Abstract
Alpharetroviruses provide a useful system for the study of the molecular mechanisms of host range and receptor interaction. These viruses can be divided into subgroups based on diverse receptor usage due to variability within the two host range determining regions, hr1 and hr2, in their envelope glycoprotein SU (gp85). In previous work, our laboratory described selection from a subgroup B avian sarcoma-leukosis virus of an extended-host-range variant (LT/SI) with two adjacent amino acid substitutions in hr1. This virus retains its ability to use the subgroup BD receptor but can also infect QT6/BD cells, which bear a related subgroup E receptor (R. A. Taplitz and J. M. Coffin, J. Virol 71:7814-7819, 1997). Here, we report further analysis of this unusual variant. First, one (L154S) of the two substitutions is sufficient for host range extension, while the other (T155I) does not alter host range. Second, these mutations extend host range to non-avian cell types, including human, dog, cat, mouse, rat, and hamster. Third, interference experiments imply that the mutants interact efficiently with the subgroup BD receptor and possibly the related subgroup E receptor, but they have another means of entry that is not dependent on these interactions. Fourth, binding studies indicate that the mutant SU proteins retain the ability to interact as monomers with subgroup BD and BDE receptors but only bind the subgroup E receptor in the context of an Env trimer. Further, the mutant SU proteins bind well to chicken cells but do not bind any better than wild-type subgroup B to QT6 or human cells, even though the corresponding viruses are capable of infecting these cells.
Alpharetroviruses, or avian sarcoma-leukosis viruses (ASLVs), display a great deal of diversity in their envelope glycoprotein (Env) sequences leading to diverse host range but, with the exception of the long terminal repeat (LTR), are nearly identical throughout the remainder of their genomes (11, 15, 16, 25-27, 29, 42). This pattern suggests a response to selective pressures to replicate in a variety of hosts. Alpharetroviruses are divided into subgroups (A to J) depending on host range, superinfection resistance patterns, and neutralizing antibody cross-reactivity. The surface (SU) subunit of the envelope glycoprotein is responsible for receptor recognition. Through a poorly characterized process, probably requiring a low-pH step (34), the SU-receptor interaction triggers the transmembrane (TM) subunit to mediate fusion between the viral envelope and the target cell membrane (10, 26, 48).
Receptors for ASLV of subgroups A, B, D, and E have been cloned. The receptor for subgroup A viruses is a low-density lipoprotein receptor-like protein and is unrelated to any other known retroviral receptor (5, 51). The receptors for B, D, and E are encoded by orthologous genes in the tumor necrosis factor receptor family (2, 3, 8, 40). Chickens have two alleles capable of acting as the receptor for these viruses. The tv-bs1 allele can serve as a receptor for all three subgroups. Infection with virus of subgroups B or D blocks superinfection by all three of these subgroups. Infection with subgroup E virus blocks superinfection by virus of subgroup E but allows subsequent infection by B or D virus (26, 48). This nonreciprocal interference probably reflects the presence of two different conformational forms of the receptor on the cell surface. Subgroup B and D viruses can recognize both forms, while subgroup E viruses can only recognize one (1). The second allele, tv-bs3, can serve as a receptor for subgroups B and D only. Quail, turkey, and some related birds have an allele for a third type of receptor, tv-bq or tv-bt, conferring susceptibility to infection by subgroup E but not subgroup B and D viruses (2, 13, 14, 26, 35, 48). Cells used for these studies are designated by the first letter of the species from which they are derived and the classical alpharetrovirus subgroups (A to E) to which the cells are resistant. For example, C/E are chicken cells that are resistant to infection by subgroup B alpharetroviruses and therefore susceptible to infection by subgroup A, B, C, and D alpharetroviruses.
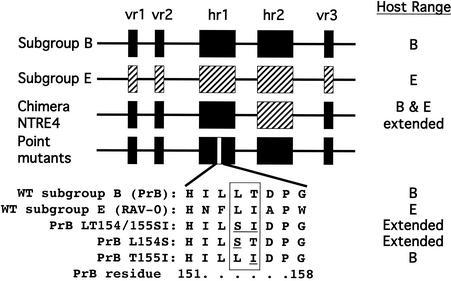
Our laboratory has previously shown that determinants of host range and receptor recognition lie predominantly within two short stretches of gp85 SU called hr1 and hr2 (15, 16) (Fig. 1). We have described a chimeric ASLV, NTRE4, which has a recombinant envelope that has a subgroup E hr2 but otherwise consists of subgroup B sequences. Unlike either parent, this virus can infect both C/E and Q/BD cells. It exhibits reciprocal interference with subgroup B viruses on C/E chicken embryo fibroblasts (CEF) and with subgroup E viruses on quail and turkey cells, indicating that it makes use of both the subgroup B and subgroup E receptors (47). Our laboratory has also described a mutant virus, LT154/155SI (S20) (43), that is derived from a subgroup B virus selected for host range extension to quail cells. It contains two amino acid substitutions in hr1 that are sufficient for this host range extension (Fig. 1). In the study reported here, we have examined these two substitutions individually for host range extension and measured binding and interference patterns among these mutants, wild-type subgroup B and E viruses, and NTRE4. Finally, we examined the ability of these viruses to infect a panel of cell lines from diverse species. We found that the L154S mutation alone suffices for the extension of host range and that virus containing this mutation is also capable of infecting cell lines of human, dog, cat, and to a lesser extent mouse, rat, and hamster origin. Surprisingly, the large increase in the ability of the mutant viruses to infect these cells was not accompanied by a detectable increase in binding of their SU proteins. However, binding to one candidate receptor, TVB-T, was detectable when the envelope glycoproteins were expressed on the cell surface. These findings suggest novel modes of interaction with receptors and perhaps entry pathways that are receptor independent.
FIG. 1.
Schematic representation of the alpharetrovirus gp85 SU. Sequences shown to be important for host range determination are represented by boxes (black for subgroup B and hashed for subgroup E). vr1, vr2, and vr3 are variable regions, and hr1 and hr2 are highly variable host-range-determining regions. NTRE4 is a chimeric virus resulting in recombination between td-PrRSV-B and RAV-0. The location of the point mutations studied here is indicated at the bottom in the hr1 rectangle. WT subgroup E (RAV-0), subgroup B (td-PrRSV-B), and the mutant amino acid sequences are listed below. The host range phenotypes are listed on the right (43, 47).
MATERIALS AND METHODS
Cells.
Cells were grown using modified Richter's medium (Tufts formulation; Irvine Scientific) supplemented with 10% fetal calf serum (FCS; Sigma), except where otherwise noted. CEF (C/E) were prepared from fertilized eggs from Lansing line 0 chickens (USDA Poultry Station, East Lansing, Mich.). QT6 cells (QT6/BD) are a quail fibrosarcoma line. Q24 cells (a gift from Jürgen Brojatsch) are QT6 cells expressing the tv-bs3 gene. Q24gfp cells additionally express the enhanced green fluorescent protein (EGFP) in the context of a packageable defective avian leukosis virus (ALV) genome derived from the plasmid pRDgfp. pRDgfp was constructed by replacing the β-galactosidase sequences flanked by BamHI sites from pRDlac (37) with EGFP (Clontech). Q24cg cells were stably transfected with pRDcg, constructed as described for pRDgfp, except that a cytomegalovirus-EGFP cassette (Clontech) was inserted. DF1 cells are a continuously dividing C/E line derived from a spontaneous transformant of Lansing line 0 CEF (4, 22, 39). 293 cells are a human embryonic kidney cell line. These cells as well as 293 cell lines stably expressing tv-bs1, tv-bs3, and tv-bT were a gift from John Young (2, 3, 8). Rat-1 is a rat fibroblast cell line. The following cell lines were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% FCS: mouse fibroblast (NIH 3T3) cells, D17 dog osteosarcoma cells, and AH927 feline embryo fibroblasts (FEF). Finally, Chinese hamster ovary (CHO-K1) cells were maintained in RPMI 1640 (Invitrogen) supplemented with 10% FCS.
Viruses and mutagenesis.
pBR322-tdPrRSV-B, LT154/155SI (formerly S20), and NTRE4 constructs are full-length viral genomes permuted at the unique SalI site in env and cloned into pBR322 (16, 43). L154S and T155I mutations were introduced into pBR322-tdPrRSV-B by QuikChange mutagenesis (Stratagene). Fragments containing these mutations were excised, ligated back into pBR322-tdPrRSV-B, and sequenced. To generate infectious virus, these constructs were digested with SalI and self-ligated, and 106 CEF were transfected with 10 μg of DNA (Lipofectamine Plus; Invitrogen). These viruses as well as RAV-60 and NTRE4 from viral stocks routinely used by our laboratory were used to infect Q24, Q24gfp, and Q24cg cells to generate the viruses used in subsequent experiments. The RCASBP(A) (ALV-A) plasmid was obtained from S. Hughes (32, 36) and used to transfect Q24, Q24gfp, and Q24cg cells (Lipofectamine Plus; Invitrogen).
Determination of viral titers.
Cells were infected in triplicate with egfp viruses for 1 h at 37°C in the presence of 1.5 μg of Polybrene (Aldrich)/ml [except cells infected with RCASBP(A)], as Polybrene does not aid, and may modestly inhibit, infection by subgroup A virus (44). Two days later, EGFP-positive and live cell counts were determined by flow cytometry using a FACSCalibur (Becton Dickinson). The titer was determined from the following formula: IU/ml = {(1/dilution) × (1/volume used to infect) × (cells per well at time of infection) × [−ln(1 − positive fraction)]}. Slight differences in live cells scored for each replicate can lead to small variability in the limit of detection for negative samples.
Interference assays.
CEF and QT6 cells were transfected or infected with our panel of viral clones or viruses as described above. Transfected and infected cells were passaged at least 5 times, and reverse transcriptase (RT) assays were used to determine that infection of the cultures was complete. Preinfected cells were superinfected in triplicate with egfp viruses and analyzed by flow cytometry to calculate titers as described above. The level of interference was obtained by dividing the titer of each virus on uninfected cells by the titer on preinfected cells.
Generation of immunoadhesin constructs.
TVA-rIgG, TVBs1-rIgG, TVBs3-rIgG, TVBT-rIgG, SUE-rIgG, and SUB-rIgG plasmids were obtained from J. Young (8, 52). PrBSU-rIgG was generated by replacing SU sequences from the SUB-rIgG with sequences from td-PrRSV-B. The XhoI site at position 390 bp in the Pr-RSV B env gene was disrupted with a silent mutation, and a BamHI site was introduced immediately downstream of the SU region by incorporating it into a PCR primer. The 1,021-bp PCR product encompassing the entire SU region was digested with XhoI, partially digested with BamHI, and ligated into the SUB-rIgG plasmid, replacing all SUB coding sequences with Pr-RSV B SU sequences. After disruption of internal XhoI sites (position 390), SU regions from NTRE4, L154S, T155I, and LT154/155SI were PCR amplified, digested with XhoI and EcoRI, and ligated into the PrBSU-rIgG to generate NTRE4SU-rIgG, L154SSU-rIgG, T155ISU-rIgG, and LT154/155SISU-rIgG. All of these constructs were verified by sequencing.
Production of purified immunoadhesin protein and binding assays.
293 cells were transfected using Lipofectamine Plus (Invitrogen). One day after transfection, cells were washed and fed with AIM-V serum-free medium (Invitrogen). Supernatants were harvested daily for 5 days, filtered (0.2-μm pore size; Millipore), and stored at 4°C; cells were fed with fresh AIM-V. Supernatants were passed over Immunopure Plus immobilized protein A columns (Pierce) and washed with phosphate-buffered saline (PBS), and the immunoadhesins were eluted in 0.1 M citric acid (pH 3). Positive fractions were pooled, neutralized with 1 M Tris (pH 9), and dialyzed (Spectrapor 2; Spectrum Laboratories) against three changes of 100 volumes of PBS. The samples were concentrated using Centricon YM-30 units (Millipore) and stored at −80°C. Protein concentrations were determined using the Bio-Rad protein microassay.
For binding assays, cells were removed from plates with 25 mM EDTA-PBS and washed with cold PBA (PBS, 1% bovine serum albumin, 0.1% NaN3). A total of 3 × 105 cells (105 cells for CEF) were incubated in triplicate with immunoadhesin (0.01, 0.1, 0.3, or 1 μg) in PBA at 4°C for 1 h. Cells were washed with cold PBA, resuspended in fluorescein isothiocyanate-conjugated swine anti-rabbit antibody (Dako) diluted 1/10 in PBA, and incubated at 4°C for 30 min. Cells were then washed with cold PBA and resuspended in PBS, and median fluorescence intensity was determined for each sample by using a FACScalibur (Becton Dickinson) and FlowJo FACS analysis software (Treestar).
RESULTS
A single mutation at Env residue 154 is sufficient for host range extension.
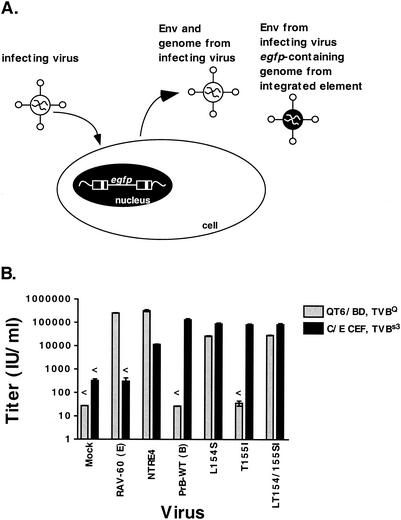
During the process of selecting the extended-host-range virus, we observed only virus containing both the L154S and the T155I mutants, suggesting that both mutations were necessary for either host range extension or for some other selected feature (43). The selected substitutions result in a conservative inversion of an amino acid containing a small hydrophobic side chain (leu) followed by a small hydroxyl-containing side chain (thr) to a pair of amino acids with the same properties in the opposite order (ser-ile). To test the role of each mutation in the extension of host range, we used site-directed mutagenesis to insert them individually into a complete viral genome. Virus derived from these constructs was used to pseudotype a vector containing the egfp gene, and the egfp viruses were used to determine the efficiency of infection of QT6 and CEF (Fig. 2A). All viruses were generated with similar efficiency, as judged by the production of approximately equal RT activity following infection or transfection of the Q24gfp viral producer cell line (data not shown). Stocks of PrB-WT, both single mutants, and the double mutant had titers around 105 IU/ml on CEF, while the titer of NTRE4 was about 10-fold less (Fig. 2B). This result shows that the mutations, alone or in combination, do not impair the ability of these viruses to replicate. As expected, PrB-WT was not capable of infecting QT6 cells. L154S and LT154/155SI, on the other hand, were able to infect QT6 cells while T155I was not, indicating that the L154S substitution is sufficient for the host range extension (Fig. 2B). Interestingly, NTRE4 had a relative titer on QT6 cells that was about 10-fold higher than that on CEF, while L154S and LT154SI infected CEF about threefold more efficiently than QT6 cells. This difference suggests that the point mutants have a preference for the subgroup B receptor, while NTRE4 has a preference for the subgroup E receptor. T155I infected the two cell types with a profile identical to that of PrB-WT in this assay.
FIG. 2.
A single mutation expands the host range of td-PrRSV-B virus. (A) Generation of pseudotyped egfp virus. Wild-type or mutant viruses were used to infect cells that contain a stably integrated replication-defective genome with the egfp gene and sequences necessary for packaging and reverse transcription. These cells then produced a mixture of viruses that bear the envelope and other viral proteins encoded by the infecting virus and contain the genome of the infecting virus, the integrated egfp element, or one copy of each. These stocks were used in subsequent experiments to score infection by egfp expression. (B) Quail cells resistant to infection by subgroup B and D alpharetroviruses (QT6/BD) and chicken cells resistant to infection by subgroup E alpharetroviruses (C/E) were infected with pseudotyped egfp viruses in triplicate, and infection was scored by flow cytometry 2 days later. Titers were determined as described in Materials and Methods. Error bars show the standard error of the mean for each determination. Titers below the limit of detection are graphed at the detection limit and marked with a “<.”
Host range extension mutants exhibit incomplete superinfection resistance.
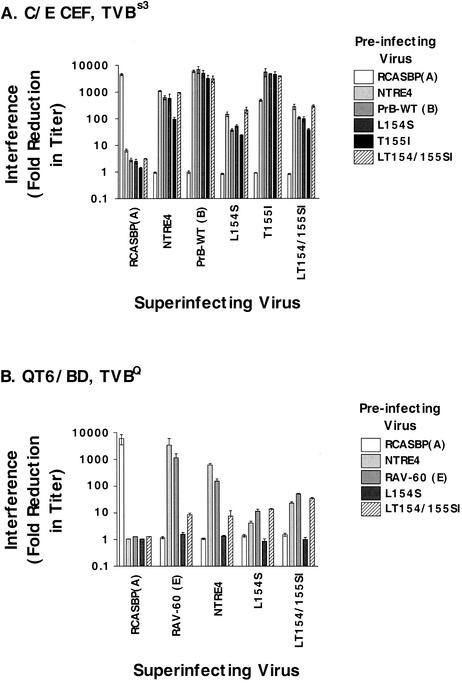
Retroviruses are able to render an infected cell resistant to subsequent infection by viruses of the same subgroup (10, 26, 48). In the simplest case, the envelope glycoprotein expressed by the infected cells interacts in cis with the receptor to prevent binding by extracellular virions. More complicated mechanisms of interference can involve removal of receptors from the cell surface (28). To examine receptor interaction with the mutant viruses in comparison to well-characterized ASLVs, we conducted interference assays on CEF and QT6 cells. Cells were infected and passaged several times to ensure complete infection. The cells were then challenged with pseudotyped egfp viruses, and infection was scored by flow cytometry 2 days later.
Figure 3A shows the results of interference assays on CEF. Infection by PrB-WT and T155I was efficiently blocked by NTRE4, PrB, and all of the PrB point mutants, but not by the subgroup A control, RCASBP(A). Infection by L154S and LT154/155SI was also blocked by all PrB mutants as well as NTRE4, but on average about 100-fold less efficiently. Interference with NTRE4 infection was intermediate to the other two groups on these cells. Preinfection with any of these viruses did not block superinfection with RCASBP(A) to any appreciable degree, confirming that the interference was specific to the subgroup B receptor on these cells (TVBs3).
FIG. 3.
Interference patterns among wild-type, recombinant, and mutant viruses. Chicken cells resistant to infection by subgroup E alpharetroviruses (C/E) and quail cells resistant to infection by subgroup B and D alpharetroviruses (QT6/BD) were infected with the viruses indicated by the shading of the bars and passaged at least 5 times, and RT assays were conducted to determine that infection was complete. They were then superinfected with pseudotyped egfp viruses in triplicate, and infection was scored by flow cytometry 2 days later. Three independent titers were determined for each virus for each preinfected group (and for uninfected cells). Interference was calculated as the titer on uninfected cells/titer on preinfected cells for each replicate. These values were then averaged and graphed as in Fig. 2. (A) CEF cells expressing the endogenous TVBs3; (B) QT6 cells expressing their endogenous TVBQ.
The interference patterns of mutant and wild-type viruses differed even more on QT6 cells (Fig. 3B). NTRE4 and a wild-type (WT) subgroup E virus (RAV-60) interfered well with each other, as has been demonstrated before (15, 43, 47). Neither virus was blocked at all by preinfection with L154S, and interference by LT154/155SI was a modest 10-fold. Interference of this magnitude is of questionable significance, given that variations as great as fivefold can often be seen among completely unrelated viruses. In the reciprocal challenge, we found that L154S infectivity was blocked no more than 10-fold by all of these viruses. Superinfection by LT154/155SI, on the other hand, was reduced as much as 20- to 50-fold. While this is not robust interference, these data do suggest that the subgroup E receptor may be involved in these infections in some manner, but the way in which these viruses use the receptor may be different from WT subgroup E virus. Further, given these data, we cannot rule out the potential involvement of other receptors in infection of QT6 cells by the mutant viruses.
Binding of SU immunoadhesins to avian cells.
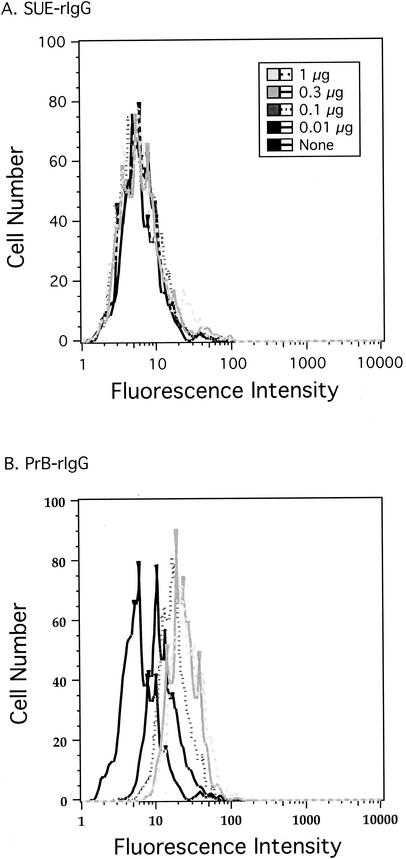
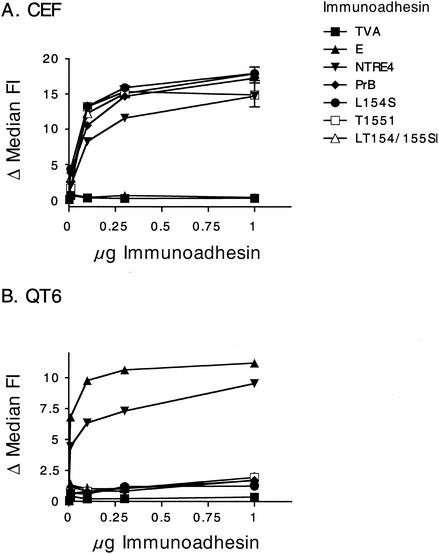
To measure the interaction of these viruses with specific cell types, we constructed a series of fusion proteins with the leader and SU region of Env fused to a rabbit immunoglobulin G (IgG) constant domain. Such constructs, known as immunoadhesins, provide a simple and reliable tool for measuring binding of retroviral Env proteins to cell surface receptors (2, 9, 23). Even though the SU domain in immunoadhesins is not in the native trimeric structure, their binding appears, in general, to accurately reflect that of the native Env protein. Binding of the immunoadhesins to cells was detected with a fluorescein-conjugated anti-rabbit IgG antibody. Receptor interaction was scored as a function of the shift in fluorescence intensity, as measured by flow cytometry. As a negative control for binding, we used TVA-rIgG, a similar immunoadhesin except with the extracellular domain of the subgroup A alpharetrovirus receptor in place of SU. This molecule only binds to cells expressing a subgroup A envelope protein. Histograms of WT subgroup E and subgroup B immunoadhesins bound to CEF are shown in Fig. 4. The fluorescence intensity of cells bound by subgroup E SU remained constant regardless of the immunoadhesin concentration (Fig. 4A). Binding of CEF by subgroup B SU immunoadhesin, on the other hand, led to a dose-dependent increase in fluorescence intensity that saturated at the two highest concentrations (Fig. 4B).
FIG. 4.
Binding of immunoadhesins to CEF. Cells were removed from plates by using EDTA (no trypsin) and bound to 0, 0.01, 0.1, 0.3, and 1 μg of purified immunoadhesin in 100 μl of PBA buffer at 4°C. They were then incubated with fluorescein-labeled secondary antibody as described in Materials and Methods and analyzed by flow cytometry. (A) Subgroup E immunoadhesin. Median fluorescence intensities were 5.69 (no immunoadhesin), 5.72 (0.01 μg), 5.73 (0.1 μg), 5.85 (0.3 μg), and 5.83 (1 μg). (B) PrB immunoadhesin. Median fluorescence intensities were 5.69 (no immunoadhesin), 11.2 (0.01 μg), 18 (0.1 μg), 22.1 (0.3 μg), and 24.5 (1 μg).
Shown in Fig. 5 are more-extensive analyses in which we examined the binding behavior of all of our immunoadhesins on QT6 and CEF cells. As expected, QT6 cells (Fig. 5B), which express a subgroup E receptor, bound both subgroup E and NTRE4 SU immunoadhesins efficiently. Binding by the control TVA was totally negative, and binding by PrB-WT and T155I was barely above background. CEF (Fig. 5A), which express the BD receptor, bound the NTRE4, PrB-WT, and T155I but not subgroup E or TVA immunoadhesins. Thus, for these viruses and cells, binding of the SU immunoadhesin exactly reflected their infectibility by the corresponding virus.
FIG. 5.
Binding of SU immunoadhesins to avian cells. Purified immunoadhesins were bound to cells in triplicate in 100 μl of PBA buffer as described in Materials and Methods. The shift in median fluorescence intensity (FI) was determined by subtracting the background median FI (no immunoadhesin) from each replicate. The graphs show the average values ± standard errors of the means. (A) CEF cells expressing the endogenous TVBs3; (B) QT6 cells expressing their endogenous TVBQ.
In contrast to the expected binding pattern obtained with the WT, T155I, and NTRE4 immunoadhesins, rather different results were obtained with L154S and LT154/155SI. Although these mutant immunoadhesins bound as efficiently as PrB to CEF, they did not bind to QT6 cells any more efficiently than PrB-WT or T155I. This result is remarkable, given that the titers of L154S and LT154/155SI were only about threefold lower on QT6 cells than on CEF. Thus, the mutation at position 154 did not appear to alter the interaction of SU with the subgroup B receptor on chicken cells, but it conferred on the virus the ability to efficiently infect QT6 cells in the absence of detectable interaction between its purified SU protein and a receptor.
Binding of SU immunoadhesins to human cells expressing defined avian receptors.
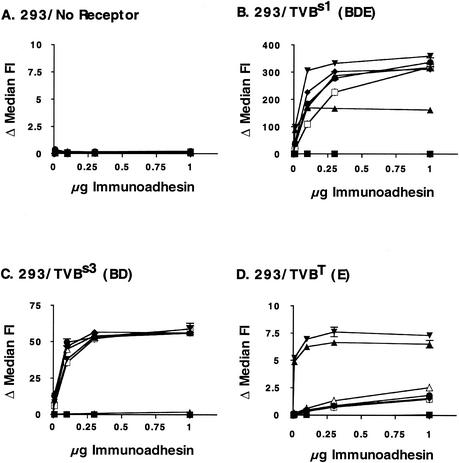
To evaluate the interaction of the Env proteins with specific ASLV receptors, we obtained a panel of 293 cell lines expressing defined avian retrovirus receptors (a generous gift from J. Young). These cells stably express TVBs1 (BDE receptor) (3), TVBs3 (BD receptor) (8), TVBT (E receptor) (2), or none of these. We bound purified SU immunoadhesin to these cells and determined the level of interaction as described in the previous section. None of the SU immunoadhesins bound to a detectable extent to 293 cells that did not express any alpharetrovirus receptors (Fig. 6A). All of them bound to cells expressing tv-bs1, in agreement with this receptor's ability to be used for infection by both subgroup B and E viruses (Fig. 6B). Interestingly, the subgroup E immunoadhesin bound well to these cells, but the fluorescence intensity leveled off at about half the value of the subgroup B derivatives and NTRE4. This result is consistent with the idea that two conformational isoforms of TVBs1 exist on the cell surface, only one of which can be recognized by subgroup E SU (1). Binding of the SU immunoadhesins to 293 cells expressing tv-bs3 (Fig. 6C) and tv-bT (Fig. 6D) recapitulated the results seen for CEF and QT6 cells. All of the immunoadhesins except for TVA and subgroup E SU bound efficiently to cells expressing the BD-specific TVBs3 receptor, while only the NTRE4 SU and subgroup E SU immunoadhesins could bind the E-specific TVBT within the limits of detection of this assay. In particular, only a very low level of binding of the extended-host-range mutants to cells expressing the subgroup E receptor was detected. Again, the L154S mutation did not enhance binding of a purified SU to the E receptor relative to that observed with PrB-WT and T155I.
FIG. 6.
Binding of SU immunoadhesins to 293 cells expressing ASLV receptors. The procedure was the same as that described for Fig. 5. (A) 293 cells, no ASLV receptors; (B) 293 cells expressing tv-bs1; (C) 293 cells expressing tv-bs3; (D) 293 cells expressing tv-bT. Immunoadhesins: ▪, TVA; ▴, E; ▾, NTRE4; ♦, PrB; •, L154S; □, T1551; ▵, LT154/155SI.
Binding of cell-surface Env protein by receptor-IgG immunoadhesins elucidates interactions between host range extension mutants and the subgroup E receptor.
We were not able to detect any significant interaction between mutant SUs and the subgroup E receptor when the receptor was located on the cell surface and the SU was supplied as part of an immunoadhesin. In this case, the SU was present as two linked monomers and the receptor was present in its native conformation on the cell surface. On virions and the surface of infected cells, on the other hand, the SU is present in the form of stable trimers (19). This trimeric conformation is not required for wild-type SU-receptor interactions as we and others have shown, but the possibility remains that it is important in the case of the host range extension mutants.
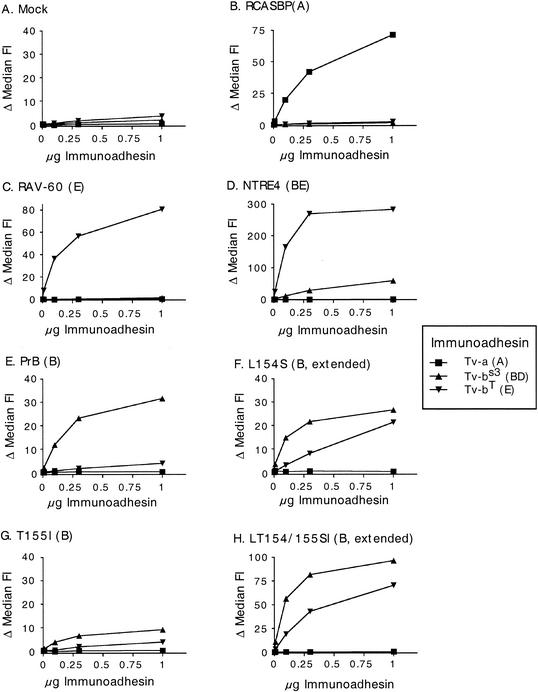
To examine the ability of receptors to bind to SU in its native form, we generated receptor-IgG immunoadhesins for TVA, TVBs3, and TVBT (a gift from J. Young). We then bound these proteins to DF1 and Q24 cells chronically infected with a panel of viruses and measured the shift in fluorescence intensity by flow cytometry. These two cell lines yielded identical results, so we have presented only one of them, Q24, in Fig. 7. Many of the interactions recapitulated the results obtained from the previous binding study. None of the immunoadhesins showed a significant interaction with mock-infected cells. TVBT bound with a higher background than the other receptor-IgGs, but the interaction was still quite low. As expected, RCASBP(A), RAV-60, and PrB envelope protein bound well to the subgroup A, E, and B receptors, respectively (Fig. 7B, C, and E). NTRE4 bound well to both receptors, but the subgroup E receptor interaction was about fivefold higher (Fig. 7D), perhaps consistent with the higher titer of this virus on QT6 cells than on CEF (Fig. 2B). T155I bound only to the subgroup B receptor, but at a very low level (Fig. 7G). This same effect was observed repeatedly, indicating that this Env protein was not expressed as efficiently on the cell surface as the others, or that less of it is competent for binding. Most striking, however, is that the subgroup E receptor immunoadhesin bound very well to cells expressing the L154S and LT154/155SI Env proteins (Fig. 7F and H). This result contrasts with that of the previous binding study, where no interaction was seen (Fig. 5 and 6), and is surprising given the interference data described in Fig. 3. The subgroup E receptor immunoadhesin exhibited a qualitatively different binding behavior on cells infected with L154S virus compared to the other interactions. Binding increased linearly throughout the range of concentrations tested and failed to saturate even at the highest concentrations tested. These characteristics may reflect a kinetics of association between this pair of proteins that is different from that of the other binding pairs. A slower association rate would account for this difference, as higher concentrations of immunoadhesin would be required to occupy all the binding sites on the cell surface. Interestingly, the different shape of the binding curve was not observed for the LT154/155SI mutant, implying that the T155I mutation in the context of the double mutant restores normal binding behavior.
FIG. 7.
Binding of receptor immunoadhesins to Q24 cells expressing alpharetrovirus Env glycoproteins. The procedure was the same as that described for Fig. 5. Q24 cells infected with the indicated viruses were bound by immunoadhesin proteins derived from TVA, TVBs3, or TVBT, and the level of binding was plotted as a function of immunoadhesin concentration. (A) Mock; (B) RCASBP(A); (C) RAV-60 (subgroup E); (D) NTRE4; (E) PrB (subgroup B); (F) L154S; (G) T155I; and (H) LT154/155SI.
Host range extension mutant viruses can infect a broad panel of hosts.
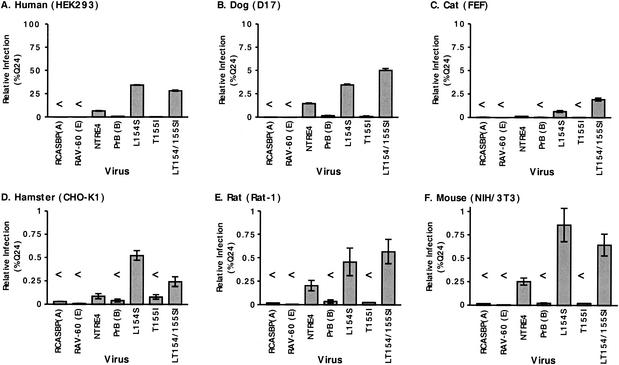
Because ASLVs of subgroup D can infect some non-avian hosts (7), we tested the ability of the subgroup B host range extension mutant viruses to infect cell lines from a diverse panel of animals. The animals tested were human (293), dog (D17), cat (FEF), mouse (NIH 3T3), hamster (CHO-K1), and rat (Rat-1). Because the ASLV LTR promoter is weak in some of these cell types, we constructed a packaging cell line that was stably transfected with an ASLV-based vector construct containing a cytomegalovirus-driven egfp flanked by two LTRs and packaging signals (similar to that shown in Fig. 2A). The diverse panel of cell types was infected 1 day after plating and scored for infection by flow cytometry 2 days later. The results (Fig. 8) indicated that all of these cell lines were infectible by NTRE4 as well as by the host range extension mutants to some degree. 293 cells could be infected almost as efficiently as avian cells. D17 and FEF cells were 1 to 2 orders of magnitude less infectible. The rodent cell lines were 2 to 3 orders of magnitude less infectible than avian cells. In all cases, infection by L154S and LT154/155SI was severalfold more efficient than by NTRE4. A low level of background infection was detected for PrB-WT and T155I in 293 and D17 cells, but this background was 30- to 50-fold lower than that for L154S and LT154/155SI infection. Expression of tv-bs1 and tv-bs3 on 293 cells increased the titers of L154S and LT154/155SI 10-fold, while expression of tv-bT had no effect (data not shown).
FIG. 8.
Host range extension mutant viruses infect a broad range of species. Infectivity is expressed as percentage of viral titer on Q24 cells (all viral stocks were approximately 105 IU/ml on these cells). (A) 293 human embryonic kidney cells; (B) D17 dog cells; (C) FEF; (D) CHO-K1 cells; (E) Rat-1 fibroblasts; and (F) NIH 3T3 mouse fibroblasts.
DISCUSSION
env genes in some retrovirus genera show remarkable variability within otherwise very similar genomes. The pattern of variability points to this region as one that is able to evolve very rapidly in comparison to the remainder of the virus, suggesting that this feature has allowed adaptation to a variety of environmental pressures in nature. While these pressures seem to disproportionately affect the env region, they do not have the same effect on all of the retroviruses characterized to date. For instance, in the case of avian (alpha-) and murine (gamma-) retroviruses, variability often reflects adaptation to a variety of receptors and hosts without greatly affecting antigenicity, while in primate lentiviruses, primary receptor usage remains constant but antigenicity and coreceptor usage are greatly modulated. In all cases, the variability is limited to well-defined regions within an otherwise conserved gene structure (6, 7, 16, 26, 31, 33, 41, 50).
Previously, our laboratory has identified env determinants of host range and cytopathogenicity in ASLV within the center third (Env residues ca. 120 to 225) of gp85 SU (16). Two regions of approximately 30 amino acids each, denoted hr1 and hr2, appear to encode the structural information necessary for receptor recognition. In further work, we identified two types of variants involving these regions that extend the host range beyond the limitations of the canonical subgroups. The first, NTRE4, is a chimeric virus that has an hr2 region from the subgroup E endogenous virus RAV-0 in an otherwise subgroup B background. Interference and infectivity studies have suggested that this envelope protein can interact with both subgroup B and E receptors (15, 16, 45, 46). The second, LT154/155SI, is a double point mutant in hr1 of gp85 SU and was isolated by repeated passage of subgroup B ALV on a mixture of CEF and QT6 cells. Like the NTRE4 recombinant, these mutations render the virus capable of infecting quail cells, and we obtained some evidence for its interaction with the subgroup E receptor (43).
In the studies reported here, we have further analyzed the basis for the extended host range of these variant viruses. We found that the NTRE4 Env protein physically associates with the BDE-specific (TVBs1), BD-specific (TVBs3), and E-specific (TVBT and, presumably, TVBQ) receptors. By contrast, SU-IgG binding and interference studies strongly implied that the L154S and double mutant viruses enter cells by a mechanism that does not involve interaction with the E-specific receptors on turkey and quail cells, although they do appear to use the BDE-specific receptor on chicken cells (Fig. 5 and 6).
Increased interaction of the mutant viruses with the subgroup E receptor could, however, be observed when their Env proteins were expressed on the cell surface and an E-specific receptor-IgG was used to detect binding (Fig. 7). The difference between the binding observed in the two cases may reflect an increased, but still low, affinity of the mutant proteins for E-specific receptors, which can only be observed as an increase in avidity when the Env protein is in the native, trimeric conformation. While this binding result may give a clue to the mechanism of entry of the host range extension mutants, the interference data, nevertheless, imply that the E receptor is not used. This point of view is bolstered by the fact that the extended-host-range viruses are capable of infecting a broad panel of cells, many of which do not express functional receptors for subgroup E virus. It must be true that either these viruses are capable of infecting cells in the absence of a specific receptor or that the receptor that they use is ubiquitous. It should be noted, however, that the interference data obtained with QT6 cells probed the interaction of proteins with quail subgroup E receptor, while receptor immunoadhesin binding studies employed the turkey subgroup E receptor. It is possible that the difference between the binding and interference results is due to a difference in these two homologous receptors. While there have been no studies to suggest that subgroup E alpharetroviruses interact with these two receptors in different ways, we cannot rule out that the host range extension mutants may only bind to the turkey subgroup E receptor. Alternatively, host range extension mutant viruses may also bind the quail subgroup E receptor but may exploit an additional route of entry in these cells, allowing infection even when robust subgroup E receptor interference is established.
By examining the point mutations separately, we identified the L154S mutation as critical for host range extension. The T155I mutation does not extend host range or allow any novel binding, but its presence in the selected LT154/155SI mutant is likely to be more than a coincidence. The rate of spread of L154S on QT6 cells is attenuated relative to LT154/155SI (data not shown). Evidence presented here suggests that the interaction between the double mutant and the subgroup E receptor may be greater than this interaction for L154S (Fig. 3B and 7H), but the effect is not large. All of these factors may have contributed to the double mutant's positive selection in the presence of QT6 cells.
Taken together, the results from this work and previous efforts (15, 16, 43, 45-47) suggest that host range extension in ASLV can develop via two nonexclusive modes. First, viruses can mutate or recombine to form determinants resulting in entry mediated by recognition of diverse receptors. Given NTRE4's high affinity for the subgroup E receptor and its ability to interfere reciprocally with a subgroup E virus, it would appear that binding is the dominant host range extension strategy employed by this virus. Second, changes in env can allow infection of diverse hosts without an evident increase in interaction with new receptors. In at least one case (the turkey subgroup E receptor), it appears that L154S and LT154/155SI can recognize another receptor, although interaction with the subgroup E receptor is probably not necessary for infection of QT6 cells, given the interference results. While it is possible that these mutant viruses recognize new receptors on all of these cell types, a simpler explanation is that the dominant mode of broad host range extension employed by this set of viruses is not strictly dependent on binding to new receptors.
This second mode of host range extension is particularly interesting because it involves mechanisms that are not yet well characterized. The SU region of retroviral envelope glycoproteins has been shown to have two distinct roles. First, it contains all of the determinants for specific receptor interaction. Second, following receptor binding it triggers a conformational change in Env protein structure and somehow signals TM to initiate events that lead to fusion (26). It is tempting to imagine that NTRE4 primarily exploits the former function of Env, while L154S and LT154/155SI primarily exploit the latter. Perhaps the mutant Env proteins are more fusogenic, and existing receptor interactions not clearly evident in the binding assays presented here are sufficient for infection by these viruses. Homologs of tv-b are present in other species (for example, DR5 in humans [3] and mice). It is possible that some of these homologs retain sufficient binding ability to render a virus with a more fusogenic envelope protein infectious. If this proves to be the case, a comparison of human and mouse DR5 could prove mechanistically informative. However, it is also possible that these envelope proteins recognize as-yet-unidentified receptors in the species that they are able to infect. For this to be true, the Env-receptor interaction must be very weak or the receptor density very low, as we did not detect any interaction between our mutant SU proteins and cells to which they have extended their host range (Fig. 5B and 6A). It is also possible that these viruses do not require any interaction with a receptor, and instead already exist in a “receptor-primed” state. These viruses would simply need to be bound to cells by nonspecific means and trafficked to a mildly acidic compartment to trigger fusion. It is also possible that these viruses do not need to bind to a primary receptor but retain the ability to interact with an as-yet-unidentified coreceptor. Mutants that can infect cells using a second signal such as coreceptor interaction, independent of the CD4 primary receptor, have been observed for human immunodeficiency virus type 1 (17, 30) and type 2 (20) and simian immunodeficiency virus (18), and this type of mechanism cannot be ruled out by the work presented here, although the putative coreceptor must be very widespread among animals.
ASLVs exhibit a striking diversity in sequence in hr1 and hr2, corresponding to the diversity in receptor usage. This diversity is most likely a consequence of selection acting on the virus due to virus-selected polymorphism in receptor genes (as exemplified by the tv-b alleles), as well as receptor blockade due to expression of endogenous proviruses (48, 49). Comparison of the different hr sequences implies that hr diversity has arisen via accumulation of point mutations, rather than by recombination (11, 12, 16, 29). Given that all wild-type ASLVs characterized to date bind their receptors with high affinity (2, 3, 8, 21, 24, 38), mutant forms like this, as well as others (7, 43, 45, 47), do not appear to be present in natural ASLV isolates. Rather, it seems more plausible that they represent intermediates in the evolutionary process by which these viruses adapt to use a new host receptor. Such viruses would be able to recognize diverse receptors and hosts and, when they find a system in which they can replicate well, could develop high-affinity binding to the receptors present as part of their adaptation to the novel host.
These considerations call into question the advantage of high-affinity receptor binding to the virus-host interaction. On the surface, it would seem advantageous to the virus to be able to infect a broad range of cells and hosts without requiring such interaction. Since such mutants must arise frequently during the course of infection of a host animal, they must confer some counterbalancing selective disadvantage to drive evolution toward the higher-affinity interaction with a specific receptor. We and others have observed at least one feature of these and similar mutants that might be relevant to this selection: they can be highly cytopathic, in at least some cell types (C. Barker, D. Negusse, G. J. A. Rainey, and J. M. Coffin, unpublished data), perhaps due to their reduced sensitivity to superinfection resistance (Fig. 3). Additionally, there may be some additional in vivo disadvantage, such as immunological effects or tissue tropism, that is not detectable in cell culture experiments.
In this work we have clarified the mechanisms of host range extension employed by viruses selected and isolated by our laboratory. Our results suggest that two modes of host range extension exist. First, viruses can broaden host range within an established repertoire of receptors. Second, viruses can extend host range by mechanisms that do not appear to involve binding of new receptors, but which make viruses more promiscuous and set up a situation in which they can explore the suitability of available hosts for replication.
Acknowledgments
We are grateful to J. A. T. Young, J. Brojatsch, and H. Adkins for cells, receptors, immunoadhesins, and helpful discussions. We thank C. Bencsics for extensive discussions and M. Bostic-Fitzgerald for administrative and editorial assistance.
This work was supported by grant R35CA 44385 from the National Cancer Institute. J.M.C. is a research professor of the American Cancer Society.
REFERENCES
- 1.Adkins, H. B., S. C. Blacklow, and J. A. Young. 2001. Two functionally distinct forms of a retroviral receptor explain the nonreciprocal receptor interference among subgroups B, D, and E avian leukosis viruses. J. Virol. 75:3520-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins, H. B., J. Brojatsch, J. Naughton, M. M. Rolls, J. M. Pesola, and J. A. Young. 1997. Identification of a cellular receptor for subgroup E avian leukosis virus. Proc. Natl. Acad. Sci. USA 94:11617-11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adkins, H. B., J. Brojatsch, and J. A. Young. 2000. Identification and characterization of a shared TNFR-related receptor for subgroup B, D, and E avian leukosis viruses reveal cysteine residues required specifically for subgroup E viral entry. J. Virol. 74:3572-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aschoff, J. M., D. Foster, and J. M. Coffin. 1999. Point mutations in the avian sarcoma/leukosis virus 3′ untranslated region result in a packaging defect. J. Virol. 73:7421-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates, P., J. A. Young, and H. E. Varmus. 1993. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74:1043-1051. [DOI] [PubMed] [Google Scholar]
- 6.Battini, J. L., J. M. Heard, and O. Danos. 1992. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J. Virol. 66:1468-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bova-Hill, C., J. C. Olsen, and R. Swanstrom. 1991. Genetic analysis of the Rous sarcoma virus subgroup D env gene: mammal tropism correlates with temperature sensitivity of gp85. J. Virol. 65:2073-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brojatsch, J., J. Naughton, M. M. Rolls, K. Zingler, and J. A. Young. 1996. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell 87:845-855. [DOI] [PubMed] [Google Scholar]
- 9.Byrn, R. A., J. Mordenti, C. Lucas, D. Smith, S. A. Marsters, J. S. Johnson, P. Cossum, S. M. Chamow, F. M. Wurm, T. Gregory, et al. 1990. Biological properties of a CD4 immunoadhesin. Nature 344:667-670. [DOI] [PubMed] [Google Scholar]
- 10.Coffin, J. M. 1996. Retroviridae: the viruses and their replication, p. 1767-1847. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Raven Press Lippincott-Raven, New York, N.Y.
- 11.Coffin, J. M., M. Champion, and F. Chabot. 1978. Nucleotide sequence relationships between the genomes of an endogenous and an exogenous avian tumor virus. J. Virol. 28:972-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coffin, J. M., P. N. Tsichlis, K. F. Conklin, A. Senior, and H. L. Robinson. 1983. Genomes of endogenous and exogenous avian retroviruses. Virology 126:51-72. [DOI] [PubMed] [Google Scholar]
- 13.Crittenden, L. B., and J. V. Motta. 1975. The role of the tvb locus in genetic resistance to RSV(RAV-O). Virology 67:327-334. [DOI] [PubMed] [Google Scholar]
- 14.Crittenden, L. B., E. J. Wendel, and J. V. Motta. 1973. Interaction of genes controlling resistance to RSV(RAV-O). Virology 52:373-384. [DOI] [PubMed] [Google Scholar]
- 15.Dorner, A. J., and J. M. Coffin. 1986. Determinants for receptor interaction and cell killing on the avian retrovirus glycoprotein gp85. Cell 45:365-374. [DOI] [PubMed] [Google Scholar]
- 16.Dorner, A. J., J. P. Stoye, and J. M. Coffin. 1985. Molecular basis of host range variation in avian retroviruses. J. Virol. 53:32-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dumonceaux, J., S. Nisole, C. Chanel, L. Quivet, A. Amara, F. Baleux, P. Briand, and U. Hazan. 1998. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J. Virol. 72:512-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edinger, A. L., J. L. Mankowski, B. J. Doranz, B. J. Margulies, B. Lee, J. Rucker, M. Sharron, T. L. Hoffman, J. F. Berson, M. C. Zink, V. M. Hirsch, J. E. Clements, and R. W. Doms. 1997. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc. Natl. Acad. Sci. USA 94:14742-14747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Einfeld, D., and E. Hunter. 1988. Oligomeric structure of a prototype retrovirus glycoprotein. Proc. Natl. Acad. Sci. USA 85:8688-8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Endres, M. J., P. R. Clapham, M. Marsh, M. Ahuja, J. D. Turner, A. McKnight, J. F. Thomas, B. Stoebenau-Haggarty, S. Choe, P. J. Vance, T. N. Wells, C. A. Power, S. S. Sutterwala, R. W. Doms, N. R. Landau, and J. A. Hoxie. 1996. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell 87:745-756. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert, J. M., P. Bates, H. E. Varmus, and J. M. White. 1994. The receptor for the subgroup A avian leukosis-sarcoma viruses binds to subgroup A but not to subgroup C envelope glycoprotein. J. Virol. 68:5623-5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Himly, M., D. N. Foster, I. Bottoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248:295-304. [DOI] [PubMed] [Google Scholar]
- 23.Holmen, S. L., and M. J. Federspiel. 2000. Selection of a subgroup A avian leukosis virus [ALV(A)] envelope resistant to soluble ALV(A) surface glycoprotein. Virology 273:364-373. [DOI] [PubMed] [Google Scholar]
- 24.Holmen, S. L., D. C. Melder, and M. J. Federspiel. 2001. Identification of key residues in subgroup A avian leukosis virus envelope determining receptor binding affinity and infectivity of cells expressing chicken or quail Tva receptor. J. Virol. 75:726-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu, S. F., M. M. Lai, and P. K. Vogt. 1978. Characterization of the env gene in avian oncoviruses by heteroduplex mapping. J. Virol. 27:667-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter, E. 1997. Viral entry and receptors, p. 71-119. In J. M. Coffin, S. H. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 27.Hunter, E., E. Hill, M. Hardwick, A. Bhown, D. E. Schwartz, and R. Tizard. 1983. Complete sequence of the Rous sarcoma virus env gene: identification of structural and functional regions of its product. J. Virol. 46:920-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jobbagy, Z., S. Garfield, L. Baptiste, M. V. Eiden, and W. B. Anderson. 2000. Subcellular redistribution of Pit-2 Pi transporter/amphotropic leukemia virus (A-MuLV) receptor in A-MuLV-infected NIH 3T3 fibroblasts: involvement in superinfection interference. J. Virol. 74:2847-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joho, R. H., M. A. Billeter, and C. Weissmann. 1975. Mapping of biological functions on RNA of avian tumor viruses: location of regions required for transformation and determination of host range. Proc. Natl. Acad. Sci. USA 72:4772-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaBranche, C. C., T. L. Hoffman, J. Romano, B. S. Haggarty, T. G. Edwards, T. J. Matthews, R. W. Doms, and J. A. Hoxie. 1999. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J. Virol. 73:10310-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacKrell, A. J., N. W. Soong, C. M. Curtis, and W. F. Anderson. 1996. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J. Virol. 70:1768-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, A. D. 1997. Development and application of retroviral vectors, p. 437-474. In J. M. Coffin, S. H. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 33.Modrow, S., B. H. Hahn, G. M. Shaw, R. C. Gallo, F. Wong-Staal, and H. Wolf. 1987. Computer-assisted analysis of envelope protein sequences of seven human immunodeficiency virus isolates: prediction of antigenic epitopes in conserved and variable regions. J. Virol. 61:570-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mothes, W., A. L. Boerger, S. Narayan, J. M. Cunningham, and J. A. Young. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103:679-689. [DOI] [PubMed] [Google Scholar]
- 35.Payne, L. N. 1992. Biology of avian retroviruses, p. 299-404. In J. A. Levy (ed.), The Retroviridae, vol. 1. Plenum Press, New York, N.Y.
- 36.Petropoulos, C. J., and S. H. Hughes. 1991. Replication-competent retrovirus vectors for the transfer and expression of gene cassettes in avian cells. J. Virol. 65:3728-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy, S. T., A. W. Stoker, and M. J. Bissell. 1991. Expression of Rous sarcoma virus-derived retroviral vectors in the avian blastoderm: potential as stable genetic markers. Proc. Natl. Acad. Sci. USA 88:10505-10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rong, L., K. Gendron, B. Strohl, R. Shenoy, R. J. Wool-Lewis, and P. Bates. 1998. Characterization of determinants for envelope binding and infection in Tva, the subgroup A avian sarcoma and leukosis virus receptor. J. Virol. 72:4552-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaefer-Klein, J., I. Givol, E. V. Barsov, J. M. Whitcomb, M. VanBrocklin, D. N. Foster, M. J. Federspiel, and S. H. Hughes. 1998. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology 248:305-311. [DOI] [PubMed] [Google Scholar]
- 40.Smith, E. J., J. Brojatsch, J. Naughton, and J. A. Young. 1998. The CAR1 gene encoding a cellular receptor specific for subgroup B and D avian leukosis viruses maps to the chicken tvb locus. J. Virol. 72:3501-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Starcich, B. R., B. H. Hahn, G. M. Shaw, P. D. McNeely, S. Modrow, H. Wolf, E. S. Parks, W. P. Parks, S. F. Josephs, R. C. Gallo, et al. 1986. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell 45:637-648. [DOI] [PubMed] [Google Scholar]
- 42.Tal, J., D. J. Fujita, S. Kawai, H. E. Varmus, and J. M. Bishop. 1977. Purification of DNA complementary to the env gene of avian sarcoma virus and analysis of relationships among the env genes of avian leukosis-sarcoma viruses. J. Virol. 21:497-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taplitz, R. A., and J. M. Coffin. 1997. Selection of an avian retrovirus mutant with extended receptor usage. J. Virol. 71:7814-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toyoshima, K., and P. K. Vogt. 1969. Enhancement and inhibition of avian sarcoma viruses by polycations and polyanions. Virology 38:414-426. [DOI] [PubMed] [Google Scholar]
- 45.Tsichlis, P. N., and J. M. Coffin. 1980. Recombinants between endogenous and exogenous avian tumor viruses: role of the C region and other portions of the genome in the control of replication and transformation. J. Virol. 33:238-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsichlis, P. N., and J. M. Coffin. 1979. Recombination between the defective component of an acute leukemia virus and Rous associated virus O, an endogenous virus of chickens. Proc. Natl. Acad. Sci. USA 76:3001-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsichlis, P. N., K. F. Conklin, and J. M. Coffin. 1980. Mutant and recombinant avian retroviruses with extended host range. Proc. Natl. Acad. Sci. USA 77:536-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss, R. A. 1993. Cellular receptors and viral glycoproteins involved in retrovirus entry, p. 1-108. In J. A. Levy (ed.), The Retroviridae, vol. 2. Plenum Press, New York, N.Y.
- 49.Weiss, R. A., and C. S. Tailor. 1995. Retrovirus receptors. Cell 82:531-533. [DOI] [PubMed] [Google Scholar]
- 50.Willey, R. L., R. A. Rutledge, S. Dias, T. Folks, T. Theodore, C. E. Buckler, and M. A. Martin. 1986. Identification of conserved and divergent domains within the envelope gene of the acquired immunodeficiency syndrome retrovirus. Proc. Natl. Acad. Sci. USA 83:5038-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young, J. A., P. Bates, and H. E. Varmus. 1993. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J. Virol. 67:1811-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zingler, K., and J. A. Young. 1996. Residue Trp-48 of Tva is critical for viral entry but not for high-affinity binding to the SU glycoprotein of subgroup A avian leukosis and sarcoma viruses. J. Virol. 70:7510-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]