Abstract
Objective
Acquiring and representing biomedical knowledge is an increasingly important component of contemporary bioinformatics. A critical step of the process is to identify and retrieve relevant documents among the vast volume of modern biomedical literature efficiently. In the real world, many information retrieval tasks are difficult because of high data dimensionality and the lack of annotated examples to train a retrieval algorithm. Under such a scenario, the performance of information retrieval algorithms is often unsatisfactory, therefore improvements are needed.
Design
We studied two approaches that enhance the text categorization performance on sparse and high data dimensionality: (1) semantic-preserving dimension reduction by representing text with semantic-enriched features; and (2) augmenting training data with semi-supervised learning. A probabilistic topic model was applied to extract major semantic topics from a corpus of text of interest. The representation of documents was projected from the high-dimensional vocabulary space onto a semantic topic space with reduced dimensionality. A semi-supervised learning algorithm based on graph theory was applied to identify potential positive training cases, which were further used to augment training data. The effects of data transformation and augmentation on text categorization by support vector machine (SVM) were evaluated.
Results and Conclusion
Semantic-enriched data transformation and the pseudo-positive-cases augmented training data enhance the efficiency and performance of text categorization by SVM.
Introduction
Acquiring and representing biomedical knowledge is a perpetual task for the biomedical informatics community. 1–3 An example of these efforts is the knowledge base maintained by the Gene Ontology (GO) consortium. 4 The GO consortium annotates the proteins from multiple organisms with a controlled vocabulary—the GO terms. One important and practical task during the annotation process is to identify and retrieve the relevant documents from the vast volume of biomedical publications efficiently and accurately. Over the last a few years, this need has prompted special conferences and challenge tasks dedicated to retrieving protein-related information from biomedical literature. 2,3,5,6 The annual Genomics Track of the Text Retrieval Conference (TREC) 7 has been dedicated to such task since 2003. 2,5
In general, information retrieval tasks can be divided into two types: ad hoc retrieval and text categorization, or filtering. 8 The ad hoc retrieval refers to retrieving text from a relatively static text collection in response to short term queries, e.g., retrieving Web pages using the Google search engine. Text categorization or filtering refers to meeting the long-term information needs of users, where users' preferences are predefined and new text is divided into groups according to such preferences. Since examples of documents labeled with preference categories are often available, the task is usually cast as a supervised classification problem. 9,10 Among many classification algorithms available, SVM 9–11 is often considered as one of the best text categorization algorithms. Application of SVM in the biomedical informatics setting spans both ends of the spectrum. For example, recent research by Aphinyanaphongs et al 12 concentrates on retrieval from the clinical science literature, where SVM outperform most other text categorization algorithms, while our task is to retrieval documents related to molecular biology. However, when trained with a small number of high dimensional training cases, SVM, like other classification methods, does not perform well 9 because a combination of high-dimensionality of the text data and scarcity of training cases makes it difficult to train classifiers that generalize well.
In this research, we address such difficulties in text categorization tasks by (1) representing the document with low-dimensional semantic-enriched features; and (2) augmenting the training data by semi-supervised learning. Our experiments show promising results, and the overall strategy should be applicable to a wide range of classification tasks.
Methods
Text Data and Preprocessing
To make our experiments reflect the real world IR tasks, we used the data from the TREC 2005 text categorization tasks. 13 The text categorization tasks of the TREC 2005 were designed based on the practical needs of the Mouse Genomics Informatics (MGI). 14 The data set for the text categorization task consisted of the full articles published during the years of 2002 ∼ 2003 from three journals: the Journal of Cell Biology, the Proceedings of the National Academy of Sciences, and the Journal of Biological Chemistry. There were four text categorization subtasks: (1) the allele subtask (A) is to retrieve documents related to allele mutation; (2) the expression subtask (E) is to retrieve documents related to gene expression during embryonic development; (3) the gene ontology subtask (G) is to retrieve documents that contain the contents which warrant GO annotations; and (4) the tumor subtask (T) is to retrieve documents related to tumors. The training data consist of 5,837 full article documents, and the numbers of positive training cases for the subtasks A, E, G, and T are 338, 81, 462 and 36, respectively.
The documents were preprocessed by removing the stop words, stemming, 15 and discarding terms that appeared less than ten times in the corpus. As a result, the size of the vocabulary (V) was trimmed to 23, 484 and a document was represented by the V-dimensional vector, d = (w 1, w 2,…,wV)′, in which an element, wi, represents the number of the times the i th word of the vocabulary occurs in the document. The elements of the vector were further weighted using a modified term frequency and inverse document frequency (tf-idf) scheme, 9 which calculates the weight wd(t) for the term t in the document d:
| (1) |
where n(t, d) is the number of times the term t occurs in the document d, D is the total number of documents in the corpus and n(t) is the number of the documents that contain t. To remove the influence of different document lengths on the weights, the document vectors were further normalized to have Euclidean norm of 1.0, as follows:
| (2) |
where w′d (t) is the normalized weight, is summation over all elements of the vector.
LDA Model
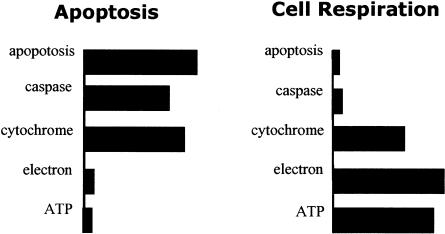
The latent Dirichlet allocation (LDA) model 16 is a member of the generative probabilistic models that use a small number of topics to describe a collection of documents. 16–19 The LDA model captures the multi-topical feature of documents and treats a document as a mixture of words from topics represented as multinomial distributions over the vocabulary. For example, when a biologist discusses the topic cell respiration, words like “electron,” “cytochrome,” and “ATP” are commonly used while, on the other hand, the words like “apoptosis,” “programmed,” “death” and “caspase” are commonly used to discuss the concept of apoptosis. Thus, a document discussing a protein located in mitochondrion and participating in apoptosis can be treated as a mixture of words from two topics. Based on such assumptions, statistical learning techniques can be applied to identify the topics from a collection of documents. The distributional representation of a concept captures the key relationship between the concept and words: a concept is conveyed by word choice, and the sense of a word depends on context. ▶ shows how a topic can be represented as word usage distribution in a probabilistic topic model. Note that the word “cytochrome” has high probability in both topics. The LDA model is capable of determining to which topic a word like this should be assigned, based on the context of the document.
Figure 1.
Representing concepts with word distributions. Two hypothetical topics are depicted. The bar lengths indicate the word-sage preference or the conditional probability of observing the a word for a given topic.
Model Specification
Let a corpus C = be a set of documents, where D denotes the number of documents in the corpus; a document di = (w 1, w 2,…, wNi) consist of a sequence of words; and let w be a word that takes a value from the vocabulary {v 1, v 2,…,vV. Let T be the number of topics of a LDA model and V be the size of the vocabulary of the corpus. The LDA model simulates the generation of a document with the following stochastic process:
• For each document, sample a topic proportion vector θ = (θ1, θ2,…θT)′ from a Dirichlet distribution with parameter α′: θ: ∼Dir(θ/α). This is equivalent to an author deciding what topics to include in a paper.
• For each word in the document, sample a topic z according to multinomial distribution governed by θ: z∼ Multi(z/θ). This can be regarded as assigning a word to a topic.
• Conditioning on z, sample a word w according to the multinomial distribution with the parameter θz: w∼ Multi(w/θz). This corresponds to picking words to represent a concept.
• The parameter θt, with t ∈ {1, 2,…T} is a V-dimensional vector that defines the multinomial word-usage distribution of a topic. It is distributed as Dirichlet with parameter β: θ∼: Dir(θ/b).
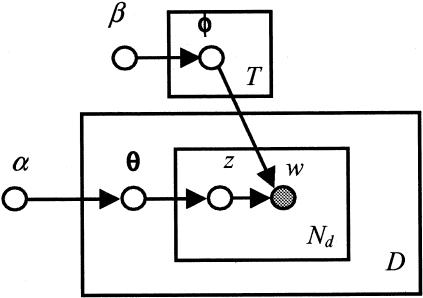
The probabilistic directed acyclic graphical representation of the LDA model is shown in ▶ in plate notation. 20 In a probabilistic graph, nodes represent random variables, and edges represent the probabilistic relationship, i.e., the conditional probability, between the variables. The shaded and un-shaded nodes represent the observed and unobserved variables, respectively. Each rectangular plate represents a replica of a data structure, e.g., a document; the variable at the bottom right of a plate indicates the number of the replicates. In this graph, each document is associated with a topic composition variable θ, and a total of Nd replicates of topic variable z and word w. The graph also shows that there are T topic word distributions.
Figure 2.
The directed acyclic graphical representation of the LDA model. Each node represents a variable, and a shaded node indicates an observed variable. Each rectangle plate represents a replica of the data structure. The variables D and Nd at the bottom right of plate indicate the number of the replicates of the structure.
Statistical Inference
The LDA model extracts the latent semantic topics by estimating the word-usage distributions and assigns each word in a corpus to a latent semantic topic through statistical inference. The exact inference of the parameters and latent variables of a LDA model is intractable. The approximate methods based on variational methods 16 and Markov chain Monte Carlo (MCMC) 21 have been developed. In this study, the Gibbs sampling inference algorithm by Griffith and Steyvers 18 was employed, because it was less likely to be trapped in local maxima as its variational counterpart. Let z denote a vector of the instances of all latent topic variables and w denote a vector of all the observed words of the corpus. The inference algorithm concentrates on the joint probability p(w, z) and applies a sampling approach to instantiate the latent topic variable for each word. Gibbs sampling is a technique to generate samples from a complex posterior distribution p(z∣w) by iteratively sampling and updating each latent variable zi according to the conditional distribution p(zi∣w, z −i) where z −i denotes the current instantiation of all the latent topic variables except zi and w denotes the vector of all observed words of the corpus. The Gibbs sampling algorithm is as follows: 1) the latent variables z are first randomly initialized; 2) each element zi of z is iteratively sampled and updated; 3) repeat the last step until the Markov chain converges to the target posterior distribution p(z ∣ w) (“burn in”); (4) samples of z can be collected from the Markov chain. The conditional distribution p(zi w, z −i) is defined as follows:
| (3) |
In Equation 3, denotes the count of the words in the corpus that are indexed by wi and assigned to the topic j, excluding the word wi; is the count of all words assigned to the topic j, excluding the word wi; is the count of words assigned to the topic j in document di which contains zi; stands for the count of all the words in the document di; and α, β are parameters defined by modeler, and, V and T were defined previously. The inference of topic labeling for each word and the word composition for each topic can be efficiently achieved by repeatedly invoking Equation 3 and updating the contents of two tables: a D×T table C DT (for nj(d)) and a T×V table C WT (for nj(w)). These two tables contain sufficient statistics for estimating parameters, θ and φ, of the model. Therefore the computational complexity of the inference algorithm is proportional to O(W ′ T′ I), where W is the total number of words in the corpus, T is the number of topics of the LDA model, and I is number of sampling sweeps during the inference. After the Markov chain converges to the posterior distribution, the rows of the table C DT can be used as a vector representing the semantic content of the documents in the topic space.
Support Vector Machine (SVM)
SVM is a well-studied, kernel-based classification algorithm that searches for a linear decision surface with the largest margin between positive and negative training cases. It is arguably one of the best benchmark classifiers for text categorization. 9,11 The purpose of our experiments was to assess the effects of semantic-preserving dimension reduction and data augmentation on the classification performance of SVM. We used a publicly available implementation, SVMlight version 6.01. 11 Following the benchmark experiments by Lewis et al., 9 we used the linear kernel and only adjusted the cost-factor parameter, “−j,” which specifies how the cost of an error on a positive training case outweighs that of a negative case, although an earlier version of SVMlight was used in the study by Lewis et al.
Training Data Augmentation with Semi-supervised Learning
To enhance the classification performance of the SVM in the scenario of sparse training cases, as in some subtasks of TREC 2005, data augmentation experiments were performed using the semi-supervised learning algorithm described by Zhu et al. 22 This algorithm is based on the theory of Gaussian random fields, which allows the labels of the training cases to be propagated to the unlabeled data probabilistically. To perform label propagation, a weighted undirected graph was constructed. The vertices of the graph are the text documents of the corpus, and the weight of an edge reflects the similarity of the two documents connected by the edge. Given two text documents represented as two vectors in the vector space form, vi and vj, the weight of the edge connecting them was determined as follows:
| (4) |
where wij is the weight of edge connecting the vertices i and j; vi′vj is the dot product of the vectors vi and vj. The term vi stands for the norm of the vector. Note that the last term in Equation 4 is the cosine of the angle between the two vectors vi and vj, which is commonly used to determine the similarity of two text documents within the vector space. In addition to the graph, the learning algorithm also requires labeled positive and negative cases. We manually selected a set of 500 training cases not belonging to any of the four categories and labeled them as negative cases. These negative cases were combined with the provided positive training cases for each category as the labeled data to perform label propagation on the rest of the training data for each subtask, respectively. The label propagation is achieved by probabilistically assigning labels to the unlabeled cases according to the harmonic function of the weighted graph, such that the free energy of the graph is minimized. The Matlab© implementation by Zhu et al. 22 was used for label propagation. For each category, 100 cases that had the highest probability of being labeled as positive for the category (pseudo-positive cases) were selected and used to augment the training data set.
Information Retrieval Evaluation
Common evaluation metrics 8 for information retrieval were used to generate the experiment results, including recall, precision, F-score, normalized utility and the receiver operating curve (ROC). 23 Let TP be the number of true positives; FP be the number of false positives, these metrics are defined as follows:
Recall (R):
| (5) |
Precision (P):
| (6) |
F-score (F):
| (7) |
Normalized utility (U norm):
| (8) |
where UTP and UFP are the task-specific utility scores for true positives (usually a positive score) and false positives (usually a negative score). The utility scores are task specific and determined by the organizer, 13 and the utility scores for the relevant documents (Ur) for A, E, T, and G categories are 16.27, 71.06, 11.63 and 161.14 respectively. These utility scores were determined by dividing the total number of negative documents in the corpus with the positive number for each subtask respectively. The score can be intuitively thought of as the number of non-relevant documents one has to read in order to find a relevant document if one manually searches for the positive cases.
Results
Classification with Vocabulary Representation
As a baseline experiment, we trained the SVMlight program on the TREC 2005 corpus with each document represented as a vector in the original vocabulary vector space, which we referred to as VocRep. To deal with the situations of unbalanced positive and negative training cases, the SVMlight implementation allows users to specify a cost-factor parameter to indicate how training errors on positive cases outweigh those of the negative cases. 24 The cost-factor is specified through the option parameter “−j” of the program. When training the SVMlight classifier on the TREC 2005 VocRep corpus, we tested different cost weights, with option “−j” set to 1, 1.5, 2, 3, 4, 6, 8, and 10. The classification performance was evaluated by the built-in leave-one-out (LOO) test of the SVMlight implementation.
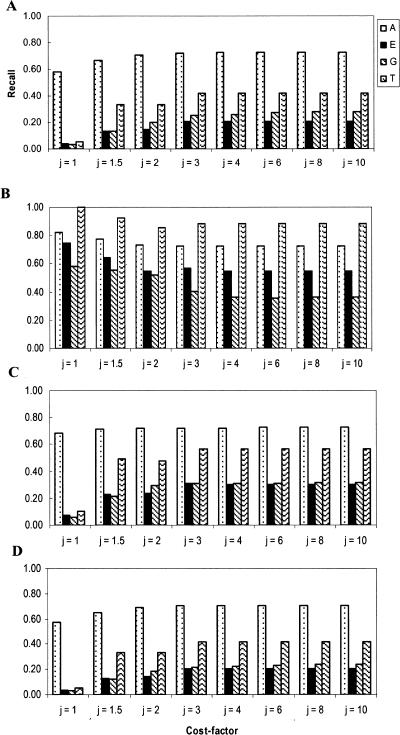
▶ shows the results of the recall (Panel A), precision (Panel B), F scores (Panel C), and normalized utility (Panel D) on the VocRep data. From this figure, it can be seen that increasing cost-factor for the positive training cases enhances the classification performance of SVM. However, the performance of the SVMlight reached plateaus when the “−j” parameter was set higher than 3. Overall, the classification performance of SVM on this data set is not very satisfactory, especially on the categories E, G, and T task. These results are not surprising as they agree with the observations from the benchmark studies by Lewis et al., 9 in which the F values for the low frequency categories in the Reuter's corpus are equivalent to our results. These results indicate that, although the SVMlight implementation is capable of alleviating the problem of skewed training data with an adjustable cost-factor parameter, the insufficient training data remain an obstacle to effective text classification. Such a problem manifests itself further when the training cases are represented as sparse vectors in a high dimension space. In such a scenario, the data points represented by these vectors tend to share few common elements and thus are far apart in the high-dimensional space. The decision surface learned under such a condition tends to over-fit the data. To determine if the SVM algorithm was over-fitting on our training data, we further evaluated the training error of the program by invoking the trained classifiers on the training data. The training errors for all these classifiers were zeros, while the LOO estimation of testing errors were around 20% ∼ 80%. Thus, these experiments indicate that, although utilizing maximal margin to protect itself from over-fitting on the high dimensional data, the SVM algorithm suffered from over-fitting in this scenario.
Figure 3.
Baseline performance of SVM. Different with different cost-factor settings were used for training SVM. Panel A: Recall; Panel B: Precision; Panel C: F scores; and Panel D: normalized utility.
Semantic-enriched Representation of Documents
Possible reasons for the relatively poor baseline classification performance are the sparseness in both the non-zero elements of the vector representation and the number of data points within a high dimensional space. The first problem potentially can be addressed by reducing the dimensionality of the data while maintaining the semantic structure of the original vector space. Recently developed probabilistic semantic analysis techniques 16,18,19 may serve as useful tools to achieve this goal. The LDA model is a full probabilistic generative model that can capture the human understandable semantic topics, which are represented as distributions over the vocabulary. By representing a document in the topic space instead of in the vocabulary space, the LDA model effectively reduces the dimension of the texts while maintaining the semantic content of the document.
We have implemented a Gibbs sampling inference algorithm for LDA in C language. 18 In a previous study, we found that the LDA models with 300 ∼ 400 topics simulated a MEDLINE© titles and abstracts corpus well. The size of that corpus is similar to the size of the TREC 2005 data set, 25 thus a LDA model with 400 topics was trained on the TREC 2005 data in the form of VocRep. Inspecting the topics extracted from the corpus, we found that most of topics were coherent biological topics while some were not specifically related to biology. ▶ shows examples of a few semantic topics and their top composition words in stemmed forms.
Table 1.
Table 1. Semantic Topics and Their High Probability Words
| Topic | Topic words |
|---|---|
| 3 | arteri smooth vascular aortic aorta atherosclerosi muscle atherosclerot lesion smc |
| 25 | endocytosi recycle transferring clathrin endocyt intern pit uptake vesicl traffick |
| 51 | raft caveolin caveola microdomain cholesterol gpi lipid cav drm anchor |
| 102 | snap snare syntaxin vamp nsf synaptobrevin complexing exocytose munc18 |
| 192 | tumor cancer carcinoma malign metastasi metastat breast tumorigen melanoma colon |
Given the high probability words of the topics shown in ▶, it is not difficult for a biologist to discern the content of a topic. This characteristic distinguishes LDA from another well-studied semantic analysis technique, latent semantic indexing (LSI). LSI applies singular value decomposition (SVD) techniques to extract the orthogonal major directions of the vector space of the term-document matrix. However, the requirement of orthogonality prevents LSI from identifying the major semantic topics that are readily understandable by humans. Free from such a restrictive requirement, the LDA model is capable of identifying the topics that agree with human representation of topics, thus maintaining the semantic structure of the original texts.
The LDA model infers the latent topic for each word in the corpus. Associating every word with a topic enables us to represent a document within a lower-dimensional vector space spanned by semantic topics, in our case the dimension of topic space T = 400, instead of the original high-dimensional vocabulary space. In this semantic space, each document can be represented by a T-dimensional (in our case 400 topics) vector, and each element, t, of the vector contains the number of words within the document that belong to the topic t. Thus, this transformation of text document serves to project the document from the vocabulary space onto the topic space. This projection addresses the problem of sparseness of the elements of original vocabulary vector space. We refer to the documents represented with the semantic-enriched features as SemRep and examined the effect of such transformation on the text categorization performance of the SVMlight.
SVM Classification on Documents Represented with Semantic-enriched Features
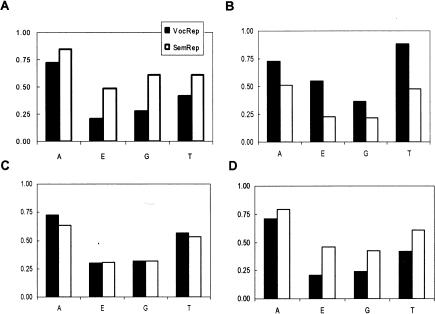
The text documents of TREC 2005 represented in SemRep form were used to train the SVMlight program, and the performance was evaluated with the LOO estimation of the test errors as in the baseline experiments. To avoid differences in the performance introduced by the different parameter settings, we fixed the cost-factor parameter “−j” to 10, a value that gave rise to the saturated performance on the baseline experiments. ▶ shows the comparison of the classification performance of SVMlight for these experiments.
Figure 4.
Semantic representation and classification. Text documents are represented in vocabulary (VocRep) and semantic space (SemRep). Panels A, B, C and D correspond to the recall, precision, F value and utility score comparisons of VocRep and SemRep.
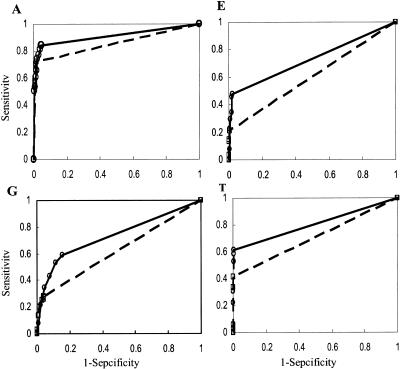
The results indicate that representing the text in the form of SemRep significantly enhances the recall and utility of the SVMlight on the SemRep data set compared to the results on the VocRep data. The increase in recall is most noticeable (more than one fold) for categories E and G, while it is also clear for categories A and T. Due to the well-known trade off between recall and precision, the precision for all categories declined to different degrees. However, the metrics that measure the overall performance of the classifier, the utility score and F values, increased for both E and G categories, indicating that SemRep produced balanced improvement for these subtasks. On the other hand, the utility scores for the subtasks A and T are significantly increased while the F values declined slightly. An F value is a balanced metric considering both recall (sensitivity) and precision (positive predictive value). Since the numbers of positive cases for these classification tasks are small, precision values are sensitive to a relatively small change of false positive number which may dominate the true positive number, which in turn influences the F values. This produces a kind of “boundary-effect” in that small changes in the TP and FP counts significantly change the values of precision and F values. To address this issue, the ROC curve, another balanced metric considering both recall and false positive rate, was used to evaluate the performance of SVMlight based on VocRep and SemRep for all four subtasks. The curves are shown in ▶ and the values of the areas under the curves (AUC) are shown in ▶. The results show that, for every subtask, representing the documents in SemRep clearly increases the AUCs, i.e., the discriminative power of SVMlight, in comparison to those for VocRep. In summary, representing the text document in the form of SemRep can significantly increase the recall (sensitivity), utility scores and the AUC of the SVMlight, thus the SemRep enhances the discriminative power of the SVM algorithm.
Figure 5.
The ROC curve analysis for the TREC text categorization. The panels correspond to the A, E, G, and T subtasks respectively. Within each panel, the ROC curve for VocRep is shown as dashed line and open boxes, while that for the SemRep is shown as solid line and open circles. Each symbol represents the sensitivity and false positive rate (1 - specificity) of the trained SVMlight classifier with a given cost-factor (-j) value.
Table 2.
Table 2. Comparison of the ROC Area under Curve for VocRep vs SemRep
| VocRep | SemRep | Changes (%) | |
|---|---|---|---|
| A | 0.856 | 0.909 | 6.19% |
| E | 0.604 | 0.730 | 20.86% |
| G | 0.622 | 0.733 | 17.85% |
| T | 0.708 | 0.804 | 13.56% |
Augmenting Training Data with Semi-supervised Learning
In addition to the sparseness of the vector element of the VocRep representation, another potential reason for the poor baseline performance may be due to a small number of training cases. A small number of training cases tends to lead a classifier to over-fitting because it is easy to find a decision rule that separates a small number of samples well. Such decision rules rarely generalize well on the new unseen data. An obvious remedy for such difficulty is to collect more training cases. However, manually annotating more training cases is an expensive process and does not scale well for the large data set in our case. To overcome the difficulty associated with scarce positive training data, we automatically augmented the training data by imputing more potentially positive cases. Such techniques are usually referred to as self-training, and have been applied in various machine learning applications. 26 The assumption underlying this approach is that there are potentially more positive training cases among a large amount of unlabeled data. We believe that this is a reasonable assumption in our case in that the positive cases in our corpus are labeled by the human annotators as they go through the corpus and many of the articles remain unlabeled. In the baseline experiments, these unlabeled articles were by default treated as negative cases for a given subtask. It is highly possible that there are more positive cases remaining unlabeled in the corpus. This condition not only subjects the classifier to scarce training data but also potentially causes confusion for classifiers.
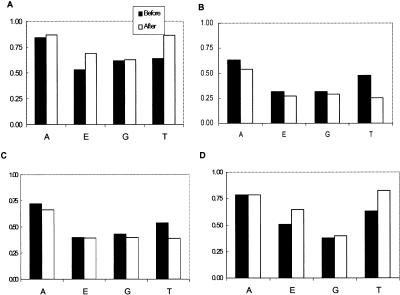
To address these problems, we applied semi-supervised learning, a learning process that combines both labeled and unlabeled data, 26 to identify more potentially positive cases so that these pseudo-positive cases could be used to augment the training set. The algorithm used for this study is based on the graphical model of Gaussian random fields and its harmonic functions, 22 and allows the information from the labeled data to propagate to the unlabeled data based on the manifold of combined data. As the output of the algorithm, the potential membership of unlabeled data is probabilistically determined. As the original data set only provided positive cases, we further manually labeled 500 randomly selected cases not belonging to any of the four subtasks as negative cases. These negative cases were combined with the provided positive cases for each subtask as the labeled data in the semi-supervised learning experiments. After applying the semi-supervised learning algorithm to the data, we extracted 100 cases that had the highest probability of belonging to one of the target categories. Then, the augmented training data, i.e., the combined set of true and pseudo positive cases represented in the SemRep form, were used to train the SVMlight program. The performance of the program on the augmented data was evaluated by a ten-fold cross-validation approach, in which the recall, precision, F values, utility scores and AUC were evaluated only based on the program's calls on the true positive case and discarded the calls on the pseudo-positive cases. Due to the relatively small number of true positive cases for some subtasks, we pooled the results from all ten cross-validation runs to calculate the metrics, equivalent to the commonly used macro-average metrics in IR. 10 The performance of the SVMlight trained with the augmented training data is shown in ▶.
Figure 6.
Effect of data augmentation. The panels A, B, C and D correspond to recall, precision, utility score and F value.
From ▶, it can be seen that the recall and utility scores for all subtasks are improved after training data augmentation. In particular, subtasks E and T, which have fewer positive training cases, benefited most significantly from this approach, with an increase in recall by 30% and 34%, respectively. The effect of data augmentation on subtasks A and G, which have more than a few hundreds positive training cases, is not as significant as in the previous two subtasks. On the other hand, the precision declined slightly for all subtasks. Again, we believe that the reason for such a decline is due to the sensitivity of the metric to the small number of positive training cases, especially in the subtasks E and T. Indeed, the results from the ROC curve analysis showed that the AUC values increased after data augmentation for all categories except the G category, as shown in ▶, which clearly demonstrated the benefit of data augmentation for the subtasks that are highly specific and have very few positive training cases, namely the subtasks E and T.
Table 3.
Table 3. The Effect of Data Augmentation on Classification Performance Evaluated by AUC
| SemRep ∗ | DataAug | Changes (%) | |
|---|---|---|---|
| A | 0.903 | 0.908 | 0.55% |
| E | 0.7533 | 0.826 | 9.65% |
| G | 0.707 | 0.677 | −4.24% |
| T | 0.8173 | 0.915 | 11.95% |
∗ AUCs for SemRep in this table are calculated only using points that match the parameter settings used in the data augmentation experiments.
Discussion
Contemporary supervised learning algorithms, e.g., SVM, usually perform well when sufficient training data are available. However, in the real world, it is not uncommon for a classifier to face a scenario of data scarcity and high-dimensionality, as in our case. In such cases, even a state of the art classifier like SVM may perform poorly as demonstrated in our baseline experiments and other benchmark studies. 9,10 A combination of high dimensionality and sparse training data almost inevitably leads to over-fitting. Such difficulty does not only occur in text categorization tasks but is a general problem for many classification tasks.
In this research, we address both aspects of the problem by reducing the dimension of the data with the LDA model and augmenting the training cases with semi-supervised learning techniques. Our experiment results indicate that the two approaches work well in the domain of text categorization using the TREC 2005 data as the test bed.
Dimension Reduction with LDA
The LDA model is an unsupervised learning algorithm that extracts the semantic topics from a text collection. By labeling each word with a topic, it allows representation of a document in the form of its semantic topic content rather than the words of vocabulary, thus achieving a significant reduction in the dimensionality of text representation, usually from tens of thousands to hundreds. Our experimental results indicate such low-dimensional semantic representation resulted in significant improvements in the classification performance of the SVMlight over the original high-dimensional representation. The enhancements are demonstrated as the cross-board increases in recall, utility and AUC, together with increased precision and F values in two of four subtasks. The benefit of SemRep may be attributed to the following reasons: (1) the semantic representation strengthens the signal contained in the original text, therefore increases the discriminative power of the representation; and (2) the decision rules learned within the low dimension space generalize better than those learned in the high dimension space. We believe that the former is likely to play a major role in improving the performance because SVM aims at finding a decision surface with maximal margins and is relatively insensitive to the dimensionality of the data in comparison to other classifiers. In fact, SVM can perform well by implicitly projecting the data into higher dimensional space using the kernel tricks.
Human annotators categorize the texts according to their semantic context rather than individual words. The LDA semantic analysis extracts the semantic concepts that are understandable to humans, thus such representation aligns well with the human categorization of concepts and texts. Therefore, the LDA model captures and retains the necessary information that is utilized for text categorization by human annotators, thus retaining the discriminative power of the original representation. Furthermore, the LDA model is capable of resolving the ambiguities, e.g., polysemy and synonymy, often associated with natural language texts. When inferring the semantic topics, it naturally groups the synonyms into topics while resolving the ambiguity caused by a polysemous word based on the semantic content of the document. Thus, projecting a document from the vocabulary space onto the semantic space serves to pool the signals of the words from the same semantic topic and strengthen the signals along the semantic directions.
There are other techniques that can be applied to reduce the dimensionality of text documents. Notably, LSI 27,28 and the recently developed generalized singular value decomposition (GSVD) 29 can be used to reduce the dimensionality of text collection. Recently, Kim et al. 29 reported the effects of dimension reduction using both LSI and GSVD on text categorization performance of SVM and other classifiers. The data set used in their study was a collection of MEDLINE data, in which each category had 500 positive training cases. Similar to LSI, the GSVD identifies the orthogonal low rank representation of the term-document matrix. In addition, if the training data are labeled, i.e., data form clusters, GSVD also strives to maintain the cluster structure of the documents. Kim et al. reported that dimension reduction with LSI worsened the classification performance of SVM in some cases, while the results by GSVD retained the classification performance, in terms of classification accuracy. However, since both LSI and GSVD are SVD-based, the requirement of orthogonality by SVD prevents these methods from identifying the human understandable semantic topics. The methods recover major directions of the term-document matrix, and such directions less likely align well with criteria used by human annotators for text categorizations in comparison to the LDA model, thus are less likely to produce similar improvements in the performance of the classifier. Furthermore, the SVD based methods are computationally more expensive compared to LDA. A SVD analysis on our corpus would require a supercomputer level computation facility that is currently unavailable to us, making it difficult for us to compare the effectiveness of dimension reduction by SVD-based approaches with LDA.
It is also noteworthy that the mechanism used by LDA and LSI to achieve dimension reduction is different from that used by another family of dimension reduction methods, namely feature selection. 30 In the latter approach, various criteria, e.g., information gain or mutual information, are used to identify informative features, and the features deemed less informative are discarded in order to reduce the dimensionality of the data. In the case of text categorization, this amounts to retaining the words from the vocabulary that are informative with respect to specific categorization tasks and discard the rest. The differences between the two approaches are three: (1) In the feature selection approach, representation of a text document remains at word-level, whereas it is transcended to topic or semantic level by LDA or LSI. (2) Feature selection discards features, i.e., the words of the vocabulary. Thus, many of original words of a document will be discarded, together with the potentially useful semantic information associated with these words. In constrast, LDA and LSI retain most of the original words in a document and pool the semantic information from these words. (3) Dimension reduction by feature selection is task specific, while that by semantic analysis is not. In the latter case, if the semantic topics align well with human perception, the semantic-enriched representation can be used for various classification tasks without re-run data processing. This characteristic was clearly demonstrated by our results where the same SemRep data set was used for all categorization subtasks and resulted in the improved performance for all the subtasks. If we were to apply a feature selection approach, we would need to select features for each subtask. It will be interesting to compare the categorization performance from these two different dimension reduction approaches in future studies.
Augmenting Training Data with Semi-supervised Learning
Another approach we have adopted to improve the performance of SVM in the sparse training data scenario is to augment the training data with semi-supervised learning. A classifier trained on a larger number of training cases is likely to generalize better than one trained with sparse data. Although the labeled data may be scarce due to lack of manpower to annotate the training data, usually ample unlabeled data are available. Recently, the techniques of tapping into the unlabeled data to enhance learning have drawn much attention in the machine learning community, see Zhu 26 for a literature review. The semi-supervised learning approach applied in the current study is based on graph theory. In this model, texts are represented as the vertices of an undirected graph and the weighted edges of the graph reflect the similarity between the vertices. The algorithm can be thought of as propagating the labels from the labeled vertices to the neighbors by random walk, and the probability distribution for the final category of an unlabeled data point is determined as the weighted average of information propagated to it from neighbors. The advantage of this approach is that the labeling is probabilistic, allowing incorporating uncertainty into the label propagation process, and labels can be sorted according to labeling confidence (probability). Our results show that the data augmentation is more effective in a subtask with very few positive training cases and becomes less effective as the number of the original training cases increases. These observations are intuitive and sensible. Furthermore, the usefulness of this method for sparse positive training cases makes the approach more valuable in the domain of applied text mining in biomedical informatics, e.g., automatic protein annotation with GO terms, where the task is commonly cast as text categorization and positive training cases are relatively sparse. That data augmentation resulted in a deteriorated performance in the G subtask is also noteworthy. This is due to the nature of the task in that the goal of the subtask G is very non-specific, i.e., finding the documents that warrant a GO annotation, which may include documents with a wide range of contents. Unlike in very specific tasks, such as subtask T, where the semi-supervised learning is more likely to find additional cases that are true positive, it is difficult for the method to identify the cases that are representative for the variety of documents in category G. Therefore, imputing pseudo-positive data may mislead the classifier, and erode the specificity and AUC. These experiments indicate that many questions remain to be further studied, such as in what scenarios one should apply the data augmentation technique, how many pseudo-positive cases to add to the training data, and what threshold to use to include a potentially positive case?
Limitations of the Present Study and Future Directions
It might be argued that the results of the current study, by some metrics, i.e., precision and in some cases F value, do not show improvements. While this can be explained as commonly observed recall-precision tradeoff, it can also result from drastic dimension reduction causing loss of information. To alleviate this problem, one obvious direction is to further improve or extend the LDA model in order to extract finer-grained semantic topics to capture more semantic information from data.
We have also applied various combinations of the study methods to the official test set of TREC 2005. 31 Therefore, the results reflect outcomes of both “good” and “bad” approaches for the evaluation. We note that those relatively better runs were all produced by combining the SemRep and data augmentation, which again provided evidence that our approach improves performance of SVMlight over its baseline results. While noting that even the best TREC genomic track results are modest, our results measured by normalized utility score, the official metric used in TREC genomics to measure system performance, are also modest. We believe the unsatisfactory recalls are due to the fact that the SVM seeks to reduce the overall risk of misclassification. This pursuit of minimum risk translates into striving to make correct calls on the class with the largest number of cases, which happens to be the negative cases in the TREC evaluation, while sacrificing the calls on the class with relatively fewer cases, which is the positive class (of greatest overall interest). Thus, the recall using the SVMlight approach on this data set was somewhat capped, and the non-probabilistic output of the SVM prevented the authors from resorting to decision theory to find an “optimal” threshold to achieve the highest possible utility score. Note that utility scores for relevant and irrelevant documents can be used to bias towards either recall or precision, while the F value is equivalent to an unbiased utility score. In this respect, our test results were not suboptimal. 32 Most importantly, the relatively unsatisfactory recall and utility scores on the TREC test results do not undermine the fact that SemRep improves the performance of SVMlight over its baseline performance—thus the value of representing data in the semantic-enriched features. We believe that the improved feature representation by SemRep can be combined with other probabilistic learning algorithms, such as Bayesian logistic regression 33 or the relevance vector machine (RVM), 34 and this will enable researchers to apply decision theory to maximize the utility.
Conclusion
In summary, we have studied two approaches for enhancing text categorization under the scenario of high dimensionality and scarce training data: (1) semantic-preserving dimension reduction with the LDA model; and (2) training data augmentation with semi-supervised learning. We find that these approaches enhance text categorization performance of the SVM algorithm, especially in terms of the recall, utility scores and AUC. Although the methods were primarily tested on text categorization tasks, the approaches embody principles generally applicable to other classification tasks.
Footnotes
Xinghua Lu is partially supported by the NIH grants 5P20 RR016434-04 and T15 LM07438-02. ChengXiang Zhai is supported by NSF grants IIS-0347933 and IIS-0428472. Atulya Velivelli is partially supported by the NSF Grant CCF 04-26627. The authors thank Dr. John Lafferty for insightful discussions, Dr. Thorsten Joachims for making SVMlight available, and the organizers of the TREC genomics track for preparing the data set. The authors also thank the anonymous reviewers for their constructive critiques and suggestions.
References
- 1.Bourne P. Will a Biological Database Be Different from a Biological Journal? Plos Computational Biology 2005;1(3):179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hersh WR, Bhuptiraju RT, Ross L, Johnson P, Cohen AM, Kreamer DF. TREC 2004 genomics track overview. 2004. Paper presented at: Text Retrieval Conference (TREC) 2004.
- 3.Hirschman L, Yeh A, Blaschke C, Valencia A. Overview of BioCreAtIvEcritical assessment of information extraction for biology. BMC Bioinformatics 2005;6(Suppl 1):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium Nat Genet 2000;25(1):25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hersh W, Bhupatiraju R. TREC genomics track overview. 2003. Paper presented at: Twelfth Text Retrieval Conference - TREC 2003.
- 6.Yeh AS, Hirschman L, Morgan AA. Evaluation of text data mining for database curationlessons learned from the KDD Challenge Cup. Bioinformatics 2003;19(Suppl 1):i331-i339. [DOI] [PubMed] [Google Scholar]
- 7.Text REtrieval Conference (TREC), National Institute of Standards and Technology. Available at: http://trec.nist.gov/. Accessed November 2005.
- 8. Modern Information Retrieval. Pearson Education Limited; Essex, England and ACM Press; New York; 1999.
- 9.Lewis DD, Yang Y, Rose TG, Li F. RCV1A new benchmark collection for text categorization research. J Mach Learn Res 2004;5:361-397. [Google Scholar]
- 10.Yang Y. An evaluation of statistical approaches to text categorization Inform Retriev 1999;1:69-90. [Google Scholar]
- 11.Joachims T. Text Categorization with Support Vector Machines. Learning with Many Relevant Features. 1998. Paper presented at: Proceedings of the European Conference on Machine Learning.
- 12.Aphinyanaphongs Y, Tsamardinos I, Statnikov A, Hardin D, Aliferis CF. Text categorization models for high-quality article retrieval in internal medicine J Am Med Inform Assoc 2005;12(2):207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.TREC Genomics Track Protocol, National Institute of Standards and Technology. Available at: http://ir.ohsu.edu/genomics/2005protocol.html. Accessed November 2005..
- 14.Eppig JT, Bult CJ, Kadin JA, et al. The Mouse Genome Database (MGD)from genes to mice—a community resource for mouse biology. Nucleic Acids Res 2005;33(Database issue):D471-D475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porter MF. An algorithm for suffix stripping Program 1980;14(3):130-137. [Google Scholar]
- 16.Blei DM, Ng AY, Jordan MI. Latent Dirichlet Allocation J Mach Learn Res 2003;3:993-1022. [Google Scholar]
- 17.Hofmann T. Probabilistic Latent Semantic Indexing. 1999. Paper presented at: the 22nd International Conference on Research and Development in Information Retrieval (SIGIR'99).
- 18.Griffiths TL, Steyvers M. Finding scientific topics Proc Natl Acad Sci U S A 2004;101(Suppl 1):5228-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng B, McLean Jr. DC, Lu X. Identifying biological concepts from a protein-related corpus with a probabilistic topic model BMC Bioinformatics 2006;7:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buntine W. Operations for learning with graphical models J Artif Intell Res 1994;3:993. [Google Scholar]
- 21.Andrieu C, Freitas Nd, Doucet A, Jordan MI. An Introduction to MCMC for Machine Learning Mach Learn 2003;50(1–2):5-43. [Google Scholar]
- 22.Zhu X, Ghahramani Z, Lafferty J. Semi-Supervised Learning Using Gaussian Fields and Harmonic Functions. 2003. Paper presented at: International Conference on Machine Learning.
- 23.Bradley AP. The use of the area under the ROC curve in the evaluation of machine learning algorithms Patt Recognit 1997;30(7):1145-1159. [Google Scholar]
- 24.Morik K, Brockhausen P, Joachims T. Combining statistical learning with a knowledge-based approach—A case study in intensive care monitoring. 1999. Paper presented at: Proceedings of the 16th International Conference on Machine Learning.
- 25.Zheng B, McLean Jr. DC, Lu X. Identifying biological concepts from a protein-related corpus with a probabilistic topic model BMC Bioinformatics. 2006;7:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu X. Semi-supervised learning literature survey. Uiversity of Wisconsin; 2005. Computer Science Technical Report 1530.
- 27.Deerwester S, Dumais ST, Landauer TK, Furnas GW, Harshman RA. Indexing by latent semantic analysis J Am Soc Inf Sci 1990;41:391-407. [Google Scholar]
- 28.Berry MW, Dumais ST, O'Brien GW. Using linear algebra for intelliegent information retrieval SIAM Review 1995;37:573. [Google Scholar]
- 29.Kim H, Howland P, Park H. Dimension reduction in text classification with support vector machine J Mach Learn Res 2005;6:37-53. [Google Scholar]
- 30.Yang Y, Pedersen JO. A comparative study on feature selection in text categorization. 1997. Paper presented at: Proceedings of the Fourteenth International Conference on Machine Learning (ICML'97).
- 31.Zhai C, Lu X, Ling X, et al. UIUC/MUSC at TREC 2005 Genomics Track. 2005. Paper presented at: The Fourteenth Text REtrieval Conference (TREC 2005).
- 32.Hersh W, Cohen A, Yang J, Bhupatiraju RT, Roberts P, Hearst M. TREC 2005 Genomics Track Overview. Paper presented at: The 14th Text REtrieval Conference, 2005; Gaithersburgh..
- 33.Dayanik A, Genkin A, Kantor P, Lewis DD, Madigan D. DIMACS at the TREC 2005 Genomics Track. 2005. Paper presented at: The 14th Text Retrieval Conference.
- 34.Tipping ME. Sparse Bayesian learning and the relevance vector machine J Mach Learn Res 2001;1:211. [Google Scholar]