Abstract
Objective
To estimate the coverage provided by SNOMED CT for clinical research concepts represented by the items on case report forms (CRFs), as well as the semantic nature of those concepts relevant to post-coordination methods.
Design
Convenience samples from CRFs developed by rheumatologists conducting several longitudinal, observational studies of vasculitis were selected. A total of 17 CRFs were used as the basis of analysis for this study, from which a total set of 616 (unique) items were identified. Each unique data item was classified as either a clinical finding or procedure. The items were coded by the presence and nature of SNOMED CT coverage and classified into semantic types by 2 coders.
Measurements
Basic frequency analysis was conducted to determine levels of coverage provided by SNOMED CT. Estimates of coverage by various semantic characterizations were estimated.
Results
Most of the core clinical concepts (88%) from these clinical research data items were covered by SNOMED CT; however, far fewer of the concepts were fully covered (that is, where all aspects of the CRF item could be represented completely without post-coordination; 23%). In addition, a large majority of the concepts (83%) required post-coordination, either to clarify context (e.g., time) or to better capture complex clinical concepts (e.g., disease-related findings). For just over one third of the sampled CRF data items, both types of post-coordination were necessary to fully represent the meaning of the item.
Conclusion
SNOMED CT appears well-suited for representing a variety of clinical concepts, yet is less suited for representing the full amount of information collected on CRFs.
Introduction
Data standards in clinical medicine generally receive greater attention than the use of standards to represent, manage, and share data in clinical research, 1 although the need for data standards in clinical research is being identified. 2 The National Institutes of Health (NIH) roadmap includes goals for the use of data standards in clinical research that are compatible with health care data standards. 3 Inherently, clinical research is on the cutting edge of medicine, often creating new terminology and standards needs. In many cases, standards have not caught up with new concepts in the various subspecialty fields. Unlike day-to-day operational clinical systems, research data are collected for cumulative analyses that require data integrity, thereby rendering the use of standards even more crucial. Since there is no widespread use of data standards in clinical research, there is little known about whether the data standards for health care delivery and clinical medicine (e.g., those standards recommended by the Consolidated Health Informatics [CHI] initiative) 4 are adequate for clinical research. Determining the extent to which concepts embedded in clinical research can be represented by current terminology standards will help illuminate unmet needs and inherent complexities that may impede semantic interoperability and effective clinical research data management.
Background
Clinical Research Data
Clinical research, as defined by NIH, is patient-oriented research conducted with human subjects (or on specimens that can be linked to an individual) with whom an investigator directly interacts. 5 Such research includes mechanisms of human disease, therapeutic interventions, clinical trials, development of new technologies, epidemiologic studies, behavioral studies, and outcomes and health services research. Clinical research, as evidenced by the content of the definition above, encompasses a broad scope of users, purposes, requirements, and concepts. Although the goals for clinical research are varied, they generally include empirical evaluations or comparisons of one or more interventions using a variety of outcome measures, which often include clinical findings or observations. Rather than focusing on all possible observations of the whole patient, as in clinical care, the data focus of clinical research is narrower, and motivated to answer one or more questions that are explicitly defined before any data are collected.
Since data collection in clinical research generally supports pre-defined analyses, coded data elements are preferable. The notion of standardized data includes shared and adopted specifications for both data fields and value sets that encode the data within these fields. Representing the breadth, depth, and overall variety of data collected in clinical research is a key challenge to identifying and properly utilizing existing data standards.
Data Standards
We define standards here as consensual specifications for the like collection, scope (i.e., content), and representation (i.e., encoding) of data from different sources or settings. These specifications can include data elements, value sets for data elements (which can be entire terminologies), survey questions and responses, 6-9 and processes for data collection and/or coding that are equivalent among multiple data collection sites. Additionally, microarray research data standards are becoming important for comparing the results of experiments. Ideally, the use of standards results in having the same data representations from different applications, and the capture of variations in data collection or representation that affect the quality, utility, and comparability of data for future, sometimes unforeseen, uses. In clinical research, standards are applicable to structured data that represent inputs [independent variables] (e.g., baseline patient status, patient description such as age and gender), interventions (e.g., medications, procedures), outcome measures [dependent variables] (e.g., signs and symptoms, test results), and study descriptors (e.g., study design, length of follow-up).
In the rheumatology domain, the Arthritis, Rheumatism, and Aging Medical Information System (ARAMIS) project 10,11 represents a long and ambitious history of using standards for patient assessment and outcomes, 9,12-14 clinical research procedures, 15,16 and data representation. The use of these standards has resulted in many advances in the diagnosis and treatment of rheumatologic and other chronic diseases. 17 Over the past 30 years, the project has constructed large longitudinal data banks of patients with chronic and rheumatic diseases, and used them to answer questions about the natural history of diseases, toxicities of medications, identification of diagnostic subgroups, prediction of risk, efficacy and safety of treatment strategies, costs of care, and the development of risk factor models. Data standards and procedures for the evaluation of patient assessment (including clinical findings and outcomes) are critical to the epidemiological study of disease and the pursuit and evaluation of various treatment strategies. 16 The ARAMIS project, the first large-scale chronic disease data bank system, illustrates the impact of data standards on care delivery, and should inspire broader efforts to address standards that apply across all domains of clinical care and research.
In the United States, the Consolidated Health Informatics (CHI) initiative is a collaborative agreement between all federal organizations that collect health care data. Representatives from multiple agencies have worked together to identify and recommend the use of the “best” data standards in a variety of areas (e.g., anatomy, laboratory, diagnoses and problem lists). Although the CHI standards have named SNOMED CT as the standard to use for diagnoses and problem lists, anatomy, and procedures, 4 there is no consensus for the use of SNOMED CT in local applications, nor proof that the Description Logics that underlie SNOMED CT concept organization are sufficient to determine equivalence across variations in coding strategy.
Federally funded clinical research has recently been charged to share data following NIH guidelines, 18 and is obligated to follow the CHI-recommended data standards, including SNOMED CT. This project was conducted to explore the adequacy and coverage of current standards in a specific clinical research domain, vasculitis, which is a part of a larger, NIH-funded clinical research network. 19 The questions undertaken by this project include: Is SNOMED CT adequate for coding clinical research data in a specific clinical research domain? Is it clear and straightforward to use? Are there SNOMED CT structure and implementation issues that are unique to clinical research applications? Following a short description of the research network that provided the setting for this research, and an overview of the current SNOMED CT terminology model, we report the results of a study conducted using a sample of data concepts from several vasculitis research studies to demonstrate both the extent of coverage and areas where more attention may be needed. Lastly, we discuss important research directions that may help inform future data standards implementation discussions in the broader clinical research arena.
The Rare Disease Clinical Research Network
The Rare Disease Clinical Research Network (RDN) consists of ten clinical research consortia, each focused on several related rare diseases. Each research consortium consists of a team of clinical investigators partnering with patient support groups and institutions (mostly within the United States). The network is funded by several NIH components, including the Office of Rare Diseases (ORD), National Center for Research Resources (NCRR), National Institute of Neurological Disorders and Stroke (NINDS), National Institute of Child Health and Human Development (NICHD), National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), and the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). 19 One goal is to accelerate the development of diagnostics and treatments across a variety of rare diseases by encouraging cooperative partnerships and data sharing among the investigators at these centers.
The ten research consortia of the RDN each focus on specific research activities in the areas of: urea cycle disorders, neurological channelopathies, bone marrow failure diseases, cholestatic liver diseases, vasculitis, genetic steroid disorders, rare thrombotic diseases, rare lung diseases, genetic diseases of mucociliary clearance, and Angelman, Rett, and Prader-Willi syndromes. Collectively, these ten consortia research over 50 rare diseases. Currently, a centralized Data and Technology Coordinating Center supports the research design, data storage, study monitoring, and analysis for more than 35 protocols at various stages of development. The RDN is committed to the use of data standards, and is storing all data related to clinical findings, procedures, and anatomy using SNOMED CT, as recommended by CHI. The study reported here emerged from these efforts.
SNOMED CT
SNOMED CT is the CHI-recommended data standard in three CHI-defined areas (procedures, anatomy, problem lists and diagnoses), and is identified as the standard terminology for several RDN data constructs. Further, because SNOMED CT is the largest and most comprehensive clinical vocabulary, there are additional areas where SNOMED content could be used (e.g., adverse events, eligibility criteria, EKG results, vital signs, family history, behavioral risk factors), although it is not a recommended standard in those areas at this time.
SNOMED CT contains approximately 800,000 terms that represent over 350,000 unique concepts, and is experiencing a period of renewed growth with an increase in access generated by the National Library of Medicine public license agreement in 2000. 20,21 While SNOMED CT is often considered the most comprehensive vocabulary, 22-24 widespread adoption has not been achieved in clinical medicine or research, and there has been little exploration about consistency and reliability of SNOMED CT “coding” across persons and institutions, especially since the expansion of the SNOMED CT terminology model in 1999. 25-27 After the merging of SNOMED RT and Read codes, SNOMED CT became a “third generation” terminology, with a robust conceptual model that allows for post-coordination (i.e., the creation of new concepts using the logical combinations of other concepts). The use of post-coordination is relatively straightforward in many areas where the SNOMED CT (conceptual) terminology model (called the clinical context model) is complete and intuitive. In some important cases, however, such as for the use of context-dependent concept qualifiers, such as negation and subject of observation, the use of post-coordination is novel and complex.
The SNOMED CT terminology model specifies a series of valid attributes for each different “axis” (or broad grouping type) of concepts, and defines legal values (i.e., groups of concepts) for each attribute using subsets of SNOMED CT concepts. As an example, ▶ illustrates the attributes whose domain is the SNOMED CT Procedures hierarchy, and the allowable ranges of SNOMED CT concepts for each attribute. There are other defined sets of valid attributes of other groups of SNOMED CT concepts (e.g., clinical findings, body sites, etc.).
Figure 1.
Valid Attributes and Ranges for Procedure Concepts in SNOMED CT: A Partial Representation of the SNOMED CT Terminology Model. Any SNOMED CT concept in the Procedure hierarchy can be modified by these defining attributes and the ranges of concepts listed as permissible values. From SNOMED CT Users Guide, January 2006.
Using the sanctioned SNOMED CT terminology model (as depicted in ▶) for the post-coordination of new or complex concepts eases terminology maintenance and makes for a more efficient terminology by reducing “combinatorial explosion.” However, in practice, there is tension between terminology management and navigation; i.e., the needs for overall efficiency of the terminology, the desired ease of coding (enhanced by offering “pre-coordinated” terms such as “fracture of the left clavicle”), and the flexibility for users to quickly create missing concepts are often competing interests. 28
Theoretically, the SNOMED CT terminology model is suited to clinical research data insofar as it has the potential to represent complex clinical concepts, including time, subject, and negation. What is not clear at this point is how much post-coordination is necessary to fully represent clinical research data, and whether inadequacies in coding reliability and validity might emerge as an inevitable consequence of complexity in coding tasks. This early examination of the types of data collected for a typical clinical research study should help to illuminate this and related issues.
Perhaps the most widely-used terminology evaluation criterion is coverage. 22,24,29-40 A second common evaluation metric is to rate existing terminologies on whether they support various attributes or desired criteria, 41,42 including post-coordination. 23,37,43 Coverage studies comparing multiple terminologies, including those that allow post-coordination in addition to enumerated concepts, have shown that those terminologies that support post-coordination (specifically SNOMED CT) have higher coverage than terminologies that do not support post-coordination. 24,37,44
Concept coverage and assessment of structural features of the terminology are obviously important and useful evaluation metrics, but activities surrounding the evaluation and adoption of standard terminology in new domains, such as clinical research, will benefit from the development of other evaluation criteria, including complexity and the resources required for accurate and reliable use of the terminology. In terminology coverage studies, various designs usually include one or more domain experts searching pre-coordinated terms. Interfaces and tools provided to testers can be considered confounding factors. 45 Because use of post-coordination is difficult and prone to variation, fewer studies thoroughly evaluate the coverage achieved with post-coordination or compare the reliability of coding across multiple individuals. 26,46
A recent review by Rosenbloom et al. 47 summarizes the problems with post-coordination succinctly as: a) the need for mechanisms or syntax to restrict post-coordination to meaningful concepts; b) the creation of duplicate concepts (or “undetected synonymy”); and c) the potential for inefficiency in creating concept expressions. These three consequences imply a need for guidance and structural features to ensure “correct” use of post-coordination. The ability to create duplicate concepts in terminologies supporting post-coordination has been noted. 46,48-50 The increased likelihood for duplicate concepts directly implies variation in coding across coders, and has implications for any other study trying to truly measure coverage in post-coordinated terminology systems.
McKnight et al. 46 used automated term composition and custom interfaces to control the view and use of the terminology model in an attempt to reduce coding burden on research coders, but despite good coverage concluded that post-coordination was too cumbersome for practical use by clinicians (although inter-coder variation was not examined). McDonald et al. 51 note a drawback of complex terminologies was limitations in clinicians' ability to document data efficiently and perhaps reliably.
Methods
Research Objectives
This study estimates the coverage provided in SNOMED CT for clinical research concepts represented by the items on case report forms (CRFs), as well as the semantic nature of those concepts relevant to post-coordination methods. We examined the data items collected from three similar longitudinal studies being conducted on three rare types of vasculitis. Our aim was to characterize requirements for the use of SNOMED CT to represent data items collected for these studies. We expected that a large majority of the concepts needed to represent the data items collected in these studies would indeed be represented to one degree or another by SNOMED CT, but hoped to identify specific issues related to the use of this terminology to code the data items collected in clinical research.
Data Source
The CRFs from several longitudinal, observational studies of vasculitis were selected. Each CRF is simply a data collection form; multiple CRFs [e.g., Eligibility, Physical Exam, Medical History] collectively comprise the data collection for a given research study. A team of rheumatologists that are experienced researchers and clinicians designed these studies and spent a period of one year developing the CRF data collection forms. Several of these instruments have been used in previous research.
A total of 17 CRFs were used as the basis of analysis for this study. The CRFs are designed to collect the data needed to accurately track the progression of the disease, and to answer specific research questions described in the three longitudinal research protocols. The assumption is that, collectively, the items on the CRFs represent all of the important variables to be used for later analyses. From these 17 CRFs, 13 contained (clinical findings and procedures) data items appropriate for SNOMED CT coding. CRFs with content covered by other CHI data standards, such as Logical Observation Identifiers Names and Codes (LOINC) for lab test names and RxNorm for clinical drugs names, were excluded from the study. The specific forms analyzed in this study are listed in ▶, along with the number of data items on each form for which SNOMED CT is the appropriate data standard.
Table 1.
Table 1. Sample of Data Items by Form Name and Construct Type
| Name of Case Report Form | Number of Unique Items | Construct ∗ |
|---|---|---|
| Baseline Medical History Form | 142 | Findings |
| Baseline Medical History Form | 5 | Procedures |
| Follow-up Medical History | 41 | Findings |
| Baseline Comorbidity Form | 79 | Findings |
| Follow-up Comorbidity Form | 52 | Findings |
| Follow-up Comorbidity Form | 1 | Procedures |
| Hospitalization Form | 6 | Findings |
| Physical Exam Form | 37 | Findings |
| Physical Exam Form | 7 | Vital Signs |
| Patient Global Assessment Form | 2 | Findings |
| Angiogram Study Form | 113 | Findings |
| Patient Assessment (LWID) | 42 | Findings |
| Eligibility Forms (3) | 35 | Findings |
| Vasculitis Damage Index Form | 54 | Findings |
| Total | 616 |
∗ Construct type assigned by informatician.
A set of (unique) items (n = 616) for which SNOMED CT is an appropriate data standard was used in this study. In an attempt to examine the items for which SNOMED CT coding was clearly the appropriate standard, the sampled items represented only the findings (99%) and procedures (1%) constructs. The sampled items included all of the findings and procedures items collected in the three longitudinal studies.
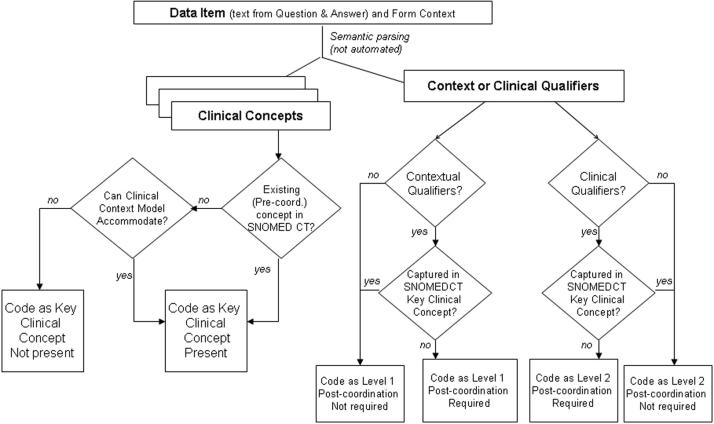
Each unique data item was identified and classified as either a clinical finding or procedure. The items were then coded into SNOMED CT concepts jointly by two coders (RR, JA) using a commercial coding tool (TermWorks, Apelon, Inc.) 52 that used term matching algorithms and hierarchy navigation to offer a list of possible SNOMED CT concepts for each item. Both coders are informaticians and familiar with the SNOMED CT terminology model. The coding tool used allowed the investigators to search existing pre-coordinated SNOMED CT terms and “descriptions” (i.e., synonyms). When not successful, a top-down approach was used, which essentially involved navigating the hierarchies (drilling down from broad conceptual groupings to more detailed conceptual groupings) until a term could be selected. Both coders' knowledge of the SNOMED CT terminology model allowed for the identification of SNOMED CT concepts that might be missed by the coding tool, but could be constructed using post-coordination. The coders did not use a shared syntax for post-coordination, but rather looked at the semantics of the data item and the SNOMED CT terminology model to determine if the concept expression could be covered by the current SNOMED CT model. ▶ describes the process that was followed. The July 2005 version of SNOMED CT was used.
Figure 2.
Process for Coding Coverage and Semantic Characteristics of Data Items
In this study, coverage refers to whether or not a concept appears explicitly in the terminology. Coverage decisions were based on a consensual agreement reached between the coders, who consulted domain experts when needed. It is important to note that the goal of this study was to determine if concepts could be represented in SNOMED CT (via existing terms or post-coordinated using the current SNOMED CT terminology model), not how they would be coded. Because of the lack of shared post-coordination syntax between coders and the collaborative nature of the coding (including the mutual development of coding and classification rules for the sample), the inter-rater agreement of coding was not analyzed.
Each coder independently recorded additional information on the nature of the semantics of the original data item while coding with SNOMED CT. For each data item, the following were recorded as either yes or no: 1) whether the primary clinical concept [defined as: the medical concept(s) that is the clinical focus/subject of the data item] was present in SNOMED CT; 2) whether all dimensions [including time, subject, and any clinical qualifiers] of the actual CRF item were adequately captured by an existing SNOMED CT concept; and 3) if post-coordination was needed to fully represent (in a SNOMED CT expression) all of the clinical concepts and dimensions that made up the CRF item. Further, for those CRF items requiring post-coordination to be covered, each item was rated on the nature of the post-coordination required: “Level 1” post-coordination refers to the need for non-clinical or context qualifiers, such as the time period or date, to fully represent the item; and “Level 2” post-coordination refers to the need for additional clinical concepts or qualifiers (e.g., site, finding due to disease, or other complex concepts) to modify the meaning of an existing clinical concept to fully represent the complete intended meaning of the data item from the original CRF.
One way to view the distinction between Level 1 (contextual) and Level 2 (clinical) post-coordination is that those concepts requiring a Level 1 post-coordination are not changing the essential clinical concept, but only the situational context (e.g., time and place dimensions). CRF data items requiring Level 2 qualifiers (e.g., “severe,” “sudden onset,” “left side”) have potential to alter the clinical meaning of the term to a domain expert. An example of the distinction between Level 1 and Level 2 post-coordination is whether a data item describing that a subject has suffered from a neurological disease is found on a Medical History CRF. If so, then Level 1 post-coordination would be required to add “History of” to the concept “Neurologic disease.” Level 2 post-coordination requires a more complex combination of SNOMED CT concepts by the coder, usually involving some clinical qualification of the clinical concept. As an example, Level 2 post-coordination is required in cases where hypertension, for example, is associated with another condition within a single data item, such as “renal vascular involvement with hypertension.” Therefore, some data items would need to be coded in such a way as to require both Level 1 and Level 2 post-coordination when both contextual and clinical qualifiers are present in the data item (e.g., “Pulmonary embolus with documented deep vein thrombosis—since last visit”).
There are many instances in SNOMED CT where particular complex concepts have been pre-coordinated, even though comparable concept expressions could be built from post-coordination. For instance, “Thrombosis of inferior vena cava” is a pre-coordinated (i.e., existing singular) concept in SNOMED CT, but a coder could use post-coordination to represent the same concept. In these instances, the sampled data items were coded as having the coverage for both the concept and the entire meaning of the entire data item. For these measures of the nature of SNOMED coverage (i.e., the types of concept qualifiers needed for post-coordination), decisions were based on consensual agreement between the authors. The authors jointly reviewed coding and/or characterization of coverage, arriving at a SNOMED CT coverage decision (covered/not covered) and a characterization of whether post-coordination was required and if so what type. Simple tabulations were then performed that are the basis of our analysis, including an examination of the semantic nature of these clinical research data items.
For open-ended data items (e.g., “Specify lung disease: ____”), we considered the item to be covered if a suitable parent concept (i.e., a concept with existing sub-concepts) was present in SNOMED CT. The post-coordination characterization for these types of questions was made based upon the wording of the item available on the CRF. Since there is no way to anticipate the concepts needed as answers to these types of items, the assumption is that the item is covered if SNOMED CT could represent a parent concept with multiple child-concepts (e.g., “lung disease” in the “Specify lung disease: ____” item example). In contrast, if an item included a structured list of value options, (e.g., Item = “Type of study performed”; selection values = “Dye,” “Catheter-directed angiogram,” “Magnetic resonance angiogram,” “Computerized-tomography angiogram”), then all of the concepts in the “answers” had to be included in SNOMED CT to be classified as having coverage for the clinical concept. Despite the use of multiple SNOMED CT concepts in these cases, the data item was still considered a single item in our analysis. This strategy was chosen to keep the focus of the study on the characteristics of CRF “data items” which consist of questions and answer values. To separate the values into different instances would give some questions (those with long answer groups) more weight in the descriptive results than others.
All items with an explicit time point or interval (e.g., current, history of, last three months, last seven days) were classified as having a need for Level 1 post-coordination; if post-coordination was also necessary to represent the key clinical concepts [defined as: the medical concept(s) that is the clinical focus/subject of the data item], then the item was classified as requiring Level 2 post-coordination as well. Because the default temporal context in the SNOMED CT clinical context model is “current,” if no time dimension was specified in the item, but a “current” time was clearly implied (e.g., from the context of a Physical Exam form, or the header “Current Findings”), we did not include these as requiring post-coordination to represent the dimension of “current.” However, when the dimension of “current” was explicitly part of the item, and other items on the same data form had other time periods specified (e.g., “past ten days”), then the item was classified as requiring Level 1 post-coordination, because it would be to distinguish a clinical concept from multiple contexts (e.g., “history of,” “past 30 days,” “current”) on a single form. Had we classified all items with a current time dimension as requiring post-coordination, then our entire sample (100%) would be classified as requiring Level 1 (contextual) post-coordination. Our strategy to highlight those items with time dimensions other than current (except for those needing an explicit specification for current) allowed us to dissect the semantic context of clinical research data items that are likely different from concepts used in clinical care delivery.
Results
A total of 616 unique data items were identified from the 13 CRFs examined. ▶ shows the overall breakdown of coverage by SNOMED CT.
Table 2.
Table 2. SNOMED CT Estimated Coverage of Sampled CRF Data Items (N = 616 Concepts)
| Features of Sampled CRF Data Items | Coverage and Type | Notes and Examples |
|---|---|---|
| Key Clinical Concepts Covered in SNOMED CT | 88% covered; 12% not covered | Examples of key clinical concept covered: |
| a.) “Hypertension” | ||
| b.) “Hypertension—since last visit” (The key clinical concept Hypertension is covered in SNOMED CT; although the time dimension requires post-coordination.) | ||
| Example of concept not covered: “Total lifetime thrombotic events.” | ||
| Full Meaning of CRF Data Item Covered | 23% full concept covered; 77% not fully covered | Example of concept not fully covered: “Lung disease-since last visit.” (The concept lung disease exists in SNOMED CT, but the “since last visit” piece does not.) |
| Post-Coordination | 83% require post-coordination; 17% do not | Note: If a pre-coordinated term was available, that term was selected and post-coordination deemed unnecessary (even if possible). Exceptions to this specified in methods. |
| Level 1 | 52% require Level 1 post-coordination | Example: Data (attribute) of Arterial thrombosis (disorder) |
| Level 2 | 67% require Level 2 post-coordination | Example: “Radial Left Pulse: Present/Absent/Unknown” |
| Note: for any items using present/absent as “answers”—we consider the notion of Present/Absent a clinical qualifier—because this qualifier is so important to the essence of the finding. | ||
| Both | 36% both Levels 1 and 2 post-coordination | Examples: “Pulmonary embolus without documented deep vein thrombosis—since last form completed” |
| “Above the knee deep vein thrombosis—Ever” | ||
| Note: Both of the above examples have contextual (time) and clinical (meaning) qualifiers. |
As expected, most clinical concepts needed for clinical research data in these studies are covered by SNOMED CT. Interestingly, however, is the inverse proportion of concepts fully covered; that is, where all aspects of the CRF item can be represented completely by existing SNOMED CT codes without post-coordination. It appears that SNOMED CT is well-suited for representing a variety of clinical concepts yet is less suited for representing the full amount of information collected on CRFs. [Note: we did not assess features of the actual language or wording of the item, but did include parts of the question text, e.g., “ever” or “history of” as important dimensions of the data item.]
An important feature of SNOMED CT is the use of a formal terminology model for post-coordination that allows for great flexibility in the creation of new concepts. A vast majority of the concepts appear to require post-coordination, either to clarify context (e.g., time) or to better capture complex clinical concepts (e.g., disease-related findings). For roughly half of the sampled CRF data items, both levels of post-coordination were necessary to fully represent the meaning of the item.
▶ illustrates the coverage and post-coordination requirements for the sample, stratified by the temporal context of each item (as determined by the informaticians). While still having high coverage, items that constituted “current” (in relation to the administration of the CRF) findings or procedures had a lower percentage (80% vs. 99%) of coverage of key clinical concepts than those items assessing historical events and observations. Although both “current” and historical data items overwhelmingly required post-coordination for the representation of their complete intended meaning, more historical items (93% vs. 25%) required Level 1 (contextual) coordination than did the “current” data items.
Table 3.
Table 3. SNOMED CT Estimated Coverage and Detail of Coding Requirements of Data Items by Temporal Context of Item
| Temporal Context | SNOMED CT Coverage of Key Concept(s) | SNOMED CT Coverage of Full Meaning of Data Item | Post-Coordination Required | Level 1 | Level 2 |
|---|---|---|---|---|---|
| Current (n = 367) | 296 (80%) | 132 (36%) | 275 (75%) | 91 (25%) | 240 (65%) |
| History (n = 249) | 246 (99%) | 11 (<1%) | 238 (96%) | 231 (93%) | 175 (70%) |
Discussion
A key finding from this investigation is that a large majority of the concepts represented in the CRF data items for this specific clinical research domain can only be partially represented by existing SNOMED CT concepts. SNOMED CT is an extensive clinical vocabulary, which also allows for complex concept construction through post-coordination. However, the more complex the nature of the concepts, the more difficult SNOMED CT is to use. Examining the semantics of the concepts and classifying the type of post-coordination needed (and therefore the expertise required) is one way to understand the complexities involved in moving a clinical data standard into the world of clinical research data. Most Level 1 post-coordination (defined by us as that post-coordination where existing clinical concepts are qualified via attributes describing only time or place) can be relatively straightforward given the numerous contextual qualifiers offered in the terminology. Perhaps this type of coding may be done by even a novice without domain expertise. These types of data items are particularly important in the context of clinical research, where clinical status is monitored at specific time intervals and in relationship to various interventions. For some diseases, the time concepts are clear and important markers of disease or disease progression, but can be quite complex—e.g., “Petechiae in last 30 days but not in past 24 hours.” It is possible that the current SNOMED CT terminology model, or instructions for post-coordination using both SNOMED CT attributes and numerical values, will need to be expanded to support these requirements (especially various time periods of follow-up), and it is not clear how much of this is part of the SNOMED CT mission. It might be more efficient to address these types of qualifiers as part of the local data model than by adding more complexity to the current SNOMED CT terminology model.
When Level 2 post-coordination (that involving clinical concept qualifiers) also is necessary, and in cases where the intended clinical meaning is at stake, both domain expertise and an intimate understanding of the SNOMED CT terminology structure will be important requirements for effective coding. Even with such expertise, the size, flexibility, and complexity of the SNOMED CT terminology model could lead to significant variance among coders, an area that requires further research. Our findings indicate that more attention is needed in the data standards' evolution regarding the representation of data items on clinical research CRFs.
Future work should examine how well the current SNOMED CT clinical context model captures the Level 1 and 2 (contextual and clinical) qualifiers (please see Appendix 1, available as a JAMIA on-line supplement at www.jamia.org) identified in this study, and whether the model needs to be expanded. For this project, we were not interested in capturing details related to the strict format (that is, the verbatim text on the CRF) and administration of the sampled data items, although others have argued that these are important attributes to capture, especially when measuring psychometric variables. 53,54 The items that we examined on the CRFs were all intended to be completed by a clinician (as opposed to the research subject) as part of standardized research protocols. While the structure of the item undoubtedly can bias the result in many examples, our intention was to evaluate the coverage of SNOMED CT on the complete intended meaning of each CRF data item. Many of these data items are unique to research, but future use of these items in clinical care is plausible; for instance, to establish risk factors or history in the context of a clinical visit. Also, because data items are the vehicle of structured data entry into electronic health record systems in clinical care, the representational needs of clinical research data items presented here are relevant to SNOMED CT implementation discussion in the milieu of health care delivery. It would be interesting to similarly explore common clinical data concepts to see whether clinical care delivery has the same or different needs from clinical research data.
Limitations
All items were sampled from CRFs from several longitudinal studies of a group of similar vasculitis diseases, and so the content is biased to the rheumatology domain. The area of rheumatology has a long history of recognizing the value of structured data and data standards. 17 These might not be representative of clinical research data, but anecdotally they appear similar in nature to items on the CRFs of other RDN longitudinal studies—Specifically, the presence of multiple temporal qualifiers and complex clinical concepts. While the sample did include all of the clinical finding data items collected for these studies, it would be interesting to see if these characterizations of clinical research data apply in other domains and study designs.
Another limitation comes from the likelihood of variability in coding between the two coders—both for coverage determination and semantic characterization. We attempted to address these by seeking consensus on all items, and consulting outside expertise to help clarify the nature of the item where needed. The coding procedures shown in ▶ were arrived at by collaboration and discussion with both coders throughout the process. Although the coding process was somewhat subjective, and the informaticians lacked clinical expertise, the purpose was to estimate, rather than measure, SNOMED CT coverage of concepts and dimensions as seen on selected CRFs, and to characterize the semantic nature and complexity of the sampled CRF data items. Future studies exploring inter-rater agreement among coders and samples from different disciplines are warranted to measure the complexity and reliability of this standard, particularly in light of the recent expansion of the SNOMED CT clinical context model.
There was no formal process for capturing context (e.g., form name, heading name), although the coders did have access to view the original forms, and the data set included the form name and heading name. A more robust description and capture of context would strengthen these results and provide insight for how to capture these contextual semantics in future SNOMED CT implementations.
Because a key objective of this study was to look at the semantics of clinical research data items and to get a gross estimate of coverage, the scope and rigor of the methods to determine SNOMED coverage differ from other coverage studies. Additionally, our coverage estimates possibly slightly over-estimate coverage, given our liberal approach to classifying coverage for open-ended items. For open-ended data items (e.g., “Specify lung disease: ____”), we considered the item to be covered if a suitable parent concept (concept with existing sub-concepts) was present in SNOMED CT. Since there was no way to anticipate the concepts needed as answers to open-ended items, our assumption was that the item is covered by SNOMED CT if an appropriate parent concept existed. This decision was forced by lack of actual subject data to evaluate the “answers” that vasculitis researchers are seeking for open-ended items. Of the 616 sampled items, 23 are open-ended and fall into this category.
The forms sampled for this study were designed by researchers for planned analyses, not for efficiency of SNOMED coding. The assumption is that these items address clinical research needs and are typical. Future studies in other clinical research domains will be required to know if these assumptions are correct. Further awareness of data semantics and coding issues as described here can facilitate standards in CRF design, which has received little attention from a data standards perspective. 17 The issues presented are not all necessarily addressed by changing the terminological standards or implementation guidance; additional standards at the “front end” of clinical research might be warranted.
Implications for Future Clinical Research
The results from this study suggest the need for improved understanding of how best to capture context in clinical research. A shared representation of context is critical in any kind of communication, but particularly in clinical settings. 55 For data to be successfully retrieved and useful, and in order to ensure a more ideal level of data integrity, some standard representation of context is necessary. Context is a complex and multi-dimensional construct. In one sense, context can be viewed as the situational attributes surrounding a core concept. Without standards or processes that capture the context indicators contained within the form itself (e.g., “Physical Exam Form,” “Family History Form”) or section headings within a form (e.g., “Current Findings,” “Co-Morbidities,” “Reason for Hospitalization,” “Maternal History”), the data items cannot be free-standing and retrieval loses meaning. This is an emerging issue for clinical care messaging standards and is the subject of several HL7 technical committees and special interest groups. We look forward to the recommendations of various groups examining the terminology model/information model interactions, particularly within the HL7 Vocabulary Technical Committees. 56 The issues of how to address overlaps and gaps between a sophisticated terminology such as SNOMED CT, and complex information models, such as HL7's RIM, mirror the challenges of inserting terminology into diverse data models that are inevitable across different research organizations. Standards activities in health care delivery have long recognized the need for information model standards within which to house and define terminological standards. 57-59 This study illustrates that discussion of standard information models and information model / terminology model interaction should be discussion topics for the clinical research data standards community.
Because the data items on CRFs could be thought of as “questions,” there are important relationships between this study and the efforts to represent structured assessments or patient response questionnaires. Others have pursued the use of existing data standards to represent questions. For instance, two previous studies 33,53 used LOINC to capture elements of questions on standardized questionnaires, yet both reported limitations on the coverage of clinical concepts within LOINC. Brandt et al. 22 stress the importance of standards for representing the content of questions and questionnaires for the maintenance and curating of data libraries that support the clinical research process. 60 They also speculate that such standards could allow intelligent aggregation and analysis of multiple question formats that attempt to measure the same construct in different settings. Although SNOMED CT does not claim to represent questions per se, it may be flexible and comprehensive enough to accommodate this unmet need.
There is no recommended CHI standard for questions on standardized or non-standardized instruments, nor is this listed as a subject area for consideration. However, many of the data standards recommended by CHI cover domains that would be found on case report forms in clinical research (e.g., diagnoses on a medical history form, findings on a physical exam form, medications on a self-reported medication form), suggesting the possible need for more than one standard to properly code CRF data items. Additionally, other areas of great interest in clinical research (e.g., quality of life, risk factors, family history) are not addressed by current CHI standards, yet are also captured via “questions” (patient directed) or data items (clinician directed) on data collection forms. A broad model for indexing questions (including the content, the exact text, the answer values, and other structural features of the question construction) might be required to represent the “context” within which other standards operate. The use of such a model in clinical research settings could encourage the item re-use, and therefore the promulgation of similar data, facilitating standardization.
Many types of clinical research data represent construct areas where CHI-recommended standards are not named (e.g., eligibility criteria, adverse events), and candidate standards need to be identified and evaluated for these areas. Coverage of concepts is typically the most quantitative evaluation metric, although several methods have been employed to test this. 24,31-33,37,45,61-67,62-68 As the SNOMED CT model becomes more comprehensive and more widely used, traditional coverage studies will need to address the issues of the nature and complexity of post-coordination that were explored in this study. We propose that additional metrics can supplement coverage studies and provide other solid data to help in making informed choices between competing data standards. The complexity of coding, type of coding required, coder requirements, and estimates of inter-rater reliability are all measurable and important metrics to explore when evaluating data standards for clinical research. A central question for determining “best” standards is identifying the future information retrieval needs and the implications of using multiple terminologies to best capture the data from various studies. While there is general agreement on the features of a “good” terminology, 42 there is little consensus or direction on how to formally and quantitatively evaluate and compare terminological standards as they apply to this domain. 43
While an organization could easily become submerged with these complex issues of the implementation of data standards, it is important to maintain a spirit of practicality and purpose. The clinical research community must clearly define the purpose of data standards, describing real “use cases” for interoperability and data sharing. An explicit and collective understanding of the purpose of data standards is critical to successful implementation and evaluation. 68 What is the purpose of standardized data in clinical research? To share CRF data items? To share concepts? To represent context? To share data sets? If the purpose is to share data items, then perhaps clinical research interests should push for standards in CRF design, and advocate for the alteration or expansion of the SNOMED CT model or standards for its use in clinical research applications. If the purpose is to make clinical research data sets “free standing,” then standards for data model design and the level of reliance on the terminology model aspects of SNOMED CT will be needed. Regardless, lobbying efforts to bring forward clinical research data needs 56,69 to relevant standards bodies are warranted and should continue. The intended nature of data sharing will determine in which standards activities clinical researchers need to be represented.
Certainly, data standards have enormous potential to impact the clinical research process and the standard of care delivery, as the ARAMIS project has demonstrated. To date, hundreds of peer-reviewed publications have been published from the ARAMIS group of investigators. The project (which involves over a dozen centers in the United States and Canada) has just been re-funded by the National Institutes of Health for years 26–30. 70 The fruitful examination of data from multiple sites and time points, as well as the diversity of research staff (clinicians, epidemiologists, biostatisticians, information scientists, health economists, and health service researchers), requires data standards and informatics tools. While most standards selection has been within the ARAMIS network, this and similar projects could benefit from an awareness of strategies to apply mainstream clinical data standards into their clinical domains. This study illustrates the issues that emerge with attempts to use standard terminologies, such as SNOMED CT, in several studies of rare vasculitis. As with the clinical concepts used in vasculitis research, the key clinical concepts used in many ARAMIS studies are likely contained in SNOMED CT. Our findings should give an appreciation for the complexities involved in applying data standards into a focused clinical research domain, and inspire future implementation and evaluation activities in other areas of clinical research.
Conclusion
The data items from clinical research CRFs contain important contextual data that are perhaps as elaborate as that collected in health care delivery, and complete coverage in SNOMED CT requires the use of post-coordination. The semantic characteristics of these data items imply the need for guidance on how to use the current SNOMED CT terminology model. The semantic nature of these CRF items presents a possible dividing line for coding tasks, and might indicate areas where non-domain experts could relieve the coding burden. Further examination of this work within other clinical domains is warranted, and further exploration of the use of data standards, such as SNOMED CT, in the context of a multi-institutional research network (NIH's Rare Disease Clinical Research Network), is ideal since many domains and study designs are represented. Additionally, standards in the area of data model design and appropriate use of SNOMED CT will be required. Once the nature of the coding required for clinical research can be described, different expertise can be used to apply different pieces of the standard, and implementation approaches can be discussed and tried, leading to interoperability of clinical research data and realizing the vision of standardized data and data sharing.
Footnotes
The project described was supported by grant number RR019259 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
The authors wish to thank Dr. Peter Merkel, Principal Investigator of the Vasculitis Clinical Research Consortium (VCRC), Boston University School of Medicine, Ann Corbo, Project Manager (VCRC), and the investigators of the VCRC (NIH NCRC grant 1 U54 RR019497) for sharing their data collection forms and expertise. The authors are grateful to David Cuthbertson, Biostatistician/Associate in Biostatistics, Pediatrics Epidemiology Center at the University of South Florida for helping to develop the Vasculitis Clinical Research Consortium case report forms and for providing background information. The authors also thank Alicia Livinski for her data management and editing contributions and Dr. Timothy Patrick, Assistant Professor, College of Health Sciences, University of Wisconsin-Milwaukee for his thoughtful reviews. The authors also wish to thank the JAMIA external reviewers for their constructive comments and assistance finalizing this manuscript.
References
- 1.Rindfleisch TC, Brutlag BL. Directions for Clinical Research and Genomic Research into the Next Decade. Implications for Informatics J Am Med Assoc 1998;5:404-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zerhouni EA. Keynote Presentation. Washington, DC: American Medical Informatics Association; 2005.
- 3.NIH NIH Roadmap. Accelerating Medical Discovery to Improve Health. National Institutes of Health; 2005. [DOI] [PubMed]
- 4.CHI CHI Executive Summaries. Consolidated Health Informatics. 2004.
- 5.NIH The NIH Director's Panel on Clinical Research Report to the Advisory Committee to the NIH Director, December, 1997. NIH Director's Panel on Clinical Research (CRP); 1997.
- 6.Bruce B, Fries JF. The Stanford Health Assessment QuestionnaireDimensions and Practical Applications. Health Qual Life Outcomes 2003;1:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fries JF, Bruce B, Cella D. The promise of PROMISusing item response theory to improve assessment of patient-reported outcomes. Clin Exp Rheumatol 2005;23(5 Suppl 39):S53-S57. [PubMed] [Google Scholar]
- 8.Bruce B, Fries JF. The Health Assessment Questionnaire (HAQ) Clin Exp Rheumatol 2005;23(5 Suppl 39):S14-S18. [PubMed] [Google Scholar]
- 9.Spitz PW, Fries JF. The present and future of comprehensive outcome measures for rheumatic diseases Clin Rheumatol 1987;6(Suppl 2):105-111. [DOI] [PubMed] [Google Scholar]
- 10.Stanford. ARAMIS (the Arthritis, Rheumatism, and Aging Medical Information System) Stanford University Medical Center; 2003..
- 11.Fries JF, McShane DJ. ARAMIS (the American Rheumatism Association Medical Information System). A prototypical national chronic-disease data bank West J Med 1986;145(6):798-804. [PMC free article] [PubMed] [Google Scholar]
- 12.Hunder GG, Arend WP, Bloch DA, et al. The American College of Rheumatology 1990 criteria for the classification of vasculitis. Introduction Arthritis Rheum 1990;33(8):1065-1067. [DOI] [PubMed] [Google Scholar]
- 13.Leavitt RY, Fauci AS, Bloch DA, et al. The American College of Rheumatology 1990 criteria for the classification of Wegener's granulomatosis Arthritis Rheum 1990;33(8):1101-1107. [DOI] [PubMed] [Google Scholar]
- 14.Singh G, Ramey DR, Morfeld D, et al. Gastrointestinal Tract Complications of Nonsteroidal Anti-inflammatory Drug Treatment in Rheumatoid ArthritisA Prospective Observational Cohort Study. Arch Int Med 1996;156(14):1530-1536. [PubMed] [Google Scholar]
- 15.Fries JF, Miller SR, Spitz PW, Williams CA, Hubert HB, Bloch DA. Toward an epidemiology of gastropathy associated with nonsteroidal antiinflammatory drug use Gastroenterology 1989;96(2 Pt 2 Suppl):647-655. [DOI] [PubMed] [Google Scholar]
- 16.Edworthy SM, Bloch DA, Brant RF, Fries JF. Detecting treatment effects in patients with rheumatoid arthritisthe advantage of longitudinal data. J Rheumatol 1993;20(1):40-44. [PubMed] [Google Scholar]
- 17.Bruce B, Fries JF. The Arthritis, Rheumatism and Aging Medical Information System (ARAMIS)Still young at 30 years. Clin Exp Rheumatol 2005;23(Suppl. 39):S163-S167. [PubMed] [Google Scholar]
- 18.NIH Final NIH Statement on Sharing Research Data, February 26, 2003, N.I.o. Health, Editor. 2003.
- 19.NIH Press Release. NIH Establishes Rare Diseases Clinical Research Network, NCRR, Editor. 2003.
- 20.CAP News Release. HHS Secretary Tommy G. Thompson Announces Access to SNOMED CT Through National Library of Medicine, S. International, Editor. Northfield, IL: SNOMED International; 2004.
- 21.NIH SNOMED Clinical Terms® To Be Added To UMLS® Metathesaurus, NLM, Editor. 2003.
- 22.Wasserman H, Wang J. An applied evaluation of SNOMED CT as a clinical vocabulary for the computerized diagnosis and problem list AMIA Annu Symp Proc. 2003:699-703. [PMC free article] [PubMed]
- 23.Dykes PC, Currie LM, Cimino JJ. Adequacy of evolving national standardized terminologies for interdisciplinary coded concepts in an automated clinical pathway J Biomed Inform 2003;36(4-5):313-325. [DOI] [PubMed] [Google Scholar]
- 24.Humphreys BL, McCray AT, Cheh ML. Evaluating the coverage of controlled health data terminologiesreport on the results of the NLM/AHCPR large scale vocabulary test. J Am Med Inform Assoc 1997;4(6):484-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burkhart L, Konicek R, Moorhead S, Androwich I. Mapping parish nurse documentation into the nursing interventions classificationa research method. Comput Inform Nurs 2005;23(4):220-229. [DOI] [PubMed] [Google Scholar]
- 26.Hasman A, de Bruijn LM, Arends JW. Evaluation of a method that supports pathology report coding Methods Inf Med 2001;40(4):293-297. [PubMed] [Google Scholar]
- 27.Baumann RP. [Medical data in pathology—evaluation of a large collection. (530,000 diagnoses coded in SNOMED II)] Rev Med Suisse Romande 1999;119(10):805-824. [PubMed] [Google Scholar]
- 28.Tuttle MS, Cole WG, Sheretz DD, Nelson SJ. Navigating to Knowledge Methods Inf Med 1995;34:214-231. [PubMed] [Google Scholar]
- 29.Henry SB, Holzemer WL, Reilly CA, Campbell KE. Terms used by nurses to describe patient problemscan SNOMED III represent nursing concepts in the patient record?. J Am Med Inform Assoc 1994;1(1):61-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henry SB, Holzemer WL. Can SNOMED International represent patients' perceptions of health-related problems for the computer-based patient record? Proc Annu Symp Comput Appl Med Care 1994:184-187. [PMC free article] [PubMed]
- 31.Campbell JR, Payne TH. A comparison of four schemes for codification of problem lists Proc Annu Symp Comput Appl Med Care 1994:201-205. [PMC free article] [PubMed]
- 32.Brown PJ, Warmington V, Laurence M, Prevost AT. Randomised crossover trial comparing the performance of Clinical Terms Version 3 and Read Codes 5 byte set coding schemes in general practice BMJ 2003;326(7399):1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakken S, Cimino JJ, Haskell R, Kukafka R, Matsumoto C, Chan GK, Huff SM. Evaluation of the clinical LOINC (Logical Observation Identifiers, Names, and Codes) semantic structure as a terminology model for standardized assessment measures J Am Med Inform Assoc 2000;7(6):529-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White MD, Kolar LM, Steindel SJ. Evaluation of Vocabularies for Electronic Laboratory Reporting to Public Health Agencies J Am Med Inform Assoc 1999;6(3):185-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinha U, Yaghmai A, Thompson L, Dai B, Taira RK, Dionisio JD, Kangarloo H. Evaluation of SNOMED3.5 in representing concepts in chest radiology reports: integration of a SNOMED mapper with a radiology reporting workstation Proc AMIA Symp. 2000:799-803. [PMC free article] [PubMed]
- 36.Ruggieri AP, Elkin P, Chute CG. Representation by standard terminologies of health status concepts contained in two health status assessment instruments used in rheumatic disease management Proc AMIA Symp. 2000:734-738. [PMC free article] [PubMed]
- 37.Campbell JR, Carpenter P, Sneiderman C, Cohn S, Chute CG, Warren J, CPRI Work Group on Codes and Structures Phase II evaluation of clinical coding schemescompleteness, taxonomy, mapping, definitions, and clarity. J Am Med Inform Assoc 1997;4(3):238-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Penz JF, Brown SH, Carter JS, Elkin PL, Nguyen VN, Sims SA, Lincoln MJ. Evaluation of SNOMED coverage of Veterans Health Administration terms Medinfo 2004;11(Pt 1):540-544. [PubMed] [Google Scholar]
- 39.Brown SH, Bauer BA, Wahner-Roedler DL, Elkin PL. Coverage of oncology drug indication concepts and compositional semantics by SNOMED-CT AMIA Annu Symp Proc. 2003:115-119. [PMC free article] [PubMed]
- 40.Strang N, Cucherat M, Boissel JP. Which coding system for therapeutic information in evidence-based medicine Comput Methods Programs Biomed 2002;68(1):73-85. [DOI] [PubMed] [Google Scholar]
- 41.Chute GC, Cohn SP, Campbell JR. A Framework for Comprehensive Health Terminology Systems in the United States J Am Med Inform Assoc 1998;5(6):503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cimino J. Desiderata for Controlled Medical Vocabularies in the Twenty-First Century Methods of Information in Medicine 1998. [PMC free article] [PubMed]
- 43.Elkin PL, Brown SH, Carter J, et al. Guideline and Quality Indicators for Development, Purchase and Use of Controlled Health Vocabularies Int J Med Inform 2002;68(1-3):175-186. [DOI] [PubMed] [Google Scholar]
- 44.Chute CG, Cohn SP, Campbell KE, Oliver DE, Campbell JR. The content coverage of clinical classifications. For The Computer-Based Patient Record Institute's Work Group on Codes & Structures J Am Med Inform Assoc 1996;3(3):224-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humphreys BL, Hole WT, McCray AT, Fitzmaurice JM. Planned NLM/AHCPR large-scale vocabulary testusing UMLS technology to determine the extent to which controlled vocabularies cover terminology needed for health care and public health. J Am Med Inform Assoc 1996;3(4):281-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKnight LK, Elkin PL, Ogren PV, Chute CG. Barriers to the Clinical Implementation of Compositionality Proc AMIA Symp. 1999:320-324. [PMC free article] [PubMed]
- 47.Rosenbloom ST, Miller RA, Johnson KB, Elkin PL, Brown SH. Interface TerminologiesFacilitating Direct Entry of Clinical Data into Electronic Health Record Systems. [Review Paper] J Am Med Inform Assoc 2006;13:277-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rector AL, Bechhofer S, Goble CA, Horrocks I, Nowlan WA, Solomon WD. The GRAIL Concept Modelling Language for Medical Terminology Artif Intell Med 1997;9:139-171. [DOI] [PubMed] [Google Scholar]
- 49.Evans DA, Rothwell DJ, Monarch IA, Lefferts RG, Cote RA. Toward representations for medical concepts Med Decis Making 1991;11(4 Suppl):S102-S108. [PubMed] [Google Scholar]
- 50.Fung KW, Hole WT, Nelson SJ, Srinivasan S, Powell T, Roth L. Integrating SNOMED CT into the UMLSan exploration of different views of synonymy and quality of editing. J Am Med Inform Assoc 2005;12(4):486-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McDonald CJ. The barriers to electronic medical record systems and how to overcome them J Am Med Inform Assoc 1997;4(3):213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Apelon I. Terminology Web Services. Apelon, Inc; 2006.
- 53.White TM, Hauan MJ. Extending the LOINC Conceptual Schema to Support Standardized Assessment Instruments J Am Med Inform Assoc 2002;9(6):586-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aday LA. Designing and Conducting Health Surveys. 2nd ed. San Francisco: Jossey-Bass; 1996. pp. 560.
- 55.Degoulet P, Fieschi M. Introduction to Clinical Informatics. 2nd ed. New York: Springer-Verlag; 1999.
- 56. HL7, Health Level Seven. Health Level Seven, Inc; 2005.
- 57.Huff S, Carter J. A Characterization of Terminology Models, Clinical Templates, Message Models, and Other Kinds of Clinical Information Models AMIA Symp. 2000.
- 58.Dampney CNG, Pegler G, Johnson M. Harmonising Health Information Models—A Critical Analysis of Current Practice. Canberra ACT, Australia: Ninth National Health Informatics Conference; 2001.
- 59.Dolin RH, Spackman KA, Markwell D. Selective retrieval of pre- and post-coordinated SNOMED concepts Proc AMIA Symp. 2002:210-214. [PMC free article] [PubMed]
- 60.Brandt CA, Cohen DB, Shifman MA, Miller PL, Nadkarni PM, Frawley SJ. Approaches and Informatics Tools to Assist in the Integration of Similar Clinical Research Questionnaires Meth Inform Med 2004;43:156-162. [PubMed] [Google Scholar]
- 61.Bakken S, Warren JJ, Lundberg C, Casey A, Correia C, Konicek D, Zingo C. An evaluation of the usefulness of two terminology models for integrating nursing diagnosis concepts into SNOMED Clinical Terms Int J Med Inform 2002;68(1-3):71-77. [DOI] [PubMed] [Google Scholar]
- 62.Brown PJ, Warmington V, Laurence M, Prevost AT. A methodology for the functional comparison of coding schemes in primary care Inform Prim Care 2003;11(3):145-148. [DOI] [PubMed] [Google Scholar]
- 63.Chiang MF, Casper DS, Cimino JJ, Starren J. Representation of ophthalmology concepts by electronic systemsadequacy of controlled medical terminologies. Ophthalmology 2005;112(2):175-183. [DOI] [PubMed] [Google Scholar]
- 64.Klimczak JC, Hahn AW, Sievert ME, Petroski G, Hewett J. Comparing clinical vocabularies using coding system fidelity Proc Annu Symp Comput Appl Med Care 1995:883-887. [PMC free article] [PubMed]
- 65.Lussier YA, Bourque M. Comparing SNOMED and ICPC retrieval accuracies using relational database models Proc AMIA Annu Fall Symp. 1997:514-518. [PMC free article] [PubMed]
- 66.Moore GW, Berman JJ. Performance analysis of manual and automated systemized nomenclature of medicine (SNOMED) coding Am J Clin Pathol 1994;101(3):253-256. [DOI] [PubMed] [Google Scholar]
- 67.Mullins HC, Scanland PM, Collins D, Treece L, Petruzzi Jr P, Goodson A, Dickinson M. The efficacy of SNOMED, Read Codes, and UMLS in coding ambulatory family practice clinical records Proc AMIA Annu Fall Symp. 1996:135-139. [PMC free article] [PubMed]
- 68.Gardner M. Why clinical information standards matter. Because they constrain what can be described BMJ 2003;326:1101-1102[editorial]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.CDISC Clinical Data Interchange Standards Consortium. CDISC; 2005.
- 70.Fries BB. The Arthritis, Rheumatism and Aging Medical Information System (ARAMIS)still young at 30 years. Clin Exp Rheumatol 2005;5(Suppl 39 (23):S163-S167. [PubMed] [Google Scholar]