Abstract
Although the HLA B*5701 class I allele is highly overrepresented among human immunodeficiency virus (HIV)-infected long-term nonprogressors (LTNPs), it is also present at the expected frequency (11%) in patients with progressive HIV infection. Whether B57+ progressors lack restriction of viral replication because of escape from recognition of highly immunodominant B57-restricted gag epitopes by CD8+ T cells remains unknown. In this report, we investigate the association between restriction of virus replication and recognition of autologous virus sequences in 27 B*57+ patients (10 LTNPs and 17 progressors). Amplification and direct sequencing of single molecules of viral cDNA or proviral DNA revealed low frequencies of genetic variations in these regions of gag. Furthermore, CD8+ T-cell recognition of autologous viral variants was preserved in most cases. In two patients, responses to autologous viral variants were not demonstrable at one epitope. By using a novel technique to isolate primary CD4+ T cells expressing autologous viral gene products, it was found that 1 to 13% of CD8+ T cells were able to respond to these cells by gamma interferon production. In conclusion, escape-conferring mutations occur infrequently within immunodominant B57-restricted gag epitopes and are not the primary mechanism of virus evasion from immune control in B*5701+ HIV-infected patients. Qualitative features of the virus-specific CD8+ T-cell response not measured by current assays remain the most likely determinants of the differential abilities of HLA B*5701+ LTNPs and progressors to restrict virus replication.
Direct evidence from the rhesus macaque model of simian immunodeficiency virus (SIV) infection and correlative data from human studies have convincingly shown that CD8+ T cells play a major role in restricting lentivirus replication and determining the rate of disease progression (28, 30, 33, 36, 49). However, the reasons why cellular immune responses ultimately fail to contain virus replication in the majority of human immunodeficiency virus (HIV)-infected patients remain unclear. The accumulation of sequence mutations within or near epitopes targeted by HIV- or SIV-specific CD8+ T cells during the acute (1, 7, 29, 47) and chronic phases of infection (4, 10, 15, 25, 29, 45) has been demonstrated and is consistent with selection pressure exerted by dominant HLA-restricted immune responses. When associated with loss of recognition by cytotoxic T lymphocytes (CTLs), as measured in standard in vitro assays, stable sequence variations are thought to represent a major mechanism of virus escape from immune control (4, 7, 8, 10, 15, 23, 25, 29, 45, 47).
To better understand the components of an effective HIV-specific immune response, we have focused our investigation on a unique group of HIV-infected long-term nonprogressors (LTNPs), who maintain normal CD4+ T-cell counts and <50 copies of HIV RNA/ml of plasma despite almost 20 years of infection. The HLA B*5701 class I allele has been found to be dramatically overrepresented in this cohort (15 of 17 thus far identified). However, the B*5701 allele is also present in 11% of patients with progressive disease, similar to the frequency in the Caucasian U.S. population (41). The virus-specific CD8+ T-cell response of LTNPs is typically highly focused on four conserved B*5701-restricted epitopes within the product of the HIV gag gene, suggesting that these epitopes and the B5701 molecule are critically involved in the restriction of viral replication in these patients. In contrast, the response in progressors is broader and includes responses restricted by other class I alleles carried by an individual patient. These observations have suggested that the difference between the HIV-specific CD8+ T-cell responses in B*5701+ LTNPs and progressors is either qualitative or due to partial or complete virus escape from immune recognition at these epitopes (20).
In this study, we investigated the prevalence and recognition of autologous viral sequence variants in patients with highly divergent abilities to restrict virus replication but with a common HLA B*5701 allele. To determine their role in escape from immune system-mediated control over virus replication, sequence changes in immunodominant HLA B57-restricted gag epitopes and the responses to these variations in 10 B*57+ LTNPs and 17 B*57+ progressors were examined. In addition, we estimated the total frequency of CD8+ T cells that respond to autologous virus by using primary CD4+ T-cell targets expressing high levels of autologous viral gene products. These results confirm that high frequencies of HIV-specific CD8+ T cells persist in most patients. They also suggest that mutations within highly immunodominant B5701-restricted gag epitopes do not result in a loss of recognition by CD8+ T cells and do not account for the vast differences in the ability to restrict virus replication between LTNPs and patients with progressive infection.
MATERIALS AND METHODS
Patients.
HIV infection in study participants was documented by HIV type 1 (HIV-1) and -2 enzyme immunoassay. All subjects signed informed-consent forms approved by the National Institute of Allergy and Infectious Diseases investigational review board. HLA class I and II typing was performed by hybridization with sequence-specific oligonucleotide probes following amplification of the corresponding genes by PCR as described elsewhere (41). CCR5 deletion mutations were detected as described previously (9). Patients 5 to 9, 12 to 14, 25, 30, 34, 35, and 101 to 107 have been described in separate reports (19, 37, 39-41).
Plasma HIV-1 virion RNA isolation and cell-associated proviral DNA extraction.
Plasma was concentrated by centrifugation at 150,000 × g for 1 h, and virion RNA was extracted from the subsequent pellet by use of the QIAamp viral RNA minikit (Qiagen, Inc., Valencia, Calif.). The isolated RNA was resuspended in RNase-free water, and then reverse transcription was done with Superscript-II reverse transcriptase (Invitrogen, Carlsbad, Calif.). Primer 5′-CTAATAGAGCTTCCTTTAGTTGCC-3′ (antisense; nucleotides 2302 to 2325 of HIV-1 HXB2) was used for cDNA synthesis of the gag gene. Proviral DNA was isolated from 106 cells by use of the PUREGENE DNA isolation kit (Gentra Systems, Minneapolis, Minn.).
Gag PCR and sequencing.
Single molecules of viral cDNA, obtained through limiting dilution, were amplified and sequenced directly, as described elsewhere (27). PCRs were performed with the Expand High Fidelity PCR system (Roche Molecular Biochemicals, Mannheim, Germany). Amplification of 1.4 kbp of the fragment encompassing the p17, p24, p2, p7, and p6 regions of the HIV-1 gag gene was done in a 50-μl reaction mixture containing 1× Expand high-fidelity buffer 3, 0.2 mM deoxynucleoside triphosphates, 2.4 mM MgCl2, 20 pmol of primers, and 1.75 U of Expand High Fidelity PCR system enzyme mixture. The forward primer was 5′-GCGAGAGCGTCAGTATTAAGC-3′ (sense; nucleotides 796 to 816), and the reverse primer was 5′-CTAATAGAGCTTCCTTTAGTTGCC-3′ (antisense; nucleotides 2302 to 2325) in a first-round reaction. Two microliters of the PCR product from the first reaction was used in a second-round reaction with the following primer pair: 5′-GGGAAAAAATTCGGTTAAGGCC-3′ (sense; nucleotides 836 to 857; forward primer) and 5′-CGAGGGGTCGTTGCCAAAGA-3′ (antisense; nucleotides 2264 to 2283; reverse primer). Each round of PCR consisted of 25 cycles, with the initial denaturation at 94°C for 2 min, followed by 25 cycles of denaturation at 94°C for 15 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min 30 s, with the final extension at 72°C for 7 min. The PCR products were purified by the QIAquick PCR purification kit (Qiagen Inc.). The DNA was sequenced with the dRhodamine terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase FS (PE Applied Biosystems, Foster City, Calif.) and analyzed with a model 377 automated sequencing system (Applied Biosystems). The nucleotide sequences of the gag region were translated and edited with the MASE (multiply aligned sequence editor) program.
Phylogenetic analysis and sequence alignment.
Phylogenetic relationships among the p24 regions of HIV-1 gag sequences were estimated by the neighbor-joining method. The neighbor-joining tree was constructed by using the HKY85 model of evolution (data not shown). Three HIV-1 reference sequences (HXB2, JRFL, and RF) were included in the analysis, and the HIV-1 U455 was used as an outgroup. The phylogenetic tree showed that gag sequences were well confined within a distinct clade with bootstrap supports of 83 to 100 and that HIV-1 reference sequences fell well outside this clade. Also, all new sequences reported in this study were scanned against sequences that are commonly used in our laboratory. The phylogenetic tree showed that newly reported sequences were free from contamination with material that was recently used in the laboratory. Thus, these sequences are free from known contaminants. Patient-specific clustering was observed.
All of the sequences obtained from the patients were aligned with HIV-1 Consensus_B as a reference. A total of 35 HIV-1 clade B sequences (obtained from the Los Alamos HIV sequence database) were also included in the alignment.
Autologous targets.
Autologous Epstein-Barr virus (EBV)-transformed B cells were infected for 16 h at 37°C with recombinant vaccinia viruses containing vVK1 (HIV-1HXB2 gag-pol gene), vP1287 (HIV-1IIIB gag), vP1289 (HIV-1IIIB p24), vP1290 (HIVIIIB p17), vP1288 (HIVIIIB pol), vPE16 (HIV-1BH10 env), vTFnef (HIV-1pNL432 nef), vP1490 (HIVIIIB rev), HIVIIIB tat, or the negative-control virus vSC8 (Escherichia coli β-galactosidase) as previously described (19, 41). The vP1287 (HIV-1IIIB gag), vP1289 (HIV-1IIIB p24), vP1290 (HIVIIIB p17), vP1288 (HIVIIIB pol), and vP1490 (HIVIIIB rev) viruses were contributed to the National Institutes of Health AIDS Research and Reference Reagent Program by Virogenetics Corp. The vTFnef virus was contributed by MedImmune Corporation, and vPE16, vVK1, HIVIIIB tat, and vSC8 viruses were contributed by Bernard Moss (16).
In experiments using HIVSF162-infected primary CD4+ T cells, autologous CD4+ T cells were positively selected from peripheral blood mononuclear cells (PBMC) by magnetic automated cell sorting (autoMACS; Miltenyi Biotec), stimulated with media containing anti-CD3 (OKT3; Coulter, Hialeah, Fla.), anti-CD28 (BD-Pharmingen, San Diego, Calif.), and human interleukin-2 (Roche Diagnostics Corp., Indianapolis, Ind.), infected with 5,000 50% tissue culture infectious doses of HIVSF162, and depleted of CD8+ T cells as described previously (17, 41). Purity (>98%) was confirmed by flow cytometry. The percentage of HIV-infected CD4+ T cells (17 to 45%) was documented by intracellular p24 staining with Kc57-fluorescein isothiocyanate (FITC; Coulter).
In experiments using autologous primary CD4+ T cells expressing autologous viral gene products, CD4+ T cells were positively selected and stimulated with blasting media as described above. On day 3, the cells were resuspended in RPMI medium containing 20% human interleukin-2 and maintained in culture for 6 days before undergoing CD8+ T-cell depletion. The CD4+ T-cell lymphoblasts were sampled daily starting on day 10 to check for intracellular p24 expression with Kc57-FITC (Coulter). The frequency of cells expressing HIV p24 (10 to 46% Kc57+ CD3+ T lymphocytes) was confirmed prior to use in 6-h stimulation assays.
CD8+ T-cell stimulation and intracellular cytokine detection assay.
PBMC were obtained by sodium diatrizoate density centrifugation (Organon-Teknika, Durham, N.C.) of apheresis donor packs and were cryopreserved in freezing medium (Gibco BRL, Grand Island, N.Y.) at −140°C. Cryopreserved PBMC were cultured overnight at 37°C in RPMI medium-10% fetal bovine serum prior to use.
Intracellular cytokine detection was performed as previously described (19, 41). Briefly, PBMC were incubated with autologous EBV-transformed B cells, either uninfected or infected with vac-β-galactosidase or recombinant vac-HIV, at an effector-to-target cell (E:T) ratio of 10:1 or with autologous primary CD4+ T-cell targets (expressing either autologous virus or HIVSF162) at an E:T ratio of 1:1. In peptide stimulation experiments, autologous EBV-transformed B cells were pulsed for 1 h in 50 μl of RPMI medium containing 10% fetal bovine serum with the relevant peptide before addition of effector PBMC at an E:T ratio of 40:1 (39, 41). The HLA B5701-restricted HIV Gag p24308-316 peptides QASQEVKNW and QASQDVKNW were purchased from Multiple Peptide Systems (San Diego, Calif.). These E:T ratios gave optimal responses with low background or bystander activation. At 2 h of incubation, brefeldin A (Sigma, St. Louis, Mo.) was added to the medium at a final concentration of 10 μg/ml to inhibit cytokine secretion. At 6 h of incubation, the cells were washed twice, fixed in 4% paraformaldehyde (Sigma), and permeabilized and blocked overnight prior to staining as described previously (19).
Flow cytometry.
Multiparameter flow cytometry was performed according to standard protocols (26). Surface and intracellular staining was performed with the following antibodies: FITC-conjugated anti-CD3 and anti-CD8; phycoerythrin (PE)-conjugated anti-CD3 and anti-CD8, mouse immunoglobulin G1 (IgG1) isotype control and anti-CD69; PerCP-conjugated anti-CD3 and anti-CD8; and allophycocyanin-conjugated anti-CD3, anti-CD4, anti-CD8, mouse IgG1 isotype control, and anti-gamma interferon (IFN-γ) (BD-Pharmingen). In addition, FITC-conjugated anti-HIV Gag p24 (Kc57) and the IgG1 isotype (Coulter) were used. By gating on CD3+ CD8+ lymphocytes, 15,000 to 200,000 events were collected. Color compensation settings were made with each round of data acquisition by using patient cells labeled with a fluorochrome-conjugated anti-CD3 antibody. Data were analyzed with FlowJo software (TreeStar, Cupertino, Calif.).
Statistical methods.
Comparisons of the mean total CD8+ T-cell responses for the two patient groups were done by Student's t test. The median total numbers of sequence mutations within B57-restricted gag epitopes in each group were compared by Wilcoxon's two-sample test. The frequencies of the E312D mutation in the two patient groups were compared by the one-sided Fisher's exact test. Adjustment of P values for multiple testing was done by the Bonferroni method.
RESULTS
Patient characteristics.
All subjects were confirmed to be HLA B*5701+ and were divided into three groups (Table 1). Patients in the LTNP group are included in a unique cohort of LTNPs that have been infected for a median of 16 years yet typically maintain normal CD4+ T-cell counts and <50 copies of HIV RNA/ml of plasma. In most of these patients, plasma virion RNA could not be isolated. In a subset of LTNPs (patients 6, 8, and 25), there was intermittent or low-level viremia, which permitted plasma virion RNA to be isolated. Only patients 12, 25, and 34 were found to be heterozygous for the 32-bp deletion within CCR5. Detailed characteristics of most LTNPs have been provided in recent reports (19, 39-41). B*57+ progressors were studied a median of 8 years after diagnosis of HIV infection. In the untreated group, patients 35, 101, 102, 105, 110, 114, and 123 were antiretrovirally naive at the time of study. Patients 14, 106, 107, 119, and 203 had not received antiretroviral drugs in the previous 6 months. To increase the sample size and include some progressors with more modest levels of plasma viral RNA, five patients (103, 104, 115, 116, and 122) with progressive disease receiving antiretroviral therapy were included.
TABLE 1.
Patient characteristics
| Patient no. | Allele(s) for HLA class I
|
Yr of diagnosis | Sample date | No. (cells/μl) of:
|
Viral load (copies/ml) | Source | QW9 5th aaa | |||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | CD4+ T cells | CD8+ T cells | ||||||
| LTNPs | ||||||||||
| 5 | 2, 24 | 57 | 6, 7 | 1985 | 9/99 | 971 | 870 | <50 | Proviral DNA | E |
| 7 | 1, 2 | 57 | 6 | 1985 | 8/98 | 371 | 551 | <50 | Proviral DNA | E |
| 9 | 23, 26 | 57, 44 | 1, 7 | 1997 | 3/99 | 1,079 | 985 | <50 | Proviral DNA | E |
| 12 | 3, 11 | 57, 7 | 6, 7 | 1986 | 3/01 | 500 | 218 | <50 | Proviral DNA | D |
| 13 | 1, 11 | 57, 35 | 4, 6 | 1986 | 4/99 | 1,016 | 767 | <50 | Proviral DNA | E |
| 34 | 1, 2 | 57, 8 | 6, 7 | 1989 | 3/01 | 1,523 | 1,111 | <50 | Proviral DNA | E |
| 30 | 31, 74 | 57, 51 | 7, 16 | 1990 | 11/00 | 450 | 828 | 56 | Proviral DNA | E |
| 6 | 11, 30 | 57, 52 | 7, 12 | 1986 | 6/99 | 737 | 998 | 152 | Virion RNA | E |
| 8 | 11, 23 | 57, 44 | 4, 6 | 1985 | 5/00 | 490 | 1,033 | 567 | Virion RNA | E |
| 25 | 3, 24 | 57, 27 | 2, 6 | 1986 | 4/00 | 541 | 727 | 1,089 | Virion RNA | E |
| Untreated progressors | ||||||||||
| 35 | 2, 3 | 57 | 6, 7 | 2000 | 8/00 | 636 | 618 | 1,149 | Virion RNA | E |
| 203 | 30, 32 | 57, 18 | 8, 14 | 1989 | 1/97 | 1,360 | 1,170 | 8,255 | Virion RNA | D |
| 106 | 3, 30 | 57, 18 | 5, 18 | 1986 | 7/00 | 546 | 1,959 | 24,245 | Virion RNA | D |
| 119 | 2, 25 | 57, 44 | 4, 6 | 1985 | 3/98 | 512 | 1,638 | 24,640 | Virion RNA | E |
| 105 | 2, 80 | 57, 8 | 2, 7 | 1992 | 10/96 | 333 | 866 | 25,040 | Virion RNA | D |
| 14 | 36, 68 | 57, 45 | 7, 16 | 1998 | 7/98 | 306 | 838 | 26,260 | Virion RNA | E |
| 107 | 3 | 57, 40 | 3, 7 | 1987 | 7/00 | 530 | 1,519 | 26,740 | Virion RNA | D |
| 123 | 1, 11 | 57, 55 | 3, 6 | 1992 | 6/93 | 189 | 852 | 52,960 | Virion RNA | E |
| 110 | 36, 68 | 57, 45 | 7, 16 | 1985 | 6/98 | 226 | 704 | 63,480 | Virion RNA | E |
| 101 | 1, 31 | 57, 51 | 6, 15 | 1986 | 11/94 | 260 | 588 | 91,910 | Virion RNA | E/D |
| 114 | 2 | 57, 51 | 6, 14 | 1989 | 5/98 | 317 | 382 | 156,700 | Virion RNA | E |
| 102 | 24, 68 | 57, 15 | 6, 7 | 1993 | 11/95 | 624 | 798 | 183,000 | Virion RNA | E |
| Treated progressors | ||||||||||
| 116 | 32, 68 | 57 | 7 | 1993 | 8/96 | 1,433 | 1,538 | 1,200 | Virion RNA | E |
| 115 | 2, 25 | 57, 44 | 4, 6 | 1995 | 8/96 | 393 | 437 | 1,245 | Virion RNA | D/E |
| 103 | 2, 11 | 57, 55 | 3, 6 | 1991 | 6/98 | 463 | 678 | 2,249 | Virion RNA | D/E |
| 104 | 2 | 57, 58 | 3, 6 | 1988 | 3/98 | 291 | 1,028 | 3,507 | Virion RNA | D |
| 122 | 1, 2 | 57, 51 | 6, 14 | 1986 | 12/96 | 595 | 984 | 5,371 | Virion RNA | E |
Amino acids (aa) in boldface indicate the E312D variation. E/D, admixture of consensus sequence and the E312D variation.
gag sequencing.
In previous studies, it has been shown that the highest frequencies of HIV-specific CD8+ T cells are typically directed against the gag gene product in both B*57+ LTNPs and B*57+ patients with progressive infection. Furthermore, peptide mapping has demonstrated that the gag-specific CD8+ T-cell response is more narrowly focused on B57-restricted epitopes in B*57+ LTNPs than in progressors, even though the frequencies of the individual peptide-specific cells for the two patient groups are similar (19, 24, 39, 41). To investigate the role that sequence mutations in HLA B57-restricted gag epitopes might play in escape from immunologic control of virus replication, plasma virion gag gene sequences in 17 B*57+ patients with progressive HIV infection and 10 B*57+ LTNPs were examined. Despite multiple attempts, 7 of 10 LTNPs with <50 copies of viral RNA/ml of plasma were reverse transcription-PCR negative. Therefore, proviral DNA sequencing was performed for these seven patients so that a comparison of viral genotypic characteristics between the LTNPs and progressors could be made (Table 1 and Fig. 1).
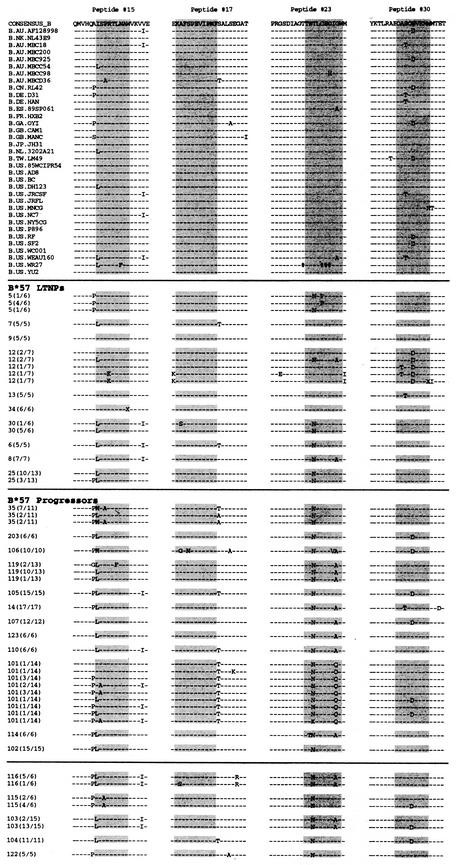
FIG.1.
gag gene sequencing results and alignment with references. Thirty-five reference sequences from the Los Alamos HIV sequence database and sequencing results from 10 B*57+ LTNPs and 17 B*57+ progressors are shown for four regions of Gag that contain known HLA B57-restricted epitopes (highlighted). Peptide numbers (15, 17, 23, and 30) correspond to 20-mers used previously to map the Gag peptide-specific CD8+ T-cell responses in B*57+ patients: 15, residues 141 to 160; 17, residues 161 to 180; 23, residues 231 to 250; 30, residues 301 to 320. Only sequence differences are shown. X, position corresponding to a stop codon; #, position of amino acid whose codon has an ambiguous base. Anchor residues within the epitope that stabilize binding with B57 are located at positions 2 (S, T, and A) and 9 to 11 (W and F).
Sequencing was performed with a single transcript in every case, and sequences were aligned with the HIV-1 clade B consensus sequence as a reference. Thirty-five other clade B sequences obtained from the Los Alamos HIV sequence database were included for comparison. In Fig. 1, sequencing results for four regions of the Gag protein that contain well-characterized HLA B57-restricted epitopes (highlighted) are shown for both patient groups.
Responses targeting the HLA B57-restricted epitope KAF11 in peptide 17 (KAFSPEVIPMF; residues 162 to 172) have been previously shown to be quantitatively immunodominant in B*57+ HIV-infected patients, including several individuals in the present study (22, 24, 39, 41). Interestingly, this sequence was completely homologous to the consensus clade B sequence in 9 of 10 LTNPs and in 15 of the 17 patients with progressive infection despite very prolonged durations of HIV infection (as late as 14 years after initial diagnosis) (Table 1; Fig. 1). The highly conserved nature of this epitope, despite a high-frequency response that likely exerts considerable immune selection pressure, may reflect the inability of the virus to tolerate changes in these positions without compromising its own fitness.
Overall, progressors had a slightly higher cumulative number of sequence variations within all four B57-restricted gag epitopes compared with the consensus clade B sequence than LTNPs (medians, 3 [range, 1 to 7] versus 2 [range, 0 to 4]), respectively; P < 0.01). However, most of these mutations were not highly overrepresented in progressors relative to LTNPs and/or in the clade B sequences. Furthermore, with the exceptions of an A162G (peptide 17) mutation in patient 106 and a W320X (peptide 30) mutation in patient 12, most of the mutations did not alter the motifs of the anchor residues required for optimal peptide binding to HLA B57: serine (S), threonine (T), or alanine (A) at position 2 and tryptophan (W) or phenylalanine (F) at positions 9 to 11 (3, 24). This result suggests that the predicted peptide sequences of a majority of these epitopes should retain the ability to bind HLA B5701.
In the QW9 epitope (QASQEVKNW; residues 308 to 316), a glutamic acid-to-aspartic acid mutation (E312D) was common at position 5 in many progressors. Although this mutation was present among clade B sequences, it tended to occur at a higher frequency in progressors than in LTNPs (8 of 17 versus 1 of 10, respectively, P = 0.06; Table 1 and Fig. 1). This sequence was of particular interest given that the percentages of CD8+ T cells targeting this particular epitope were previously found to be more variable in five B*57+ progressors than in eight B*57+ LTNPs (39, 41). Although the E312D mutation should not affect binding to HLA B5701, sequence mutations that spare the anchoring residues might still lead to virus evasion from immune control by other mechanisms such as impaired recognition of the peptide-HLA complex by the T-cell receptor (10, 13, 15). For these reasons, we examined CD8+ T-cell recognition of this variant epitope in these patients.
HIV-specific CD8+ T-cell responses.
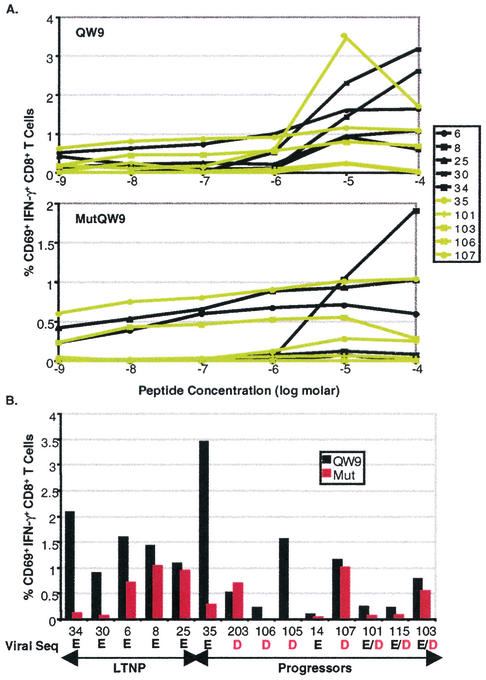
The ability of CD8+ T cells to respond to either the mutated (E312D) or consensus sequence peptides over a broad range of concentrations was explored. The CD8+ T-cell responses to these synthetic peptides for five B*57+ LTNPs and five progressors are shown in Fig. 2A. Although the frequencies of CD8+ T cells that upregulate CD69 expression and produce IFN-γ in response to the mutated peptide were lower in almost every case, the concentration at which the highest frequency of IFN-γ+ cells was observed for both peptides was approximately 10 μM. Consistent with our previous observations, the CD8+ T-cell responses directed against this B57-restricted epitope were more variable in B*57+ progressors than in LTNPs (Fig. 2B) (39, 41). Both virus variants were recognized, to various degrees, by CD8+ T cells of five progressors whose autologous viruses contained either the consensus sequence or both the mutated and consensus sequences (patients 35, 14, 101, 115, and 103). Of the four patients in whom the E312D sequence was the only sequence found in the plasma (patients 203, 106, 105, and 107), the CD8+ T cells of only two patients, 106 and 105, did not recognize this variant.
FIG. 2.
The percentages of CD69+ IFN-γ+ CD8+ T cells responding to autologous B cells pulsed with either the consensus sequence (QW9) or mutated (mutQW9) peptides corresponding to codons 308 to 316 in gag. (A) Peptide titration using the consensus sequence peptide (top) and mutated peptide (bottom) in five B*57+ LTNPs (black) and five B*57+ progressors (lime) revealed that the responses to the consensus sequence peptide are typically higher but that the optimal concentration for both peptides is 10 μM. (B) Summary of the CD8+ T-cell responses to QW9 (black) and the mutated peptide (red) for five B*57+ LTNPs and nine B*57+ progressors. Background activity against autologous B cells without the peptide has been subtracted. The results shown are representative of three experiments. Patients are listed in order of increasing plasma viral RNA levels. The autologous sequence at position 312 (position 5 within the epitope) is listed below the patient numbers.
In addition to sequence changes within a given epitope, changes in amino acid sequences that surround it within the protein, which might affect processing or presentation, may affect CD8+ T-cell recognition (10, 12, 14). For this reason, the potential effect of the alanine-to-proline change in the flanking sequence of autologous virus on recognition of peptide 15 was also investigated (Fig. 1). For two patients (106 and 35) in whom this change was found in all clones, the ability of B5701-peptide 15-tetramer+ CD8+ T cells to produce IFN-γ in response to autologous virus was measured. In both cases B5701-peptide 15-tetramer+ CD8+ T cells produced IFN-γ whether they were stimulated with CD4+ T cells expressing autologous gene products or HIVSF162 (not shown). Taken together, these data suggest that the majority of progressor patients maintained recognition of the mutated and/or consensus clade B peptide(s) that corresponded to their autologous virus sequences yet were still unable to restrict virus replication to low levels.
Total response to heterologous and autologous viral sequences.
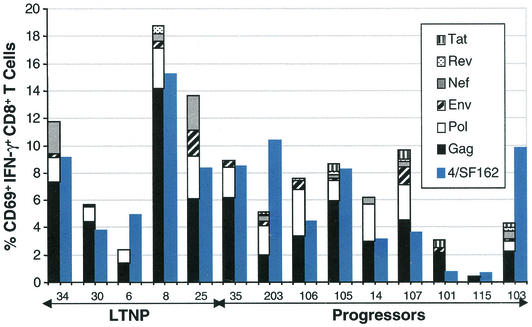
Although responses to the conserved Gag or variant E312D peptides in LTNPs and progressors were not consistently different, it remained possible that differences may lie in the frequency of CD8+ T cells specific for other peptides or gene products. We and others have previously shown that the response to a single peptide is not an accurate representation of the patient's global HIV-specific CD8+ T-cell response (5, 6, 19, 39). As shown in Fig. 3, the total frequencies of CD8+ T cells that produce IFN-γ in response to EBV-transformed B cells infected with vaccinia virus-HIV recombinants (means, 10.48% ± 2.91% versus 5.97% ± 1.02% for LTNPs and progressors, respectively; P = 0.5) and primary CD4+ T cells infected with HIVSF162 (means, 8.26% ± 2.01% versus 5.49% ± 1.27% for LTNPs and progressors, respectively; P = 0.2) were not statistically different between the two patient groups. Consistent with previous results, high frequencies of HIV-specific CD8+ T cells targeting multiple gene products were detected in all patients, even those with poor restriction of virus replication (5, 11, 19, 32, 39, 41). Although the responses to heterologous virus in the two patient groups are not different, it remained possible that substantial differences may be apparent when the total response to autologous virus is examined.
FIG. 3.
A summary of the total HIV-specific CD8+ T-cell responses by using two techniques of antigen presentation. The percentages of CD69+ IFN-γ+ CD8+ T cells responding to autologous EBV-transformed B cells infected with vaccinia virus-HIV recombinants (stacked-bar histograms) and autologous primary CD4+ T cells infected with HIVSF162 (blue histograms) for five B*57+ LTNPs and nine B*57+ progressors are shown. Background activity against B cells infected with the control virus vaccinia virus-β-galactosidase or uninfected autologous primary CD4+ T-cell lymphoblasts, respectively, has been subtracted. These results are representative of at least three experiments.
An estimate of the frequencies of CD8+ T cells specific for autologous virus has not been well characterized thus far. To address this, we adapted a technique to culture autologous virus in autologous CD4+ T-cell lymphoblasts. These cells were then used to stimulate effector PBMC to examine the global CD8+ T-cell responses to a patient's autologous virus (Fig. 4). This approach offers a number of theoretical advantages over other techniques in that culture in heterologous cells or synthesis of large numbers of peptides is not required. In addition, this system maintains surface major histocompatibility complex (MHC) expression and peptide-MHC stoichiometry that may be closer to the in vivo situation.
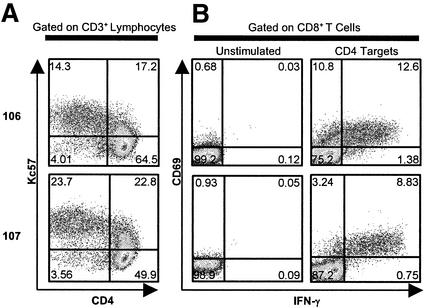
FIG. 4.
CD8+ T-cell responses to autologous primary CD4+ T cells expressing autologous virus. (A) By gating on CD3+ lymphocytes, the percentages of primary CD4+ T-cell targets expressing autologous virus are determined by intracellular p24 (Kc57) staining and estimated as the sums of values in the upper left and upper right quadrants. Representative data are shown for two B*57+ patients. (B) The percentages of CD69+ IFN-γ+ CD8+ T cells (upper right quadrant) in response to autologous primary CD4+ T-cell targets expressing autologous virus are shown for the same two B*57+ patients.
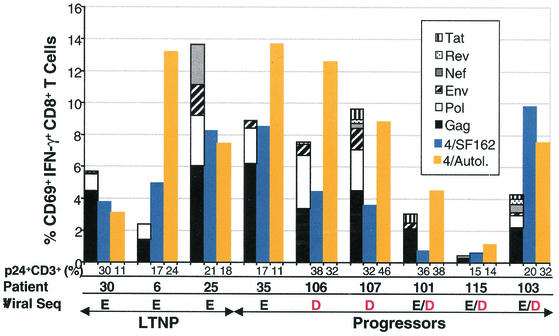
This response was measured in six progressors and three LTNPs from whom high levels of autologous virus could be cultured (Fig. 5). Between approximately 1 and 13% of peripheral blood CD8+ T cells produced IFN-γ in response to autologous virus. The frequencies of CD8+ T cells responding to CD4+ T cells expressing autologous virus for LTNPs and progressors were similar (means, 7.93% ± 2.92% versus 8.02% ± 1.97%, respectively; P > 0.5) and were similar to or exceeded the responses to CD4+ T cells infected with HIVSF162 in most cases. In addition, some patients with modest responses to the QW9 peptide or E312D variant had strong total responses to the autologous virus. For example, although patient 106 did not have a detectable response to the E312D variant that predominates in the plasma, 12.6% of peripheral blood CD8+ T cells were specific for autologous virus. These results suggest that even in those patients with modest responses to QW9 or the variant E312D peptide sequences, a high frequency of CD8+ T cells capable of recognizing autologous viral sequences persists in the peripheral blood. They also suggest that the difference in the abilities of these patients to restrict viral replication does not lie simply in the total number of cells capable of responding to autologous virus.
FIG. 5.
Summary of the total HIV-specific CD8+ T-cell responses by using three techniques of antigen presentation. The percentages of CD69+ IFN-γ+ CD8+ T cells responding to autologous EBV-transformed B cells infected with vaccinia virus-HIV recombinants (stacked-bar histograms), autologous primary CD4+ T cells infected with HIVSF162 (blue histograms), and autologous primary CD4+ T cells expressing high levels of autologous virus (gold histograms) for three B*57+ LTNPs and six B*57+ progressors are shown. p24+ CD3+ percentages are the percentages of CD4+ T cells infected with and expressing HIVSF162 or expressing autologous p24 in each experiment. These results are representative of at least three experiments.
DISCUSSION
These results provide some additional insight into the potential factors that might underlie the differential ability of LTNPs and progressors to restrict HIV replication. Our previous work has indicated that the extraordinary ability of B*5701+ LTNPs to restrict HIV replication compared to B*5701+ or B*5701− progressors is not simply explained by the epitopes restricted by this allele (39, 41). B*5701+ progressors that target these conserved epitopes do not appear to restrict HIV replication any better than B*5701− progressors. Some of the B*5701+ patients described in this report have some of the highest plasma viral RNA levels in our untreated cohorts. In addition, B*5701+ LTNPs and progressors maintain similar conserved-epitope-specific or total CD8+ T-cell responses to heterologous HIV viruses yet have vast differences in the ability to restrict HIV replication during the chronic phase of infection. These results suggested that differences may lie either in qualitative aspects of the CD8+ T-cell response or, alternatively, in the ability to recognize autologous viral variants.
In the present study, differences in epitope sequences and responses to these changes were examined in a cohort matched for the B*5701 allele but with very disparate abilities to restrict HIV replication. Differences between these patient groups in the predicted sequences of three of four immunodominant epitopes were not observed. A single mutation within the QW9 epitope (E312D) was overrepresented in patients with progressive disease. Consistent with our previous results, responses to the consensus sequence QW9 peptide were more variable in progressors, suggesting their CD8+ T cells may not recognize HIV at this epitope. However, this mutation did not reliably confer loss of recognition on the E312D variant. In addition, most progressor patients maintained high-frequency total responses to autologous virus. Thus, high-frequency epitope- and total-HIV-specific responses to autologous viruses persisted in B*5701+ progressors. These results then suggest that differences between LTNPs and progressors in restriction of virus replication reside not in the loss of immune system recognition of autologous virus but rather in qualitative differences in the CD8+ T-cell response that are not accounted for in current assays (40).
A number of additional lines of evidence suggest that the E312D change within the QW9 epitope does not represent a true escape mutation that permits poorly restricted viral replication in progressor patients. Changes in epitopes resulting in true escape should result in impaired binding by MHC or loss of recognition by CD8+ T cells (4, 7, 8, 10, 13, 15, 23, 25, 45, 47, 48). In addition, the mutant epitope should be rapidly selected for by the immune response and the epitopes at which this selection pressure is effective should be relatively well conserved. However, the E312D change within QW9 does not have these characteristics since it does not result in impaired binding or a reliable loss of recognition by CD8+ T cells. It was found at a frequency similar to that for the clade B consensus sequence in the plasma virus of three patients and thus does not appear to be under strong immune selection pressure. It was also the dominant sequence in the plasma virus quasispecies of one LTNP. In addition, it occurred at a relatively high frequency in B clade viruses. Taken together, these results suggest that this change represents a relatively common variant and not selection pressure-driven escape from the CD8+ T-cell response.
The results from the present study contrast with the well-characterized role that escape mutations have been shown to play in progressive infection in HLA B*2705+ patients (23, 25, 29, 38, 43-45). Similar to HLA B*5701, B*2705 is a Bw4 allele that has also been associated with slower progression of HIV disease. In most B*27+ patients, the immunodominant CTL response is directed against highly conserved epitope KK10 (KRWIILGLNK; residues 263 to 272) within Gag p24 (25, 43). The substitution of lysine (K), glycine (G), or threonine (T) for arginine (R) at the anchoring position 264 has been shown to result in epitopes that have diminished binding with HLA B2705 (23, 25, 29). In two patients, the R264K mutation did not appear until late in the course of infection and coincided with progressive disease (25, 29). In four other B*27+ adults and in four B*27+ pediatric patients, sequence variations involving the critical 264 anchor position were detected at the time of clinical immunodeficiency and/or elevated plasma HIV RNA levels (23, 25, 29). Even though these cases associate the presence of sequence mutations and disease progression, at least three other B*27+ patients that did not carry these mutations yet experienced declining CD4+ T-cell counts and high levels of plasma viremia have been reported (25, 29). One possible explanation for the different roles of escape mutations in B*2705+ and B*5701+ patients might be the existence of several codominant B57-restricted epitopes within the Gag protein (and possibly another in the Nef protein) compared with only a single highly immunodominant B27-restricted gag epitope. As discussed previously, the dominant virus-specific CD8+ T-cell responses in B*57+ LTNPs are typically focused on three or four B57-restricted gag epitopes (24, 39, 41). Similar to observations made in the murine lymphocytic choriomeningitis virus infection model (18, 42, 46, 53), loss of CTL recognition at one of multiple immunodominant B5701-restricted epitopes would be expected to confer minimal survival advantage on the virus variants if the other dominant responses maintained restriction of virus replication. Conversely, loss of recognition at a single immunodominant epitope in B*2705+ patients may have a more profound effect (44).
Based on associations with poor immunologic control, assigning causality to changes within viral sequences of lentiviruses is particularly difficult. These host-virus dynamics are extraordinarily complex given the large number of permutations of viral epitopes, timing of gene expression during the viral replication cycle, and complement of MHC class I alleles. The host CTL response is constrained by the ability of the MHC class I alleles to bind to various viral epitopes, while the virus is constrained by the degree to which an escape mutation impairs viral replicative capacity or “fitness.” In addition, the CD8+ T-cell response to each of the viral proteins in the context of each of a single patient's MHC alleles is very broad. Although escape mutations may be found within a single epitope, it is likely that other conserved epitopes within the same gene remain as targets (48). In addition, although some responses may appear to be quantitatively larger, this does not necessarily mean they are immunodominant with regard to restriction of viral replication (51, 52). Thus, it remains uncertain whether mutations found in cross-sectional or longitudinal studies of lentivirus infection cause true escape from immune system control or whether escape mutations at single epitopes simply occur in the context of high levels of virus replication. Although changes within HIV or SIV sequences that alter MHC binding or CD8+ T-cell recognition clearly occur, formal proof of the precise role played by these changes in the restriction of viral replication is likely to come only from challenge studies using cloned variant SIV viruses in rhesus macaques carrying the allele of interest.
The results of the present study confirm and extend some of our previous work on the frequency of HIV-specific CD8+ T cells in the peripheral blood. We and others have previously shown that up to 22% of the peripheral blood CD8+ T cells are specific for heterologous HIV viruses (41) (5, 19, 32, 39). These frequencies are commonly present in LTNPs but also in progressors with high levels of plasma viral RNA and without HIV-specific CD4+ proliferative responses. Given that only 20 to 80% of HIV-specific monoclonal expansions or tetramer+ cells express IFN-γ following stimulation with antigen, the true frequency might be severalfold above these frequencies (2, 19, 21, 31, 32, 34, 50). However, an estimate of the total response to autologous virus has not been well characterized thus far. In one recent report, diminished frequencies of CD8+ T cells that recognize autologous viral gene products were observed in some patients with advanced disease (35). However, under the present experimental conditions and in patients with less-advanced disease, the response to autologous virus was occasionally higher than the response to heterologous virus, albeit only modestly in some cases. Nonetheless, relatively high frequencies of CD8+ T cells specific for autologous virus are present in most patients. Thus far, our understanding of how such high frequencies of CD8+ T cells persist in the absence of restriction of viral replication during the chronic phase of infection remains incomplete.
At present, a further definition of the relative role of viral escape mutations or qualitative abnormalities within the HIV-specific immune response is of particular importance. The majority of vaccines currently in preclinical or clinical trials are thought to rely on CD8+ T-cell responses that may alter disease progression but that are unlikely to prevent infection. In this report, the ability of relatively high frequencies of HIV-specific CD8+ T cells of infected patients to cross recognize autologous and heterologous viruses is encouraging. However, the ability of vaccines to induce restriction of HIV replication and the durability of this effect in humans remain unclear at present. Thus a better understanding of the relative roles of viral escape mutations or qualitative changes in the CD8+ T-cell response in control of viral replication may provide important information on how the effects of these changes may be avoided or possibly reversed by prophylactic or therapeutic vaccines or immunotherapies.
REFERENCES
- 1.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 2.Appay, V., D. F. Nixon, S. M. Donahoe, G. M. Gillespie, T. Dong, A. King, G. S. Ogg, H. M. Spiegel, C. Conlon, C. A. Spina, D. V. Havlir, D. D. Richman, A. Waters, P. Easterbrook, A. J. McMichael, and S. L. Rowland-Jones. 2000. HIV-specific CD8+ T cells produce antiviral cytokines but are impaired in cytolytic function. J. Exp. Med. 192:63-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barber, L. D., L. Percival, K. L. Arnett, J. E. Gumperz, L. Chen, and P. Parham. 1997. Polymorphism in the alpha 1 helix of the HLA-B heavy chain can have an overriding influence on peptide-binding specificity. J. Immunol. 158:1660-1669. [PubMed] [Google Scholar]
- 4.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 5.Betts, M. R., D. R. Ambrozak, D. C. Douek, S. Bonhoeffer, J. M. Brenchley, J. P. Casazza, R. A. Koup, and L. J. Picker. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75:11983-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betts, M. R., J. P. Casazza, B. A. Patterson, S. Waldrop, W. Trigona, T. M. Fu, F. Kern, L. J. Picker, and R. A. Koup. 2000. Putative immunodominant human immunodeficiency virus-specific CD8+ T-cell responses cannot be predicted by major histocompatibility complex class I haplotype. J. Virol. 74:9144-9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Z. W., A. Craiu, L. Shen, M. J. Kuroda, U. C. Iroku, D. I. Watkins, G. Voss, and N. L. Letvin. 2000. Simian immunodeficiency virus evades a dominant epitope-specific cytotoxic T lymphocyte response through a mutation resulting in the accelerated dissociation of viral peptide and MHC class I. J. Immunol. 164:6474-6479. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, O. J., S. Paolucci, S. M. Bende, M. Daucher, H. Moriuchi, M. Moriuchi, C. Cicala, R. T. Davey, Jr., B. Baird, and A. S. Fauci. 1998. CXCR4 and CCR5 genetic polymorphisms in long-term nonprogressive human immunodeficiency virus infection: lack of association with mutations other than CCR5-Δ32. J. Virol. 72:6215-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couillin, I., B. Culmann-Penciolelli, E. Gomard, J. Choppin, J. P. Levy, J. G. Guillet, and S. Saragosti. 1994. Impaired cytotoxic T lymphocyte recognition due to genetic variations in the main immunogenic region of the human immunodeficiency virus 1 NEF protein. J. Exp. Med. 180:1129-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalod, M., M. Dupuis, J. C. Deschemin, D. Sicard, D. Salmon, J. F. Delfraissy, A. Venet, M. Sinet, and J. G. Guillet. 1999. Broad, intense anti-human immunodeficiency virus (HIV) ex vivo CD8+ responses in HIV type 1-infected patients: comparison with anti-Epstein-Barr virus responses and changes during antiretroviral therapy. J. Virol. 73:7108-7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Val, M., H. J. Schlicht, T. Ruppert, M. J. Reddehase, and U. H. Koszinowski. 1991. Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell 66:1145-1153. [DOI] [PubMed] [Google Scholar]
- 13.Douek, D. C., M. R. Betts, J. M. Brenchley, B. J. Hill, D. R. Ambrozak, K. L. Ngai, N. J. Karandikar, J. P. Casazza, and R. A. Koup. 2002. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J. Immunol. 168:3099-3104. [DOI] [PubMed] [Google Scholar]
- 14.Eisenlohr, L. C., J. W. Yewdell, and J. R. Bennink. 1992. Flanking sequences influence the presentation of an endogenously synthesized peptide to cytotoxic T lymphocytes. J. Exp. Med. 175:481-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans, D. T., D. H. O'Connor, P. Jing, J. L. Dzuris, J. Sidney, J. da Silva, T. M. Allen, H. Horton, J. E. Venham, R. A. Rudersdorf, T. Vogel, C. D. Pauza, R. E. Bontrop, R. DeMars, A. Sette, A. L. Hughes, and D. I. Watkins. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270-1276. [DOI] [PubMed] [Google Scholar]
- 16.Falkner, F. G., T. R. Fuerst, and B. Moss. 1988. Use of vaccinia virus vectors to study the synthesis, intracellular localization, and action of the human immunodeficiency virus trans-activator protein. Virology 164:450-457. [DOI] [PubMed] [Google Scholar]
- 17.Ferrari, G., W. Humphrey, M. J. McElrath, J. L. Excler, A. M. Duliege, M. L. Clements, L. C. Corey, D. P. Bolognesi, and K. J. Weinhold. 1997. Clade B-based HIV-1 vaccines elicit cross-clade cytotoxic T lymphocyte reactivities in uninfected volunteers. Proc. Natl. Acad. Sci. USA 94:1396-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gairin, J. E., H. Mazarguil, D. Hudrisier, and M. B. Oldstone. 1995. Optimal lymphocytic choriomeningitis virus sequences restricted by H-2Db major histocompatibility complex class I molecules and presented to cytotoxic T lymphocytes. J. Virol. 69:2297-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gea-Banacloche, J. C., S. A. Migueles, L. Martino, W. L. Shupert, A. C. McNeil, L. Ehler, C. Prussin, R. Stevens, L. Lambert, J. Altman, C. W. Hallahan, J. C. de Quiros, and M. Connors. 2000. Maintenance of large numbers of virus-specific CD8+ T cells in HIV-infected progressors and long-term nonprogressors. J. Immunol. 165:1082-1092. [DOI] [PubMed] [Google Scholar]
- 20.Gillespie, G. M., R. Kaul, T. Dong, H. B. Yang, T. Rostron, J. J. Bwayo, P. Kiama, T. Peto, F. A. Plummer, A. J. McMichael, and S. L. Rowland-Jones. 2002. Cross-reactive cytotoxic T lymphocytes against a HIV-1 p24 epitope in slow progressors with B*57. AIDS 16:961-972. [DOI] [PubMed] [Google Scholar]
- 21.Goepfert, P. A., A. Bansal, B. H. Edwards, G. D. Ritter, Jr., I. Tellez, S. A. McPherson, S. Sabbaj, and M. J. Mulligan. 2000. A significant number of human immunodeficiency virus epitope-specific cytotoxic T lymphocytes detected by tetramer binding do not produce gamma interferon. J. Virol. 74:10249-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goulder, P. J., M. A. Altfeld, E. S. Rosenberg, T. Nguyen, Y. Tang, R. L. Eldridge, M. M. Addo, S. He, J. S. Mukherjee, M. N. Phillips, M. Bunce, S. A. Kalams, R. P. Sekaly, B. D. Walker, and C. Brander. 2001. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J. Exp. Med. 193:181-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 24.Goulder, P. J., M. Bunce, P. Krausa, K. McIntyre, S. Crowley, B. Morgan, A. Edwards, P. Giangrande, R. E. Phillips, and A. J. McMichael. 1996. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res. Hum. Retroviruses 12:1691-1698. [DOI] [PubMed] [Google Scholar]
- 25.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 26.Holmes, K., B. Fowlkes, I. Schmid, and J. Giorgi. 1995. Preparation of cells and reagents for flow cytometry, p. 5.3.1-5.3.23. In J. Coligan, A. Kruisbeek, D. Margulies, E. Sheevac, and W. Strober (ed.), Current protocols in immunology, vol. 1. Green Publishing Assoc., New York, N.Y. [DOI] [PubMed]
- 27.Imamichi, H., K. A. Crandall, V. Natarajan, M. K. Jiang, R. L. Dewar, S. Berg, A. Gaddam, M. Bosche, J. A. Metcalf, R. T. Davey, Jr., and H. C. Lane. 2001. Human immunodeficiency virus type 1 quasi species that rebound after discontinuation of highly active antiretroviral therapy are similar to the viral quasi species present before initiation of therapy. J. Infect. Dis. 183:36-50. [DOI] [PubMed] [Google Scholar]
- 28.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein, M. R., C. A. van Baalen, A. M. Holwerda, S. R. Kerkhof-Garde, R. J. Bende, I. P. Keet, J. K. Eeftinck-Schattenkerk, A. D. Osterhaus, H. Schuitemaker, and F. Miedema. 1995. Kinetics of Gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J. Exp. Med. 181:1365-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kostense, S., S. A. Otto, G. J. Knol, E. H. Manting, N. M. Nanlohy, C. Jansen, J. M. Lange, M. H. van Oers, F. Miedema, and D. van Baarle. 2002. Functional restoration of human immunodeficiency virus and Epstein-Barr virus-specific CD8+ T cells during highly active antiretroviral therapy is associated with an increase in CD4+ T cells. Eur. J. Immunol. 32:1080-1089. [DOI] [PubMed] [Google Scholar]
- 32.Kostense, S., K. Vandenberghe, J. Joling, D. Van Baarle, N. Nanlohy, E. Manting, and F. Miedema. 2002. Persistent numbers of tetramer+ CD8+ T cells, but loss of interferon-gamma+ HIV-specific T cells during progression to AIDS. Blood 99:2505-2511. [DOI] [PubMed] [Google Scholar]
- 33.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andres, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune response with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larsson, M., X. Jin, B. Ramratnam, G. S. Ogg, J. Engelmayer, M. A. Demoitie, A. J. McMichael, W. I. Cox, R. M. Steinman, D. Nixon, and N. Bhardwaj. 1999. A recombinant vaccinia virus based ELISPOT assay detects high frequencies of Pol-specific CD8 T cells in HIV-1-positive individuals. AIDS 13:767-777. [DOI] [PubMed] [Google Scholar]
- 35.Lee, S. K., Z. Xu, J. Lieberman, and P. Shankar. 2002. The functional CD8 T cell response to HIV becomes type-specific in progressive disease. J. Clin. Investig. 110:1339-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matano, T., R. Shibata, C. Siemon, M. Connors, H. C. Lane, and M. A. Martin. 1998. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J. Virol. 72:164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNeil, A. C., W. L. Shupert, C. A. Iyasere, C. W. Hallahan, J. Mican, R. T. Davey, Jr., and M. Connors. 2001. High-level HIV-1 viremia suppresses viral antigen-specific CD4+ T cell proliferation. Proc. Natl. Acad. Sci. USA 98:13878-13883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyerhans, A., G. Dadaglio, J. P. Vartanian, P. Langlade-Demoyen, R. Frank, B. Asjo, F. Plata, and S. Wain-Hobson. 1991. In vivo persistence of a HIV-1-encoded HLA-B27-restricted cytotoxic T lymphocyte epitope despite specific in vitro reactivity. Eur. J. Immunol. 21:2637-2640. [DOI] [PubMed] [Google Scholar]
- 39.Migueles, S. A., and M. Connors. 2001. Frequency and function of HIV-specific CD8+ T cells. Immunol. Lett. 79:141-150. [DOI] [PubMed] [Google Scholar]
- 40.Migueles, S. A., A. C. Laborico, W. L. Shupert, M. S. Sabbaghian, R. Rabin, C. W. Hallahan, D. Van Baarle, S. Kostense, F. Miedema, M. McLaughlin, L. Ehler, J. Metcalf, S. Liu, and M. Connors. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 3:1061-1068. [DOI] [PubMed] [Google Scholar]
- 41.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 97:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moskophidis, D., and R. M. Zinkernagel. 1995. Immunobiology of cytotoxic T-cell escape mutants of lymphocytic choriomeningitis virus. J. Virol. 69:2187-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nixon, D. F., A. R. Townsent, J. G. Elvin, C. R. Rizza, J. Gallwey, and A. J. McMichael. 1988. HIV-1 gag-specific cytotoxic T lymphocytes defined with recombinant vaccinia virus and synthetic peptides. Nature 336:484-487. [DOI] [PubMed] [Google Scholar]
- 44.Nowak, M. A., R. M. May, R. E. Phillips, S. Rowland-Jones, D. G. Lalloo, S. McAdam, P. Klenerman, B. Koppe, K. Sigmund, C. R. Bangham, et al. 1995. Antigenic oscillations and shifting immunodominance in HIV-1 infections. Nature 375:606-611. [DOI] [PubMed] [Google Scholar]
- 45.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, et al. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453-459. [DOI] [PubMed] [Google Scholar]
- 46.Pircher, H., D. Moskophidis, U. Rohrer, K. Burki, H. Hengartner, and R. M. Zinkernagel. 1990. Viral escape by selection of cytotoxic T cell-resistant virus variants in vivo. Nature 346:629-633. [DOI] [PubMed] [Google Scholar]
- 47.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puglielli, M. T., A. J. Zajac, R. G. van der Most, J. L. Dzuris, A. Sette, J. D. Altman, and R. Ahmed. 2001. In vivo selection of a lymphocytic choriomeningitis virus variant that affects recognition of the GP33-43 epitope by H-2Db but not H-2Kb. J. Virol 75:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 50.Shankar, P., M. Russo, B. Harnisch, M. Patterson, P. Skolnik, and J. Lieberman. 2000. Impaired function of circulating HIV-specific CD8+ T cells in chronic human immunodeficiency virus infection. Blood 96:3094-3101. [PubMed] [Google Scholar]
- 51.van der Most, R. G., R. J. Concepcion, C. Oseroff, J. Alexander, S. Southwood, J. Sidney, R. W. Chesnut, R. Ahmed, and A. Sette. 1997. Uncovering subdominant cytotoxic T-lymphocyte responses in lymphocytic choriomeningitis virus-infected BALB/c mice. J. Virol 71:5110-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Most, R. G., A. Sette, C. Oseroff, J. Alexander, K. Murali-Krishna, L. L. Lau, S. Southwood, J. Sidney, R. W. Chesnut, M. Matloubian, and R. Ahmed. 1996. Analysis of cytotoxic T cell responses to dominant and subdominant epitopes during acute and chronic lymphocytic choriomeningitis virus infection. J. Immunol. 157:5543-5554. [PubMed] [Google Scholar]
- 53.Weidt, G., W. Deppert, O. Utermohlen, J. Heukeshoven, and F. Lehmann-Grube. 1995. Emergence of virus escape mutants after immunization with epitope vaccine. J. Virol. 69:7147-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]