minor group HRVs (K224 in HRV2) is shown in Fig. 3. It revealed no obvious conservation of any other residue. It was thus of interest to determine which of the amino acids in the other serotypes lying within the region analogous to the receptor footprint on HRV2 were exposed at similar locations, possibly resulting in a conserved charge pattern.
FIG. 3.
Structural alignment of HRV VP1 sequences of the BC, DE, and HI loops, taking into account all amino acid residues within 12 Å (shaded) from K224 in HRV2 and from the equivalent amino acid residues in the other HRVs. K224 conserved in all minor group HRVs and the corresponding residues in major group HRVs are highlighted. Residues within the footprint of V3 on HRV2 (Neumann et al., submitted) are underlined. Note that residues of the neighboring VP1 are also close to K224 but are in fact not in the footprint of the single repeat.
Ligands of LDLR, VLDLR, LRP, and other members of the LDLR family all exhibit a pattern of positive charges at one face of the molecule (e.g., apo-E [42] and lactoferrin [2]), and ionic interactions between acidic residues of the receptor and basic residues of the ligands have been implicated in binding (43). Therefore, we wondered whether positive charges would also be conserved within minor group HRVs in the receptor footprint region. First, we estimated the quality of three-dimensional models built with SwissModel. Structural predictions of all HRV serotypes, whose structures are known, were carried out, each time with omission of the respective target structure from the data set. As seen in Table 2, the RMS deviation between the experimentally determined structures and the calculated structures of VP1 was equal to or below 3 Å for the backbone and below 3.7 Å when all atoms were included. As expected from the phylogenetic distance of HRV3 and HRV14 from all other rhinovirus serotypes (Fig. 2), their prediction was poor. This is also in line with their belonging to species B, whereas the other HRVs belong to species A. The position of the amino acid residues was fairly correct, so we believe that the models reflect the approximate location of the charges rather well. This allowed calculation of the approximate positions of surface-exposed amino acid residues and investigation of whether charge patterns are conserved within the minor group. However, the accuracy of the resulting models should not be overinterpreted, as the RMS implies that a major portion of the error lies in the less conserved loop region.
TABLE 2.
RMS deviationsa
| Model | RMS (all atoms) | RMS (backbone) |
|---|---|---|
| HRV1A | 2.05 | 1.58 |
| HRV2 | 1.47 | 0.77 |
| HRV3 | 3.30 | 2.67 |
| HRV14 | 3.67 | 3.01 |
| HRV16 | 2.21 | 1.62 |
VP1 models of the HRVs were superimposed onto the corresponding X-ray structures with Swiss-PdbViewer and were subjected to magic fit with all atoms, and the RMS was calculated.
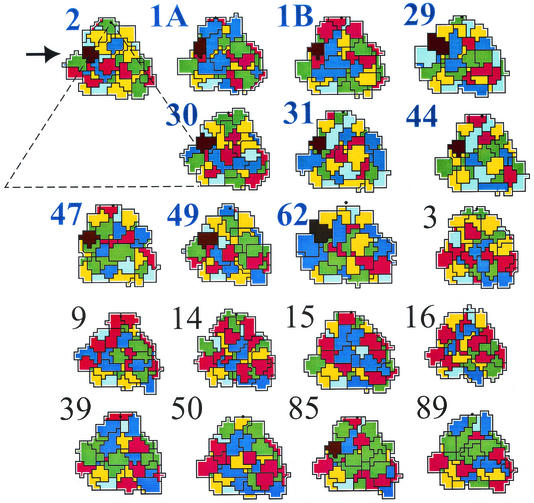
For those serotypes whose atomic structures were not available, three-dimensional models of VP1 were thus calculated and road maps were drawn from all HRVs. The program ROADMAP (6) plots a projection of an asymmetrical icosahedral unit, taking into account the surface exposure of each amino acid residue; amino acids were color coded according to their properties. As the footprint of VLDLR on HRV2 covers residues contributed by VP1 only and as sequences of the remaining capsid proteins were not available, we felt it legitimate to omit VP2, -3, and -4. Again, we first estimated the quality of the models. Comparison of the road maps based on the X-ray coordinates with those drawn by using calculated model coordinates gave a rather good overall agreement of the patterns (data not shown). As seen in Fig. 4, in minor group HRVs there are invariably several closely spaced, positively charged residues (blue) on the left (west) side of the fivefold axis with the conserved lysine playing a central role. These basic groups are largely replaced by uncharged or acidic ones in major group HRVs. There are two interesting exceptions: HRV85 has a lysine as well at a position equivalent to that of the conserved lysine in minor group HRVs. However, except for K132, no other basic amino acids are in its vicinity. On the other hand, HRV15 has a glutamic acid instead of the conserved lysine but a large number of basic residues, which are, however, rather in the middle than at the left side of the asymmetric unit.
FIG. 4.
Road maps of all minor group HRVs (HRV1A, -1B, -2, -29, -30, -31, -44, -47, -49, and -62), indicated in blue and boldface, and 9 major group HRV2 (HRV3, -9, -14, -15, -16, -50, -85, and -89), indicated in black, including VP1 amino acid residues 35 Å from the fivefold axis. Blue, basic; red, acidic; yellow, hydrophobic; and green, hydrophilic uncharged; His is colored light blue. For orientation, the size of the entire asymmetric unit is indicated with a triangle in HRV2. The position of the lysine in the HI loop of minor group HRVs is colored black and is specified with an arrow. Note the presence of a lysine in the HI loop of HRV85.
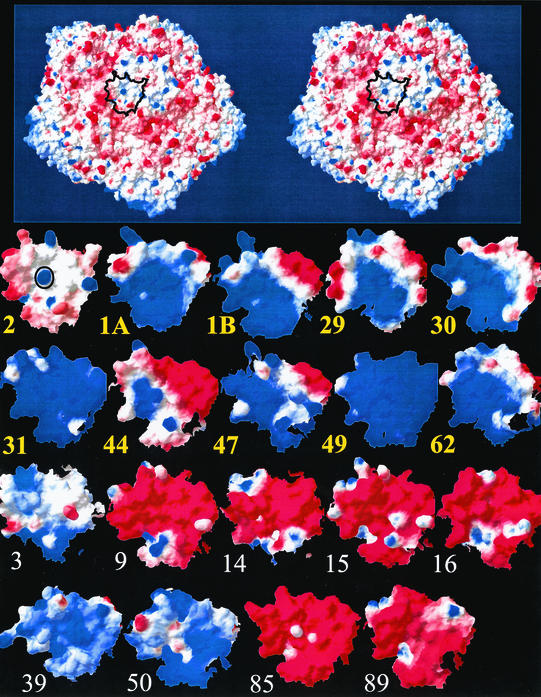
The footprint of V3 on HRV2 extends down into the canyon and covers amino acid residues at the canyon wall (Neumann et al., submitted). However, ROADMAP produces a projection down the z axis that might result in an underestimation of the exposure of residues at the canyon wall. Therefore, in order to more closely examine in all HRVs the charge properties at the position equivalent to the receptor binding site in HRV2, the surface potentials were calculated and displayed as a side view centered on K224, in HRV2, or on the corresponding residues. The approximate dimension (N terminus to C terminus) of a single repeat is about 24 Å, and the footprint of a single repeat is roughly 18 by 10 Å; therefore, the amino acid residues within 12 Å from the lysine conserved in the minor group (and from the equivalent residues in the major group) were selected (Fig. 3), and the potentials at this site are depicted in Fig. 5. From this picture it becomes even clearer that all minor group HRVs exhibit a strong positive charge at this particular position. However, it also shows that the basic amino acids in HRV15, as seen in the road map (Fig. 4), is remote from the minor group receptor binding site, that K224 in HRV85 is not sufficient to overwhelm the negative charges in its vicinity, and finally, that HRV3, HRV39, and HRV50 have positive charge densities similar to those in minor group HRVs; nevertheless, K224 is absent and thus appears to be indispensable for binding to minor group receptors.
FIG. 5.
Surface potential of 10 minor group (bold yellow lettering) and 9 major group HRVs (plain white lettering) centered on the position of binding of V3 on HRV2 as delineated in the stereo drawing of an HRV2 pentamer (top). Surface potentials were calculated with Swiss-PdbViewer by using the available X-ray coordinates and the coordinates predicted by SwissModel. For the models the same scale (blue, 2.5, positive; and red, −2.5, negative) was used. Amino acids within 12 Å from K224 in HRV2 (indicated by O) or from the amino acids at the equivalent position (see structural alignment in Fig. 3) are displayed. The view is from the side onto the north wall of the canyon and is centered on K224 as indicated on the top. This view avoids the underestimation of the exposed surfaces by the road map due to the steep descent at the canyon.
DISCUSSION
Although the overall structures of five HRV serotypes, two belonging to the minor and three to the major receptor group, are rather similar, these viruses recognize exclusively and specifically their respective receptors (1, 26). For example, not a single plaque was seen after a challenge of HeLa cells with HRV14 at high multiplicity of infection under conditions of the ICAM-1 was blocked with a monoclonal antibody (8). Therefore, at least for HRV14, a single mutation, which occurs with a frequency of about 1 in 104 to 1 in 105 (38), is certainly not sufficient to change the receptor specificity. This is also in line with recent results for HRV89, where a number of mutations had to accumulate for the virus to bind to a different receptor. However, the novel specificity was not directed toward a member of the LDLR family (32).
Differences of major group HRVs with respect to their affinity for human ICAM-1 were noted (8) and mutations of canyon residues in HRV14 led to a decrease or an increase in affinity toward ICAM-1 (9). On the other hand, amino acid residues in ICAM-1 involved in the interaction with HRV14 were characterized by systematically exchanging those of the mouse homologue, which does not bind, for those present in the human homologue, which does bind (31). Although a somewhat higher conservation of amino acid residues within the canyon than for more exposed sites was noted (7), alignment of the now-available sequences did not give us any hint as to which amino acids might invariably take part in the recognition of ICAM-1. Nevertheless, it is very likely that charge patterns also participate in receptor recognition by major group HRVs (24). As VP2 and VP3 residues are also involved, the situation is, however, more complicated.
We observed a large difference in the affinity of HRV1A for human and mouse LDLR. Unexpectedly, HRV1A binds much better to the mouse homologue, whereas HRV2 binds equally well to LDLR of both species (33). Therefore, serotypes belonging to the same groups bind to the same receptor with different affinity.
The individual repeats in LDLR are differently implicated in ligand binding; repeat 5 is predominantly involved in recognition of LDL (via apo-B), and any one of repeats 2 to 7 is required for recognition of β-VLDL (via apo-E) (12, 36). By the same token, LRP binds its collection of different ligands via distinct domains (41). It is thus possible that nonidentical ligand binding repeats and/or combinations thereof recognize different minor group serotypes in keeping with their amino acid differences except from the conserved lysine in the HI loop. It is interesting that a ClustalW analysis on parts of the BC, DE, and HI loop comprising roughly 70 amino acid residues within 12 Å from the central Lys-224 (Fig. 3) still resulted in the same clusters of serotypes as those obtained upon analyzing the entire VP1 sequences. Again, minor and major group HRVs could not be discriminated (Fig. 6). It is possible that closely related minor group serotypes preferentially bind to the same combinations of the repeats. Preliminary data on the interaction of artificial concatemers of individual repeats with the different serotypes suggest that this assumption might be correct.
FIG. 6.
Neighbor-joining tree of 10 minor and 9 major group HRVs based on the VP1 amino acid sequences within a sphere around K224 (HRV2) with a 12-Å radius. Sequence fragments were joined and treated as single sequences comprising parts of the BC, the DE, and the HI loops contributed from the two symmetry-related VP1 molecules. The analysis was carried out with ClustalW. Note that the clusters of serotypes HRV1A and -B; -29, -31, -44, -47, and -62; and -2, -30, and -49 are the same as in Fig. 2, where the entire VP1 was analyzed. Minor group HRVs are shown in boldface; classification as species A and B is also indicated.
The interaction between the LDLR family and their ligands is most probably of ionic nature. Nevertheless, the search for conserved charge patterns in the ligands failed to yield any particular sequence but rather indicated the involvement of a local concentration of positive charges. Similar to our finding, a single lysine conserved in vitellogenin of a number of species was found to be strongly but not exclusively involved in binding to the oviparous homologue of VLDLR (25). In this case, repeats 123 of the receptor were sufficient for high-affinity interaction and further deletion of one repeat strongly reduced binding. This compares well with HRV2 binding to VLDLR, where the same repeats (i.e., V123) bind well, whereas repeats 23 (V23) exhibit much weaker binding and attachment of single repeat 2 or 3 is almost undetectable (unpublished data). This is in keeping with the small area of contact of a single repeat with the viral surface and makes it difficult to explain why only repeat 3 was seen in cryoelectron microscopy images in contact with the viral surface in a complex between MBP-V23 (a maltose binding protein fusion to V23) and HRV2 (Neumann et al., submitted). We thus believe that a second repeat contributes by a small, by this method almost undetectable, extent to the interaction with the virus. Furthermore, these data also indicate that amino acid residues, which are not adjacent in sequence but come close to each other in the context of the three-dimensional structure, are involved in receptor recognition. Structural changes like those occurring during uncoating (17) apparently destroy this arrangement, and receptor binding is lost (29).
The finding that HRV15 has a glutamic acid instead of a lysine in the HI loop but has a number of other exposed basic residues, whereas HRV85 possesses a lysine but lacks other basic residues, lets us assume that these serotypes might require fewer changes to switch receptor specificity or at least to acquire affinity towards LDLR. However, the overall negative charges in HRV85 might prevent binding, even when a lysine is introduced at the appropriate site. On the other hand, HRV39 and HRV50 appear to have the correct basic environment and exchange of a few amino acid residues might be sufficient to allow binding to LDLR, in addition to ICAM-1. This is even more so, as there are no major deletions or insertions in the loops with respect to HRV2 (Fig. 3). Experiments along these lines are currently being carried out in our laboratory.
Acknowledgments
This work was funded by the Virology Foundation and the Austrian Science Foundation grant no. P-14503-MOB. Support for M.V. by an EMBO fellowship is acknowledged.
REFERENCES
- 1.Abraham, G., and R. J. Colonno. 1984. Many rhinovirus serotypes share the same cellular receptor. J. Virol. 51:340-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, B. F., H. M. Baker, G. E. Norris, D. W. Rice, and E. N. Baker. 1989. Structure of human lactoferrin: crystallographic structure analysis and refinement at 2.8 A resolution. J. Mol. Biol. 209:711-734. [DOI] [PubMed] [Google Scholar]
- 3.Andries, K., B. Dewindt, J. Snoeks, L. Wouters, H. Moereels, P. J. Lewi, and P. A. J. Janssen. 1990. Two groups of rhinoviruses revealed by a panel of antiviral compounds present sequence divergence and differential pathogenicity. J. Virol. 64:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieri, S., J. T. Djordjevic, N. L. Daly, R. Smith, and P. A. Kroon. 1995. Disulfide bridges of a cysteine-rich repeat of the LDL receptor ligand-binding domain. Biochemistry 34:13059-13065. [DOI] [PubMed] [Google Scholar]
- 5.Blomqvist, S., C. Savolainen, L. Raman, M. Roivainen, and T. Hovi. 2002. Human rhinovirus 87 and enterovirus 68 represent a unique serotype with rhinovirus and enterovirus features. J. Clin. Microbiol. 40:4218-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman, M. S. 1993. Mapping the surface properties of macromolecules. Protein Sci. 2:459-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman, M. S., and M. G. Rossmann. 1993. Comparison of surface properties of picornaviruses: strategies for hiding the receptor site from immune surveillance. Virology 195:745-756. [DOI] [PubMed] [Google Scholar]
- 8.Colonno, R. J., P. L. Callahan, and W. J. Long. 1986. Isolation of a monoclonal antibody that blocks attachment of the major group of human rhinoviruses. J. Virol. 57:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colonno, R. J., J. H. Condra, S. Mizutani, P. L. Callahan, M. E. Davies, and M. A. Murcko. 1988. Evidence for the direct involvement of the rhinovirus canyon in receptor binding. Proc. Natl. Acad. Sci. USA 85:5449-5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, C. G., J. L. Goldstein, T. C. Sudhof, R. G. Anderson, D. W. Russell, and M. S. Brown. 1987. Acid-dependent ligand dissociation and recycling of LDL receptor mediated by growth factor homology region. Nature 326:760-765. [DOI] [PubMed] [Google Scholar]
- 11.Duechler, M., S. Ketter, T. Skern, E. Kuechler, and D. Blaas. 1993. Rhinoviral receptor discrimination: mutational changes in the canyon regions of human rhinovirus types 2 and 14 indicate a different site of interaction. J. Gen. Virol. 74:2287-2291. [DOI] [PubMed] [Google Scholar]
- 12.Esser, V., L. E. Limbird, M. S. Brown, J. L. Goldstein, and D. W. Russell. 1988. Mutational analysis of the ligand binding domain of the low density lipoprotein receptor. J. Biol. Chem. 263:13282-13290. [PubMed] [Google Scholar]
- 13.Fass, D., S. Blacklow, P. S. Kim, and J. M. Berger. 1997. Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature 388:691-693. [DOI] [PubMed] [Google Scholar]
- 14.Guex, N., A. Diemand, and M. C. Peitsch. 1999. Protein modelling for all. Trends Biochem. Sci. 24:364-367. [DOI] [PubMed] [Google Scholar]
- 15.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 16.Herz, J. 2001. Deconstructing the LDL receptor—a rhapsody in pieces. Nat. Struct. Biol. 8:476-478. [DOI] [PubMed] [Google Scholar]
- 17.Hewat, E., E. Neumann, and D. Blaas. 2002. The concerted conformational changes during human rhinovirus 2 uncoating. Mol. Cell 10:317-326. [DOI] [PubMed] [Google Scholar]
- 18.Hewat, E. A., E. Neumann, J. F. Conway, R. Moser, B. Ronacher, T. C. Marlovits, and D. Blaas. 2000. The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view. EMBO J. 19:6317-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofer, F., M. Gruenberger, H. Kowalski, H. Machat, M. Huettinger, E. Kuechler, and D. Blaas. 1994. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc. Natl. Acad. Sci. USA 91:1839-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howell, B. W., and J. Herz. 2001. The LDL receptor gene family: signaling functions during development. Curr. Opin. Neurobiol. 11:74-81. [DOI] [PubMed] [Google Scholar]
- 21.Hughes, P. J., C. North, C. H. Jellis, P. D. Minor, and G. Stanway. 1988. The nucleotide sequence of human rhinovirus 1B: molecular relationships within the rhinovirus genus. J. Gen. Virol. 69:49-58. [DOI] [PubMed] [Google Scholar]
- 22.Kim, S., T. J. Smith, M. S. Chapman, M. G. Rossmann, D. C. Pevear, F. J. Dutko, P. J. Felock, G. D. Diana, and M. A. McKinlay. 1989. Crystal structure of human rhinovirus serotype-1A (Hrv1A). J. Mol. Biol. 210:91-111. [DOI] [PubMed] [Google Scholar]
- 23.King, A. M. Q., F. Brown, P. Christian, T. Hovi, T. Hyypiä, N. J. Knowles, S. M. Lemon, P. D. Minor, A. C. Palmenberg, T. Skern, and G. Stanway. 2000. Family Picornaviridae, p. 657-678. In M. H. V. van Regenmortel, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 24.Kolatkar, P. R., J. Bella, N. H. Olson, C. M. Bator, T. S. Baker, and M. G. Rossmann. 1999. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 18:6249-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, A., M. Sadasivam, and J. L. Ding. 2003. Receptor-ligand interaction between vitellogenin receptor (VtgR) and vitellogenin (Vtg), implications on low density lipoprotein receptor and alipoprotein B/E. The first three ligand-binding repeats of VtgR interact with the amino-terminal region of Vtg. J. Biol. Chem. 278:2799-2806. [DOI] [PubMed] [Google Scholar]
- 26.Lonberg-Holm, K., R. L. Crowell, and L. Philipson. 1976. Unrelated animal viruses share receptors. Nature 259:679-681. [DOI] [PubMed] [Google Scholar]
- 27.Mikhailenko, I., W. Considine, K. M. Argraves, D. Loukinov, B. T. Hyman, and D. K. Strickland. 1999. Functional domains of the very low density lipoprotein receptor: molecular analysis of ligand binding and acid-dependent ligand dissociation mechanisms. J. Cell Sci. 112:3269-3281. [DOI] [PubMed] [Google Scholar]
- 28.Neels, J. G., B. M. M. van den Berg, A. Lookene, G. Olivecrona, H. Pannekoek, and A. J. van Zonneveld. 1999. The second and fourth cluster of class A cysteine-rich repeats of the low density lipoprotein receptor-related protein share ligand-binding properties. J. Biol. Chem. 274:31305-31311. [DOI] [PubMed] [Google Scholar]
- 29.Noble, J. N., and K. Lonberg-Holm. 1973. Interactions of components of human rhinovirus type 2 with HeLa cells. Virology 51:270-278. [DOI] [PubMed] [Google Scholar]
- 30.Oberste, M. S., K. Maher, M. R. Flemister, G. Marchetti, D. R. Kilpatrick, and M. A. Pallansch. 2000. Comparison of classic and molecular approaches for the identification of untypeable enteroviruses. J. Clin. Microbiol. 38:1170-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Register, R. B., C. R. Uncapher, A. M. Naylor, D. W. Lineberger, and R. J. Colonno. 1991. Human-murine chimeras of ICAM-1 identify amino acid residues critical for rhinovirus and antibody binding. J. Virol. 65:6589-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reischl, A., M. Reithmayer, G. Winsauer, R. Moser, I. Gosler, and D. Blaas. 2001. Viral evolution toward change in receptor usage: adaptation of a major group human rhinovirus to grow in ICAM-1-negative cells. J. Virol. 75:9312-9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reithmayer, M., A. Reischl, L. Snyers, and D. Blaas. 2002. Species-specific receptor recognition by a minor-group human rhinovirus (HRV): HRV serotype 1A distinguishes between the murine and the human low-density lipoprotein receptor. J. Virol. 76:6957-6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudenko, G., L. Henry, K. Henderson, K. Ichtchenko, M. S. Brown, J. L. Goldstein, and J. Deisenhofer. 2002. Structure of the LDL receptor extracellular domain at endosomal pH. Science 298:2353-2358. [DOI] [PubMed] [Google Scholar]
- 35.Rueckert, R. R. 1996. Picornaviridae: the viruses and their replication, p. 609-654. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, Pa.
- 36.Russell, D. W., M. S. Brown, and J. L. Goldstein. 1989. Different combinations of cysteine-rich repeats mediate binding of low density lipoprotein receptor to two different proteins. J. Biol. Chem. 264:21682-21688. [PubMed] [Google Scholar]
- 37.Savolainen, C., S. Blomqvist, M. N. Mulders, and T. Hovi. 2002. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J. Gen. Virol. 83:333-340. [DOI] [PubMed] [Google Scholar]
- 38.Sherry, B., and R. Rueckert. 1985. Evidence for at least two dominant neutralization antigens on human rhinovirus 14. J. Virol. 53:137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uncapher, C. R., C. M. Dewitt, and R. J. Colonno. 1991. The major and minor group receptor families contain all but one human rhinovirus serotype. Virology 180:814-817. [DOI] [PubMed] [Google Scholar]
- 40.Vash, B., N. Phung, S. Zein, and D. DeCamp. 1998. Three complement-type repeats of the low-density lipoprotein receptor-related protein define a common binding site for RAP, PAI-1, and lactoferrin. Blood 92:3277-3285. [PubMed] [Google Scholar]
- 41.Willnow, T. E., K. Orth, and J. Herz. 1994. Molecular dissection of ligand binding sites on the low density lipoprotein receptor-related protein. J. Biol. Chem. 269:15827-15832. [PubMed] [Google Scholar]
- 42.Wilson, C., M. R. Wardell, K. H. Weisgraber, R. W. Mahley, and D. A. Agard. 1991. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science 252:1817-1822. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto, T., C. G. Davis, M. S. Brown, W. J. Schneider, M. L. Casey, J. L. Goldstein, and D. W. Russell. 1984. The human LDL receptor: a cysteine-rich protein with multiple Alu sequences in its mRNA. Cell 39:27-38. [DOI] [PubMed] [Google Scholar]