Abstract
The wnt signaling pathway is constitutively activated in colon tumors by mutations in the adenomatous polyposis coli and β-catenin genes. We have modified the minute virus of mice (MVM) P4 promoter to make it responsive to wnt signaling by inserting binding sites for the heterodimeric β-catenin/Tcf transcription factor. In luciferase assays we can see up to 20-fold selectivity of Tcf mutant P4 promoters for cells with activated wnt signaling. Hybrid MVM/H-1 viruses containing Tcf mutant promoters were tested for NS1 expression, viral DNA replication, virus replication, and cytopathic effect on colon, lung, kidney, and cervical cancer cell lines. Activation of the wnt pathway by expression of ΔN-β-catenin increased NS1 expression and viral burst size in 293T and H1299 lung cancer cells, showing that the Tcf mutant P4 promoter can respond to wnt signals in the context of the virus. Compared to the parental virus, the burst size of the Tcf mutant viruses was reduced at least 1,000-fold in H1299, 293T, NB324K, and HeLa cells, which have inactive wnt signaling pathways. The burst size and cytopathic effect of the Tcf viruses was near wild-type levels in SW480 and Isreco1 colon cancer cell lines, which have high Tcf activity. The high specificity of these viruses should permit the development of H-1 virus-based vectors which combine high safety and greater efficacy in cancer therapy.
Several replicating adenoviruses have now been tested in clinical trials for the treatment of cancer (2). The strength of this approach is that the therapeutic effect of the injected virus is augmented by that of the virus produced within the tumor. Since the acute side effects of treatment are related to the amount of virus injected (39), the ability to inject a smaller amount of virus is an important advantage of replicating viruses. Deliberate release of replicating viruses designed to kill human cells raises biosafety concerns, but with adenovirus the immune system limits the number of cycles of virus replication so effectively that poor efficacy is a much more important barrier to the widespread use of these viruses for cancer therapy than safety issues. The genetic instability of tumors is almost certain to produce broad-spectrum resistance to adenoviruses, at least in a minority of patients. To overcome this resistance it would be useful to have alternative agents drawn from an entirely different virus family (2). Most current targeting exploits tumor-specific defects in the regulation of cellular DNA replication or transcription. Several RNA viruses have been proposed for use as cancer therapies, but all share the problem that it is difficult to rationally design tumor specificity into an RNA virus. Among DNA viruses, most are known or suspected to cause serious diseases, such as progressive multifocal leucoencephalopathy in AIDS patients by JC virus or mesothelioma by simian virus 40. Other DNA viruses are so large that the consequences of modification of the viral genome are difficult to predict. These viruses, such as herpesviruses, often produce latent infections which would be a source of concern in cured patients. Still other DNA viruses are either difficult to produce, like papillomaviruses, or replicate in the cytoplasm, like poxviruses. The most interesting remaining candidates are members of the autonomous parvovirus family.
B19 virus is the only parvovirus known to cause human disease. Minute virus of mice (MVM) and H-1 parvoviruses have several properties that make them interesting candidates for development as cancer therapies. They have been shown to reduce the incidence of spontaneous and chemically induced tumors in laboratory animals (14, 18, 43). They possess intrinsic oncotropism and show selective toxicity to tumor cells in culture (8, 9, 43). The viral particles are small, which should favor spread within tumors (15). There is little or no preexisting neutralizing antibody and no reason to expect cross-resistance to adenovirus. The drawbacks of autonomous parvoviruses are their inability to induce S phase and their limited potential for expressing transgenes. Although they are currently difficult to produce in large quantities, intensive efforts to produce other parvoviruses for gene therapy are likely to overcome this obstacle in the near future. Despite their inherent advantages, H-1 virus and MVM are too restricted in their tropism to be useful without modification. For example, careful analysis of MVM host range mutants has identified blocks at multiple levels, including decapsidation, amplification, and postencapsidation, in human cell lines (32, 37).
Artificially removing barriers to replication of wild-type viruses raises biosafety concerns. These issues were addressed with adenovirus by attenuating the virus before attempting to increase the toxicity or modify the tropism of the virus. We have chosen to adopt the same strategy with autonomous parvovirus. To build an additional layer of safety into H-1 virus we have inserted binding sites for Tcf family transcription factors into the P4 promoter, which controls expression of the NS1 and NS2 proteins. NS1 is a multifunctional phosphoprotein (10) that plays essential roles in viral replication (35), viral promoter transactivation (13, 21, 25), and virus toxicity (5). Tcf regulation of NS1 expression should restrict virus replication to colon tumors because constitutive activation of the wnt signaling pathway is a universal causal oncogenic defect in colon tumors (36). In normal cells, Tcf's recruit Groucho and other corepressors to prevent transcription. Activation of wnt signaling, either by binding of wnt ligand to Frizzled receptors or through mutation of the adenomatous polyposis coli (APC) and β-catenin genes in colon tumors, results in activation of transcription from promoters containing Tcf binding sites. We describe here the properties of hybrid H-1/MVM parvoviruses with four Tcf sites inserted into the P4 promoter.
MATERIALS AND METHODS
Cell lines.
HT29 and 293T cells were supplied by the American Type Culture Collection (ATCC). Isreco1 and SW480 cells were provided by B. Sordat. HeLa cells were supplied by the Imperial Cancer Research Fund cell production lab. The H1299-derived cell line H24 containing the Tet-VP16 transactivator was provided by C. Prives (6). NB324K cells were supplied by P. Beard. CR2 cells were derived from 293T cells by infection with a lentivirus expressing Myc-tagged ΔN-β-catenin (19, 31, 42). cMM1 cells were derived from H24 cells by stable transfection with pMM1, which contains a Myc-tagged ΔN-β-catenin cDNA (42) cloned into pUHD10-3 (20). H24 cells were cotransfected with 10 μg of pMM1 and 1 μg of pBabe-puro (29) and were grown in medium containing 2 μg of puromycin per ml and 1 μg of tetracycline per ml. Transformants were single cell cloned and screened for inducible ΔN-β-catenin expression by Western blotting and in situ staining using the anti-Myc antibody 9E10 (16) and the anti-β-catenin antibody C19220 (Transduction Laboratories, Basel, Switzerland). ΔN-β-catenin expression was induced by removing tetracycline from the medium.
Parvovirus mutagenesis.
The P4xLuc series of plasmids has been described previously (11). P4mut25Luc was cut with PmeI and self-ligated to eliminate a duplication of the NcoI-AflIII fragment within the P4 promoter. P4mut22Luc, P4mut23Luc, and P4mut24Luc contain a BglII linker which replaces the E2F, ets, and Sp1 sites, respectively. P4mut19Luc and P4mut25Luc contain the linker before the E2F and after the Sp1 sites, respectively. A double-stranded oligonucleotide containing four Tcf binding sites (GATC-TCCTTTGATCTTAATCCCTTTGATCTGGATCCCTTTGATCTCCAACCCTTT-GATC) was cloned in both orientations into the BglII site of the P4xLuc plasmids to give plasmids pMM23 and -24 (mut19), pMM25 and -26 (mut22), pMM27 and -28 (mut23), pMM29 and -30 (mut24), and pMM33 and -34 (mut25), where pMM23, -25, -27, -29, and -33 have the C/T-rich strand of the Tcf site on the viral genomic strand and the rest have the Tcf sites reversed. The P4-Tcf promoters obtained were inserted into phH1, a plasmid containing the MVM left hairpin and P4 promoter in the H-1 virus genome (22). To achieve this, the 3,298-bp NdeI-EcoRI fragment of phH1 was blunted and self-ligated to give pMM39, in which the vector AflIII site was destroyed by self-ligation. The 240-bp AflIII-NcoI fragment of the P4-Tcf-Luc plasmids (pMM24, -26, -28, -30, and -34) was cloned into the AflIII-NcoI sites of pMM39 to give pMM42, -44, -46, -48, and -50. Finally, the 4,565-bp SpeI-SspI fragment of phH1 was cloned into the SpeI-SspI sites of pMM42 to -50 to give pMM66 to -74, the plasmids used to make viruses vMM66 to -74.
Parvovirus amplification and titration.
Virus was produced by cotransfection of phH1-derived plasmids and pXNS1 (33) into cR2 cells. At 3 days posttransfection cells were harvested by scraping, washed once in phosphate-buffered saline, and resuspended in 50 mM Tris-0.5 mM EDTA, pH 8.7. Virus was released by five rounds of freeze-thawing, cell debris was pelleted by centrifugation, and the virus-containing supernatant was stored at 4°C. Viral titers were estimated by measuring the amount of viral DNA in HT29 cells 24 h after infection in the presence of hydroxyurea. This approach was used to avoid underestimating the titer by plaque assay or infectious center assay on nonpermissive indicator cells such as NB324K cells. None of the viruses replicates in HT29 cells in the presence of hydroxyurea. The amount of viral DNA was measured by quantitative PCR and expressed as the number of genome copies per milliliter.
Quantitative PCR assays.
Cells were harvested and DNA extraction was performed with a Dneasy tissue kit (Qiagen, Basel, Switzerland) according to the manufacturer's instructions. Ten nanograms of DNA was used for each quantitative PCR. PCR was performed by use of a TaqMan Universal PCR master mix (Perkin-Elmer, Rotkreuz, Switzerland), 800 nM primers (Invitrogen, Basel, Switzerland), and 500 nM TaqMan probe (MWG Biotech AG, Munchenstein, Switzerland) in a PE5700 PCR machine (Perkin-Elmer). The primers and probe lie in the NS1 coding sequence as follows: forward primer, CCACACTCAAAGAGTTGGTACATAA; reverse primer, CACCTGGTTGAGCCATCAT; and probe, AACTGTCTGGCTGCATCATCATCCA.
Luciferase assays.
Dual luciferase reporter assays were performed according to the manufacturer's instructions (Promega, Madison, Wis.) in a Biocounter (Lumac bv, Landgraaf, The Netherlands). cMM1 cells were seeded at 3.5 × 105 cells per 35-mm-diameter well 24 h before transfection in medium with or without 1 μg of tetracycline per ml, incubated with Lipofectamine (Invitrogen) for 18 h with 100 ng of reporter plasmid and 1 ng of control Renilla luciferase plasmid (pRL-CMV; Promega), and harvested 48 h later. SW480 cells were seeded at 4 × 105 cells per 35-mm-diameter well; incubated with Lipofectamine for 18 h with 100 ng of reporter plasmid, 500 ng of ΔN-Tcf4 plasmid (23), and 1 ng of pRL-CMV; and harvested 48 h later. 293T cells were seeded at 4 × 105 cells per 35-mm-diameter well 24 h before transfection; transfected using a calcium phosphate protocol for 6 h with 100 ng of reporter plasmid, 50 ng of β-catenin Δ45S plasmid (30), 50, 100, or 200 ng of ΔN-Tcf4 plasmid, and 5 ng of Renilla plasmid (pRL-TK; Promega); and harvested 24 h later. Each value is normalized to the activity of the Renilla control.
Western blotting.
Cells were infected at a multiplicity of 500 viral genome copies/cell for 1 h in serum-free Dulbecco's modified Eagle medium (DMEM), after which the medium was replaced with DMEM containing 10% fetal bovine serum (FBS) (Invitrogen). Cells were harvested 24 h later. NS1 expression was detected with SP8 rabbit polyclonal antibody (3, 17). The Myc-tagged ΔN-β-catenin was detected with anti-β-catenin antibody C19220 (Transduction Laboratories).
Virus replication assay.
Cells were infected at a multiplicity of 500 viral genome copies/cell for 1 h in serum-free DMEM, after which the medium was replaced with DMEM containing 10% FBS (Invitrogen). Cells were harvested 48 h later and lysed by five cycles of freeze-thawing. The viral titer in the supernatant was tested on HT29 cells as described above.
Cytopathic effect assays.
Cells were seeded in six-well plates at a density of 50% the day before infection. Infection was performed in DMEM containing 10% FBS (Invitrogen) with serial 10-fold dilutions of virus suspension, starting at a multiplicity of 1,000 viral genome copies/cell. At 8 days postinfection cells were fixed with 4% formaldehyde in PBS and stained with 1% crystal violet in 20% methanol.
RESULTS
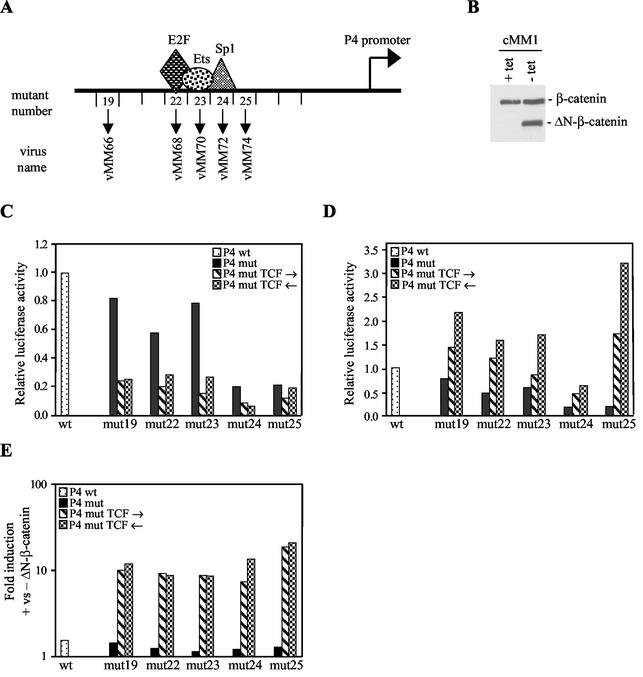
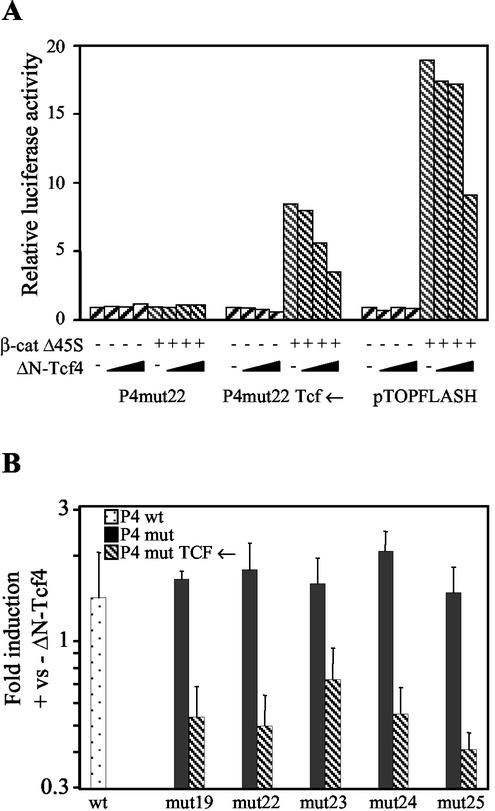
The P4 promoter has previously been characterized by BglII linker scanning mutagenesis in a hybrid MVM/H-1 virus construct (11). We inserted four Tcf binding sites, in both orientations, into the BglII linker of plasmids with mutations outside the hairpin (Fig. 1A). The first mutant nucleotide in mut19 is 12 nucleotides downstream of the NS1 nick site required for DNA replication. To test the inducibility of the promoters by wnt pathway activation, H1299 human lung cancer cells were engineered to express an active β-catenin mutant (ΔN-β-catenin) from a tetracycline-regulated promoter. The N-terminal deletion in the mutant removes the destruction box. The wnt pathway is inactive in parental H1299 cells. Western blotting showed that the resulting cell line (cMM1) expresses the mutant β-catenin only after removal of tetracycline from the medium (Fig. 1B). The upper band represents endogenous β-catenin, and the lower band represents the exogenous active mutant. Despite being present in reasonable amounts, the endogenous protein does not normally activate transcription of Tcf target genes in the absence of wnt ligand. Transfection of P4-luciferase reporters into cMM1 cells in the presence of tetracycline was performed to test the basal activity of the Tcf-P4 promoters (Fig. 1C). Deletion of the Sp1 site (mut24) or insertion of a BglII site immediately after the Sp1 site (mut25) reduced the luciferase activity to about 20% of the wild-type promoter activity. BglII insertion alone in mut19, -22, and -23 had smaller effects. Insertion of Tcf sites reduced the luciferase activity of these mutants to about 20% of the wild-type promoter activity and further reduced the already low activity of the mut24 promoter. This suggests that Tcf recruits corepressors to the promoter, consistent with published reports that Tcf binds to Groucho and C-terminal binding protein (CtBP) in the absence of a wnt signal (36). Induction of ΔN-β-catenin expression by tetracycline removal activated all of the Tcf-P4 promoters, except the mut24 version, to wild-type or supra-wild-type levels (Fig. 1D). The inducibility of the Tcf-P4 promoters by ΔN-β-catenin was 10- to 20-fold for all of the mutants (Fig. 1E, which combines the data shown in Fig. 1C and D). Low basal activity (Fig. 1C), together with wild-type-induced activity (Fig. 1D), are sensible prerequisites for producing a selective and active virus. To demonstrate that the Tcf sites confer responsiveness to Tcf4, 293T cells were cotransfected with luciferase reporters, a constitutively active β-catenin mutant (Δ45S), and increasing amounts of a dominant-negative Tcf4 mutant (ΔN-Tcf4) (Fig. 2A). The ΔN-Tcf4 mutant lacks the amino-terminal β-catenin binding site but can still bind to Groucho and CtBP (36). The pTOPFLASH reporter, which contains multiple Tcf sites, is widely used to test activation of the wnt pathway (23). Δ45S β-catenin had no effect on the reporters lacking Tcf sites, but it activated the Tcf-P4 promoter 9-fold and the pTOPFLASH reporter 19-fold (Fig. 2A). The lower inducibility of the Tcf-P4 promoter relative to pTOPFLASH may reflect the greater complexity of the Tcf-P4 promoter, which contains additional transcription factor binding sites not present in pTOPFLASH. ΔN-Tcf4 had no effect on the P4mut promoter lacking the Tcf site but inhibited the transactivation of Tcf reporters by β-catenin. Activation of both pTOPFLASH and the Tcf-P4 reporter by Δ45S β-catenin was reduced to a similar extent by ΔN-Tcf4. To confirm that Tcf contributes to the activity of the Tcf-P4 promoters in colon cancer cells, SW480 cells were transfected with the luciferase reporters and the dominant-negative ΔN-Tcf4 mutant. Unlike the wild-type and P4mut promoters, the Tcf-P4 promoters were repressed two- to fourfold by ΔN-Tcf4 expression (Fig. 2B). Promoter activity was reduced but not abolished by ΔN-Tcf4 in both 293T and SW480 cells, suggesting that this Tcf4 mutant does not act as a strong dominant negative. Taken together, the luciferase assays represented in Fig. 1 and 2 show that insertion of Tcf sites into the parvovirus P4 promoter confers responsiveness to activation of the prototypic wnt signaling pathway.
FIG. 1.
Tcf-P4 promoters respond to activation of the wnt pathway. (A) Promoter map showing the position of the Tcf sites. The Tcf insertions at positions 22, 23, and 24 replace the E2F, ets, and Sp1 sites. (B) Western blot showing expression of ΔN-β-catenin (lower band) after removal of tetracycline from the medium. The upper band corresponds to endogenous β-catenin. (C to E) Activity of the Tcf-P4 promoters determined by luciferase assays in cMM1 cells in a basal state (plus tetracycline) (C) and an induced state (minus tetracycline) (D) and fold induction by tetracycline removal (E). P4 mut, plasmids with BglII linkers at the sites shown in panel A; wt, wild type. The Tcf oligonucleotide was inserted in both possible orientations: “P4 mut Tcf →” indicates plasmids with the C/T-rich strand of the Tcf site on the genomic strand, and “P4 mut Tcf ←” indicates plasmids with the C/T-rich strand on the coding strand. The vMM viruses have the C/T-rich strand of the Tcf sites on the viral coding (antigenomic) strand.
FIG. 2.
Luciferase assays showing repression of Tcf-P4 promoters by dominant-negative Tcf4 (ΔN-Tcf4). (A) 293T cells were transfected with pTOPFLASH or P4 luciferase reporter, mutant β-catenin expression vector (β-cat Δ45S), and 50, 100, or 200 ng of ΔN-Tcf4 expression vector. (B) SW480 cells were transfected with P4 luciferase reporters and 500 ng of ΔN-Tcf4 expression vector. wt, wild type.
The constructs with the C/T-rich strand of the Tcf sites on the viral coding strand were selected for further study because they gave the highest absolute activity (Fig. 1D). The Tcf promoters were transferred to a viral vector (phH1 [22]) for production of virus. The viruses are called vMM66, -68, -70, -72, and -74 (Fig. 1A). The producer cell line (cR2) was made by infection of 293T cells with a lentivirus expressing ΔN-β-catenin to activate Tcf-dependent transcription. To further reduce the risk of selecting suppressor mutations in the P4 promoter, the producer cells were cotransfected with phH1 vectors and a plasmid expressing NS1 (pXNS1 [33]). Conventional indicator cell lines used for infectious center assays and plaque assays are not permissive for the Tcf viruses. To titer the Tcf viruses, HT29 cells were infected in the presence of hydroxyurea and DNA was harvested after 24 h. Preliminary tests with wild-type virus showed that the viral DNA content of HT29 cells was the same after 24 h in the presence of hydroxyurea as after 4 h in the absence of hydroxyurea, suggesting that the assay measures unreplicated DNA that is stably associated with the cell in a nuclease-resistant form. The number of copies of viral DNA was measured by quantitative PCR. NB324K cells are highly permissive for H-1 virus and form large, easily scored plaques. For phH1, a titer of 700 genome copies/ml on HT29 cells corresponds to a titer of 1 PFU/ml on NB324K cells.
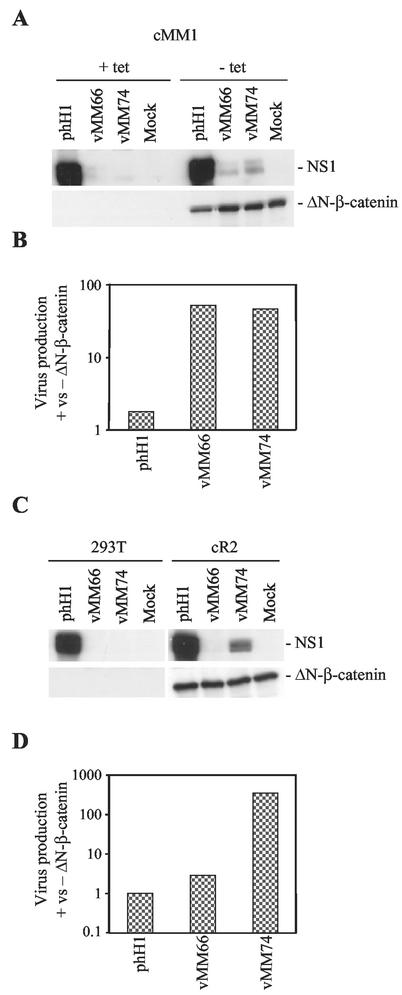
To determine whether the Tcf-P4 promoters can respond to activation of the wnt pathway in the context of the virus, cMM1 cells were infected with parental virus (phH1) or the Tcf versions of mut19 and -25 (vMM66 and vMM74) in the presence or absence of tetracycline (Fig. 3A). Induction of β-catenin expression resulted in an increase in NS1 expression from both Tcf viruses, albeit not to wild-type levels, showing that the Tcf-P4 promoter can respond to activation of the wnt signaling pathway in the context of the virus. This is consistent with previous studies showing that heterologous genes can be expressed from modified P4 promoters in recombinant LuIII parvoviruses (27, 28). Differences in NS1 level observed by Western blotting at 24 h may reflect not only differences in promoter activity but also changes in template copy number following virus replication, which is itself caused by NS1 expression. To test whether promoter activation is accompanied by virus replication, the infections were repeated and burst assays were performed. Compared to parental phH1 virus, vMM66 and -74 showed a 50-fold increase in burst size following expression of the β-catenin mutant (Fig. 3B). The small increase in phH1 replication following ΔN-β-catenin expression could be due to an increase in S-phase fraction caused by the oncogene. As a further test of the Tcf-P4 response to artificial activation of the wnt pathway, 293T cells and ΔN-β-catenin-expressing derivatives (cR2 cells) were infected with vMM66 and -74. Neither Tcf virus gave detectable NS1 expression in parental 293T cells, whereas expression could again be detected from vMM74 in cR2 cells (Fig. 3C). In parental 293T cells, vMM74 virus production was reduced 4,000-fold compared to phH1. Consistent with the increase in NS1 expression, there was a 300-fold increase in vMM74 virus production in the cells expressing mutant β-catenin (Fig. 3D). We conclude that artificial activation of the wnt pathway by exogenous ΔN-β-catenin can activate Tcf-P4 promoters in the context of the virus and that this leads to virus production.
FIG. 3.
ΔN-β-catenin renders nonpermissive cells permissive for Tcf virus replication. (A and C) Western blots done at 24 h postinfection for NS1 and ΔN-β-catenin in cells infected with phH1, vMM66, and vMM74. (B and D) Burst assays measuring virus production at 48 h postinfection with phH1, vMM66, and vMM74, expressed as the ratio of virus production in the presence and absence of ΔN-β-catenin. (A and B) ΔN-β-catenin expression was induced in cMM1 cells by tetracycline removal. (C and D) 293T and cR2 cells (293T derivatives expressing ΔN-β-catenin).
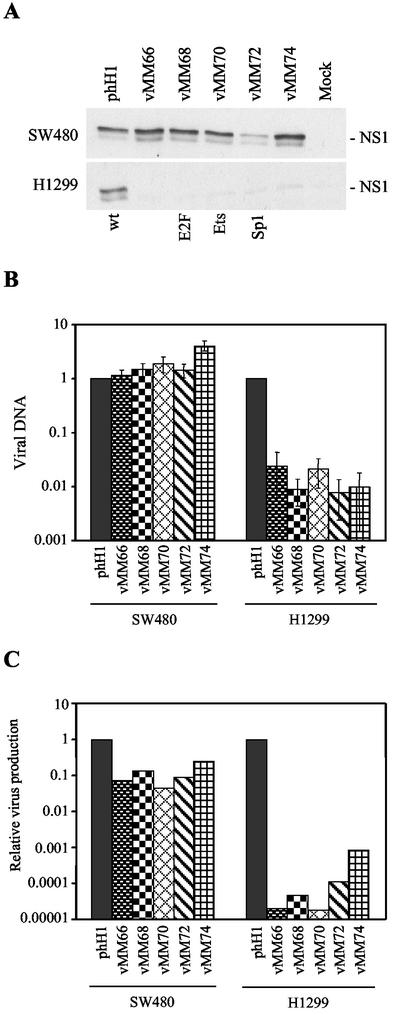
The differences between vMM66 and -74 in cMM1 and cR2 cells indicated that the site of Tcf insertion in the promoter might have large effects on the behavior of the Tcf viruses in other cell lines. Western blotting 24 h after infection of SW480 colon cancer cells, in which the wnt pathway is activated by mutation of APC, showed that NS1 is expressed normally from all of the Tcf viruses except vMM72, the virus with Tcf sites replacing the Sp1 site (Fig. 4A, upper panel). Deletion of the Sp1 site is known to reduce the activity of the basal P4 promoter (11). Normal NS1 expression from the other Tcf mutant viruses demonstrates that the Tcf mutations do not interfere with decapsidation or conversion of the viral DNA to the monomeric replicative form in these cells. In control H1299 lung cancer cells, the phH1 parental virus was able to express NS1, but all of the Tcf viruses were defective in NS1 expression (Fig. 4A, lower panel). This suggests that the insertion of Tcf sites into the P4 promoter may confer selectivity for colon cancer cells, as expected from the luciferase assays. To determine whether the level of regulation of NS1 expression is sufficient to modulate viral DNA replication, the amount of DNA in cells 24 h after infection was measured by quantitative PCR (Fig. 4B). All of the Tcf viruses gave wild-type levels of DNA replication in SW480 cells. In contrast, the Tcf viruses gave 100-fold less viral DNA than the parental virus in H1299 cells. To test whether actual virus is produced from the replicated DNA, burst assays were performed (Fig. 4C). The titers were calculated by HT29 assay because NB324K indicator cells are nonpermissive for the Tcf viruses. Tested in this way, the absolute burst size of phH1 was 0.04 and 2 in SW480 and H1299 cells, respectively. In control experiments, the burst size of phH1 in NB324K cells was 5 by HT29 assay and 19 by PFU titer on NB324K cells, indicating that the HT29 assay may underestimate the true burst size. The results for the Tcf viruses shown in Fig. 3C are expressed relative to phH1 to eliminate the confounding effect of differences in the permissiveness of individual cell lines for autonomous parvoviruses. The relative burst size of the Tcf mutant viruses was reduced ∼10-fold in SW480 cells but was reduced >1,000-fold in H1299 cells (Fig. 4C). We conclude that the insertion of Tcf sites in the P4 promoter modifies the host range of the virus and most likely confers colon cell line and wnt pathway-specific viral replication.
FIG.4.
Tcf virus infection of SW480 and H1299 cells. (A) Western blots done at 24 h postinfection showing expression of NS1 in the colon cancer cell line SW480 (upper panel) and the lung cancer cell line H1299 (lower panel). (B) Viral DNA content at 24 h postinfection of SW480 and H1299 cells, as determined by quantitative PCR. The values are normalized to the parental phH1 results. (C) Burst assays in SW480 and H1299 cells measuring virus production at 48 h postinfection. The values are normalized to the phH1 results.
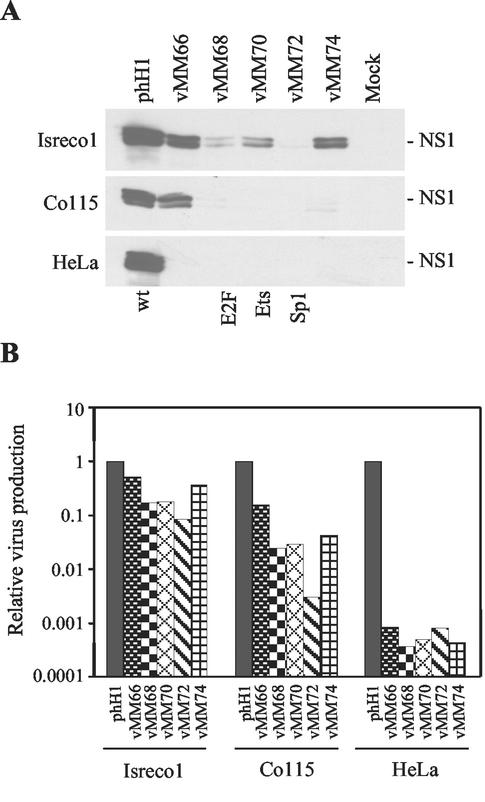
We have previously shown that adenoviruses with early promoters regulated by the wnt pathway have wild-type activity in about half of the colon cancer cell lines tested (4). This is partly due to differences in the absolute level of Tcf activity in the different cell lines but could also reflect intrinsic limitations of adenoviruses as tumor targeting vectors. To determine whether the Tcf-regulated parvoviruses show similar cell line specificity, a panel of cell lines was tested (Fig. 5). The absolute burst size of phH1 calculated by HT29 assay was 0.08, 4, and 7 in Isreco1, Co115, and HeLa cells, respectively. Isreco1 and Co115 are colon tumor cell lines with high and intermediate Tcf activity levels, respectively (unpublished data). Only the parvoviruses with the Tcf sites in positions 19 and 25 (vMM66 and -74) expressed NS1 well and had a near normal burst size in Isreco1 cells (Fig. 5). Only vMM66 was able to express NS1 in Co115 cells (Fig. 5A). A possible explanation for the differences in NS1 expression by the Tcf viruses in SW480, Isreco1, and Co115 cells is that the contribution of the ets and E2F proteins to overall P4 activation depends on the state of the relevant signaling pathways in the different cells. The burst size of all the Tcf mutant viruses was reduced in Co115, ranging from a 10-fold reduction with vMM66 to a 300-fold reduction with vMM72, in which the Tcf site replaces the Sp1 site. These differences were all smaller than the ∼1,000-fold reduction in burst size of the Tcf viruses in HeLa cells (Fig. 5B). vMM66 and -74 were also tested in NB324K cells, which are highly permissive for H-1 virus replication and have inactive wnt signaling. Compared to phH1, the burst size was reduced 1,000-fold for vMM66 and 6,000-fold for vMM74 in NB324K cells.
FIG. 5.
Tcf virus infection of Isreco1, Co115, and HeLa cells. (A) Western blots done at 24 h postinfection showing expression of NS1 in the colon cancer cell lines Isreco1 and Co115 (upper and middle panels, respectively) and in HeLa cells (lower panel). (B) Burst assays in Isreco1, Co115, and HeLa cells measuring virus production at 48 h postinfection. The values are normalized to the phH1 results.
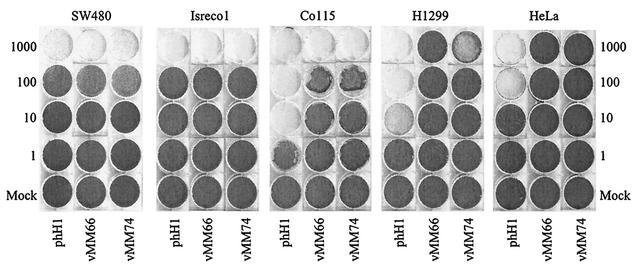
To determine the toxicity of the viruses, we performed cytopathic effect assays on H1299, HeLa, SW480, Isreco1, and Co115 cells (Fig. 6). There were substantial differences in the sensitivities of the cell lines to the parental virus (Fig. 6, phH1 wells). In SW480 and Isreco1 cells, the Tcf mutant viruses vMM66 and vMM74 were as toxic as phH1. In Co115 cells, phH1 was 100-fold more active than the Tcf mutant viruses. H1299 cells were sensitive to phH1 and to the largest amount of vMM74 used but were resistant to vMM66. HeLa cells were sensitive to phH1 but were resistant to both vMM66 and vMM74. These results correlate with those obtained in burst assays (Fig. 4 and 5), except those for Co115, in which vMM74 was less active than vMM66. The reason for the difference is unknown, but it could reflect the longer duration of the cytopathic effect assay. We conclude that the Tcf-regulated parvoviruses show selective toxicity for some cell lines with activation of the wnt pathway.
FIG. 6.
Cytopathic effect assays on SW480, Isreco1, Co115, HeLa, and H1299 cells infected with phH1, vMM66, or vMM74. Cells were stained with crystal violet at 8 days postinfection. The multiplicity of infection is expressed as genome copies per cell based on the HT29 titer.
DISCUSSION
The viruses we have produced are restricted at multiple levels. In addition to the intrinsic oncotropism of H-1 parvovirus, our viruses can only replicate in cells with activated wnt signaling. The chance that these viruses will replicate in normal tissues is thus remote, because the few normal cells with active wnt signaling, like colon crypt stem cells, are unlikely to fulfill the other requirements for parvovirus replication.
Previous attempts to restrict autonomous parvovirus replication were confounded by positive feedback of NS1 on its own expression (26). This is mainly due to the increase in copy number of the viral genome following DNA replication, which is induced by NS1 (24). This change in copy number, rather than direct transcriptional regulation by Tcf, explains the very large differences in NS1 expression detected by Western blotting at 24 h in many of our experiments. Previous studies on the phH1 virus with a BglII linker replacing the E2F site in the P4 promoter showed that E2F activity is essential for virus production (12). Exogenous NS1 was able to complement the defect in trans, showing that the mutation did not interfere with other essential functions. Presumably, there is a threshold level of NS1 below which positive feedback is not triggered. By recruiting Tcf to the promoter, we repress P4 transcription in normal cells and thereby maintain NS1 levels below this threshold. Once the threshold is exceeded, differences in promoter activity may have little effect. The fact that the Tcf viruses did not replicate better than wild-type virus in SW480 cells indicates that NS1 expression from the wild-type promoter is not limiting for replication in these cells.
We have not directly compared our Tcf mutant viruses with the parental viruses containing only BglII linkers at the corresponding sites in the P4 promoter. The fact that the virus with Tcf sites in place of the E2F site had a relatively normal burst size on SW480 cells suggests that Tcf can substitute for the normally essential E2F and transactivate the promoter in the context of the virus. Tcf could partially compensate for deletion of the Sp1 site, but overall this virus was still the least active, consistent with previous data showing that Sp1 is important for basal promoter activity (11). In principle, the combination of Tcf and ets sites is an ideal choice to target colon tumors, because these tumors typically have activated wnt and ras signaling pathways. The role of E2F is more controversial, because mutation of the proximal players in the retinoblastoma (Rb) pathway (cyclin D, p16, cdk4, and Rb) is strangely absent in colon cancer (1). Indeed, one study reported that Rb is the most strongly overexpressed gene in colon cancer (38). Indirect inactivation of Rb, for example, by Tcf-dependent transactivation of the cyclin D1 promoter, is a possible explanation (41). One potential drawback of vMM66 and -74 is that loss of the Tcf sites could revert the P4 promoter to the wild type. We have not seen changes in the promoter sequence after sequential passage of vMM74 on Co115 cells (V. Perrin and R. Iggo, unpublished data), but this is clearly an issue which should be addressed.
Tumor-specific replication is only the first requirement for converting H-1 parvovirus for colon cancer therapeutic use. The burst size of the parental phH1 virus was 0.04 in SW480 cells and 0.08 in Isreco1 cells. This is very low and clearly incompatible with the therapeutic goal of disseminating new viruses within a tumor. The phH1 burst size was higher but still low (range, 2 to 7) in H1299, HeLa, and Co115 cells. It is possible that the low burst size in some way reflects a defect in the HT29 assay. The phH1 burst size measured by PFU in NB324K cells was 19, compared with 5 measured by the HT29 assay, lending some credence to this view. The lack of a good cell line for performing plaque or infectious center assays with the Tcf viruses obliged us to measure the viral titer by HT29 assay, which is based on PCR of infected indicator cells. Like all PCR assays, it can detect defective viruses with deletions outside the PCR product. The conclusion that the viruses show selectivity for cells with active wnt signaling is based on comparing the burst size of the phH1 virus with that of the Tcf viruses in individual cell lines. By expressing the result in this way we sidestep the issue of differences in absolute burst size and can detect at least 1,000-fold decreases in burst size for the Tcf viruses in H1299, 293T, NB324K, and HeLa cells. Parvoviruses are unusually dependent on host functions to permit virus replication. Hence, it is not surprising that there should be differences between cell lines. To produce a colon cancer therapeutic virus will require viruses that are more broadly active in colon cancer cells, and in cells like SW480 this will require changes that are unrelated to wnt signaling.
The small size of the virus means it is not possible to express large prodrug-activating enzymes from replication-competent parvoviruses. Hence, cell killing must rely heavily on the intrinsic properties of the virus. Several toxic effects of parvoviruses have been described. The viral hairpins may trigger checkpoints which selectively kill p53 mutant cells (34, 40). NS1 expression is also toxic, particularly to tumor cells (5). Repression of the Tcf-P4 promoter by Groucho and CtBP should protect normal cells from the toxic effects of NS1. Several mutations in NS1 have been described which alter the balance between replication and cytotoxicity (7). By combining these with modifications that alter the strength of the P4 promoter it may be possible to produce a more effective replicating parvovirus.
In summary, we have developed new autonomous parvoviruses whose replication is tightly restricted to tumor cells with activated wnt signaling. The ease of manipulation of the parvovirus genome makes these viruses ideal tools to develop and test new cancer-targeting viruses.
Acknowledgments
We thank N. Salomé, D. Trono, B. Sordat, H. Clevers, and C. Prives for providing reagents. We thank V. Perrin, P. Beard, L. Deleu, and J. P. F. Nüesch for helpful discussions.
We thank the Swiss National Science Foundation and EMBO for financial support.
REFERENCES
- 1.Bartkova, J., M. Thullberg, P. Slezak, E. Jaramillo, C. Rubio, L. H. Thomassen, and J. Bartek. 2001. Aberrant expression of G1-phase cell cycle regulators in flat and exophytic adenomas of the human colon. Gastroenterology 120:1680-1688. [DOI] [PubMed] [Google Scholar]
- 2.Biederer, C., S. Ries, C. H. Brandts, and F. McCormick. 2002. Replication-selective viruses for cancer therapy. J. Mol. Med. 80:163-175. [DOI] [PubMed] [Google Scholar]
- 3.Brockhaus, K., S. Plaza, D. J. Pintel, J. Rommelaere, and N. Salome. 1996. Nonstructural proteins NS2 of minute virus of mice associate in vivo with 14-3-3 protein family members. J. Virol. 70:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunori, M., M. Malerba, H. Kashiwasaki, and R. Iggo. 2001. Replicating adenoviruses that target tumors with constitutive activation of the wnt signaling pathway. J. Virol. 75:2857-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caillet-Fauquet, P., M. Perros, A. Brandenburger, P. Spegelaere, and J. Rommelaere. 1990. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J. 9:2989-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, X., L. J. Ko, L. Jayaraman, and C. Prives. 1996. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 10:2438-2451. [DOI] [PubMed] [Google Scholar]
- 7.Corbau, R., V. Duverger, J. Rommelaere, and J. P. Nuesch. 2000. Regulation of MVM NS1 by protein kinase C: impact of mutagenesis at consensus phosphorylation sites on replicative functions and cytopathic effects. Virology 278:151-167. [DOI] [PubMed] [Google Scholar]
- 8.Cornelis, J. J., P. Becquart, N. Duponchel, N. Salome, B. L. Avalosse, M. Namba, and J. Rommelaere. 1988. Transformation of human fibroblasts by ionizing radiation, a chemical carcinogen, or simian virus 40 correlates with an increase in susceptibility to the autonomous parvoviruses H-1 virus and minute virus of mice. J. Virol. 62:1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis, J. J., N. Spruyt, P. Spegelaere, E. Guetta, T. Darawshi, S. F. Cotmore, J. Tal, and J. Rommelaere. 1988. Sensitization of transformed rat fibroblasts to killing by parvovirus minute virus of mice correlates with an increase in viral gene expression. J. Virol. 62:3438-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotmore, S. F., and P. Tattersall. 1986. The NS-1 polypeptide of the autonomous parvovirus MVM is a nuclear phosphoprotein. Virus Res. 4:243-250. [DOI] [PubMed] [Google Scholar]
- 11.Deleu, L., F. Fuks, D. Spitkovsky, R. Horlein, S. Faisst, and J. Rommelaere. 1998. Opposite transcriptional effects of cyclic AMP-responsive elements in confluent or p27KIP-overexpressing cells versus serum-starved or growing cells. Mol. Cell. Biol. 18:409-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deleu, L., A. Pujol, S. Faisst, and J. Rommelaere. 1999. Activation of promoter P4 of the autonomous parvovirus minute virus of mice at early S phase is required for productive infection. J. Virol. 73:3877-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doerig, C., B. Hirt, P. Beard, and J. P. Antonietti. 1988. Minute virus of mice non-structural protein NS-1 is necessary and sufficient for trans-activation of the viral P39 promoter. J. Gen. Virol. 69:2563-2573. [DOI] [PubMed] [Google Scholar]
- 14.Dupressoir, T., J. M. Vanacker, J. J. Cornelis, N. Duponchel, and J. Rommelaere. 1989. Inhibition by parvovirus H-1 of the formation of tumors in nude mice and colonies in vitro by transformed human mammary epithelial cells. Cancer Res. 49:3203-3208. [PubMed] [Google Scholar]
- 15.Enger, P. O., F. Thorsen, P. E. Lonning, R. Bjerkvig, and F. Hoover. 2002. Adeno-associated viral vectors penetrate human solid tumor tissue in vivo more effectively than adenoviral vectors. Hum. Gene Ther. 13:1115-1125. [DOI] [PubMed] [Google Scholar]
- 16.Evan, G. I., G. K. Lewis, G. Ramsay, and J. M. Bishop. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5:3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faisst, S., S. R. Faisst, T. Dupressoir, S. Plaza, A. Pujol, J. C. Jauniaux, S. L. Rhode, and J. Rommelaere. 1995. Isolation of a fully infectious variant of parvovirus H-1 supplanting the standard strain in human cells. J. Virol. 69:4538-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faisst, S., D. Guittard, A. Benner, J. Y. Cesbron, J. R. Schlehofer, J. Rommelaere, and T. Dupressoir. 1998. Dose-dependent regression of HeLa cell-derived tumours in SCID mice after parvovirus H-1 infection. Int. J. Cancer 75:584-589. [DOI] [PubMed] [Google Scholar]
- 19.Fuerer, C., and R. Iggo. 2002. Adenoviruses with Tcf binding sites in multiple early promoters show enhanced selectivity for tumour cells with constitutive activation of the wnt signalling pathway. Gene Ther. 9:270-281. [DOI] [PubMed] [Google Scholar]
- 20.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson, N. D., and S. L. D. Rhode. 1991. Parvovirus NS1 stimulates P4 expression by interaction with the terminal repeats and through DNA amplification. J. Virol. 65:4325-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kestler, J., B. Neeb, S. Struyf, J. Van Damme, S. F. Cotmore, A. D'Abramo, P. Tattersall, J. Rommelaere, C. Dinsart, and J. J. Cornelis. 1999. cis requirements for the efficient production of recombinant DNA vectors based on autonomous parvoviruses. Hum. Gene Ther. 10:1619-1632. [DOI] [PubMed] [Google Scholar]
- 23.Korinek, V., N. Barker, P. J. Morin, D. van Wichen, R. de Weger, K. W. Kinzler, B. Vogelstein, and H. Clevers. 1997. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science 275:1784-1787. [DOI] [PubMed] [Google Scholar]
- 24.Legendre, D., and J. Rommelaere. 1994. Targeting of promoters for trans activation by a carboxy-terminal domain of the NS-1 protein of the parvovirus minute virus of mice. J. Virol. 68:7974-7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorson, C., L. R. Burger, M. Mouw, and D. J. Pintel. 1996. Efficient transactivation of the minute virus of mice P38 promoter requires upstream binding of NS1. J. Virol. 70:834-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maxwell, I. H., and F. Maxwell. 1999. Control of parvovirus DNA replication by a tetracycline-regulated repressor. Gene Ther. 6:309-313. [DOI] [PubMed] [Google Scholar]
- 27.Maxwell, I. H., F. Maxwell, S. L. Rhode, 3rd, J. Corsini, and J. O. Carlson. 1993. Recombinant LuIII autonomous parvovirus as a transient transducing vector for human cells. Hum. Gene Ther. 4:441-450. [DOI] [PubMed] [Google Scholar]
- 28.Maxwell, I. H., A. L. Spitzer, C. J. Long, and F. Maxwell. 1996. Autonomous parvovirus transduction of a gene under control of tissue-specific or inducible promoters. Gene Ther. 3:28-36. [PubMed] [Google Scholar]
- 29.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morin, P. J., A. B. Sparks, V. Korinek, N. Barker, H. Clevers, B. Vogelstein, and K. W. Kinzler. 1997. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 275:1787-1790. [DOI] [PubMed] [Google Scholar]
- 31.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 32.Previsani, N., S. Fontana, B. Hirt, and P. Beard. 1997. Growth of the parvovirus minute virus of mice MVMp3 in EL4 lymphocytes is restricted after cell entry and before viral DNA amplification: cell-specific differences in virus uncoating in vitro. J. Virol. 71:7769-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pujol, A., L. Deleu, J. P. Nuesch, C. Cziepluch, J. C. Jauniaux, and J. Rommelaere. 1997. Inhibition of parvovirus minute virus of mice replication by a peptide involved in the oligomerization of nonstructural protein NS1. J. Virol. 71:7393-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raj, K., P. Ogston, and P. Beard. 2001. Virus-mediated killing of cells that lack p53 activity. Nature 412:914-917. [DOI] [PubMed] [Google Scholar]
- 35.Rhode, S. L. D. 1989. Both excision and replication of cloned autonomous parvovirus DNA require the NS1 (Rep) protein. J. Virol. 63:4249-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roose, J., and H. Clevers. 1999. TCF transcription factors: molecular switches in carcinogenesis. Biochim. Biophys. Acta 1424:M23-M37. [DOI] [PubMed] [Google Scholar]
- 37.Rubio, M. P., S. Guerra, and J. M. Almendral. 2001. Genome replication and postencapsidation functions mapping to the nonstructural gene restrict the host range of a murine parvovirus in human cells. J. Virol. 75:11573-11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takemasa, I., H. Higuchi, H. Yamamoto, M. Sekimoto, N. Tomita, S. Nakamori, R. Matoba, M. Monden, and K. Matsubara. 2001. Construction of preferential cDNA microarray specialized for human colorectal carcinoma: molecular sketch of colorectal cancer. Biochem. Biophys. Res. Commun. 285:1244-1249. [DOI] [PubMed] [Google Scholar]
- 39.Tao, N., G. P. Gao, M. Parr, J. Johnston, T. Baradet, J. M. Wilson, J. Barsoum, and S. E. Fawell. 2001. Sequestration of adenoviral vector by Kupffer cells leads to a nonlinear dose response of transduction in liver. Mol. Ther. 3:28-35. [DOI] [PubMed] [Google Scholar]
- 40.Telerman, A., M. Tuynder, T. Dupressoir, B. Robaye, F. Sigaux, E. Shaulian, M. Oren, J. Rommelaere, and R. Amson. 1993. A model for tumor suppression using H-1 parvovirus. Proc. Natl. Acad. Sci. USA 90:8702-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tetsu, O., and F. McCormick. 1999. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422-426. [DOI] [PubMed] [Google Scholar]
- 42.van de Wetering, M., R. Cavallo, D. Dooijes, M. van Beest, J. van Es, J. Loureiro, A. Ypma, D. Hursh, T. Jones, A. Bejsovec, M. Peifer, M. Mortin, and H. Clevers. 1997. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88:789-799. [DOI] [PubMed] [Google Scholar]
- 43.Van Pachterbeke, C., M. Tuynder, J. P. Cosyn, L. Lespagnard, D. Larsimont, and J. Rommelaere. 1993. Parvovirus H-1 inhibits growth of short-term tumor-derived but not normal mammary tissue cultures. Int. J. Cancer 55:672-677. [DOI] [PubMed] [Google Scholar]