Abstract
Objective
Natural language processing (NLP) approaches have been explored to manage and mine information recorded in biological literature. A critical step for biological literature mining is biological named entity tagging (BNET) that identifies names mentioned in text and normalizes them with entries in biological databases. The aim of this study was to provide quantitative assessment of the complexity of BNET on protein entities through BioThesaurus, a thesaurus of gene/protein names for UniProt knowledgebase (UniProtKB) entries that was acquired using online resources.
Methods
We evaluated the complexity through several perspectives: ambiguity (i.e., the number of genes/proteins represented by one name), synonymy (i.e., the number of names associated with the same gene/protein), and coverage (i.e., the percentage of gene/protein names in text included in the thesaurus). We also normalized names in BioThesaurus and measures were obtained twice, once before normalization and once after.
Results
The current version of BioThesaurus has over 2.6 million names or 2.1 million normalized names covering more than 1.8 million UniProtKB entries. The average synonymy is 3.53 (2.86 after normalization), ambiguity is 2.31 before normalization and 2.32 after, while the coverage is 94.0% based on the BioCreAtive data set comprising MEDLINE abstracts containing genes/proteins.
Conclusion
The study indicated that names for genes/proteins are highly ambiguous and there are usually multiple names for the same gene or protein. It also demonstrated that most gene/protein names appearing in text can be found in BioThesaurus.
Introduction
Natural language processing (NLP) approaches have been explored to manage and mine information recorded in biological literature. 1–15 One critical step for the development of NLP applications in the biomedical domain is biological named entity tagging (BNET) that identifies names mentioned in text and normalizes them with entries in biological databases. 16,17 For example, in EDGAR, 2 which extracted relationships between cancer-related drugs and genes from the literature, terms for genes need to be identified before the extraction of the relationships. In various pathway construction NLP systems, 8–11 the identification of biological entity terms such as genes/proteins is the crucial component in order to construct molecular pathways from the text.
Methods for biological entity name identification can be categorized in two ways: one is to use a dictionary and a mapping method 12,18–20 and the other is to mark up terms in the text according to contextual cues or specific verbs. 21–23 The performance of systems relying on dictionaries depends on the coverage of the dictionary and a method to disambiguate gene/protein names from other biomedical terms or general English words. Another requirement is the ability to associate these names with corresponding entries in biomedical databases in order to be used by other automated systems for literature mining. 14,20 This task is called biological entity name normalization, which requires a knowledge base that associates names identified in text with entries in databases. We refer to the task that identifies entity terms in the text and associates them with entries in the database as biological named entity tagging (BNET). BNET is not a trivial task because of several characteristics associated with biological entity names, namely: synonymy (i.e., different terms refer to the same database entry), ambiguity (i.e., one term is associated with different entries), and novelty (i.e., entity terms or entities are not present in databases or knowledge bases). Both tasks (i.e., biological entity name identification and biological entity name normalization) were tackled by the researchers in the community (i.e., Task 1A and Task 1B) in the first BioCreAtive workshop (Critical Assessment of Information Extraction in Biology) (see http://www.pdg.cnb.uam.es/BioLINK/BioCreAtive.eval.html and http://www.mitre.org/public/BioCreAtive/). 14 The performance of the systems participating in the workshop was measured using precision, recall, and F-Measure. A system’s precision is defined as the ratio of the number of names (or entities) correctly identified (or normalized) to the total number of names (or entities) being identified (or normalized). A system’s recall is defined as the ratio of the number of names (or entities) correctly identified (or normalized) to the total number of names (or entities) in the test set. An F-Measure of a system is defined as the harmonic mean of its precision and recall. In the following, we provide a brief summary of the workshop related to the above two tasks.
For the biological entity name identification task (Task 1A), the data were prepared using a gene/protein name tagger, AbGene, 22 and manually curated by domain experts. 17,24 There were 15 teams in Task 1A and a number of teams achieved F-Measure of 80%. Most systems in Task 1A fell into the following two machine learning approaches: Statistical modeling 25–27 and Support vector machine. 27–29 Besides machine learning approaches, a rule-based system, Text Detective, was using a combination of handcrafted rules with lexical knowledge sources to identify genes mentioned in the text. 30 The teams that achieved an F-Measure of 90% or more tended to use Statistical modeling together with post-processing methods, various features, and external knowledge sources. The data for the normalization task of BioCreAtive (Task 1B) 31 were prepared using a gene list associated with the full journal articles found in the model organism databases, i.e., SGD (yeast), 32 MGD (mouse), 33 and Flybase (fly). 34 As described by Hirschman et al., 14 the normalization task can be divided into several steps: i) identifying gene occurrences in the text; ii) associating gene occurrences to one or more unique gene identifiers; iii) selecting the correct identifier in case of ambiguity; and iv) assembling the final gene list for each abstract. Eight systems participated in the evaluation and a variety of approaches were adapted for the above steps. 19,30,35 Generally, identifying gene occurrences in text can be classified into two groups: i) matching against the lexical resource 36,37 ; and ii) using the results obtained in Task 1A. The second step was simply a table look-up. The methods used to select unique identifiers fell into two categories: prune the lexical resource by removing ambiguous lexicon, or perform word sense disambiguation. Most systems used thresholds to select final lists and one system applied a maximum entropy classifier for removing bad matches. The precision and recall rates reported for Task 1B ranged from a maximum of 92% F-Measure for yeast to 79% for mouse. We participated in Task 1B and used an extensive list of synonyms obtained from online resources to perform biological named entity identification and normalization. The system achieved the best recall for mouse and yeast while the precision needs to be improved. Incorporating more synonyms into the system could improve recall while word sense disambiguation would be critical to improve the precision. 12
In this paper, we present a quantitative assessment of the complexity of dictionary-based protein named entity tagging where a protein named entity thesaurus, BioThesaurus, was assessed on synonymy, ambiguity and coverage. In the following, we first present background and related work. The assessment method is presented next. We then provide a discussion and conclude our work.
Background and Related Work
In this section, we first present a brief description of BioThesaurus. Background information about synonymy, ambiguity, and coverage is described next. We then provide background description of online resources used in the study. Examples of synonymy and ambiguity as well as related work are then given next.
BioThesaurus
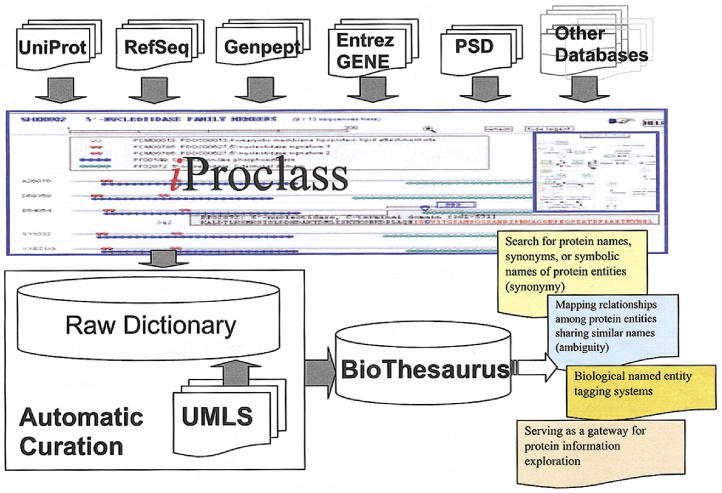
BioThesaurus 38 is a Web-based system designed to map a comprehensive collection of protein and gene names to protein entries in the UniProt Knowledgebase (UniProtKB), a knowledge base of protein sequence and function created by UniProt—the Universal Protein Resource. 39 UnitProtKB combined information contained in three databases: Swiss-Prot (a curated protein sequence database), TrEMBL (a computer-annotated supplement of Swiss-Prot, translated from nucleotide sequence database EMBL), and PIR-PSD (PIR-International Protein Sequence Database). Currently covering more than two million proteins, BioThesaurus consists of over 2.8 million names extracted from multiple molecular biological databases according to the database cross-references in iProClass, an integrated database that provides rich links with executive summaries to over 90 biological databases. 40 An overview of BioThesaurus construction is shown in ▶. The thesaurus was designed to provide comprehensive gene/protein names for all protein entries in UniProtKB. An extensive collection of names was extracted from many online resources according to database cross-references in iProClass. A parser was developed to automatically gather names from the underlying sources and parse individual names from annotation fields that contain multiple names separated by parentheses or other delimiters such as semicolons or commas. A raw thesaurus was then compiled, associating names with the corresponding UniProtKB entries. The raw thesaurus was further filtered to remove highly ambiguous and nonsensical names. The “name filter” was compiled based on frequency counts of names in UniProtKB entries and by curator judgment as “nonsensical.” Examples of filtered names include “novel protein,” “fragment,” and “hypothetical protein.” We also mapped names in the thesaurus to concepts in the UMLS (Unified Medical Language System), 41 a biomedical knowledge resource. Note that mapping names to UMLS concepts can be helpful for further curating the thesaurus. Some usages of semicolons and parentheses in annotation fields were not for separating synonyms but for comments, while our parser could not distinguish them and extract those comments as names. For example, the usages of parentheses in the description field for UniProtKB entry Q727V5, “precorrin-6Y C5,15-methyltransferase (decarboxylating)” and that for entry Q5JBL9, “TraA fimbrial protein precursor (pilin)” are different where the latter one is used to denote a synonym while the former one is not.
Figure 1.
The construction of BioThesaurus. Annotation fields from Genpept, PSD, RefSeq, Entrez GENE, Swiss-Prot and TrEMBL were extracted and associated with iProClass entries. Several other databases were also included including several model organism databases, HUGO, and ENZYME etc. Terms obtained from the annotation fields comprised the Raw Dictionary. An automatic curation process was performed using the UMLS. We also manually inspected high ambiguous entries in the raw dictionary and removed nonsensical terms. After curation, we obtained BioThesaurus, where terms were associated with entities from iProClass. BioThesaurus could be used for extensive information retrieval, investigating relationships among entities sharing the same name, biological named entity tagging, and serving as a gateway for protein information exploration.
Characteristics Associated with Biological Entity Names
There are several characteristics associated with biological entity names: synonymy (also referred to as alias), ambiguity (or referred to as homonymy) and coverage (or novelty), which are detailed in the following.
Synonymy—Different terms can refer to the same entity. The synonymous pairs can be classified into two types: derivable and non-derivable. A pair is considered derivable if one can be derived from the other algorithmically (i.e., can be unified through ignoring cases or punctuations, normalizing morphological difference, or ignoring word order). Another type of derivable is abbreviation-derivable. For example, the pair (“cAMP receptor protein,” “cyclic AMP receptor protein”) is derivable because “cAMP” is an abbreviation for “cyclic AMP.” In contrast, the synonymous pair (“cyclic AMP receptor protein,” “catabolite activator protein”) is non-derivable. Different methods have been used to recognize synonyms. One way to obtain synonyms is to use various online knowledge resources. For example, synonymous relations that are specified in the UMLS can be used to recognize synonym pairs (e.g., “cyclic AMP receptor protein” and “catabolite activator protein” were associated with the same UMLS Concept Unique Identifier, C0007364). For abbreviation-derivable pairs, methods such as mapping abbreviations to full names using contextual cues (e.g., parentheses) have been developed. Wren et al. provided a review of a list of abbreviation databases obtained by extracting full names for abbreviations from text using either patter-based methods or supervised machine learning methods that are available to the public. 42 Non-derivable synonymous pairs can be detected using contextual cues, such as “also called” or “also known as.” 43 For example, the synonymous pair “IL-8,” “neutrophil-activating peptide 1” can be detected in the sentence “IL-8 (also known as neutrophil-activating peptide 1) is recognized as a potent effector of neutrophil functions.”
Ambiguity—One term can be associated with different entities as well as other concepts. The ambiguity of biological entity terms can be classified into four different types:
• Systematic ambiguity—terms that represent concepts that are closely related. For example, gene products (e.g., mRNA or proteins) are usually represented by names that refer to genes (note that such ambiguity has been referred to as class ambiguity in the literature).44 Homologous genes are usually represented by the same name. For example, “CAP” refers to rat cystinyl aminopeptidase protein and also human cystinyl aminopeptidase protein.
• Entity-specific ambiguity—terms that represent multiple un-related proteins/genes. For example, the term “CAP” refers to biological entities, such as capsid protein, cystine aminopeptidase, catabolite gene-activator protein, cyclase-associated protein, and calcium activated protease.
• Cross-medicine ambiguity—terms that refer to clinical terms as well as protein/gene terms. For example, the term “CAP” also refers to the following clinical concepts: community acquired pneumonia, congenital alveolar proteinosis, cochlear action potential, carotid artery pressure, and compound action potentials.
• Cross-general ambiguity—symbols that are also general English words. For example, the following are common English words and also biological entity terms (“not,” “can,” “bad,” and “for”).
Several researchers have started to investigate the ambiguity issues. Hatzivassiloglou et al. considered systematic ambiguity (i.e., classifying names to three different classes: gene, protein, or mRNA) and applied machine learning techniques to disambiguate the semantic context of a term with precision up to 85%. 44 However, in the same paper, they also reported a pair-wise agreement between experts is around 77.5%. In several previous studies, 45–47 we demonstrated that classifiers could be constructed in an unsupervised way for disambiguation of frequently occurring ambiguous biomedical abbreviations with a precision over 95%. A similar method was also proposed by Gaudan et al. to resolve abbreviations. 48 Schijvenaar et al. 49 proposed a thesaurus-based disambiguation method for ambiguous human gene symbols. They claimed that an overall accuracy of the disambiguation algorithm was up to 92.7% on a test set automatically generated from MEDLINE. However, they did not mention the recall of the disambiguation algorithm. The work of Chang et al., GAPSCORE, handles the synonymy and ambiguity of terms based on a statistical model of gene names that quantifies their appearance, morphology and context. 50 The method requires a human-annotated corpus and when evaluating against the Yapex data set, 51 the method achieved an F-measure of 82.5% for partial matching and 57.6% for exact matching. In the BioCreAtive workshop, various methods have been proposed for disambiguation. For example, Hanisch et al. used a multi-stage process that included correlating abbreviations with their full names and also a filter for abstracts based on organism specificity. 19 Our system in the BioCreAtive task used features derived from online resources to create feature vectors used in word sense disambiguation. 37 Crim et al. used maximum entropy classifier to disambiguate. 36 Currently, no work has been reported that disambiguates the cross-general ambiguous biomedical terms.
Coverage—Novelty or coverage refer to entity names in text that are not present in databases or knowledge sources. There are two cases of novelty. One case concerns a new term for an existing biological entity, which may be due to the discovery of a new function or to a new derivable synonym or text variants of an existing term that could not be detected automatically. Such novelty is term novelty (or term coverage). The other case concerns a term for a new biological entity that we call conceptual coverage. Currently, there are very few studies on the coverage of names in databases of knowledge sources.
UniRef Databases
In order to assess systematic ambiguity caused by homologous entries, we used the UniProtKB Non-redundant Reference (UniRef) databases which combine similar sequences into a single record based on sequence similarities of UniProtKB entries. 39 Three UniRef data sets (UniRef100, UniRef90 and UniRef50) are available for download: UniRef100 combines identical sequences and sub-fragments into a single UniRef entry; and UniRef90 and UniRef50 are built by clustering UniRef100 sequences into clusters based on the CD-HIT algorithm such that each cluster is composed of sequences that have at least 90% or 50% sequence similarity, respectively, to the representative sequence. 52 Since homologous entries in UniProtKB can share sequence similarities ranging from over 90% for close homologs to well below 50% for remote homologs, utilizing UniRef databases, we could assess systematic ambiguity caused by homologous proteins.
Examples of Ambiguity and Synonymy in BioThesaurus
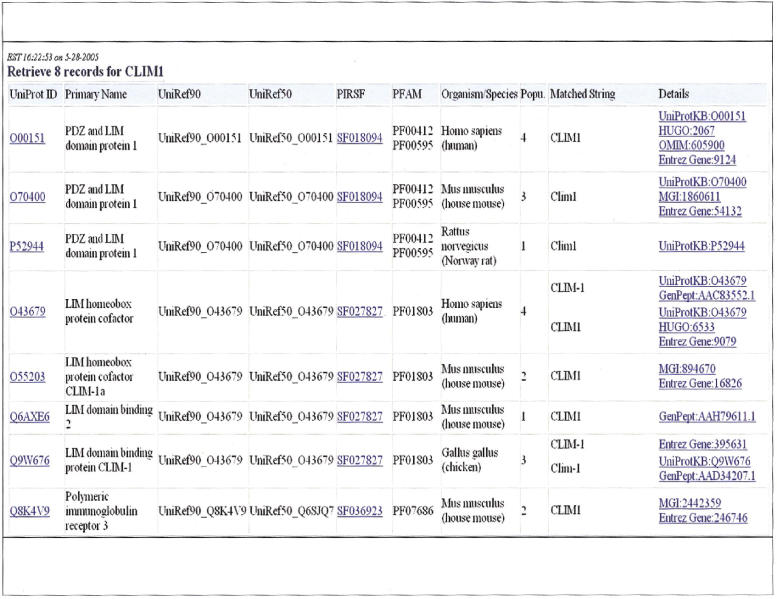
▶ shows 17 synonymous protein names from multiple data sources for the human PDZ and LIM domain protein 1 (UniProt entry O00151). The names and synonyms can be normalized based on case and morphological variations and the number of synonyms decreases to 11 after normalization. ▶ shows both entity-specific and systematic ambiguities of protein names. The eight UniProtKB entries associated with “CLIM1” demonstrate both systematic ambiguity resulting from protein homology and name overloading from gene symbols that represent different proteins. Indeed there are three heterogeneous groups of proteins as indicated by their different assignments in UniRef90/50 sequence clusters as well as in distinct families in PIRSF (PIR SuperFamily), a protein family classification system at PIR that classifies proteins based on global sequence similarities of full-length proteins to reflect their evolutionary relationships. 56 The “CLIM1” example represents entity-specific ambiguity among the three PIRSF families as well as systematic ambiguity within each PIRSF family. Examination of the source of protein name ambiguities revealed that “CLIM1” was derived from “human 36-Kda carboxyl terminal LIM domain protein (hCLIM1),” a cytoskeleton regulator. 53 Independently, “cofactors of LIM homeodomain proteins,” transcriptional activators associated with the LIM homeoproteins, were abbreviated as “CLIM,” and two forms of CLIMs were named as “CLIM1” and “CLIM2,” respectively. 54 Clearly, the ambiguity could arise from independent naming of different proteins. However, it is not clear how “CLIM1” was annotated as a synonym of “polymeric immunoglobulin receptor” found in MGD.
Figure 2.
Synonymous protein names from multiple data sources (UniProtKB: O00151). Names/synonyms of the protein entry are listed based on their rank of frequency (in parentheses) of unique sources (Source Attribute) from which the names are derived. Names with higher frequency may correlate with their more popular usage than those with lower frequency. Textual variants provide the name as appeared from the source of its origin.
Figure 3.
Protein name ambiguities using query of “CLIM1” from BioThesaurus. The query name “CLIM1” corresponds to eight UniProtKB entries (ambiguity of 8); each is displayed with corresponding UniRef clusters (90 and 50), as well as PIRSF families and Pfam domains. The UniRef cluster and PIRSF family information can be used to estimate the name ambiguity disregarding the homologous proteins, in this case, the ambiguity drops to 4 based on UniRef90 or 50, or to 3 based on PIRSF. In fact, the eight proteins belong to three functionally heterogeneous groups of proteins.
Related Work
In Chen et al., 55 gene information associated with 21 organisms was obtained and quantified, naming ambiguity with species, across species, with English words and with medical terms, was measured. The purpose of Chen’s work was to study gene name ambiguity associated with eukaryotic nomenclatures that were obtained from each specific organism database, where the assessment of ambiguity did not distinguish systematic ambiguity from other types of ambiguity.
Here, we assessed the complexity of dictionary-based biological named entity tagging through several aspects related to biological entity names. According to our knowledge, there is no related work that provides a detailed analysis of synonymy, ambiguity and coverage of a thesaurus for all protein records in UniProtKB with respect to biological named entity tagging even though most researchers are aware of them. 12
Methods
In the study, we first generated the gene/protein name thesaurus using the iProClass release 2.67. Noticing the ambiguous usages of parentheses and semicolons in annotation fields of online resources, we post-processed BioThesaurus to remove obvious non-protein names obtained from the online resources such as species names using the UMLS that is described in the following. We then evaluated BioThesaurus with respect to three characteristics: synonymy, ambiguity, and coverage.
Data Preparation
A version of BioThesaurus was constructed using the iProClass release 2.67 (May 15, 2005). As we have presented earlier in this paper, BioThesaurus associates terms derived from various online resources with protein records in UniProtKB. To prepare data for our analysis, BioThesaurus was computationally curated using the UMLS to remove terms that are obviously not genes/proteins such as cells, small molecules, or organisms (▶; also see Background section for rationale). To do this, we first identified the UMLS semantic categories that correspond to genes/proteins. For each UMLS semantic category, we counted the number of terms in the raw dictionary with seven or more letters that can be found in the UMLS using strict string matching while ignoring care differences. The reason for restricting to terms with seven or more letters in the process is that abbreviations especially with less than seven letters are used frequently for medical concepts as well as for genes/proteins; including them in the identification process will favor categories that contain many abbreviations (e.g., body part). Based on the acquired frequency information and the UMLS semantic category definition, we derived a list of UMLS semantic categories containing genes/proteins. We considered terms with a semantic category not in this list as ones representing other types of concepts instead of genes/proteins. After removing terms representing non-gene or non-protein concepts and those that are nonsensical, we acquired the final thesaurus for assessment.
Assessment
Gene/protein names in BioThesaurus were assessed according to the three major characteristics of biological entity names—synonymy, ambiguity, and coverage. The assessment was conducted under two circumstances: i) using names before or after normalization, and ii) including or removing systematic ambiguity. Name normalization is to convert names to lower cases, to remove punctuation marks, and to unify morphological variants. The normalization was done because most mapping methods used by biological named entity tagging systems can handle derivable synonyms, such as case variants (e.g., “arsC” vs. “ARSC”), punctuation variants (e.g., “IL12A” vs. “IL-12A”), and morphological variants (e.g., “superoxide dismutase” vs. “superoxide dismutases”).
Removing systematic ambiguity (the same name for homologous proteins across species) can help estimate the extent of name ambiguity for entities representing different functions (entity-specific) and concepts from different domains (cross-medicine) in the thesaurus. Systematic ambiguity can be removed based on a full-scale family classification system with curated families of homologous proteins. While the PIRSF family classification provides manually curated families of full-length proteins, the curated families of the system have not covered the complete UniProtKB (coverage ∼50%–75%). 56 Instead, we used the UniRef clusters as a way for removing systematic ambiguity. UniRef clusters provide full-scale automatic clustering of all UniProtKB entries based on sequence identity, and the clusters even at 50% sequence identity level (UniRef50) were found to well correspond to one PIRSF family, even to a subfamily within the PIRSF family. To remove systematic ambiguity, we grouped protein entries belonging to the same UniRef90 or UniRef50 clusters, where proteins shared at least 90% or 50% sequence identity, respectively. We manually inspected several UniRef50 clusters and found all proteins in the same cluster belong to the same PIRSF family.
The synonymy is a measure of the number of names per UniProtKB entry (i.e., entity). The average synonymy was measured by the total number of entity-name pairs (EID, NAME) in BioThesaurus divided by the total number of entities with one or more names, where EID is UniProtKB entry ID and Name is gene/protein name. We counted the number of entities in eight synonymy ranges: 1, 2, 3–4, 5–8, 9–16, 17–32, 33–64, and >64 (i.e., [2n -1, 2n+1] for n from 1 to 5). We also measured the synonymy considering systematic ambiguity by using UniRef90 and UniRef50 to group UniProtKB entries.
The ambiguity is a measure of the number of UniProtKB entries per name. The average ambiguity was measured by the total number of entity-name pairs (EID, NAME) divided by the total number of names. We counted the number of names in eight ambiguity ranges: 1, 2, 3–4, 5–8, 9–16, 17–32, 33–64, and >64. The ambiguity of BioThesaurus with biomedical concepts was assessed based on names that were mapped to the UMLS concepts with semantic categories that were not highly related to gene or proteins. The ambiguity with general English words was assessed by matching single-word names in BioThesaurus with a common English word list we assembled using frequent word lists generated by Waring (http://www1.harenet.ne.jp/∼waring/vocab/wordlists/vocfreq.html).
We evaluated the term coverage using the test set of Task 1B provided by the 2003 BioCreAtive Workshop for three model organisms (yeast, mouse, and fly). The following is a brief summary of the test set used in BioCreAtive for Task1B in detail. In the test set, genes that are described in the associated papers were identified and mapped to corresponding identifiers in the associated model organism databases by human experts with a total of 3,375 pairs of (GENE, PAPER). Among them, the number of pairs identified by human experts as GENE that could be inferred explicitly from the abstract of PAPER was 1,593. The remaining 1,782 pairs that were judged as either GENEs were inferred explicitly from full paper or were inferred implicitly from the paper. Note that systems participating in Task1B of the 2003 BioCreAtive Workshop were evaluated using the 1,593 pairs only. When assessing the coverage, we also eliminated 250 pairs from the 1,593 because GENEs were not mapped directly to UniProtKB protein entries, thus had no corresponding BioThesaurus records. Note that when considering the whole 3,375 pairs, a total of 905 pairs did not have corresponding BioThesaurus records. The final identifier list consisted of 1,343 identifiers. For each gene GENE, names associated with GENE were extracted from BioThesaurus and, if there was at least one occurrence of any of those names in the associated abstract, we considered GENE was covered by BioThesaurus. Note that when identifying the occurrences of those terms, we used the normalized form of the terms. We computed the percentage of identifiers for each model organism that were covered by BioThesaurus.
To map to the correct entity, biological named entity tagging systems need to resolve ambiguity of terms that occur in the text. We assessed the ambiguity using the following methods. For each pair (GENE, ABSTRACT), there might be multiple terms associated with GENE occurring in ABSTRACT, and they may have various ambiguities. Tagging systems need to at least resolve the ambiguity of the least ambiguous term (that is, the term with the least number of gene identifiers associated with it) in order to map to the right entity. We considered the ambiguity of the least ambiguous term as the ambiguity that BNET systems need to resolve. We measured the ambiguity that BNET systems need to resolve twice: once considering systematic ambiguity and once ignoring systematic ambiguity according to UniRef90.
Results
The version of BioThesaurus used here consisted of 1.8 million UniProtKB entries with a total of 3.2 million names.
A total of 34,598 names in BioThesaurus were mapped to concepts from 120 (out of 134) UMLS semantic categories when ignoring cases. Among them, we considered 18 categories highly related to genes/proteins based on the UMLS semantic category definitions. ▶ lists these 18 categories with the number of mapped names. The top seven UMLS semantic categories: Amino Acid, Peptide, or Protein (T116), Gene or Genome (T028), Enzyme (T126), Biological Active Substance (T123), Receptor (T192), Immunological Factor (T129), and Pharmacologic Substance (T121), each had over one thousand matched names. Note that some categories listed in ▶ are not concepts for genes/proteins per se but are highly related. For example, disease or syndrome (T047) and lab procedure (T059) are highly related to genes/proteins because disease terms are often used to name disease genes/proteins (e.g., disease name “Wolman disease” used to name the associated protein with UniProtKB identifier Q5T073) and lab procedures involving genes/proteins may be named after them (e.g., enzyme name Leucine aminopeptidase used to name the associated lab procedure with UMLS identifier C0202118). Several UMLS semantic categories had more than 100 matched names in BioThesaurus but were not deemed highly related to genes/proteins based on their definitions, such as Plant (T002), cell component (T026), Body Part, Organ or Organ Component (T023), and a few other categories (e.g., Quantitative Concept (T081) and Intellectual Product (T170)).
Table 1.
Table 1 UMLS Semantic Categories Highly Related to Genes/Proteins Concepts
| Semantic Category ID | Semantic Type Definition | # Matched Concepts |
|---|---|---|
| T116 | Amino acid, peptide, or protein | 24,471 |
| T028 | Gene or genome | 16,029 |
| T126 | Enzyme | 11,125 |
| T123 | Biologically active substance | 7,615 |
| T192 | Receptor | 2,345 |
| T129 | Immunologic factor | 2,025 |
| T121 | Pharmacologic substance | 1,585 |
| T109 | Organic chemical | 986 |
| T044 | Molecular function | 709 |
| T047 | Disease or syndrome | 422 |
| T125 | Hormone | 326 |
| T131 | Hazardous or poisonous substance | 323 |
| T124 | Neuroreactive substance or biogenic amine | 259 |
| T118 | Carbohydrate | 200 |
| T114 | Nucleic acid, nucleoside, or nucleotide | 174 |
| T119 | Lipid | 92 |
| T059 | Laboratory procedure | 77 |
| T045 | Genetic function | 57 |
As shown in ▶, the average synonymy of each BioThesaurus record is 3.53 (i.e., one protein entry is represented by an average of 3.53 names). After normalization, there were 2,106,222 names with an average synonymy 2.32. After grouping homologous entries and combining their names to remove systematic ambiguity according to UniRef90 and UniRef50, the number of entries is reduced to 1,015,207 and 580,956 respectively, and the synonymy was increased to 4.41 and 6.13 respectively before normalization and 3.61 and 4.90 respectively after.
Table 2.
Table 2 The synonymy assessment result of BioThesaurus, where Original denotes measures before removing systematic ambiguity, UniRef90 shows measures after grouping entities according to UniRef90, and UniRef50 for measures after grouping entities according to UniRef50. We measured the synonymy using eight ranges (1, 2, 3–4, 5–8, 9–16, 17–32, 33–64, and >64). The average synonymy as well as the total number of entities in the dictionary are also provided. The percentage in parentheses shows the percentage of entities with synonymy in the range to the total number of entities.
| # of Synonyms | Original |
UniRef90 |
UniRef50 |
|||
|---|---|---|---|---|---|---|
| Before Norm. | After Norm. | Before Norm. | After Norm. | Before Norm. | After Norm. | |
| 1 | 154,444 | 368,141 | 82,167 | 143,594 | 50,049 | 70,018 |
| (8.6%) | (20.4%) | (8.1%) | (14.1%) | (8.6%) | (12.1%) | |
| 2 | 523,523 | 887,875 | 270,976 | 432,730 | 141,410 | 216,890 |
| (29.1%) | (49.3%) | (26.7%) | (42.6%) | (24.3%) | (37.3%) | |
| 3–4 | 808,714 | 299,472 | 373,322 | 216,730 | 177,196 | 118,454 |
| (44.9%) | (16.6%) | (36.8%) | (21.3%) | (30.5%) | (20.4%) | |
| 5–8 | 222,083 | 175,789 | 182,712 | 146,388 | 106,232 | 92,761 |
| (12.3%) | (9.8%) | (18.0%) | (14.4%) | (18.3%) | (16.0%) | |
| 9–16 | 76,031 | 57,598 | 83,169 | 61,331 | 65,823 | 55,209 |
| (4.2%) | (3.2%) | (8.2%) | (6.0%) | (11.3%) | (9.5%) | |
| 17–32 | 14,761 | 11,034 | 20,234 | 13,046 | 28,828 | 20,784 |
| (8.2%) | (0.6%) | (2.0%) | (1.3%) | (5.0%) | (3.6%) | |
| 33–64 | 908 | 673 | 2,387 | 1,271 | 8,971 | 5,560 |
| (<0.1%) | (<0.1%) | (0.2%) | (0.1%) | (1.5%) | (1.0%) | |
| >64 | 41 | 23 | 240 | 117 | 2,447 | 1,280 |
| (<0.1%) | (<0.1%) | (<0.1%) | (<0.1%) | (0.4%) | (0.2%) | |
| Total entities | 1,800,505 | 1,015,207 | 580,956 | |||
| Average synonymy | 3.53 | 2.86 | 4.41 | 3.64 | 6.13 | 4.90 |
The ambiguity result of BioThesaurus shows that the majority of names (83.7%) were associated with a single UniProtKB entry, with an average ambiguity of 2.31 for the 2,665,968 names (▶). After normalization, the average ambiguity of the 2,106,222 names is slightly increased to 2.32. When systematic ambiguity was removed according to UniRef90 and UniRef50, the ambiguity dropped to 1.77 and 1.46 respectively before normalization and 1.79 and 1.49 respectively after.
Table 3.
Table 3 The ambiguity assessment result of BioThesaurus. Refer to ▶ for notations. The ambiguity was measured using eight ranges (1, 2, 3–4, 5–8, 9–16, 17–32, 33–64, and >64). The total number of terms and the average ambiguity in the dictionary are also provided. The percentage in parentheses shows the percentage of terms with ambiguity in the range to the total number of terms.
| Before Normalization |
After Normalization |
|||||
|---|---|---|---|---|---|---|
| Ambiguity | Original | UniRef90 | UniRef50 | Original | UniRef90 | UniRef50 |
| 1 | 2,233,471 | 2,233,471 | 2,233,471 | 1,810,435 | 1,810,435 | 1,810,435 |
| (83.7%) | (83.7%) | (83.7%) | (84.0%) | (84.0%) | (84.0%) | |
| 2 | 223,698 | 270,826 | 303,400 | 176,041 | 211,427 | 237,711 |
| (8.4%) | (10.2%) | (11.4%) | (8.2%) | (9.8%) | (11.0%) | |
| 3–4 | 109,701 | 94,164 | 80,654 | 84,206 | 76,375 | 66,536 |
| (4.1%) | (3.5%) | (3.0%) | (3.9%) | (3.5%) | (3.1%) | |
| 5–8 | 53,535 | 35,528 | 26,968 | 44,717 | 29,490 | 21,585 |
| (2%) | (1.3%) | (1%) | (2.1%) | (1.4%) | (1.0%) | |
| 9–16 | 21,936 | 15,757 | 11,113 | 19,301 | 12,894 | 9,190 |
| (0.8%) | (0.6%) | (0.4%) | (0.9%) | (0.6%) | (0.4%) | |
| 17–32 | 10,841 | 7,579 | 5,579 | 9,007 | 6,208 | 4,600 |
| (0.4%) | (0.3%) | (0.2%) | (0.4%) | (0.3%) | (0.2%) | |
| 33–64 | 5,646 | 4,183 | 3,134 | 4,532 | 3,472 | 2,700 |
| (0.2%) | (0.2%) | (0.1%) | (0.2%) | (0.2%) | (0.1%) | |
| >64 | 7,140 | 4,460 | 1,649 | 5,996 | 3,934 | 1,478 |
| (0.3%) | (0.2%) | (<0.1%) | (0.3%) | (0.2%) | (<0.1%) | |
| Total names | 2,665,968 | 2,106,222 | ||||
| Average ambiguity | 2.31 | 1.77 | 1.46 | 2.32 | 1.79 | 1.49 |
The cross-biomedical domain ambiguity was evaluated based on BioThesaurus names that were mapped to terms in UMLS semantic categories using exact string match. Among 34,598 matched BioThesaurus names, only 1,483 names (4.3%) were mapped to UMLS semantic categories not considered highly related to genes/proteins (i.e., those not listed in ▶). After normalization, 2,143 (4.0%) out of 53,052 matched normalized names were mapped to concepts with categories not considered highly related to genes/proteins. Examples include “Juvenile,” “minute,” and “purple.” The common English collection contained 8,090 words, all in lower case. Among them, 367 were also single-word names in BioThesaurus; when case difference was ignored, the number increased to 594 words. Examples of gene/protein names in common English words include “dare,” “air,” “all” and “ago.”
The coverage result is shown in ▶. The coverage of BioThesaurus is 94.0%, i.e., there are 1,262 out of 1,343 pairs of (GENE, ABSTRACT) where at least one name associated with GENE in BioThesaurus occurred in ABSTRACT. Additionally, 119 out of 1,127 (i.e., 3,375–905–1,343) pairs of (GENE, ABSTRACT), which were originally curated as a GENE that was not explicitly mentioned in ABSTRACT, were found to be errors since names associated with GENE in BioThesaurus were actually found to occur in ABSTRACT.
Table 4.
Table 4 The coverage assessment for BioThesaurus using the test set of the BioCreAtive workshop. The percentage in the first column shows the percentage of terms in BioCreAtive text present in BioThesaurus. The percentages in the second, third, and last columns were acquired by inversing the ambiguity which refer to the precisions of a base system that randomly picks an associated entity for those ambiguous terms.
| Organisms | The Coverage of Genes in the Evaluation Set Present in BioThesaurus |
The Ambiguity of Matched Terms in BioThesaurus |
||
|---|---|---|---|---|
| Mentioned in Abstracts | Including Systematic Ambiguity | Ignoring Systematic Ambiguity | Limited to Specific Organism | |
| Yeast | 557/595 (93.6%) | 55.7 (1.8%) | 23.8 (4.2%) | 1.23 (81.3%) |
| Mouse | 346/378 (91.5%) | 28.0 (3.6%) | 13.4 (7.5%) | 1.13 (88.5%) |
| Fly | 359/370 (98.1%) | 50.7 (2.0%) | 17.7 (5.6%) | 7.34 (13.6%) |
| Total | 1,262/1,343 (94.0%) | 46.9 (2.1%) | 19.1 (5.3%) | 3.02 (33.1%) |
The ambiguity that a dictionary-based BNET system needs to resolve according to BioCreAtive text was in the range of 28–55 when including systematic ambiguity. It dropped to the range of 13–23 when ignoring systematic ambiguity. If we ignore other types of ambiguity (i.e., cross-general ambiguity and cross-medicine ambiguity), a base system which randomly picks an entity for those ambiguous terms could achieve a precision in the range of 1.8%–3.4% when including systematic ambiguity and of 4.5%–7.5% when ignoring systematic ambiguity (i.e., when we only consider entity-specific ambiguity, a base dictionary-based BNET system achieved a precision in the range of 4.5%–7.5% and a recall in the range of 91.5%–98% for BioCreAtive Task1B test set).
Discussion and Conclusion
In this paper we have assessed BioThesaurus regarding its ambiguity, synonymy, and coverage to reveal the complexity of dictionary-based biological named entity tagging. Most existing NLP systems currently do not associate terms appearing in the text with database entries and they do not need to resolve entity-specific ambiguity (i.e., a term represents multiple database entries). The dictionary we constructed links terms with protein entries in UniProtKB. Using the comprehensive cross-reference information stored in iProClass, entities (i.e., concepts) in the dictionary can be tailored for systems that use records in other databases, such as SGD and MGD, cross-referenced by iProClass.
Because of the incomplete coverage of all UniProtKB records by the curated classification systems such as PIRSF, we used UniRef clusters to assess the synonymy and ambiguity when ignoring systematic ambiguity. In fact, UniRef clusters provide tighter sequence groupings than PIRSF, i.e., one PIRSF family can map to one or more UniRef50 clusters. It is known that functions of some homologous proteins can be well conserved from higher to lower organisms, while those of others may only be conserved in close lineage of organisms (e.g., among mammals). Moreover, even homologous proteins including paralogs sharing high degree of sequence identity may differ in functions due to changes of key residues such as active sites (including gaining or losing functions). It is not known the percentage of such proteins in UniRef clusters, which is beyond the scope of this paper. Nonetheless, we feel that using UniRef90 and 50 is still a reasonable way to estimate the level of name ambiguity when disregarding systematic ambiguity of homologous proteins, albeit the true level of this ambiguity could be higher. One advantage of estimating the name ambiguity by removing the systematic ambiguity is that it could help identify protein name misnomers and name annotation inconsistency in databases. For example, human EGFR (epidermal growth factor receptor) was incorrectly assigned as gene name of the human EGF entry (UniProtKB: Q8NDU8), thus, giving the name “EGFR” higher ambiguity.
The assessment of synonymy and ambiguity provided in the result section was derived disregarding the fact that some terms may never appear in literature. For terms that actually occur in the text, the average ambiguity for them tends to be higher. We could not assess them directly. We plan to explore ways to assess the real ambiguity in the text. Even though there are not many terms in BioThesaurus that are common English words, but they occur frequently in the literature where most of these occurrences hold meanings other than biological entities (e.g., all or to). We plan to investigate further for disambiguating these terms in text. In the assessment of synonymy, we did not include abbreviation-derivable in the assessment. It is because most gene/protein symbols are actually abbreviation-derivable. For example in ▶, the symbol “CLIM1” is abbreviated from “C-TERMINAL LIM DOMAIN PROTEIN 1.” However, it is not easy to handle abbreviation-derivable synonyms in entity tagging systems because of their ambiguities. We plan to further study abbreviation-derivable synonyms.
During the coverage assessment, we removed 250 pairs of (GENE, ABSTRACT) with no corresponding UniProtKB records associated with them. Most of them were from fly (with 63 pairs) and mouse (with 168 pairs). Among these pairs, almost all were associated with records of non-coding genomic regions or gene clusters/complexes. For example, there are over 60 pairs of (GENE, ABSTRACT) in MGI having a type as DNA segment (e.g., “D2Mit6” with identifier MGI:92196), over 10 pairs having a type as complex (e.g., “Hbb” with identifier MGI:96020). Most of such pairs in fly were associated with transposable elements instead of coding genes. Pairs in yeast that were not associated with UniProtKB records are also caused by non-coding gene records. For example, “HSX1” with identifier S000006707 in SGD has a type tRNA.
During the assessment, we also found that UniProtKB has finer granularity than model organism databases for genes that were cross-referenced by iProClass. For example, 25,514 UniProtKB entries were associated with 16,553 FlyBase records. One reason is that while entries in model organism databases are gene-centric, multiple protein products (isoforms) from alternative splicing of the same gene can be represented by multiple entries in UniProtKB. Another factor is that multiple protein fragments of the same gene could be represented in UniProtKB, which may have derived from independent labs, and sometimes may be given different names. For example, there are two UniProtKB entries (i.e., O76923 for isoform A and O96378 for isoform B) associated with the Ap-2 gene (FBgn00023417). In mouse, there are eight UniProtKB entries (e.g., Q08501, Q8C7G1, Q3UPQ1), including spliced variants and fragments, associated with one mouse gene Prlr (MGI:97763). Because of this many-to-one mapping of UniProtKB to the model organism databases, utilizing the rich cross-reference information supplied by iProClass, BioThesaurus can be tailored to dictionaries for entities in other databases (e.g., MGD) by providing a richer source of protein names and synonyms.
During the coverage assessment study, most terms (94.0%) identified by human experts in abstract of the BioCreAtive test set were included in BioThesaurus. It indicates that a BNET system which simply annotates a term with all associated entities can achieve a high recall (as high as 94.0%). However, the precision of such system would be very low since there are a lot of false positives caused by the ambiguities (as low as 2.1% when considering systematic ambiguity and entity-specific ambiguity and 5.3% when only considering entity-specific ambiguity). If we already know the corresponding organism for the paper, we can limit entities to a specific organism. The last column of ▶ shows the result of the ambiguity when we tailored BioThesaurus to specific organisms. From ▶, the ambiguity (including systematic ambiguity and entity-specific ambiguity) for mouse (or yeast) decreased to 1.23 (or 1.12) which implies that the precision for a BNET system which randomly picks an entity from entities associated with an ambiguous term was 81.3% (or 88.5%). This feature is helpful in literature-based protein database annotation and indeed is already used for semi-automatic mapping of gene/protein names to UniProtKB entries from a rule-based text mining system RLIMS-P for protein phosphorylation. 57 However, the real precision measure for such a system would be relatively lower because there are problems with tokenization, cross-general ambiguities, and cross-medicine ambiguities.
In conclusion, biological entity tagging is an essential task for NLP systems in the biomedical domain. We assessed BioThesaurus through several perspectives: ambiguity, synonymy, and coverage. Based on the assessment, we quantified the complexity of dictionary-based biological named entity tagging systems. The study demonstrated that most gene/protein names appearing in text can be found in BioThesaurus. The study confirmed that biological named entity tagging is a non-trivial task because of the high ambiguity of gene and protein names as well as multiple names corresponding to the same gene/protein. While classifying ambiguity into different categories and applying different approaches to resolve these ambiguities, we believe the performance of dictionary-based biological named entity tagging could be improved.
Footnotes
This research was supported by grant no. IIS-0430743 from the National Science Foundation.
References
- 1.Humphreys K, Demetriou G, Gaizauskas R. Two applications of information extraction to biological science journal articlesenzyme interactions and protein structures. Pac Symp Biocomput 2000:505-516. [DOI] [PubMed]
- 2.Rindflesch TC, Tanabe L, Weinstein JN, Hunter L. EDGARextraction of drugs, genes and relations from the biomedical literature. Pac Symp Biocomput 2000:517-528. [DOI] [PMC free article] [PubMed]
- 3.Yakushiji A, Tateisi Y, Miyao Y, Tsujii J. Event extraction from biomedical papers using a full parser Pac Symp Biocomput 2001:408-419. [DOI] [PubMed]
- 4.Andrade MA, Valencia A. Automatic extraction of keywords from scientific textapplication to the knowledge domain of protein families. Bioinformatics 1998;14(7):600-607. [DOI] [PubMed] [Google Scholar]
- 5.Craven M, Kumlien J. Constructing biological knowledge bases by extracting information from text sources Proc Int Conf Intell Syst Mol Biol 1999:77-86. [PubMed]
- 6.Chiang JH, Yu HC. MeKEdiscovering the functions of gene products from biomedical literature via sentence alignment. Bioinformatics 2003;19:1417-1422. [DOI] [PubMed] [Google Scholar]
- 7.Chiang JH, Yu HC, Hsu HJ. GISa biomedical text-mining system for gene information discovery. Bioinformatics 2004;20(1):120-121. [DOI] [PubMed] [Google Scholar]
- 8.Rzhetsky A, Iossifov I, Koike T, et al. GeneWaysa system for extracting, analyzing, visualizing, and integrating molecular pathway data. J Biomed Inform 2004;37(1):43-53. [DOI] [PubMed] [Google Scholar]
- 9.Daraselia N, Yuryev A, Egorov S, Novichkova S, Nikitin A, Mazo I. Extracting human protein interactions from MEDLINE using a full-sentence parser Bioinformatics 2004;20(5):604-611. [DOI] [PubMed] [Google Scholar]
- 10.Yuryev A, Mulyukov Z, Kotelnikova E, et al. Automatic pathway building in biological association networks BMC Bioinformatics 2006;7:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JC, Kim HS, Kim JJ. Bidirectional incremental parsing for automatic pathway identification with combinatory categorial grammar Pac Symp Biocomput 2001:396-407. [PubMed]
- 12.Hirschman L, Colosimo M, Morgan A, Yeh A. Overview of BioCreAtIvE task 1Bnormalized gene lists. BMC Bioinformatics 2005;6(Suppl 1):S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirschman L, Park JC, Tsujii J, Wong L, Wu CH. Accomplishments and challenges in literature data mining for biology Bioinformatics 2002;18(12):1553-1561. [DOI] [PubMed] [Google Scholar]
- 14.Hirschman L, Yeh A, Blaschke C, Valencia A. Overview of BioCreAtIvEcritical assessment of information extraction for biology. BMC Bioinformatics 2005;6(Suppl 1):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shatkay H, Feldman R. Mining the biomedical literature in the genomic eraan overview. J Comput Biol 2003;10(6):821-855. [DOI] [PubMed] [Google Scholar]
- 16.Krauthammer M, Nenadic G. Term identification in the biomedical literature J Biomed Inform 2004;37(6):512-526. [DOI] [PubMed] [Google Scholar]
- 17.Yeh A, Morgan A, Colosimo M, Hirschman L. BioCreAtIvE task 1Agene mention finding evaluation. BMC Bioinformatics 2005;6(Suppl 1):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanisch D, Fluck J, Mevissen HT, Zimmer R. Playing biology’s name gameidentifying protein names in scientific text. Pac Symp Biocomput 2003:403-414. [PubMed]
- 19.Hanisch D, Fundel K, Mevissen HT, Zimmer R, Fluck J. ProMinerrule-based protein and gene entity recognition. BMC Bioinformatics 2005;6(Suppl 1):S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenssen TK, Laegreid A, Komorowski J, Hovig E. A literature network of human genes for high-throughput analysis of gene expression Nat Genet 2001;28(1):21-28. [DOI] [PubMed] [Google Scholar]
- 21.Fukuda K, Tamura A, Tsunoda T, Takagi T. Toward information extractionidentifying protein names from biological papers. Pac Symp Biocomput 1998:707-718. [PubMed]
- 22.Tanabe L, Wilbur WJ. Tagging gene and protein names in biomedical text Bioinformatics 2002;18(8):1124-1132. [DOI] [PubMed] [Google Scholar]
- 23.Sekimizu T, Park HS, Tsujii J. Identifying the Interaction between Genes and Gene Products Based on Frequently Seen Verbs in Medline Abstracts Genome Inform Ser Workshop Genome Inform 1998;9:62-71. [PubMed] [Google Scholar]
- 24.Tanabe L, Xie N, Thom LH, Matten W, Wilbur WJ. GENETAGa tagged corpus for gene/protein named entity recognition. BMC Bioinformatics 2005;6(Suppl 1):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finkel J, Dingare S, Manning CD, Nissim M, Alex B, Grover C. Exploring the boundariesgene and protein identification in biomedical text. BMC Bioinformatics 2005;6(Suppl 1):S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald R, Pereira F. Identifying gene and protein mentions in text using conditional random fields BMC Bioinformatics 2005;6(Suppl 1):S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou G, Shen D, Zhang J, Su J, Tan S. Recognition of protein/gene names from text using an ensemble of classifiers BMC Bioinformatics 2005;6(Suppl 1):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hakenberg J, Bickel S, Plake C, et al. Systematic feature evaluation for gene name recognition BMC Bioinformatics 2005;6(Suppl 1):S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitsumori T, Fation S, Murata M, Doi K, Doi H. Gene/protein name recognition based on support vector machine using dictionary as features BMC Bioinformatics 2005;6(Suppl 1):S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamames J. Text detectivea rule-based system for gene annotation in biomedical texts. BMC Bioinformatics 2005;6(Suppl 1):S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colosimo ME, Morgan AA, Yeh AS, Colombe JB, Hirschman L. Data preparation and interannotator agreementBioCreAtIvE task 1B. BMC Bioinformatics 2005;6(Suppl 1):S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balakrishnan R, Christie KR, Costanzo MC, et al. Fungal BLAST and Model Organism BLASTP Best Hitsnew comparison resources at the Saccharomyces Genome Database (SGD). Nucleic Acids Res 2005;33(Database issue):D374-D377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blake JA, Eppig JT, Bult CJ, Kadin JA, Richardson JE. The Mouse Genome Database (MGD)updates and enhancements. Nucleic Acids Res 2006;34(Database issue):D562-D567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drysdale RA, Crosby MA. FlyBasegenes and gene models. Nucleic Acids Res 2005;33(Database issue):D390-D395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fundel K, Guttler D, Zimmer R, Apostolakis J. A simple approach for protein name identificationprospects and limits. BMC Bioinformatics 2005;6(Suppl 1):S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crim J, McDonald R, Pereira F. Automatically annotating documents with normalized gene lists BMC Bioinformatics 2005;6(Suppl 1):S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H, Wu C, Friedman C. BioTagger: a biological entity tagging system. Paper presented at: BioCreAtive Workshop, 2004; Spain.
- 38.Liu H, Hu ZZ, Zhang J, Wu C. BioThesaurusa web-based thesaurus of protein and gene names. Bioinformatics 2006;22(1):103-105. [DOI] [PubMed] [Google Scholar]
- 39.Wu CH, Apweiler R, Bairoch A, et al. The Universal Protein Resource (UniProt)an expanding universe of protein information. Nucleic Acids Res 2006;34(Database issue):D187-D191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu CH, Huang H, Nikolskaya A, Hu Z, Barker WC. The iProClass integrated database for protein functional analysis Comput Biol Chem Feb 2004;28(1):87-96. [DOI] [PubMed] [Google Scholar]
- 41.Bodenreider O. The Unified Medical Language System (UMLS)integrating biomedical terminology. Nucleic Acids Res 2004;32(Database issue):D267-D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wren JD, Chang JT, Pustejovsky J, Adar E, Garner HR, Altman RB. Biomedical term mapping databases Nucleic Acids Res 2005;33(Database issue):D289-D293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu H, Agichtein E. Extracting synonymous gene and protein terms from biological literature Bioinformatics 2003;19(Suppl 1):i340-i349. [DOI] [PubMed] [Google Scholar]
- 44.Hatzivassiloglou V, Duboue PA, Rzhetsky A. Disambiguating proteins, genes, and RNA in texta machine learning approach. Bioinformatics 2001;17(Suppl 1):S97-S106. [DOI] [PubMed] [Google Scholar]
- 45.Liu H, Lussier YA, Friedman C. Disambiguating ambiguous biomedical terms in biomedical narrative textan unsupervised method. J Biomed Inform 2001;34(4):249-261. [DOI] [PubMed] [Google Scholar]
- 46.Liu H, Teller V, Friedman C. A multi-aspect comparison study of supervised word sense disambiguation J Am Med Inform Assoc 2004;11(4):320-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu H, Johnson SB, Friedman C. Automatic resolution of ambiguous terms based on machine learning and conceptual relations in the UMLS J Am Med Inform Assoc 2002;9(6):621-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaudan S, Kirsch H, Rebholz-Schuhmann D. Resolving abbreviations to their senses in Medline Bioinformatics 2005;21(18):3658-3664. [DOI] [PubMed] [Google Scholar]
- 49.Schijvenaars BJ, Mons B, Weeber M, Schuemie MJ, van Mulligen EM, Wain HM, Kors JA. Thesaurus-based disambiguation of gene symbols BMC Bioinformatics 2005;6:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang JT, Schutze H, Altman RB. GAPSCOREfinding gene and protein names one word at a time. Bioinformatics 22 2004;20(2):216-225. [DOI] [PubMed] [Google Scholar]
- 51.Franzen K, Eriksson G, Olsson F, Asker L, Liden P, Coster J. Protein names and how to find them Int J Med Inform 4 2002;67(1–3):49-61. [DOI] [PubMed] [Google Scholar]
- 52.Li W, Jaroszewski L, Godzik A. Clustering of highly homologous sequences to reduce the size of large protein databases Bioinformatics 2001;17(3):282-283. [DOI] [PubMed] [Google Scholar]
- 53.Kotaka M, Ngai SM, Garcia-Barcelo M, Tsui SK, Fung KP, Lee CY, Waye MM. Characterization of the human 36-kDa carboxyl terminal LIM domain protein (hCLIM1) J Cell Biochem 1999;72(2):279-285. [DOI] [PubMed] [Google Scholar]
- 54.Ueki N, Seki N, Yano K, Ohira M, Saito T, Masuho Y, Muramatsu M. Isolation and chromosomal assignment of human genes encoding cofactor of LIM homeodomain proteins, CLIM1 and CLIM2 J Hum Genet 1999;44(2):112-115. [DOI] [PubMed] [Google Scholar]
- 55.Chen L, Liu H, Friedman C. Gene name ambiguity of eukaryotic nomenclatures Bioinformatics 2004. Aug 27. [DOI] [PubMed]
- 56.Wu CH, Nikolskaya A, Huang H, et al. PIRSFfamily classification system at the Protein Information Resource. Nucleic Acids Res 2004;32(Database issue):D112-D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan X, Hu ZZ, Wu HT, et al. An online literature mining tool for protein phosphorylation Bioinformatics 2006. Apr 27. [DOI] [PubMed]