Abstract
The chemokine receptors CCR5 and CXCR4 are promising non-virus-encoded targets for human immunodeficiency virus (HIV) therapy. We describe a selection procedure to isolate mutant forms of RANTES (CCL5) with antiviral activity considerably in excess of that of the native chemokine. The phage-displayed library of randomly mutated and N-terminally extended variants was screened by using live CCR5-expressing cells, and two of the selected mutants, P1 and P2, were further characterized. Both were significantly more potent HIV inhibitors than RANTES, with P2 being the most active (50% inhibitory concentration of 600 pM in a viral coat-mediated cell fusion assay, complete protection of target cells against primary HIV type 1 strains at a concentration of 10 nM). P2 resembles AOP-RANTES in that it is a superagonist of CCR5 and potently induces receptor sequestration. P1, while less potent than P2, has the advantage of significantly reduced signaling activity via CCR5 (30% of that of RANTES). Additionally, both P1 and P2 exhibit not only significantly increased affinity for CCR5 but also enhanced receptor selectivity, retaining only trace levels of signaling activity via CCR1 and CCR3. The phage chemokine approach that was successfully applied here could be adapted to other chemokine-chemokine receptor systems and used to further improve the first-generation mutants reported in this paper.
Despite the success of highly active antiretroviral therapy, new human immunodeficiency virus type 1 (HIV-1) inhibitors are still needed and among the most promising new approaches is the blockade of viral entry into target cells (20). HIV-1 entry into target cells is initially dependent on the interaction of its envelope glycoproteins with CD4 and a coreceptor, with the chemokine receptors CCR5 and CXCR4 being by far the most commonly used by HIV-1 (5).
HIV entry is inhibited by the natural chemokine ligands of the coreceptors, including MIP-1α (CCL3), MIP-1β (CCL4), and RANTES (CCL5) for CCR5 (8) and SDF-1 (CXCL12) for CXCR4 (6, 23). Certain N-terminal modifications have been shown to increase the anti-HIV activity of native chemokines (21, 32, 33, 36), and the most potent of these molecules owe their anti-HIV activity to their ability to induce prolonged intracellular sequestration of coreceptors (18, 31).
Up until now, chemokine structure-activity relationships have been studied via either scanning or truncation mutagenesis (14, 16, 19, 24), peptide scanning of primary sequence (22), or semirational design of chemokine analogues (21, 32, 36). A more-rapid, bioengineering-based approach for the selection of useful chemokine variants has yet to be described.
We decided to apply current knowledge of the structure-activity relationship of chemokines and the mechanism by which they inhibit HIV entry (2, 7, 18) to the design of a phage display strategy for the discovery of N-terminally mutated RANTES variants with improved anti-HIV activity. Selection led to the isolation of around 40 clones that exhibited a consensus sequence, and two clones were chosen for further evaluation. Both show greatly enhanced anti-HIV-1 activity compared to RANTES as well as increased selectivity for CCR5.
MATERIALS AND METHODS
Reagents.
Chemokines were prepared by total chemical synthesis, essentially as described in (35). The aminooxypentane (AOP)-RANTES used in this study was from the batch described in reference 32. The purity and authenticity of the chemokines were verified by analytical high-performance liquid chromatography and mass spectrometry (data not shown), and their concentrations in solution were determined by measurement of absorbance at 280 nm. The 1D2 anti-RANTES antibody and the 2D7 phycoerythrin-conjugated anti-CCR5 antibody were obtained from Pharmingen (San Diego, Calif.).
Cells.
CHO-K1 cells were provided by BioWhittaker. CHO-CCR5 cells were kindly provided by T. Schwartz (Panum Institute, Copenhagen, Denmark), HEK-CCR5 (9) and HEK-CX3CR1 (10) cells were provided by C. Combadiere (Hôpital Pitié-Salpétrière, Paris, France). HEK-CCR1 and HEK-CCR3 cells were stably transduced by using retroviral vectors derived from the appropriate pBABE expression constructs (obtained from the National Institutes of Health AIDS Reagent Program). Human peripheral blood mononuclear cells (PBMC), isolated on Ficoll gradients (Pharmacia Biotech) from the buffy coats of healthy donors seronegative for HIV, were cultured for 72 h in RPMI 1640 medium supplemented as described above. PBMC were stimulated with 1 μg of phytohemagglutinin (Murex Diagnostics, Dartford, United Kingdom)/ml, for 72 h. The cells were then cultured in the presence of 500 U of interleukin-2 (Chiron)/ml for 24 h prior to viral challenge. Monocyte-derived macrophages (MDM) were derived from freshly isolated PBMC. Adherent cells were purified after overnight culture of PBMC and then cultured for 7 days in Teflon bags in medium supplemented with 15% human AB serum. At the end of the procedure, cells were >98% MDM as assessed by morphology and flow cytometry (CD14+, CD16+).
Library construction.
The DNA sequence coding for human RANTES, amplified from cDNA prepared from activated human PBMCs, was used as the scaffold for library construction. PCR mutagenesis of the gene's 5′ region was performed by using a specific downstream primer (TGGGGCCCCTCTAGACATCTCCAAAGAGTTGATGTACTC) and a degenerate upstream primer (CTCGCGGCCCAGCCGGCCATGGCCNNKTCCNCANNKTCCTCGNNKNCCNCANCCTGCTGCTTTGCCTACATTGCGCGGCCGCTGCCCCGTGCCCACATC) (N represents any of the four bases; K represents either G or T). The product was cut with NcoI and PspOMI and inserted into phagemid pHEN1 (17) (kindly provided by G. Winter, Cambridge University, Cambridge, United Kingdom), previously linearized with NcoI and NotI. The ligated DNA was electroporated into Escherichia coli TG1. Colonies were PCR screened before selection, and their DNA inserts were sequenced with an automatic sequencer ABI 377 (Perkin-Elmer). In this way, the complexity of the RANTES library was determined to be at least 5 × 106 clones, thus exceeding its theoretical diversity (2 × 106). Phage stocks were prepared essentially as described in reference 17.
Panning on live cells.
Phage stocks used for selection were supplemented with 1.5% (wt/vol) bovine serum albumin (BSA) prior to their use. Phage (1010 CFU) was directly incubated with 5 × 106 adherent cells growing in six-well plates (Nunc, Roskilde, Denmark) at 37°C at 10% CO2 in 5 ml of supplemented RPMI 1640 medium. After 1 h, cells were washed 10 times at room temperature with 10 ml of phosphate-buffered saline (PBS) and then scraped from the plate into 10 ml of PBS-1% (wt/vol) BSA. Cells were pelleted (538 × g; 10 min; 4°C), resuspended in 1 ml of elution buffer (0.1 M glycine-HCl, pH 3), and kept on ice for 10 min. The cell suspension was then neutralized with 1 M Tris-Cl (pH 8) and pelleted. Cells were then resuspended in Tris (30 mM, pH 8) and EDTA (1 mM) and lysed by three cycles of freezing (−80°C) and thawing (37°C). The total cell lysate was prewarmed to 37°C and mixed with log-phase E. coli TG1 for the production and purification of phage (as described above) for use in further rounds of selection.
Competitive radioligand binding on CCR5.
Assays were carried out essentially as described in reference 32. Competitive binding was performed on CHO-CCR5 cells at 4°C by using 12 pM 125I-MIP-1β or 125I-RANTES (Amersham) plus variable amounts of unlabeled ligand. After incubation, cells were washed and lysed and the lysates were counted with a Beckman Gamma 4000 scintillation counter. Determinations were performed in duplicate, and 50% inhibitory concentrations (IC50) were derived from monophasic curves (competition binding; one-site) fitted with Prism software (GraphPad).
Steady-state receptor down-modulation experiments.
CHO-CCR5 cells (105) were incubated for 30 min at 37°C in supplemented culture medium containing various concentrations (0 to 100 nM) of chemokines. After 5 washes with 10 ml of cold PBS, cells were scraped from the plates and resuspended in 0.5 ml of elution buffer, kept on ice for 1 min (in order to elute cell surface-bound chemokines), and then neutralized (see “Panning on live cells” above for buffers). Cells were resuspended in 100 μl of PBS-0.5% BSA, incubated for 30 min on ice with 0.5 μg of 2D7-phycoerythrin, washed two times, fixed with 4% paraformaldehyde, and analyzed with a FACScan (Becton Dickinson, San Jose, Calif.). At least 10,000 events were accumulated for each sample.
Calcium mobilization assay.
Experiments were carried out on 96-well plates using a FLEXstation machine (Molecular Devices). Fluorescence measurements were carried out on HEK-293 cells that were loaded with Fluo-4 (Molecular Probes) according to the manufacturer's recommendations and maintained at 37°C. Fluorescence readings were converted into cytosolic Ca2+ concentrations by the method of Grynkiewicz et al. (15), and dose curves were fitted to the data by using Prism software.
Cell fusion assay.
Envelope-mediated cell fusion assays were carried out as described in reference 31 with the HeLa-P5L (32) and HeLa-Env-ADA (25) cell lines. Briefly, HeLa-P5L cells were seeded in 96-well plates (104 cells per well). Twenty-four hours later, medium was removed and replaced with medium containing 104 HeLa-Env-ADA cells per well plus chemokines. After a further 24 h, cells were washed once in PBS, lysed, and assayed for β-galactosidase activity by the addition of the colorigenic substrate CPRG (chlorophenol red-β-d-galactopyranoside). Results are expressed as 100 × (mean absorbance [treated] − mean absorbance [no envelope cells])/(mean absorbance [no chemokine] − mean absorbance [no envelope cells]). Experiments were performed in triplicate, and dose-inhibition curves were fitted by using Prism software (GraphPad).
Infectivity assay.
Activated PBMC (105) in triplicate wells were treated for 30 min at 37°C with medium alone or medium containing serial dilutions of chemokines. HIV-1 BX08 (multiplicity of infection, 2 × 10−5) or HIV-1BaL at a multiplicity of infection of 10−4 (data not shown) was then added. After overnight incubation, cells were washed extensively and cultured in medium containing appropriate concentrations of chemokines. A quantitative HIV-1 p24 antigen-capture enzyme-linked immunosorbent assay (Beckman Coulter, Villepinte, France) was performed on the supernatants. Cell viability was not affected by the chemokines used in this assay. For single-round infectivity assays, we used the pNL-Luc-E−R+ vector (11). The HIV-1BaL envelope expression vector is the pSV7d plasmid bearing the R5 HIV-1BaL env gene. Both constructs were kindly provided by T. Dragic and N. Landau (ADARC, New York, N.Y.). MDM (105), pretreated with chemokines for 30 min at 37°C, were incubated overnight with HIV-1BaL pseudotypes at a concentration of 50 ng of p24/ml. MDM were then washed extensively, and medium was replenished but not chemokine supplemented. After 48 h, luciferase activity was assayed in lysates according to the manufacturer's instructions (Promega) by using a Berthold LB9501 luminometer. All assays were performed in triplicate.
RESULTS
Library design.
Having verified by enzyme-linked immunosorbent assay and Western blotting that RANTES-g3p fusion proteins are incorporated into phage (data not shown), we set out to construct a phage library of RANTES mutants. Since a number of chemokine analogues featuring hydrophobic N-terminal extensions have been reported as having enhanced anti-HIV activity (21, 32, 36), a one-residue N-terminal extension (position 0) was incorporated into our library design. Additionally, partial or complete randomization was introduced into positions 2, 3, 6, 7, 8, and 9. Hence, the library of RANTES mutants had the following sequence XS#XSSX###-RANTES (10 to 68), where X represents any amino acid and # represents either A, P, S, or T (Table 1).
TABLE 1.
Sequences of clones from RANTES mutant librarya
| N-terminal sequence of selected cloneb | No. of clones used for selection from cell line:
|
Total | ||
|---|---|---|---|---|
| HEK-CCR5 | CHO-CCR5 | HEK-CCR5 and CHO-CCR5 | ||
| LSPVSSQSSA (P1) | 6 | 4 | 10 | |
| FSPLSSQSSA (P2) | 6 | 6 | ||
| LSPMSSQSPA | 6 | 1 | 4 | 11 |
| WSPLSSQSPA | 1 | 1 | 2 | |
| WSPLSSQSSP | 2 | 2 | ||
| LSPQSSLSSS | 1 | 1 | ||
| ASSGSSQSTS | 1 | 1 | ||
| ISAGSSELAA | 1 | 1 | ||
| RSPMSSQSSP | 1 | 1 | ||
| YSPSSSLAPA | 1 | 1 | ||
| MSPLSSQASA | 1 | 1 | ||
| ASPLSSQSSS | 1 | 1 | ||
| QSPLSSQAST | 1 | 1 | ||
| QSPLSSTASS | 1 | 1 | ||
| LSPLSSQSAA | 1 | 1 | ||
| GSSSSSQTPA | 1 | 1 | ||
| YSPLSSQSSP | 1 | 1 | ||
| FSSVSSQSSS | 1 | 1 | ||
| Total | 20 | 12 | 12 | 44 |
Sequences of clones selected from the library of phage-displayed RANTES mutants. Three independent selections, with the indicated target cells, were carried out. Cell-based screening led to the definition of a consensus sequence (shown in bold), which was different from that obtained via conventional panning on an anti-RANTES antibody (see below). Symbols: *, L, M, or V; ‡, A, P, or S, no T; #, A, P, S, or T.
LSP*SSQSSA, Consensus sequence (selection on CCR5+ cells); RSPPSSR‡AS, Consensus sequence (panning on 1D2 antibody); SPYSSDTTP, wild-type RANTES sequence; XS#XSSX###, RANTES library sequence.
Selection of RANTES mutants.
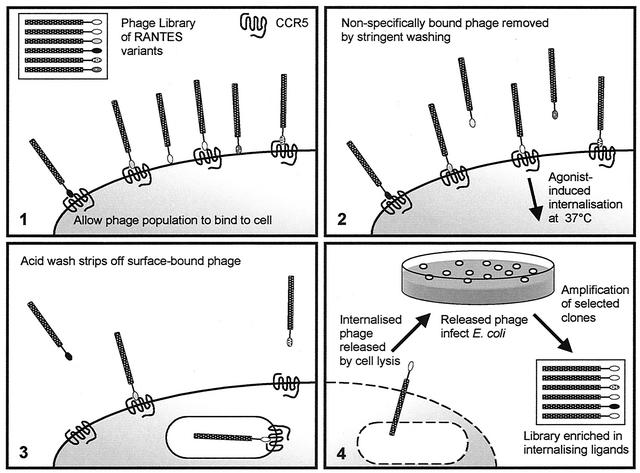
Our selection strategy involved panning against CCR5 presented on the surface of living cells (Fig. 1). To allow for the influence of cell type-dependent variations in the quantity and type of cellular components such as cell surface proteoglycan or the intracellular proteins involved in receptor endocytosis, we chose to use two different cellular backgrounds, human embryonic kidney (HEK) and Chinese hamster ovary (CHO). Three independent experiments were carried out, each of which featured three rounds of selection. In two cases, a single cell background was used in each of three rounds (HEK-CCR5 or CHO-CCR5); our third strategy involved alternating the cell background between rounds (HEK-CCR5 then CHO-CCR5). Comparison of the selected sequences enabled us to define a consensus sequence, LSP*SSQSSA, where * represents either L, M, or V (Table 1).
FIG. 1.
Strategy used to select for phage-displayed RANTES variants that are internalized by cells expressing CCR5. Phage particles are represented as cylinders (encapsulated phage genome) with shaded heads (pIII-RANTES mutant fusion). (1) The library of phage-displayed RANTES mutants is allowed to bind to cells expressing CCR5. (2) Stringent washing is used to remove nonspecifically bound phage, and the cells are incubated at 37°C to permit ligand-induced internalization of phage particles. (3) A low-pH wash is used to remove surface-associated phage, and (4) internalized phage are released by cell lysis. Eluted phage are allowed to infect E. coli, and stocks of selected phage are prepared. Steps 1 to 4 constitute a round of selection.
In order to ascertain whether the selection of this consensus sequence was CCR5-dependent, we performed three subsequent experiments. Firstly, we panned the library by using an anti-RANTES antibody that does not interact with the N terminus of RANTES. This would be expected to select for variants that have advantages of stability and/or high expression while conserving the epitope recognized by the antibody. Clones isolated in this way yielded a strikingly different consensus sequence from that obtained via selection on cells, notably with arginine and proline ubiquitous in positions 0 and 3, respectively (Table 1). Secondly, the same selection procedure was performed on untransfected HEK or CHO cells. This approach would be expected to favor mutants with an enhanced ability to interact with cell surface components other than CCR5 and, by default, those merely endowed with a growth advantage. Here, sequence analysis revealed that the RANTES gene had been partially or entirely deleted in the majority of selected phage (data not shown). Finally, we confirmed that clones expressing the consensus sequence engage cells in a ligand- and CCR5-dependent manner by flow cytometry, by using one of the selected phage clones (P2), which, unlike irrelevant phage, binds to CHO-CCR5 and HEK-CCR5 cells but does not bind to the untransfected parent cell lines (data not shown).
It is noteworthy that the cell background used appeared to influence the outcome of selection (Table 1). With this in mind, we chose to conduct further investigations with two mutant proteins, P1, which was selected on HEK-CCR5 cells, and P2, which was selected on CHO-CCR5 cells.
Enhanced anti-HIV activity.
To evaluate the anti-HIV activity of these proteins, we deliberately used very stringent assays, in which AOP-RANTES shows measurable activity but RANTES is either partially or completely inactive over the concentration ranges used. We had previously validated the anti-HIV activity of the RANTES preparation used in these experiments by performing a replication assay with HIV-1BaL, in which 10 nM RANTES achieved 100% inhibition (data not shown). Firstly, we compared the anticoreceptor activity of P1 and P2 with that of both RANTES and AOP-RANTES in an R5-tropic envelope-mediated cell fusion assay (Fig. 2A). In this assay, both P1 and P2 had extremely potent activities (IC50 of 7 nM (95% confidence limits, 3 to 15) and 0.6 nM (95% confidence limits, 0.4 to 0.9), respectively), which compare favorably with that of AOP-RANTES (IC50 of 3 nM [95% confidence limits, 2 to 6]), while RANTES did not achieve 50% inhibition at the concentrations used. We then measured the ability of the RANTES mutants to inhibit entry into MDM in a single-round infectivity assay by using HIV-1 particles pseudotyped with the R5 HIV-1BaL envelope. Although 10 nM wild-type RANTES achieved only relatively inefficient protection of macrophages from R5-tropic HIV (Fig. 2B), the same concentration efficiently protected PBMCs from HIV-1BaL challenge (data not shown). Both P1 and P2, in contrast, showed significant inhibitory activity, especially P2, which was active in the low nanomolar range and exhibited a greater protective effect than AOP-RANTES (Fig. 2B). Finally, we tested the capacity of the selected mutants to inhibit replication of R5-tropic HIV-1 in activated PBMCs. P2 exhibited potency equivalent or superior to that of AOP-RANTES against both the laboratory-adapted HIV-1BaL (data not shown) and the primary isolate, BX08 (Fig. 2C). Significantly, 50% inhibition of baseline HIV replication was not reached with wild-type RANTES under the conditions used (data not shown).
FIG. 2.
Improved anti-HIV activity of RANTES variants. (A) R5-tropic envelope-dependent cell fusion assay. P1 and P2 prevent R5 envelope-mediated cell fusion more efficiently than RANTES, which only begins to show activity in this assay at concentrations greater than 1 μM. P2 is more effective than AOP-RANTES (IC50 of 0.6 nM [range, 0.4 to 0.9 nM] and 3 nM [range, 2 to 6 nM], respectively). (B) Infection of macrophages by HIV-1 particles pseudotyped with the BaL envelope. RANTES does not achieve 50% inhibition in the concentration range used, and P1 and P2 achieve 50% inhibition between 10−8 to 10−7 M and 10−9 to 10−8 M, respectively. The corresponding value for AOP-RANTES is approximately 10−8 M. (C) Replication of the field isolate, BX08, in activated PBMCs. While P1 is active at concentrations above 10−8 M, both AOP-RANTES and P2 strongly inhibit replication at concentrations above 10−9 M. Error bars indicate 95% confidence intervals.
Mechanism underlying improved anti-HIV activity. (i) Affinity for CCR5.
Two mechanisms have been put forward to explain how chemokines inhibit HIV entry: steric blockade of the interaction between HIV envelope and cell surface coreceptors (4, 32) and reduction of cell surface coreceptor concentration via the induction of receptor down-modulation (1, 2, 18). In order to evaluate the relative contributions of these two mechanisms towards the anti-HIV activity of P1 and P2, we compared the affinity of P1 and P2 for CCR5 with that of RANTES and AOP-RANTES in competition binding assays using both MIP-1β and RANTES as radiolabeled tracers (Fig. 3).
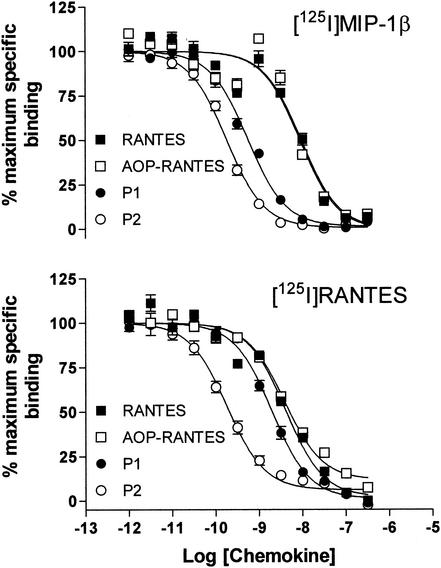
FIG. 3.
Affinity of RANTES variants for CCR5 measured by competition binding assay on CHO-CCR5 cells. The selected mutants P1 and P2 have significantly higher affinity for CCR5 than either RANTES or AOP-RANTES. With [125I]MIP-1β used as a tracer (top panel), IC50 of 9.0, 9.5, 0.56, and 0.18 nM were observed for RANTES, AOP-RANTES, P1, and P2, respectively. With [125I]RANTES used as a tracer (bottom panel), the respective IC50 were 4.1, 3.9, 2.0, and 0.18 nM.
Although RANTES inhibits binding of iodinated MIP-1β in the low nanomolar range, in agreement with previously reported work (30), we find that the apparent affinity of AOP-RANTES is indistinguishable from that of RANTES with either tracer. The selected mutants, on the other hand, particularly P2, show substantially increased affinity for CCR5. P2 shows a 50-fold increase in affinity when MIP-1β is used as a tracer and a 20-fold increase when RANTES is used as a tracer. The increase in affinity of P1 is more modest, with a 16-fold increase in affinity measured against MIP-1β and a 2-fold increase in affinity against RANTES. The detectable gain in affinity for both P1 and P2 over native RANTES is more pronounced when MIP-1β, rather than RANTES is used as a tracer. A possible explanation for this observation would be that RANTES and MIP-1β have overlapping but nonidentical binding sites on CCR5 and that the modifications introduced into P1 and P2 have more of an impact on competition with MIP-1β than with RANTES.
(ii) Enhanced capacity to sequester CCR5.
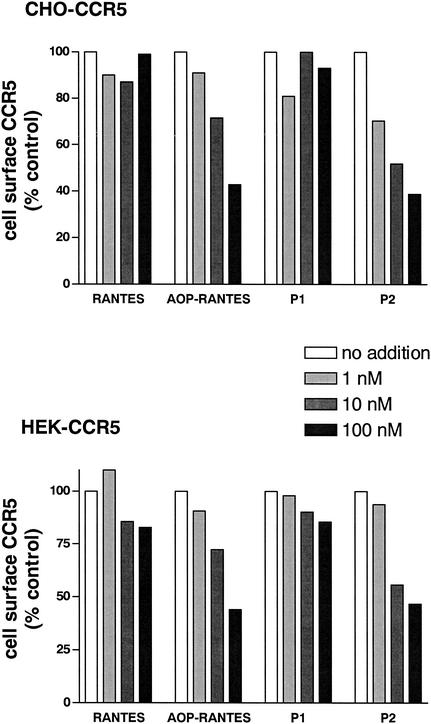
Since it is known that phage can be efficiently endocytosed by mammalian cells in a receptor-dependent manner (27), and panning was carried out at 37°C, it is possible that the capacity of some of the phage clones to induce their own endocytosis may have played a role in the selection process. The proteins encoded by these phage might then have improved antiviral activity owing to an increased capacity to induce CCR5 sequestration, as has been previously shown for a number of N-terminally modified chemokines with enhanced anti-HIV activity (7, 18, 31, 36). With this in mind, we measured the capacity of P1 and P2 to induce steady-state down-modulation of CCR5 in both of the cell lines that were used for panning (Fig. 3). Under the assay conditions used, RANTES did not reduce steady-state surface CCR5 concentration below control levels. This observation is in agreement with previous work with nonleukocytic cell lines, where it is necessary to increase intracellular levels of GRKs and arrestins in order to obtain detectable levels of internalization of heterologously expressed CCR5 by RANTES (3). AOP-RANTES, on the other hand, induced significant, dose-dependent down-modulation of CCR5, as previously described (18). P1, like RANTES, did not induce detectable down-modulation of CCR5, whereas P2 induced down-modulation with a capacity comparable to that of AOP-RANTES (Fig. 4).
FIG. 4.
CCR5 internalization induced by RANTES and RANTES variants. P2 and AOP-RANTES induce dose-dependent receptor down-modulation. Steady-state levels of surface CCR5 on the indicated cells (HEK- or CHO-CCR5) were measured by flow cytometry after incubation with chemokines. Levels are expressed as percentages of that of the control (medium, no chemokine). These data are representative of three independent experiments performed under identical conditions.
Agonist activity and receptor selectivity.
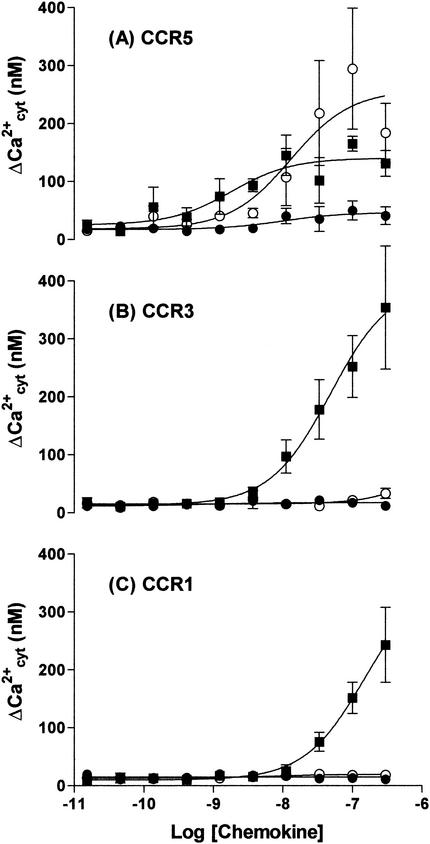
Since GPCR down-modulation is generally held to be closely linked with receptor activation (13), we chose to compare the capacity of RANTES with that of the selected RANTES mutants to elicit cytosolic calcium flux via CCR5 (Fig. 5). P2 is clearly a stronger agonist than RANTES; while both compounds have a similar EC50 values, lying in the range 1 to 10 nM, the maximum response (measured at concentrations above 100 nM) elicited by P2 is 180% of that of RANTES (Fig. 5A). On the other hand, P1 is a much weaker agonist than RANTES, with a maximal response of only 30% of that of the native chemokine (Fig. 5A). It is also noteworthy that P1 and P2 appear to be endowed not only with an increased affinity for CCR5 but also with increased receptor selectivity. Indeed, while wild-type RANTES also signals via both CCR1 and CCR3, the mutants P1 and P2 have negligible signaling activity via these two receptors (Fig. 5B and C).
FIG. 5.
Increased selectivity of selected mutant chemokines P1 and P2 for CCR5. Cytosolic calcium-dependent fluorescence changes in response to various concentrations of RANTES (▪), P1 (•), and P2 (○) via CCR5 (A), CCR3 (B), and CCR1 (C) were determined by using stably transfected HEK cells that had been loaded with Fluo-4 dye.
DISCUSSION
Chemokine structure-activity relationships have until now been investigated with limited numbers of chemokine variants that must be produced and purified separately before screening. Here, we show that by applying appropriate selection pressure to a library of chemokine variants displayed on phage, it is possible to isolate mutants endowed with predefined characteristics. As an example, we opted for the directed improvement of the anti-HIV activity of the chemokine RANTES. Directed evolution gave us rapid access to ligands not only having increased affinity and selectivity for CCR5 but also showing significantly enhanced anti-HIV activity compared to native RANTES. The structure-activity data obtained in this study would not have been accessible by conventional truncation or scanning mutagenesis.
We have shown that, as would be expected from the inhibitory mechanism of chemokines, the enhanced anti-HIV activity of P1 and P2 is due to the potency with which they achieve coreceptor blockade (Fig. 2A). Notably, given previous reports of the inefficiency with which RANTES protects macrophages (32), we show that P1 and P2 achieve inhibition of macrophage infection at low nanomolar concentrations (Fig. 2B). Furthermore, both mutants showed much greater potency than RANTES in a replication assay with a field isolate, BX08 (Fig. 2C). To our knowledge, P2 represents the most potent nonchemically modified anti-HIV chemokine so far described. P1 and P2 are the products of a single cycle of mutagenesis and selection. It is conceivable that further cycles, involving retention of the consensus sequence and mutation of neighboring residues, would result in the isolation of even more active compounds.
The gain in antiviral activity seen with P2 is most probably due in part to its increased ability to compete with the viral envelope for a common binding site on the receptor, as shown by its increased affinity for CCR5 in the competition binding assays, and in part to its increased capacity to induce intracellular sequestration of the receptor, a property it shares with other potent anti-HIV chemokines (7, 18, 31). The more modest improvement in antiviral activity seen with P1, which does not bring about CCR5 sequestration, would then be entirely due to its improved affinity for CCR5.
Since P1 and P2 were chosen from a group of selected mutants exhibiting an N-terminal consensus sequences (Table 1), it is likely that their increased antiviral activity is owed to the presence of a structural motif that enhances the ability of RANTES to engage CCR5. The key features of this motif are hydrophobic and aromatic residues at position 0, hydrophobic, but not aromatic, residues at position 3, which is a tyrosine residue in wild-type RANTES, and ubiquitous selection of proline and glutamine at positions 2 and 6, respectively. While the selected hydrophobic residues at position 0 would correspond to N-terminal extensions in Met-RANTES (29), Leu-RANTES (26), and AOP-RANTES (32) and the conservation of proline at position 2 of chemokines has been previously documented (34), the presence of glutamine at position 6 would not have been expected from previously published data. Since the mutants were selected by phage display, residue selection at some of the positions may have occurred as a result of parameters related to either phage life cycle or protein stability, rather than to functional interaction with CCR5. This is unlikely in the case of position 6, since in the control selection on the anti-RANTES antibody, arginine was exclusively selected at this position (Table 1). It is important to note that modified residues in each selected mutant may have improved receptor engagement either individually or in concert. In order to evaluate the contribution to functional engagement with CCR5 made by individual residues in the selected consensus sequence, it will be necessary to test the effect on activity of individually substituted residues in a wild-type RANTES background.
P1 and P2, unlike native RANTES, are unable to signal via either CCR1 or CCR3. Tighter selectivity of this kind could be the consequence of either reduced binding or loss of signaling activity via these receptors. While previous work with both AOP-RANTES and Met-RANTES suggest that N-terminal modifications have an impact on receptor signaling rather than receptor specificity (28, 29), competition binding studies with P1 and P2 on CCR1 and CCR3 would be necessary in order to establish whether the N-terminal structures selected by phage display affect address or message function, or both. Such studies would resolve whether P1 and P2 have lost their signaling activity because they no longer bind to CCR1 and CCR3 or whether the modifications at their N-termini have produced receptor antagonists. Whatever the explanation turns out to be, reduced signaling via these receptors makes P1 and P2 more attractive as potential anti-HIV agents, since the capacity to activate these receptors would increase the potential for undesirable, proinflammatory effects.
Given the broad similarities in structure activity that have been noted across the chemokine family, the approach described here for RANTES (CCL5)/CCR5 might be applied to the study of other chemokine-receptor couples. An obvious example in the HIV field would be the selection of SDF-1 (CXCL12) mutants with enhanced activity against CXCR4-tropic HIV strains. Additionally, it may be possible to exploit the high level of homology across the chemokine gene family by using a DNA shuffling approach (12), in which pools of sequences from different chemokines would be used to construct libraries of hybrid mutants. Phage display might be applied in this way to rapidly obtain fresh structure activity ideas, leading to the development of novel inhibitors of chemokine receptor drug targets, not only those for which inhibitors have already been described but also those for which inhibitors are currently unavailable.
Acknowledgments
O.H. and K.D. contributed equally to this work.
We thank Annie David (Institut Pasteur) for assistance in the viral challenge assays; Alain Thierry (Montpellier) and Don Mosier (TSRI, La Jolla, Calif.) for critical reading of the manuscript; and Christophe Parizot (Paris, France), Gabriel Künzi, and Astrid Melotti (Geneva, Switzerland) for expert technical assistance.
G.G. was supported by the Fondation pour la Recherche Médicale (SIDACTION), Paris, France, by the European Commission (contract QLK3-1999-01262), and by a Bristol-Myers Squibb research grant. O.H. and R.E.O. were funded by the Swiss National Science Foundation AIDS Commission.
REFERENCES
- 1.Alkhatib, G., M. Locati, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1997. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor down-modulation. Virology 234:340-348. [DOI] [PubMed] [Google Scholar]
- 2.Amara, A., S. L. Gall, O. Schwartz, J. Salamero, M. Montes, P. Loetscher, M. Baggiolini, J. L. Virelizier, and S. F. Arenzana. 1997. HIV coreceptor downregulation as antiviral principle: SDF-1alpha-dependent internalization of the chemokine receptor CXCR4 contributes to inhibition of HIV replication. J. Exp. Med. 186:139-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aramori, I., S. S. Ferguson, P. D. Bieniasz, J. Zhang, B. Cullen, and M. G. Cullen. 1997. Molecular mechanism of desensitization of the chemokine receptor CCR-5: receptor signaling and internalization are dissociable from its role as an HIV-1 co-receptor. EMBO J. 16:4606-4616. (Erratum, 16:6055.) [DOI] [PMC free article] [PubMed]
- 4.Arenzana-Seisdedos, F., J. L. Virelizier, D. Rousset, I. Clark-Lewis, P. Loetscher, B. Moser, and M. Baggiolini. 1996. HIV blocked by chemokine antagonist. Nature 383:400.. [DOI] [PubMed] [Google Scholar]
- 5.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 6.Bleul, C. C., M. Farzan, H. Choe, C. Parolin, I. Clark-Lewis, J. Sodroski, and T. A. Springer. 1996. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature 382:829-833. [DOI] [PubMed] [Google Scholar]
- 7.Brandt, S. M., R. Mariani, A. U. Holland, T. J. Hope, and N. R. Landau. 2002. Association of chemokine-mediated block to HIV entry with coreceptor internalization. J. Biol. Chem. 277:17291-17299. [DOI] [PubMed] [Google Scholar]
- 8.Cocchi, F., A. L. DeVico, D. A. Garzino, S. K. Arya, R. C. Gallo, and P. Lusso. 1995. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811-1815. [DOI] [PubMed] [Google Scholar]
- 9.Combadiere, C., S. K. Ahuja, H. L. Tiffany, and P. M. Murphy. 1996. Cloning and functional expression of CC CKR5, a human monocyte CC chemokine receptor selective for MIP-1(alpha), MIP-1(beta), and RANTES. J. Leukoc. Biol. 60:147-152. [DOI] [PubMed] [Google Scholar]
- 10.Combadiere, C., K. Salzwedel, E. D. Smith, H. L. Tiffany, E. A. Berger, and P. M. Murphy. 1998. Identification of CX3CR1. A chemotactic receptor for the human CX3C chemokine fractalkine and a fusion coreceptor for HIV-1. J. Biol. Chem. 273:23799-23804. [DOI] [PubMed] [Google Scholar]
- 11.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 12.Crameri, A., S. Cwirla, and W. P. Stemmer. 1996. Construction and evolution of antibody-phage libraries by DNA shuffling. Nat. Med. 2:100-102. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson, S. S. 2001. Evolving concepts in G protein-coupled receptor endocytosis: the role in receptor desensitization and signaling. Pharmacol. Rev. 53:1-24. [PubMed] [Google Scholar]
- 14.Gong, J. H., M. Uguccioni, B. Dewald, M. Baggiolini, and I. Clark-Lewis. 1996. RANTES and MCP-3 antagonists bind multiple chemokine receptors. J. Biol. Chem. 271:10521-10527. [DOI] [PubMed] [Google Scholar]
- 15.Grynkiewicz, G., M. Poenie, and R. Y. Tsien. 1985. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 260:3440-3450. [PubMed] [Google Scholar]
- 16.Hemmerich, S., C. Paavola, A. Bloom, S. Bhakta, R. Freedman, D. Grunberger, J. Krstenansky, S. Lee, D. McCarley, M. Mulkins, B. Wong, J. Pease, L. Mizoue, T. Mirzadegan, I. Polsky, K. Thompson, T. M. Handel, and K. Jarnagin. 1999. Identification of residues in the monocyte chemotactic protein-1 that contact the MCP-1 receptor, CCR2. Biochemistry 38:13013-13025. [DOI] [PubMed] [Google Scholar]
- 17.Hoogenboom, H. R., A. D. Griffiths, K. S. Johnson, D. J. Chiswell, P. Hudson, and G. Winter. 1991. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 19:4133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mack, M., B. Luckow, P. J. Nelson, J. Cihak, G. Simmons, P. R. Clapham, N. Signoret, M. Marsh, M. Stangassinger, F. Borlat, T. N. Wells, D. Schlondorff, and A. E. Proudfoot. 1998. Aminooxypentane-RANTES induces CCR5 internalization but inhibits recycling: a novel inhibitory mechanism of HIV infectivity. J. Exp. Med. 187:1215-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizoue, L. S., S. K. Sullivan, D. S. King, T. N. Kledal, T. W. Schwartz, K. B. Bacon, and T. M. Handel. 2001. Molecular determinants of receptor binding and signaling by the CX3C chemokine fractalkine. J. Biol. Chem. 276:33906-33914. [DOI] [PubMed] [Google Scholar]
- 20.Moore, J. P., and M. Stevenson. 2000. New targets for inhibitors of HIV-1 replication. Nat. Rev. Mol. Cell Biol. 1:40-49. [DOI] [PubMed] [Google Scholar]
- 21.Mosier, D. E., G. R. Picchio, R. J. Gulizia, R. Sabbe, P. Poignard, L. Picard, R. E. Offord, D. A. Thompson, and J. Wilken. 1999. Highly potent RANTES analogues either prevent CCR5-using human immunodeficiency virus type 1 infection in vivo or rapidly select for CXCR4-using variants. J. Virol. 73:3544-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nardese, V., R. Longhi, S. Polo, F. Sironi, C. Arcelloni, R. Paroni, C. DeSantis, P. Sarmientos, M. Rizzi, M. Bolognesi, V. Pavone, and P. Lusso. 2001. Structural determinants of CCR5 recognition and HIV-1 blockade in RANTES. Nat. Struct. Biol. 8:611-615. [DOI] [PubMed] [Google Scholar]
- 23.Oberlin, E., A. Amara, F. Bachelerie, C. Bessia, J. L. Virelizier, S. F. Arenzana, O. Schwartz, J. M. Heard, I. Clark-Lewis, D. F. Legler, M. Loetscher, M. Baggiolini, and B. Moser. 1996. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature 382:833-835. [DOI] [PubMed] [Google Scholar]
- 24.Pakianathan, D. R., E. G. Kuta, D. R. Artis, N. J. Skelton, and C. A. Hebert. 1997. Distinct but overlapping epitopes for the interaction of a CC-chemokine with CCR1, CCR3 and CCR5. Biochemistry 36:9642-9648. [DOI] [PubMed] [Google Scholar]
- 25.Pleskoff, O., C. Treboute, A. Brelot, N. Heveker, M. Seman, and M. Alizon. 1997. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science 276:1874-1878. [DOI] [PubMed] [Google Scholar]
- 26.Polo, S., V. Nardese, C. D. Santis, C. Arcelloni, R. Paroni, F. Sironi, A. Verani, M. Rizzi, M. Bolognesi, and P. Lusso. 2000. Enhancement of the HIV-1 inhibitory activity of RANTES by modification of the N-terminal region: dissociation from CCR5 activation. Eur. J. Immunol. 30:3190-3198. [DOI] [PubMed] [Google Scholar]
- 27.Poul, M. A., and J. D. Marks. 1999. Targeted gene delivery to mammalian cells by filamentous bacteriophage. J. Mol. Biol. 288:203-211. [DOI] [PubMed] [Google Scholar]
- 28.Proudfoot, A. E., R. Buser, F. Borlat, S. Alouani, D. Soler, R. E. Offord, J. M. Schroder, C. A. Power, and T. N. Wells. 1999. Amino-terminally modified RANTES analogues demonstrate differential effects on RANTES receptors. J. Biol. Chem. 274:32478-32485. [DOI] [PubMed] [Google Scholar]
- 29.Proudfoot, A. E., C. A. Power, A. J. Hoogewerf, M. O. Montjovent, F. Borlat, R. E. Offord, and T. N. Wells. 1996. Extension of recombinant human RANTES by the retention of the initiating methionine produces a potent antagonist. J. Biol. Chem. 271:2599-2603. [DOI] [PubMed] [Google Scholar]
- 30.Raport, C. J., J. Gosling, V. L. Schweickart, P. W. Gray, and I. F. Charo. 1996. Molecular cloning and functional characterization of a novel human CC chemokine receptor (CCR5) for RANTES, MIP-1beta, and MIP-1alpha. J. Biol. Chem. 271:17161-17166. [DOI] [PubMed] [Google Scholar]
- 31.Sabbe, R., G. R. Picchio, C. Pastore, O. Chaloin, O. Hartley, R. Offord, and D. E. Mosier. 2001. Donor- and ligand-dependent differences in C-C chemokine receptor 5 reexpression. J. Virol. 75:661-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simmons, G., P. R. Clapham, L. Picard, R. E. Offord, M. M. Rosenkilde, T. W. Schwartz, R. Buser, T. N. C. Wells, and A. E. Proudfoot. 1997. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science 276:276-279. [DOI] [PubMed] [Google Scholar]
- 33.Townson, J. R., G. J. Graham, N. R. Landau, B. Rasala, and R. J. Nibbs. 2000. Aminooxypentane addition to the chemokine macrophage inflammatory protein-1alpha P increases receptor affinities and HIV inhibition. J. Biol. Chem. 275:39254-39261. [DOI] [PubMed] [Google Scholar]
- 34.Vanhoof, G., F. Goossens, I. De Meester, D. Hendriks, and S. Scharpe. 1995. Proline motifs in peptides and their biological processing. FASEB J. 9:736-744. [PubMed] [Google Scholar]
- 35.Wilken, J., D. Hoover, D. A. Thompson, P. N. Barlow, H. McSparron, L. Picard, A. Wlodawer, J. Lubkowski, and S. B. Kent. 1999. Total chemical synthesis and high-resolution crystal structure of the potent anti-HIV protein AOP-RANTES. Chem. Biol. 6:43-51. [DOI] [PubMed] [Google Scholar]
- 36.Yang, O. O., S. L. Swanberg, Z. Lu, M. Dziejman, J. McCoy, A. D. Luster, B. D. Walker, and S. H. Herrmann. 1999. Enhanced inhibition of human immunodeficiency virus type 1 by met-stromal-derived factor 1β correlates with down-modulation of CXCR4. J. Virol. 73:4582-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]