Abstract
The core promoter mutants of hepatitis B virus (HBV) emerge as the dominant viral population at the late HBeAg and the anti-HBe stages of HBV infection, with the A1762T/G1764A substitutions as the hotspot mutations. The double core promoter mutations were found by many investigators to moderately enhance viral genome replication and reduce hepatitis B e antigen (HBeAg) expression. A much higher replication capacity was reported for a naturally occurring core promoter mutant implicated in the outbreak of fulminant hepatitis, which was caused by the neighboring C1766T/T1768A mutations instead. To systemically study the biological properties of naturally occurring core promoter mutants, we amplified full-length HBV genomes by PCR from sera of HBeAg+ individuals infected with genotype A. All 12 HBV genomes derived from highly viremic sera (5 × 109 to 5.7 × 109 copies of viral genome/ml) harbored wild-type core promoter sequence, whereas 37 of 43 clones from low-viremia samples (0.2 × 107 to 4.6 × 107 copies/ml) were core promoter mutants. Of the 11 wild-type genomes and 14 core promoter mutants analyzed by transfection experiments in human hepatoma cell lines, 6 core promoter mutants but none of the wild-type genomes replicated at high levels. All had 1762/1764 mutations and an additional substitution at position 1753 (T to C), at position 1766 (C to T), or both. Moreover, these HBV clones varied greatly in their ability to secrete enveloped viral particles irrespective of the presence of core promoter mutations. High-replication clones with 1762/1764/1766 or 1753/1762/1764/1766 mutations expressed very low levels of HBeAg, whereas high-replication clones with 1753/1762/1764 triple mutations expressed high levels of HBeAg. Experiments with site-directed mutants revealed that both 1762/1764/1766 and 1753/1762/1764/1766 mutations conferred significantly higher viral replication and lower HBeAg expression than 1762/1764 mutations alone, whereas the 1753/1762/1764 triple mutant displayed only mild reduction in HBeAg expression similar to the 1762/1764 mutant. Thus, core promoter mutations other than those at positions 1762 and 1764 can have major impact on viral DNA replication and HBeAg expression.
The hepatitis B virus (HBV) core gene is divided into the precore region (29 amino acid codons) and the core region (181 codons) by two in-frame initiating ATG codons. The heterogeneity at the 5′ end of the core gene transcript enables initiation of translation from either the precore or core ATG codon to express two related proteins. The major core gene transcript (pregenomic RNA) has the 5′ end downstream of the precore ATG codon and thus can express core (nucleocapsid) protein only, whereas a subset of transcript (precore mRNA) has its 5′ end located upstream of the precore region to express a longer protein form, the precursor to hepatitis B e antigen (HBeAg). Efficient translational initiation from precore ATG codon prevents core protein expression from this subset of mRNA species. Maturation of HBeAg requires two proteolytic cleavage events en route the secretory pathway. The N-terminal 19 residues of this 210-amino-acid (i.e., 29 plus 181 amino acids) protein target the nascent protein to the endoplasmic reticulum, where it is cleaved off. The C-terminal arginine rich sequence of 34 residues is removed subsequently by a furin-like protease during passage through the Golgi apparatus. Thus, the mature HBeAg protein differs from core protein by 10 extra residues at the N terminus and lacks of the C-terminal DNA-binding sequence. Formation of intramolecular disulfide bond between two cysteine residues (precore residue 26 and core residue 61) generates the unique secondary structure of HBeAg distinct from core protein (35, 53).
Although multiple copies of core protein assemble to form nucleocapsid essential for the packaging of pregenomic RNA, as well as for virion formation, HBeAg is not required for HBV replication in vitro (50). HBeAg homologue is also dispensable for infectivity of related duck and woodchuck hepatitis viruses, although it is necessary for persistent infection in woodchucks (7, 8, 43). Expression of HBeAg during perinatal infection has been proposed to promote immune tolerance (33). By sharing antigenic epitopes with the core protein, HBeAg may serve as a decoy to buffer anti-core protein immune response, which develops soon after infection. On the other hand, once the host develops an anti-HBe immune response, HBV-infected hepatocytes are destroyed through membrane-bound HBeAg. Thus, seroconversion from HBeAg to anti-HBe is usually associated with a 2-log reduction in the viremia titer. At the same time, the anti-HBe immune pressure provides a strong selective force for the emergence of viral variants that express less or no HBeAg.
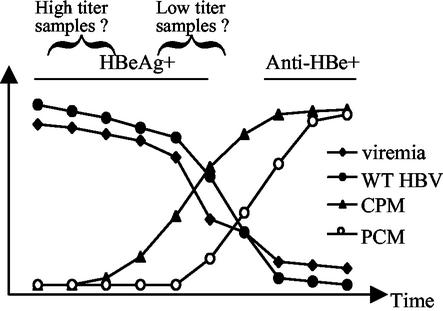
Thus far, two types of HBeAg variants have been described: the precore mutants and the core promoter mutants. HBeAg expression is abolished in the precore mutants at the translational level by nonsense or frameshift mutation, or mutated initiation codon in the precore region, a region outside the core protein coding sequence. The core promoter mutants were discovered by Okamoto et al. (37), which harbor mutations (mostly substitutions) in the basic core promoter region. These mutations are clustered within the region from positions 1750 to 1770 of the HBV genome, with an A-to-T mutation at 1762 and a G-to-A mutation at 1764 being the most common (1, 10-12, 17, 19, 23, 32, 37, 44, 48). When introduced into wild-type HBV genomes, the double mutation indeed decreased HBeAg expression and, surprisingly, also enhanced viral genome replication of about twofold (5). Later work from others has confirmed this finding (3, 34, 42). The reduction of HBeAg expression is apparently mediated by reduced precore mRNA transcription, while the mechanism of enhanced replication might be complex, involving both transcription factor binding and mutated HBx protein (25). The differential effect of the naturally occurring mutations on the transcription of precore versus pregenomic RNA is supported by the similar impact of some artificial mutations (55). Consistent with their different degrees of HBeAg downregulation, core promoter mutations are detectable at the late HBeAg phase of infection, whereas the precore mutations are found later, at the height of anti-HBe immune response (Fig. 1). However, the core promoter mutations are usually maintained after the rise of precore mutations, implying a function other than modulating HBeAg expression.
FIG. 1.
Diagram of the dynamic change of viral populations during seroconversion from HBeAg to anti-HBe. The seroconversion is accompanied by rapid drop in viremia titer. The wild-type (WT) HBV population declines and eventually disappears as a result of the selective pressure of anti-HBe immunity, whereas the core promoter mutants (CPM) and precore mutants (PCM) arise sequentially to replace the wild-type HBV. We propose that the highly viremic samples used in the present study were at an early HBeAg+ stage of infection, whereas the low-viremia samples were close to seroconversion.
Isolated analysis on the effect of two hotspot mutations in the core promoter region may have limitations, since many different combinations of core promoter mutations are present in patients (1, 10-12, 17, 19, 23, 32, 37, 44, 48). In this regard, a core promoter mutant that caused an outbreak of fulminant hepatitis replicated in human hepatoma cell lines at a level 10 times higher than did a wild-type control (18, 28). Interestingly, two less-common core promoter mutations at 1766 (C to T) and 1768 (T to A) were implicated in the high-replication phenotypes (2, 3). Considering the importance of core promoter mutants as the predominant viral species in the late stage of HBV infection, we performed comparative transfection experiments of 14 naturally occurring core promoter mutants with 11 wild-type clones and analyzed viral replication, virion secretion, and expression of HBeAg and HBsAg.
MATERIALS AND METHODS
Serum samples and PCR.
Serum samples were collected retrospectively from 13 French patients positive for HBeAg (based on the Abbott assay). Six individuals had ca. 5 × 109 copies of genome/ml, and seven others had ca. 0.2 × 107 to 4.6 × 107 copies/ml as determined by branched DNA assay (Bayer Diagnostics). The alanine aminotransferase levels were not significantly different between the two groups (Table 1). When rechecked with the enzyme immunoassay kit from Diasorin and 40-μl serum samples, the HBeAg titer was found to be low for the sample derived from patient 4 and borderline positive for the serum from patient 8 (Table 1). The serum samples were diluted in TEN buffer (10 mM Tris [pH 8.0], 1 mM EDTA, 100 mM NaCl) and digested at 37°C for 2 h with proteinase K (0.5 mg/ml) in the presence of sodium dodecyl sulfate (SDS; 0.5%). DNA was extracted with phenol-chloroform-isoamyl alcohol (25:24:1) and precipitated with ethanol. Purified DNA was resuspended in water. Full-length HBV genomes were amplified according to the method of Gunther et al. (16). For efficient directional cloning of the PCR products, we modified sense primer into 5′-CCGGAAAGCTTATGCTCTTCTTTTTCACCTCTGCCTAATCATC-3 (the HindIII site is underlined) and antisense primer into 5′-CCGGAGAGCTCATGCTCTTCAAAAAGTTGCATGGTGCTGGTG-3′ (the SacI site is underlined). Forty cycles of amplification were performed with the Expand high-fidelity PCR system (Roche).
TABLE 1.
Correlation between viremia titer and prevalence of core promoter mutantsa
| Patient no. | Viremia level (106/ml) | HBeAgb titer | Transaminase (ULN) | No. of clones with CPM/total no. of clones tested |
|---|---|---|---|---|
| 1 | 7 | 15,956 | 1.54 | 7/8 |
| 2 | 33 | 12,891 | 2.12 | 2/3 |
| 3 | 2 | ND | 1.54 | 2/5 |
| 4 | 43 | 528 | 1.5 | 6/6 |
| 8 | 32 | 97 | 0.82 | 6/7 |
| 9 | 46 | 16,739 | 2.54 | 11/11 |
| 14 | 22 | 17,824 | ND | 3/3 |
| 5 | 5,700 | 15,585 | 1.33 | 0/2 |
| 6 | 5,700 | 14,611 | 1.17 | 0/2 |
| 7 | 5,700 | 12,452 | 3.12 | 0/2 |
| 11 | 4,960 | 20,648 | 2.33 | 0/2 |
| 12 | 5,700 | 13,468 | ND | 0/2 |
| 13 | 5,700 | 15,243 | 1.8 | 0/2 |
Abbreviations: CPM, core promoter mutations; ULN, upper limit of normal levels; ND, not determined.
The value for the negative control was 150.
Cloning and dimer construction.
The PCR products were cloned into the HindIII/SacI sites of pUC18 vector. For HBV transcription to proceed under the control of endogenous core promoter, tandem dimers were made. The HBV insert was released from pUC18 vector by digestion with SapI and BglI, circularized with T4 DNA ligase, and relinearized with EcoRI. Next, HBV DNA with EcoRI ends were ligated to EcoRI-digested, dephosphorylated pUC18 DNA at an insert/vector ratio of at least 10:1. After transformation, colonies harboring tandem HBV dimers were screened with an oligonucleotide probe spanning the tail-to-head junction as described previously (51). The oligonucleotide has the sequence 5′-GGCCATGCAGTGGAATTCCACWRCYTTCCA-3′ (W = A + T; R = A + G; Y = C + T).
Site-directed mutants.
Mutations in the core promoter region were introduced by overlap extension PCR, by using Expand high-fidelity PCR system and fewer than 25 PCR cycles. The PCR products were digested with RsrII and ApaI restriction enzymes to exchange with the cognate fragment in clone 2A. The entire PCR-derived fragment was sequenced to guarantee the lack of unwanted mutations. Tandem dimeric version of the mutants was prepared.
Transfection and detection of HBV replication.
Dimeric HBV DNA was transfected into the Huh7 and HepG2 human hepatoma cells by a calcium phosphate transfection kit (5 Primer-3 Primer or Promega), with 15 μg of HBV dimer per 10-cm dish or 8 μg per 6-cm dish. After incubation of the cells with DNA precipitates for 4 to 6 h, cells were washed and replaced with fresh medium. Cells were harvested by trypsin or scrapping at 5 days posttransfection with no interim medium change, and culture supernatant was collected. HBV core particles were obtained from cell lysate and treated with nuclease as described previously (52). Cells were lysed with 300 μl of buffer containing 10 mM HEPES (pH 7.5), 100 mM NaCl, 1 mM EDTA, and 1% NP-40. The lysate was supplemented with 10 mM CaCl2-12 mM MgCl2 and digested at 37°C for 15 min with DNase I (1.5 U) and mung bean nuclease (20 U) to degrade transfected HBV DNA. Core particles were precipitated with 110 μl of polyethylene glycol (PEG) solution (1.2 M NaCl, 60 mM EDTA, 30% sucrose, 26% PEG), resuspended in 100 μl of solution containing 10 mM Tris (pH 7.5)-6 mM MgCl2-8 mM CaCl2, and digested with 1.5 U of DNase I and 2.5 U of mung bean nuclease at 37°C for 10 min to further degrade transfected DNA. After the addition of 270 μl of protease digestion buffer (25 mM Tris [pH 7.5], 10 mM EDTA, 100 mM NaCl, 0.5% SDS), samples were digested at 37°C for 2 h with proteinase K (0.5 mg/ml). DNA was extracted with phenol and precipitated with ethanol. Purified DNA was subjected to Southern blot analysis with a highly pure full-length HBV probe (obtained by two rounds of PCR amplification). Gel electrophoresis was either performed in the absence of ethidium bromide or in its presence both in the gel and in the running buffer, which greatly accelerated migration of the single-stranded HBV genome.
Strand-specific HBV probes.
Single-stranded RNA probes were used to confirm the nature of HBV genomes associated with intracellular core particles and extracellular particles. We have recently sequentially subcloned a 2.2-kb EcoRV-EcoRI fragment and a 2.6-kb EcoRI-ApaI fragment (with ApaI end flushed) of clone 3.4 into the SmaI-XhoI (flushed) sites of pBluescript vector for studies on virion secretion. This subclone was digested to completion with EcoRI and then transcribed with T7 polymerase to generate 2.6-kb negative-stranded riboprobe or with T3 polymerase to generate 2.2-kb positive-stranded RNA probe (Riboprobe In Vitro Transcription Systems; Promega), with [α-32P]CTP (3,000 Ci/mmol) as a radioactive tracer. Template DNA was subsequently degraded by RNase-free DNase, and the probe was purified by phenol-chloroform extraction and Sephadex G-50 column chromatography. Diethyl pyrocarbonate-treated solutions were used during hybridization and washing steps.
Analysis of extracellular viral particles.
Culture supernatant was prespun at 4,000 rpm for 10 min to remove cell debries and loaded on top of 10 and 20% sucrose (in TEN buffer). Samples were spun at 39,000 rpm for 18 h with a Sorvall SW41 rotor to pellet extracellular particles. It should be noted that this procedure collects both Dane particles and naked core particles. Transfected HBV DNA was digested away at 37°C for 15 min by 1 U of DNase I and 1.5 U of mung bean nuclease in the presence of 8 mM CaCl2 and 6 mM MgCl2. The proteinase K digestion buffer (270 μl) was added, and samples were digested at 37°C for 2 h with proteinase K (0.5 mg/ml). DNA was extracted with phenol and precipitated with ethanol in the presence of 20 μg of glycogen as a carrier. HBV DNA was detected by Southern blot. Gel electrophoresis was performed in the presence or absence of ethidium bromide.
Further separation of Dane particles from naked core particles was achieved by sedimentation through CsCl gradient as previously described (50). After concentration of viral particles from culture supernatant through a 10 and 20% sucrose cushion, particles in the pellet fraction were resuspended in 4.5 ml of TEN solution. CsCl (1.5 g) was added and dissolved, and samples were spun at 12°C and 46,000 rpm for 48 h or longer in a Sorvall SW65 rotor. Fractions of 400 μl were taken from the top and weighed to obtain density values. After dialysis against TEN buffer, an aliquot from each fraction (10 to 15 μl) was used for HBsAg measurement, and the remaining solution was used for DNA extraction. Each sample was supplemented with 8 mM CaCl2 and 6 mM MgCl2 and digested with 1 U of DNase I and 1.5 U of mung bean nuclease at 37°C for 10 min. Tris (pH 7.5), EDTA, and SDS were added to final concentrations of 25 mM, 10 mM, and 0.5%, respectively, and proteins were digested with 0.5 mg of proteinase K/ml. After phenol extraction and ethanol precipitation, DNA was analyzed in Southern blot.
HBsAg and HBeAg.
The HBsAg secreted into the culture supernatant was measured by using the Auszyme kit from Abbott Laboratories. Secreted HBeAg was detected by the enzyme immunoassay kit from Diasorin. The samples were diluted four times or more to avoid signal saturation.
RESULTS
Experimental approaches.
Our original goal was to identify naturally occurring HBV genomes with high- and low-replication capacities, with the long-term prospect of mapping the trans- or cis-acting elements that regulate viral replication levels. HBeAg+ samples were selected, and the full-length HBV genomes were amplified. Assuming a positive correlation between viral titers and intrinsic replication capacities of the viral strains, we studied samples with either highest or lowest viremia levels (Table 1). The patients infected with the same genotype were selected so as to avoid potential genotypic difference in replication capacity. Genotype A was chosen since it is the most common HBV genotype in HBeAg+ patients in France (15, 26). The limited sequence variability within this particular genotype (usually <2%) would simplify the subsequent identification of underlying mutations. The patients were not under antiviral therapy at the time of sample collection. Although HBV replication can proceed (to various extents) from different DNA constructs, including vector-free circularized or linear genome (16), vector-linked tandem dimer (52), and a DNA copy of pregenomic RNA under foreign (cytomegalovirus) promoter, we selected to pursue the tandem dimer approach. Such a DNA form allows HBV replication to proceed under the endogenous core promoter rather than an artificial cytomegalovirus promoter and thus permits the expression of HBeAg. This turned out to be judicious, since all of the high-replication genomes identified in the present study are core promoter mutants. Core promoter mutations modulate HBV replication and HBeAg expression at the transcriptional level (5, 34, 42).
Due to the quantitative nature of the work, measures were taken to ensure the accuracy of the results. HBV dimers were purified with the same plasmid DNA purification kit, and DNA concentrations determined by a spectrometer were validated by gel electrophoresis after HindIII digestion. Transfection efficiency was monitored by cotransfection with genes encoding glycine decarboxylase (27) or luciferase. Each panel of constructs was tested at least three times, and similar results were obtained. For site-directed core promoter mutants derived from clone 2A, levels of HBsAg expression (which is not expected to be modulated by core promoter mutations) were used as an internal control for the transfection efficiency.
Frequent isolation of core promoter mutants from low-viremia sera positive for HBeAg.
We initially sequenced the core promoter region of a total of 12 full-length HBV genomes derived from six patients: three (patients 5, 6, and 7) with high levels (5.7 × 109 copies/ml) and three (patients 2, 3, and 4) with low levels of viremia (0.2 × 107 to 4 × 107 copies/ml) (Table 1). Of the six clones derived from low-viremia sera (2A, 2B, 3.4, 3.10, 4B, and 4D), all but 2A and 3.10 had core promoter mutations (Fig. 2). In contrast, none of the six clones derived from highly viremic serum (5.2, 5.4, 6.1, 6.2, 7.2, and 7.4) had core promoter mutations. The prevalence of core promoter mutants in less-viremic samples suggests that these patients may be close to seroconversion to anti-HBe, which is associated with both the emergence of core promoter mutants and a decrease in viremia (Fig. 1). To substantiate this finding, we sequenced 8 more clones from patients 2 to 4, 29 clones from four more low-viremia samples (patients 1, 8, 9, and 14), and 6 clones from three additional high-viremic patients (patients 11, 12, and 13) (Table 1). All six new clones from highly viremic samples also possessed a wild-type sequence in the core promoter, whereas 33 of 37 genomes from low-titer samples had core promoter mutations (Table 1 and Fig. 2). Taken together, all 12 clones derived from six highly viremic sera were of the wild-type sequence, whereas 37 of 43 clones derivd from the seven sera with low viremia levels had core promoter mutations.
FIG. 2.
Core promoter sequences found in HBV clones derived from HBeAg+ serum samples with high- or low-viremia titers. (A) Sequence patterns at 1751 to 1777 of the basic core promoter region. Pattern 1 represents the wild-type sequence. Hyphens indicate a lack of nucleotides at these positions. Patterns 2 to 8 contained only substitutions. Insertions were present in patterns 9 to 11, and deletions occurred in sequences under patterns 12 and 13. The most common substitutions at 1762 and 1764 are marked by asterisks. (B) HBV clones with such divergent sequence patterns. For each clone, the number at the beginning identifies the patient (clone 1.2 was derived from patient 1, whereas clone 2A was from patient 2). It is evident that dominant viral populations from patients 1, 4, and 9 had patterns 3, 4, and 7, respectively.
Some core promoter mutants have very high replication potential.
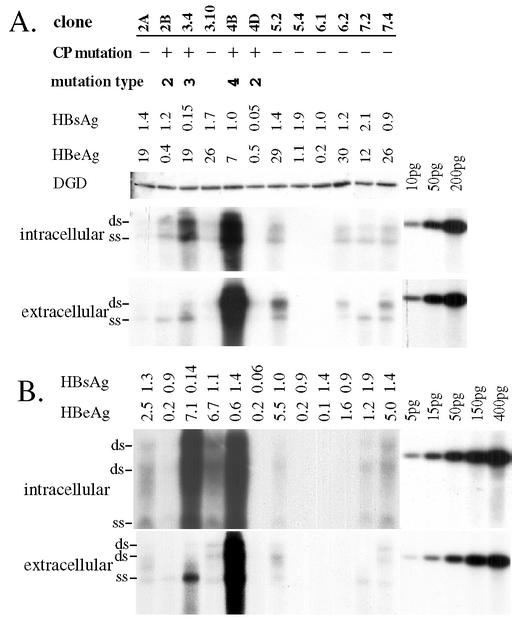
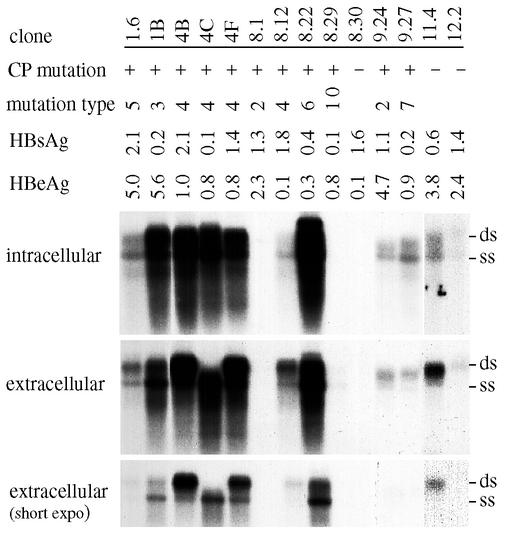
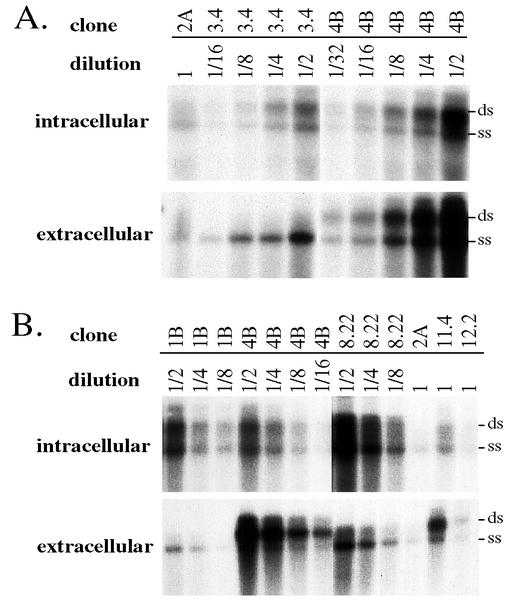
We initially tested 12 HBV genomes (4 core promoter mutants and 8 wild-type clones) for replication capacity in transfected Huh7 cells. Three clones (4D, 5.4, and 6.1) failed to replicate or to express HBeAg (Fig. 3A), and sequencing of the core gene revealed deletion of a single nucleotide (2088 for 5.4 and 6.1; 2324 for 4D). Clone 4B and, to a lesser extent, clone 3.4 replicated at much higher levels than the other genomes (Fig. 3A, panel of intracellular particles). Similar results were obtained in repeat experiments (data not shown) and in another HCC cell line, HepG2 (Fig. 3B, panel of intracellular particles). Both clones contained the 1762T 1764A common core promoter mutations. In addition, clone 3.4 harbored the 1753C mutation, whereas clone 4B had 1753C and 1766T mutations. To correlate the replication capacity with the type of core promoter mutations, we prepared 10 additional tandem dimers of core promoter mutants and 3 dimers of wild-type clones. The 10 core promoter mutants covered seven different types of core promoter mutations (Fig. 2 and 4). Four additional highly replicating genomes were identified: 1B, 4C, 4F, and 8.22 (Fig. 4, panels of intracellular particles). Of these, 1B contained the same triple core promoter mutations as clone 3.4 (1753C 1762T 1764A), 4C and 4F had the same four point mutations (1753C 1762T 1764A 1766T) as clone 4B derived from the same individual, and 8.22 harbored 1762T 1764A 1766T triple mutations (three of the four substitutions as observed in 4B). Clone 8.22 also had two additional point mutations further upstream: T1636G/C1678T. A quantitative analysis in Huh7 cells revealed that 4B replicated >8-fold higher than 2A, a genome with wild-type core promoter sequence, whereas 3.4 replicated at least 4-fold more efficiently than 2A (Fig. 5A, panel of core particles). In another experiment involving 4B, 1B, and 8.22, the replication capacities of clone 1B and 4B were estimated to be eightfold higher than for clone 11.4 and even higher than for clone 12.2 (Fig. 5B). Furthermore, clone 8.22 replicated at an even higher level. Both clones 11.4 and 12.2 had a wild-type core promoter sequence.
FIG. 3.
Transfection of 12 HBV genomes in Huh7 cells (A) and HepG2 cells (B). Cells grown in 10-cm dishes were transfected with 15 μg of HBV dimer and 15 μg of duck glycine decarboxylase (DGD) cDNA (24) and then harvested 5 days later. From the cell lysates, duck glycine decarboxylase expression was determined by Western blotting, and HBV DNA replication was detected by Southern blot from core particles. From the culture supernatant, HBsAg (shown as the optical density at 490 nm) and HBeAg (shown as counts per minute [103]) were measured by commercial kits after 1:4 and 1:5 dilution of samples, respectively. Extracellular viral particles (both Dane particles and naked core particles) were concentrated by ultracentrifugation through sucrose cushions and analyzed by Southern blotting. Various amounts of 3.2-kb linear HBV DNA were run in parallel as a size marker and for quantification. CP mutation, core promoter mutations; ds, double-stranded viral genome; ss, single-stranded viral genome. Gels were run in the absence of ethidium bromide (A) or in its presence (B). The two “ds” bands in panel B may represent relaxed circular DNA and duplex linear DNA, respectively, whereas the “ds” band in panel A appears to be duplex linear.
FIG. 4.
Transfection of 14 HBV genomes in Huh7 cells. Cells were harvested at day 5 posttransfection. For released viral particles, a short exposure is also given to better visualize the bands corresponding to single- and double-stranded viral genomes. ds,: double- stranded viral genome; ss, single-stranded viral genome.
FIG. 5.
Quantitative analysis of the relative replication and secretion capacities of different HBV clones. (A) Comparison of clones 2A, 3.4, and 4B. (B) Comparison of clones 1B, 2A, 4B, 8.22, 11.4, and 12.2. Huh7 cells were transfected with various HBV constructs and harvested at day 5 posttransfection. HBV DNA from intracellular core particles and extracellular viral particles were analyzed, with the agarose gels run in the absence of ethidium bromide. The entire samples of 2A, 11.4, and 12.2 were loaded into single wells, while serially diluted samples of 1B, 3.4, 4B, and 8.22 were loaded into separate lanes. ds, double-stranded viral genome; ss, single-stranded viral genome. Please note that clones 3.4 and 1B and, to a lesser extent, clone 8.22, released viral particles with predominantly single-stranded viral genome despite a similar ratio of the two DNA forms inside intracellular core particles.
Core promoter mutants vary greatly in their rates of virion secretion.
HBV DNA replication is asymmetric. The negative strand is synthesized first from the pregenomic RNA template and, after degradation of the RNA template, positive-strand DNA is synthesized by using the negative-strand DNA as a template (13, 47). Only core particles containing such double-stranded viral genome are enveloped and secreted into serum (13), although human hepatoma cell lines transfected with HBV DNA may release naked core particles containing single-stranded genome (14, 54, 57). Parallel to the detection of replicating viral DNA within transfected cells, we concentrated extracellular viral particles (both enveloped virus particles and naked core particles) by ultracentrifugation and analyzed particle-associated HBV DNA by Southern blot. Clone 4B was found to release many more viral particles than did other HBV genomes, especially clone 3.4, by comparing the relative intensities of extratracellular viral DNA with intracellular viral DNA (Fig. 3A). This point is better demonstrated by serial dilution of both intracellular and extracellular viral DNA derived from clones 3.4 and 4B (Fig. 5A). Similar degrees of difference can be found between 4B and two other high-replication clones, 1B and 8.22 (Fig. 5B). On the other hand, two wild-type genomes with low replication capacities displayed elevated extracellular viral DNA signals compared to most other clones (clone 5.2 in Fig. 3 and clone 11.4 in Fig. 4 and 5). Therefore, release of particles to culture medium did not correlate with core promoter mutations (Table 2).
TABLE 2.
Properties of some naturally occurring genotype A strains
| Clone | Core promoter pattern | DNA replication | Virion secretiona | HBeAg expression | HBsAg expression | Full-length sequence |
|---|---|---|---|---|---|---|
| 2A | 1 | + | + | ++++ | ++++ | Yes |
| 5.2 | 1 | + | ++ | ++++ | ++++ | No |
| 7.2 | 1 | + | +/− | ++++ | ++++ | No |
| 11.4 | 1 | + | +++ | ++++ | +++ | No |
| 9.24b | 2 | + | + | ++++ | ++ | No |
| 1B/3.4 | 3 | +++ | +/− | ++++c | +/− | Yes |
| 4B | 4 | ++++ | +++ | ++ | ++++ | Yes |
| 4C | 4 | ++++ | +/− | ++ | − | No |
| 8.12 | 4 | + | +++ | −d | ++++ | No |
| 1.6 | 5 | + | + | ++++ | ++++ | No |
| 8.22 | 6 | ++++ | + | + | + | Yes |
| 9.27b | 7 | + | + | ++ | +/− | No |
Based on the intensity the of extracellular double-stranded DNA relative to intracellular double-stranded DNA.
South African subgroup of genotype A, with precore codons 17 and 25 mutated to TTT and GGA, respectively.
Even higher in HepG2 cells.
Contains a TAG nonsense mutation at precore codon 28.
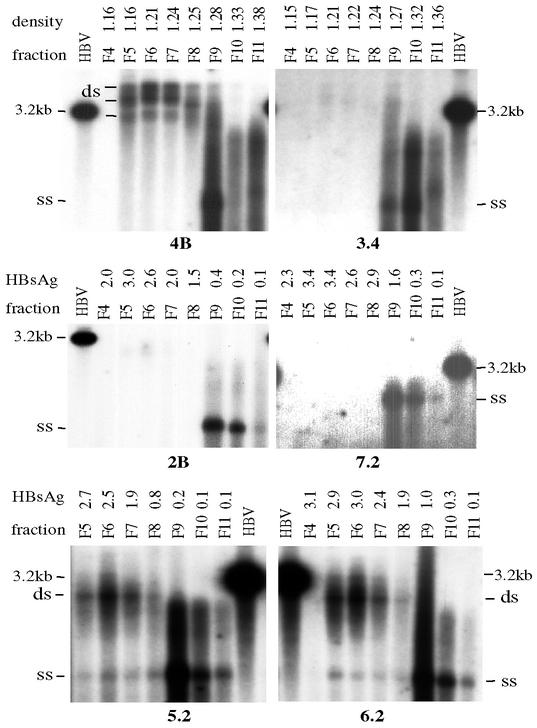
Ultracentrifugation through 10 to 20% sucrose will pellet down both Dane particles and naked core particles. Surprisingly, extracellular particles produced from clones 1B, 2B, 3.4, 4C, 7.2, and 8.22 displayed mainly the single-stranded form, whereas both DNA forms were produced by many other clones (Fig. 3 to 5). Since naked core particles released by human hepatoma cell lines usually contain single-stranded viral genome (14, 54, 57), we separated core particles from Dane particles through CsCl gradient. Indeed, clones such as 4B, 5.2, and 6.2 released two types of particles, with a double-stranded genome inside lighter Dane particles and a single-stranded genome inside heavier core particles (Fig. 6). In contrast, clones 2B, 3.4, and 7.2, which had primarily singled-stranded DNA in the culture supernatant (Fig. 3), released essentially core particles with single-stranded DNA (Fig. 6). Since the release of naked core particles with single-stranded genome is an artifact of hepatoma cells, our results suggest high virion secretion efficiency of clones 4B and 5.2 and much reduced virion secretion by clones 1B, 2B, 3.4, 7.2, and 8.22.
FIG. 6.
CsCl gradient separation of naked core particles from Dane particles. Culture supernatants from 10-cm dishes of Huh7 or HepG2 cells were subjected to ultracentrifugation in 10 to 20% sucrose gradient to pellet the viral particles, which were further spun in CsCl gradient. Fractions of 400 μl were collected, dialyzed, and DNA extracted for Southern blot analysis with an HBV DNA probe (for samples with a high-secretion phenotype, only fractions of DNA samples were used for Southern blotting). The results shown for clones 3.4 and 4B were from an experiment in HepG2 cells, whereas data for the other clones were obtained from transfected Huh7 cells. Except for clone 7.2, gels were run in the presence of ethidium bromide to better separate the single-stranded genome from the double-stranded genome. For clones 3.4 and 4B, the density values of the CsCl fractions were determined. The HBsAg values were measured for CsCl fractions of other samples. Note that clones 3.4, 7.2, and 2B secreted very few Dane particles. HBV, 3.2-kb HBV genome; ss, single-stranded viral genome.
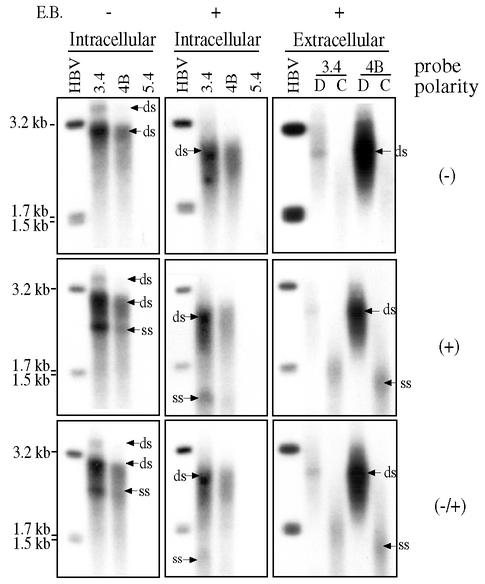
The two major HBV DNA bands were closely spaced if separated in agarose gels lacking ethidium bromide (compare panels A and B in Fig. 3; clone 7.2 against other clones in Fig. 6). To confirm the fast-migrating band as negative-stranded HBV genome, we separated intracellular and extracellular viral DNA of clones 4B and 3.4 in triplicate and hybridized strips of blots with negative-stranded riboprobe, positive-stranded riboprobe, and double-stranded DNA probe, respectively. The extracellular viral particles were resolved into naked core particles and Dane particles by CsCl gradient. Linear HBV DNA of 3.2, 1.7, and 1.5 kb were included as size markers. Considering that the positive strand is present only in double-stranded genomes, the negative-stranded probe should hybridize to double-stranded DNA only. On the other hand, the positive-stranded riboprobe and the double-stranded DNA probe will hybridize to both DNA species. This was indeed the case (Fig. 7). The single-stranded DNA migrated much slowly in the absence of ethidium bromide (to a position above 2 kb) than in its presence (to <1.5 kb). The double-stranded DNA was primarily linear (ca. 3.2 kb) in Huh7 cells, whereas more relaxed circular DNA (running at around 4kb position) was generated in HepG2 cells (Fig. 3B versus A; clone 4B in Fig. 6 versus right panels of Fig. 7). Whether derived from HBV 3.4 or 4B, extracellular naked core particles contained primarily negative-stranded HBV DNA, whereas Dane particles had only double-stranded DNA (Fig. 7). This is consistent with the presence of a “maturation signal” for virion formation and secretion (47).
FIG. 7.
Detection of intracellular and extracellular HBV DNA by strand-specific probes. Huh7 cells were transfected with HBV clones 3.4, 4B, and 5.4 (as a negative control). DNA was isolated from intracellular core particles, triplicate DNA samples were separated in agarose gels in the presence or absence of ethidium bromide as indicated, and hybridized with one of the three probes. Extracellular viral particles were separated by CsCl gradient, and fractions with densities of Dane particles (D) and core particles (C) were pooled. DNA was separated in an agarose gel in the presence of ethidium bromide and hybridized with one of the three probes. HBV DNA linearized with EcoRI (3.2 kb) and EcoRI plus RsrII (1.7 plus 1.5 kb) were run in parallel as size markers. More DNA samples of clone 3.4 were added than 4B. ds, double stranded; ss, single stranded.
Sustained HBeAg expression by some core promoter mutants.
Besides viral DNA detection, we also measured secretion of HBsAg and HBeAg to culture medium. Several clones, e.g., 4C, 4D, 8.29, 14.3, 1B, 3.4, and 9.27, expressed no or little HBsAg (Fig. 3 and 4). Clones 4D, 5.4, 6.1, and 8.30 were defective in HBeAg expression (Fig. 3 and 4) owing to a frameshift mutation in the core gene, whereas the lack of HBeAg expression by clones 2B and 8.12 was attributable to a precore region deletion (at position 1827) and a TAG nonsense mutation (codon 28), respectively. Several core promoter mutants (8.22, 4B, 4C, 4F, 8.29, and 9.27) expressed very low levels of HBeAg (Fig. 3 and 4). However, clones 1B and 3.4 and, to a lesser extent, clones 1.6 and 9.24 expressed high levels of HBeAg (Fig. 3 and 4 and Table 2). Clones derived from patient 9 belonged to the South African subgroup of genotype A as previously described (4).
Limited sequence divergence among HBV genomes with different replication and secretion phenotypes.
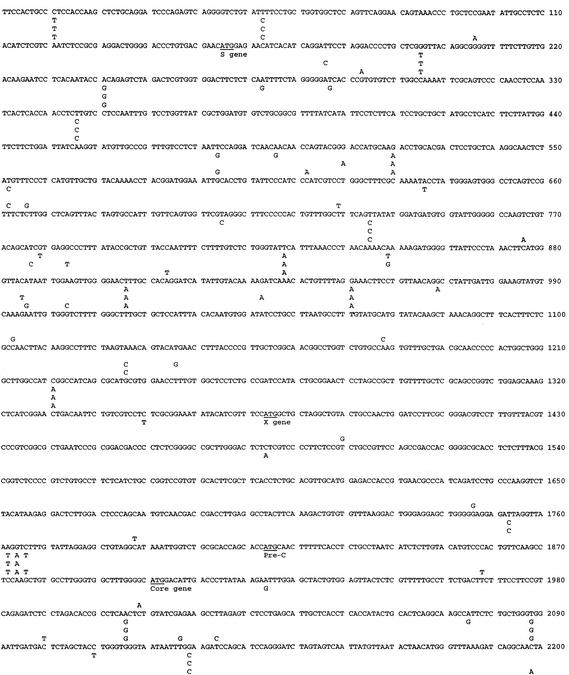
As the initial step toward understanding the structural basis for the diverse replication, secretion, and HBeAg expression phenotypes, we determined the complete nucleotide sequences of five HBV genomes. Clone 2A has a wild-type core promoter sequence and replicated at a low level. In contrast, clones 1B, 3.4, 4B, and 8.22 are core promoter mutants with high replication capacities. Among these, clones 1B and 3.4 have low-virion-secretion and high-HBeAg-expression phenotypes; 4B has high-virion-secretion and low-HBeAg-expression phenotypes; 8.22 has low-secretion and low-HBeAg phenotypes. All five clones contained a genome size of 3,221 nucleotides characteristic of genotype A (Fig. 8). Sequence differences were only 36 nucleotides (1.1%) between clones 2A and 3.4, 46 nucleotides (1.4%) between clones 2A and 4B, 54 nucleotides (1.7%) between clones 2A and 8.22, 43 nucleotides (1.3%) between clones 3.4 and 4B, 50 nucleotides (1.6%) between clones 3.4 and 8.22, and 56 nucleotides (1.7%) between clones 4B and 8.22. Clone 1B turned out to have the same sequence as clone 3.4. These two clones have mutated pre-S2 initiation codon and will not express the middle envelope protein.
FIG. 8.
Comparison of the complete nucleotide sequences of HBV clones 2A (top), 4B (second), 3.4 (third), and 8.22 (bottom). These sequences are available in GenBank under accession numbers AF536524, AF537372, AF537371, and AY152726. The translational start sites for envelope, core, HBx, and polymerase protein are indicated.
Cumulative effect of core promoter mutations on viral replication and HBeAg expression.
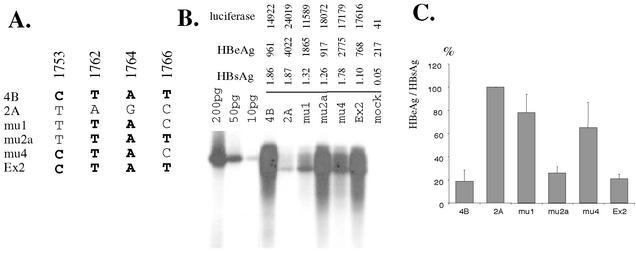
In order to determine the extent to which different core promoter mutations contribute to the different replication and HBeAg expression phenotypes of naturally occurring mutants, we generated four site-directed mutants from clone 2A (Fig. 9A). After transfection into Huh7 cells, viral DNA replication and HBeAg expression were determined. Since the mutations are not expected to affect HBsAg expression, we used HBeAg/HBsAg ratios (after subtraction of values from nontransfected cells) to accurately determine the effect of various core promoter mutations on HBeAg expression. Results from seven independent experiments revealed a moderate effect of 1762/1764 hotspot mutations on HBeAg expression (mu1). The expression of HBeAg was further reduced by the addition of the 1753 mutation (mu4) and, more potently, by the 1766 mutation (mu2a). Introduction of all four point mutations (Ex2) reduced HBeAg expression to the level of 4B, the naturally occurring core promoter mutant (Fig. 9C). Similarly, viral DNA replication was enhanced by the hotspot mutations (mu1), augmented by the 1753/1762/1764 mutations (mu4) and, more effectively, by the 1762/1764/1766 mutations (mu2a) or the four point mutations together (Ex2) (Fig. 9B).
FIG. 9.
Effect of different combinations of core promoter mutations on HBV genome replication and HBeAg expression. (A) Site-directed mutants. Clone 4B contains four point mutations in the core promoter compared with clone 2A. Mu1, mu2a, mu4, and Ex2 are clone 2A-based site-directed mutants containing different mutation combinations. Mutated nucleotides are shown in boldface. (B) Results from one transfection experiment. Huh7 cells grown in 6-cm dishes were transfected with 10 μg of HBV DNA and 2 μg of luciferase plasmid and then harvested 5 days later. The values of luciferase were determined from cell lysate. HBsAg and HBeAg values were determined from culture supernatant. (C) HBeAg expression profiles of the mutants. We used HBeAg/HBsAg ratios to minimize the effect of variation in transfection efficiency. The graph is based on results from seven independent experiments in Huh7 cells. Mu2a and Ex2 displayed highest degree of viral DNA replication and the lowest level of HBeAg expression. Their replication and HBeAg expression phenotypes are close to those of the naturally occurring core promoter mutant, clone 4B.
DISCUSSION
The core promoter mutants are the predominant viral species at the late HBeAg+ phase and the anti-HBe stage of infection. Characterization of their biological and pathogenic properties is essential for therapeutic interventions. Extensive clinical studies have attempted to correlate core promoter mutants with HBeAg status or level, viremia titer, liver diseases, and response to antiviral therapy (1, 6, 11, 19, 20, 23, 30). Due to the presence of viral quasispecies, the complexity of the host immune system, and the difficulty of assessing the cause-effect relationship, conflicting results have been reported. In this regard, functional characterization of the core promoter mutants in human hepatoma cell lines permits the evaluation of their biological properties. Up to now, most investigators have taken the approach of introducing the core promoter mutations into the background of a wild-type genome and have focused on the 1762T 1764A hotspot mutations (5, 34, 42, 49, 56). These mutations reduce HBeAg expression and slightly increase viral DNA replication. In contrast, a naturally occurring core promoter mutant associated with fulminant hepatitis outbreak displayed a 10-fold-higher replication capacity than the wild-type clone, as a result of 1766T 1768A core promoter mutations, rather than the common 1762T 1764A mutations (2, 3, 18, 28). The present study sought to characterize large number of naturally occurring core promoter mutants, together with naturally occurring wild-type isolates. A complex pattern of viral DNA replication, viral secretion, and HBeAg and HBsAg expression was observed for these core promoter mutants (see Table 2 for summary).
Viremia titer and prevalence of core promoter mutants.
Consistent with reports in the literature (1, 6, 11, 20, 37), we could detect core promoter mutants from HBeAg+ patients. Such mutants are highly prevalent in genotype A strains owing to their reduced tendency to develop the HBeAg-minus precore mutation (6, 26, 29, 31, 40). Interestingly, distribution of core promoter mutants segregated with viremia titer: no such mutants were found in highly viremic sera, whereas a high prevalence of such mutants was detectable in samples with much-reduced viremia (Table 1). Similar observations, though not as clearcut as ours, were made with HBeAg+ samples covering the entire spectrum of viremia titer (11, 30). It is well established that the level of viremia declines over the course of HBV infection, especially during the period of seroconversion from HBeAg to anti-HBe (Fig. 1). We propose that the highly viremic samples were still at the early HBeAg+ stage of infection, while the low-titer samples were already approaching the period of seroconversion (Fig. 1).
Effect of less-common core promoter mutations on viral replication and HBeAg expression.
Eight HBV genomes with wild-type core promoter sequence (2A, 3.10, 5.2, 6.2, 7.2, 7.4, 11.4, and 12.2) showed evidence of DNA replication, but none replicated at high levels. In contrast, six core promoter mutants (4B, 4C, 4F, 3.4, 1B, and 8.22) replicated at much higher rates, and all harbored the common A1762T/G1764A mutations. In addition, there was a T1753C mutation in clones 1B and 3.4 and a C1766T mutation in clone 8.22. Both mutations were found in 4B, 4C, and 4F genomes (clone 8.12 failed to replicate at high level despite identical mutations as found in 4B). Site-directed mutagenesis not only confirmed the role of 1762/1764 common mutations on viral DNA replication but also demonstrated the importance of the less-common mutations, especially the C1766T mutation (Fig. 8B, mu2a and Ex2). This result is consistent with previous work with the fulminant hepatitis strain, although for that strain the 1766 mutation alone enhanced replication by twofold only (2, 3). We did not test the effect of 1766 mutation alone in the present study. Based on the results of site-directed mutants, we conclude that the high replication capacity of clones 4B (4C and 4F) and 8.22 is at least partly contributed by the combined core promoter mutations but is not due to the 1762/1764 mutations alone.
Findings with the site-directed mutants may also partly explain why naturally occurring core promoter mutants varied greatly in HBeAg expression. The 1762/1764 hotspot mutations (mu1) and 1753/1762/1764 triple mutations (mu4) reduced HBeAg expression only moderately in genotype A, whereas the 1762/1764/1766 mutations (mu2a) and 1753/1762/1764/1766 quadruple mutations (Ex2) markedly suppressed HBeAg production (Fig. 9A). In agreement with these findings, the HBeAg expression level was high for 9.24 (1762/1764) and 1B/3.4 (1753/1762/1764) but much lower for 4B (1753/1762/1764/1766) and 8.22 (1762/1764/1766). Consistent with these observations, only serum samples from patients 4 and 8 showed low or no HBeAg expression (Table 1). The dominance of 1753/1762/1764/1766 quadruple mutations in patient 4 was evidenced by their presence in clones 4B, 4C, 4F, 4G, and 4H (though not in 4D).
What is the mechanism whereby the core promoter mutations in clones 4B (or Ex2) and 8.22 (or mu2a) markedly enhance viral replication and reduce HBeAg expression? The 1762/1764/1766 mutations (shared by Ex2 and mu2a) are located in the nuclear receptor-binding site of the core promoter, which is part of X gene coding sequence. In this regard, the 1762/1764 hotspot mutations cause a double amino acid change in the HBx protein, which suppresses the transcription of both precore and pregenomic RNA (25). The double mutation also creates a new HNF1 binding site, which specifically enhances pregenomic RNA transcription (25). The net effect is specific reduction in the transcription of precore but not pregenomic RNA. Recent studies from Tang et al. (49) found that RXRα-PPARα binding and enhancement of viral transcription and/or replication are abrogated by the double mutations, whereas the transcription-enhancing effect of HNF4 is maintained and even augmented. A weak effect of the HNF1 on transcription was also demonstrated. Since the HNF1-binding site created by the 1762/1764 double mutation is imperfect (GGTTAATGATC) compared to the 1762/1764/1766 triple mutations (GGTTAATGATT), the stronger replication and lower HBeAg expression by the triple mutant could be caused by a better response to HNF1. Indeed, other types of core promoter sequences shown in Fig. 2, including those with both 1766 and 1768 mutations, do not completely conform to this optimal HNF1 consensus sequence. Interestingly, naturally occurring HBV strains with HNF1-binding sites (as a result of insertions) have been identified and found to possess enhanced pregenome transcription and viral replication (17, 39). We plan to formally test this hypothesis in the near future.
The paradox of enhanced in vitro replication and reduced in vivo viremia.
Contrary to our initial hypothesis, high replicating viral strains were isolated from patients with diminished viremia titer, while serum with a high viral titer contained viral strains with limited replication capacity in hepatoma cell lines. Nevertheless, a high replication capacity of core promoter mutants is consistent with their frequent association with fulminant hepatitis (12, 21, 28, 32, 36, 41, 44, 45), and one such genome associated with fulminant hepatitis was found to replicate at a very high level (2, 18). Our results suggest that very high replication potential is a feature shared by many naturally occurring core promoter mutants rather than limited to core promoter mutants implicated in fulminant hepatitis. Thus, the real question is why patients infected with core promoter mutants fail to produce high viremia. We cannot exclude the possibility that viral replication in hepatoma cell lines might be quantitatively different from that in normal hepatocytes, considering that transcription factors such as RXRα-PPARα and HN4 promote the replication of wild-type HBV and core promoter mutants differentially (49, 56). One may also argue that the high replicating clone may constitute a minor fraction of viral quasispecies in vivo. However, most clones derived from patient 4 had the identical core promoter mutations (1753C 1762T 1764A 1766T) associated with high replication capacity. A third possibility is a decline in virion secretion, as exemplified by core promoter mutants 1B, 3.4, and 8.22. Finally, we believe reduced viremia associated with core promoter mutants could be explained by enhanced destruction of viral particles by the host immune system. Consistent with the high replication capacity of core promoter mutants, genotype C of HBV causes more severe liver diseases than genotype B and develops core promoter mutations much more frequently than genotype B (9, 22, 30, 38, 46).
Core promoter mutants differ greatly in virion secretion.
Comparison among different clones revealed great variability in the intensity of extracellular HBV DNA relative to intracellular DNA. Although most clones released both double- and single-stranded genomes, clones such as 3.4 failed to release double-stranded genomes. CsCl gradient centrifugation and strand-specific probes revealed an association of extracellular single (negative)-strand DNA with core particles and of double-stranded DNA with Dane particles. The fact that naked core particles were detected from virtually all of the clones suggests constitutive nature of its release in Huh7 cells, either as a result of cell death or through a specific release mechanism (considering similar amounts of double- and single stranded HBV DNA intracellularly for most clones). Since the release of naked core particles does not occur in vivo, virion secretion efficiency can be easily estimated by comparing the double-stranded DNA species inside cells and in culture medium. Thus, clones such as 4B, 11.4, and 8.12 displayed much higher secretion efficiencies than clones 3.4, 1B, and 8.22 (Fig. 5), although we do not know whether 4B has an increased secretion capacity, whether 3.4 has a decreased secretion capacity, or whether both are true. It is anticipated that high viral replication coupled with limited viral secretion in the long-living hepatocytes will cause marked viral retention and may trigger severe liver diseases.
The molecular basis for the altered secretion capacities remains to be determined. Recently, Le Pogam et al. (24) reported reduction in virion secretion by naturally occurring substitutions at the core protein (Pro5Thr and Leu60Val). In another study, amino acid substitutions in the small envelope protein reduced secretion of both viral and subviral particles (21). Taken together, these results highlight the importance of core-envelope interaction in virion formation and secretion. The mutations described by these investigators are absent in clone 3.4, although this clone does express a reduced level of HBsAg (Fig. 3 and 4). However, our ongoing mapping experiment suggests that the defects in HBsAg secretion and virion secretion are genetically separable (N. Khan et al., unpublished data). In summary, the present study establishes much higher replication capacities of some naturally occurring core promoter mutants compared to their wild-type counterparts. Such highly replicating core promoter mutants differ greatly in the efficiency of HBeAg and HBsAg expression and virion secretion. The replication and HBeAg expression profiles are controlled, at least in part, by the overall pattern of their core promoter mutations. In particular, increased viral DNA replication and reduced HBeAg expression brought by the hotspot 1762T 1764A mutations are augmented by the C1766T mutation.
Acknowledgments
This work was supported by grants CA-35711, AA-20169, and p20RR15578 from the National Institutes of Health and from Life Span Research Funds. This work was also supported in part by a grant from the Brain Korea 21 Project for Medical Science (to S.H.A.), a Student Research Fellowship from American Liver Foundation (to A.T.) and an Alpha Omega Alpha Student Research Fellowship (to N.K.). J.L. is a Liver Scholar from the American Liver Foundation.
REFERENCES
- 1.Baptista, M., A. Kramvis, and M. Kew. 1999. High prevalence of 1762T 1764A mutations in the basic core promoter of hepatitis B virus isolated from Black Africans with hepatocellular carcinoma compared with asymptomatic carriers. Hepatology 29:946-953. [DOI] [PubMed] [Google Scholar]
- 2.Baumert, T., S. Rogers, K. Hasegawa, and T. Liang. 1996. Two core promoter mutations identified in a hepatitis B virus strain associated with fulminant hepatitis result in enhanced viral replication. J. Clin. Investig. 98:2268-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumert, T., A. Marrone, J. Vergalla, and T. Liang. 1998. Naturally occurring mutations define a novel function of the hepatitis B virus core promoter in core protein expression. J. Virol. 72:6785-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowyer, S., L. van Staden, M. Kew, and J. Sim. 1997. A unique segment of the hepatitis B virus group A genotype identified in isolates from South Africa. J. Gen. Virol. 78:1719-1729. [DOI] [PubMed] [Google Scholar]
- 5.Buckwold, V., Z. Xu, M. Chen, T. Yen, and J. Ou. 1996. Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J. Virol. 70:5845-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, H., M. Hussin, and A. Lok. 1999. Different hepatitis B virus genotypes are associated with different mutations in the core promoter and precore regions during hepatitis B e antigen seroconversion. Hepatology 29:976-984. [DOI] [PubMed] [Google Scholar]
- 7.Chang, C., G. Enders, R. Sprengel, N. Peters, H. Varmus, and D. Ganem. 1987. Expression of the precore region of an avian hepatitis B virus is not required for viral replication. J. Virol. 61:3322-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, H., M. Kew, W. Hornbuckle, B. Tennant, P. Cote, J. Gerin, R. Purcell, and R. Miller. 1992. The precore gene of the woodchuck hepatitis virus genome is not essential for viral replication in the natural host. J. Virol. 66:5682-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu, C., and A. Lok. 2002. Clinical significance of hepatitis B virus genotypes. Hepatology 35:1274-1276. [DOI] [PubMed] [Google Scholar]
- 10.Chun, Y., J. Kim, H. Woo, S. Oh, I. Kang, J. Ha, and S. Kim. 2000. No significant correlation exists between core promoter mutations, viral replication, and liver damage in chronic hepatitis B infection. Hepatology 32:1154-1162. [DOI] [PubMed] [Google Scholar]
- 11.Erhardt, A., U. Reineke, D. Blondin, W. Gerlich, O. Adams, T. Heintges, C. Niederau, and D. Haussinger. 2000. Mutations of the core promoter and response to interferon treatment in chronic replicative hepatitis B. Hepatology 31:716-725. [DOI] [PubMed] [Google Scholar]
- 12.Friedt, M., P. Gerner, E. Lausch, H. Trubel, B. Zabel, and S. Wirth. 1999. Mutations in the basic core promotor and the precore region of hepatitis B virus and their selection in children with fulminant and chronic hepatitis B. Hepatology 29:1252-1258. [DOI] [PubMed] [Google Scholar]
- 13.Ganem, D., and R. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 2923-2970. In D. Knipe and P. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 14.Gerelsaikhan, T., J. Tavis, and V. Bruss. 1996. Hepatitis B virus nucleocapsid envelopment does not occur without genomic DNA synthesis. J. Virol. 70:4269-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grandjacques, C., P. Pradat, L. Stuyver, M. Chevallier, P. Chevallier, C. Pichoud, M. Maisonnas, C. Trepo, and F. Zoulim. 2000. Rapid detection of genotypes and mutations in the pre-core promoter and the pre-core region of hepatitis B virus genome: correlation with viral persistence and disease severity. J. Hepatol. 33:430-439. [DOI] [PubMed] [Google Scholar]
- 16.Gunther, S., B. Li, S. Miska, D. Kruger, H. Meisel, and H. Will. 1995. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J. Virol. 69:5437-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunther, S., N. Piwon, A. Iwanska, R. Schilling, H. Meisel, and H. Will. 1996. Type, prevalence, and significance of core promoter/enhancer II mutations in hepatitis B viruses from immunosuppressed patients with severe liver disease. J. Virol. 70:8318-8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa, K., J. Huang, S. Rogers, H. Blum, and T. Liang. 1994. Enhanced replication of a hepatitis B virus mutant associated with an epidemic of fulminant hepatitis. J. Virol. 68:1651-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honda, A., O. Yokosuka, T. Ehata, M. Tagawa, F. Imazeki, and H. Saisho. 1999. Detection of mutations in the enhancer 2/core promoter region of hepatitis B virus in patients with chronic hepatitis B virus infection: comparison with mutations in precore and core regions in relation to clinical status. J. Med. Virol. 57:337-344. [PubMed] [Google Scholar]
- 20.Hou, J., G. Lau, J. Cheng, C. Cheng, and W. Carman. 1999. T1762/A1764 variants of the basal core promoter of hepatitis B virus; serological and clinical correlations in Chinese patients. Liver 19:411-417. [DOI] [PubMed] [Google Scholar]
- 21.Kalinina, T., A. Riu, L. Fisher, H. Will, and M. Sterneck. 2001. A dominant hepatitis B virus population defective in virus secretion because of several S-gene mutations from a patient with fulminant hepatitis. Hepatology 34:385-394. [DOI] [PubMed] [Google Scholar]
- 22.Kao, J., P. Chen, M. Lai, and D. Chen. 2003. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology 124:327-334. [DOI] [PubMed] [Google Scholar]
- 23.Kidd-Ljunggren, K., M. Oberg, and A. Kidd. 1997. Hepatitis B virus X gene 1751 to 1764 mutations: implications for HBeAg status and disease. J. Gen. Virol. 78:1469-1478. [DOI] [PubMed] [Google Scholar]
- 24.Le Pogam, S., T. Yuan, G. Sahu, S. Chatterjee, and C. Shih. 2000. Low-level secretion of human hepatitis B virus virions caused by two independent, naturally occurring mutations (P5T and L60V) in the capsid protein. J. Virol. 74:9099-9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, J., V. Buckwold, M. Hon, and J. Ou. 1999. Mechanism of suppression of hepatitis B virus precore RNA transcription by a frequent double mutation. J. Virol. 73:1239-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, J., S. Tong, Y. Wen, L. Vitvitski, Q. Zhang, and C. Trepo. 1993. Hepatitis B virus genotype A rarely circulates as an HBe-minus mutant: possible contribution of a single nucleotide in the precore region. J. Virol. 67:5402-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, J., S. Tong, and J. Wands. 1999. Identification and expression of glycine decarboxylase (p120) as a duck hepatitis B virus pre-S envelope-binding protein. J. Biol. Chem. 274:27658-27665. [DOI] [PubMed] [Google Scholar]
- 28.Liang, T., K. Hasegawa, N. Rimon, J. Wands, and E. Ben-Porath. 1991. A hepatitis B virus mutant associated with an epidemic of fulminant hepatitis. N. Engl. J. Med. 324:1705-1709. [DOI] [PubMed] [Google Scholar]
- 29.Lindh, M., Y. Furuta, A. Vahlne, G. Norkrans, and P. Horal. 1995. Emergence of precore TAG mutation during hepatitis B e seroconversion and its dependence on pregenomic base pairing between nucleotides 1858 and 1896. J. Infect. Dis. 172:1343-1347. [DOI] [PubMed] [Google Scholar]
- 30.Lindh, M., C. Hannoun, A. Dhillon, G. Norkrans, and P. Horal. 1999. Core promoter mutations and genotypes in relation to viral replication and liver damage in East Asian hepatitis B virus carriers. J. Infect. Dis. 179:775-782. [DOI] [PubMed] [Google Scholar]
- 31.Lok, A., U. Akarca, and S. Greene. 1994. Mutations in the pre-core region of hepatitis B virus serve to enhance the stability of the secondary structure of the pre-genome encapsidation signal. Proc. Natl. Acad. Sci. USA 91:4077-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMillan, A., D. Bowen, P. Angus, G. McCaughan, and S. Locarnini. 1996. Mutations in the hepatitis B virus precore/core gene and core promoter in patients with severe recurrent disease following liver transplantation. Hepatology 24:1371-1378. [DOI] [PubMed] [Google Scholar]
- 33.Milich, D., J. Jones, J. Hughes, J. Price, A. Raney, and A. McLachlan. 1990. Is a function of the secreted hepatitis B e antigen to induce immunologic tolerance in utero? Proc. Natl. Acad. Sci. USA 87:6599-6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moriyama, K., H. Okamoto, F. Tsuda, and M. Mayumi. 1996. Reduced precore transcription and enhanced core-pregenome transcription of hepatitis B virus DNA after replacement of the precore-core promoter with sequences associated with e antigen-seronegative persistent infections. Virology 226:269-280. [DOI] [PubMed] [Google Scholar]
- 35.Nassal, M., and A. Rieger. 1993. An intramolecular disulfide bridge between cys7 and cys61 determines the structure of the secretory core gene product (e antigen) of hepatitis B virus. J. Virol. 67:4307-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogata, N., R. Miller, K. Ishak, and R. Purcell. 1993. The complete nucleotide sequence of a pre-core mutant of hepatitis B virus implicated in fulminant hepatitis and its biological characterization in chimpanzees. Virology 194:263-276. [DOI] [PubMed] [Google Scholar]
- 37.Okamoto, H., F. Tsuda, Y. Akahane, Y. Sugai, M. Yoshiba, K. Moriyama, T. Tanaka, Y. Miyakawa, and M. Mayumi. 1994. Hepatitis B virus with mutations in the core promoter for an e antigen-negative phenotype in carriers with antibody to e antigen. J. Virol. 68:8102-8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orito, E., M. Mizokami, H. Sakugawa, K. Michitaka, K. Ishikawa, T. Ichida, T. Okanoue, H. Yotsuyanagi, and S. Iino. 2001. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Hepatology 33:218-223. [DOI] [PubMed] [Google Scholar]
- 39.Pult, I., T. Chouard, S. Wieland, R. Klemenz, M. Yaniv, and H. Blum. 1997. A hepatitis B virus mutant with a new hepatocyte nuclear factor binding site emerging in transplant-transmitted fulminant hepatitis B. Hepatology 25:1507-1515. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez-Frias, F., M. Buti, R. Jardi, M. Cotrina, L. Viladomiu, R. Esteban, and J. Guardia. 1995. Hepatitis B virus infection: precore mutants and its relation to viral genotypes and core mutations. Hepatology 22:1641-1647. [DOI] [PubMed] [Google Scholar]
- 41.Sato, S., K. Suzuki, Y. Akahane, K. Akamatsu, K. Akiyama, K. Yunomura, F. Tsuda, T. Tanaka, H. Okamoto, Y. Miyakawa, and M. Mayumi. 1995. Hepatitis B virus strains with mutations in the core promoter in patients with fulminant hepatitis. Ann. Intern. Med. 122:241-248. [DOI] [PubMed] [Google Scholar]
- 42.Scaglioni, P., M. Melegari, and J. Wands. 1997. Biological properties of hepatitis B viral genomes with mutations in the precore promoter and precore open reading frame. Virology 233:374-381. [DOI] [PubMed] [Google Scholar]
- 43.Schlicht, H., J. Salfeld, and H. Schaller. 1987. The duck hepatitis B virus pre-C region encodes a signal sequence which is essential for synthesis and secretion of processed core proteins but not for virus formation. J. Virol. 61:3701-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sterneck, M., S. Gunther, T. Santantonio, L. Fischer, C. Broelsch, H. Greten, and H. Will. 1996. Hepatitis B virus genomes of patients with fulminant hepatitis do not share a specific mutation. Hepatology 24:300-306. [DOI] [PubMed] [Google Scholar]
- 45.Stuyver, L., S. De Gendt, F. Cadranel, C. Van Geyt, G. Van Reybroeck, R. Dorent, I. Gandjbachkh, M. Rosenheim, F. Charlotte, P. Opolon, J. Huraux, and F. Lunel. 1999. Three cases of severe subfulminant hepatitis in heart-transplanted patients after nosocomial transmissiion of a mutant hepatitis B virus. Hepatology 29:1876-1883. [DOI] [PubMed] [Google Scholar]
- 46.Sumi, H., O. Yokosuka, N. Seki, M. Arai, F. Imazeki, T. Kurihara, T. Kanda, K. Fukai, M. Kato, and H. Saisho. 2003. Influence of hepatitis B virus genotypes on the progression of chronic type B liver disease. Hepatology 37:19-26. [DOI] [PubMed] [Google Scholar]
- 47.Summers, J., and W. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi, K., Y. Ohta, K. Kanai, Y. Akahane, Y. Iwasa, K. Hino, N. Ohno, H. Yoshizawa, and S. Mishiro. 1999. Clinical implications of mutations C-to-T1663 and T-to-C/A/G1753 of hepatitis B virus genotype C genome in chronic liver disease. Arch. Virol. 144:1299-1308. [DOI] [PubMed] [Google Scholar]
- 49.Tang, H., A. Raney, and A. McLachlan. 2001. Replication of the wild type and a natural hepatitis B virus nucleocapsid promoter variant is differentially regulated by nuclear hormone receptors in cell culture. J. Virol. 75:8937-8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong, S., C. Diot, P. Gripon, L. Vitvitski, C. Trepo, and C. Guguen-Guillouzo. 1991. In vitro replication competence of a cloned hepatitis B virus variant with a nonsense mutation in the distal pre-C region. Virology 181:733-737. [DOI] [PubMed] [Google Scholar]
- 51.Tong, S., J. Li, L. Vitvitski, S. Benjelloun, and C. Trepo. 1991. Rapid screening for bacterial colonies harbouring tandem hepatitis B virus sequences by an oligonucleotide probe. J. Virol. Methods 32:109-114. [DOI] [PubMed] [Google Scholar]
- 52.Tong, S., J. Li, L. Vitvitski, and C. Trepo. 1992. Replication capacities of natural and artificial precore stop codon mutants of hepatitis B virus: relevance of pregenome encapsidation signal. Virology 191:237-245. [DOI] [PubMed] [Google Scholar]
- 53.Wasenauer, G., J. Kock, and H. Schlicht. 1993. Relevance of cysteine residues for biosynthesis and antigenicity of human hepatitis B virus e protein. J. Virol. 67:1315-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei, Y., J. Tavis, and D. Ganem. 1996. Relationship between viral DNA synthesis and virion envelopment in hepatitis B viruses. J. Virol. 70:6455-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu, X., and J. Mertz. 1996. Promoters for synthesis of the pre-C and pregenomic mRNAs of human hepatitis B virus are genetically distinct and differentially regulated. J. Virol. 70:8719-8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu, X., and J. Mertz. 2001. Critical roles of nuclear receptor response elements in replication of hepatitis B virus. J. Virol. 75:11354-11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan, T., G. Sahu, W. Whitehead, R. Greenberg, and C. Shih. 1999. The mechanism of an immature secretion phenotype of a highly frequent naturally occurring missense mutation at codon 97 of human hepatitis B virus core antigen. J. Virol. 73:5731-5740. [DOI] [PMC free article] [PubMed] [Google Scholar]